Abstract
Introgressive hybridization is widespread in wild plants and has important consequences. However, frequent hybridization between species makes the estimation of the species’ phylogeny challenging, and little is known about the genomic landscape of introgression as it results from complex interactions of multiple evolutionary processes. Here, we reconstructed the phylogeny of ten wild diploid strawberries with whole genome resequencing data and then investigated the influence of recombination rate variation on phylogeny and introgression. We found that genomic regions with low recombination showed reduced levels of incomplete lineage sorting and introgression, and concentrated phylogenetic signals, thus contributing to the most likely species tree of wild diploid strawberries. We revealed complex and widespread introgression across the genus Fragaria, with an average proportion of approximately 4.1% of the extant genome. Introgression tends to be retained in the regions with high recombination rates and low gene density. Furthermore, we identified four SLF genes under selective sweeps that may play potential roles in the possible regain of self-incompatibility by ancient introgression. Altogether, our study yielded novel insights into the evolutionary history and genomic characteristics of introgression in wild diploid strawberries and provides evidence for the role of introgression in plant mating system transitions.
Keywords: Fragaria, introgression, mating system, phylogenomics, recombination rate, wild diploid strawberry
Introduction
Hybridization is quite common and an important force in plant evolution, with many plant species showing hybrid origins (Mallet 2005; Soltis and Soltis 2009). One of its outcomes is introgression—the transfer of genetic material between or within species by hybridization and repeated backcrossing. This process is believed to be widespread in nature, as revealed by recent genome-scale sequence data (Mallet et al. 2016; Edelman and Mallet 2021; Moran et al. 2021). As such, introgressive hybridization has been detected in many groups of species, including Arabidopsis (Arnold et al. 2016), monkeyflowers (Stankowski and Streisfeld 2015), tomatoes (Pease et al. 2016), Drosophila (Suvorov et al. 2022), butterflies (Martin et al. 2013; Edelman et al. 2019; Kozak et al. 2021), lizards (Finger et al. 2022), birds (Ellegren et al. 2012; Singhal et al. 2021), mammals (Jones et al. 2018; Shi and Yang 2018; Ferreira et al. 2021), and hominins (Nielsen et al. 2017). These studies have considerably improved our understanding of the role of hybridization and introgression in nature (Payseur and Rieseberg 2016; Martin and Jiggins 2017; Edelman and Mallet 2021). However, it remains a challenge to characterize the genomic landscape of introgression as it results from complex interactions of multiple evolutionary processes (e.g., selection, demography, and recombination) (Martin and Jiggins 2017).
Following hybridization, recombination can break up the linkage between alleles at different loci, thereby generating novel combinations across loci that can be exposed to selection (Schumer et al. 2018). As a result, regions of the genome with high recombination can more rapidly decouple neutral and adaptive introgressed alleles from deleterious alleles and, therefore, tend to harbor a higher proportion of introgression, whereas a localized reduction in introgression is expected within low recombination regions due to the increased linkage between introgressed loci and neighboring selected variants (Nachman and Payseur 2012; Schumer et al. 2018; Martin et al. 2019). Therefore, recombination rate variation is expected to play an important role in mediating the efficacy of selection and shaping patterns of introgression across the genome. However, how recombination rate variation impacts the genomic landscape of introgression remains an open question despite the increasing attention it has received in the last few years (Martin and Jiggins 2017; Kim et al. 2018; Schumer et al. 2018; Leitwein et al. 2019; Martin et al. 2019; Owens et al. 2022).
Reconstructing the history of hybridization and introgression in any evolutionary radiation requires a robust backbone of phylogenetic relationships among species. The recent emergence of phylogenomic data sets with hundreds or thousands of loci provides unprecedented opportunities to resolve the evolutionary history of species. However, phylogenetic trees based on genome-wide sequence data may not always represent the true, species-level relationships. Gene tree heterogeneity is widespread across the genome and often poses significant challenges for phylogenetic inference as a result of two primary processes, incomplete lineage sorting (ILS) and gene flow (Degnan and Rosenberg 2009). Although several methods have been developed and can explicitly accommodate ILS as the source of discordance (Edwards et al. 2016; Mirarab et al. 2016; Xu and Yang 2016), identifying and accounting for other processes such as gene flow in empirical datasets remain challenging (Li et al. 2019), particularly for lineages with an extensive history of hybridization and introgression, which in turn makes it difficult to infer the history of hybridization and introgression. As a key genetic parameter that influences assessments of gene flow, ILS, and genetic diversity, recombination rate variation plays a critical role in shaping the distribution of phylogenetic signals. The recombination rate interacts with the effects of natural selection to influence how genealogical histories are distributed across the genome. As mentioned above, low recombination regions of the genome are generally depleted in signatures of hybridization and are typically enriched for the most likely species tree (Pease and Hahn 2013). However, recombination rate variation among markers is not considered in most phylogenetic studies, and only a few empirical studies have attempted to directly quantify the impact of recombination rate variation on phylogenetic inferences (Pease and Hahn 2013; Edelman et al. 2019; Li et al. 2019; Martin et al. 2019; Hennelly et al. 2021; Owens et al. 2022). Therefore, we still have a poor understanding of the impact of recombination on species tree reconstructions.
The genus Fragaria contains approximately 25 species in five even-ploidy levels, ranging from diploid to decaploid with a basic chromosome number of seven (Folta and Davis 2006; Hummer and Hancock 2009; Liston et al. 2014). Wild Fragaria have a distribution spanning the Northern Hemisphere with the center of diversity being within China, where the majority of diploid (9 out of 12) and all five tetraploid species of the genus occur (Liston et al. 2014; Lei et al. 2017). The mating system varies substantially across species in Fragaria, from self-compatibility (SC), through self-incompatibility (SI), to dioecy (Liston et al. 2014). Moreover, Fragaria species are known to have small genomes (200–300 Mb for diploid species) and are amenable to propagation in tissue culture and genetic transformation. These characteristics render Fragaria an emerging model system for studies of sexual system evolution, polyploidization, and evolutionary genomics (Liston et al. 2014).
The phylogenetic relationships among species of Fragaria remain unresolved despite numerous efforts using chloroplast genomes (Njuguna et al. 2013), mitochondrial genomes (Fan et al. 2022), target capture sequencing (Kamneva et al. 2017), RNASeq (Qiao et al. 2016), multi-locus genes (Yang and Davis 2017), and the sequencing of whole genomes (Feng et al. 2021; Qiao et al. 2021). A major challenge in reconstructing the Fragaria phylogeny is the rapid radiation process, which is accompanied by both extensive ILS and hybridization between nascent lineages, leading to extensive gene-tree heterogeneity across the genome. For example, Feng et al. (2021) reconstructed a genome-wide phylogeny of five diploid species with 8,663 single-copy genes and revealed extensive gene–tree discordance, due to both extensive ILS and interspecific hybridization. More recently, Qiao et al. (2021) conducted the phylogenetic analysis of ten diploid species based on concatenation analysis of 1,007 single-copy genes with a maximum likelihood (ML) method, but the effect of neither ILS nor gene flow on phylogenetic reconstruction was accounted for in the analysis. As a result, our understanding of the full speciation history of Fragaria remains limited. Furthermore, natural hybridization and introgression between species are well-documented within Fragaria (Bringhurst and Khan 1963; Bringhurst and Senanayake 1966; Staudt 1999; Lei et al. 2005). However, the spatial landscape of introgression across the genome and its association with recombination and gene density are poorly understood.
The aim of the present study is to reconstruct phylogenetic relationships of wild diploid strawberries and characterize the genomic landscape of introgression among species. To this end, we generated whole-genome resequencing data of 68 wild diploid strawberries representing ten Fragaria species and reconstructed phylogenetic trees with partitioned recombination rates to investigate the relationships between recombination rates and frequencies of particular topologies. We then integrated a series of population genetic and phylogenetic approaches to assess hybridization patterns and the rate of gene flow between taxa. In particular, we performed a detailed characterization of genomic introgression in terms of recombination rate and gene density. Finally, we carried out a selective sweep analysis to identify regions and genes under selection in wild diploid strawberries and discuss their potential role in mating system transitions.
Results
Phylogeny and Population Structure
We sequenced and analyzed the genomes of 68 wild diploid strawberries representing ten Fragaria species, F. chinensis, F. nipponica, F. nubicola, F. pentaphylla, F. nilgerrensis, F. daltoniana, F. iinumae, F. viridis, F. vesca, and F. mandshurica, with a mean depth of approximately 54× per individuals covering an average of approximately 86% of the reference genome (supplementary table S1, Supplementary Material online). Along with one sample from the outgroup Potentilla microphylla (Buti et al. 2018), we identified approximately 14 million high-quality single-nucleotide polymorphisms (SNPs) within and across species.
We created a concatenation ML tree and a coalescent-based summary ASTRAL tree at the whole genome level, chromosome scales, and genomic windows with partitioned recombination rate, respectively (fig. 1a, supplementary fig. S1, Supplementary Material online and supplementary table S2, Supplementary Material online). In addition, we estimated a site-based coalescent SVDQuartets tree with reduced SNPs across the genome and built the whole chloroplast genome tree (supplementary fig. S1, Supplementary Material online). Although ML, ASTRAL, and SVDQuartets trees had strong support (100%) for nodes that separated species groups, these phylogenetic reconstructions were inconsistent with each other (supplementary fig. S1, Supplementary Material online) and showed strong discordance with the phylogenetic relationship of species trees from previous studies (supplementary fig. S2, Supplementary Material online; Njuguna et al. 2013; Kamneva et al. 2017; Qiao et al. 2016, 2021; Edger et al. 2019; Liston et al. 2020; Fan et al. 2022). All these phylogenetic trees support that F. vesca and F. mandshurica comprise one clade, while in most studies, several species native to Southwest China, F. chinensis, F. nubicola, F. pentaphylla, F. nilgerrensis, and F. daltoniana, are grouped into another clade (designated as Clade Southwest China). The main discordance of these phylogenies involves the phylogenetic relationships of F. iinumae and F. viridis. The phylogenetic trees inferred by Kamneva et al. (2017), Edger et al. (2019), and Liston et al. (2020), and this study clearly indicates that F. iinumae is sister to the Clade Southwest China (supplementary fig. S2, Supplementary Material online), while the species tree inferred from genome-wide orthologous single-copy genes (Qiao et al. 2021) and mitochondrial genome (Fan et al. 2022) support F. iinumae as the first-diverging lineage among diploid strawberry (supplementary fig. S2, Supplementary Material online). Additionally, two coalescent trees (ASTRAL and SVDQuartets tree) inferred from the whole genome and ASTRAL/ML tree inferred from low recombination regions in this study illustrate that F. viridis is a sister to the Clade Southwest China and F. iinumae (supplementary figs. S1 and S2, Supplementary Material online), while the ML tree constructed by concatenation sites across the whole genome, plastome trees (Njuguna et al. 2013), mitochondrial genome tree (Fan et al. 2022), and several other studies (Kamneva et al. 2017; Edger et al. 2019; Liston et al. 2020; Qiao et al. 2021) show that F. viridis is sister to the Clade of F. vesca and F. mandshurica (supplementary figs. S1 and S2, Supplementary Material online). Furthermore, the relative phylogenetic relationships among F. chinensis, F. nipponica, F. pentaphylla, and F. nubicola are heterogeneous (supplementary figs. S1 and S2, Supplementary Material online).
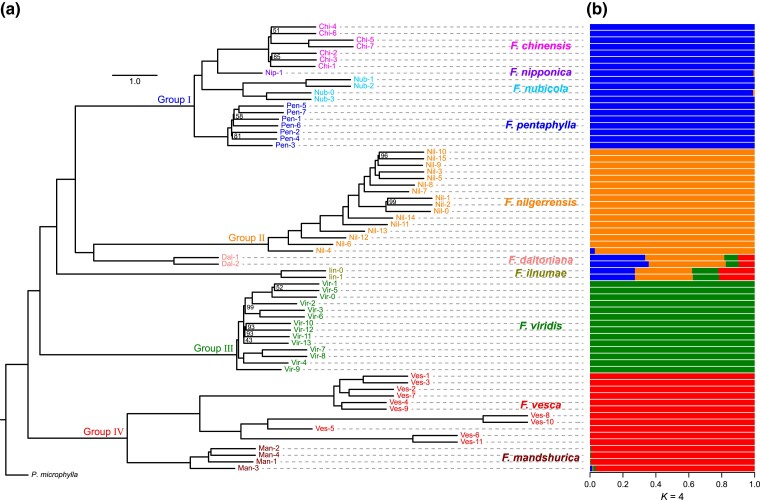
Fig. 1.
The phylogenetic tree and genetic structure of the ten wild diploid strawberries. (a) The ASTRAL tree for the 68 individuals representing the ten wild diploid strawberry species from low recombination regions, setting P. microphylla as the outgroup. The tree is constructed using 3,859 ML trees from SNPs of 10 kb nonoverlapping sliding windows with the 20% lowest recombination rate across the genome. The bootstrap support values less than 100% are shown along the nodes. (b) The optimal genetic structure of the 68 wild diploid strawberries detected by the Structure analysis (K = 4) based on genome-wide unlinked SNPs.
We investigated population structure in Fragaria using STRUCTURE and principal component analysis (PCA) analyses (fig. 1b and supplementary fig. S3, Supplementary Material online). The ΔK analysis indicates that the best K value was K = 4 (supplementary fig. S3, Supplementary Material online). For K = 4, the individuals were generally separated into four groups corresponding to the four phylogenetic lineages, while two species, F. daltoniana and F. iinumae, showed a high degree of admixture (fig. 1b), indicating potential pervasive hybridization or introgression. The population genetic structure with K ranging from 2 to 5 is also shown in supplementary figure S3, Supplementary Material online to fully explore population subdivision. The individual ancestry assignment estimated by STRUCTURE (K = 4) is highly consistent with the PCA results (supplementary fig. S3, Supplementary Material online), where the first three principal components contributed nearly half (46.9%) of total genetic variance and clearly distinguished these four groups and two admixed species (F. daltoniana and F. iinumae). For example, group III (F. viridis) and group IV (F. mandshurica and F. vesca) were independent of each other along the PC2, while group II (F. nilgerrensis) separated from other groups according to PC3 (supplementary fig. S3, Supplementary Material online).
Topology Weighting Reveals Phylogenetic Discordance
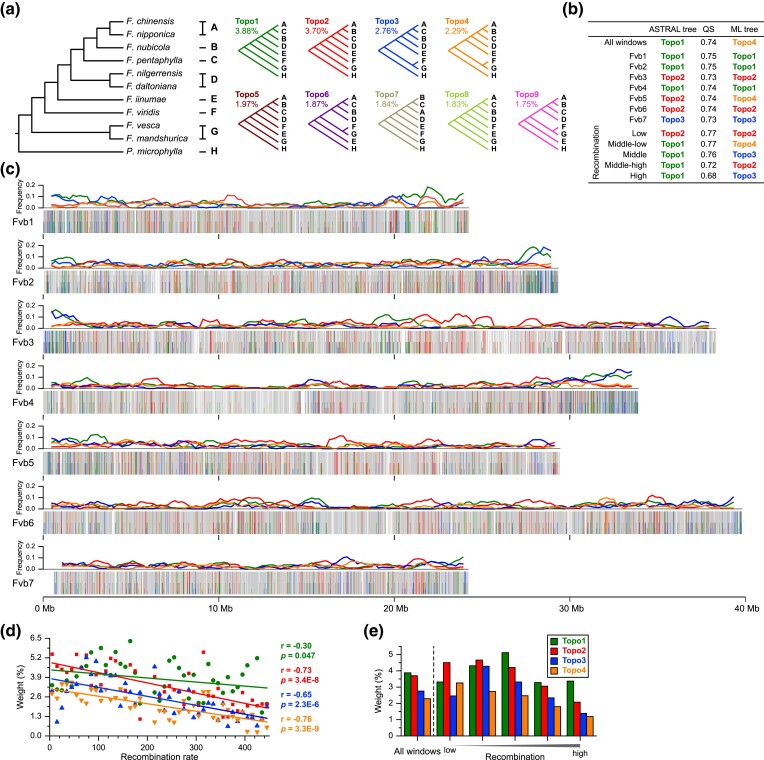
To explore phylogenetic conflicts along the chromosomes, we calculated the frequency of topology under eight taxa combinations (see Materials and Methods) for each of the 10 kb nonoverlapping sliding windows (supplementary table S3, Supplementary Material online). There are nine main topologies with occurrence frequency over 1.5% at the whole genome level, the top four of which have topology weighting ranging from 2.2% to 3.9% (fig. 2a and supplementary table S4, Supplementary Material online). Topo1 and Topo2 are the phylogenetic hypotheses inferred from ASTRAL with the whole genome and low recombination windows, respectively (fig. 2a and b, supplementary fig. S1, Supplementary Material online and supplementary table S4, Supplementary Material online). Topo3 and Topo4 correspond to Topo1 and Topo2, respectively, with the sole change in the phylogenetic position of F. viridis as the sister group of F. vesca and F. mandshurica, instead of the sister group of Clade Southwest China and F. iinumae (fig. 2a). The main topologies do not appear to be randomly distributed across the genome. Topo1 and Topo3 tend to be located at the end of chromosomes, while Topo2 is enriched in the center of chromosomes (fig. 2c). It is worth noting that Topo2 has the highest topology weighting and is consistent with both the ASTRAL and ML trees inferred with low recombination windows and the two longest chromosomes (Fvb3 and Fvb6; fig. 2b and supplementary table S4, Supplementary Material online). We further investigated the genomic characteristics of the main topologies and found that the weightings of the top four abundant topologies were significantly reduced with the increase of recombination rate (fig. 2d). Although Topo1 has a slightly higher average frequency compared to Topo2 at the whole genome level, Topo 2 occurs more frequently in low recombination regions (fig. 2e and supplementary table S4, Supplementary Material online).
Fig. 2.
Variation of topology weighting within and among chromosomes reveals widespread phylogenetic discordance and is correlated with recombination rate. (a) The main possible topologies, with occurrence frequency over 1.5%, for the eight taxa groups. (b) Topology for ASTRAL trees and ML tree at the whole genome level, chromosome scales, and groups with partitioned recombination rate. The normalized quartet score (QS) values reflect the level of phylogenetic discordance among gene trees inferred from corresponding windows. (c) Frequency distribution of four main topologies (colors as in panel a) for 500 kb sliding windows with the step size of 100 kb (Upper line chart), and weighting for the four main possible topologies (colors as in panel a) plotted across 10 kb nonoverlopping sliding windows (Lower bar chart) along the chromosomes, where grey and white columns indicate the windows belonging to other topologies, and the windows with unknown topology, respectively. (d) Average weightings for the top four possible topologies (colors as in panel a) binned according to their recombination rate and the linear regression curve. The r (Pearson Correlation Coefficient) and P values are shown as the colors of the corresponding topology in panel (a). (e) Frequency of the top four possible topologies at the whole genome level and five groups with partitioned recombination rate.
The normalized quartet score is relatively high (0.77) for the species tree inferred from low/medium-low recombination windows compared to that for high recombination regions (0.68), indicating a lower level of gene tree discordance in regions of low recombination. Additionally, the MSCquartets analyses show less blue circles plotted close to centroids of the simplexes in the trees inferred from low recombination windows, compared to the trees from high recombination regions (supplementary fig. S4, Supplementary Material online), which indicates that low recombination regions tend to contain less ILS. As a result, we expect that the topology resulting from low recombination regions should be more likely to reflect the true phylogeny of the wild diploid strawberries.
Widespread Introgression Among Species
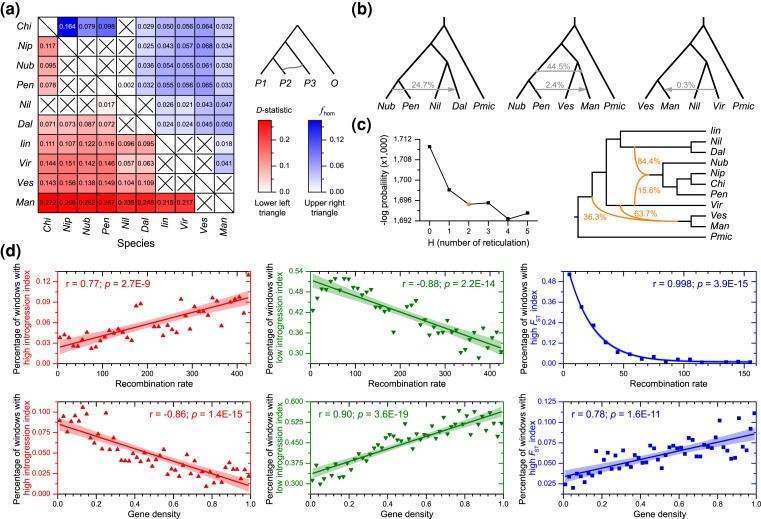
ABBA-BABA tests revealed that about 79% (95/120) of tested four-taxon phylogenies had a significant signal of introgression (P < 0.01 after Benjamini–Hochberg correction) (fig. 3a and supplementary table S5, Supplementary Material online), indicating widespread hybridization and introgression history in wild diploid Fragaria species. The D statistic is widely used in detecting introgression but is unable to estimate the proportion of the genome with evidence of introgression (Martin et al. 2015; Malinsky et al. 2021; Morales-Cruz et al. 2021). To further determine the proportion of the genome with evidence of introgression, we calculated fhom value of 95 trios with significant introgression signals, as an alternative estimator of genome-wide fraction of admixture (Pulido-Santacruz et al. 2020). As the estimates of fhom value for the same P2-P3 species pairs varied on different P1 species (supplementary table S5, Supplementary Material online), we used their maximum values to reflect the genomic proportion of introgression between P2 and P3 species, remaining maximum fhom values of 49 P2-P3 species pairs (supplementary table S5, Supplementary Material online). Overall, the average extent of introgression in Fragaria is approximately 4.1% of the genome, comprising up to approximately 16.4% of the extant genome in comparisons between F. nipponica (P2) and F. chinensis (P3) (fig. 3a and supplementary table S5, Supplementary Material online). The extensive genomic introgression detected between F. nipponica and F. chinensis could explain the closer phylogenetic relationship of the two species.
Fig. 3.
The widespread introgression across wild diploid strawberry genomes and its association with recombination rate and gene density. (a) Heatmap of significantly elevated Dmin score (Lower left triangular matrix) and genomic properties of introgression (fhom value, Upper right triangular matrix) between P2 and P3. The color of the corresponding heatmap cell represents the most significant Dmin score with adjusted P value <0.01 and the maximum fhom value across all possible species in P1. (b) Signals of introgression in Fragaria inferred by DFOIL analyses based on five-taxon phylogeny (((P1, P2), (P3, P4)), O), where the divergence time between P3 and P4 was earlier than that between P1 and P2. The horizontal line with double-ended dots and lines with single-ended arrows indicate ancient introgression and post-speciation gene flow, respectively. The numbers above the lines show the proportion of individual combinations that detected significant signals of introgression for the corresponding species combinations. (c) Signals of gene flow in wild diploid Fragaria species inferred by PhyloNet. The numbers next to the lines indicate inheritance probabilities for corresponding edges. (d) Average percentage of windows with high ingression index, low ingression index, and high genetic differentiation (FST) index binned according to their recombination rate and gene density. The r (Pearson Correlation Coefficient) and P values are shown along the linear regression curve. Chi, F. chinensis; Nip, F. nipponica; Nub, F. nubicola; Pen, F. pentaphylla; Nil, F. nilgerrensis; Dal, F. daltoniana; Iin, F. iinumae; Vir, F. viridis; Ves, F. vesca; Man, F. mandshurica; Pmic, P. microphylla.
We performed DFOIL to evaluate 34 alternative symmetric five-taxon phylogenies and found signals for introgression at three species combinations, including ancient introgression and three instances of post-speciation gene flow (fig. 3b). The ancient introgression signal was detected between F. mandshurica and the ancestor of F. nubicola and F. pentaphylla (i.e., the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla) at a relatively high proportion (44.5%) of individual combinations (fig. 3b). Since a small number of ancient introgression events could create the impression of more widespread recent introgression, widespread introgression inferred by D statistic may be over-inflated by possible effects of early ancient introgression. In addition, nearly a quarter (24.7%) of individual combinations showed significant signals for post-speciation gene flow from F. nubicola to F. daltoniana (fig. 3b), which is consistent with the high degree of admixture of F. daltoniana in the STRUCTURE analysis (fig. 1b). We also estimated the substitution rates along the phylogeny at low recombination regions and observed relatively low variation among species, and no significant difference (P = 0.36) between SI and SC species (supplementary fig. S5, Supplementary Material online). Nevertheless, further research is needed to better understand the impact of substitution rate variation on the assessment of introgression in wild diploid strawberries.
We further applied two phylogeny-based approaches to assess the influence of hybridization and introgression on topological discordance. PhyloNet detected at least two ancient hybridizations/introgression in Fragaria, showing F. viridis may have contributed to hybrid origination of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla, and/or the lineage of F. vesca and F. mandshurica (fig. 3c), which could explain the conflicting phylogenetic positions of F. viridis (fig. 2a and b, supplementary figs. S1 and S2, Supplementary Material online, and supplementary tables S2 and S3, Supplementary Material online) and mixture state of F. viridis when K = 2 and 3 in STRUCTURE result (supplementary fig. S3, Supplementary Material online). Limited by the requirement of a symmetric five-taxon phylogeny, partial ancient introgression events cannot be discovered in DFOIL software, for example, the possibility of ancient introgression between F. viridis and the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla would be omitted, because F. viridis cannot be set as P3/P4 in respective species combinations. However, our PhyloNet analyses hypothesize ancient introgression from F. viridis to the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla (fig. 3c), which provides an informative supplement to the DFOIL analyses. The PhyloNetworks also identified complex hybridization and introgression in the lineage of F. chinensis, F. nipponica, F. pentaphylla, and F. nubicola (supplementary fig. S6, Supplementary Material online), which is consistent with phylogenetic discordance in our above analyses (supplementary figs. S1 and S2, Supplementary Material online). Together, these results support complex and widespread introgression in the Fragaria genus, and introgression substantially contributed to phylogenetic discordance.
Genomic Landscape of Introgression is Shaped by Recombination Rate and Gene Density
We calculated the fdM of each 10 kb nonoverlapping window for 95 trios with significant introgression signals and detected 15 to 2,749 putatively introgression regions (pIR) from each trio (supplementary table S6, Supplementary Material online), yielding 873 high introgression windows and 7,528 low introgression windows, respectively (supplementary table S7, Supplementary Material online), according to introgression index that was estimated by the proportion of trios with pIRs for each window (see Materials and Methods). Additionally, we identified 1,151 high genetic differentiation windows (introgression barrier regions) (supplementary table S8, Supplementary Material online), on a basis of the FST index of each window that was calculated as the proportion of combinations with top 5% FST values (see Materials and Methods). The genomic distribution of high/low introgression regions is a mosaic across the genome (supplementary fig. S7, Supplementary Material online), whereas the high genetic differentiation regions tend to be concentrated in the center of chromosomes (supplementary fig. S8, Supplementary Material online). Nevertheless, genomic regions with high differentiation (FST) mainly overlapped with low and middle-low introgression regions (supplementary fig. S9, Supplementary Material online). The genes in high introgression regions are enriched for transcription regulation (supplementary table S9, Supplementary Material online), while genes in low introgression regions and high differentiation regions largely comprise specific enzyme activity (supplementary tables S10 and S11, Supplementary Material online). Worth noting, we observed that the introgression proportion of the Fragaria genome has an extremely strong positive relationship with recombination rate and a highly significant negative correlation with gene density (fig. 3d). Genomic windows of high differentiation exhibited similar characteristics with genomic windows of low introgression in that they showed a significantly negative and positive correlation with recombination rate and gene density, respectively (fig. 3d).
In addition, the main topology weighting reduced dramatically in the regions of high introgression and increased continually in the low introgression windows (supplementary table S12, Supplementary Material online), which indicates that introgression could influence topology weighing and phylogeny. More specifically, Topo2 has a much higher frequency than Topo1 in high genetic differentiation regions (introgression barriers) (supplementary table S12, Supplementary Material online), further providing evidence that Topo2 might reflect the most likely species tree of the Fragaria genus.
Mating System Transitions May be Associated With Selective Sweeps and Introgressed Regions
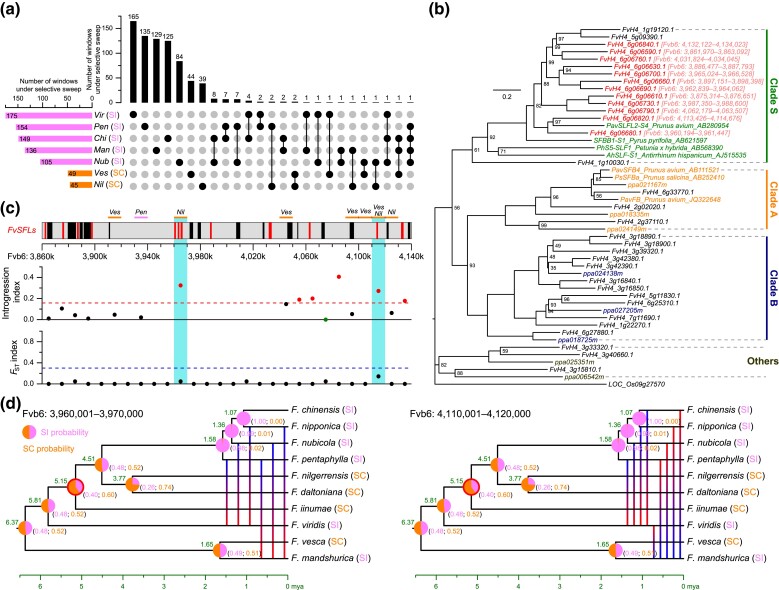
Positive selection enhances adaptive evolution and leaves distinct signatures across the genome. We identified genomic regions under selective sweeps by using selscan (Szpiech and Hernandez 2014) and RAiSD (Alachiotis and Pavlidis 2018) approaches for each Fragaria species separately, leaving the overlapped regions regarded as the selective sweep regions for the corresponding strawberry. We screened a total of 765 windows under selective sweep for the seven wild diploid strawberries (supplementary table S13, Supplementary Material online). Two SC species, F. nilgerrensis and F. vesca, have 45 and 49 selective sweeps, respectively, which was much smaller than the number of the selective sweeps in SI species, ranging from 105 to 175 (fig. 4a and supplementary table S13, Supplementary Material online). Of the 765 windows, approximately 94.2% were identified under selective sweeps in a single species, while approximately 5.8% were shared by at least two species (fig. 4a and supplementary table S13, Supplementary Material online), indicating relatively unique adaptive patterns for each species. This conclusion is also supported by differentiated GO enrichment (specific metabolic process and enzyme activity) for genes located in selective sweeps of different diploid strawberries (supplementary table S14, Supplementary Material online).
Fig. 4.
Four S locus-related FvSLFs, which are located in selective sweep and high introgression regions, may influence mating system transitions in wild diploid strawberries. (a) The upset plot of windows under selective sweeps of the seven Fragaria species. (b) ML tree of SLF/SLF-like F-box genes from the F. vesca genome and partial F-box genes from peach (prefix of “ppa”) and cherry. The bootstrap support values less than 100% are shown along the nodes. Clades A, B, and S are defined according to the previous phylogenetic tree of the peach genome (Akagi et al. 2016). (c) Gene structure, introgression, and FST statistics along the 280 kb putative S locus containing 12 FvSLFs in tandem duplications. The distribution of FvSLFs and other genes in this region is signed with red and black columns, respectively. Eight selective sweeps (10 kb window) in specific species were marked with the horizontal lines, with the species names shown above the line. (d) The probability of ancestral mating system state across Fragaria genus. The numbers in the brackets indicate the relative probabilities of SI and SC. The vertical lines indicate the introgression between P3 and P2 species in two windows. Vir, F. viridis; Pen, F. pentaphylla; Chi, F. chinensis; Man, F. mandshurica; Nub, F. nubicola; Ves, F. vesca; Nil, F. nilgerrensis.
Wild diploid strawberries have served as an important model for studying mating system transitions, due to their relatively recent origin and variation in SI and SC (Njuguna et al. 2013; Liston et al. 2014). Model comparison (symmetric vs. asymmetric) of mating system transition rates reveals that bidirectional evolution may give more reliable results (supplementary table S15, Supplementary Material online). To evaluate this, we reconstructed the probability of the ancestral type of mating system across the Fragaria genus by BayesTraits in RASP 4 (Yu et al. 2020). The common ancestral state of the lineages of F. chinensis, F. nipponica, F. nubicola, F. pentaphylla, F. nilgerrensis, F. daltoniana, and F. iinuma was SC with a probability of 60% (fig. 4d). Although this probability is not very high due to the small number of species involved in this study, it still indicates the possible regain of SI along the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla. To reveal the genetic mechanism of mating system transition in wild diploid strawberries, we focused on the S locus, including S-RNase and SLF genes, the key regions/genes that regulate the reproduction system of Fragaria. Here, we identified 33 members of SLF/SLF-like F-box genes from the F. vesca strawberry genome, 12 of which were clustered in Clade S and located within one end of the S locus of F. vesca in tandem duplications (Fvb6: 3,860–4,140k) (fig. 4b and c). This 280-kb region contains three and five 10-kb windows with evidence of selective sweep in F. vesca and F. nilgerrensis, respectively (fig. 4c). In particular, four FvSLFs were located in two selective sweeps (Fvb6: 3,960–3,970k; Fvb6: 4,110–4,120k) of F. nilgerrensis, while the latter window (containing one FvSLF) was also identified in selective sweeps of F. vesca (fig. 4c). Meanwhile, we screened six S-RNase/S-RNase-like genes in F. vesca, two of which were clustered in Class III and located at the other end of the S locus (Fvb6: 4,630–4,651k; supplementary fig. S10, Supplementary Material online), approximately 500 kb away from the region of tandem duplicated SLFs. However, no S-RNase/S-RNase-like genes overlapped with either of the selective sweeps for all wild diploid strawberries (supplementary table S13, Supplementary Material online). These findings indicated that the FvSLFs may contribute potential roles in the mating system transitions in wild diploid strawberries.
Furthermore, these two windows fall in high introgression regions even with the top 1% highest introgression index (fig. 4c and supplementary table S7, Supplementary Material online). We also detected introgression signals between F. mandshurica/F. viridis (P3) and F. nipponica/F. pentaphylla/F. nubicola (P2) at the window Chr6: 3,960–3,970k, and signals between F. viridis (P3) and F. chinensis/F. nipponica/F. pentaphylla/F. nubicola (P2) at the window Chr6: 4,110–4,120k (fig. 4d and supplementary table S6, Supplementary Material online), but none was found between two SC species for either of these two windows. Combining with evidence of ancient introgression by DFOIL and PhyloNet (fig. 3b and c), we hypothesize that these two windows experienced ancient introgression from F. viridis (and/or F. mandshurica) to the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla, which may directly contribute to the mating system transition from SC (the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, F. pentaphylla, F. nilgerrensis, F. daltoniana, and F. iinumae) to SI (the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla) (fig. 4d).
Discussion
Recombination-aware Phylogenomics of the Wild Diploid Strawberries
The phylogenetic relationships among diploid Fragaria species have not been resolved even in large multilocus datasets (Edger et al. 2019; Liston et al. 2020; Feng et al. 2021; Qiao et al. 2021; Fan et al. 2022). Previous phylogenomic studies based on genome-wide loci demonstrated conflicting topological positions for many diploid species (supplementary fig. S1, Supplementary Material online). Although these discordances can result from artifacts arising from tree inference methods, both gene flow and ILS accompanying rapid radiation or recently diverged lineages can overwhelm the genealogical signal of the original population branching pattern, and lead to gene trees that contradict the true species tree (Shen et al. 2017; Irisarri et al. 2018; Li et al. 2019). However, despite the rapid development of more sophisticated approaches (Edwards 2009; Edwards et al. 2016; Mirarab et al. 2016; Xu and Yang 2016), estimating species tree that explicitly account for gene flow and ILS remains challenging for lineages with an extensive history of hybridization and introgression.
In the present study, by partitioning genomic windows into different levels of recombination, we show that topology weightings are significantly correlated with the recombination rate (fig. 2d and e). Specifically, we found that the genomic windows with low recombination exhibited lower levels of introgression and ILS compared to the medium to high recombination regions (fig. 3d and supplementary fig. S4, Supplementary Material online). As a result, the low recombination regions tend to harbor a greater proportion of topologies reflecting speciation and, therefore, are more likely to give a topology matching the species tree (Topo2 in fig. 2a). These results are consistent with previous reports of enriched true species trees in genomic regions with low and no recombination (Pease and Hahn 2013; Fontaine et al. 2015; Edelman et al. 2019; Li et al. 2019; Martin et al. 2019; Hennelly et al. 2021; Owens et al. 2022). For example, Pease and Hahn (2013) found genomic regions with low or no recombination showed significantly stronger support for the putative species tree in the presence of ILS, because low recombination regions have lower effective population size (Ne) and, therefore, less retention of shared ancestral polymorphisms than higher-recombination regions. Recent phylogenomic studies of Heliconius butterflies (Edelman et al. 2019; Martin et al. 2019), the cat family Felidae (Li et al. 2019), grey wolves (Hennelly et al. 2021), and sunflowers (Owens et al. 2022) showed that the most likely species tree is notably enriched within low recombination regions of sex chromosomes in the presence of gene flow, owing to the strong linkage in low recombination regions which leads to the more effective removal of deleterious alleles introduced through hybridization (Nachman and Payseur 2012; Schumer et al. 2018). Together, these findings indicate that phylogenomic parameters inferred from whole genome data may be misleading and highlight the consideration of taking into account recombination rate variation in phylogenetic reconstruction, especially for lineages that experienced extensive hybridization.
Genomic Landscape of Introgression Across Wild Diploid Fragaria Species
Genomic analyses have proved mounting evidence that introgression seems to be more common than previously thought, and that mosaic genomes are more common than homogeneous genomes (Taylor and Larson 2019). However, the genomic proportion and landscape of introgression are not well understood. Most previous studies are focused on the hybridization and gene flow between single or few number pairs of taxa, which makes it hard to infer general patterns at a broad scale. In this study, we applied D-statistic and fhom values to detect and quantify introgression among ten wild diploid strawberries and observed that 0.2% to 16.4% of the genomes were introgressed ancestry (fig. 3a and supplementary table S5, Supplementary Material online). The genomic proportion of introgression in Fragaria is similar to 2.0–8.0% among six North American wild grapes (Morales-Cruz et al. 2021) and 0.4–10.7% among the seven species from section Populus of the genus Populus (Liu et al. 2022), but higher than that reported for wild tomato species (0.2–2.5%; Hamlin et al. 2020). However, it is noteworthy that ancient introgression events can affect the estimation of genomic introgression among extant species (Malinsky et al. 2018). Therefore, our detected high genomic proportion of introgression in Fragaria might be overestimated due to ancient introgression and should be interpreted with caution. Nevertheless, the similar levels of genomic introgression observed in Fragaria and other species suggest that introgression shapes substantial genomic variation of wild plant genomes.
The fate of any given introgressed allele will depend not only on the adaptive value of its associated variants but also on the local genomic landscape in terms of recombination rates and the nature of linked genes (Martin and Jiggins 2017). In this study, we found that genomic windows with high introgression tend to be retained in the regions of high recombination rate, whereas low introgression regions are concentrated in genomic regions with low recombination rates (fig. 3d). Such a strong positive correlation between introgression and recombination rate has been observed in genomic studies of a range of taxa, including butterfly (Martin et al. 2019), alpine bumblebees (Christmas et al. 2021), maize (Calfee et al. 2021), wild grapes (Morales-Cruz et al. 2021), Populus species (Liu et al. 2022), and sunflowers (Owens et al. 2022). These findings are consistent with theoretical expectations that introgressed genomic alleles (mostly deleterious mutations) would break down rapidly during subsequently repeated backcrossing following initial hybridization and tend to be retained due to the selection against deleterious foreign alleles within high recombination regions (Schumer et al. 2018; Martin et al. 2019; Morales-Cruz et al. 2021). Given the fact that gene regions have relatively low recombination rate (Martin et al. 2019), it is not unexpected for the observed negative relationship between gene density and introgressed ancestry (fig. 3d). However, it is difficult to test whether such regions directly shape barriers to introgression (Martin et al. 2019). Nevertheless, we found a stronger correlation between introgression and gene density, than introgression and recombination rate (fig. 3d), which might support the direct contribution of gene density on the genomic landscape, because gene-rich foreign regions contain numerous deleterious mutations, and thus are less likely to be introgressed (Morales-Cruz et al. 2021). However, introgression of large genomic regions is known to occur (Dasmahapatra et al. 2012). Furthermore, we also found extremely low levels of recombination rate in high genetic differentiation (FST) regions, in addition to a negative relationship between FST and recombination rate (fig. 3d), which supports a role for linked selection (Burri 2017). Overall, our findings demonstrate that recombination rate variation is an important factor in shaping the landscape of genomic introgression and differentiation among wild diploid Fragaria species.
Selective Sweep of SLFs May Contribute to Mating System Transitions
SI is a widespread genetic system in angiosperms and occurs in approximately 39% of plant species (Igic et al. 2008). SI systems fall into two major classes: gametophytic self-incompatibility (GSI) and sporophytic self-incompatibility (SSI) (Hiscock and McInnis 2003). GSI is quite common in families such as Rosaceae, Solanaceae, and Plantaginaceae, and SI is regarded as the ancestral state (Igic and Kohn 2001). The Fragaria genus provides an exceptional model to study mating system transitions, not only because it underwent a full range of sexual systems from hermaphroditism (diploid) to dioecy that is associated with all increases in ploidy in the genus, and back again from subdioecy of a wild octoploid to hermaphroditism of commercial strawberry along domestication, but also because of variation in SI/SC of diploid species (Liston et al. 2014). Combining the results of model comparisons and ancestral mating system reconstruction, we propose the possible transition from SC to SI along the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla (fig. 4d). However, Chen et al. (2022) considered that wild diploid Fragaria evolved multiple losses of SI, according to a phylogeny inferred by orthologous single-copy genes. Although the loss of SI is believed to be more frequently than the transition from SC to SI (Igic et al. 2008), bidirectional transitions were inferred in the Asteraceae and the Brassicaceae (Ferrer and Good-Avila 2007; Sherman-Broyles and Nasrallah 2008). In particular, Njuguna et al. (2013) also proposed that both loss and gain of SI are possible in Fragaria according to phylogeny estimated from the chloroplast genome.
Introgression from closely related species can introduce adaptive genetic variation that may affect traits. However, linking adaptive traits with signatures of introgression is still rare and remains a challenge in most systems (Taylor and Larson 2019; Morales-Cruz et al. 2021). As well known, the GSI system contains the female S determinant (S-RNase) in the pistil, and the male S determinant (SLF or called SFB, SFBB, SLFL) in pollen, while the SLF genes were usually derived from tandem duplications (Akagi et al. 2016). The deletion/mutation of specific SLF in SC haplotypes could be responsible for the loss of SI in the tribe Maleae and Amygdaleae (Ushijima et al. 2004; Sonneveld et al. 2005; Kakui et al. 2011; Ashkani and Rees 2016). However, the knowledge about the SI/SC mechanism for Fragaria is only limited to the identification of S-RNase genes (Bošković et al. 2010; Du et al. 2021; Chen et al. 2022). Here, we identified a roughly 280 kb region in the S-locus of F. vesca (Fvb6: 3,860–4,140k), which contained 27 genes including 12 FvSLFs and several windows under selective sweep (fig. 4b and c), and may potentially play a role in mating system transitions. More interesting, the two selective sweep regions (Fvb6: 3,960–3,970k; Fvb6: 4,110–4,120k) that contain four FvSLFs exhibited ancient introgression from F. viridis (and/or F. mandshurica) to the ancestor of the lineage of F. chinensis, F. nipponica, F. nubicola, and F. pentaphylla, which could further explain the regain of SI in this lineage, following the initial transitions from SI to SC (figs. 3b,3c, and 4d and supplementary table S7, Supplementary Material online). Our finding on the mechanism of mating system transitions is important for genetic breeding and germplasm innovation in strawberries, and also provides a novel example of adaptive introgression related to mating system transitions in plants.
Materials and Methods
Plant Material and Genome Resequencing
We collected 64 samples from ten wild diploid Fragaria species in the China National Germplasm Repository for Strawberry (Nanjing) at the Jiangsu Academy of Agricultural Sciences (Nanjing, China), with detailed sampling information provided in supplementary table S1, Supplementary Material online. Genomic DNA was extracted from fresh leaves with the plant DNA extraction kit (AU31111, Bioteke, China). Libraries were constructed with a short insert size of 350 for each sample and then subjected to sequence (PE150) on Illumina NovaSeq 6000 platform.
SNP Calling and Genotyping
We downloaded genome survey data of F. viridis (designated as Vir-0), F. nubicola (designated as Nub-0), F. nilgerrensis (designated as Nil-0), F. iinumae (designated as Iin-0), and Potentilla microphylla (designated as Pmic) from SRA database under SRR11833746, SRR11833747, SRR11833748, SRR9217951, and PRJEB18433, respectively. The first three datasets were generated from our previous study (Feng et al. 2021). After filtering low-quality reads of these five samples, plus 64 newly sequenced wild strawberries, according to pipeline QC_pe (Feng et al. 2017), we mapped the clean data of each individual to the F. vesca Genome v4.0 (https://www.rosaceae.org/species/fragaria_vesca/genome_v4.0.a1; Edger et al. 2017) by BWA (Li and Durbin 2009) and then conducted SNP calling using SAMtools v0.1.19 (Li et al. 2009), Picard tool v1.119 (http://broadinstitute.github.io/picard/), and GATK v4.1.4.0 (DePristo et al. 2011). Only sites with a quality score above 30 were kept in HaplotypeCaller. Furthermore, we joined genotype files (gVCF) of all individuals together and filtered the variant sites using GATK VariantFiltration with the expression “QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < -12.5 || ReadPosRankSum < −8.0.” Additionally, we removed the indels and kept the sites for downstream analyses, according to minQ 30, max-missing 0.67, max-alleles 2, max-meanDP 500, min-meanDP 10.
Finally, we retained 69 genotypes (including the outgroup Pmic), and 14,175,029 high-quality SNPs, plus 68,550,711 invariant sites (Signed as Dataset 1). Meanwhile, we excluded minor allele frequency sites (maf 0.05) by VCFtools (Danecek et al. 2011) to generate Dataset 2 that contains 5,751,545 SNPs and 69 genotypes. Additionally, we excluded genotypes of outgroup Pmic and removed highly correlated SNPs by PLINK (Purcell et al. 2007) with the parameter indep-pairwise of 50 5 0.2, yielding Dataset 3 (459,031 unlinked SNPs, 68 genotypes).
Population Structure Analyses
To investigate population structure in Fragaria, we performed admixture analyses according to Dataset 3, by using STRUCTURE (Pritchard et al. 2000) and PCA. Firstly, we ran eight independent STRUCTURE with different K values ranging from 2 to 11 hypothetical ancestral populations, setting the length of burn-in and number of MCMC replications after burn-in as 20,000 and 100,000, respectively. Then, we calculated the best K value by the ΔK method using STRUCTURE HARVESTER (Earl and vonHoldt 2012) and averaged the cluster assignment from eight replications by CLUMPP (Jakobsson and Rosenberg 2007). Also, we conducted PCA for 68 individuals from the 10 diploid strawberry species with GCTA (Yang et al. 2011) and displayed the result between PC1 and PC2/PC3.
Recombination Rate and Gene Density
To estimate the recombination rate along the strawberry genome, we calculated the population-scaled recombination rate for each 10 kb nonoverlapping sliding window across the ten wild diploid strawberries. We phased each chromosome according to unlinked SNPs of 68 Fragaria individuals (Dataset 3) by using Beagle (Browning and Browning 2009). Then, we used FastEPRR (Gao et al. 2016) to estimate the recombination rate (Rho) per 10 kb windows by combining all the species together. FastEPRR is a widely used R package for the rapid and accurate estimation of population recombination rates from DNA polymorphisms. To investigate the gene density across seven strawberry chromosomes, we calculated the proportion of gene regions at each 10 kb nonoverlapping sliding window based on gff files of Fragaria vesca v4.0.a1.
Phylogenetic Tree Estimation
We applied four approaches to reconstruct phylogenetic relationships among wild diploid Fragaria species. Firstly, we divided the genome (Dataset 1) into 10 kb nonoverlapping sliding windows but only focused on the windows with at least 200 sites and 20 parsimony-informative sites (PIS), and then constructed ML trees for each window by using IQ-TREE (Nguyen et al. 2015). Furthermore, we also removed the trees with the average bootstrap support value less than 60%, resulting in 19,295 ML trees/windows. We used a coalescent-based summary method (ASTRAL; Mirarab and Warnow 2015) to construct the species tree based on 19,295 ML trees (Approach 1). Meanwhile, we adopted a concatenation approach to infer ML trees by using IQ-TREE (Nguyen et al. 2015) according to concatenation nucleotide sites from 19,295 windows (Approach 2). Additionally, we rebuilt ASTRAL trees and ML trees at chromosomal scales (Fvb1–Fvb7), and for five groups that were equally classified by recombination rate of corresponding windows (low, medium–low, medium, medium–high, and high recombination), respectively. In addition, we inferred divergence times and substitution rates for 68 accessions representing the 10 wild diploid strawberries by using r8s (Sanderson 2003) with parameter “smooth” of 0.01, according to ML tree reconstructed with low recombination regions, setting a secondary age calibration, the crown age of genus Fragaria at 6.37 Ma (Feng et al. 2021).
The third approach is that we used a site-based coalescent method (SVDQuartets; Chifman and Kubatko 2014) to infer species tree according to Dataset 2. Lastly and the fourth approach, we assembled the chloroplast genome for each of 69 individual by NOVOplasty (Dierckxsens et al. 2017) and built the chloroplast ML tree by using MAFFT (Katoh and Standley 2013) and IQ-TREE (Nguyen et al. 2015).
Topology Weighting
To explore evolutionary relationships among wild diploid strawberries, we carried out Twisst (Martin and Van Belleghem 2017) to quantify topology weighting among and within chromosomes. First, we generated 19,295 species trees from each ML tree of 10 kb nonoverlapping sliding windows by ASTRAL (Mirarab and Warnow 2015) under the map of the relations between individuals and species, setting P. microphylla as the outgroup. The software Twisst does not scale to more than eight taxa combinations; therefore, we combined monophyletic clade into one taxa combination (e.g., group A contains F. chinensis and F. nipponica, group D contains F. nilgerrensis and F. daltoniana, group G contains F.vesca and F. mandshurica), according to the variations of Fragaria phylogeny in previous and this study (supplementary figs. S1 and S2, Supplementary Material online). For eight taxa combinations, there is a maximum of 10,395 possible rooted, bifurcating tree topologies. Here, we applied Twisst (Martin and Van Belleghem 2017) to classify 19,295 species trees into 3,441 possible topologies, yielding weightings for each topology. Moreover, we focused on the main topologies with occurrence frequency of over 1.5%, and drew the distribution of the four main topologies along chromosomes. Lastly, we analyzed average weightings for the main possible topologies at chromosomal scales and groups with partitioned recombination rates.
Admixture and Introgression Analyses
To evaluate the contribution of hybridization and ILS on the discordance of topology across the genome, we performed MSCquartets analyses (Rhodes et al. 2021), estimated quartet score (Mirarab and Warnow 2015), and detected the signals for introgression (Solis-Lemus et al. 2017; Wen et al. 2018). Firstly, we carried out quartetTreeTest with the “T3 model” in the MSCquartets package (Rhodes et al. 2021) based on the 19,295 species trees inferred from 10 kb nonoverlapping sliding windows (same as the input file of topology analyses), with the rejection level of 1e−6. The package would generate a series of plots for all four-taxon subsets with a red triangle (reject tree) or blue circle (fail to reject tree). More blue circles lying closer proximity to the centroid represent substantial ILS, while closer to the vertex represent little ILS. Secondly, we used “q option” of ASTRAL (Mirarab and Warnow 2015) to calculate the normalized quartet score (i.e., the proportion of quartet trees in the input trees that are satisfied by the ASTRAL tree) at the whole genome (19,295 species trees), chromosomal levels (ranging from 2,110 to 3,590 trees), and groups with partitioned recombination rate (3,859 trees for each group). The score is a number between zero and one, the lower this number indicates the more discordant the input trees. Thirdly, we performed DFOIL analyses (Pease and Hahn 2015) to detect introgression based on a five-taxon phylogeny (((P1, P2), (P3, P4)), O), where O is the outgroup (Pmic) and P1 to P4 are the ingroups, and the divergence between P3 and P4 should be earlier than that between P1 and P2. DFOIL analysis could distinguish the ancient introgression and post-speciation gene flow, and also estimate the direction of post-speciation gene flow. Here, we applied the software DFOIL to examine the introgression signals in 55,207 individual combinations from 34 species combinations, according to the species tree inferred from low recombination regions. Fourthly, we adopted the Infer_Network_MPL model (Wen et al. 2018) to detect gene flow, according to 19,295 rooted species trees, by setting maximum reticulations of 5. Meanwhile, we applied SNPs2CF (https://github.com/melisaolave/SNPs2CF/) to compute concordance factors from unlinked SNPs of 19 represented individuals (Chi-3, Chi-7, Nip-1, Nub-0, Nub-1, Pen-3, Pen-7, Nil-0, Nil-4, Dal-1, Dal-2, Iin-0, Iin-1, Vir-0, Vir-9, Ves-1, Ves-11, Man-2, and Man-3), which could greatly reduce the computational complexity. Then, we compared and estimated the hypothetical hybridization with hman setting from 0 to 10, by using PhyloNetworks (Solis-Lemus et al. 2017).
To further detect and characterize introgression across the Fragaria genome, we used Dsuite (Malinsky et al. 2021) and ABBABABAwindows.py script (Martin et al. 2015). Firstly, we applied the Dmin model of program Dtrios in Dsuite (Malinsky et al. 2021) to perform the ABBA-BABA test and calculate the overall D statistic and associate P value for 120 (.) four-taxa trios, 95 trios of which had adjusted P value < 0.01, and were defined as trios with significant introgression signals. Furthermore, we also followed the formula, fhom = S (P1, P2, P3, O)/S (P1, P3, P3, O) = (ABBA1-BABA1)/(ABBA2-BABA2) (Martin et al. 2015; Pulido-Santacruz et al. 2020), to calculate fhom value for 95 trios with significant introgression signals. Then, we counted the fdM values in each 10 kb nonoverlapping sliding window for each of the 95 trios by using ABBABABAwindows.py script (Martin et al. 2015), removing the windows containing less than 10 SNPs. We defined putatively introgression regions (pIR) as windows with the highest x% of fdM values, where x was determined for each trio by the fhom estimate, and then calculated an introgression index for each window as the proportion of trio with pIRs. The windows that have the top approximately 5% highest introgression index were defined as high introgression regions (the threshold value is 0.1578 in this study), while the windows having an introgression index equal to 0 were defined as low introgression regions.
Similarly, we used VCFtools (Danecek et al. 2011) to calculate the genetic differentiation (FST) value for each 10 kb nonoverlapping sliding window of 21 (.) combinations that were pairwise of the seven wild species containing at least three individuals (i.e., F. chinensis, F. nubicola, F. pentaphylla, F. nilgerrensis, F. viridi, F. vesca, and F. mandshurica). For each combination, we marked the window with the top 5% of FST value as the FST outlier and defined the FST index of each window as the proportion of the combination with the FST outlier. We then ranked the windows by FST index and classified the windows with the highest approximately 5% FST index (>0.3333) as high FST regions (i.e., introgression barrier regions). Lastly, we analyzed the average percentage of windows with high introgression and low introgression/introgression barrier regions, binned according to their recombination rate and gene density.
Detection of Selective Sweeps
We used a combination of a haplotype-based method (selscan; Szpiech and Hernandez 2014) and a composite evaluation approach (RAiSD; Alachiotis and Pavlidis 2018) to screen the regions under positive selection for the seven Fragaria species containing at least three individuals (like FST analysis), following our previous pipeline (Hu et al. 2022). Firstly, we calculated raw XP-nSL scores for each chromosome on all 42 comparisons of seven species by using selscan v1.3.0 (Szpiech and Hernandez 2014) and then normalized the XP-nSL value across 10 kb nonoverlapping windows using norm v1.3.0 (Szpiech and Hernandez 2014). For each species, we collected the windows with the top 1% extreme score, setting either one of six other species as a reference, respectively. After removing redundancy, the remaining windows were used for downstream analysis. Secondly, we applied RAiSD (Alachiotis and Pavlidis 2018) to generate raw μ value across the genome for each of the seven species and then calculated the average μ value for 10 kb nonoverlapping windows. Here, we defined the windows with the highest 5% values as candidate regions. Finally, the overlap regions from the above two approaches were regarded as the highly confident selective sweep regions for each wild diploid strawberry. We compared the shared and specific windows under selective sweep regions of the seven Fragaria species and drew the Upset diagram by TBtools (Chen et al. 2020).
GO Enrichment
We applied the topGO script (http://bioconductor.uib.no/2.7/bioc/vignettes/topGO/), following our previous pipeline (Feng et al. 2020), to analyze the gene ontology enrichment of genes in the high introgression and low introgression/introgression barrier regions across the wild diploid strawberries, as well as genes from the selective sweep regions of the seven Fragaria species, setting all F. vesca genes as background. Only the lowest-level GO terms in MF and BP, with a P value <0.05, were retained as enriched terms, whereas the P value was calculated according to a “classic” algorithm under Fisher's test.
Identification of SLFs and S-RNases in F. vesca Genome
Following the pipeline of Akagi et al. (2016), we scanned F-box genes in F. vesca genome (v4.0) by using BLASTP against SFB gene (AB111521) or SLF2 gene (AB280954) from Prunus avium with the cutoff of 1e−19 and further removed the genes that were absent of the F-box domain (PF00646.33) by Pfam search (Mistry and Finn 2007). Furthermore, we used IQ-TREE (Nguyen et al. 2015) to construct the ML tree of SLF/SLF-like F-box genes from the strawberry genome and partial F-box genes from peach (prefix of “ppa’), cherry, etc., based on CDS sequences guided by the alignment of protein sequences by using the PAL2NAL script (Suyama et al. 2006). Finally, we classified these F-box genes into Clades A, B, S, and Others, according to the standard of Akagi et al. (2016), whereas the genes in Clade S were considered as SLFs that contribute to pollen S function. Similarly, we followed the pipeline of Morimoto et al. (2015) to identify S-RNase-like genes according to S3-RNase gene (AB010306) from P. avium and ribonuclease T2 Pfam domain (PF00445.18). According to the standard of Morimoto et al. (2015), the members in Class III were regarded as candidate S-RNases which may have pistil S function. The region containing at least a single pistil S determinant S-RNase and several pollen S-related SLFs in tandem duplications was defined as the S locus in F. vesca.
Estimation of Ancestral Mating System Type in Wild Diploid Strawberries
We simulated the probability of ancestral mating system states at each ancestral node using BayesTraits implemented in RASP 4 (Yu et al. 2020), according to phylogenetic relationship at low recombination regions with divergence time, and current mating systems (SI or SC) of the ten wild diploid strawberries described by Njuguna et al. (2013). With the ape package in phytools (Revell 2012), two models (symmetric and asymmetric transition rates) in ML analyses were compared to determine whether the gain rate (transition rate from SC to SI) is significantly different from the loss rate (transition rate from SI to SC).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32270396 and 32072533), Youth Innovation Promotion Association, Chinese Academy of Sciences (2021348), and South China Botanical Garden, Chinese Academy of Sciences (QNXM-07).
Contributor Information
Chao Feng, Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China; Guangdong Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China.
Jing Wang, Institute of Pomology, Jiangsu Academy of Agricultural Sciences/Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, Nanjing, China.
Aaron Liston, Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR.
Ming Kang, Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China; Guangdong Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The genome resequencing data has been deposited to the SRA database at NCBI under BioProject PRJNA879993.
References
- Akagi T, Henry IM, Morimoto T, Tao R. 2016. Insights into the Prunus-specific S-RNase-based self-incompatibility system from a genome-wide analysis of the evolutionary radiation of S locus-related F-box genes. Plant Cell Physiol. 57(6):1281–1294. [DOI] [PubMed] [Google Scholar]
- Alachiotis N, Pavlidis P. 2018. RAiSD detects positive selection based on multiple signatures of a selective sweep and SNP vectors. Commun Biol. 1:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MD, Gruber C, Flokova K, Miersch O, Strnad M, Novak O, Wasternack C, Hause B. 2016. The recently identified isoleucine conjugate of cis-12-oxo-phytodienoic acid is partially active in cis-12-oxo-phytodienoic acid-specific gene expression of Arabidopsis thaliana. PLoS One 11(9):e0162829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkani J, Rees DJG. 2016. A comprehensive study of molecular evolution at the self-incompatibility locus of Rosaceae. J Mol Evol. 82(2–3):128–145. [DOI] [PubMed] [Google Scholar]
- Bošković RI, Sargent DJ, Tobutt KR. 2010. Genetic evidence that two independent S-loci control RNase-based self-incompatibility in diploid strawberry. J Exp Bot. 61(3):755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringhurst RS, Khan DA. 1963. Natural pentaploid Fragaria chiloensis-F. vesca hybrids in coastal California and their significance in polyploid Fragaria evolution. Am J Bot. 50(7):658–661. [Google Scholar]
- Bringhurst RS, Senanayake YDA. 1966. The evolutionary significance of natural Fragaria chiloensis × F. vesca hybrids resulting from unreduced gametes. Am J Bot. 53(10):1000–1006. [Google Scholar]
- Browning BL, Browning SR. 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 84(2):210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R. 2017. Dissecting differentiation landscapes: a linked selection's perspective. J Evol Biol. 30(8):1501–1505. [DOI] [PubMed] [Google Scholar]
- Buti M, Moretto M, Barghini E, Mascagni F, Natali L, Brilli M, Lomsadze A, Sonego P, Giongo L, Alonge M, et al. 2018. The genome sequence and transcriptome of Potentilla micrantha and their comparison to Fragaria vesca (the woodland strawberry). Gigascience 7(4):giy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee E, Gates D, Lorant A, Perkins MT, Coop G, Ross-Ibarra J. 2021. Selective sorting of ancestral introgression in maize and teosinte along an elevational cline. PLoS Genet. 17(10):e1009810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 13(8):1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen W, Wan H, Liu F, Du HY, Zhang CJ, Fan WS, Zhu AD. 2022. Rapid evolution of T2/S-RNase genes in Fragaria linked to multiple transitions from self-incompatibility to self-compatibility. Plant Divers. 10.1016/j.pld.2022.04.003. [DOI] [PMC free article] [PubMed]
- Chifman J, Kubatko L. 2014. Quartet inference from SNP data under the coalescent. Bioinformatics 30(23):3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmas MJ, Jones JC, Olsson A, Wallerman O, Bunikis I, Kierczak M, Peona V, Whitley KM, Larva T, Suh A, et al. 2021. Genetic barriers to historical gene flow between cryptic species of alpine bumblebees revealed by comparative population genomics. Mol Biol Evol. 38(8):3126–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra KK, Walters JR, Briscoe AD, Davey JW, Whibley A, Nadeau NJ, Zimin AV, Hughes DST, Ferguson LC, Martin SH, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487(7405):94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 24(6):332–340. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du JK, Ge CF, Li TT, Wang SH, Gao ZH, Sassa H, Qiao YS. 2021. Molecular characteristics of S-RNase alleles as the determinant of self-incompatibility in the style of Fragaria viridis. Hortic Res. 8(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, Vonholdt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 4(2):359–361. [Google Scholar]
- Edelman NB, Frandsen PB, Miyagi M, Clavijo B, Davey J, Dikow RB, Garcia-Accinelli G, Van Belleghem SM, Patterson N, Neafsey DE, et al. 2019. Genomic architecture and introgression shape a butterfly radiation. Science 366(6465):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman NB, Mallet J. 2021. Prevalence and adaptive impact of introgression. Annu Rev Genet. 55:265–283. [DOI] [PubMed] [Google Scholar]
- Edger PP, Poorten TJ, VanBuren R, Hardigan MA, Colle M, McKain MR, Smith RD, Teresi SJ, Nelson ADL, Wai CM, et al. 2019. Origin and evolution of the octoploid strawberry genome. Nat Genet. 51(3):541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, VanBuren R, Colle M, Poorten TJ, Wai CM, Niederhuth CE, Alger EI, Ou S, Acharya CB, Wang J, et al. 2017. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7(2):gix124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV. 2009. Is a new and general theory of molecular systematics emerging? Evolution 63(1):1–19. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Xi ZX, Janke A, Faircloth BC, McCormack JE, Glenn TC, Zhong BJ, Wu SY, Lemmon EM, Lemmon AR, et al. 2016. Implementing and testing the multispecies coalescent model: a valuable paradigm for phylogenomics. Mol Phylogenet Evol. 94(A):447–462. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Smeds L, Burri R, Olason PI, Backstrom N, Kawakami T, Kunstner A, Makinen H, Nadachowska-Brzyska K, Qvarnstrom A, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491(7426):756–760. [DOI] [PubMed] [Google Scholar]
- Fan WS, Liu F, Jia QY, Du HY, Chen W, Ruan JW, Lei JJ, Li DZ, Mower JP, Zhu AD. 2022. Fragaria mitogenomes evolve rapidly in structure but slowly in sequence and incur frequent multinucleotide mutations mediated by micro-inversions. New Phytol. 236(2):745–759. [DOI] [PubMed] [Google Scholar]
- Feng C, Wang J, Harris AJ, Folta KM, Zhao MZ, Kang M. 2021. Tracing the diploid ancestry of the cultivated octoploid strawberry. Mol Biol Evol. 38(2):478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Wang J, Wu L, Kong H, Yang L, Feng C, Wang K, Rausher M, Kang M. 2020. The genome of a cave plant, Primulina huaijiensis, provides insights into adaptation to limestone karst habitats. New Phytol. 227(4):1249–1263. [DOI] [PubMed] [Google Scholar]
- Feng C, Xu MZ, Feng C, von Wettberg EJB, Kang M. 2017. The complete chloroplast genome of Primulina and two novel strategies for development of high polymorphic loci for population genetic and phylogenetic studies. BMC Evol Biol. 17:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MS, Jones MR, Callahan CM, Farelo L, Tolesa Z, Suchentrunk F, Boursot P, Mills LS, Alves PC, Good JM, et al. 2021. The legacy of recurrent introgression during the radiation of hares. Syst Biol. 70(3):593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer MM, Good-Avila SV. 2007. Macrophylogenetic analyses of the gain and loss of self-incompatibility in the Asteraceae. New phytol. 173(2):401–414. [DOI] [PubMed] [Google Scholar]
- Finger N, Farleigh K, Bracken J, Leache A, Francois O, Yang Z, Flouri T, Charran T, Jezkova T, Williams D, et al. 2022. Genome-scale data reveal deep lineage divergence and a complex demographic history in the Texas horned lizard (Phrynosoma cornutum) throughout the Southwestern and Central USA. Genome Biol Evol. 14(1):evab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Davis TM. 2006. Strawberry genes and genomics. Crit Rev Plant Sci. 25(5):399–415. [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347(6217):1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Ming C, Hu WJ, Li HP. 2016. New software for the fast estimation of population recombination rates (FastEPRR) in the genomic era. G3 (Bethesda). 6(6):1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin JAP, Hibbins MS, Moyle LC. 2020. Assessing biological factors affecting postspeciation introgression. Evol Lett. 4(2):137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennelly LM, Habib B, Modi S, Rueness EK, Gaubert P, Sacks BN. 2021. Ancient divergence of Indian and Tibetan wolves revealed by recombination-aware phylogenomics. Mol Ecol. 30(24):6687–6700. [DOI] [PubMed] [Google Scholar]
- Hiscock SJ, McInnis SM. 2003. The diversity of self-incompatibility systems in flowering plants. Plant Biol. 5(1):23–32. [Google Scholar]
- Hu YX, Feng C, Yang LH, Edger P, Kang M. 2022. Genomic population structure and local adaptation of the wild strawberry Fragaria nilgerrensis. Hortic Res. 9:uhab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer KE, Hancock J. 2009. Strawberry genomics: botanical history, cultivation, traditional breeding, and new technologies. In: Folta K, Gardiner S, editors. Genetics and genomics of Rosaceae. New York: Springer. p. 413–435. [Google Scholar]
- Igic B, Kohn JR. 2001. Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci U S A. 98(23):13167–13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B, Lande R, Kohn JR. 2008. Loss of self-incompatibility and its evolutionary consequences. Int J Plant Sci. 29(9):521–530. [Google Scholar]
- Irisarri I, Singh P, Koblmuller S, Torres-Dowdall J, Henning F, Franchini P, Fischer C, Lemmon AR, Lemmon EM, Thallinger GG, et al. 2018. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat Commun. 9:3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806. [DOI] [PubMed] [Google Scholar]
- Jones MR, Mills LS, Alves PC, Callahan CM, Alves JM, Lafferty DJR, Jiggins FM, Jensen JD, Melo-Ferreira J, Good JM. 2018. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360(6395):1355–1358. [DOI] [PubMed] [Google Scholar]
- Kakui H, Kato M, Ushijima K, Kitaguchi M, Kato S, Sassa H. 2011. Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). Plant J. 68(6):1028–1038. [DOI] [PubMed] [Google Scholar]
- Kamneva OK, Syring J, Liston A, Rosenberg NA. 2017. Evaluating allopolyploid origins in strawberries (Fragaria) using haplotypes generated from target capture sequencing. BMC Evol Biol. 17(1):180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Huber CD, Lohmueller KE. 2018. Deleterious variation shapes the genomic landscape of introgression. PLoS Genet. 14(10):e1007741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak KM, Joron M, McMillan WO, Jiggins CD. 2021. Rampant genome-wide admixture across the Heliconius radiation. Genome Biol Evol. 13(7):evab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei JJ, Li YH, Du GD, Dai HP, Deng MQ. 2005. A natural pentaploid strawberry genotype from the Changbai Mountains in northeast China. HortScience. 40(5):1194–1195. [Google Scholar]
- Lei JJ, Xue L, Guo RX, Dai HP. 2017. The Fragaria species native to China and their geographical distribution. Acta Hortic. 1156:37–46. [Google Scholar]
- Leitwein M, Cayuela H, Ferchaud AL, Normandeau E, Gagnaire PA, Bernatchez L. 2019. The role of recombination on genome-wide patterns of local ancestry exemplified by supplemented brook charr populations. Mol Ecol. 28(21):4755–4769. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Figueiro H, Eizirik E, Murphy WJ. 2019. Recombination-aware phylogenomics reveals the structured genomic landscape of hybridizing cat species. Mol Biol Evol. 36(10):2111–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Cronn R, Ashman T-L. 2014. Fragaria: a genus with deep historical roots and ripe for evolutionary and ecological insights. Am J Bot. 101(10):1686–1699. [DOI] [PubMed] [Google Scholar]
- Liston A, Wei N, Tennessen JA, Li J, Dong M, Ashman T-L. 2020. Revisiting the origin of octoploid strawberry. Nat Genet. 52(1):2–4. [DOI] [PubMed] [Google Scholar]
- Liu SY, Zhang L, Sang YP, Lai Q, Zhang XX, Jia CF, Long ZQ, Wu JL, Ma T, Mao KS, et al. 2022. Demographic history and natural selection shape patterns of deleterious mutation load and barriers to introgression across Populus genome. Mol Biol Evol. 39(2):msac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Matschiner M, Svardal H. 2021. Dsuite – Fast D-statistics and related admixture evidence from VCF files. Mol Ecol Resour. 21(2):584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Svardal H, Tyers AM, Miska EA, Genner MJ, Turner GF, Durbin R. 2018. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat Ecol Evol. 2(12):1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. 2005. Hybridization as an invasion of the genome. Trends Ecol Evol. 20(5):229–237. [DOI] [PubMed] [Google Scholar]
- Mallet J, Besansky N, Hahn MW. 2016. How reticulated are species? Bioessays. 38(2):140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD. 2015. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 32(1):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Salazar C, Jiggins CD. 2019. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17(2):e2006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Jiggins CD. 2017. Interpreting the genomic landscape of introgression. Curr Opin Genet Dev. 47:69–74. [DOI] [PubMed] [Google Scholar]
- Martin SH, Van Belleghem SM. 2017. Exploring evolutionary relationships across the genome using topology weighting. Genetics 206(1):429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Bayzid MS, Warnow T. 2016. Evaluating summary methods for multilocus species tree estimation in the presence of incomplete lineage sorting. Syst Biol. 65(3):366–380. [DOI] [PubMed] [Google Scholar]
- Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31(12):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Finn R. 2007. Pfam: a domain-centric method for analyzing proteins and proteomes. Methods Mol Biol. 396:43–58. [DOI] [PubMed] [Google Scholar]
- Morales-Cruz A, Aguirre-Liguori JA, Zhou YF, Minio A, Riaz S, Walker AM, Cantu D, Gaut BS. 2021. Introgression among North American wild grapes (Vitis) fuels biotic and abiotic adaptation. Genome Biol. 22(1):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran BM, Payne C, Langdon Q, Powell DL, Brandvain Y, Schumer M. 2021. The genomic consequences of hybridization. eLife. 10:e69016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto T, Akagi T, Tao R. 2015. Evolutionary analysis of genes for S-RNase-based self-incompatibility reveals S locus duplications in the ancestral Rosaceae. Horticult J. 84(3):233–242. [Google Scholar]
- Nachman MW, Payseur BA. 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos Trans R Soc Lond B Biol Sci. 367(1587):409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E. 2017. Tracing the peopling of the world through genomics. Nature 541(7637):302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njuguna W, Liston A, Cronn R, Ashman T-L, Bassil N. 2013. Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Mol Phylogenet Evol. 66(1):17–29. [DOI] [PubMed] [Google Scholar]
- Owens GL, Huang K, Todesco M, Rieseberg LH.. 2022. Re-evaluating homoploid reticulate evolution in the annual sunflowers. bioRxiv. doi: 10.1101/2022.10.14.512273. [DOI]
- Payseur BA, Rieseberg LH. 2016. A genomic perspective on hybridization and speciation. Mol Ecol. 25(11):2337–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Haak DC, Hahn MW, Moyle LC. 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14(2):e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Hahn MW. 2013. More accurate phylogenies inferred from low-recombination regions in the presence of incomplete lineage sorting. Evolution 67(8):2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Hahn MW. 2015. Detection and polarization of introgression in a five-taxon phylogeny. Syst Biol. 64(4):651–662. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephans M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Santacruz P, Aleixo A, Weir JT. 2020. Genomic data reveal a protracted window of introgression during the diversification of a neotropical woodcreeper radiation. Evolution 74(5):842–858. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Edger PP, Xue L, Qiong L, Lu J, Zhang YC, Cao Q, Yocca AE, Platts AE, Knapp SJ, et al. 2021. Evolutionary history and pan-genome dynamics of strawberry (Fragaria spp). Proc Natl Acad Sci U S A. 118(45):e2105431118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Xue L, Wang Q, Sun H, Zhong Y, Huang J, Lei J, Zhang T. 2016. Comparative transcriptomics of strawberries (Fragaria spp.) provides insights into evolutionary patterns. Front Plant Sci. 7:1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. [Google Scholar]
- Rhodes JA, Banos H, Mitchell JD, Allman ES. 2021. MSCquartets 1.0: quartet methods for species trees and networks under the multispecies coalescent model in R. Bioinformatics 37(12):1766–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. R8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19(2):301–684. [DOI] [PubMed] [Google Scholar]
- Schumer M, Xu CL, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, Sankararaman S, Andolfatto P, Rosenthal GG, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 360(6389):656–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Hittinger CT, Rokas A. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 1(5):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman-Broyles S, Nasrallah JB. 2008. Self-incompatibility and evolution of mating systems in the Brassicaceae. In: Franklin-Tong VE, editor. Self-Incompatibility in flowering plants: evolution, diversity, and mechanisms. Berlin: Springer-Verlag. pp. 123–147. [Google Scholar]
- Shi C, Yang Z. 2018. Coalescent-based analyses of genomic sequence data provide a robust resolution of phylogenetic relationships among major groups of gibbons. Mol Biol Evol. 35(1):159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal S, Derryberry GE, Bravo GA, Derryberry EP, Brumfield RT, Harvey MG. 2021. The dynamics of introgression across an avian radiation. Evol Lett. 5(6):568–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Lemus C, Bastide P, Ane C. 2017. Phylonetworks: a package for phylogenetic networks. Mol Biol Evol. 34(12):3292–3298. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2009. The role of hybridization in plant speciation. Annu Rev Plant Biol. 60:561–588. [DOI] [PubMed] [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP. 2005. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell. 17(1):37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankowski S, Streisfeld MA. 2015. Introgressive hybridization facilitates adaptive divergence in a recent radiation of monkeyflowers. Proc R Soc B. 282(1814):154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt G. 1999. Systematics and geographic distribution of the American strawberry species: taxonomic studies in the genus Fragaria (Rosaceae: Potentilleae). Berkeley, University of California Press. [Google Scholar]
- Suvorov A, Kim BY, Wang J, Armstrong EE, Peede D, D′Agostino ER, Price DK, Wadell PJ, Lang M, Courtier-Orgogozo V, et al. 2022. Widespread introgression across a phylogeny of 155 Drosophila genomes. Curr Biol. 32(1):111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(SI):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD. 2014. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 31(10):2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Larson EL. 2019. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat Ecol Evol. 3(2):170–177. [DOI] [PubMed] [Google Scholar]
- Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AF, Tao RT. 2004. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 39(4):573–586. [DOI] [PubMed] [Google Scholar]
- Wen DQ, Yu Y, Zhu JF, Nakhleh L. 2018. Inferring phylogenetic networks using PhyloNet. Syst Biol. 67(4):735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Yang ZH. 2016. Challenges in species tree estimation under the multispecies coalescent model. Genetics 204(4):1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Davis TM. 2017. A newperspective on polyploid Fragaria (strawberry) genome composition based on large-scale, multi-locus phylogenetic analysis. Genome Biol Evol. 9(12):3433–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. 2011. GCTA: a tool for genome wide complex trait analysis. Am J Hum Genet. 88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Blair C, He XJ. 2020. RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol Biol Evol. 37(2):604–606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome resequencing data has been deposited to the SRA database at NCBI under BioProject PRJNA879993.