Abstract
Large genomic data sets are becoming the new normal in phylogenetic research, but the identification of true orthologous genes and the exclusion of problematic paralogs is still challenging when applying commonly used sequencing methods such as target enrichment. Here, we compared conventional ortholog detection using OrthoFinder with ortholog detection through genomic synteny in a data set of 11 representative diploid Brassicaceae whole-genome sequences spanning the entire phylogenetic space. Then, we evaluated the resulting gene sets regarding gene number, functional annotation, and gene and species tree resolution. Finally, we used the syntenic gene sets for comparative genomics and ancestral genome analysis. The use of synteny resulted in considerably more orthologs and also allowed us to reliably identify paralogs. Surprisingly, we did not detect notable differences between species trees reconstructed from syntenic orthologs when compared with other gene sets, including the Angiosperms353 set and a Brassicaceae-specific target enrichment gene set. However, the synteny data set comprised a multitude of gene functions, strongly suggesting that this method of marker selection for phylogenomics is suitable for studies that value downstream gene function analysis, gene interaction, and network studies. Finally, we present the first ancestral genome reconstruction for the Core Brassicaceae which predating the Brassicaceae lineage diversification ∼25 million years ago.
Keywords: ancestral genome, Angiosperms353, OrthoFinder, ortholog, paralog, phylogenomics
Significance.
The identification of orthologs, homologous genes originated through speciation, has become a crucial first step in phylogenomic studies. Many phylogenomic data sets are therefore restricted to highly conserved single-copy genes, limiting their use for further downstream analyses. We used synteny to identify reliable orthologs across 11 diploid Brassicaceae species covering all major evolutionary lineages and compared them to orthologs identified with conventional methods. The obtained phylogenetic trees showed some differences between sets, highlighting the need to carefully select genes for species tree reconstruction. Our syntenic gene sets comprised a large number of genes with diverse gene functions, making this approach suitable for studies combining phylogenomics with comparative genomics and trait evolution.
Introduction
Sequencing data used for molecular phylogenetics has increased manifold over the last 30 years. Recent studies often use target enrichment, a cost-efficient method to simultaneously obtain nucleotide sequences of multiple genes from many taxa (Cronn et al. 2012; Weitemier et al. 2014). The bait sets for target enrichment may contain hundreds of genes and can be used for diverse sets of taxa, such as for a specific group like the Brassicaceae (Nikolov et al. 2019) or for larger groups such as all angiosperms (Johnson et al. 2019). However, orthology, not paralogy of each gene (i.e., the homologous gene copy in each taxon is related by linear decent [Fitch 1970]), is required for many downstream analyses to obtain an accurate species tree, for example, using ASTRAL (Zhang et al. 2018). Apart from obtaining an incorrect tree topology, the inclusion of paralogs for species tree inference can also lead to erroneous older divergence time estimates (Siu-Ting et al. 2019; Zhou et al. 2022). Orthology is not trivial to achieve in target enrichment studies. Software such as orthoMCL (Li et al. 2003) or OrthoFinder (Emms and Kelly 2019) is often used to cluster genes from a few available whole-genome sequences or transcriptomes in the study species into groups of presumed orthologous genes (orthogroups) based on sequence similarity, and subsequently single-copy genes are selected for phylogenetic reconstruction (Johnson et al. 2019; One Thousand Plant Transcriptomes Initiative 2019; Baker et al. 2022). The orthology of the obtained sequences is then inferred from the nucleotide or amino acid sequence, ideally with additional filtering criteria (e.g., based on sequence identity and length) to exclude paralogs (Johnson et al. 2016). However, few studies assess whether the orthogroups reliably only contain orthologs and no paralogs and what the effects of mixed alignments on inferred species trees are.
Gene and genome duplications, gene loss, and variable rates of molecular evolution are all potential sources of error for ortholog detection. Error rates are highest for genes with high evolutionary rates, in particular for those with high between site rate heterogeneity, leading to the clustering of many incomplete orthogroups (Natsidis et al. 2021). Single-copy gene families are thus predominantly comprised of genes with conserved nucleotide sequence and gene function (De Smet et al. 2013; Li et al. 2016) such as those involved in photosynthesis and cell cycle, whereas genes involved in local adaptation, such as those related to response to stimulus or signal transduction, are underrepresented (Li et al. 2017). This limits the subsequent use of phylogenomic target enrichment data sets. However, more and more research projects aim to investigate trait evolution in a phylogenomic context. For such studies, larger gene sets of reliable orthologs covering a wider range of gene functions would be highly advantageous, so that they can be used both to reconstruct the phylogenetic tree and subsequently identify the molecular basis of trait differences between taxa.
When whole-genome sequences are available, additional information apart from nucleotide or amino acid sequence can be used to detect orthologous genes. Synteny, the collinearity of genes across genomes, could help in identifying reliable orthologs, because conserved gene order is expected between orthologous blocks. Additionally, many paralogs are derived from transposition to different locations in the genome. Notable exceptions are paralogous blocks that originated from ancient whole-genome duplication (WGD), a phenomenon common in plants (e.g., Bowers et al. 2003; Vanneste et al. 2014; One Thousand Plant Transcriptomes Initiative 2019). The duplicated genes may subsequently undergo fractionation, through which one or the other gene copy is lost, or subfunctionalization, which may be linked to relaxed selection (Cheng et al. 2018). Both may hinder the correct identification of one-to-one orthologs, since a WGD-derived paralog may be wrongly added to a group of orthologous genes following the loss of the respective ortholog, or clustering may add multiple paralogs from some taxa. However, syntenic paralog blocks derived from WGDs can easily be identified based on synonymous substitution rate (Ks).
The Brassicaceae are a relatively large angiosperm family with ∼4,000 species in 350 genera (Walden, German, et al. 2020). With genome sequences available for the model plant Arabidopsis thaliana and many important crop species such as cabbages and rapeseed, the family has become a model system for genome evolution in recent years. However, the phylogeny of this family has so far proved difficult to resolve despite considerable efforts. The branching order of the main lineages differs between plastid-based phylogenies (Walden, German, et al. 2020; Hendriks et al. 2022) and phylogenies based on nuclear genes (Huang et al. 2016; Kiefer et al. 2019; Nikolov et al. 2019; Hendriks et al. 2022), and support values for deeper nodes can be low. All Brassicaceae share the At-α WGD (Bowers et al. 2003; Schranz and Mitchell-Olds 2006) that occurred after the divergence from the sister family Cleomaceae ∼40 million years ago (Edger et al. 2015), and many Brassicaceae tribes have undergone additional meso- and neopolyploidizations (Hohmann et al. 2015; Mandáková, Li, et al. 2017). The genome structure of the family has been studied in detail in many species of different tribes, and a system of 22 genomic blocks A-X has been established based on the synteny of the genomes of A. thaliana, Arabidopsis lyrata, Capsella rubella, and Brassica rapa (Schranz et al. 2006; Lysak et al. 2016). With many genome sequences now available, the Brassicaceae provide an ideal study system to investigate whether the use of synteny information for ortholog identification could be beneficial for future phylogenomic studies.
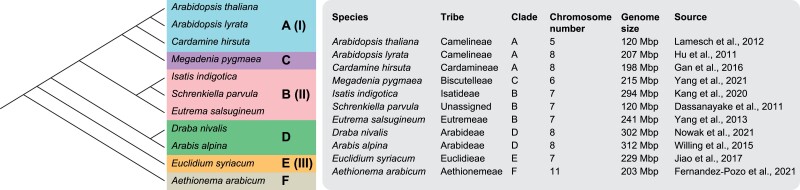
We selected 11 diploid Brassicaceae species with whole-genome sequences available covering all major evolutionary lineages (fig. 1). We compared orthologs identified through synteny with single-copy orthogroups from OrthoFinder regarding the total number of genes identified, bootstrap support, ASTRAL quartet scores, and the resulting species tree topology. Furthermore, we assessed differences between both methods using subsets of available target enrichment gene sets for angiosperms (Johnson et al. 2019) and Brassicaceae (Nikolov et al. 2019). We evaluated the use of larger gene sets for studying trait evolution by comparing the number of gene ontology (GO) terms and protein classes found in every gene set. Finally, we showed that the usefulness of synteny does not end with ortholog detection for phylogenomics and reconstructed ancestral genomes at nine crucial nodes in the evolutionary history of the Brassicaceae family.
Fig. 1.
Brassicaceae species included in the study. The phylogenetic tree following Huang et al. (2016) with clades (A–F) (and lineage names I–III according to Walden, German et al. 2020) is given on the left. Placement of Megadenia as sister to clade A followed Guo et al. (2021). Information on genome sequences for the studied species, including tribe and clade, chromosome number, genome size of the genome sequence, and citation for the genome sequence are given in the right panel.
Results
Syntenic Mapping Reliably Identifies Many Orthologous Genes
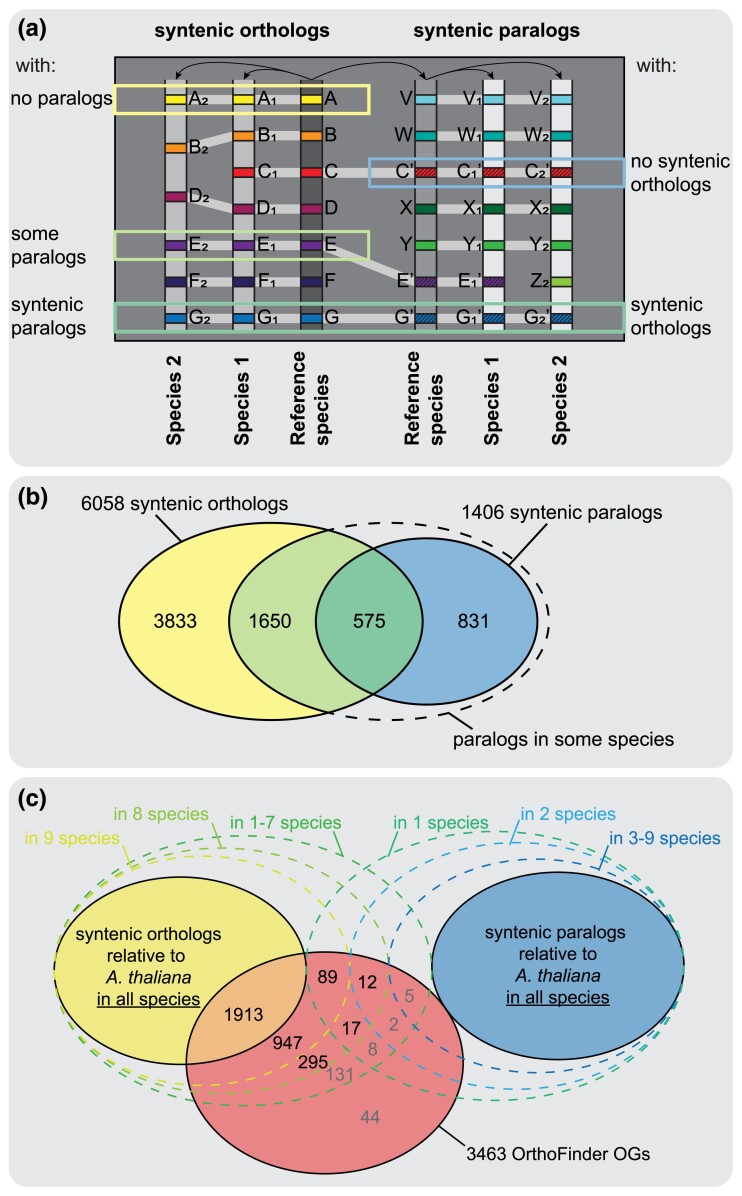
To reliably identify orthologs across the Brassicaceae, we first used syntenic mapping of our selected ten diploid genomes (fig. 1) to A. thaliana. We identified 21,221 genes with an ortholog in synteny in at least one of our ten other study species, and 7,825 genes with a syntenic paralog retained from the At-α WGD. Our further analyses were restricted to the 6,058 orthologs and 1,406 At-α paralogs (hereafter simply termed “paralogs”) that were found in synteny across all taxa, and we split them into five groups illustrated in figure 2a: syntenic orthologs without paralogs (“no paralogs”), syntenic paralogs retained only in some species (“some paralogs”) or syntenic paralogs in all species (“syntenic paralogs”), and syntenic paralogs with syntenic orthologs retained only in some species (“no syntenic orthologs”) or with syntenic orthologs across all species (“syntenic orthologs”). As paralogs were only detected when orthologs were present, there was no group containing paralogs without orthologs. Only 9.5% of orthologs (575 genes, 40.9% of paralogs) were also retained in synteny in paralogs in all species (fig. 2b); an additional 1,650 orthologs had paralogs retained in synteny in some species (27.2% of orthologs); for 3,833 orthologs (63.3%), we did not detect a paralog in any species. Of the 1,406 syntenic paralogs, 831 (59.1%) did not have orthologous copies kept in synteny in all species.
Fig. 2.
Distribution of syntenic orthologs/paralogs and OrthoFinder orthologs. (a) Schematic drawing of syntenic mappings of two species against a reference. The following categories of orthologs/paralogs were analyzed: Syntenic orthologs across all species without syntenic paralogs (“no paralogs”), with syntenic paralogs found in some species (“some paralogs”), or with syntenic paralogs retained in all species (“syntenic paralogs”); syntenic paralogs without syntenic orthologs detected in all species (“no syntenic orthologs”) and syntenic paralogs with a complete set of syntenic orthologs (“syntenic orthologs”). (b) Number of syntenic orthologs/paralogs found across the 11 species of our study displayed as a Venn diagram. (c) Venn diagram showing the number of single-copy OrthoFinder orthogroups found for each combination of orthologs/paralogs in synteny to Arabidopsis thaliana and orthologs not in synteny. Numbers of orthogroups in the subsets we analyzed in detail (i.e., those with syntenic orthologs in at least eight species) are highlighted in black, others are given in gray. Few orthogroups had less than eight syntenic orthologs and are thus summarized in one category.
Many OrthoFinder Orthogroups are Syntenic and few Contain Paralogs
For comparison, we used OrthoFinder to cluster homologous genes from all 11 species by sequence similarity. This resulted in 46,838 orthogroups, 3,463 of which strict single-copy orthogroups having a single gene copy in each taxon, which were the focus of our analysis. Using the information gained from syntenic ortholog and paralog identification, we assessed the composition of these single-copy orthogroups. As each orthogroup contains a single A. thaliana sequence, we searched for the genes from the other ten species among all syntenic orthologs and paralogs identified relative to A. thaliana. Genes that were not found may instead represent other paralogs such as transposed duplicates. No orthogroups were comprised solely of paralogs, but only 1,913 (55%) had syntenic orthologs relative to A. thaliana in all ten species (fig. 2c); this group represents the most reliable orthologs among OrthoFinder orthogroups. Most other orthogroups (1,242 or 35.9%) were comprised of syntenic orthologs in eight or nine species without any detected syntenic At-α paralogs, that is, the other one or two genes in the orthogroup must be homologs located in a different genomic position. The two species in the highest number of orthogroups, not in synteny, were Draba nivalis and Aethionema arabicum (supplementary fig. S1, Supplementary Material online); generally, taxa more closely related to the reference species A. thaliana had more syntenic orthologs. Only 44 orthogroups (1.27%) contained no syntenic genes. The remaining 133 orthogroups (3.8%) were comprised of intermediate numbers of syntenic and other orthologs and paralogs, including paralogs retained from At-α but potentially also transposed duplicates or older paralogs.
We also investigated the overlap between our syntenic orthologs and paralogs with orthogroups from OrthoFinder, including multi-copy gene families. Most syntenic orthologs (5,807 or 95.9%) were contained within only a single orthogroup (supplementary fig. S2, Supplementary Material online), indicating that across the ca. 30 million years of divergence among Brassicaceae, splitting orthologs into multiple orthogroups does not seem to be a widespread problem, at least when only considering genes with conserved genomic position. However, for syntenic paralogs, we detected 562 (40%) across multiple orthogroups, in line with the idea that sequence similarity-based clustering algorithms are more prone to errors for faster evolving gene copies such as paralogs, which may underlie less strict evolutionary pressure. In both groups, syntenic homologs were found across up to four different OrthoFinder orthogroups.
Evaluation of Gene Sets for Phylogenomics
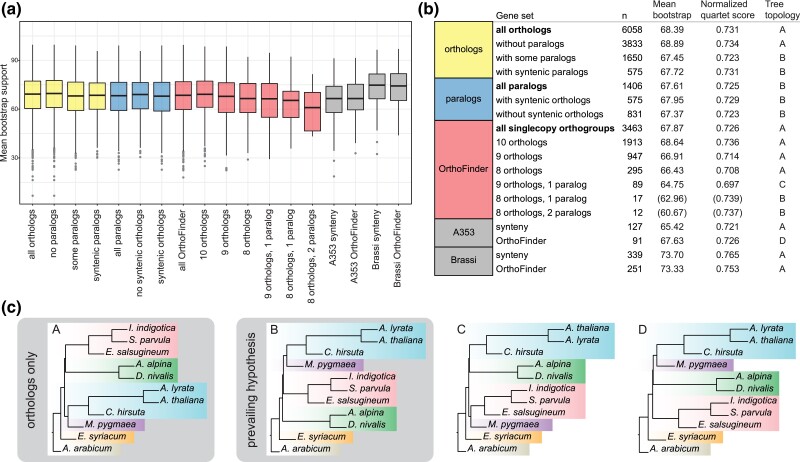
To assess the impact of the two ortholog detection methods on inferred species trees, we first compared mean maximum-likelihood (ML) bootstrap support as a proxy for well-resolved gene trees. In addition to the five synteny gene sets from above, we also analyzed six of the larger sets of OrthoFinder orthogroups with eight, nine, or ten syntenic orthologs and two, one, or no paralogs. The mean bootstrap support among gene sets was in the range of 60.7–68.9%, with the highest value in the set of syntenic orthologs without paralogs, followed by OrthoFinder orthogroups containing only syntenic orthologs (fig. 3aandb). The lowest mean bootstrap support values were found in the sets of OrthoFinder orthologs with only eight orthologs and one or two paralogs; however, the sample size was small for these two sets. The subsets of syntenic and OrthoFinder orthogroups from the two target enrichment gene sets showed contrasting patterns: Gene trees from the Angiosperms353 set had relatively low mean bootstrap support (65.4% and 67.6%, respectively), whereas support was very high among gene trees from the Brassicaceae set (73.7% and 73.3%, respectively). The latter is likely due to the implementation of stringent filtering criteria.
Fig. 3.
Evaluation of phylogenetic trees reconstructed from syntenic orthologs/paralogs and OrthoFinder orthogroups. (a) Boxplot with mean bootstrap support for all trees of the respective gene sets. (b) Information on gene set size, mean bootstrap support, normalized quartet score of the ASTRAL analysis against the species tree and the four different tree topologies (A–D) identified when running ASTRAL without constraint. (c) The four tree topologies (A–D).
We also tested the effect of orthogroup selection on inferred species trees. First, we reconstructed species trees using ASTRAL (Zhang et al. 2018) for all gene sets separately. Gene sets with only orthologs showed a different branching order (topology A) from those that included paralogs (topology B, fig. 3bandc). Interestingly, the syntenic ortholog subset of Angiosperms353 and Brassicaceae sets as well as the OrthoFinder subset of the latter also followed topology A, although the use of the entire Brassicaceae gene set in the respective study resulted in topology B (Nikolov et al. 2019), as did the combination of both sets (Hendriks et al. 2022). Additionally, we ran ASTRAL for all gene sets against the Brassicaceae phylogeny following the prevailing species tree hypothesis (also shown in fig. 1) derived from three recent comprehensive phylogenomic studies (Huang et al. 2016; Nikolov et al. 2019; Hendriks et al. 2022) and compared the normalized quartet scores. The scores roughly followed the same trend as mean bootstrap support, with data sets comprised mainly of orthologs having higher values than those comprised of or including paralogs (with the exception of the two smallest gene sets, for which no reliable estimate could be obtained).
Syntenic Orthologs Cover a Large Range of Gene Functions
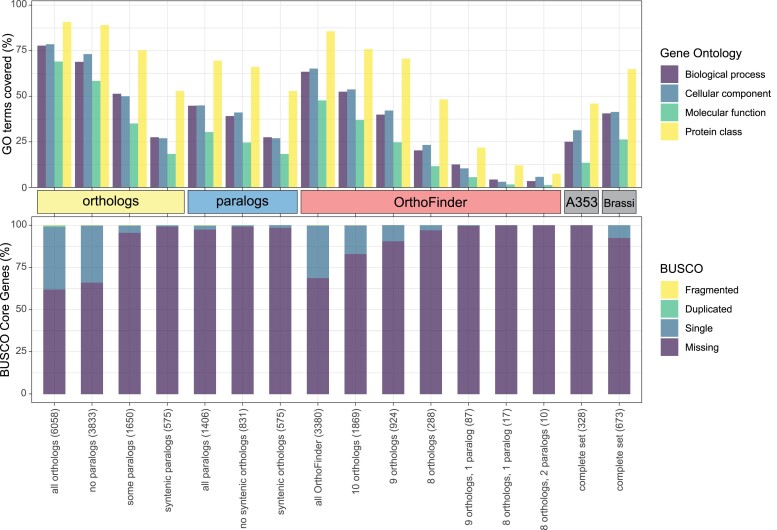
Next, we assessed the coverage of different gene functions among the gene sets using GO terms and Benchmarking Universal Single-Copy Orthologs (BUSCO; Manni et al. 2021). We evaluated the coverage of GO terms and protein classes for each gene set relative to the complete annotation of the A. thaliana genome. The number of GO terms and protein classes in each gene set was related to gene number, with syntenic orthologs having the highest number of biological process, cellular component, and molecular function–related genes as well as the most different protein classes (fig. 4). Completeness of BUSCO showed a similar trend, though to a lesser degree, with the highest completeness in syntenic orthologs (37.9%), followed by syntenic orthologs without paralogs (33.9%), OrthoFinder orthologs (31.2%), and syntenic OrthoFinder orthologs (17%); all other gene sets had >90% missing markers (fig. 4).
Fig. 4.
Fraction of GO terms and protein classes and BUSCO completeness per gene set. GO terms and protein classes were identified using PANTHER, fraction is given relative to the total number of GO terms and protein classes annotated in Arabidopsis thaliana. BUSCO completeness was assessed relative to the BUSCO Brassicales database. Gene set size is given in parentheses.
When analyzing GO overrepresentation, we found that fold enrichment was generally low (<3-fold) for significantly overrepresented terms in larger gene sets, whereas small gene sets (e.g., Angiosperms353 or Brassicaceae) showed enrichment up to 50-fold, matching the expectation that the smaller gene sets comprise genes with a restricted subset of gene functions. The overrepresented terms also supported this idea; for example, plastid-related cellular component terms were highly overrepresented in the Angiosperms353 set but less so in synteny-based gene sets (supplementary fig. S3, Supplementary Material online). In line with previous studies on angiosperm core genes (Li et al. 2016), transcription factor–related terms in the molecular function category as well as the protein class were underrepresented in both the Angiosperms353 and Brassicaceae set, whereas they were slightly overrepresented in most synteny and OrthoFinder sets (supplementary fig. S4, Supplementary Material online). Similarly, different terms related to stimulus response were highly underrepresented in the Angiosperms353 set in the biological process category (supplementary fig. S5, Supplementary Material online). Interestingly, this set was also highly enriched for terms related to tRNA and rRNA processing. The larger gene sets, however, showed underrepresentation of defense related protein classes and MADS-box transcription factors (supplementary fig. S6, Supplementary Material online).
Synteny Data Can Serve as Input for Ancestral Gene Order Reconstruction
Finally, we used the syntenic blocks as input for ancestral genome reconstruction. Our marker-based ancestral genome reconstruction used 336 markers containing blocks of syntenic genes present and in synteny in all 11 species across the Brassicaceae and consisted of 1–82 genes per marker, in total of 3,392 genes. The average number of genes was 10 per marker, spanning 93 kb on average but ranging from 0.5 to 783.0 kb (supplementary fig. S7, Supplementary Material online). As markers were required to be present in all species, the quality of the genome assemblies played a major role in marker coverage. For example, no synteny to A. thaliana could be detected for some sections of the Euclidium syriacum genome with the settings used here, and thus no markers were available for these parts of the genome (supplementary fig. S8, Supplementary Material online). As a result, three of the shorter ABC blocks (G, P, and T) are not covered in our reconstructions.
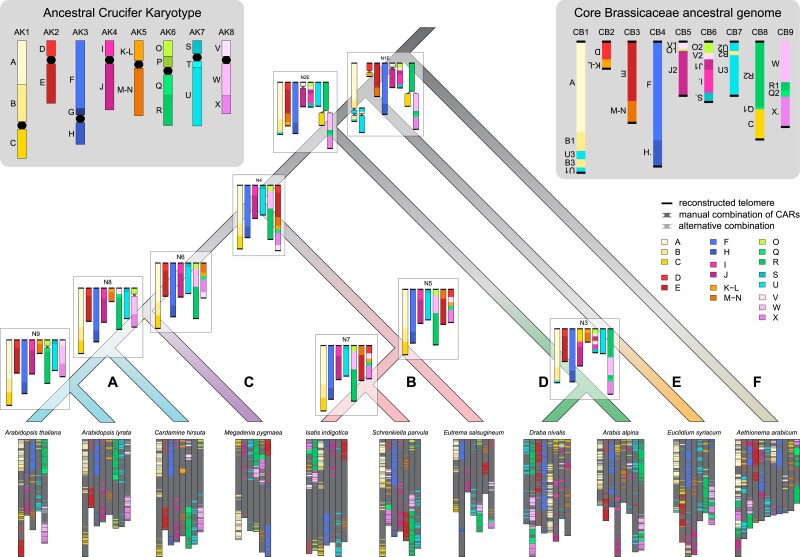
We reconstructed the ancestral marker order for all nine internal nodes (N1–N9) of the Brassicaceae phylogeny, using either E. syriacum or tribe Arabideae as the first diverging lineage (supplementary fig. S9, Supplementary Material online) in the input phylogeny guiding the analysis, resulting in 18 reconstructions in total. As telomere position was inferred in the analysis, the number of chromosomes could be deduced from the number of telomeres present in each reconstruction. Between 12 and 17, telomeres were inferred, indicating chromosome numbers between seven and nine. Six to 15 Contiguous Ancestral Regions (CARs) were reconstructed by the algorithm, and where more CARs than inferred chromosomes were reconstructed, we combined them manually to obtain a likely version of the ancestral genome at the respective node (fig. 5). Reconstructions at nodes N3–N9 were identical for both input phylogenies. At five nodes (N3–N7), the reconstruction only contained CARs with telomeres at both ends, indicating complete chromosomes; at two nodes (N8 and N9), two CARs contained only one telomere and were combined. In contrast, different results were obtained using the two input phylogenies at nodes N1 and N2 (supplementary fig. S10, Supplementary Material online). At node N2, 15 telomeres were reconstructed for both analyses, with 10 CARs for N2E and 11 for N2A. Two possible combinations of CARs were found for N2A, with the lower half of chromosome 7 being either the same as in N2E or the unmatched CAR from N2E. At node N1, 17 telomeres were inferred for N1E, whereas 15 were found for N1A; distinct differences between the two reconstructions included the number of B-U adjacencies found on CB1/7 and the combination of CB2/3 into a single chromosome in N1A.
Fig. 5.
Schematic illustrations of ancestral genomes along the Brassicaceae phylogeny. Ancestral genome reconstruction was guided by an input phylogeny with Euclidium syriacum as the first diverging lineage (see fig. 1). Markers are represented by horizontal bars colored by ABC blocks (Schranz et al., 2006; Lysak et al., 2016), black bars represent telomeres reconstructed by ANGES. CARs were combined manually for reconstructions containing CARs with less than two telomeres. Where more than one chromosome needed manual assembly, CARs were combined to retain marker adjacencies also found in related extant genomes; when multiple combinations were possible, the one found in E. syriacum is displayed in dark gray, whereas the alternative combination is displayed in light gray. Note that the displayed length of chromosomes here is relative to marker number, not nucleotide length or gene number except for the extant genomes. The Ancestral Crucifer Karyotype (ACK; Schranz et al., 2006; Lysak et al., 2016) is shown in the top left corner for comparison; black circles represent centromeres. The Core Brassicaceae ancestral genome as reconstructed here is highlighted in the top right corner.
Ancestral genome reconstructions at more recent nodes closely resembled extant genomes from the respective clades: nodes N3 (Arabideae, clade D), N5/N7 (clade B), and N8/N9 are all identical to extant genomes or showed at most a single rearrangement. Notably, the reconstructed ancestor of Arabidopsis (N9) was identical to the Ancestral Crucifer Karyotype (ACK; fig. 5). Chromosomes AK1, AK3, AK4, and AK7 from the ACK were conserved throughout clades A, B, and C with only minor inversions and are also found in the reconstructed genome of their most recent common ancestor. At the next deeper node, however, only AK3 was conserved as a unit, whereas all others showed some rearrangement or fusion. Common rearrangements between chromosomes involved AK6/8, AK2/5, and AK1/7, and all of them can be seen in the Core Brassicaceae ancestral genome (fig. 5). In this most recent common ancestor of all Brassicaceae but tribe Aethionemeae, 7 of the 22 ABC blocks are broken into 2 pieces, or 4 in the case of block U, and are found on different chromosomes, whereas 12 blocks are intact relative to the ACK. To highlight the rearrangements relative to the ancestral genome of Core Brassicaceae, we show genomic blocks colored by CAR in N1E in supplementary figure S11, Supplementary Material online.
Discussion
Paralogs are homologous gene copies that originate from gene or genome duplication (Fitch 1970). For phylogenomic studies, the inclusion of paralogs in gene trees gives problems that hinder the reconstruction of accurate species trees. Gene and genome duplications often lead to neo- or subfunctionalization of gene function, fueled by the relaxed constraints on sequence mutation that can arise with the existence of a second gene copy (Cheng et al. 2018; Birchler and Yang 2022). However, duplicated genes may subsequently be lost, and this phenomenon occurs even after millions of years (Johri et al. 2022). Phylogenetic trees reconstructed from genes where one taxon is represented by a paralog derived from gene duplication with subsequent loss of the ortholog may not follow the species tree topology, and divergence times estimation from this gene may not be accurate either (Siu-Ting et al. 2019; Zhou et al. 2022). Most species tree reconstruction models thus require and assume orthology, and phylogenomic studies only select orthologs a priori.
Potential paralogs can be excluded at different stages of phylogenomic analysis. Many target enrichment studies begin by selecting only single- or low-copy genes during probe design (e.g., Johnson et al. 2019; Nikolov et al. 2019). However, strict exclusion of all genes with paralogs is rarely possible or limits the number of potential target genes considerably when the taxa of interest have undergone WGDs in their recent history, which is common for plants, and many genes have paralogs (Ufimov et al. 2022). Subsequent filtering for paralogs during analysis of the sequencing data is often conducted, for example, in one of the most commonly used target enrichment analysis pipelines, HybPiper (Johnson et al. 2016), based on sequence length and similarity. Paralogs can also be detected based on phylogenetic trees (Kocot et al. 2013), and the problem of hidden paralogy through early gene duplication followed by late gene loss can be overcome to some extent by filtering gene trees for the presence of known monophyletic clades (Siu-Ting et al. 2019). On the other hand, some more recent approaches explicitly use paralogs for species tree reconstruction. The inclusion of genes consisting of both orthologs and paralogs may result in accurate species tree reconstruction when methods accounting for incomplete lineage sorting (Yan et al. 2021) or gene duplications (Zhang et al. 2020) are used. Alternatively, detection of paralogs can be conducted based on patterns of sequence divergence between alleles and paralogs within a sample to obtain separate alignments for orthologs and paralogs from older WGDs, which may contain phylogenetic signal and thus contribute to overall better phylogenetic resolution at the species tree level by including more gene trees (Ufimov et al. 2022).
In recent years, synteny has emerged as a source of genomic information for comparative genomics (Zhao et al. 2017; Zhao and Schranz 2017, 2019; Conover et al. 2021; Lovell et al. 2022) and phylogenomics (Zhao et al. 2021). Here, we make use of positional information to identify reliable orthologs for species tree reconstruction and evaluate the resulting gene sets with respect to gene numbers, functional annotations, gene and species trees, as well as their further use in comparative genomics. One of the most important advantages of using synteny is that paralogs can be easily identified. In the context of whole-genome sequences, they can either be identified by position (transposed paralogs are located in a different genomic context) or by sequence divergence (paralogs retained from polyploidy events have higher Ks). We thus found a large number of orthologs, representing 22.3% of all genes annotated in A. thaliana, and were also able to reliably identify many At-α paralogs as well as filter out other paralogs. Interestingly, both the Angiosperms353 and Brassicaceae set of target enrichment genes performed well regarding the exclusion of paralogs. This is likely due to strict downstream filtering, leading to the selection of highly conserved genes that are preferentially retained in single-copy and are also under strong selective constraints, and thus have low substitution rates.
The genes in our gene sets encoded different protein classes, were involved in a wide variety of biological processes, had many molecular functions, and were located in various cellular components. Overrepresentation analysis also showed generally low fold enrichment (<3-fold) of many GO terms, suggesting in turn a high coverage of many functional categories. Few categories were underrepresented in synteny sets; interestingly, type 1 MADS box transcription factors were among the underrepresented protein classes for two sets of syntenic ortholog genes (“all orthologs” and “orthologs without paralogs”), in line with a recent study showing that some MADS-domain transcription factors are frequently transposed in the Brassicaceae (Madrid et al. 2021). In contrast to the diverse gene functional annotations in our synteny gene sets, single-copy genes and thus gene sets selected for phylogenomics using conventional methods often have a conserved nucleotide sequence and gene function, such as photosynthesis and DNA metabolism (Li et al. 2017, 2016).
We compared our Brassicaceae synteny gene set with a previously published set of target enrichment genes designed for phylogenomic analysis of the Brassicaceae (Nikolov et al. 2019). The Brassicaceae gene set contained 673 genes (2.5% of A. thaliana), thus unsurprisingly, a smaller fraction of functional categories was covered. Interestingly, different GO categories were over- and underrepresented in the two sets, likely due to the strict filtering criteria used to exclude paralogs in the Brassicaceae set (including single-copy status, long branch score, patristic distance, Robinson–Foulds distance and saturation). Specifically, more conserved genes, such as those involved in tRNA modification, were overrepresented in the Brassicaceae set, and less conserved transcription factor related terms were underrepresented. This pattern was even more striking when comparing our synteny gene sets with the Angiosperms353 set, which was specifically designed to include only single-copy genes and avoid genes with potential paralogs, since accurate filtering for paralogs may not always be feasible for large-scale analyses with such a broad taxonomic focus. Genes localized to the plastid were among the most highly overrepresented in the Angiosperms353 set, in line with previous studies showing enrichment for organellar localization among single-copy genes (Han et al. 2014). We thus conclude that the use of larger gene sets and different gene selection criteria for phylogenomics may provide genomic information allowing for downstream identification of traits under selection from target enrichment data.
Our analysis of gene and species tree resolution and topology showed surprisingly few differences between gene sets. Bootstrap values were not significantly different between most sets, with the exception of the Brassicaceae set, which had considerably higher mean bootstrap values, likely due to strict filtering criteria, as well as higher quartet scores compared with other gene sets. Species tree topologies generally fall into two groups. Trees based solely on orthologs had clades A and C as sisters to tribe Arabideae and clade B, whereas trees based on genes with paralogs had tribe Arabideae as sister to a clade containing clades A, B, and C. The differences in topology are likely caused by generally low quartet scores at deeper nodes in the Brassicaceae phylogeny, where first and second topology can be almost equally common (supplementary table S1, Supplementary Material online). Due to the high number of genes used to reconstruct the species tree, these nodes can have high posterior probabilities despite similar quartet scores for main and alternate topologies. The selection of genes may nonetheless influence the topology. We found that although the Brassicaceae species tree based on the complete Brassicaceae set as used in the original publication supports Arabideae as sister to clades A, B, and C (Nikolov et al. 2019), the tree based on the syntenic subset we analyzed here followed the same topology we detected for other gene sets without paralogs, with clades A and C as sisters to Arabideae and clade B. Interestingly, a recent genus-level phylogeny of Brassicaceae also found different topologies at deep nodes depending on filtering level, with the species tree inferred from the complete combined Angiosperms353 and Brassicaceae gene sets supporting topology B and stricter filtering of potential paralogs resulting in a species tree more similar to our topology A (Hendriks et al. 2022). Altogether, we conclude that the use of synteny for selecting genes for phylogenomics does not necessarily perform better than more established methods when it comes to the topology of the resulting species tree with regards to tree resolution. For taxa with difficult to resolve relationships, the method may aid in obtaining large sets of reliable orthologs. However, although including synteny information in the selection of orthologs can avoid the influence of paralogs on species tree reconstruction, it should be noted that other processes can obfuscate phylogenetic analyses. For example, in the presence of high levels of incomplete lineage sorting or ancient gene flow, phylogenetic networks may be better suited than methods resulting in bifurcating trees to reconstruct and visualize the evolutionary history of such clades (Cai et al. 2021; Stull et al. 2022). It would be interesting to compare our results to another species group with a difficult backbone phylogeny. Furthermore, future studies should consider including synteny in their selection of genes for phylogenomic reconstruction.
Our approach was aimed at identifying a large number of reliable orthologs for species tree reconstruction through conserved gene position. As it relies on high quality genome assemblies, its use for now is limited to taxa where such data is available. We restricted sampling to diploid genomes to provide a proof of concept for our approach. However, the evolutionary history of the Brassicaceae is highly influenced by repeated cycles of polyploidization, with all Brassicaceae sharing the At-α event and preceding WGDs (Bowers et al. 2003; Schranz and Mitchell-Olds 2006), mesopolyploidization in at least 11 tribes (Walden, German, et al. 2020) and a neopolyploid rate as high as 43% (Hohmann et al. 2015). Both meso- and neopolyploid species were not considered here, because only a few chromosome-level genome assemblies are available for such species, and those are restricted to a few positions in the phylogeny, such as the tribe Brassiceae. However, we demonstrated that it is possible to even identify syntenic paralogs derived from the more ancient At-α event, and methodology for the identification of younger WGD-derived paralogs should work analogously, in particular in species with distinct subgenomes. As new genome assemblies become available, polyploid genomes may also be included in similar analyses.
Finally, we made use of the synteny blocks identified for phylogenomic analyses and reconstructed ancestral genomes for the Brassicaceae. Previous ancestral genome reconstructions in the Brassicaceae were limited by the availability of high-quality genome sequences. As only genomes from the two largest clades, A and B, were available, reconstructions should not be considered ancestral to the entire family; however, genomes from the earlier diverging lineages have become available over the past few years, now allowing us to reconstruct ancestral genomes from deeper nodes of the family. The ACK with n = 8 (Schranz et al. 2006; Lysak et al. 2016) was the first published ancestral Brassicaceae genome, reconstructed from the genome structures of A. thaliana, A. lyrata, C. rubella, and B. rapa. A later ancestral genome reconstruction added Schrenkiella parvula, but the reconstructed genome still greatly resembled the ACK (Murat, Louis, et al. 2015). In our study, the ancestor of Arabidopsis strongly resembles the ACK, whereas the ancestor of clade A has an additional translocation. This difference is likely due to our inclusion of Megadenia pygmaea as sister to clade A, as well as further outgroups showing similar adjacencies. Due to the lack of a non-Brassicaceae outgroup, we cannot obtain the ancestral genome of all Brassicaceae, including the first diverging tribe Aethionemeae; however, for the first time, we reconstruct the genome of the MRCA of Core Brassicaceae from ∼25 million years ago (Walden, German, et al. 2020). Our newly reconstructed ancestral genome of Core Brassicaceae had a haploid chromosome number of n = 9. Interestingly, the ancestral chromosome number for Brassicaceae was estimated to be n = 7 (Carta et al. 2020) in an angiosperm wide context. It should be noted that the estimated number may be strongly impacted by the studied taxa and outgroups. This is likely true for the ACK with its strong influence from clade A, where n = 8 is the base chromosome number in most tribes, as well as in our study, where the chromosome numbers of representatives from earlier diverging lineages ranged from n = 7 in E. syriacum to n = 11 in A. arabicum. Furthermore, telomeres here were reconstructed based on telomere position in our 11 extant species, not based on sequence data. The addition of other genomes may thus change the number of chromosomes in reconstruction. Despite the uncertainties that arise from the reconstruction method itself, evidence for the correctness of the results can be found in extant genomes. For example, ancestral adjacencies of blocks B-U (Core Brassicaceae chromosome CB1) and O-V-J (CB6) are also found within paralogous blocks retained from At-α (Walden, Nguyen, et al. 2020) and can thus even be dated back to before this WGD event. In the future, the availability of a (diploid) genome from sister family Cleomaceae would allow us to potentially reconstruct the ancestral genome of all Brassicaceae including first diverging lineage Aethionemeae and further advance our understanding of genome evolution in the family.
Materials and Methods
Taxon Sampling and Genomic Resources
We selected diploid species with available high-quality genome assemblies representing the major taxonomic lineages of Brassicaceae following recent nuclear phylogenies (Huang et al. 2016; Nikolov et al. 2019): Aethionema arabicum (Fernandez-Pozo et al. 2021) from first-diverging tribe Aethionemeae (clade F), which was used as an outgroup here; Euclidium syriacum (Jiao et al. 2017) from clade E; Arabis alpina (Willing et al. 2015) and Draba nivalis (Nowak et al. 2021) from clade D; Eutrema salsugineum (Yang et al. 2013), Isatis indigotica (Kang et al. 2020), and Schrenkiella parvula (Dassanayake et al. 2011) from clade B; Megadenia pygmaea from clade C (Yang et al. 2021); and Cardamine hirsuta (Gan et al. 2016), Arabidopsis lyrata (Hu et al. 2011), and Arabidopsis thaliana (Lamesch et al. 2012) from clade A. For the phylogenetic position of M. pygmaea from the tribe Biscutelleae, we followed Guo et al. (2021) and placed it as sister to clade A. More information on genomic resources can be found in figure 1.
Assembly of Pseudochromosomes
The published genome sequences of E. syriacum, E. salsugineum, and S. parvula were not assembled at the chromosome level yet. For S. parvula, assignment of the largest scaffolds to seven pseudochromosomes was already available (Dassanayake et al. 2011), whereas for the other two species, we first manually generated pseudochromosomes by combining the longest contigs from the published genome assemblies using evidence from chromosome painting data. We obtained pairwise syntenic blocks of both species with A. thaliana using SynMap in CoGe (https://genomevolution.org/coge/; Lyons et al. 2008; Nelson et al. 2018) with default settings and assigned them to ABC blocks. We then used information from chromosome painting (Mandáková and Lysak 2008; Mandáková, Hloušková, et al. 2017) to infer the position and orientation of scaffolds. Finally, the scaffolds were combined into pseudochromosomes (separated by 100 Ns) and new, matching annotation files were generated for downstream analyses for all three species. The newly generated assemblies are available at CoGe (https://genomevolution.org/coge/) under accessions 61751 (E. syriacum), 61750 (E. salsugineum), and 61748 (S. parvula).
Synteny Detection and Grouping of Orthologs and Paralogs
Pairwise synteny between A. thaliana and the other ten species was detected using SynMap with default parameters; synonymous substitution rates (Ks) were calculated using the CodeML program from the PAML package (Yang 2007) implemented in CoGe. The resulting syntenic blocks were combined into blocks of genes present and in synteny in all species using R 4.0.5 (R Core Team 2022). Bad syntenic matches (Ks > 5) and resulting short blocks (block length < 5) were removed. Blocks were then split into orthologs, paralogs derived from At-α WGD (“paralogs” from here on), and all other blocks. Only orthologs and paralogs mapping to the main chromosomes were retained for further analysis. Filtering for paralogs was performed by median block Ks determined for each species manually (see supplementary fig. S12, Supplementary Material online). For A. arabicum and E. syriacum, where the Ks peaks for orthologs and paralogs overlap slightly, assignments were adjusted manually for blocks with intermediate Ks using Ks, gene content, and duplication status. Syntenic orthologs and paralogs were then assigned to their A. thaliana counterparts, and gene groups containing all 11 species were selected; by restricting our analysis to orthologs present in all species, we reduced potential bias toward the A. thaliana genome that served as our reference for synteny detection. A subset of orthologous genes had an At-α derived paralog. We also ran OrthoFinder version 2.4.0 (Emms and Kelly 2019) using the longest transcripts of each gene for all species as input to independently obtain single-copy orthogroups. Using the gene names, we then counted the number of orthologs and paralogs identified in synteny analysis in OrthoFinder orthogroups.
We compared our synteny and OrthoFinder gene sets to two target enrichment gene sets: (1) The Angiosperms353 set (Johnson et al. 2019) is meanwhile widely used for phylogenomic studies within and across angiosperm families and contains 353 genes. (2) A Brassicaceae-specific gene set (Nikolov et al. 2019) was recently developed to resolve the phylogeny of the family and comprises 673 genes. From each of these two gene sets, we selected syntenic orthologs and single-copy OrthoFinder orthogroups for comparison with our other gene sets.
Sequence Alignment and Phylogenetic Inference
For each orthogroup from synteny analysis or OrthoFinder, we first aligned the coding sequences of all 11 species using MACSE version 2.05 (Ranwez et al. 2018) to obtain a codon-aware nucleotide alignment. The nucleotide sequence was then curated using Gblocks version 0.91b (Castresana 2000) using codon mode and discarding nonconserved blocks. ML trees were reconstructed using RAxML version 8.2.12 (Stamatakis 2014) with GTR+Γ model of rate heterogeneity and rapid bootstrap inference with 1,000 replicates followed by a thorough ML search.
A species tree reconstruction was performed using ASTRAL version 5.7.8 (Zhang et al. 2018). To compare gene sets obtained through synteny and orthology detection, we analyzed syntenic orthologs, paralogs, and OrthoFinder gene sets separately. We also split the sets further to study whether the presence of a paralogous gene copy in syntenic orthologs or a paralog in an OrthoFinder orthogroup influenced the reconstructed phylogeny. For all sets, we ran ASTRAL first without any constraints, and then tested the data versus the hypothesized species tree (fig. 1) to obtain comparable normalized quartet scores.
Analysis of Gene Function
GO and protein class overrepresentation analysis was conducted using PANTHER v. 16.0 (Mi et al. 2021, released 12-01-2020). Overrepresentation was tested for each synteny and OrthoFinder gene set and additionally for the entire Angiosperms353 and Brassicaceae gene set, including genes that were not found in synteny or in OrthoFinder orthogroups. All A. thaliana genes were used as background for synteny, Angiosperms353 and Brassicaceae gene sets, and all A. thaliana genes assigned to an orthogroup for OrthoFinder gene sets. Significance was tested using Fisher's exact test using FDR < 0.05.
We furthermore ran BUSCO v. 5.4.2 (Manni et al. 2021) using protein mode on A. thaliana orthologs with the BUSCO Brassicales data set to assess the gene set completeness of all gene sets.
Ancestral Genome Reconstruction
The ancestral gene order was reconstructed with ANGES (Jones et al. 2012), as this software has previously been successfully used to reconstruct the ancestral genomes of eukaryotes, both animals (Neafsey et al. 2015) and plants (Murat et al. 2014; Murat, Zhang, et al. 2015). In short, the method rearranges user-specified orthologous markers such as genes or larger genomic regions found across species into CARs and is guided by a bifurcating phylogenetic tree. As small rearrangements (e.g., transpositions) are common across many plant genomes and also among our selected species, the use of orthologous genes as markers does not result in long, contiguous output at the chromosome or chromosome-arm level. Instead, we thus used longer syntenic blocks of genes present in all species. Markers were generated using pairwise synteny with A. thaliana. Syntenic maps were created using SynMap with parameters optimized for the following analysis steps. Default parameters were used except for the following: DAGChainer maximum distance between two matches was set to 10, DAGChainer minimum number of aligned pairs was set to 20, and synonymous substitution rates (Ks) were calculated. The resulting syntenic blocks were combined into blocks of genes present and in synteny in all species using R v4.0.5 (R Core Team 2022). Only orthologous blocks mapping to the main chromosomes were considered. Filtering for paralogs and block optimization were performed as above. This resulted in 336 blocks containing 3,392 genes found in the same block in all species. The genomic coordinates of the blocks in each species were then used as input for ancestral genome reconstruction. To assess the extent to which the input phylogeny influenced the reconstructed ancestral genomes, we used two different input trees representing different hypotheses for the nuclear phylogeny: The representative of the first diverging lineage after tribe Aethionemeae was either E. syriacum (clade E, see fig. 1) or the clade containing A. alpina and D. nivalis (clade D). The latter was chosen due to the proposed phylogenetic position of Arabideae or clade D as first diverging lineage based on similarities between the genome structures of A. alpina and A. arabicum (Walden, Nguyen, et al. 2020). Markers were set as unique and universal, doubled markers were utilized to infer marker direction, and telomeres were added after greedy heuristic C1P optimization. For each of the two phylogenies, we reconstructed the ancestral genomes at all nine nodes, resulting in a total of 18 reconstructions.
Combination of CARs with Ancestral Genomes
Ancestral genomes were manually assembled from the CARs obtained from ANGES following three rules: (1) Chromosomes end with telomeres. A CAR with reconstructed telomeres at both ends was thus considered a complete chromosome, whereas a CAR with one telomere was located at the chromosome end and could be combined with another one-telomere CAR to obtain a complete chromosome. (2) If a genome contained more than one chromosome made from two CARs, marker adjacencies in closely related extant lineages were considered, and CARs were combined in such a way that at least one extant adjacency was present in the ancestral reconstruction. (3) In cases where the combination of CARs was ambiguous, we followed adjacencies in the respective first diverging lineage; however, alternative adjacencies are shown in supplementary figure S10, Supplementary Material online. For details, see supplementary methods, Supplementary Material online.
The system of ABC blocks (Schranz et al. 2006; Lysak et al. 2016) that has been used to visualize Brassicaceae genomes for the past 15 years is based solely on the genomes of clades A and B. Here, we reconstruct the ancestral genome of Core Brassicaceae, the ancestor of all major lineages except tribe Aethionemeae (clade F). To display the chromosomal rearrangements relative to this ancestor, we assigned a new coloring scheme based on the CARs and chromosomes of the ancestral genome at node N1E (supplementary fig. S11, Supplementary Material online).
Supplementary Material
Acknowledgment
This work was supported by the German Research Foundation [HO 6443/1 to N.W.].
Contributor Information
Nora Walden, Biosystematics Group, Wageningen University, Wageningen, The Netherlands; Centre for Organismal Studies, Heidelberg University, Heidelberg, Germany.
Michael Eric Schranz, Biosystematics Group, Wageningen University, Wageningen, The Netherlands.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Author Contributions
N.W. and M.E.S. designed the study, N.W. performed the analysis with input from M.E.S., and N.W. drafted the manuscript with input from M.E.S.
Data availability
The newly generated pseudochromosome assemblies are available at CoGe (https://genomevolution.org/coge/) under accessions 61751 (E. syriacum), 61750 (E. salsugineum) and 61748 (S. parvula). Data and code used for the analyses are available at Zenodo (https://doi.org/10.5281/zenodo.7573131).
Literature Cited
- Baker WJ, et al. . 2022. A comprehensive phylogenomic platform for exploring the angiosperm tree of life. Syst Biol. 71:301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yang H. 2022. The multiple fates of gene duplications: deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 34:2466–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438. [DOI] [PubMed] [Google Scholar]
- Cai L, et al. . 2021. The perfect storm: gene tree estimation error, incomplete lineage sorting, and ancient gene flow explain the most recalcitrant ancient angiosperm clade, Malpighiales. Syst Biol. 70:491–507. [DOI] [PubMed] [Google Scholar]
- Carta A, Bedini G, Peruzzi L. 2020. A deep dive into the ancestral chromosome number and genome size of flowering plants. New Phytol. 228:1097–1106. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cheng F, et al. . 2018. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. 4:258–268. [DOI] [PubMed] [Google Scholar]
- Conover JL, Sharbrough J, Wendel JF. 2021. pSONIC: ploidy-aware syntenic orthologous networks identified via collinearity. G3 GenesGenomesGenetics 11:jkab170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R, et al. . 2012. Targeted enrichment strategies for next-generation plant biology. Am J Bot. 99:291–311. [DOI] [PubMed] [Google Scholar]
- Dassanayake M, et al. . 2011. The genome of the extremophile crucifer Thellungiella parvula. Nat Genet. 43:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, et al. . 2013. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci U S A. 110:2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, et al. . 2015. The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci U S A. 112:8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, et al. . 2021. Aethionema arabicum genome annotation using PacBio full-length transcripts provides a valuable resource for seed dormancy and Brassicaceae evolution research. Plant J. 106:275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. 1970. Distinguishing homologous from analogous proteins. Syst Zool. 19:99–113. [PubMed] [Google Scholar]
- Gan X, et al. . 2016. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants. 2:16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, et al. . 2021. Linked by ancestral bonds: multiple whole-genome duplications and reticulate evolution in a Brassicaceae tribe. Mol Biol Evol. 38:1695–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Peng Y, Xu L, Xiao P. 2014. Identification, characterization, and utilization of single copy genes in 29 angiosperm genomes. BMC Genomics. 15:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks KP, et al. . 2022. Global phylogeny of the Brassicaceae provides important insights into gene discordance. bioRxiv 2022.09.01.506188. [Google Scholar]
- Hohmann N, Wolf EM, Lysak MA, Koch MA. 2015. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27:2770–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, et al. . 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, et al. . 2016. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol Biol Evol. 33:394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W-B, et al. . 2017. Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 27:778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, et al. . 2016. Hybpiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl Plant Sci. 4:1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, et al. . 2019. A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst Biol. 68:594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P, Gout J-F, Doak TG, Lynch M. 2022. A population-genetic lens into the process of gene loss following whole-genome duplication. Mol Biol Evol. 39:msac118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BR, Rajaraman A, Tannier E, Chauve C. 2012. ANGES: reconstructing ANcestral GEnomeS maps. Bioinformatics 28:2388–2390. [DOI] [PubMed] [Google Scholar]
- Kang M, et al. . 2020. A chromosome-scale genome assembly of Isatis indigotica, an important medicinal plant used in traditional Chinese medicine. Hortic Res. 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, et al. . 2019. Interspecies association mapping links reduced CG to TG substitution rates to the loss of gene-body methylation. Nat Plants. 5:846–855. [DOI] [PubMed] [Google Scholar]
- Kocot KM, Citarella MR, Moroz LL, Halanych KM. 2013. Phylotreepruner: a phylogenetic tree-based approach for selection of orthologous sequences for phylogenomics. Evol Bioinforma. 9:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, et al. . 2012. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. . 2016. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 28:326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. . 2017. Single-copy genes as molecular markers for phylogenomic studies in seed plants. Genome Biol Evol. 9:1130–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JT, et al. . 2022. GENESPACE tracks regions of interest and gene copy number variation across multiple genomes. eLife 11:e78526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, et al. . 2008. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with Rosids. Plant Physiol. 148:1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Mandáková T, Schranz ME. 2016. Comparative paleogenomics of crucifers: ancestral genomic blocks revisited. Curr Opin Plant Biol. 30:108–115. [DOI] [PubMed] [Google Scholar]
- Madrid E, et al. . 2021. Transposition and duplication of MADS-domain transcription factor genes in annual and perennial Arabis species modulates flowering. Proc Natl Acad Sci U S A. 118:e2109204118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Hloušková P, German DA, Lysak MA. 2017. Monophyletic origin and evolution of the largest crucifer genomes. Plant Physiol. 174:2062–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Li Z, Barker MS, Lysak MA. 2017. Diverse genome organization following 13 independent mesopolyploid events in Brassicaceae contrasts with convergent patterns of gene retention. Plant J. 91:3–21. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2008. Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20:2559–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, et al. . 2021. PANTHER Version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49:D394–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F, et al. . 2014. Shared subgenome dominance following polyploidization explains grass genome evolutionary plasticity from a seven protochromosome ancestor with 16K protogenes. Genome Biol Evol. 6:12–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F, Louis A, et al. . 2015. Understanding Brassicaceae evolution through ancestral genome reconstruction. Genome Biol 16:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F, Zhang R, et al. . 2015. Karyotype and gene order evolution from reconstructed extinct ancestors highlight contrasts in genome plasticity of modern Rosid crops. Genome Biol. Evol 7:735–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsidis P, Kapli P, Schiffer PH, Telford MJ. 2021. Systematic errors in orthology inference and their effects on evolutionary analyses. iScience 24:102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, et al. . 2015. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347:1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ADL, Haug-Baltzell AK, Davey S, Gregory BD, Lyons E. 2018. EPIC-CoGe: managing and analyzing genomic data. Bioinformatics 34:2651–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov LA, et al. . 2019. Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 222:1638–1651. [DOI] [PubMed] [Google Scholar]
- Nowak MD, et al. . 2021. The genome of Draba nivalis shows signatures of adaptation to the extreme environmental stresses of the Arctic. Mol Ecol Resour. 21:661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative . 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. 2018. MACSE V2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35:2582–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2022. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/. [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. 2006. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11:535–542. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mitchell-Olds T. 2006. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18:1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu-Ting K, et al. . 2019. Inadvertent paralog inclusion drives artifactual topologies and timetree estimates in phylogenomics. Mol Biol Evol. 36:1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull G, Pham K, Soltis P, Soltis D. 2023. Deep reticulation: the long legacy of hybridization in vascular plant evolution. Plant J. tpj.16143. [DOI] [PubMed] [Google Scholar]
- Ufimov R, et al. . 2022. Utilizing paralogs for phylogenetic reconstruction has the potential to increase species tree support and reduce gene tree discordance in target enrichment data. Mol Ecol Resour. 22:3018–3034. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y. 2014. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the cretaceous–paleogene boundary. Genome Res. 24:1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden N, German DA, et al. . 2020. Nested whole-genome duplications coincide with diversification and high morphological disparity in Brassicaceae. Nat Commun. 11:3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden N, Nguyen T-P, Mandáková T, Lysak MA, Schranz ME. 2020. Genomic blocks in Aethionema arabicum support Arabideae as next diverging clade in Brassicaceae. Front Plant Sci. 11:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitemier K, et al. . 2014. Hyb-Seq: combining target enrichment and genome skimming for plant phylogenomics. Appl Plant Sci. 2:1400042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing E-M, et al. . 2015. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants. 1:14023. [DOI] [PubMed] [Google Scholar]
- Yan Z, Smith ML, Du P, Hahn MW, Nakhleh L. 2022. Species tree inference methods intednded to deal with incomplete lineage sorting are robust to the presence of paralogs. Syst Biol. 71:367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang R, et al. . 2013. The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci. 4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, et al. . 2021. The chromosome-level genome sequence and karyotypic evolution of Megadenia pygmaea (Brassicaceae). Mol Ecol Resour. 21:871–879. [DOI] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Scornavacca C, Molloy EK, Mirarab S. 2020. ASTRAL-Pro: quartet-based species-tree inference despite paralogy. Mol Biol Evol. 37:3292–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, et al. . 2017. Phylogenomic synteny network analysis of MADS-box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell tpc 00312:2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, et al. . 2021. Whole-genome microsynteny-based phylogeny of angiosperms. Nat Commun. 12:3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Schranz ME. 2017. Network approaches for plant phylogenomic synteny analysis. Curr Opin Plant Biol. 36:129–134. [DOI] [PubMed] [Google Scholar]
- Zhao T, Schranz ME. 2019. Network-based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes. Proc Natl Acad Sci U S A. 116:2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Soghigian J, Xiang Q-Y. 2022. A new pipeline for removing paralogs in target enrichment data. Syst Biol. 71:410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The newly generated pseudochromosome assemblies are available at CoGe (https://genomevolution.org/coge/) under accessions 61751 (E. syriacum), 61750 (E. salsugineum) and 61748 (S. parvula). Data and code used for the analyses are available at Zenodo (https://doi.org/10.5281/zenodo.7573131).