ABSTRACT
Staphylococcus aureus generates biofilms during many chronic human infections, which contributes to its growth and persistence in the host. Multiple genes and pathways necessary for S. aureus biofilm production have been identified, but knowledge is incomplete, and little is known about spontaneous mutations that increase biofilm formation as infection progresses. Here, we performed in vitro selection of four S. aureus laboratory strains (ATCC 29213, JE2, N315, and Newman) to identify mutations associated with enhanced biofilm production. Biofilm formation increased in passaged isolates from all strains, exhibiting from 1.2- to 5-fold the capacity of parental lines. Whole-genome sequencing identified nonsynonymous mutations affecting 23 candidate genes and a genomic duplication encompassing sigB. Six candidate genes significantly impacted biofilm formation as isogenic transposon knockouts: three were previously reported to impact S. aureus biofilm formation (icaR, spdC, and codY), while the remaining three (manA, narH, and fruB) were newly implicated by this study. Plasmid-mediated genetic complementation of manA, narH, and fruB transposon mutants corrected biofilm deficiencies, with high-level expression of manA and fruB further enhancing biofilm formation over basal levels. This work recognizes genes not previously identified as contributing to biofilm formation in S. aureus and reveals genetic changes able to augment biofilm production by that organism.
KEYWORDS: Staphylococcus aureus, biofilm, evolution, genomics, infection, adaptation, cystic fibrosis
INTRODUCTION
Staphylococcus aureus is a prevalent clinical pathogen worldwide that is capable of causing acute and chronic disease in a variety of organ systems (1–5). Morbidity and mortality attributable to S. aureus infection have increased substantially over the past decades, due in part to the rise and dissemination of community-acquired methicillin-resistant S. aureus (MRSA) clones and attendant challenges in treatment (6).
Among the many virulence factors S. aureus deploys during chronic infection is the production of biofilms or biofilm-like aggregates (7, 8). Biofilm formation has been implicated in the pathogenesis of S. aureus osteomyelitis, indwelling device infections, wound infections, endocarditis, and other medical conditions (7) and is believed to promote persistence through several complementary mechanisms: staphylococcal biofilms impair action of the innate immune system by expressing factors that disable immune cells and circumvent inflammatory responses (9, 10), reducing host-mediated clearance. They similarly provide resistance to antibiotic-mediated killing (11), blunting the impact of a broad range of therapeutic agents. Dispersion of cells from mature biofilms facilitates the continuance and progression of chronic infection, including the colonization of new body sites (8).
Substantial work has been conducted to identify regulatory and effector genes essential to S. aureus biofilm formation using various methods including examination of orthologs with relevant function in other species (12), saturation transposon mutagenesis screens (13), and transcriptomic (14, 15) or proteomic (16, 17) analyses. These efforts have provided an increasingly detailed picture of the pathways and genes relevant to biofilm formation in this organism, but even so, the factors involved remain incompletely enumerated (7, 8, 10, 18). Moreover, chronically infecting S. aureus populations are known to undergo mutational adaptation during the course of disease that selects for increased fitness in the host, including changes that promote biofilm formation phenotypes (19–24). Mutations enhancing S. aureus biofilm production in vivo are therefore likely of relevance to chronic pathogenesis, but comparatively little is known about what genomic changes are able to elevate biofilm formation beyond basal levels (25), and systematic studies to investigate that phenomenon have not yet been performed.
Here, we utilized in vitro selection and whole-genome sequencing to identify spontaneous chromosomal mutations associated with enhanced biofilm formation in four laboratory S. aureus strains (ATCC 29213, JE2, N315, and Newman). Using a combination of isogenic transposon mutant knockouts (26) and genetic complementation studies, we provide functional validation of genes not previously described as contributing to S. aureus biofilm formation.
RESULTS
In vitro selection of S. aureus for enhanced biofilm formation.
We performed selection of S. aureus for spontaneous mutations conferring increased capacity for biofilm production. We serially passaged replicate liquid cultures of four S. aureus laboratory strains (ATCC 29213, JE2, N315, and Newman) and selectively harvested the population adhering to an abiotic surface after each growth period. Thirty-seven serial passages were conducted using five separate replicates of each strain, after which single colonies were isolated from evolved populations for subsequent analysis.
Baseline phenotypes of the parental S. aureus strains differed significantly by crystal violet biofilm absorbance assays, with N315 having the greatest prepassaging biofilm formation capacity and Newman the least (Fig. 1A). Phenotyping of isolates from evolved populations (Fig. 1B) revealed statistically significant increases in biofilm-forming capacity relative to their matched parental strains for 18 of 20 replicates tested. The exceptions were N315 replicate 3, which showed slightly reduced biofilm formation, and N315 replicate 5, which did not change significantly. N315 exhibited the smallest relative gains in biofilm formation during passaging, with isolates having a maximum of 1.2 times the biofilm formation of their progenitor, while derivatives of other strains achieved maxima of 4- to 5-fold increases relative to their respective parents.
FIG 1.
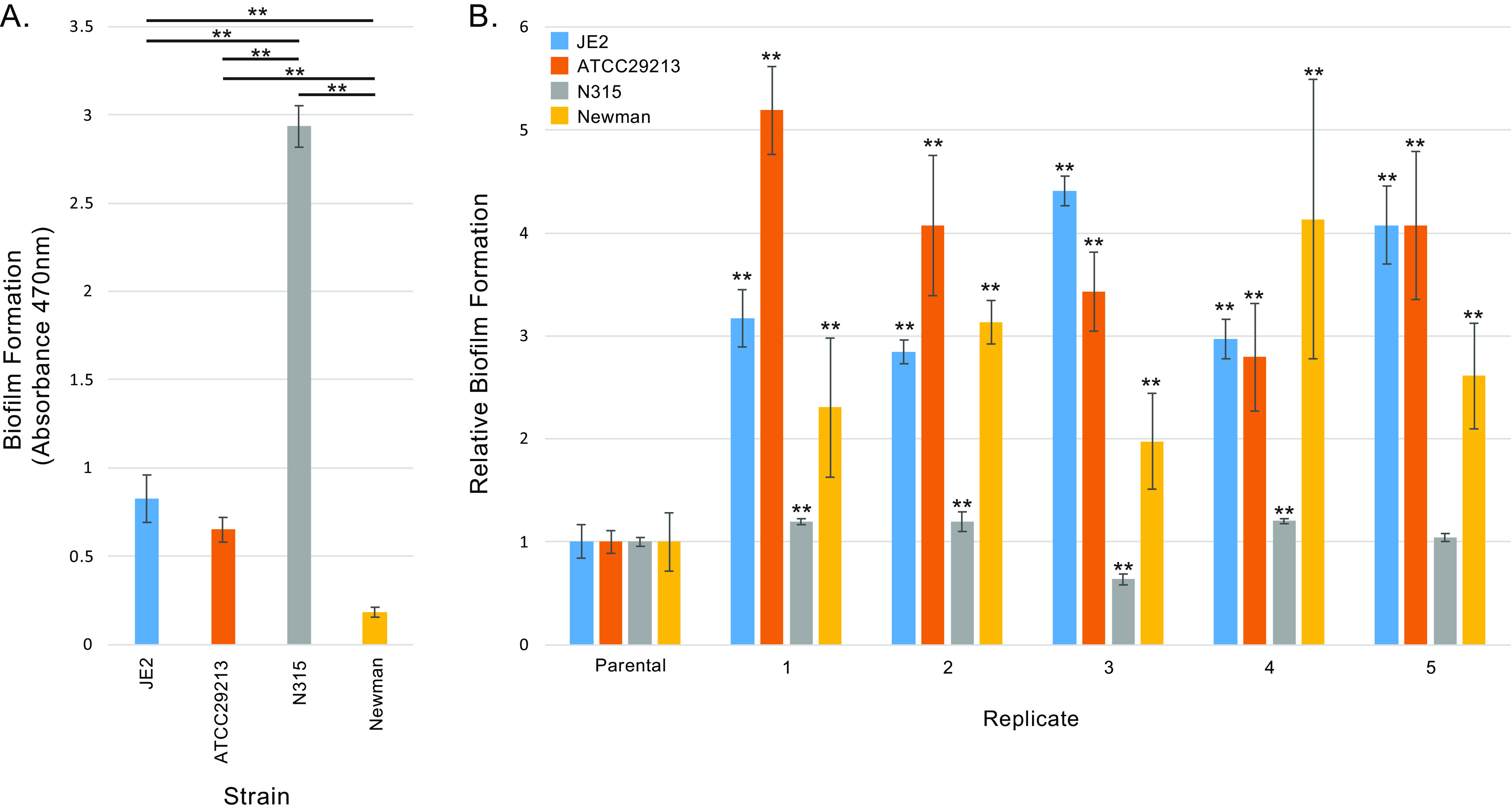
Biofilm formation of S. aureus laboratory strains and passaged derivatives. (A) Biofilm-forming capacity of S. aureus laboratory strains. Error bars indicate standard deviation for at least four separate measurements. ** represents a P value of <0.01 by two-tailed t test for the indicated comparison between strains (black lines). (B) Biofilm-forming capacity of laboratory strain derivatives obtained from serial passaging for increased adherence. Results are normalized to biofilm formation rates of the matched parental strain. Error bars show standard deviation from at least four different measurements. ** indicates a P value of <0.01 by two-tailed t test against the parental strain.
Mutations associated with biofilm formation in vitro.
Whole-genome sequencing of passaged isolates having enhanced biofilm formation identified nonsynonymous coding sequence mutations in 23 different genes (Table 1 and see Table S2 in the supplemental material). Most genes (n = 19) were altered in a single isolate each, while three genes (spdC, icaR, and codY) were recurrently mutated in multiple isolates from different strains and one (eap/map) was mutated in two independently evolved replicates of the Newman strain. At least one gene mutation with unequivocally inactivating consequences (stop-gain, frameshift, and/or start codon loss) consistent with loss-of-function effects was noted in spdC, a hypothetical 5′-nucleotidase of the lipoprotein e(P4) family (pangenome gene identifier [27] SAUPAN001228000), icaR, and a HAD-family hydrolase (pangenome gene identifier SAUPAN002478000). Mutations in the remaining genes exclusively comprised missense variants or in-frame indels.
TABLE 1.
Gene mutations associated with increased biofilm formation in vitro
| Pangenome gene identifier | Common gene name | Gene function | Mutant count per selected strain (total replicates) |
|||
|---|---|---|---|---|---|---|
| Newman (n = 5) | JE2 (n = 5) | N315 (n = 3) | ATCC 29213 (n = 5) | |||
| SAUPAN000916000 | sbnC | IucA/IucC family siderophore biosynthesis protein | 1 | 0 | 0 | 0 |
| SAUPAN005823000 | spdC | Lysostaphin resistance protein A | 0 | 2 | 3 | 0 |
| SAUPAN006410000 | icaR | Biofilm operon icaADBC HTHa-type negative transcriptional regulator | 2 | 5 | 0 | 0 |
| SAUPAN005927000 | narH | Nitrate reductase subunit beta | 1 | 0 | 0 | 0 |
| SAUPAN003492000 | rpoZ | DNA-directed RNA polymerase subunit omega | 0 | 1 | 0 | 0 |
| SAUPAN004777000 | DUF445 domain-containing protein | 1 | 0 | 0 | 0 | |
| SAUPAN003350000 | Hypothetical protein | 1 | 0 | 0 | 0 | |
| SAUPAN003016000 | Nitronate monooxygenase | 0 | 0 | 0 | 1 | |
| SAUPAN003553000 | codY | GTP-sensing pleiotropic transcriptional regulator | 3 | 5 | 3 | 3 |
| SAUPAN005440000 | manA | Mannose-6-phosphate isomerase | 0 | 0 | 1 | 0 |
| SAUPAN001024000 | argC | N-Acetyl-gamma-glutamyl-phosphate reductase | 0 | 0 | 1 | 0 |
| SAUPAN006300000 | Amino acid permease | 0 | 0 | 0 | 1 | |
| SAUPAN003545000 | dprA | DNA processing protein | 1 | 0 | 0 | 0 |
| SAUPAN002478000 | HAD family hydrolase | 0 | 0 | 0 | 1 | |
| SAUPAN006252000 | gbaA | TetR family transcriptional regulator | 0 | 0 | 1 | 0 |
| SAUPAN001228000 | 5′-Nucleotidase, lipoprotein e(P4) family | 0 | 1 | 0 | 0 | |
| SAUPAN006369000 | isaB | Immunodominant antigen B | 0 | 0 | 0 | 1 |
| SAUPAN002607000 | ABC transporter ATP-binding protein | 1 | 0 | 0 | 0 | |
| SAUPAN002583000 | fruB | 1-Phosphofructokinase | 1 | 0 | 0 | 0 |
| SAUPAN000959000 | Bifunctional metallophosphatase/5′-nucleotidase | 1 | 0 | 0 | 0 | |
| SAUPAN000382000 | dusB | tRNA-dihydrouridine synthase | 0 | 0 | 1 | 0 |
| SAUPAN005009000 | eap/map | MAP domain-containing protein | 2 | 0 | 0 | 0 |
| SAUPAN000011000 | gyrB | DNA gyrase subunit B | 0 | 1 | 0 | 0 |
HTH, helix-turn-helix.
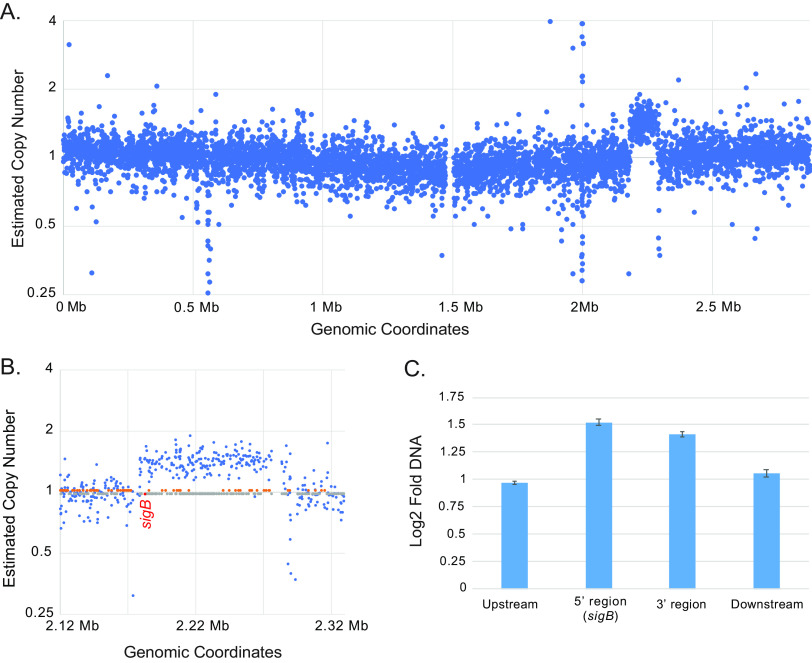
Structural analysis of whole-genome sequencing reads additionally revealed an ~110-kb region of copy number gain (approximate positions 2,183,746 to 2,293,258 bp of the JE2 reference genome) in a single passaged isolate, JE2 replicate 3 (Fig. 2A). The affected region encompassed 110 genes, notably including the sigB locus (Fig. 2B and Table S3). Verification of the copy number alteration was achieved using real-time PCR to measure DNA content within the presumptive event (sigB at the 5′ end and a second locus at the 3′ terminus) and genomic regions flanking the event and was consistent with a duplication event occurring in this region (Fig. 2C).
FIG 2.
Copy number gain of the sigB locus in JE2 replicate 3. (A) Plot of estimated copy number for regions across the JE2 reference genome. (B) Enlargement of the region affected by copy number gain. Locations of genes carried in the forward orientation (orange dots) and reverse orientation (gray dots) are indicated along the central axis, with the location of sigB (reverse orientation) highlighted in red. (C) Confirmation of copy number gain by real-time PCR. Analysis was performed using primers designed upstream and downstream of the presumptive amplification and within both ends of the amplification. DNA copy number was estimated by the ΔΔCT method, using an uninvolved region of the genome as a comparator. Error bars indicate standard deviation from three separate measurements.
Validation of biofilm-associated mutations by transposon mutagenesis.
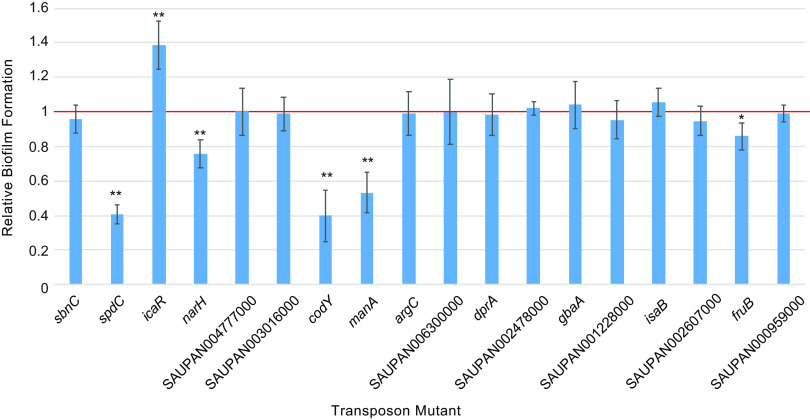
We next tested the contributions of candidate gene mutations to biofilm formation using a defined, isogenic transposon knockout mutant library of strain JE2 (26). Eighteen of the 23 candidate genes had corresponding transposon mutants available for testing (Fig. 3). Of the mutants examined, six gene disruptions significantly (P < 0.05 by two-tailed t test) altered biofilm formation relative to the parental strain. These comprised knockouts of spdC (15), icaR (28), and codY (13), each previously reported to influence biofilm formation in S. aureus, and three newly implicated genes, narH, manA, and fruB. Transposon knockouts of spdC, narH, codY, manA, and fruB showed reduced biofilm capacity. The transposon mutant of icaR uniquely enhanced biofilm formation, consistent with its loss-of-function promoting that virulence phenotype.
FIG 3.
Biofilm formation of isogenic transposon mutant strains. Relative biofilm formation of isogenic JE2 transposon knockouts in the indicated genes, normalized to biofilm production of the parental JE2 strain (red line). Error bars indicate standard deviation from at least four separate measurements. * indicates a P value of <0.05 and ** indicates a P value of <0.01 by two-tailed t test against the parental strain.
Complementation of newly identified biofilm-associated genes.
The demonstration that specific, isogenic transposon mutants measurably impacted biofilm formation is consistent with the impacted genes being involved in that phenotype. However, it is possible that additional, unlinked chromosomal mutations or polar effects resulting from transposition could have affected biofilm production (13).
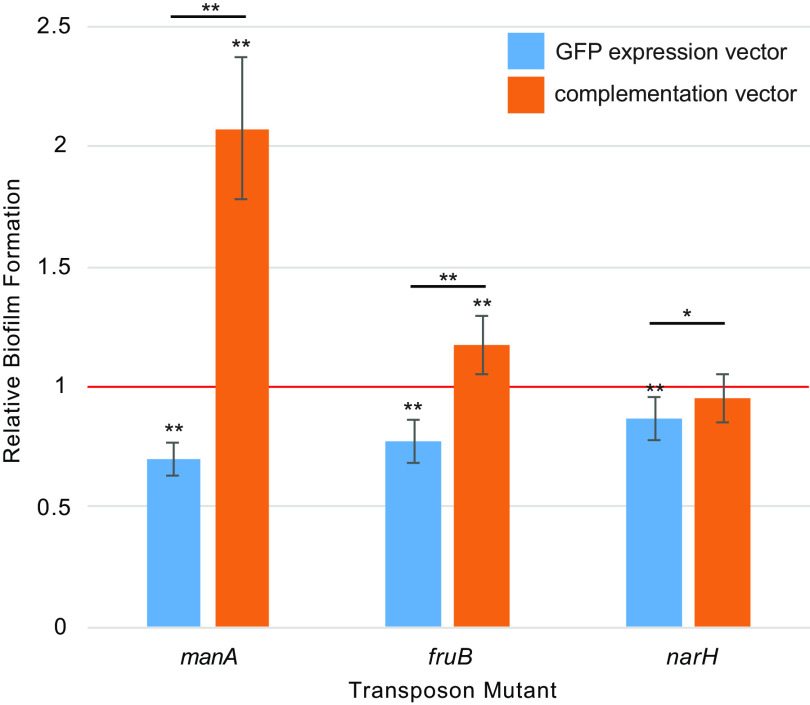
To address this uncertainty, we performed genetic complementation of the three biofilm-associated genes that were newly implicated by our study (narH, manA, and fruB). Each gene was cloned into an expression plasmid under the control of a strong, constitutive promoter and introduced into the corresponding transposon knockout mutant. To serve as a comparator for the complemented mutants, we generated an analogous vector expressing green fluorescent protein (GFP) and transformed that plasmid into the same genetic backgrounds.
Transposon mutant strains transformed with GFP expression vectors recapitulated the expected deficiencies in biofilm mutation we observed earlier (Fig. 4). In all cases, transformation of transposon mutants with plasmids expressing the corresponding gene fully compensated for those deficiencies. Moreover, complementation with manA and fruB under regulation by a strong constitutive promoter resulted in significantly increased biofilm formation levels relative to those of the original parental strain.
FIG 4.
Biofilm formation of complemented transposon mutant strains. Biofilm formation of isogenic JE2 transposon mutants after complementation with the corresponding gene or a control vector expressing GFP. Results are normalized to biofilm production of the parental JE2 strain carrying the control vector (red line). Error bars indicate standard deviation from at least four separate measurements. * indicates a P value of <0.05 and ** indicates a P value of <0.01 by two-tailed t test against the parental strain (displayed above each bar plot) or for the indicated comparisons between transposon mutants carrying control or complementation vectors (black lines).
Collectively, these results provide strong evidence that narH, manA, and fruB contribute to biofilm formation in S. aureus and that high-level constitutive expression of manA and fruB can promote enhanced biofilm formation in that organism.
DISCUSSION
S. aureus biofilms are composed of an extracellular matrix that incorporates a heterogeneous mixture of proteins, polysaccharides, and extracellular DNA and that follows a stereotyped pattern of development from initial formation to maturation and eventual dispersal (18, 29). The two major biochemical pathways responsible for aggregating S. aureus into biofilm matrices (7) comprise one dependent on the production of polysaccharide intercellular adhesin (PIA), which is biosynthesized by the ica operon (12), and a second, PIA-independent pathway mediated by adhesion of particular cell wall-associated proteins (7, 30). A number of genes directly contributing to, regulating, or providing accessory functions supporting those pathways have been catalogued, but the process of biofilm formation is complex (18), and additional effectors, including those involved in adaptive responses during chronic infection (19–24), have yet to be uncovered (7, 10, 18).
The most appropriate systems for modeling S. aureus biofilm growth in vivo are unknown (31), but studies (20) have indicated that clinical isolates from chronically infected cystic fibrosis patients show enhanced biofilm formation using standard microtiter plate assays (32), suggesting that the models employed in our study reasonably emulate relevant in vivo evolutionary pressures. Despite their initial, basal differences in biofilm formation capacity (Fig. 1A), all strains tested were able to evolve enhanced biofilm formation capacity with serial passaging (Fig. 1B). As reflected in our data, strain Newman has limited inherent capacity for biofilm formation due to a saeS mutation that actively represses multiple factors relevant to that phenotype (33). Interestingly, saeS reversion mutations were not recovered in any Newman mutants exhibiting enhanced biofilm formation, indicating that compensatory changes in other genes are more readily selected. N315, the strain with the greatest initial biofilm capacity, achieved the smallest relative gains in biofilm formation with passaging, suggesting that phenotype may already be close to a biological maximum for N315.
In vitro selection for mutants with enhanced biofilm production (Table 1) identified eight affected genes previously documented as involved in S. aureus biofilm formation (sigB, argC, icaR, gbaA, rpoZ, eap/map, spdC, and codY), three of which (icaR, spdC, and codY) significantly impacted biofilm formation as isogenic transposon mutants (Fig. 3). Expression of the virulence regulator sigB has previously been shown to drive biofilm formation in a dose-dependent fashion (34, 35). However, to the best of our knowledge, prior to this study spontaneous chromosomal duplication of the sigB locus (Fig. 2) has not been associated with enhanced S. aureus biofilm formation. Loss of icaR or gbaA, each a regulator of the ica polysaccharide intracellular adhesin operon (28, 36), results in overexpression of the canonical enzymes that manufacture major constituents of PIA-based biofilms (37) and correspondingly enhance biofilm production (28, 36). Although it was not possible to functionally assess argC, eap/map, or rpoZ in this study, prior work has linked these genes to biofilm phenotypes (38–41).
The remaining two genes, spdC and codY, are pleiotropic regulatory factors that indirectly influence biofilm formation (15, 42) and whose contributions are thereby multifactorial. Point mutations in codY were the most frequently selected genetic changes in passaged isolates, observed in a total of 11 replicates and in all four strains. All codY variants identified were single amino acid substitutions, most clustering within the helix-turn-helix motif of the DNA binding domain (residues 203 to 227 [43]) and three involving one of four essential DNA binding residues (44) (see Table S2 in the supplemental material). In contrast, only unequivocally inactivating mutations in spdC (two JE2 isolates with start-loss mutations and one N315 isolate with a stop-gain mutation) were recovered after in vitro selection for enhanced biofilm formation. Nevertheless, transposon disruption of both genes contrarily reduced biofilm formation (Fig. 2), a finding consistent with prior reports (13, 15). Together, these results suggest that mutations of spdC and codY could exert biofilm-enhancing effects when occurring concordantly with other relevant mutations or regulatory changes but could impair biofilm formation when introduced as isogenic mutants. This model is supported by our observation that all clearly inactivating mutations in spdC occur in concert with mutations in other candidate genes (Table S2) and would also explain inconsistencies reported in the phenotypic consequences of codY mutation, which in some studies have reduced biofilm formation and in others augmented it (13, 42, 45).
Excitingly, our in vitro evolution strategy newly identified and permitted functional validation of manA, narH, and fruB as genes involved in S. aureus biofilm formation (Fig. 3 and 4). Mannose-6-phosphate isomerase manA supports biofilm generation in other species by contributing to extracellular polysaccharide production (46), and our results are consistent with the gene serving a similar role in S. aureus. The contributory mechanisms of either fruB, encoding fructose-1-phosphate kinase, or narH, encoding nitrate reductase subunit beta, are less clear. However, we note that expression of both fruB and narH differs significantly between S. aureus grown in stationary phase and in biofilms (14) and that both genes contribute to biofilm production in other species (47, 48), providing orthologous support for their involvement in that phenotype. Both genes support anaerobic respiration, and it is plausible that their action may reflect the affinity of S. aureus to generate biofilms during anaerobic growth (49). Inactivation of manA, narH, and fruB by transposon mutagenesis each resulted in biofilm deficiencies, inconsistent with a loss-of-function model promoting biofilm production. Nevertheless, genetic complementation of those mutant strains (Fig. 4) overcame those deficiencies and in the case of manA and fruB indicates that constitutive, high-level expression can elevate biofilm production beyond basal levels. It is therefore possible that mutations observed in these genes after selection result in greater protein expression or activity, by means of derepression, increased stability, or elevated kinetics.
In summary, this effort demonstrates that S. aureus readily accumulates spontaneous genomic mutations in multiple genes that affect capacity for biofilm formation and that high-level expression of several novel factors results in enhanced biofilm production. Although our study has identified and validated three previously undescribed factors relevant to biofilm formation in S. aureus, it nonetheless remains subject to several limitations. First, it is possible that some candidate genes which failed functional validation legitimately contribute to biofilm formation. Given the complex epistatic interactions that govern many virulence traits in S. aureus (50), some variants may exert effects only when occurring in the presence of other genetic variants or in specific strain backgrounds. Effector genes whose contributions to biofilm production are not quantitatively large may also have been overlooked by our conservative functional validation criteria. Second, although we have considered four laboratory strains from different phylogenomic backgrounds, our work does not explore the scope of causative changes that could impact biofilm formation across all S. aureus lineages. Similarly, biofilm mutant screening is influenced by the conditions used and the substrate tested (51, 52), and any single experiment cannot identify all factors which could potentially impact that phenotype under particular circumstances. Third, our work has limited power to identify effector mutations that occur infrequently. Future work will seek to illuminate the mechanisms by which newly implicated genes influence biofilm formation and will further probe the range of mutations and mechanisms by which S. aureus is able to augment its capacity for biofilm formation as an adaptive phenotype during chronic infection.
MATERIALS AND METHODS
Strains and growth conditions.
S. aureus strain ATCC 29213 was purchased from the American Type Culture Collection. Strains JE2 and N315 were obtained from the Biodefense and Emerging Infections Research Resources Repository (BEI Resources). JE2 transposon mutants in defined genes (26) were also obtained from BEI Resources. S. aureus strain Newman was a generous gift from Lucas Hoffman (University of Washington).
Strains were routinely maintained at 37°C using tryptic soy broth (TSB) medium, except for those transformed with temperature-sensitive complementation vectors, which were cultured at 32°C with the addition of 10 μg/mL chloramphenicol to select for retention of plasmids.
Selection for biofilm formation and biofilm formation assays.
Passaging to select for mutants with enhanced biofilm formation was conducted using modifications of existing protocols (53, 54). Briefly, overnight cultures of S. aureus were grown in tryptic soy broth (TSB), diluted 1:100 in fresh medium, and deposited in 450-μL aliquots into 48-well tissue culture plates containing sterile glass coverslips (German glass round 5-mm coverslips, supplied by VWR, Radnor, PA) (54). Following overnight incubation, coverslips were removed with sterile forceps and rinsed with sterile Dulbecco’s phosphate-buffered saline (DPBS), and the biofilm was dispersed into 1 mL fresh TSB by vortexing. Cells were diluted to an optical density at 600 nm (OD600) of 0.07 and were applied to fresh culture plates with coverslips to initiate the next round of passaging.
To quantitatively assess biofilm formation, overnight cultures were grown in TSB, diluted to an OD600 of 0.063 in 450 μL TSB, and deposited into the wells of a 48-well tissue culture plate. Following 18 h of incubation, biofilm formation was assessed by crystal violet staining as described elsewhere (53). At least four replicates of each specimen were used to quantitate biofilm formation levels, with readings normalized to in-batch controls of the appropriate parental strain.
Whole-genome sequencing and genomic analysis.
Whole-genome sequencing and analysis were performed as previously described (55) using a MiSeq platform with 300 cycle chemistries (Illumina, San Diego, CA). Briefly, variant calling utilized reference genomes identified as those most closely matching the de novo assemblies of sequenced isolates. Mutations were manually verified using the Integrative Genomics Viewer (56) prior to annotation of sequence features. Common gene names and pangenome identifiers were established using BLAST (57) searches of target gene sequences against S. aureus reference genomes and by cross-correlating gene identifiers against the AureoWiki database (27). Chromosomal copy number analysis was performed using CNOGpro (58).
Real-time PCR for copy number verification.
Real-time PCR was performed to verify a presumptive copy number alteration in one evolved strain. Primers (see Table S1 in the supplemental material) targeted 5′ and 3′ regions within the alterations, regions ~1 kb upstream and downstream of the alteration, and a chromosomal locus distant to the copy number change which was used as a comparator for copy number estimation using the threshold cycle (ΔΔCT) method. All oligonucleotides used in this study were synthesized by IDT (Coralville, IA).
Genetic complementation.
Genes of interest and an S. aureus codon-optimized SuperfolderGFP gene (59) (serving as a control) were synthesized (gBlocks; IDT) under the control of the strong, constitutive S. aureus Pcap promoter (60) (Table S1). The erythromycin resistance determinant of temperature-sensitive shuttle vector pCN50-ermc-E194ts (61) was replaced with the chloramphenicol resistance cassette of pCN50-BSAI (61) to yield pCNpE194ts-cat, and each expression cassette was subsequently cloned upstream of that vector’s transcriptional terminator to yield the final complementation vectors. Complementation vectors and the GFP-expression control were transformed into matched JE2 transposon mutant strains as described elsewhere (61). Biofilm formation of complemented mutants was measured as described above, with the addition of 10 μg/mL chloramphenicol to all culture media, with readings normalized to in-batch controls of the parental JE2 strain transformed with the GFP-expression control vector.
Data availability.
Sequence data generated from this study are available from the NCBI Sequence Read Archive under accession PRJNA860616.
ACKNOWLEDGMENTS
This work was supported by grants from Vertex Pharmaceuticals, the Cystic Fibrosis Foundation (SINGH19R0 to S.J.S.), and NIH (P30 DK089507 to S.J.S., K23 AR080209 to D.R.L.). The funders of this study had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Stephen J. Salipante, Email: stevesal@uw.edu.
Victor J. Torres, New York University Grossman School of Medicine
REFERENCES
- 1.Conlon BP. 2014. Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells: an investigation of persister cells, their formation and their role in S. aureus disease. Bioessays 36:991–996. 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- 2.Shah PL, Mawdsley S, Nash K, Cullinan P, Cole PJ, Wilson R. 1999. Determinants of chronic infection with Staphylococcus aureus in patients with bronchiectasis. Eur Respir J 14:1340–1344. 10.1183/09031936.99.14613409. [DOI] [PubMed] [Google Scholar]
- 3.Garzoni C, Kelley WL. 2009. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol 17:59–65. 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Clement S, Vaudaux P, Francois P, Schrenzel J, Huggler E, Kampf S, Chaponnier C, Lew D, Lacroix J-S. 2005. Evidence of an intracellular reservoir in the nasal mucosa of patients with recurrent Staphylococcus aureus rhinosinusitis. J Infect Dis 192:1023–1028. 10.1086/432735. [DOI] [PubMed] [Google Scholar]
- 5.Kalinka J, Hachmeister M, Geraci J, Sordelli D, Hansen U, Niemann S, Oetermann S, Peters G, Löffler B, Tuchscherr L. 2014. Staphylococcus aureus isolates from chronic osteomyelitis are characterized by high host cell invasion and intracellular adaptation, but still induce inflammation. Int J Med Microbiol 304:1038–1049. 10.1016/j.ijmm.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Copin R, Sause WE, Fulmer Y, Balasubramanian D, Dyzenhaus S, Ahmed JM, Kumar K, Lees J, Stachel A, Fisher JC, Drlica K, Phillips M, Weiser JN, Planet PJ, Uhlemann A-C, Altman DR, Sebra R, van Bakel H, Lighter J, Torres VJ, Shopsin B. 2019. Sequential evolution of virulence and resistance during clonal spread of community-acquired methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA 116:1745–1754. 10.1073/pnas.1814265116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459. 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister JL, Horswill AR. 2014. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol 4:178. 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherr TD, Heim CE, Morrison JM, Kielian T. 2014. Hiding in plain sight: interplay between staphylococcal biofilms and host immunity. Front Immunol 5:37. 10.3389/fimmu.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilcher K, Horswill AR. 2020. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol Mol Biol Rev 84:e00026-19. 10.1128/MMBR.00026-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin H-J, Yang S, Lim Y. 2021. Antibiotic susceptibility of Staphylococcus aureus with different degrees of biofilm formation. J Anal Sci Technol 12:41. 10.1186/s40543-021-00294-2. [DOI] [Google Scholar]
- 12.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433. 10.1128/IAI.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, Kelley WL. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect Immun 75:1079–1088. 10.1128/IAI.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. 2004. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186:4665–4684. 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poupel O, Proux C, Jagla B, Msadek T, Dubrac S. 2018. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog 14:e1006917. 10.1371/journal.ppat.1006917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brady AJ, Laverty G, Gilpin DF, Kearney P, Tunney M. 2017. Antibiotic susceptibility of planktonic- and biofilm-grown staphylococci isolated from implant-associated infections: should MBEC and nature of biofilm formation replace MIC? J Med Microbiol 66:461–469. 10.1099/jmm.0.000466. [DOI] [PubMed] [Google Scholar]
- 17.Resch A, Leicht S, Saric M, Pásztor L, Jakob A, Götz F, Nordheim A. 2006. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6:1867–1877. 10.1002/pmic.200500531. [DOI] [PubMed] [Google Scholar]
- 18.Moormeier DE, Bayles KW. 2017. Staphylococcus aureus biofilm: a complex developmental organism: molecular mechanisms of S. aureus biofilm development. Mol Microbiol 104:365–376. 10.1111/mmi.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabryszewski SJ, Wong Fok Lung T, Annavajhala MK, Tomlinson KL, Riquelme SA, Khan IN, Noguera LP, Wickersham M, Zhao A, Mulenos AM, Peaper D, Koff JL, Uhlemann A-C, Prince A. 2019. Metabolic adaptation in methicillin-resistant Staphylococcus aureus pneumonia. Am J Respir Cell Mol Biol 61:185–197. 10.1165/rcmb.2018-0389OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan X, Coureuil M, Ramond E, Euphrasie D, Dupuis M, Tros F, Meyer J, Nemazanyy I, Chhuon C, Guerrera IC, Ferroni A, Sermet-Gaudelus I, Nassif X, Charbit A, Jamet A. 2019. Chronic Staphylococcus aureus lung infection correlates with proteogenomic and metabolic adaptations leading to an increased intracellular persistence. Clin Infect Dis 69:1937–1945. 10.1093/cid/ciz106. [DOI] [PubMed] [Google Scholar]
- 21.Schwartbeck B, Birtel J, Treffon J, Langhanki L, Mellmann A, Kale D, Kahl J, Hirschhausen N, Neumann C, Lee JC, Götz F, Rohde H, Henke H, Küster P, Peters G, Kahl BC. 2016. Dynamic in vivo mutations within the ica operon during persistence of Staphylococcus aureus in the airways of cystic fibrosis patients. PLoS Pathog 12:e1006024. 10.1371/journal.ppat.1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suligoy CM, Lattar SM, Noto Llana M, González CD, Alvarez LP, Robinson DA, Gómez MI, Buzzola FR, Sordelli DO. 2018. Mutation of Agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front Cell Infect Microbiol 8:18. 10.3389/fcimb.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. 2011. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS One 6:e24301. 10.1371/journal.pone.0024301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Awan F, Ali MM, Mushtaq MH, Ijaz M. 2021. Genetic diversity in Staphylococcus aureus and its relation to biofilm production. In Aqib A (ed), Insights into drug resistance in Staphylococcus aureus. IntechOpen Limited, London, United Kingdom. [Google Scholar]
- 25.Liu H, Shang W, Hu Z, Zheng Y, Yuan J, Hu Q, Peng H, Cai X, Tan L, Li S, Zhu J, Li M, Hu X, Zhou R, Rao X, Yang Y. 2018. A novel SigB(Q225P) mutation in Staphylococcus aureus retains virulence but promotes biofilm formation. Emerg Microbes Infect 7:72. 10.1038/s41426-018-0078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs S, Mehlan H, Bernhardt J, Hennig A, Michalik S, Surmann K, Pané-Farré J, Giese A, Weiss S, Backert L, Herbig A, Nieselt K, Hecker M, Völker U, Mäder U. 2018. Aureo Wiki-The repository of the Staphylococcus aureus research and annotation community. Int J Med Microbiol 308:558–568. 10.1016/j.ijmm.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Conlon KM, Humphreys H, O’Gara JP. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol 184:4400–4408. 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Idrees M, Sawant S, Karodia N, Rahman A. 2021. Staphylococcus aureus biofilm: morphology, genetics, pathogenesis and treatment strategies. Int J Environ Res Public Health 18:7602. 10.3390/ijerph18147602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penadés JR, Lasa I. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol 191:832–843. 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staudinger BJ, Muller JF, Halldórsson S, Boles B, Angermeyer A, Nguyen D, Rosen H, Baldursson O, Gottfreðsson M, Guðmundsson GH, Singh PK. 2014. Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am J Respir Crit Care Med 189:812–824. 10.1164/rccm.201312-2142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp (47):2437. 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cue D, Junecko JM, Lei MG, Blevins JS, Smeltzer MS, Lee CY. 2015. SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS One 10:e0123027. 10.1371/journal.pone.0123027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bateman BT, Donegan NP, Jarry TM, Palma M, Cheung AL. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect Immun 69:7851–7857. 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachid S, Ohlsen K, Wallner U, Hacker J, Hecker M, Ziebuhr W. 2000. Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J Bacteriol 182:6824–6826. 10.1128/JB.182.23.6824-6826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray A, Edmonds KA, Palmer LD, Skaar EP, Giedroc DP. 2020. Staphylococcus aureus glucose-induced biofilm accessory protein A (GbaA) is a monothiol-dependent electrophile sensor. Biochemistry 59:2882–2895. 10.1021/acs.biochem.0c00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson KM, Abraham N, Jefferson KK. 2010. Staphylococcus aureus extracellular adherence protein contributes to biofilm formation in the presence of serum. FEMS Microbiol Lett 305:143–147. 10.1111/j.1574-6968.2010.01918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonemoto K, Chiba A, Sugimoto S, Sato C, Saito M, Kinjo Y, Marumo K, Mizunoe Y. 2019. Redundant and distinct roles of secreted protein Eap and cell wall-anchored protein SasG in biofilm formation and pathogenicity of Staphylococcus aureus. Infect Immun 87:e00894-18. 10.1128/IAI.00894-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss A, Moore BD, Tremblay MHJ, Chaput D, Kremer A, Shaw LN. 2017. The ω subunit governs RNA polymerase stability and transcriptional specificity in Staphylococcus aureus. J Bacteriol 199:e00459-16. 10.1128/JB.00459-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu B, Ge X, Stone V, Kong X, El-Rami F, Liu Y, Kitten T, Xu P. 2017. ciaR impacts biofilm formation by regulating an arginine biosynthesis pathway in Streptococcus sanguinis SK36. Sci Rep 7:17183. 10.1038/s41598-017-17383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol 190:2257–2265. 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han A-R, Kang H-R, Son J, Kwon DH, Kim S, Lee WC, Song HK, Song MJ, Hwang KY. 2016. The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism. Nucleic Acids Res 44:9483–9493. 10.1093/nar/gkw775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joseph P, Ratnayake-Lecamwasam M, Sonenshein AL. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J Bacteriol 187:4127–4139. 10.1128/JB.187.12.4127-4139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mlynek KD, Bulock LL, Stone CJ, Curran LJ, Sadykov MR, Bayles KW, Brinsmade SR. 2020. Genetic and biochemical analysis of CodY-mediated cell aggregation in Staphylococcus aureus reveals an interaction between extracellular DNA and polysaccharide in the extracellular matrix. J Bacteriol 202:e00593-19. 10.1128/JB.00593-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amos MR, Sanchez-Contreras M, Jackson RW, Muñoz-Berbel X, Ciche TA, Yang G, Cooper RM, Waterfield NR. 2011. Influence of the Photorhabdus luminescens phosphomannose isomerase gene, manA, on mannose utilization, exopolysaccharide structure, and biofilm formation. Appl Environ Microbiol 77:776–785. 10.1128/AEM.02326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangalea MR, Borlee BR. 2022. The NarX-NarL two-component system regulates biofilm formation, natural product biosynthesis, and host-associated survival in Burkholderia pseudomallei. Sci Rep 12:203. 10.1038/s41598-021-04053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakraborty B, Zeng L, Burne RA. 2022. The fruB gene of Streptococcus mutans encodes an endo-levanase that enhances growth on levan and influences global gene expression. Microbiol Spectr 10:e0052222. 10.1128/spectrum.00522-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mashruwala AA, van de Guchte A, Boyd JM. 2017. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife 6:e23845. 10.7554/eLife.23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laabei M, Recker M, Rudkin JK, Aldeljawi M, Gulay Z, Sloan TJ, Williams P, Endres JL, Bayles KW, Fey PD, Yajjala VK, Widhelm T, Hawkins E, Lewis K, Parfett S, Scowen L, Peacock SJ, Holden M, Wilson D, Read TD, van den Elsen J, Priest NK, Feil EJ, Hurst LD, Josefsson E, Massey RC. 2014. Predicting the virulence of MRSA from its genome sequence. Genome Res 24:839–849. 10.1101/gr.165415.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiltunen AK, Savijoki K, Nyman TA, Miettinen I, Ihalainen P, Peltonen J, Fallarero A. 2019. Structural and functional dynamics of Staphylococcus aureus biofilms and biofilm matrix proteins on different clinical materials. Microorganisms 7:584. 10.3390/microorganisms7120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cassat JE, Lee CY, Smeltzer MS. 2007. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol 391:127–144. 10.1007/978-1-59745-468-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40:175–179. 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 54.Dassanayake RP, Falkenberg SM, Stasko JA, Shircliff AL, Lippolis JD, Briggs RE. 2020. Identification of a reliable fixative solution to preserve the complex architecture of bacterial biofilms for scanning electron microscopy evaluation. PLoS One 15:e0233973. 10.1371/journal.pone.0233973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Werth BJ, Ashford NK, Penewit K, Waalkes A, Holmes EA, Bryan A, Salipante SJ. 2022. Evolution of cefiderocol resistance in Stenotrophomonas maltophilia using in vitro serial passage techniques. JAC Antimicrob Resist 4:dlac011. 10.1093/jacamr/dlac011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 58.Brynildsrud O, Snipen L-G, Bohlin J. 2015. CNOGpro: detection and quantification of CNVs in prokaryotic whole-genome sequencing data. Bioinformatics 31:1708–1715. 10.1093/bioinformatics/btv070. [DOI] [PubMed] [Google Scholar]
- 59.Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 60.Schwendener S, Perreten V. 2015. New shuttle vector-based expression system to generate polyhistidine-tagged fusion proteins in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol 81:3243–3254. 10.1128/AEM.03803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Penewit K, Holmes EA, McLean K, Ren M, Waalkes A, Salipante SJ. 2018. Efficient and scalable precision genome editing in Staphylococcus aureus through conditional recombineering and CRISPR/Cas9-mediated counterselection. mBio 9:e00067-18. 10.1128/mBio.00067-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download iai.00538-22-s0001.xlsx, XLSX file, 0.01 MB (11.7KB, xlsx)
Table S2. Download iai.00538-22-s0002.xlsx, XLSX file, 0.01 MB (14.1KB, xlsx)
Table S3. Download iai.00538-22-s0003.xlsx, XLSX file, 0.01 MB (13.7KB, xlsx)
Data Availability Statement
Sequence data generated from this study are available from the NCBI Sequence Read Archive under accession PRJNA860616.