Abstract
The metagenomics approach accelerated the study of genetic information from uncultured microbes and complex microbial communities. In silico research also facilitated an understanding of protein-DNA interactions, protein–protein interactions, docking between proteins and phyto/biochemicals for drug design, and modeling of the 3D structure of proteins. These in silico approaches provided insight into analyzing pathogenic and nonpathogenic strains that helped in the identification of probable genes for vaccines and antimicrobial agents and comparing whole-genome sequences to microbial evolution. Artificial intelligence, more precisely machine learning (ML) and deep learning (DL), has proven to be a promising approach in the field of microbiology to handle, analyze, and utilize large data that are generated through nucleic acid sequencing and proteomics. This enabled the understanding of the functional and taxonomic diversity of microorganisms. ML and DL have been used in the prediction and forecasting of diseases and applied to trace environmental contaminants and environmental quality. This review presents an in-depth analysis of the recent application of silico approaches in microbial genomics, proteomics, functional diversity, vaccine development, and drug design.
Keywords: Artificial intelligence, Deep learning, Machine learning, Metagenomics, Microbiology
Introduction
The discipline of microbiology means exploring the structure and function, interrelationships, and mechanisms within communities of microorganisms and their interactions with the immediate environments or hosts. Microscopy has been the key technique for the identification of microbes, which is complementarily followed by culture techniques to elucidate their physiology, genetic constructs, metabolism, and pathogenicity. However, these procedures are time-consuming and labor-intensive. The incorporation of advanced techniques such as high-throughput sequencing and next-generation sequencing in the field of microbiology has presented a plethora of genomic data. This accumulation of data from various domains of microbial genomics has enabled the development of new diagnostic and genotyping tools, deciphered microbial genetic diversity, and identified virulence and resistance mechanisms. Additionally, in silico methods assist in gathering genetic information that can be used to identify therapeutic targets, investigate host–pathogen interactions, and establish mechanisms of antibiotic resistance and virulence.
Therefore, an optimal analysis and interpretation of these large intricate data is the next challenge to achieve these promising advances. This task is beyond human expertise with a high risk of errors involved and calls out for advanced computational techniques that can detect meaningful patterns from the heaps of data. Artificial intelligence can fill these gaps with techniques such as machine learning, which uses structured data and recognizes meaningful patterns with supervised and unsupervised learning methods.
Bioinformatics, an application of information technology, helps in the processing and analysis of the data generated in biological research and experiments by applying computer-based algorithms. It helps in DNA barcoding and designing the patterns of disease outbreaks and new biological products. Proteomics also facilitates the study of protein structures and the identification of protein–protein interaction sites (Rao et al. 2014). In the study of metabolomics, dynamics in cell and cellular interactions are possible with the help of bioinformatics (Kushwaha et al. 2017). Bioinformatics has not only helped in genome sequencing and presented accomplishments in gene allocations but also helped to draw phylogenetic relationships and detect transcription factor-binding sites of the genes. Microarray data analysis is made possible by bioinformatics tools. Biological data are growing exponentially due to the availability of low-cost sequencing technologies. The enormous amount of data generated has led to the development of databases of nucleic acid sequences, protein sequences, and their structures. For example, Swiss-Prot and PIR for protein sequences, GenBank and DDBJ for genome sequences and protein structures, and protein databanks are established primary databases. Various software and tools that could be helpful in microbiological studies are summarized in Table 1.
Table 1.
Useful software and tools for microbial studies
| S. no | Software name | Usage | Description | URL | References |
|---|---|---|---|---|---|
| 1 | Prodigal | Gene annotation | Software tool for protein-coding gene prediction of bacterial and archaeal genomes | https://github.com/hyattpd/prodigal/wiki | Hyatt et al. (2010) |
| 2 | Prokka | Functional annotation of bacterial genomes produces standards-compliant output file | https://github.com/tseemann/prokka | Seemann (2014) | |
| 3 | RAST | Automated annotation tool for bacterial and archaeal genomes | https://rast.nmpdr.org/ | Aziz et al. (2008) | |
| 4 | MicroScope | A detailed analytical platform for annotation and analysis of bacterial genomes | https://mage.genoscope.cns.fr/microscope/home/index.php | Vallenet et al. (2009) | |
| 5 | NCBI prokaryotic genome annotation pipeline (PGAP) | Automated genome annotation pipeline for bacteria, combination of ab initio gene prediction algorithms with homology-based methods | https://www.ncbi.nlm.nih.gov/genome/annotation_prok/NCBI Pathogen Detection | Tatusova et al. (2016) | |
| 6 | HarvestA | Genome alignments | Core genome alignment and visualization tools for quick and high-throughput analysis of intraspecific bacterial genomes | http://harvest.readthedocs.io/en/latest/ | Treangen et al. (2014) |
| 7 | Roary | Homology clustering and association studies |
Aligner for comparative analysis of full bacterial genomes High speed stand-alone pangenome pipeline for bacterial genomes |
http://sanger-pathogens.github.io/Roary/ | Page et al. (2015) |
| 8 | Scoary Neptune | Pangenome wide association studies using Roary output software designed for detecting genomic signatures within bacterial populations | Brynildsrud et al. (2016); Marinier et al. (2017) | ||
| 9 | RAxML | Phylogenetic inference | Sequential and parallel maximum-likelihood phylogeny estimation based on nucleotide and protein sequence alignments | https://cme.h-its.org/exelixis/software.html | Alexey et al. Kozlov et al. (2019) |
| 10 | Fast Tree | Maximum likelihood phylogenetic trees from large nucleotide or protein multiple sequence alignments | http://www.microbesonline.org/fasttree/ | Price et al. (2010) | |
| 11 | Gubbins | Compute maximum likelihood from alignment after removing regions containing elevated densities of base substitution | https://github.com/sangerpathogens/gubbins | Croucher et al. (2015) | |
| 12 | Clonal Frame | MLA maximum-likelihood implementation of clonal frame designed for genomes sequences | www.github.com/xavierdidelot/ClonalFrameML | Didelot and Wilson (2015) | |
| 13 | PHYLOViZ | Online web-based tool for phylogenetic analysis and sequence-based typing methods to generate allelic profiles and associated epidemiologic data | www.online.phyloviz.net/index | Francisco et al. (2012) | |
| 14 | PHYLOViZ 2.0 | Java software used for integration and visualization of multiple phylogenetic inference methods, also used for analysis of sequence-based typing methods that generate allelic profiles and their associated epidemiologic data | www.phyloviz.net/ | Nascimento et al. (2017) | |
| 15 | Microreact | Visualization tool | Browser based application for visualization and sharing of genomic epidemiology data | https://microreact.org/ | Argimón et al. (2016) |
| 16 | Phandango | Online tool for quick evaluation of large-scale population genomics datasets that combines output from multiple genomic analysis methods | https://github.com/jameshadfield/phandango | Hadfield et al. (2018) | |
| 17 | iTOL | Online application used to manage, display, and annotate phylogenetic trees | https://itol.embl.de/ | Letunic and Bork (2021) | |
| 18 | GenGIS 2 | Applications including 3-D graphical and Python interfaces that allow users to combine sequences and digital map data | https://beikolab.cs.dal.ca/gengis/Main_Page | Parks et al. (2013) |
In silico approaches for microbial genomics
Metagenomics is an approach of advanced genomics techniques to study the microbial communities directly from their natural environments without cultivation in the lab and isolation of individual species (DeLong 2002; Riesenfeld et al. 2004a, b; Handelsman 2004; Rodriguez-Valera 2004; Streit and Schmitz 2004; Edwards and Rohwer 2005). This is the culture-independent approach for retrieval of 16S rRNA genes, established two decades ago by Pace and colleagues (Olsen et al. 1986). In 2002, Hugenholtz reported that until that time, 99% of microbial species had not been cultivated due to limitations but metagenomics approaches, revolutionized microbiology by eliminating the need for clonal isolates (Hugenholtz 2002; Rappe and Giovannoni 2003; Singh and Porwal 2021).
Metagenomic assembly facilitates gene prediction and annotation and is therefore considered a significant step when studying the functional constitution and size of microbiomes (Van der Walt et al. 2017).
To facilitate microbial identification studies, various techniques have emerged. DNA pyrosequencing, also known as sequencing by synthesis, was developed in the mid-1990s (Ronaghi et al. 1996). The major limitation of this method is its inability to read the long stretches of DNA sequence (sequences hardly exceed 100–200 base pairs with first- and second-generation pyrosequencing chemistries) (Joseph et al. 2009).
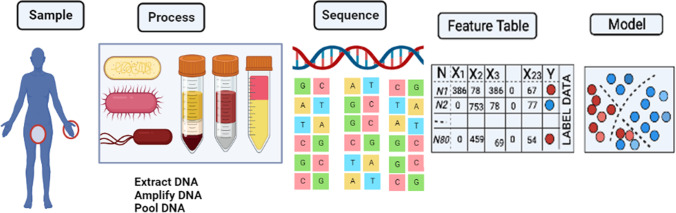
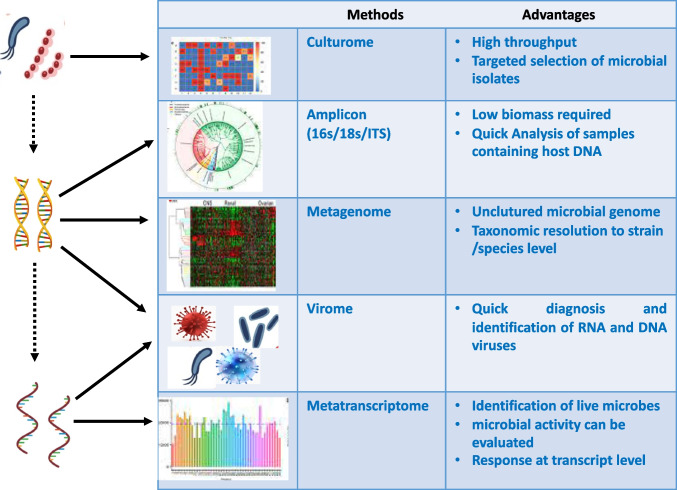
With the advent of sequencing technology, next-generation sequencing (NGS) has emerged as a rapid and reliable method for the identification of bacterial pathogens. NGS has evolved as a molecular microscope, expanding its applications into every field of microbial research (Buermans and den Dunnen 2014). The application of NGS in the microbial world includes both wet lab and bioinformatics tools/computational methods (Fig. 1) (Ghannam and Techtmann 2021). The first step of this technique is the molecular profiling of the microbial community that incorporates collection of sample (from the patient or environment), nucleic acid extraction, and library preparation. Several biases could be introduced with wet lab methods (Hazen et al. 2013). After sequencing, the primary analysis was performed using bioinformatics tools. Several studies have taken place on the processing of sequencing reads. This includes methods for binning marker genes into operational taxonomic units (OTUs) and is representative of biologically meaningful categories (Edgar 2010). Liu et al. (2021) have elaborated the step wise analysis methods used for high throught put analysis of microbiome. The collected samples are first diluted and then distributed in microtiter plate of 96 wells. The wells are then subjected to amplicon sequencing and selected as candidate. The candidates are further subjected to 16rDNA full length Sangers sequencing (Fig. 2).
Fig. 1.
Schematic representation of the next-generation sequencing approach used for the investigation of microbial communities through a pipeline that comprises collection of samples, nucleic acid extraction from hosts or the environment and preparation of libraries for sequencing (figure recreated in Biorender.com)
Fig. 2.
Advantages and HTS methods for different levels of microbiome analysis. At the molecule level, microbiome studies are divided into three types: microbe, DNA, and mRNA. The corresponding research techniques include culturome, amplicon, metagenome, metavirome, and metatranscriptome analyses. And corresponding advantages of various HTS methods used for analysis
Peker et al. compared the three methods for NGS data analysis for speed and diagnostic accuracy: de novo assembly followed by the Basic Local Alignment Search Tool (BLAST), operational taxonomic unit (OTU) for clustering and an in house developed database (16S–23S rRNA encoding region). They directly used the patient samples to perform NGS of the 16S and 23S rRNA encoding regions for reliable identification of pathogens. Although NGS data analysis is tedious and laborious, a database for the complete 16S–23S rRNA coding region is not obtainable. The study suggested and recommended de novo assembly followed by BLAST as a better method. This method showed the shortest turnaround time (2 h and 5 min), which is two hours less than OTU clustering and 4.5 h less than mapping, with a sensitivity of 80%. This analysis concluded that the blend of de novo assembly and BLAST seems to be the best approach for the analysis of data (Peker et al. 2019). Additionally, comprehension of protein-DNA interactions, protein–protein interactions, docking between proteins and phyto/biochemicals for drug design, and modelling of the three-dimensional structure of proteins were made possible by in silico research (Qiu et al. 2020; Bryant et al. 2022; Baig et al. 2016; Ali et al. 2021; Fatoki et al. 2021).
Machine learning for metagenomic data analysis
With the evolution of technology and machine learning (ML) models, metagenomics has become a popular field of bioinformatics. One can create more competent models to address the problems of DNA sequencing and genome classification. As the technology is becoming more sophisticated, new more precise DNA sequencing techniques have been developed, and the enhanced computational power of modern computers has helped to achieve that. As a result, much larger quantities of data can now be processed and trained with more complex machine learning models that were earlier not feasible due several limitations. The advantage of ML is that it can fully appreciate the depth of data generated while microbiome studies and build predictive models based on outcomes for the data achieved from the microbial community (Ghannam and Techtmann 2021). ML approaches use several forms, involving unsupervised, semisupervised, reinforced, or supervised learning (Kumar et al. 2018; Saxena et al. 2019; Sathya and Abraham 2013: Zitnik et al. 2019) (Fig. 2). The model that uses a training set falls under supervised learning (Stoter et al. 2019). Statistical classification and regression analysis come under common supervised learning algorithms (Kumar et al. 2011). Clustering, also known as unsupervised learning, implements k-means to determine a centriole and reduces error by iteration and descent to achieve classification (Omer et al. 2014).
The progression of ML has led to the use of this technique in various fields of research (Chen et al. 2016; Li et al. 2016; Zou et al. 2016; Ding et al. 2017; Feng et al. 2017; Yu et al. 2017; Zeng et al. 2017; Pan et al. 2018; Liu et al. 2018; He et al. 2019; Kumar et al. 2021; Zhang et al. 2019). Such exemplary applications are drug repurposing (Yu et al. 2016, 2017), discovery of new antibiotics (Steele et al. 2009), identification of novel biocatalysts, personalized medicine (Virgin and Todd 2011; Pires et al. 2020a, b; Villasana et al. 2020), identification of disease-related microRNAs (Chen and Huang 2017; Zhao et al. 2018), identification of disease-related noncoding RNAs (Chen and Yan 2013; Hu et al. 2017, 2018), and bioremediation of agricultural, industrial, and domestic wastes (Mani and Kumar 2014; Pires et al. 2020a, b). Oudah and Henschel defined the four key stages of ML algorithm development (Oudah and Henschel 2018): The first step of the ML method, which is also a critical stage, addresses the extraction of the features (Liu et al. 2015) and then OTUs, which are obtained by clustering. Then, the significant features that are responsible for enhancing the precision and proficiency are selected, and the final step is training the dataset that is used to train an algorithm and fit the dataset. After that, a test set is used for the evaluation of the model.
Machine learning for disease prediction and classification
Various normal microflora residing in the gut play vital roles in human health. Disturbances in intestinal microorganisms may cause inflammatory diseases of the intestine (Chen et al. 2017a, b, c), such as colorectal cancer, tumors, diabetes, ulcerative colitis, and obesity. Consequently, it becomes essential to interpret the relationship of microbes, a disease, better clinical prognostic tests, and the development of new drugs (Yu et al. 2015, 2016; Shi et al. 2016; Su et al. 2018, Fan et al. 2019, Arango-Argoty et al. 2018, Steiner et al. 2020).
For the analysis of microiome–host interactions in the context of disease, an approach was given by Fan et al. (2019) that combines several data sources of the human microbiome–host disease consortium with HeteSim scores. Initially, they constructed heterogenicity networks and then conducted microbe–disease pair weighting with the standardized HeteSim measurement method. This was followed by the integration of the microbes–disease–disease pathway with HeteSim scores of the microbe‒microbe–disease pathway and finally calculation of the corresponding scores of probable microgenome associations.
Amgarten et al. (2018) proposed a new tool, MARVEL, for the prediction of the double-stranded DNA sequence of bacteriophages in metagenomics. MARVEL uses a random forest (RF) approach with a large dataset containing 1247 phage genomes and 1029 bacterial genomes along with a test dataset consisting of 335 bacterial and 177 phage genomes. Six features were proposed for the identification of phages, and then, RF was exercised for the selection of features. Finally, three features were established, which provided more information (Grazziotin et al. 2017).
Over the last few years, many studies have explored and scrutinized the role of microbiome communities in the prediction of diseases. Later, researchers incorporated complete genome sequencing and entire transcriptome sequencing data of 33 types of cancer from The Cancer Genome Atlas (TCGA) to examine the potential of microbial signatures as cancer predictors by using variation boosting ML models (Poore et al. 2020). The ML models successfully discriminated different cancer types and distinguished between cancer and normal tissues, suggesting that the microbiome is exclusive to each cancer type and cancer stage. The authors concluded that the proposed model could serve as a potential tool in microbiome-based cancer diagnosis. A similar study investigated the role of the vaginal microbial community based on bacterial signatures in the prediction of cervical intraepithelial neoplasia (CIN) using a random forest model (Lee et al. 2020). Sequencing data of the V3 region of 16S rRNA from vaginal swabs of 66 subjects were investigated for its taxonomic composition. A set of 33 bacterial species were obtained as marker communities differentiating between the CIN1 and CIN2 groups, with 0.952 area under curve (AUC). This finding validates the potential of the RF model in the prediction of CIN staging and VM as a biomarker.
Cai et al. (2019) focused on investigating the underlying mechanism of pathogenesis in human diseases using genomics with the help of in silico applications. They used a novel ML-based approach and recognized two genes, OTOF and SOCS1, that contribute to the pathogenesis mechanism of rhinovirus (Xu et al. 2019). The expression levels of these two genes could potentially determine the infected or noninfected state of an individual. Alongside depicting the significance of these two genes in rhinovirus pathogenesis, this study also demonstrated the effectiveness of in silico applications in studying the pathogenesis mechanisms. Wang et al. in 2019, proposed a spectral rotation method based on the triplet periodicity property to solve planted motif finding problems (Wang et al. 2019). The proposed method gives genes with several substitutions that can be detected from arbitrarily generated background sequences. The results of the experiment based on the genomic dataset of Saccharomyces cerevisiae showed that genes could be visually distinguished. The authors suggested that genes having approximately 50% mutations could be easily identified in background sequences.
Several studies have explored viral genomics with the help of in silico approaches. Remita et al. developed a machine learning-based virus classification tool called CASTOR and used different datasets of hepatitis B virus, human papillomaviruses (HPV), and HIV-1 as testing datasets (Remita et al. 2017). The model imitates the restriction fragment length polymorphism (RFLP) technique in silico and stimulates fragmention of genomic material by different restriction endonucleases. The authors noted positive cases of 99% for HPV alpha species, 99% for HBV genotyping, and 98% for HIV-1 M subtyping. They concluded that this model is a great fit to achieve accurate large-scale virus studies owing to its generality and robustness (Lebatteux et al. 2019). Ren et al. proposed VirFinder (a novel k-mer-based tool) for the identification of viral sequences from collected metagenomic data (Ren et al. 2017). This model identifies viral sequences based on the differences in k-mer signatures of viruses and hosts. The model was trained on sequences of host and viral genomes that were sequenced before January 1, 2014, and evaluated on sequences attained after January 1, 2014. When compared to the current gene-based virus classification tool VirSorter (Roux et al. 2015), the proposed model had better TPRs (true positive rates), and it also works comparatively better for small viral contigs. The authors concluded that the proposed model is an effective tool to improve viral sequence identification, especially for viral metagenomic data.
Through their intricate multilayered learning models, deep neural networks have been shown to be a promising approach for the analysis of feature-rich and high-dimensional omics data with their complex multilevel structure. Various studies have developed deep learning-based computational models for the analysis of complex genomic and metagenomic datasets. Arangp-Argoty et al. proposed DeepARG networks to analyze a metagenomic dataset to envisage antibiotic resistance genes (ARGs) (Arango-Argoty et al. 2018). This network constitutes two models, DeepARG-LS for short-read sequences and DeepARG-SS for full-length sequences. The models were trained using 30 ARG categories and showed extreme accuracy (> 0.97) and recall (> 0.90) when evaluated on different databases (Berglund et al. 2017; Lakin et al. 2017). On the basis of the results, the authors concluded that DeepARG facilitates the identification of a wide range of ARGs.
Quang et al. developed a model named deleterious annotation of genetic variants using neural networks (DANN), based on a deep neural network, to annotate the pathogenicity of coding and noncoding genetic variants while also capturing nonlinear relationships among the features (Quang et al. 2015). Trained using the same feature set and training data as combined annotation-dependent depletion (CADD), the support vector machine (SVM)–based model DANN was found to outperform CADD’s SVM by a 19% decrease in the error rate with a 14% increase in the area under the receiver opreting characteristic curve.
Artificial intelligence in microbial proteomics
The protein–protein interactions among hosts and pathogens are capable of providing insights into the host‒pathogen relationship. However, there is a lack of experimental research regarding this context, and the computational approach is proven to be of great significance in this context. Emamjomeh et al. established a collective learning method to interpret proton pump inhibitors (PPIs) between humans and the hepatitis C virus (Emamjomeh et al. 2014). They used six different descriptors to encode human and HCV proteins as feature vectors. The benchmark dataset for validation comprises confident positive and negative PPIs. Tenfold cross-validation was carried out, and the method achieved 83% accuracy and 94% specificity. This method exhibited better performance than the existing approaches and was concluded to be appropriate for future use in the interpretation of the host‒pathogen relationship. In a similar study, the group of authors used SVM models to interpret human proteins that interact with HPV proteins and HCV proteins (Kim et al. 2017). Their model achieved an average accuracy of 66.9% for HPV-human PPIs and 75% for HCV-human PPIs for independent datasets in each case.
Application of artificial intelligence in drug and vaccine development
Secondary metabolites isolated from bacteria, fungi, plants, and marine organisms are important sources of antibiotics, immunosuppressants, anticancer drug/agent herbicides, and insecticides. In microorganisms, the biosynthesis of these secondary metabolites takes place via metabolic pathways. The biosynthetic pathways of specific/secondary metabolites are governed by enzymes, and these enzymes are encoded by clustered genes (Singh et al. 2021a). Earlier, it was very tedious to find the metabolic pathways of specific metabolites, but with the increased availability of high-end gene sequencing combined with powerful bioinformatics tools, it helped to identify metabolic gene clusters (Chavali and Rhee 2018). These bioinformatics tools are primarily focused on the identification of gene clusters of bacteria and fungi. They can identify “signature enzymes,” named nonribosomal peptide synthetase (NRPS), polyketide synthase (PKS), and hybrid NRPS-PKS (Osbourn 2010).
Antimicrobial peptides are short innate immunity peptides and may belong to a wide range of diverse sequence families. These are popular candidates for the development of antimicrobials owing to their ability to disrupt the target by several mechanisms, such as DNA interference, damage to the cell membrane, or signaling for adaptive immune responses (Wimley and Hristova 2011). The search for this component even in wet labs is turning toward computational approaches. In 2018, Veltri et al. implemented a DNN model with convolutional and recurrent layers to allow the model to extract features on its own (Veltri et al. 2018). The dataset is used for the training and testing of the model that reveals the latest available antibacterial peptide data from an updated APD version 3. This model has identified approximately 98% of the AMPs that are listed and available in APD vr.3 as active against gram-positive and gram-negative bacteria. Su et al., in 2019, used a multiscale CNN with multiple layers. The proposed DNN model attained 92.4% accuracy and 94% specificity, outperforming existing DNN models by 1.3% and 1.5%, respectively (Su et al. 2019).
The amphiphilicity of AMPs is a membrane disruptive factor, but they often become hemolytic in human red blood cells (Nguyen et al. 2011; Baeriswyl et al. 2019). Most ML applications do not consider such issues while searching for AMPs. To address this issue, Capecchi et al. recently published their study, where they used ML models for AMP design, taking both the activity and hemolysis of AMPs into consideration by training their model’s sets of active, inactive, hemolytic, and nonhemolytic sequences obtained from reported activity data (Capecchi et al. 2021). They further trained the RNN classifier using data from the Antimicrobial Activity database and peptide structure to design short nonhemolytic AMPs. The authors successfully managed to identify eight new nonhemolytic AMPs against Pseudomonas aeruginosa, Acinetobacter baumannii, and methicillin-resistant Staphylococcus aureus (MRSA) and showed that ML could be used to design therapeutically safe new nonhemolytic AMPs.
Stokes et al. (2020) used DNN and trained it by using a dataset of approved antimicrobials as input to interpret the growth inhibitory activity of Escherichia coli (Stokes et al. 2020). They used a pool of 2335 molecules to train the neural network model that hampered E. coli growth. Furthermore, the model was applied to multiple chemical libraries containing > 107 million molecules to identify the best leading molecule against E. coli. The candidates were ranked according to the model’s predicted score, and finally, the list of potential candidates was selected. This dataset showed antimicrobial activity within an acceptable toxicity range in humans. This technique led to the discovery of halicin, a novel broad-spectrum antimicrobial that was effective against a wide range of MDR microbes. However, there are several drawbacks of this algorithm that need to be acknowledged. Although this algorithm will lead to a molecule with a low toxicity level, the slightest change in the amino acid sequence can cause a drastic shift in the toxicity level (Maritan et al. 2020).
Drug discovery is a long and complex pipeline and a significant stage for the identification of new compounds targeted to specific characteristics of microorganisms. This is designed to prevent/control the disease/infection either by obstructing vital microbial processes or by preventing microorganism multiplication (Singh et al. 2021b). Vamathevan et al. (2019) explained the application of ML in drug discovery and different developmental stages (Vamathevan et al. 2019).
The in silico vaccine discovery pipeline consists of computational tools to discover potential candidates that may stimulate a protective immune response in the host or typically to predict protein characteristics (Goodswen et al. 2021). Therefore, the primary aim of implementing machine learning in vaccine development should be to minimize the number of false candidates. Goodswen et al. (2013) trained their ML algorithms by using protein datasets from Toxoplasma gondii, Plasmodium sp., and Caenorhabditis elegans (Goodswen et al. 2013). They concluded that their proposed model was more effective in identifying false candidates than laboratory validation.
Data visualization is of critical importance in elucidating results and communicating knowledge among researchers. Chen et al. (2022) developed a data visualization web server Image GP to visualize and analyze data in easier and efficient way specially designed for biology and chemistry data visualization. They have used R code for plotting which is open sourced, and supplemented with 26 parameters to fulfil tailored requirements (Chen et al. 2022).
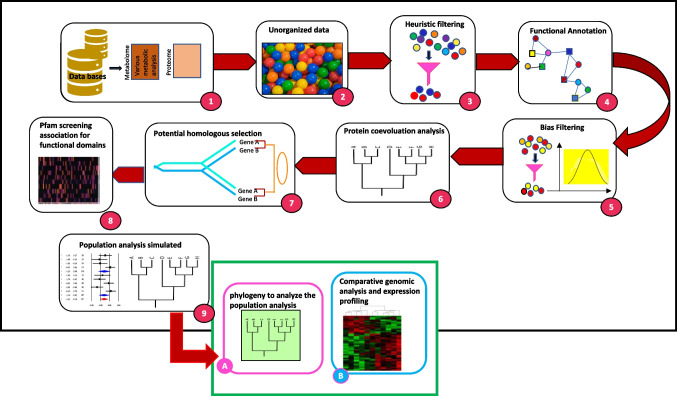
With the enormous increase and heavily utilization of high-throughput technologies including genomics, proteomics, metabolomics, a large data is generated. Now, scientists need to understand and analyze the data very precisely and also need to bridge the gap between genotype and phenotype on gigantic scale (Davis-Turak et al. 2017; O’Donoghue 2021; O’Donoghue et al. 2018). This requires either a trained professional setup to jeopardize dataset quality. But manual set up alone cannot deal with the huge amount of data and also chances of errors can occur. To deal with this, the pipelines have been developed. A pipeline is a process of an automated workflow of a complete machine learning task. This is performed by facilitating a sequence of data that has to be transformed and corelated in a model and further could be analyzed to get the output. A pipeline comprises raw data input, features, outputs, model parameters, machine learning models, and predictions. A pipeline consists of multiple sequential steps for data processing, modelling, and deployment. The flow of pipeline is depicted in Fig. 3. In the pipeline, each step is designed as an independent module and all these modules are tied together to get the final result (https://www.javatpoint.com/machine-learning-pipeline). Various computational pipelines used for microbial genomics, proteomics, and functional diversity are summerizes in Table 2.
Fig. 3.
Flow of filter of the bioinformatics pipeline. (1) Database extrapolation of data from two defined groups. (2) Unorganized raw data. (3) Heuristic filtering scripts. (4) Functional annotation retrieved from Uniprot Swiss/TrEMBL. (5) Bias filtering Section (6) “Raw” phylogenetic analysis. (7) Potential homologous selection. (8) Pfam screening association for functional domains. (9) Population analysis simulated — statistics score applied; reference sequences from selected database (A) phylogeny to analyze the population analysis result coupled with the previous raw phylogenetic analysis results; new database extrapolation of input data through API (B) comparison of my pipeline with previous findings (Pelosi 2022)
Table 2.
Various computational pipelines used for microbial genomics, proteomics, and functional diversity
| S. no | Pipeline name | Usage | Description | URL | References |
|---|---|---|---|---|---|
| 1 | MicrobioLink | A computational pipeline to analyze microbiome–host interactions at a cellular levelusing network and systems biology approaches |
MicrobioLink analyzes microbial proteins in a certain context which influences cellular processes by modulating gene or protein expression MicrobioLink facilitates to evaluate an entire microbial community or even a single microorganism, either a commensal or pathogen that can interfere with host processes via protein-mediated signal transduction |
Andrighetti et al. (2020). https://doi.org/10.3390/cells9051278 | |
| 2 | BIOCOM-PIPE | A flexible and independent suite of tools for processing data from high-throughput sequencing technologies, |
BIOCOM-PIPE is focused on the diversity of archaeal, bacterial, fungal, and photosynthetic microeukaryote amplicons It is a new pipeline designed to characterize microbial diversity from environmental DNA metabarcoding data |
https://doi.org/10.5281/zenodo.3678129) | Djemiel et al. (2020). https://doi.org/10.1186/s12859-020-03829-3 |
| 3 | Bactopia | provide efficient comparative genomic analyses for bacterial species or genera |
Bactopia is based on Nextflow workflow software make efficient use of large clusters and cloud-computing environments to process the many thousands of genomes that are currently being generated. For users that are not familiar with bacterial genomic tools and/or who require a standardized pipeline, Bactopia is a one-stop shop that can be easily deployed using conda, Docker, and Singularity containers. For researchers with particular interest in individual species or genera, BaDs can be highly customized with taxon-specific databases Running multiple tasks on a single platform standardizes the underlying data quality used for gene and variant calling between projects run in different laboratories |
https://www.github.com/bactopia/bactopia |
Robert and Timothy (2020) |
| 4 | Bacteria Genome Pipeline | An automated and scalable pipeline built on the Snakemake framework |
This pipeline will be useful for researchers in low-to-middle income countries and people with little or no bioinformatics skills in analyzing raw genomics data BAGEP for monomorphic bacteria that performs quality control on FASTQ paired end files, scan reads for contaminants using a taxonomic classifier, maps reads to a reference genome of choice for variant detection, detects antimicrobial resistant (AMR) genes, constructs a phylogenetic tree from core genome alignments, and provides interactive short nucleotide polymorphism (SNP) visualization across core genomes in the data set. The objective of our research was to create an easy-to-use pipeline from existing bioinformatics tools that can be deployed on a personal computer |
Olawoye et al. (2020) https://doi.org/10.7717/peerj.10121 | |
| 5 | MetaPhage |
Automated Pipeline for Analyzing, Annotating, and Classifying Bacteriophages in Metagenomics Sequencing Data The pipeline is implemented in Nextflow |
To assist the nonspecialist in the decision-making process and facilitate workflow management, we present here MetaPhage (MP), a fully automated computational pipeline for quality control, assembly, and phage detection as well as classification and quantification of these phages in metagenomics data. The pipeline is modular and enables the user to skip some of the steps and recover analysis in the event of execution errors. To guarantee scalability and reproducibility, | https://github.com/MattiaPandolfoVR/MetaPhage | Pandolfo et al. (2022). https://doi.org/10.1128/msystems.00741-22 |
| 6 | Virus-seeker | The VS-Virome pipeline is controlled by a master Perl script VirusSeeker-Virome. A pipeline for novel virus discovery and virome composition analysis | This pipeline helps in quick identification of candidate viral sequences by alignment to virus only databases. It also removes false positives by alignment. Detects multiple and diverse group of RNA and DNA viruses |
Zhao et al. (2017) |
|
| 7 | MetaFlow/mics | Reproducible nextflow pipeline for the analysis of Microbiome marker data |
It is a comprehensive pipelne for the analysis of microbiome marker data The pipeline produces a detailed account of the number of reads assigned to each sample and further breaks down the results by indicating whether the index matches the barcode perfectly, as well as the number of indexes containing errors. The pipeline provides a visualization of the overall read quality distribution as well as the log-transformed distribution of the number of sequences in the total samples Seamlessly scalable, interoperable, and extensible |
https://github.com/hawaiidatascience/metaflowmics |
Arisdakessian et al. (2020) |
| 8 | ASA3P | An automatic pipeline used for assembly, annotation and higher level analysis of closely related bacterial isolates | This pipeline conducts comprehensive genome characterizations and analyses like detection of antibiotic resistance gene, identification of virulence factors, and taxonomic classification | (https://github.com/oschwengers/asap) |
Schwengers et al. (2020) |
| 9 | SURPI | Sequence-based ultrarapid pathogen identification | This pipeline chelps to identify pathogen from complex NGS data generated from clinical samples. It provides extensive classification of reads against viral and bacterial databases in fast mode. SURPI pipeline consists of a set of fixed external software and database dependencies and user-defined custom parameters | http:// chiulab.ucsf.edu/surpi |
Naccache et al. (2014) |
| 10 | Diagno Top | A computational pipeline for discriminating bacterial pathogens without database search | This pipeline differentiates the spectral clusters found in top-down proteomics data sets that is been used for microbial diagnostics without database search. A promising tool for clinical microbiology and biomarker discovery | http://patternlabforproteomics.org/diagnotop/ |
Lima et al. (2021) |
| 11 | ViroMatch | Computational pipeline for the detection of viral sequences from complex metagenomic data | It is an automated pipeline where metagenomic sequences are screened for putative viral reads by neucleotide mapping and translated mapping | https://github.com/twylie/viromatch |
Wylie and Wyile (2021) |
| 12 | V-pipe | For assessment of viral genetic diversity from high-throughput sequencing data | This computational pipeline is a combination of statistical models and computational tools for end-to end analyses of raw sequencing reads | https:// https://github.com/cbg-ethz/V-pipe |
Posada-Céspedes et al. (2021) |
| 13 | IDseq | Cloud-based pipeline for metagenomic pathogen detection and monitoring | This pipeline is a cloud based metagenomics pipeline for pathogen detection and monitoring. It accepts raw mNGS data, exhibits host and quality filtration steps to finally result into reads and contigs for taxonomic categorization. It is specifically designed for detection of novel pathogens | https://idseq.net |
Katrina et al. (2020) |
| 14 | HAYSTAC | This is based on novel Bayesian framework | The pipeline High AccuracY and scalable Taxonomic Assignment of MetagenomiC data (HAYSTAC) is developed as a robust and rapid species identification from high throughput sequencing data. It can easily handle the ancient and modern DNA data and also the incomplete reference databases | https://github.com/antonisdim/HAYSTAC |
Dimopoulos et al. (2022) |
| 15 | SeqScreen | This is for accurate and sensitive functional screening of pathogenic sequences | This pipeline accurately characterize short nucleotide sequences by using taxonomic and functional labels and customized set of curated Functions of sequences of Concern (FunSoCs) specific to microbial pathogenesis. It is a combination of machine learling classifiers, alignment based tools, curated databases and curation-based labelling of protein sequences along with custom functions for accurate identification of pathogen | www.gitlab.com/treangenlab/seqscreen |
Balaji et al. (2022) |
| 16 | VAPiD | This is a portable and lightweight command line tool for annotation and GenBank deposition of viral genomes | Tha pipeline VAPiD is developed to facilitate the viral genome annotations. It can handle batch submission of multiple viruses without prior knowledge of viral species, correctly annotates RNA editing and ribosomal and runs with simple one line command slippage, and handles submission of metadata | https://github.com/rcs333/VAPiD |
Shean et al. (2019) |
Application of ML in antimicrobial resistance
Human mortality worldwide faces the widespread spread of infectious diseases. There is a major challenge for health workers for the prevention and treatment of such diseases. To address any such threat, accurate identification and characterization of pathogens are the foremost requirement and require expertise along with high-end equipment and facilities. Machine learning can automate this with precision and accuracy with the help of image and metagenomics data (Goodswen et al. 2021).
Drug-resistant tuberculosis (TB) poses major health concerns worldwide. Earlier, the identification of drug resistance was based on single nucleotide polymorphisms (SNPs). Currently, research is based on the association between genetic variants and multivariate variants (Zhang et al. 2013; Walker et al. 2015). Yang et al. (2018) studied the multivariate association with different ML models, such as RF, SVM, and LR, for the classification of multidrug resistance against eight anti-TB drugs. The reported SVM was the best model that derived the data from 1839 TB samples. Another similar study was conducted by Kouchaki et al. (2019), with 13,402 samples and tested against 11 drugs. In this study, LR performed best and indicated that ML algorithms function differently with different training datasets.
Antimalarial resistance in Plasmodium falciparum is the greatest challenge in Africa. The efficacy of antimalarial therapy was assessed by genotyping malaria parasites once the infection was identified and treated (Plucinski et al. 2015; Talundzic et al. 2016; Halsey et al. 2017), followed by parasite genotyping from the same patient if reinfected with malaria by sequencing a well-defined set of microsatellite repeats. This microsatellite comparison enables us to understand whether the patient is infected with a new strain or reoccureance is due to failure in treatment (Plucinski et al. 2015). For this study, an unsupervised Bayesian classifier was developed (Slater et al. 2005), as the manual prediction of these profiles is difficult and prone to bias. Jones et al. (2020) evaluated this approach and proposed that the Bayesian approach was immensely specific and catered to the precise assessment of treatment failure rates in comparison to manual analysis (Jones et al. 2020).
Limitations and conclusions
As indicated by the performance metrics of the research listed above, AI algorithms have shown excellent gains in microbial studies. However, large-scale clinical applications outside of limited clinical investigations are needed. This could aid in obtaining government regulatory approval for clinical applications of AI-based models in conventional patient treatment, which is currently absent.
Multiple variables must be addressed before AI may be used in regular healthcare procedures on a wide scale, such as model training, high-quality data/images, data labeling, and model validation methodologies.
In general, AI models necessitate correctly annotated genomes and proteomics data. Otherwise, the study could lead to AI model bias. AI models that are based on a single source and nonblinded microbiological data frequently produce incorrect results. AI models are typically built in a single institution with a specific patient population, which can limit the models’ ability to be applied outside of the institutional clinical setting. The accuracy achieved in AI-assisted microbiological research may not always imply efficacy in clinical practice. Furthermore, ethical concerns are likely due to biassed AI models and exaggerated accuracy, which could result in unintended misidentification or predictions with false negatives and positives. The majority of AI-driven microbiological technologies are largely research-based and not in widespread use. While several research groups strive to make AI technologies easier to integrate and implement with traditional software systems, this requires additional formal training for microbiologists and technical employees. Moreover, microbial institutions must develop uniform standards for the use of AI in relevant settings. All of these drawbacks must be addressed before regulatory organizations provide final permission for the use of AI-based technology in microbiology research.
Predictive models for metagenomics studies, disease prediction and classification, and microbial proteomics studies could be extremely beneficial, not only in the case of early disease detection and improved patient survival rates but also in terms of gaining a better understanding of pathogenic and beneficial microorganisms. During the previous decade, AI algorithms’ prediction performance improved considerably. Similarly, modern microbiological study predictive models are improving. However, to take advantage of advances in AI algorithms for data mining and building valuable patterns for better decision support, we must appropriately utilize data collected from microbial research. These prediction models are not intended to replace traditional microbiological research but rather to provide an additional layer of protection for disease detection and treatment. Additionally, these AI-based systems are capable of extracting key information with predictive significance. In regard to a tangible benefit, only models with knowledge-driven approaches provide a genuine difference when compared to traditional techniques. Fair restrictions from relevant authorities, as well as the adoption of AI approaches in microbial metagenomics, proteomics, and disease predictions, are necessary conditions for incorporating AI technology into the current healthcare environment.
Author contribution
All authors contributed to the article and approved the submitted version.
All authors contributed to the article and approved the submitted version.
Rachana Singh and Rajnish Kumar were involved in the design, conception, and critical revision of the manuscript for intellectual content. Garima Yadav was involved in the compilation of the manuscript and incorporation of important relevant information. Mohammed Kuddus and Ghulam Md Ashraf critically examined and revised the manuscript.
Data availability
NA
Declarations
Ethics approval and consent to participate
NA
Consent for publication
All the authors have given their consent to publish this review.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ali M, Aurongzeb M, Rashid Y. (2021) In-silico three dimensional structure prediction of important Neisseria meningitidis proteins. Pak J Pharm Sci. 2021;34(2):553–560. [PubMed] [Google Scholar]
- Amgarten D, Braga LPP, da Silva AM, Setubal JC. MARVEL, a tool for prediction of bacteriophage sequences in metagenomic bins. Front Genet. 2018;9:304. doi: 10.3389/fgene.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrighetti T, Bohar B, Lemke N, Sudhakar P, Korcsmaros T. MicrobioLink: an integrated computational pipeline to infer functional effects of microbiome–host interactions. Cells. 2020;9:1278. doi: 10.3390/cells9051278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango-Argoty G, Garner E, Pruden A, Heath LS, Vikesland P, Zhang L. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 2018;6:23. doi: 10.1186/s40168-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2(11):e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisdakessian,C., Sean, B., Cleveland, and Belcaid, M. (2020). MetaFlow|mics: scalable and reproducible nextflow pipelines for the analysis of micro-biome marker data. In Practice and Experience in Advanced Research Computing (PEARC ’20). 26–30. 10.1145/3311790.3396664
- Aziz RK, Bartels D, Best AA, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeriswyl S, Gan BH, Siriwardena TN, Visini R, Robadey M, Javor S, Stocker A, Darbre T, Reymond JL. X-ray crystal structures of short antimicrobial peptides as Pseudomonas aeruginosa lectin B complexes. ACS Chem Biol. 2019;14:758–766. doi: 10.1021/acschembio.9b00047. [DOI] [PubMed] [Google Scholar]
- Baig MH, Ahmad K, Roy S, Ashraf JM, Adil M, Siddiqui MH, Khan S, Kamal MA, Provazník I, Choi I. Computer aided drug design: success and limitations. Curr Pharm Des. 2016;22(5):572–581. doi: 10.2174/1381612822666151125000550. [DOI] [PubMed] [Google Scholar]
- Balaji A, Kille B, Kappell AD, et al. SeqScreen: accurate and sensitive functional screening of pathogenic sequences via ensemble learning. Genome Biol. 2022;23:133. doi: 10.1186/s13059-022-02695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund F, Marathe NP, Österlund T, Bengtsson-Palme J, Kotsakis S, Flach CF, Larsson DGJ, Kristiansson E. Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome. 2017;5:134. doi: 10.1186/s40168-017-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant P, Pozzati G, Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun. 2022;13(1):1265. doi: 10.1038/s41467-022-28865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17(1):238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermans HPJ, den Dunnen JT. Next generation sequencing technology: advances and applications. Biochimica et Biophysica Acta (BBA) – Mol Basis of Dis. 2014;1842:1932–1941. doi: 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- Cai L, Wu, Y, Gao J (2019) DeepSV: accurate calling of genomic deletions from high-throughput sequencing data using deep convolutional neural network. BMC Bioinformatics 20(1):665. 10.1186/s12859-019-3299-y [DOI] [PMC free article] [PubMed]
- Capecchi A, Cai X, Personne H, Köhler T, van Delden C, Reymond JL. Machine learning designs nonhemolytic antimicrobial peptides. Chem Sci. 2021;12:9221–9232. doi: 10.1039/d1sc01713f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali AK, Rhee SY. Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Briefings in Bioinfo. 2018;19:1022–1034. doi: 10.1093/bib/bbx020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Huang L. LRSSLMDA: laplacian regularized sparse subspace learning for MiRNA-disease association prediction. PLoS Comput Biol. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yan GY. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinfo. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- Chen XX, Tang H, Li WC, Wu H, Chen W, Ding H, et al. Identification of bacterial cell wall lyases via pseudo amino acid composition. Biomed Res Int. 2016 doi: 10.1155/2016/1654623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Guo MY, Li SM, Liu B. ProtDec-LTR2.0: an improved method for protein remote homology detection by combining pseudo protein and supervised Learning to Rank. Bioinfo. 2017;33:3473–3476. doi: 10.1093/bioinformatics/btx429. [DOI] [PubMed] [Google Scholar]
- Chen X, Huang Y-A, You Z-H, Yan G-Y, Wang X-S. A novel approach based on KATZ measure to predict associations of human microbiota with non-infectious diseases. Bioinfo. 2017;33:733–739. doi: 10.1093/bioinformatics/btw715. [DOI] [PubMed] [Google Scholar]
- Chen X, Huang YA, You ZH, Yan GY, Wang XS. A novel approach based on KATZ measure to predict associations of human microbiota with diseases. Bioinfo. 2017;33:733–739. doi: 10.1093/bioinformatics/btw715. [DOI] [PubMed] [Google Scholar]
- Chen T, Liu Y-X, Huang L. ImageGP: an easy-to-use data visualization web server for scientific researchers. iMeta. 2022;1:e5. doi: 10.1002/imt2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Turak J, Courtney SM, Hazard ES, Glen WB, Jr, da Silveira WA, et al. Genomics pipelines and data integration: challenges and opportunities in the research setting. Expert Rev Mol Diagn. 2017;17:225–237. doi: 10.1080/14737159.2017.1282822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. Microbial population genomics and ecology. Curr Opin Microbiol. 2002;5:520–524. doi: 10.1016/s1369-5274(02)00353-3. [DOI] [PubMed] [Google Scholar]
- Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11(2):e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos EA, Carmagnini A, Velsko IM, Warinner C, Larson G, Frantz LAF, et al. HAYSTAC: a Bayesian framework for robust and rapid species identification in high-throughput sequencing data. PLoS Comput Biol. 2022;18:e1010493. doi: 10.1371/journal.pcbi.1010493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YJ, Tang JJ, Guo F. Identification of drug-target interactions via multiple information integration. Inf Sci. 2017;418:546–560. doi: 10.1016/j.ins.2017.08.045. [DOI] [Google Scholar]
- Djemiel C, Dequiedt S, Karimi B, et al. BIOCOM-PIPE: a new user-friendly metabarcoding pipeline for the characterization of microbial diversity from 16S, 18S and 23S rRNA gene amplicons. BMC Bioinformatics. 2020;21:492. doi: 10.1186/s12859-020-03829-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinfo. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Emamjomeh A, Goliaei B, Zahiri J, Ebrahimpour R. Predicting protein–protein interactions between human and hepatitis C virus via an ensemble learning method. Mol Biosyst. 2014;12:3147–3154. doi: 10.1039/c4mb00410h. [DOI] [PubMed] [Google Scholar]
- Fan CY, Lei XJ, Guo L, Zhang AD. Predicting the associations between microbes and diseases by integrating multiple data sources and path-based HeteSim scores. Neurocomput. 2019;323:76–85. doi: 10.1016/j.neucom.2018.09.054. [DOI] [Google Scholar]
- Fatoki TH, Ibraheem O, Ogunyemi IO, Akinmoladun AC, Ugboko HU, Adeseko CJ, Awofisayo OA, Olusegun SJ, Enibukun JM. Network analysis, sequence and structure dynamics of key proteins of coronavirus and human host, and molecular docking of selected phytochemicals of nine medicinal plants. J Biomol Struct Dyn. 2021;39(16):6195–6217. doi: 10.1080/07391102.2020.1794971. [DOI] [PubMed] [Google Scholar]
- Feng PM, Zhang JD, Tang H, Chen W, Lin H. Predicting the organelle location of noncoding RNAs using pseudo nucleotide compositions. Interdiscip Sci Comput Life Sci. 2017;9:540–544. doi: 10.1007/s12539-016-0193-4. [DOI] [PubMed] [Google Scholar]
- Francisco AP, Vaz C, Monteiro PT, et al. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics. 2012;13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannam RB, Techtmann SM. Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Compu and Struc Biotech J. 2021;19:1092–1107. doi: 10.1016/j.csbj.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodswen SJ, Kennedy PJ, Ellis JT. A novel strategy for classifying the output from an in silico vaccine discovery pipeline for eukaryotic pathogens using machine learning algorithms. BMC Bioinfo. 2013;14:315. doi: 10.1186/1471-2105-14-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodswen SJ, Barratt JLN, Kennedy PJ, Kaufer A, Calarco L, Ellis JT. Machine learning and applications in microbiology. FEMS Micro Rev. 2021;45:fuab015. doi: 10.1093/femsre/fuab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazziotin AL, Koonin EV, Kristensen DM. Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation. Nucleic Acids Res. 2017;45:491–498. doi: 10.1093/nar/gkw975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2018;34(2):292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey ES, Venkatesan M, Plucinski MM et al (2017) Capacity development through the US President’s malaria initiative-supported antimalarial resistance monitoring in Africa Net- work. Emerg Infect Dis 23. 10.3201/eid2313.170366 [DOI] [PMC free article] [PubMed]
- Handelsman J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–684. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TC, Rocha AM, Techtmann SM. Advances in monitoring environmental microbes. Curr Opin Biotech. 2013;24:526–533. doi: 10.1016/j.copbio.2012.10.020. [DOI] [PubMed] [Google Scholar]
- He WY, Jia CZ, Zou Q. 4mCPred: machine learning methods for DNA N-4-methylcytosine sites prediction. Bioinfo. 2019;35:593–601. doi: 10.1093/bioinformatics/bty668. [DOI] [PubMed] [Google Scholar]
- Hu H, Zhu CY, Ai HX, Zhang L, Zhao J, Zhao Q, et al. LPI-ETSLP: lncRNA-protein interaction prediction using eigenvalue transformation-based semi-supervised link prediction. Mol Biosyst. 2017;13:1781–1787. doi: 10.1039/c7mb00290d. [DOI] [PubMed] [Google Scholar]
- Hu H, Zhang L, Ai HX, Zhang H, Fan YT, Zhao Q, et al. HLPI-Ensemble: prediction of human lncRNA-protein interactions based on ensemble strategy. RNA Biol. 2018;15:797–806. doi: 10.1080/15476286.2018.1457935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P (2002) Exploring prokaryotic diversity in the genomic era. Genome Biol 3. 10.1186/gb-2002-3-2-reviews0003 [DOI] [PMC free article] [PubMed]
- Hyatt D, Chen GL, LoCascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Plucinski M, Kay K et al (2020) A computer modelling approach to evaluate the accuracy of microsatellite markers for classification of recurrent infections during routine monitoring of antimalarial drug efficacy. Antimicrob Agents Chemother 64. 10.1128/AAC.01517-19 [DOI] [PMC free article] [PubMed]
- Joseph RM, Devineni AV, King IF, Heberlein U (2009) Oviposition preference for and positional avoidance of acetic acid provide a model for competing behavioral drives in Drosophila. Proc Natl Acad Sci U S A 106(27):11352–11357. 10.1073/pnas.0901419106 [DOI] [PMC free article] [PubMed]
- Katrina LK, Tiago C, Charles, de Bourcy FA, Dimitrov B, Dingle G, Egger R, et al. IDseq—an open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. GigaScience. 2020;9:giaa111. doi: 10.1093/gigascience/giaa111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Alguwaizani S, Zhou X, Huang DS, Park B, Han K. An improved method for predicting interactions between virus and human proteins. J Bioinform Comput Biol. 2017;15:1650024. doi: 10.1142/S0219720016500244. [DOI] [PubMed] [Google Scholar]
- Kouchaki S, Yang Y, Walker TM, et al. Application of machine learning techniques to tuberculosis drug resistance analysis. Bioinfo. 2019;35:2276–2282. doi: 10.1093/bioinformatics/bty949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Sharma A, Varadwaj P, Ahmad A, Ashraf GM. Classification of oral bioavailability of drugs by machine learning approaches: a comparative study. J Comp Int Sci. 2011;2:1–18. doi: 10.6062/JCIS.2011.02.03.0045. [DOI] [Google Scholar]
- Kumar R, Sharma A, Siddiqui MH, Tiwari RK. Promises of machine learning approaches in prediction of absorption of compounds. Mini Rev Med Chem. 2018;18(3):196–207. doi: 10.2174/1389557517666170315150116. [DOI] [PubMed] [Google Scholar]
- Kumar R, Sharma A, Srivastava JK, Siddiqui MH, Uddin MS, Aleya L. Hydroxychloroquine in COVID-19: therapeutic promises, current status, and environmental implications. Environ Sci Pollut Res Int. 2021;28(30):40431–40444. doi: 10.1007/s11356-020-12200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha UKS, Deo I, Jaiswal JP, Prasad B (2017) Role of bioinformatics in crop improvement. GJSFR 17(1):13–23
- Lakin SM, Dean C, Noyes NR, Dettenwanger A, Ross AS, Doster E, Rovira P, Abdo Z. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017;45:574–580. doi: 10.1093/nar/gkw1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebatteux D, Remita AM, Diallo AB. Toward an alignment-free method for feature extraction and accurate classification of viral sequences. J Comput Biol. 2019;26:519–535. doi: 10.1089/cmb.2018.0239. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kang GU, Jeon SY, Tagele SB, Pham HQ, Kim MS, Ahmad S, Jung DR, Park YJ, Han HS, et al. Vaginal microbiome-based bacterial signatures for predicting the severity of cervical intraepithelial neoplasia. Diagnostics (basel) 2020;10:1013. doi: 10.3390/diagnostics10121013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tang JJ, Guo F. Learning from real imbalanced data of 14–3-3 proteins binding specificity. Neurocomput. 2016;217:83–91. doi: 10.1016/j.neucom.2016.03.093. [DOI] [Google Scholar]
- Lima DB, Dupré M, Santos MDM, Carvalho PC, Chamot-Rooke J. DiagnoTop: a computational pipeline for discriminating bacterial pathogens without database search. J Am Soc Mass Spectrom. 2021;32:1295–1299. doi: 10.1021/jasms.1c00014. [DOI] [PubMed] [Google Scholar]
- Liu B, Liu F, Wang X, Chen J, Fang L, Chou KC. Pse-in- One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Res. 2015;43:65–71. doi: 10.1093/nar/gkv458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Jiang S, Zou Q. HITS-PR-HHblits: protein remote homology detection by combining PageRank and Hyperlink-Induced Topic Search. Brief Bioinform. 2018;2018:bby104. doi: 10.1093/bib/bby104. [DOI] [PubMed] [Google Scholar]
- Liu YX, Qin Y, Chen T, et al. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell. 2021;12:315–330. doi: 10.1007/s13238-020-00724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani D, Kumar C. Biotechnological advances in bioremediation of heavy metals contaminated ecosystems: an overview with special reference to phytoremediation. Int J Environ Sci Technol. 2014;11:843–872. doi: 10.1007/s13762-013-0299-8. [DOI] [Google Scholar]
- Marinier E, Zaheer R, Berry C, Weedmark KA, Domaratzki M, Mabon P, Knox NC, Reimer AR, Graham MR, Chui L, Patterson-Fortin L, Zhang J, Pagotto F, Farber J, et al. Neptune: a bioinformatics tool for rapid discovery of genomic variation in bacterial populations. Nucleic Acids Res. 2017;45(18):e159. doi: 10.1093/nar/gkx702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritan M, Romeo M, Oberti L, Sormanni P, Tasaki M, Russo R, Ambrosetti A, Motta P, Rognoni P, Mazzini G, Barbiroli A, et al. Inherent biophysical properties modulate the toxicity of soluble amyloidogenic light chains. J Mol Biol. 2020;432:845–860. doi: 10.1016/j.jmb.2019.12.015. [DOI] [PubMed] [Google Scholar]
- Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- O’Donoghue, Seán I. Grand challenges in bioinformatics data visualization. Front Bioinformatics. 2021;1:13. doi: 10.3389/fbinf.2021.669186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue SI, Baldi BF, Clark SJ, Darling AE, Hogan JM, Kaur S, Maier-Hein L, et al. Visualization of biomedical data. Annu Rev Biomed Data Sci. 2018;1:275–304. doi: 10.1146/annurev-biodatasci-080917-013424. [DOI] [Google Scholar]
- Olawoye IB, Frost SDW, Happi CT. The Bacteria Genome Pipeline (BAGEP): an automated, scalable workflow for bacteria genomes with Snakemake. Peer J. 2020;8:e10121. doi: 10.7717/peerj.10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Omer A, Singh P, Yadav NK, Singh RK. An overview of data mining algorithms in drug induced toxicity prediction. Mini Rev Med Chem. 2014;14:345–354. doi: 10.2174/1389557514666140219110244. [DOI] [PubMed] [Google Scholar]
- Osbourn A. Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Oudah M, Henschel A. Taxonomy-aware feature engineering for microbiome classification. BMC Bioinformatics. 2018;19:227. doi: 10.1186/s12859-018-2205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan GF, Jiang LM, Tang JJ, Guo F. A novel computational method for detecting DNA methylation sites with DNA sequence information and physicochemical properties. Int J Mol Sci. 2018;19:E511. doi: 10.3390/ijms19020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfo M, Telatin A, Lazzari G, Adriaenssens EM, Vitulo D (2022) MetaPhage: an automated pipeline for analyzing, annotating, and classifying bacteriophages in metagenomics sequencing data. mSystems 7. 10.1128/msystems.00741-22 [DOI] [PMC free article] [PubMed]
- Parks DH, Mankowski T, Zangooei S, Porter MS, Armanini DG, Baird DJ, et al. GenGIS 2: geospatial analysis of traditional and genetic biodiversity, with new gradient algorithms and an extensible plugin framework. PLoS ONE. 2013;8(7):e69885. doi: 10.1371/journal.pone.0069885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peker N, Garcia-Croes S, Dijkhuizen B, Wiersma HH, van Zanten E, Wisselink G, Friedrich AW, Kooistra-Smid M, Sinha B, Rossen JWA, Couto N. A comparison of three different bioinformatics analyses of the 16S–23S rRNA encoding region for bacterial identification. Front Microbiol. 2019;10:620. doi: 10.3389/fmicb.2019.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi B. Developing a bioinformatics pipeline for comparative protein classification analysis. BMC Genom Data. 2022;23:43. doi: 10.1186/s12863-022-01045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires I, Souza G, Junior J. An analysis of the relation between garbage pickers and women’s health risk. Acta Sci Agric. 2020;4:12–16. [Google Scholar]
- Pires IM, Marques G, Garcia NM, Flórez-Revuelta F, Ponciano V, Oniani S. A research on the classification and applicability of the mobile health applications. J Pers Med. 2020;10:11. doi: 10.3390/jpm10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plucinski MM, Morton L, Bushman M, et al. Robust algorithm for systematic classification of malaria late treatment failures as recrudescence or reinfection using microsatellite geno-typing. Antimicrob Agents Chemother. 2015;59:6096–6100. doi: 10.1128/AAC.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, Kanbar J, Miller-Montgomery, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada-Céspedes S, Seifert D, Topolsky I, Jablonski KP, Metzner KJ, Beerenwinkel N. V-pipe: a computational pipeline for assessing viral genetic diversity from high-throughput data. Bioinformatics. 2021;37:1673–1680. doi: 10.1093/bioinformatics/btab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLOS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Li X, He X, Pu J, Zhang J, Lu S. Computational methods-guided design of modulators targeting protein-protein interactions (PPIs) Eur J Med Chem. 2020;207:112764. doi: 10.1016/j.ejmech.2020.112764. [DOI] [PubMed] [Google Scholar]
- Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinfo. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VS, Srinivas K, Sujini GN, Kumar GN. Protein-protein interaction detection: methods and analysis. Int J Proteomics. 2014;2014:147648. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe M, Giovannoni S. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Remita MA, Halioui A, Malick Diouara AA, Daigle B, Kiani G, Diallo AB. A machine learning approach for viral genome classification. BMC Bioinformatics. 2017;18:208. doi: 10.1186/s12859-017-1602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Ahlgren NA, Lu YY, Fuhrman JA, Sun F. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome. 2017;5:69. doi: 10.1186/s40168-017-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004;6:981–989. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- Riesenfeld CS, Schloss P, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- Robert AP III, Timothy DR (2020) Bactopia: a flexible pipeline for complete analysis of bacterial genomes. mSystems. 5(4). 10.1128/mSystems.00190-20 [DOI] [PMC free article] [PubMed]
- Rodriguez-Valera F. Environmental genomics, the big picture? FEMS Microbiol Lett. 2004;231:153–158. doi: 10.1016/S0378-1097(04)00006-0. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. Peer J. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathya R, Abraham A. Comparison of supervised and unsupervised learning algorithms for pattern classification. Int J Adv Res Artif Intell. 2013;2:34–8. doi: 10.14569/IJARAI.2013.020206. [DOI] [Google Scholar]
- Saxena D, Sharma A, Siddiqui MH, Kumar R. Blood brain barrier permeability prediction using machine learning techniques: an update. Curr Pharm Biotechnol. 2019;20(14):1163–1171. doi: 10.2174/1389201020666190821145346. [DOI] [PubMed] [Google Scholar]
- Schwengers O, Hoek A, Fritzenwanker M, Falgenhauer L, Hain T, et al. ASA3P: An automatic and scalable pipeline for the assembly, annotation and higher-level analysis of closely related bacterial isolates. PLOS Comput Biol. 2020;16:e1007134. doi: 10.1371/journal.pcbi.1007134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Shean RC, Makhsous N, Stoddard GD, et al. VAPiD: a lightweight cross-platform viral annotation pipeline and identification tool to facilitate virus genome submissions to NCBI GenBank. BMC Bioinformatics. 2019;20:48. doi: 10.1186/s12859-019-2606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JY, Li JX, Lu HM. Predicting existing targets for new drugs base on strategies for missing interactions. BMC Bioinfo. 2016;17:282. doi: 10.1186/s12859-016-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Singh PK, Kumar R, Kabir MT, Kamal MA, Rauf A, Albadrani GM, Sayed AA, Mousa SA, Abdel-Daim MM, Uddin MS. Multi-omics approach in the identification of potential therapeutic biomolecule for COVID-19. Front Pharm. 2021;12:652335. doi: 10.3389/fphar.2021.652335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Porwal P (2021) Innovative technologies for enzyme production from extremophilic microbes. Md Kuddus (Ed), Microbial extremozymes: novel sources and industrial applications. 30–37. Elsevier Academic Press. ISBN: 978–0–12–822945–3. 10.1016/B978-0-12-822945-3.00009-9
- Singh R, Chuhan N, Kuddus Md (2021a) Exploring the therapeutic potential of marine-derived bioactive compounds against COVID-19. Env Sci Pol Res 1–12. 10.1007/s11356-021-16104-6 [DOI] [PMC free article] [PubMed]
- Slater M, Kiggundu M, Dokomajilar C, et al. Distinguishing recrudescences from new infections in antimalarial clinical trials: major impact of interpretation of genotyping results on estimates of drug efficacy. Am J Trop Med Hyg. 2005;73:256–262. doi: 10.4269/ajtmh.2005.73.256. [DOI] [PubMed] [Google Scholar]
- Steele HL, Jaeger KE, Daniel R, Streit WR. Advances in recovery of novel biocatalysts from metagenomes. J Mol Microbiol Biotechnol. 2009;16:25–37. doi: 10.1159/000142892. [DOI] [PubMed] [Google Scholar]
- Steiner MC, Gibson KM, Crandall KA. Drug resistance prediction using deep learning techniques on HIV-1 sequence data. Viruses. 2020;12(5):560. doi: 10.3390/v12050560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes JM, Yang K, Swanson K, Jin W, Cubillos-Ruiz A, Donghia NM, MacNair CR, French S, Carfrae LA, Bloom-Ackermann Z, Tran VM, et al. A deep learning approach to antibiotic discovery. Cell. 2020;181:475–483. doi: 10.1016/j.cell.2020.01.021. [DOI] [PubMed] [Google Scholar]
- Stoter FR, Chakrabarty S, Edler B, Habetse EAP. CountNet: estimating the number of concurrent speakers using supervised learning. IEEE/ACM Trans Audio Speech Lang Process. 2019;27:268–282. doi: 10.1109/taslp.2018.2877892. [DOI] [Google Scholar]
- Streit WR, Schmitz RA. Metagenomics—the key to the uncultured microbes. Curr Opin Microbiol. 2004;7:492–498. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Su R, Wu H, Xu B, Liu X, Wei L. Developing a multi-dose computational model for drug-induced hepatotoxicity prediction based on toxicogenomics data. IEEE/ACM Trans Comput Biol Bioinform. 2018 doi: 10.1109/tcbb.2018.2858756. [DOI] [PubMed] [Google Scholar]
- Su X, Xu J, Yin Y, Quan X, Zhang H. Antimicrobial peptide identification using multiscale convolutional network. BMC Bioinfo. 2019;20:730. doi: 10.1186/s12859-019-3327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talundzic E, Plucinski MM, Biliya S, et al. Advanced molecular detection of malarone resistance. Antimicrob Agents Chemother. 2016;60:3821–3823. doi: 10.1128/AAC.00171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, Rouy Z, Roche D, Salvignol G, Scarpelli C, Médigue C. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009;2009:bap021. doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M, Zhao S. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;6:463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt A, van Goethem M, Ramond JB, et al. Assembling metagenomes, one community at a time. BMC Geno. 2017;18:521. doi: 10.1186/s12864-017-3918-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri D, Kamath U, Shehu A. Deep learning improves antimicrobial peptide recognition. Bioinfo. 2018;34:2740–2747. doi: 10.1093/bioinformatics/bty179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasana MV, Pires IM, Sá J, Garcia NM, Zdravevski E, Chorbev I, Lameski P, Flórez-Revuelta F. Promotion of healthy nutrition and physical activity lifestyles for teenagers: a systematic literature review of the current methodologies. J Pers Med. 2020;10:12. doi: 10.3390/jpm10010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TM, Kohl TA, Omar SV. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect. 2015;18:21–31. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang S, Song T. A spectral rotation method with triplet periodicity property for planted motif finding problems. Comb Chem High Throughput Screen. 2019;22:683–693. doi: 10.2174/1386207322666191129112433. [DOI] [PubMed] [Google Scholar]
- Wimley WC, Hristova K. Antimicrobial peptides: successes, challenges and unanswered questions. J Membr Biol. 2011;239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie TN, Wyile KM. ViroMatch: a computational pipeline for the detection of viral sequences from complex metagenomic data. Microbiol Resour Announc. 2021;10:e01468–e1520. doi: 10.1128/MRA.01468-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang YH, Li J, Pan XY, Huang T, Cai YD. New computational tool based on machine-learning algorithms for the identification of rhinovirus infection-related genes. Comb Chem High Throughput Screen. 2019;22:665–674. doi: 10.2174/1386207322666191129114741. [DOI] [PubMed] [Google Scholar]
- Yang Y, Niehaus KE, Walker TM, et al. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinfo. 2018;34:1666–1671. doi: 10.1093/bioinformatics/btx801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Huang JB, Ma ZX, Zhang J, Zou YP, Gao L. Inferring drug-disease associations based on known protein complexes. BMC Med Genomics. 2015;8:S2. doi: 10.1186/1755-8794-8-s2-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Wang BB, Ma XK, Gao L. The extraction of drug-disease correlations based on module distance in incomplete human interactome. BMC Syst Biol. 2016;10:111. doi: 10.1186/s12918-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Zhao J, Gao L. Drug repositioning based on triangularly balanced structure for tissue-specific diseases in incomplete interactome. Artif Intell Med. 2017;77:53–63. doi: 10.1016/j.artmed.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Zeng XX, Ding NX, Rodriguez-Paton A, Zou Q. Probability- based collaborative filtering model for predicting gene-disease associations. BMC Med Genomics. 2017;10:76. doi: 10.1186/s12920-017-0313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HT, Li DF, Zhao LL, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resis- tance. Nat Genet. 2013;45:1255–1217. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zou Q, Rodriguez-Paton A, Zeng XX. Meta-path methods for prioritizing candidate disease miRNAs. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:283–291. doi: 10.1109/tcbb.2017.2776280. [DOI] [PubMed] [Google Scholar]
- Zhao G, Wu G, Lim ES, Droit L, Krishnamurthy S, Barouch DH, Virgin HW, Wang D. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology. 2017;503:21–30. doi: 10.1016/j.virol.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Yu H, Ming Z, Hu H, Ren G, Liu H. The bipartite network projection-recommended algorithm for predicting long noncoding RNA-protein interactions. Mol Ther Nucleic Acids. 2018;13:464–471. doi: 10.1016/j.omtn.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitnik M, Nguyen F, Wang B, Leskovec J, Goldenberg A, Hoffman MM. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Int J Inf Fusion. 2019;50:71–91. doi: 10.1016/j.inffus.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Li JJ, Song L, Zeng XX, Wang GH. Similarity computation strategies in the microRNA-disease network: a survey. Brief Funct Genomics. 2016;15:55–64. doi: 10.1093/bfgp/elv024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NA