Abstract
The Nuclear Casein and Cyclin-dependent Kinase Substrate 1 (NUCKS1) protein is highly conserved in vertebrates, predominantly localized to the nucleus and one of the most heavily modified proteins in the human proteome. NUCKS1 expression is high in stem cells and the brain, developmentally regulated in mice and associated with several diverse malignancies in humans, including cancer, metabolic syndrome, and Parkinson’s disease. NUCKS1 function has been linked to modulating chromatin architecture and transcription, DNA repair and cell cycle regulation. In this review, we summarize and discuss the published information on NUCKS1 and highlight the questions that remain to be addressed to better understand the complex biology of this multifaceted protein.
Introduction
The human NUCKS1 protein, originally designated P1, was discovered by Østvold et al. in 1985 [1]. The gene was cloned in 2001, and the names of both the gene and the protein were changed to NUCKS1 (Nuclear Casein and Cyclin-dependent Kinase Substrate 1; HGNC:29923) by the HUGO Gene Nomenclature Committee [2].
Using HeLa cells, we showed early on that the NUCKS1 protein is phosphorylated at ~25 different residues, and that its apparent molecular mass, as determined by 15% SDS PAGE, is ~50 kD [1, 3]. However, after the NUCKS1 gene was cloned it became clear that this gene encodes a protein of 243 amino acids (Fig. 1A), corresponding to a predicted molecular mass of 27 kD [2]. Analysis of the protein isolated from rat liver by mass spectrometry revealed a mass of 28.4 kD [4]. Hence, we concluded that human NUCKS1 migrates anomalously in SDS PAGE.
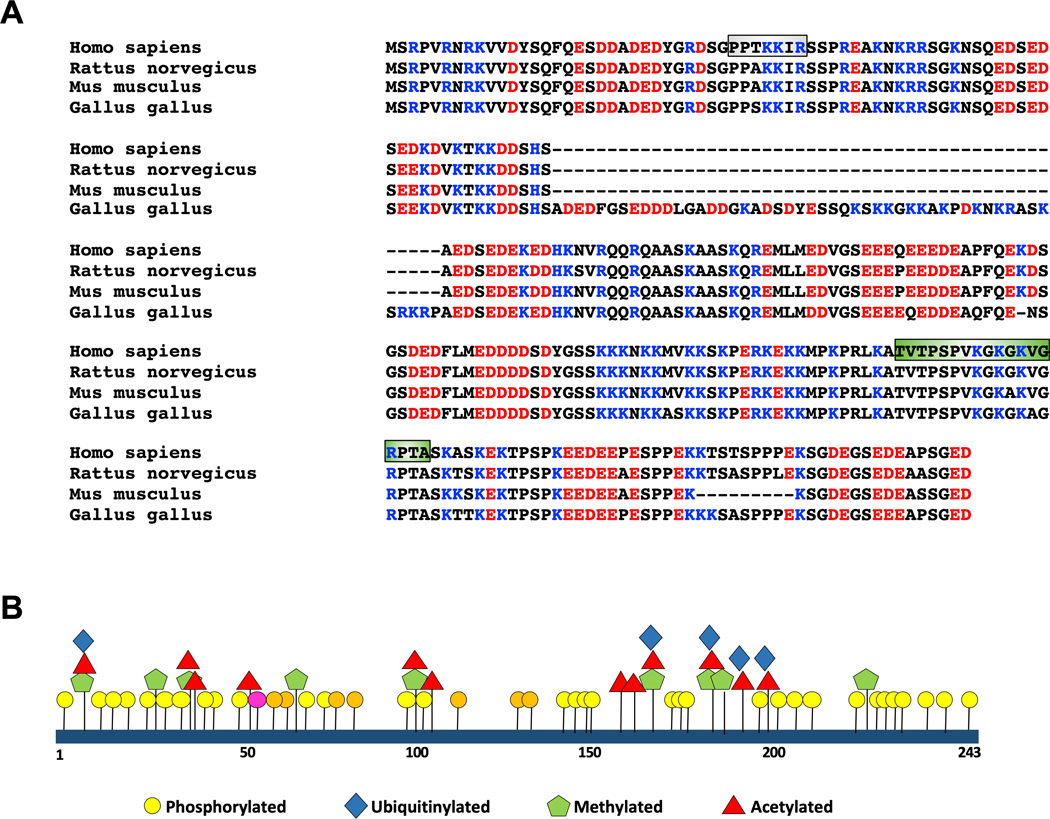
Figure 1. NUCKS1 orthologues and posttranslational modifications.
(A) ClustalW sequence alignment of the primary structures of NUCKS1 from human, rat, mouse, and chicken, with acidic residues highlighted in red and basic residues in blue. The NLS and the DNA binding domain are shown in grey and green boxes, respectively. (B) Schematic to present a summary of the identified posttranslational NUCKS1 modifications (https://www.phosphosite.org). ATM and CK2 target sites identified by us [21] are shown in pink and orange, respectively.
We went on to show that NUCKS1 has no defined structure in solution, a very low content of α-helices and β-sheets, but a relatively high content of β-turns [2]. In cells, NUCKS1 is predominantly localized to the nucleus and contains two nuclear localization signals, one of which (i.e., NLS1; Fig. 1A) physically interacts with both importin α3 and α5 [5]. NUCKS1 binds to single-stranded (ss) and double-stranded (ds)DNA, and to displacement loops (D-loops) and contains an atypical AT-hook in its DNA-binding domain (Fig. 1A; [6–8]). NUCKS1 also binds RNA [9]. These attributes are expected to be major determinants of NUCKS1 function. Yet, many of the molecular details of NUCKS1 biology remain to be determined.
The NUCKS1 gene in different vertebrate species
The NUCKS1 gene is vertebrate-specific and present in all organisms from cartilaginous fishes to man [4, 7]. In humans, the gene is located on chromosome 1 (1q32.1), consists of seven exons and six introns, and is distributed over a region of about 37 kb [4]. One of its introns (intron 1) is exceptionally long (> 20 kb). There is also a NUCKS1 pseudogene on chromosome 1q, containing both exons and introns. However, several non-sense mutations are found within its exons, and this pseudogene is probably not expressed. In the rat, one Nucks1 gene containing 7 exons and 6 introns is located on chromosome 13, and a processed retro-pseudogene is localized on chromosome 6. Apart from five residues, the open reading frame of this retro-pseudogene encodes a protein with a primary amino acid sequence that is nearly identical to the rat NUCKS1 protein encoded on chromosome 13 [2]. However, the putative promoter region of the rat Nucks1 pseudogene is very different from the promotor sequences in the mouse and human NUCKS1 genes. Moreover, the putative promoter region (600 bp upstream of the transcription start site (TSS)) of the rat Nucks1 gene on chromosome 13 is more than 80% conserved with the human and mouse promoter sequences. Furthermore, the rat Nucks1 gene on chromosome 13 is located between Elk4 and Rab7L1, which is also the case for NUCKS1 in other mammals. Collectively, these findings suggest that in rat the functional Nucks1 gene is indeed the one located on chromosome 13.
The Nucks1 gene in mice has a deletion in exon 7, giving rise to loss of nine amino acids in the C-terminal region of the protein compared to human NUCKS1 (corresponding residues: 219–227; Fig. 1A), while the Nucks1 genes in birds, amphibians and fish (zebrafish) all show an inserted sequence in exon 5, leading to an extension of the protein of ~50 residues (Fig. 1A; [4]). To date, little information with respect to the regulation of NUCKS1 transcription is available. However, we predict that the transcription of NUCKS1 must be tightly regulated.
Transcription of the NUCKS1 gene
The factors regulating the transcription of NUCKS1 are largely unknown. The promotor of the mammalian NUCKS1 gene does not contain a TATA box but contains two transcriptional initiator (Inr) elements and likely two alternative TSS. In addition, one E2F and two putative SP1 sites are also present [4]. The promoter and the 5´ UTR are GC-rich and contain CpG islands, suggesting that NUCKS1 expression could be regulated epigenetically by cytosine methylation. Northern blot experiments have shown that at least three NUCKS1 mRNA species are expressed in mammals. The longest species is ~7 kb [2] and likely due to an extraordinarily long 3´ UTR, a feature also found in HMGA genes [10].
The transcript sequences of human NUCKS1 are defined by more than 800 Genebank accessions. These sequences also indicate that at least five different mRNAs are produced, including alternatively spliced variants and one non-spliced form. One splice variant lacking the functional NLS has been isolated from human and chimpanzee cells [5], and, although mainly localized to the nucleus, cytoplasmic staining for NUCKS1 protein has been observed in some human cancer cells [11, 12]. Whether this is due to the lack of the functional NLS or a consequence of diminished nuclear membrane integrity, as reported for some cancer cell types [13], should be one aspect of future studies.
NUCKS1 transcript is present at relatively high levels in most tissues. Interestingly, human NUCKS1 mRNA is highest in the brain, the thyroid gland and skeletal muscle (The Human Protein Atlas, https://www.proteinatlas.org; [2]). NUCKS1 mRNA also is present in mammalian stem cells, and the NUCKS1 promotor has been shown to be co-occupied and co-transcriptionally regulated by OCT4, SOX2 and NANOG in human embryonic stem cells [14–16]. Northern blot analyses and data from The Mouse Atlas of Gene Expression Project (https://www.mouseatlas.org) show that Nucks1 mRNA is present in fertilized mouse oocytes and in 3.5 days post coitum (dpc) blastocysts, and that Nucks1 expression is dramatically increased during the first days of development (Mouse Gene Expression Database:1934811 (Tissue × Stage Matrix)). Dependent on the tissue, highest expression of both mouse Nucks1 mRNA (and protein) is observed between E11.5 and 14.5 dpc [17]. While its expression remains high in mouse brain, Nucks1 mRNA decreases at 15 dpc in most fetal tissues [17]. In Xenopus tropicalis, Nucks1 mRNA is highest at NF stage 14 at the onset of neurulation (https://www.xenbase.org). Interestingly, in Xenopus, Nucks1 mRNA overexpression negatively affects mesoderm development and leads to a shortened anterior-posterior axis [18], suggesting that Nucks1 expression must be tightly regulated during patterning of the early Xenopus embryo. In invertebrates, transcripts with close homology to vertebrate NUCKS1 appear to be absent [17].
The NUCKS1 protein
The NUCKS1 protein shares many characteristics with high mobility group (HMG) proteins, a superfamily of abundantly expressed, nuclear proteins that bind to DNA and nucleosomes and induce structural changes within chromatin [19]. The canonical HMG proteins are soluble in 0.35 M NaCl or 2% trichloroacetic acid (TCA), contain a high fraction of basic amino acids (≥ 25%), a high fraction of acidic amino acids (20–30%) and a relatively high fraction of proline (≥ 7%). The HMG proteins are soluble in 5% perchloric acid (PCA) and can be extracted directly into this acid [20].
Like the HMG proteins, NUCKS1 is soluble in 5% PCA and 2% TCA. It is high in basic (21.4%) and acidic amino acids (25.9%), has a high content of proline, serine and threonine residues, and binds to DNA (Fig. 1A). As such, NUCKS1 could be classified as an HMG-like protein. However, unlike what is observed for the HMG proteins, the mobility of NUCKS1 is decreased rather than increased when analyzed by SDS PAGE. Furthermore, except for the single, AT-hook like sequence and a carboxy-terminus rich in acidic amino acids, NUCKS1 shares no amino acid sequence homology with any of the HMG proteins. Instead, NUCKS1 shares sequence homology (28% identical, and 11% similar) with RAD51AP1 (a RAD51-interacting protein involved in homologous recombination DNA repair), and NUCKS1 and RAD51AP1 can be considered paralogues [8].
In human cancer cell lines, the copy number of NUCKS1 molecules per nucleus varies from 3.2×105 to 1.4×106 [21]. As such, NUCKS1 is about as abundant as other transcription factors. For example, ER copy numbers per cell/nucleus range from ~1.1×104 to 2.6×105, Tcf-1 copy numbers are ~3.9×106 and p53 copy numbers per cell/nucleus range from 2.1×104 to 1.6×105 [22–25]. In adult mice, the copy number of the NUCKS1 protein varies from 1.2×104 to 1.5×105 copies per nucleus [21].
One recent study looked at the effects of cell cycle stage on NUCKS1 protein abundance in human cells. In synchronized RPE-hTERT cells, NUCKS1 protein is expressed at low level during G0/G1-phase; NUCKS1 protein gradually increases during S-phase and remains high in S/G2-phase [26]. Although likely, it has not been directly tested if the abundance of NUCKS1 protein is cell cycle regulated in other cell types. However, it is known that many of its posttranslational modifications depend on cell cycle stage [27].
Posttranslational modification of the NUCKS1 protein
The most striking feature of the NUCKS1 protein is its multitude of posttranslational modifications (Fig. 1B). Per residue, NUCKS1 likely is the highest posttranslationally modified protein in the mammalian proteome (https://www.phosphosite.org; [27]). NUCKS1 can be phosphorylated on 41 sites of a total of 43 serine, threonine and tyrosine residues (https://www.phosphosite.org), and significant changes in the extent of NUCKS1 phosphorylation are observed between asynchronous, G1 phase, and mitotically arrested HeLa cells [27]. Compared to HeLa cells, fewer phosphorylated residues were identified in NUCKS1 purified from breast cancer tissues [27], although a basal level of NUCKS1 phosphorylation is necessary for the ability of the protein to bind DNA [7].
Little is known about the signals leading to the phosphorylation of specific residues in NUCKS1 and of the kinases involved. NUCKS1 is constitutively phosphorylated on several residues in cells [3]. Many of the constitutively phosphorylated sites are consensus phosphorylation sites for Casein Kinas 2 (CK2), and NUCKS1 is indeed a substrate for CK2 in vitro and in cells [3, 21]. Phosphorylation experiments utilizing synthetic NUCKS1-derived peptides and purified CK2 showed that 11 residues can be phosphorylated by CK2 in vitro. Mass spectrometry analysis of NUCKS1 isolated from untreated or HeLa cells treated with a specific CK2 inhibitor showed that at least seven of these residues also are phosphorylated by CK2 in cells [21].
NUCKS1 is a target of cyclin-dependent kinases in vitro and probably also in cells. Phosphorylation of Ser181 by CDK1 in vitro abolishes binding of NUCKS1 to DNA, and NUCKS1 isolated from metaphase-arrested cells exhibits no affinity for DNA [6, 7], leading us to predict that in human NUCKS1 Ser181 may be phosphorylated at the beginning of mitosis. In support of this prediction, NUCKS1 is exclusively localized to the cytoplasm in metaphase-arrested human cells [5, 6].
At least four residues in NUCKS1 (Ser14, Ser54, Ser181 and Ser214) are phosphorylated in response to induced DNA damage [28–30]. In 293T cells and after ionizing radiation, NUCKS1-Ser14 was identified as a substrate of either the ataxia telangiectasia mutated serine/threonine-protein kinase (ATM) or the ATM and Rad3-related serine/threonine-protein kinase (ATR) [30]. Ser54 and Ser181 are phosphorylated by ATM upon exposure of cells to ionizing radiation or neocarzinostatin [8, 21, 29]. Interestingly, inhibition of ATM only partially abrogated Ser54 phosphorylation in HeLa cells treated with the DNA inter-strand crosslinking agent mitomycin C (MMC), suggesting that both ATM and ATR can phosphorylate this residue. MMC challenges DNA repair by homologous recombination (HR), a DNA repair pathway regulated by NUCKS1 [8, 31]. Yet, if NUCKS1 activity in HR is regulated by Ser54 phosphorylation remains to be tested. The kinase(s) phosphorylating Ser214 in cells are presently unknown. However, both DNA-activated protein kinase (DNA-PK) and ATM can phosphorylate Ser14 in vitro [21].
Each of the three tyrosines in NUCKS1 (Tyr13, Tyr26, and Tyr146) can be phosphorylated in cells ((https://www.phosphosite.org), [27]). Tyr13 is a substrate of SYK1 (Spleen associated tyrosine kinase 1), a non-receptor tyrosine kinase with oncogenic and tumor suppressor-like characteristics in both a B-cell line (DG75) and a breast cancer cell line (MD-MAB-231) [32]. In vitro, NUCKS1 also is a substrate for second messenger-activated kinases, such as cyclic AMP-dependent protein kinase and calcium/calmodulin-dependent protein kinase II, and for members of the protein kinase C family [33, 34], indicative of several signal transduction pathways the NUCKS1 protein is involved with [35]. NUCKS1 also was identified as one component of the AKT1 interactome [36]. However, NUCKS1 does not appear to contain an AKT1 consensus phosphorylation site, and it is not known if NUCKS1 can directly be phosphorylated by this kinase.
Many of the phosphorylation sites in NUCKS1 are unrelated to any known kinase consensus sites, and the kinases phosphorylating these sites remain to be identified. However, many of these phosphorylation sites are conserved in mammals, birds, amphibians, and fish, suggesting that they likely play important roles in regulating NUCKS1 function. The NUCKS1 protein also is acetylated, methylated and formylated (https://www.phosphosite.org; Fig. 1B; [27]), but the enzymes acetylating, methylating and formylating NUCKS1 have not yet been identified. Compared to HeLa cells, however, NUCKS1 purified from human breast cancer tissues showed additional sites of lysine acetylation, formylation and mono-methylation [27], potentially indicative of altered NUCKS1 regulation.
It comes as no surprise that ubiquitinylated forms of NUCKS1 (at Lys9, Lys175, Lys184, Lys188, and Lys196) also have been identified (Fig. 1B; [37–42]). NUCKS1 physically interacts with MDM2 and is oligo-ubiquitinated by this E3 ligase in vitro [38], although the respective residues were not identified. Lys175, Lys188 and Lys196 are ubiquitinated in HEK293 cells in response to UV irradiation [42]. After γ-irradiation, we detected poly-ubiquitinated NUCKS1 in a human breast cancer cell line (Wiese et al. unpublished). Poly-ubiquitination through Lys48-linked ubiquitin chains generally leads to proteasomal degradation, while ubiquitin chains linked through Lys63 serve as non-proteolytic signals in intracellular trafficking, signal transduction and DNA repair [43, 44]. How NUCKS1 poly-ubiquitination may be linked to its role in DNA damage repair via the HR pathway is one focus of research in our laboratory.
Biology of the NUCKS1 protein
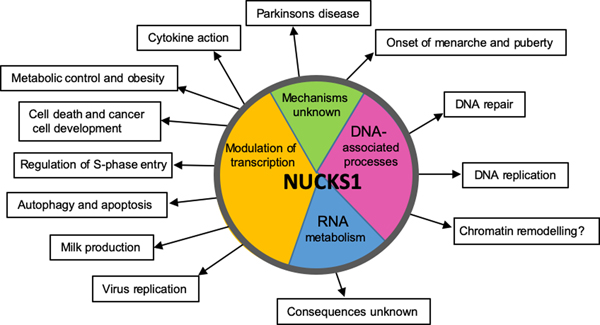
The lack of NUCKS1 orthologues in lower eukaryotes has complicated our understanding of its biological functions. Collectively, however, current knowledge suggests that NUCKS1 is a multifaceted protein involved in the regulation of several diverse biological processes and signal transduction pathways (Fig. 2).
Figure 2.
Pie chart summary of the physiological processes and pathways, and the diseases that human NUCKS1 affects through modulation of transcription (orange), affinity to different DNA (pink) and RNA (blue) substrates, and through mechanism that are currently unknown (green).
NUCKS1 functions as a transcriptional regulator
Several lines of evidence suggest that NUCKS1 is a chromatin remodeling protein involved in the regulation of gene expression. For example, NUCKS1 functions as a chromatin modifier and transcriptional regulator of insulin signaling components, including the insulin receptor (IR), and of cytokines in endocrine cells and mice [35, 45]. NUCKS1 also was identified as a Tat transcriptional coactivator and shown to play a crucial role in HIV-1 replication [46]. Interestingly, although an atypical AT-hook is present within the putative DNA binding region in NUCKS1, NUCKS1-bound DNA is enriched in GC-containing sequences, such as present in SP1 and STAT1/STAT3 binding motifs [35]. In support of these data, results from our laboratory based on SELEX experiments, in which we used a NUCKS1-derived peptide containing the DNA binding domain (including the AT-hook) and a library composed of dsDNA fragments with random DNA sequence, also show that NUCKS1 prefers binding to GC-rich DNA sequences (Østvold et al. unpublished).
NUCKS1 is involved in the regulation of cytokine expression. For example, suppressed expression of inflammatory cytokines was observed in alkali-burned corneas of Nucks1 knockout mice, and corneal epithelial cells showed reduced NF-κB activation and NF-κB-mediated cytokine expression upon silencing of Nucks1 [47]. NUCKS1 also affects the secretion of growth factors and cytokines associated with inflammation, cell proliferation and cell death in bone marrow-derived mesenchymal stem cells (BM-MSCs), and the increase in VEGFa in culture media of NUCKS1-deficient BM-MSC cells was more than three-fold [48]. These results suggest that NUCKS1 may negatively affect cell survival and angiogenesis during myocardial infarction in mice [48], which is in contrast to the positive effects on cell survival and proliferation that the NUCKS1 protein is shown to have in human cells [8, 26, 31, 49].
NUCKS1 and the regulation of metabolism
As mentioned above, NUCKS1 was identified as a positive transcriptional regulator of the insulin signaling pathway [35, 45]. Nucks1 knockout mice kept on a high-fat diet express reduced insulin receptor 1 (IR1) in adipose tissues and liver, gain weight, and develop fatty liver disease, glucose intolerance and insulin resistance [35]. Mechanistically, chromatin accessibility at the TSS of the IR1 promoter was diminished, interfering with the recruitment of RNA Polymerase II in mouse primary hepatocytes with NUCKS1 knockdown. Similarly, mice with hypothalamus-specific Nucks1 deletion are obese and insulin resistant [45]. These mice developed mild glucose intolerance on regular chow. When kept on a high-fat diet, however, obesity and insulin resistance phenotypes developed. These results show that in mice the NUCKS1 protein affects glucose homeostasis and insulin signaling through the hypothalamus, and via the regulation of IR1 expression.
In humans, NUCKS1 protein levels show an inverse correlation with body mass index and HOMA-IR (homeostatic model assessment for insulin resistance) [35]. Overweight individuals displayed > 40% reduction in NUCKS1 protein compared to lean individuals [35]. Low NUCKS1 protein also was observed in omental fat tissues of obese African American women [50]. In this regard it is interesting to note that upregulated p53 in adipose tissue has directly been linked to the development of insulin resistance [51]. As p53 can downregulate NUCKS1 [26], p53 upregulation may be one mechanism leading to the reduced expression of NUCKS1 protein in adipose cells.
NUCKS1 and regulation of cell cycle progression
Recently, NUCKS1 was shown to control the progression of cells from G1 to S phase [26]. Mechanistically, a NUCKS1-SKP2-p21/p27 checkpoint pathway was identified. The SKP2 protein is part of the SCFSKP2 complex and directs the degradation of the CDK inhibitors p21 and p27. Loss of NUCKS1 reduced SKP2 expression in both human normal and cancer cell lines and led to increased p21 and p27, demonstrating that NUCKS1 controls the SKP2-p21/p27 axis in cells with wild type p53 [26]. In response to DNA damage, p53 transcriptionally downregulates NUCKS1 by a mechanism that remains to be determined. Correspondingly, in p53 mutant mice Nucks1 was upregulated following total-body X-irradiation [52], and in a variety of human tumor types, loss of p53 or loss-of-function mutations in p53 correlate with overexpressed NUCKS1 and SKP2, and with p21/p27 degradation [26]. As such, cell proliferation may be sustained in the absence of mitogens or in the context of DNA damage [26], pointing to the importance of the NUCKS1-SKP2-p21/p27 axis in counteracting cell transformation.
In cow mammary epithelial cells, the NUCKS1 protein is upregulated in response to extracellular stimuli such as hormones (estrogen and prolactin) and amino acids (Met, Leu), and positively regulates the synthesis of milk proteins, milk fat and lactose [53]. In this model system, NUCKS1 overexpression increased both cell number and viability, and, consistent with the results presented by Hume and collaborators [26], increased the percentage of cells in S phase, although through the mTOR-SREBP-1c/Cyclin D1 signaling pathway. Similarly, NUCKS1 overexpression in gastric cancer cell lines led to enhanced cell proliferation and activation of mTOR through the PI3K/Akt/mTOR axis, potentially via enhancement of IGF-R1 transcription [54]. We surmise that NUCKS1 expression is tightly regulated to balance G1/S cell cycle arrest, cell proliferation and tissue homeostasis in healthy cells.
NUCKS1 in DNA repair
In accord with the NUCKS1 protein playing a role in stimulating the entry of cells into S phase [26, 53], we uncovered a role for NUCKS1 in regulating DNA repair by HR [8, 31], a pathway that operates in S/G2 phase and is suppressed in G1 [55]. Based on amino acid homology between NUCKS1 and RAD51AP1, a stimulator of the RAD51 recombinase [56, 57], we tested if NUCKS1 affects HR capability in human cells [8]. In HeLa and U2OS cells, knockdown of NUCKS1 phenocopies knockdown of RAD51AP1. NUCKS1 is epistatic with both RAD51AP1 and XRCC3, a second important protein in HR [58], and human cells depleted for NUCKS1 are sensitized to inter-strand DNA crosslinking agents and other drugs that induce barriers to replicant fork progression [8]. In a follow-up investigation, we identified the NUCKS1 protein as an important new regulator of the spatiotemporal events in HR [31]. We showed that NUCKS1 is required for the timely formation of HR protein complexes, that NUCKS1 interacts with DNA motor protein RAD54L in vitro and in cells, that NUCKS1 stimulates the RAD54L ATPase, and functions with RAD54L in RAD51-mediated joint molecule formation. As such, NUCKS1 appears to share attributes with other proteins, including SKP2, that regulate both G1/S transition and DNA repair [26, 59, 60].
NUCKS1 and apoptosis
Tissue homeostasis requires a balance between cell proliferation, DNA repair and cell death. Cell death by apoptosis plays a major role in organismal developmental and is deregulated in many diseases, including cancer [61, 62]. Few studies have initiated investigations to elucidate a link between NUCKS1 and apoptosis. For example, in rat development, the levels of NUCKS1 correlate with those of the pro-apoptotic BAX protein and with activated caspase-3 in all tissues and developmental stages analyzed [17]. Moreover, NUCKS1 protein is upregulated in rat cerebellar granule neurons stimulated to undergo apoptosis just prior to caspase-3 activation ([17]; (Østvold et al. unpublished)). These findings could suggest that NUCKS1 affects the initiation of the apoptotic program. Accordingly, NUCKS1 is a substrate for both caspase-3 and caspase-7 in vitro and in cells [63], and knockdown of NUCKS1 in gastric cancer cell lines induces apoptosis, while NUCKS1 overexpression has no effect [49, 54]. Collectively, these results point to some involvement of the NUCKS1 protein in affecting the apoptotic program in rodent and in human cells, although the molecular mechanisms of this link and its relevance for tissue homeostasis and NUCKS1-related pathologies remain to be determined.
Diseases associated with NUCKS1
NUCKS1 and cancer
Many studies have shown that both NUCKS1 protein and message are overexpressed in different cancer types, leading the authors to suggest that NUCKS1 could serve as a biomarker for different forms of neoplastic disease. Early on, large-scale expression data sets pointed towards a functional module of co-expressed genes in mammalian breast and ovarian cancers [64]. This module contains six genes that were classified as involved in either transcriptional regulation (PDEF, H2AFO, NUCKS1) or the ubiquitin proteasome pathway (PSMD7, SQSTMI, FLJ10111). Quantitative measurements of mRNA in cell lines derived from breast tissues confirmed that these genes are co-expressed. In addition to breast cancer, this module also is co-expressed at elevated levels in lung cancer, oligodendroglioma, fetal brain and in some stem cells [64].
Liu and collaborators described a group of differentially expressed genes as the 186-gene “invasiveness” gene signature (IGS) [65]. These genes were differently expressed between tumorigenic breast cancer cells and normal breast epithelium, predicting the likelihood of a tumor to metastasize (hence named “invasiveness”). The IGS, which NUCKS1 is part of, is associated with the risk for death and metastasis not only in breast cancer patients, but also in lung and prostate cancer patients, and in patients with medulloblastoma [65].
Several groups found NUCKS1 expressed at elevated level in breast carcinoma when compared to the adjacent healthy tissue [66–69]. This can be explained by copy number gains at chromosome 1q32.1, which – in one study – were observed in 66% of the primary breast tumors and in 78% of the breast cancer cell lines investigated [70]. Increased NUCKS1 protein also is associated with lymph node involvement and the formation of distant metastases [68], suggesting that NUCKS1 may be a predictor of the clinically aggressive phenotype. Testing for a correlation between NUCKS1 protein and other tumor subtype markers, NUCKS1 was positively associated with Ki67 and cytokeratin 5/6, but not with ER, PR or HER2, leading the authors to suggest that NUCKS1 could be a novel prognostic marker in the histopathological evaluation of invasive breast carcinoma of no special type [69]. Recently, NUCKS1 also has been identified as a direct target of miR-641 in a number of breast cancer cell lines, supporting the notion that miR-641 may function as a tumor suppressor, targeting the oncogenic effects of NUCKS1 mediated through the NUCKS1/PI3K/AKT axis [71].
NUCKS1 message is elevated in grade III ovarian serous papillary carcinoma, an aggressive cancer that affects the uterus/endometrium, peritoneum, and ovary [72], and NUCKS1 protein is increased in ~43% of ovarian cancer tissues [12]. Interestingly, NUCKS1 mRNA level gradually increases from non-metastatic to metastatic lesions, and is significantly associated with FIGO stage, histological grade, lymph node metastasis, response to chemotherapy and recurrence [12]. Hence, these investigators suggest that NUCKS1 contributes to disease progression and poor prognosis and is a significant and independent prognostic marker for overall and disease-free survival of ovarian cancer patients [12]. Similarly, elevated NUCKS1 transcript and protein are risk factors for poor prognosis and recurrence of endometrial cancer [11], and NUCKS1 protein also positively correlates with advanced FIGO stage, poor histological grade, large tumor size, parametrical involvement, deep stromal infiltration, lymph node metastasis and recurrence of cervical squamous cell carcinoma [73].
Prostate cancer cells show overexpressed NUCKS1 mRNA in metastatic samples compared to primary tumor samples [74], and NUCKS1 mRNA is significantly upregulated in drug resistant prostate cancer cell lines [75]. As in breast cancer, copy number changes, including gain of chromosome 1q32, are frequent in prostate cancer, potentially contributing to elevated NUCKS1 in these cells [76].
Elevated NUCKS1 also has also been observed in tumors of the gastrointestinal tract, skin, and liver. For example, serial analysis of gene expression showed that NUCKS1 is among the twenty most upregulated genes in esophageal squamous cell carcinoma [77]. Gains within chromosome 1q32.1 and elevated NUCKS1 were detected in colorectal cancer (CRC) cells, and NUCKS1 protein levels are linked to significantly worse overall and relapse-free survival in CRC patients, suggesting that NUCKS1 may be an independent risk factor of recurrent disease [78]. Elevated NUCKS1 protein in gastric adenocarcinoma correlates with the depth of invasion, TNM classification, Ki67 and adverse prognosis [79]. Mechanistically, NUCKS1 was shown to promote gastric cancer cell aggressiveness via upregulation of IGF-1R and subsequent activation of the PI3K/AKT/mTOR signaling pathway [49]. Recently it was also shown that NUCKS1 promotes cell proliferation and suppresses autophagy through the mTOR-Beclin1 pathway in gastric cancer [54]. Downregulation of NUCKS1 significantly upregulates Beclin1 levels, reduces phosphorylated (at Ser2448) and total mTOR, and induces autophagy and apoptosis. In accord with the results presented by Hume et al. [26], this study also shows that downregulation of NUCKS1 induces cell cycle arrest and the upregulation of p21 in gastric cancer cell lines [54].
Elevated NUCKS1 protein (in comparison to keratoacanthoma) was detected in both squamous and basal cell carcinoma [80]. In this study, the amounts of NUCKS1 exceeded those of Ki67, and the authors proposed that NUCKS1 could be a novel marker for the pathological evaluation of skin tumor biopsies. Downregulation of NUCKS1 through Ese-3 (Epithelium-specific ETS protein 3) in HaCaT human keratinocytes inhibits cell proliferation, migration, and invasion [81], indicative of high NUCKS1 as one potential determining factor of cancer development in skin.
In hepatocellular carcinoma (HCC), copy number gains within chromosome 1q32.1 are associated with vascular invasion and elevated NUCKS1 transcript [82]. Western blot analyses showed that, compared to adjacent non-tumor liver tissue, the NUCKS1 protein was significantly increased in >50% of the HCCs analyzed. Higher amounts of NUCKS1 protein also were observed in >85% of the HCC tissues in situ when compared to the surrounding normal liver. Moreover, siRNA-mediated knockdown of NUCKS1 resulted in a significant decrease in the number of viable Hep3B cells after treatment with cisplatin and led to significantly suppressed tumor growth in nude mice [82]. In HCC patients, elevated NUCKS1 expression closely is associated with tumor differentiation, TNM stage, vascular invasion and metastasis, and correlates with poor prognosis, indicative of NUCKS1 representing a novel biomarker in the prognosis of patients with HCC [83].
MiR-137, which inhibits tumor growth and increases the sensitivity to paclitaxel and cisplatin in lung cancer patients, also regulates the expression of NUCKS1. In fact, NUCKS1 is a direct target of miR-137 [84]. NUCKS1 is elevated in human lung cancer tissues and inversely correlates with that of miR-137. Restoration of miR-137 expression in tumor cells led to the inhibition of cell proliferation, suggesting that the tumor suppressive role of miR-137 is mediated by its downregulation of NUCKS1 [84–86]. Of note, lung cancer patients with low miR-137 showed shorter disease-free survival, in support of a link between elevated NUCKS1 and poor prognosis [84]. NUCKS1 also is among the nine top-ranked transcription factors significantly associated with smoking-induced lung cancer [87], and up-regulation of NUCKS1 protein in lung adenocarcinoma is associated with a poor prognosis [88].
Little is known about an association between NUCKS1 expression and tumors of the central nervous system. However, what is known is that NUCKS1 is overexpressed in cell lines derived from glioblastoma multiforme and, most prominently, in drug-resistant T98G cells [89]. This may stem from the low abundance of miR-137 in these cells [89]. Loss of miR-137 occurs in different tumor types, as stated above. However, miR-137 likely has hundreds of targets, of which only some have been identified. For example, one consequence of miR-137 expression is G1 cell cycle arrest via suppression of CDK6 and RB1 phosphorylation [86], for which – to the best of our knowledge – no evidence of NUCKS1 involvement has been identified.
Recent studies have described reciprocity among diverse RNA species, including protein-coding messenger RNAs, non-coding RNAs such as long non-coding RNAs (lncRNAs), and circular RNAs [90]. Mechanistically, these RNA species can function as competing endogenous RNA (ceRNA), sponging microRNAs and impacting upon gene expression. Accumulating evidence suggests that NUCKS1 expression is regulated by microRNA sponges. For example, in osteosarcoma cells lncRNA CBR3-AS1 sponges miR-140–5p leading to the upregulation of NUCKS1, whereby stemness and epithelial-mesenchymal transition are promoted [91]. In non-small cell lung cancer cells and tissues, circRNA (RNA circ_0008037) sequesters miR-433–3p, leading to elevated NUCKS1, cell proliferation, migration and invasion [92]. In a similar manner, chromosome 1q gains and the associated overexpression of three mRNAs with ceRNA activity (CEP170, NUCKS1, ZC3H11A) lead to melanoma progression and metastasis through sequestration of distinct miRNAs species with tumor suppressor function [93]. Collectively, these results show that the regulation of NUCKS1 transcription is complex, and that NUCKS1 can be mis-regulated post-transcriptionally in cancerous cells.
We surmise, compared to normal cells and tissues, NUCKS1 message and protein are elevated in many different tumor types. Moreover, the results from cell-based studies are in support of the NUCKS1 protein driving cell survival, DNA repair and cell proliferation, and counteracting cell cycle arrest, apoptosis, and autophagy [8, 26, 31, 49, 54, 91, 92]. Tumor cells are fueled by uncontrolled cell growth, upregulated DNA damage repair and evasion from apoptotic signals [94–96]. Autophagy, however, plays a more complex role in tumor development [97]. As autophagy functions as a tumor suppressor in early stages of tumor development [97], NUCKS1 may function during this stage to inhibit autophagy.
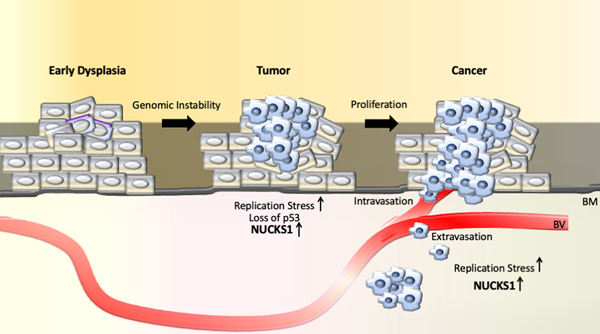
In support of a link between elevated Nucks1 and accelerated tumor initiation, promotion and/or progression (Fig. 3), we detected elevated Nucks1 transcript and loss of the remaining Trp53 wild type allele in the majority of mice that had developed thymic lymphoma (TL) in response to whole-body X-irradiation [52]. Mechanistically, elevated NUCKS1 protein may drive cell cycle progression and/or DNA damage repair in this murine model of TL [8, 26, 30, 31, 52]. As overexpression of DNA repair genes has been associated with metastasis and drug resistance in cancer cells [95, 96], and as NUCKS1 protects cycling cells in culture from replication stress [8], we speculate that NUCKS1 also may protect early neoplastic in situ (Fig. 3).
Figure 3. Model to explain the upregulation of the NUCKS1 protein as part of the activated DNA damage response network and in response to replication stress during tumor development [52, 120].
BM: basement membrane; BV: blood vessel.
Apart from copy number gains at 1q32 [76, 78, 98–101], there are few additional data on cancer-associated mutations within the NUCKS1 gene. More than 900 single nucleotide variants (SNVs) associated with cancer have been reported for NUCKS1 (ICGC data portal, COSMIC, Catalogue of Somatic Mutations in Cancer). Approximately 90 of these are located within exons, leading to non-sense, frameshift, and missense mutations, and to potentially deleterious changes in the NLS, the DNA binding domain, or in phosphorylation consensus sites in NUCKS1. For example, exchange of Pro182 to Lys182 destroys the CDK1 phosphorylation site, which regulates binding of NUCKS1 to DNA [6]. In the future, it will be important to test the phenotype of this and other NUCKS1 variants in cell culture experiments and animal models to understand if they are responsible for a change in the biology of normal cells.
NUCKS1 and Parkinson’s disease
Parkinson’s disease (PD) is a long-term degenerative disorder/synucleinopathy of the central nervous system and the second most common neurodegenerative disorder after Alzheimer’s disease [102]. Several candidate genes and susceptibility loci causing monogenic familial forms of PD have been identified [103], and a significant association of expression and transcription level of NUCKS1 with PD has been observed [104, 105]. For example, studying 2,011 PD cases and 18,381 controls from Japan, Satake et al. identified a susceptibility locus for PD on chromosome 1q32, designated PARK16, and seven small nucleotide polymorphisms (SNPs; rs16856139, rs823118, rs823122, rs947211, rs823156, rs708730, rs1240572) significantly (P < 10−7) linked to PD [104]. These SNPs are located within several linkage disequilibrium blocks that contain the SLC45A3, NUCKS1, RAB7L1, SLC41A1 and PM20D1 genes. Rs947211 and ten tightly linked HapMap SNPs positively associated with NUCKS1 transcript, and these PD-susceptibility variants were the principle genetic determinants of NUCKS1 expression [104].
Philstrøm et al. showed that rs823118, located between NUCKS1 and RAB7L1, is one of several risk loci for PD in a Caucasian population [106]. In addition, strong support for rs823118 as a susceptibility locus for sporadic PD in Han Chinese was obtained [107], although another study failed to detect this association in a Scandinavian population [106].
A meta-analysis of 13,708 PD cases and 95,282 controls from Europe showed that rs823118 is both an expression and a methylation quantitative trait locus for RAB7L1, a gene that encodes a small guanosine triphosphate-binding protein involved in the regulation of exo- and endocytotic pathways [108], and that the rs823118 risk allele is associated with decreased NUCKS1 and increased expression of RAB7L1 [105].
In a more recent study, significant associations between a rs823093 risk allele and PD and between rs823093 and late onset Alzheimer’s disease were observed [103]. Rs823093 is located within one of the introns of the NUCKS1 gene and in a region predicted to be a putative enhancer. However, NUCKS1 transcription was shown to be low in PD patients with the rs823093 risk variant [103]. In contrast, NUCKS1 positively correlated with regional atrophy in PD brains, possibly related to its involvement in cell-autonomous pathology in the context of synuclein-mediated DNA damage [109]. In this regard it is important to note that accumulated DNA double-strand breaks (DSBs) are intermediaries of alpha-synuclein-induced pathogenesis in PD [110], and that NUCKS1 functions in DSB repair [8, 31] and the DNA damage response [30]. We conclude that strong evidence has been accumulated in support of NUCKS1 impacting upon several neuronal genes and pathways. The molecular details, however, of how NUCKS1 contributes to the development of neurodegenerative disease remain to be elucidated.
NUCKS1 and other pathophysiological processes
Ulcerative colitis (UC) is a chronic inflammatory bowel disease characterized by chronic inflammation, oxidative stress, and genome instability. In patients with non-dysplastic UC, the NUCKS1 protein is overexpressed within the colonic mucosa [111]. More specifically, non-dysplastic UC lesions were shown to have ~4-fold higher NUCKS1 than sporadic CRC tissue, indicative of DSB repair operating at higher levels in UC than in CRC tissue [111]. Interestingly, in the same tissues the expression of p53 was suppressed, in support of an inverse correlation between p53 and NUCKS1 [26]. However, high NUCKS1 protein in the colonic mucosa of UC patients could also be linked to the elevated expression of inflammatory cytokines, as NUCKS1 previously was identified as a transcriptional regulator of their expression [35, 47].
Superwarfarins are highly potent vitamin K antagonist anticoagulants that are used as rodenticides. Brodifacoum (BDF), one of the world’s the most widely used superwarfarin, also induces neuroinflammation and neuronal death [112], suggesting that BDF exerts damaging effects on the rodent CNS independent of its anticoagulant actions. Proteomic analyses showed altered expression of more than six hundred proteins in cerebellar lysates from adult rats treated with BDF compared to controls. The protein most significantly increased was NUCKS1, suggesting that NUCKS1 may play a role in certain types of tissue injury [47], including BDF-induced neuroinflammation and neuronal death [112].
The age at which menarche occurs is affected by environmental and genetic factors. Robust evidence for an association between NUCKS1 expression and the age of menarche has been obtained in a meta-analysis of GWAS data encompassing 182,416 women from 57 studies. In this study, rs951366 in NUCKS1 was identified as being linked to the onset of menarche [113]. In another study, NUCKS1 was found to be associated with pubertal height, and it was hypothesized that late onset of menarche is linked to a higher prevalence of adolescent idiopathic scoliosis (AIS), indicative of a linkage between menarche age and predisposition of idiopathic scoliosis [114]. In more recent work, an association between rs951366, located in the 3úntranslated region of the NUCKS1 gene, and AIS was detected [115]. In this study, AIS patients showed remarkably decreased and allele-specific expression of NUCKS1. However, how NUCKS1 regulates puberty and puberty height growth remains to be elucidated.
Plasma leakage and hemorrhage are hallmarks of dengue fever, and antibodies directed against the dengue virus NS1 protein can react with endothelial cells. Suppression-subtractive hybridization experiments demonstrated that exposing human microvascular endothelial cells to anti-dengue virus type 2NS1 antibodies leads to the upregulation of a set of genes including NUCKS1. However, the significance of this upregulation is not known [116].
Concluding remarks and remaining questions
NUCKS1 is chromatin-associated, hyper-modified and nuclear protein that shares many biophysical characteristics with HMG proteins. Overall, the amino acid composition of NUCKS1 is very similar to that of the HMG proteins. Its primary amino acid sequence, however, shows no homology to any of the HMG proteins, but instead shares homology with RAD51AP1 [8], a vertebrate-specific protein regulating DNA repair via the HR pathway.
Unlike RAD51AP1, but like the HMGA family of proteins, NUCKS1 has one single DNA binding domain containing the GRP core sequence of an AT-hook. NUCKS1 weakly binds to ssDNA and, as RAD51AP1, more strongly to dsDNA and D-loops and, despite of its AT-hook, shows preferential binding to GC-rich sequences [8, 35]. The affinity of NUCKS1 for DNA is regulated by its phosphorylation status, which depends on cell cycle stage [6, 7]. NUCKS1 also drives cell proliferation and cell cycle entry into the S-phase, and through p53-mediated transcriptional repression NUCKS1 downregulation leads to G1 arrest [26] and inhibition of HR DNA repair [8]. Although the molecular mechanisms by which NUCKS1 regulates the HR pathway had remained elusive, important new mechanistic insights on its functional role in HR have recently been elucidated [31]. Yet, it remains unclear if NUCKS1 directly binds to damaged DNA and/or can regulate other DNA repair proteins in addition to RAD54L [31]. What is known is that NUCKS1 can bind to the promoter elements of several DNA repair genes [35], potentially regulating their expression in a cell cycle-dependent manner. Of note, the minimally essential domains in NUCKS1 affecting DNA repair and/or transcriptional control are unknown but could be identified by testing the expression of NUCKS1 deletion constructs in NUCKS1 knockout cells. Moreover, RAD51AP1 drives HR in transcriptionally active regions of the genome [117], and this may also be the case for NUCKS1.
NUCKS1 does not have a transcription activation site but might cooperate with other transcription factors to induce transcription of target genes. As such, it is of significant interest to investigate the molecular mechanisms of NUCKS1 in transcriptional control and the impact of its posttranslational modifications.
NUCKS1 lacks three-dimensional structure in solution [2]. However, a disordered to ordered structural transition is likely to take place after NUCKS1 associates with proteins, DNA and/or chromatin, interactions that frequently are regulated by posttranslational modification. With 42 annotated phosphorylation sites, phosphorylation is the most prominent modification of NUCKS1. To date, however, the responsible kinases for only 12 of these sites have been identified. Although it is a challenging task to experimentally validate the kinases and phosphatases involved, it is of substantial interest to identify the putative signal transduction pathways affected by NUCKS1 phosphorylation status. For example, NUCKS1-Ser79 phosphorylation correlates with an increased expression of cyclin D and the recurrence of nonfunctioning pituitary adenomas [118]. As such, we expect that much is to be learned with respect to the pathways affected by NUCKS1 and their relevance to disease.
The organismal consequences of NUCKS1 loss have been studied in mice with different genetic backgrounds. In C57BL/6 mice, loss of Nucks1 can lead to obesity and insulin resistance [35]. In 129SV mice, Nucks1 deficiency and Trp53 loss lead to genome instability and an increased susceptibility to radiation carcinogenesis [52]. Although some of the links between the insulin-IGF1-PI3K-AKT and DNA damage response pathways have been elucidated [30, 119], clearly additional studies are needed to better understand how NUCKS1 affects organismal health under unperturbed conditions and upon exposure to environmental mutagens.
In conclusion, the available literature suggests that NUCKS1 is a multi-functional protein involved in regulating several diverse biological processes and signal transduction pathways. Further elucidating the biology of NUCKS1 will make important contributions to a better understanding of this highly modified chromatin-associated protein and of prevalent threats to human health, including cancer, neurodegenerative disease, and metabolic syndrome.
Abbreviations
- AIS
adolescent idiopathic scoliosis
- ATM
ataxia telangiectasia mutated
- BDF
Brodifacoum
- CK2
Casein Kinase 2
- CRC
colorectal cancer
- HCC
hepatocellular carcinoma
- HMG
high mobility group
- HR
homologous recombination
- IGS
‘invasiveness’ gene signature
- IR1
insulin receptor 1
- MMC
mitomycin C
- NUCKS1
Nuclear Casein and Cyclin-dependent Kinase Substrate 1
- PCA
perchloric acid
- PD
Parkinson’s disease
- SNPs
small nucleotide polymorphisms
- TCA
trichloroacetic acid
- TL
thymic lymphoma
- TSS
transcription start site
- UC
ulcerative colitis
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Ostvold AC, Holtlund J. and Laland SG (1985) A novel, highly phosphorylated protein, of the high-mobility group type, present in a variety of proliferating and non-proliferating mammalian cells. Eur J Biochem. 153, 469–475 [DOI] [PubMed] [Google Scholar]
- 2.Ostvold AC, Norum JH, Mathiesen S, Wanvik B, Sefland I. and Grundt K. (2001) Molecular cloning of a mammalian nuclear phosphoprotein NUCKS, which serves as a substrate for Cdk1 in vivo. Eur J Biochem. 268, 2430–2440 [DOI] [PubMed] [Google Scholar]
- 3.Maelandsmo GM, Ostvold AC and Laland SG (1989) Phosphorylation of the high-mobility-group-like protein P1 by casein kinase-2. Eur J Biochem. 184, 529–534 [DOI] [PubMed] [Google Scholar]
- 4.Grundt K, Haga IV, Aleporou-Marinou V, Drosos Y, Wanvik B. and Ostvold AC (2004) Characterisation of the NUCKS gene on human chromosome 1q32.1 and the presence of a homologous gene in different species. Biochem Biophys Res Commun. 323, 796–801 [DOI] [PubMed] [Google Scholar]
- 5.Grundt K, Haga IV, Huitfeldt HS and Ostvold AC (2007) Identification and characterization of two putative nuclear localization signals (NLS) in the DNA-binding protein NUCKS. Biochim Biophys Acta. 1773, 1398–1406 [DOI] [PubMed] [Google Scholar]
- 6.Grundt K, Skjeldal L, Anthonsen HW, Skauge T, Huitfeldt HS and Ostvold AC (2002) A putative DNA-binding domain in the NUCKS protein. Arch Biochem Biophys. 407, 168–175 [DOI] [PubMed] [Google Scholar]
- 7.Ostvold AC, Hullstein I. and Laland SG (1992) The phosphate groups of the high mobility group like protein P1 strengthens its affinity for DNA. Biochem Biophys Res Commun. 185, 1091–1097 [DOI] [PubMed] [Google Scholar]
- 8.Parplys AC, Zhao W, Sharma N, Groesser T, Liang F, Maranon DG, Leung SG, Grundt K, Dray E, Idate R, Ostvold AC, Schild D, Sung P. and Wiese C. (2015) NUCKS1 is a novel RAD51AP1 paralog important for homologous recombination and genome stability. Nucleic Acids Res. 43, 9817–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C. and Landthaler M. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 46, 674–690 [DOI] [PubMed] [Google Scholar]
- 10.Johnson KR, Lehn DA, Elton TS, Barr PJ and Reeves R. (1988) Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y). J Biol Chem. 263, 18338–18342 [PubMed] [Google Scholar]
- 11.Liu T, Tan S, Xu Y, Meng F, Yang C. and Lou G. (2015) Increased NUCKS expression is a risk factor for poor prognosis and recurrence in endometrial cancer. Am J Cancer Res. 5, 3659–3667 [PMC free article] [PubMed] [Google Scholar]
- 12.Shi C, Qin L, Gao H, Gu L, Yang C, Liu H. and Liu T. (2017) NUCKS nuclear elevated expression indicates progression and prognosis of ovarian cancer. Tumour Biol. 39, 1010428317714631 [DOI] [PubMed] [Google Scholar]
- 13.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K. and Lammerding J. (2016) Nuclear envelope rupture and repair during cancer cell migration. Science. 352, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R. and Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SS, Liu YH, Tseng CN, Chung TL, Lee TY and Singh S. (2006) Characterization and gene expression profiling of five new human embryonic stem cell lines derived in Taiwan. Stem Cells Dev. 15, 532–555 [DOI] [PubMed] [Google Scholar]
- 16.Player A, Wang Y, Bhattacharya B, Rao M, Puri RK and Kawasaki ES (2006) Comparisons between transcriptional regulation and RNA expression in human embryonic stem cell lines. Stem Cells Dev. 15, 315–323 [DOI] [PubMed] [Google Scholar]
- 17.Drosos Y, Kouloukoussa M, Ostvold AC, Havaki S, Katsantoni E, Marinos E. and Aleporou-Marinou V. (2014) Dynamic expression of the vertebrate-specific protein Nucks during rodent embryonic development. Gene expression patterns : GEP. 14, 19–29 [DOI] [PubMed] [Google Scholar]
- 18.Chen JA, Voigt J, Gilchrist M, Papalopulu N. and Amaya E. (2005) Identification of novel genes affecting mesoderm formation and morphogenesis through an enhanced large scale functional screen in Xenopus. Mech Dev. 122, 307–331 [DOI] [PubMed] [Google Scholar]
- 19.Hock R, Furusawa T, Ueda T. and Bustin M. (2007) HMG chromosomal proteins in development and disease. Trends Cell Biol. 17, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin GH and Johns EW (1973) Isolation and characterisation of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur J Biochem. 40, 215–219 [DOI] [PubMed] [Google Scholar]
- 21.Grundt K, Thiede B. and Ostvold AC (2017) Identification of kinases phosphorylating 13 sites in the nuclear, DNA-binding protein NUCKS. Biochim Biophys Acta Proteins Proteom. 1865, 359–369 [DOI] [PubMed] [Google Scholar]
- 22.Ma B, Pan Y, Gunasekaran K, Keskin O, Venkataraghavan RB, Levine AJ and Nussinov R. (2005) The contribution of the Trp/Met/Phe residues to physical interactions of p53 with cellular proteins. Phys Biol. 2, S56–66 [DOI] [PubMed] [Google Scholar]
- 23.Ma L, Wagner J, Rice JJ, Hu W, Levine AJ and Stolovitzky GA (2005) A plausible model for the digital response of p53 to DNA damage. Proc Natl Acad Sci U S A. 102, 14266–14271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van de Wetering M, Castrop J, Korinek V. and Clevers H. (1996) Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol Cell Biol. 16, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reese JC and Katzenellenbogen BS (1992) Examination of the DNA-binding ability of estrogen receptor in whole cells: implications for hormone-independent transactivation and the actions of antiestrogens. Mol Cell Biol. 12, 4531–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hume S, Grou CP, Lascaux P, D’Angiolella V, Legrand AJ, Ramadan K. and Dianov GL (2021) The NUCKS1-SKP2-p21/p27 axis controls S phase entry. Nat Commun. 12, 6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wisniewski JR, Zougman A, Kruger S, Ziolkowski P, Pudelko M, Bebenek M. and Mann M. (2008) Constitutive and dynamic phosphorylation and acetylation sites on NUCKS, a hypermodified nuclear protein, studied by quantitative proteomics. Proteins. 73, 710–718 [DOI] [PubMed] [Google Scholar]
- 28.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J. and Andersen JS (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 9, 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R. and Shiloh Y. (2010) ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal. 3, rs3 [DOI] [PubMed] [Google Scholar]
- 30.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP and Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 31.Maranon DG, Sharma N, Huang Y, Selemenakis P, Wang M, Altina N, Zhao W. and Wiese C. (2020) NUCKS1 promotes RAD54 activity in homologous recombination DNA repair. J Cell Biol. 219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue L, Wang WH, Iliuk A, Hu L, Galan JA, Yu S, Hans M, Geahlen RL and Tao WA (2012) Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc Natl Acad Sci U S A. 109, 5615–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marin TL, Gongol B, Martin M, King SJ, Smith L, Johnson DA, Subramaniam S, Chien S. and Shyy JY (2015) Identification of AMP-activated protein kinase targets by a consensus sequence search of the proteome. BMC Syst Biol. 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walaas SI, Ostvold AC and Laland SG (1989) Phosphorylation of P1, a high mobility group-like protein, catalyzed by casein kinase II, protein kinase C, cyclic AMP-dependent protein kinase and calcium/calmodulin-dependent protein kinase II. FEBS Lett. 258, 106–108 [DOI] [PubMed] [Google Scholar]
- 35.Qiu B, Shi X, Wong ET, Lim J, Bezzi M, Low D, Zhou Q, Akincilar SC, Lakshmanan M, Swa HL, Tham JM, Gunaratne J, Cheng KK, Hong W, Lam KS, Ikawa M, Guccione E, Xu A, Han W. and Tergaonkar V. (2014) NUCKS is a positive transcriptional regulator of insulin signaling. Cell reports. 7, 1876–1886 [DOI] [PubMed] [Google Scholar]
- 36.Duggal S, Jailkhani N, Midha MK, Agrawal N, Rao KVS and Kumar A. (2018) Defining the Akt1 interactome and its role in regulating the cell cycle. Sci Rep. 8, 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akimov V, Barrio-Hernandez I, Hansen SVF, Hallenborg P, Pedersen AK, Bekker-Jensen DB, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM, Kratchmarova I, Kelstrup CD, Olsen JV and Blagoev B. (2018) UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol. 25, 631–640 [DOI] [PubMed] [Google Scholar]
- 38.Guo Z, Wang X, Li H. and Gao Y. (2013) Screening E3 substrates using a live phage display library. PLoS One. 8, e76622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD and Carr SA (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 10, 634–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner SA, Beli P, Weinert BT, Scholz C, Kelstrup CD, Young C, Nielsen ML, Olsen JV, Brakebusch C. and Choudhary C. (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics. 11, 1578–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto-Sato E, Fujimori S, Ishizaka M, Hirai N, Masuoka K, Saito R, Ozawa Y, Hino K, Washio T, Tomita M, Yamashita T, Oshikubo T, Akasaka H, Sugiyama J, Matsumoto Y. and Yanagawa H. (2010) A comprehensive resource of interacting protein regions for refining human transcription factor networks. PLoS One. 5, e9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boeing S, Williamson L, Encheva V, Gori I, Saunders RE, Instrell R, Aygun O, Rodriguez-Martinez M, Weems JC, Kelly GP, Conaway JW, Conaway RC, Stewart A, Howell M, Snijders AP and Svejstrup JQ (2016) Multiomic Analysis of the UV-Induced DNA Damage Response. Cell reports. 15, 1597–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deshaies RJ and Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 44.Rona G. and Pagano M. (2019) Mixed ubiquitin chains regulate DNA repair. Genes Dev. 33, 1615–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu B, Shi X, Zhou Q, Chen HS, Lim J, Han W. and Tergaonkar V. (2015) Hypothalamic NUCKS regulates peripheral glucose homoeostasis. Biochem J. 469, 391–398 [DOI] [PubMed] [Google Scholar]
- 46.Kim HY, Choi BS, Kim SS, Roh TY, Park J. and Yoon CH (2014) NUCKS1, a novel Tat coactivator, plays a crucial role in HIV-1 replication by increasing Tat-mediated viral transcription on the HIV-1 LTR promoter. Retrovirology. 11, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poon MW, Jiang D, Qin P, Zhang Y, Qiu B, Chanda S, Tergaonkar V, Li Q, Wong IY, Yu Z, Tse HF, Wong DS and Lian Q. (2017) Inhibition of NUCKS Facilitates Corneal Recovery Following Alkali Burn. Sci Rep. 7, 41224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Chiu S, Liang X, Chai YH, Qin Y, Wang J, Li X, Qiu B, Tergaonkar V, Tse HF and Lian Q. (2017) Absence of NUCKS augments paracrine effects of mesenchymal stem cells-mediated cardiac protection. Exp Cell Res. 356, 74–84 [DOI] [PubMed] [Google Scholar]
- 49.Huang YK, Kang WM, Ma ZQ, Liu YQ, Zhou L. and Yu JC (2019) NUCKS1 promotes gastric cancer cell aggressiveness by upregulating IGF-1R and subsequently activating the PI3K/Akt/mTOR signaling pathway. Carcinogenesis. 40, 370–379 [DOI] [PubMed] [Google Scholar]
- 50.Doumatey AP, Xu H, Huang H, Trivedi NS, Lei L, Elkahloun A, Adeyemo A. and Rotimi CN (2015) Global Gene Expression Profiling in Omental Adipose Tissue of Morbidly Obese Diabetic African Americans. J Endocrinol Metab. 5, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minamino T, Orimo M, Shimizu I, Kunieda T, Yokoyama M, Ito T, Nojima A, Nabetani A, Oike Y, Matsubara H, Ishikawa F. and Komuro I. (2009) A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 15, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 52.Yue Y, Leung SG, Liu Y, Huang Y, Grundt K, Ostvold AC, Jen KY, Schild D, Mao JH and Wiese C. (2016) Nucks1 synergizes with Trp53 to promote radiation lymphomagenesis in mice. Oncotarget. 7, 61874–61889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X, Zhang M, Ao J, Zhen Z, Gao X. and Li M. (2019) NUCKS1 is a novel regulator of milk synthesis in and proliferation of mammary epithelial cells via the mTOR signaling pathway. J Cell Physiol [DOI] [PubMed] [Google Scholar]
- 54.Zhao E, Feng L, Bai L. and Cui H. (2020) NUCKS promotes cell proliferation and suppresses autophagy through the mTOR-Beclin1 pathway in gastric cancer. J Exp Clin Cancer Res. 39, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, Xia B, Peter M. and Durocher D. (2015) A mechanism for the suppression of homologous recombination in G1 cells. Nature. 528, 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R. and Kanaar R. (2007) RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Molecular cell. 28, 468–481 [DOI] [PubMed] [Google Scholar]
- 57.Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P. and Schild D. (2007) Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Molecular cell. 28, 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pierce AJ, Johnson RD, Thompson LH and Jasin M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes & development. 13, 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA and Dynlacht BD (2002) E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J, Zhang X, Zhang L, Wu CY, Rezaeian AH, Chan CH, Li JM, Wang J, Gao Y, Han F, Jeong YS, Yuan X, Khanna KK, Jin J, Zeng YX and Lin HK (2012) Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1. Mol Cell. 46, 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raj S, Jaiswal SK and DePamphilis ML (2022) Cell Death and the p53 Enigma During Mammalian Embryonic Development. Stem Cells. 40, 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandey MK, Prasad S, Tyagi AK, Deb L, Huang J, Karelia DN, Amin SG and Aggarwal BB (2016) Targeting Cell Survival Proteins for Cancer Cell Death. Pharmaceuticals (Basel). 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J, Helsens K, Impens F, Wejda M, Schymkowitz J, Rousseau F, Madder A, Vandekerckhove J, Declercq W, Gevaert K. and Vandenabeele P. (2009) Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol Cell Proteomics. 8, 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson HG, Harris JW, Wold BJ, Quake SR and Brody JP (2002) Identification and confirmation of a module of coexpressed genes. Genome Res. 12, 1517–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K. and Clarke MF (2007) The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 356, 217–226 [DOI] [PubMed] [Google Scholar]
- 66.Drosos Y, Kouloukoussa M, Ostvold AC, Grundt K, Goutas N, Vlachodimitropoulos D, Havaki S, Kollia P, Kittas C, Marinos E. and Aleporou-Marinou V. (2009) NUCKS overexpression in breast cancer. Cancer cell international. 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soliman NA, Zineldeen DH and El-Khadrawy OH (2014) Effect of NUCKS-1 overexpression on cytokine profiling in obese women with breast cancer. Asian Pac J Cancer Prev. 15, 837–845 [DOI] [PubMed] [Google Scholar]
- 68.Symonowicz K, Dus-Szachniewicz K, Wozniak M, Murawski M, Kolodziej P, Osiecka B, Jurczyszyn K. and Ziolkowski P. (2014) Immunohistochemical study of nuclear ubiquitous casein and cyclin-dependent kinase substrate 1 in invasive breast carcinoma of no special type. Experimental and therapeutic medicine. 8, 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziolkowski P, Gamian E, Osiecka B, Zougman A. and Wisniewski JR (2009) Immunohistochemical and proteomic evaluation of nuclear ubiquitous casein and cyclin-dependent kinases substrate in invasive ductal carcinoma of the breast. J Biomed Biotechnol. 2009, 919645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, Zaks TZ and Weber BL (2005) High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 7, R1186–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Wei D, Zhang J, Deng R, Tang J. and Su D. (2022) miR-641 Inhibited Cell Proliferation and Induced Apoptosis by Targeting NUCKS1/PI3K/AKT Signaling Pathway in Breast Cancer. Comput Math Methods Med. 2022, 5203839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S, Skomedal H, Tu IP, Hernandez-Boussard T, Johnson SW, O’Dwyer PJ, Fero MJ, Kristensen GB, Borresen-Dale AL, Hastie T, Tibshirani R, van de Rijn M, Teng NN, Longacre TA, Botstein D, Brown PO and Sikic BI (2003) Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 14, 4376–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu L, Xia B, Zhong L, Ma Y, Liu L, Yang L. and Lou G. (2014) NUCKS1 overexpression is a novel biomarker for recurrence-free survival in cervical squamous cell carcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 35, 7831–7836 [DOI] [PubMed] [Google Scholar]
- 74.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M. and Monzon FA (2007) Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 7, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Linn DE, Yang X, Sun F, Xie Y, Chen H, Jiang R, Chen H, Chumsri S, Burger AM and Qiu Y. (2010) A Role for OCT4 in Tumor Initiation of Drug-Resistant Prostate Cancer Cells. Genes Cancer. 1, 908–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bermudo R, Abia D, Benitez D, Carrio A, Vilella R, Ortiz AR, Thomson TM and Fernandez PL (2010) Discovery of genomic alterations through coregulation analysis of closely linked genes: a frequent gain in 17q25.3 in prostate cancer. Ann N Y Acad Sci. 1210, 17–24 [DOI] [PubMed] [Google Scholar]
- 77.Sakamoto N, Oue N, Noguchi T, Sentani K, Anami K, Sanada Y, Yoshida K. and Yasui W. (2010) Serial analysis of gene expression of esophageal squamous cell carcinoma: ADAMTS16 is upregulated in esophageal squamous cell carcinoma. Cancer Sci. 101, 1038–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kikuchi A, Ishikawa T, Mogushi K, Ishiguro M, Iida S, Mizushima H, Uetake H, Tanaka H. and Sugihara K. (2013) Identification of NUCKS1 as a colorectal cancer prognostic marker through integrated expression and copy number analysis. Int J Cancer. 132, 2295–2302 [DOI] [PubMed] [Google Scholar]
- 79.Yang M, Wang X, Zhao Q, Liu T, Yao G, Chen W, Li Z, Huang X. and Zhang Y. (2014) Combined evaluation of the expression of NUCKS and Ki-67 proteins as independent prognostic factors for patients with gastric adenocarcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 35, 7505–7512 [DOI] [PubMed] [Google Scholar]
- 80.Zduniak K, Agrawal S, Symonowicz K, Jurczyszyn K. and Ziolkowski P. (2014) The comparison of nuclear ubiquitous casein and cyclin-dependent kinases substrate (NUCKS) with Ki67 proliferation marker expression in common skin tumors. Polish journal of pathology : official journal of the Polish Society of Pathologists. 65, 48–54 [DOI] [PubMed] [Google Scholar]
- 81.Zhang C, Li J, Guo Y, Wang R, Hua L, Gan D, Zhu L, Ma P, Yang J, Li H, Li S, Song Y. and Su H. (2021) Ese-3 Inhibits the Proliferation, Migration, and Invasion of HaCaT Cells by Downregulating PSIP1 and NUCKS1. Ann Clin Lab Sci. 51, 470–486 [PubMed] [Google Scholar]
- 82.Cheong JY, Kim YB, Woo JH, Kim DK, Yeo M, Yang SJ, Yang KS, Soon SK, Wang HJ, Kim BW, Park JH and Cho SW (2016) Identification of NUCKS1 as a putative oncogene and immunodiagnostic marker of hepatocellular carcinoma. Gene. 584, 47–53 [DOI] [PubMed] [Google Scholar]
- 83.Zhang X, Zhang X, Li X, Bao H, Li G, Li N, Li H. and Dou J. (2021) NUCKS1 Acts as a Promising Novel Biomarker for the Prognosis of Patients with Hepatocellular Carcinoma. Cancer Biother Radiopharm [DOI] [PubMed] [Google Scholar]
- 84.Shen H, Wang L, Ge X, Jiang CF, Shi ZM, Li DM, Liu WT, Yu X. and Shu YQ (2016) MicroRNA-137 inhibits tumor growth and sensitizes chemosensitivity to paclitaxel and cisplatin in lung cancer. Oncotarget. 7, 20728–20742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giunti L, Da Ros M, De Gregorio V, Magi A, Landini S, Mazzinghi B, Buccoliero AM, Genitori L, Giglio S. and Sardi I. (2019) A microRNA profile of pediatric glioblastoma: The role of NUCKS1 upregulation. Mol Clin Oncol. 10, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y, Chen R, Zhou X, Guo R, Yin J, Li Y. and Ma G. (2020) miR-137: A Novel Therapeutic Target for Human Glioma. Mol Ther Nucleic Acids. 21, 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.El-Aarag SA, Mahmoud A, Hashem MH, Abd Elkader H, Hemeida AE and ElHefnawi M. (2017) In silico identification of potential key regulatory factors in smoking-induced lung cancer. BMC Med Genomics. 10, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma H, Xu J, Zhao R, Qi Y, Ji Y. and Ma K. (2021) Upregulation of NUCKS1 in Lung Adenocarcinoma is Associated with a Poor Prognosis. Cancer Invest. 39, 435–444 [DOI] [PubMed] [Google Scholar]
- 89.Giunti LDR,M; Iorio AL Magi A; Mazzinghi B; Giglio S; Sardi I. (2017) Gene-03: MicroRNAs Profile in Paediatric GBMS. Neuro-Oncology. 19, iv18 [Google Scholar]
- 90.Tay Y, Rinn J. and Pandolfi PP (2014) The multilayered complexity of ceRNA crosstalk and competition. Nature. 505, 344–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yao W, Hou J, Liu G, Wu F, Yan Q, Guo L. and Wang C. (2022) LncRNA CBR3-AS1 promotes osteosarcoma progression through the network of miR-140–5p/DDX54-NUCKS1-mTOR signaling pathway. Molecular Therapy Oncolytics. 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu J, Zhang H, Zhao C, Li G, Zhang Y. and Sun Y. (2022) CircRNA circ_0008037 facilitates tumor growth and the Warburg effect via upregulating NUCKS1 by binding to miR-433–3p in non-small cell lung cancer. Thorac Cancer. 13, 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu X, Wang K, Vera O, Verma A, Elemento O, Yu X. and Karreth FA (2021) Chromosome 1q amplification perturbs a ceRNA network to promote melanoma metastasis. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanahan D. and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell. 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 95.Sarasin A. and Kauffmann A. (2008) Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutation research. 659, 49–55 [DOI] [PubMed] [Google Scholar]
- 96.Baxter JS, Zatreanu D, Pettitt SJ and Lord CJ (2022) Resistance to DNA repair inhibitors in cancer. Mol Oncol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chavez-Dominguez R, Perez-Medina M, Lopez-Gonzalez JS, Galicia-Velasco M. and Aguilar-Cazares D. (2020) The Double-Edge Sword of Autophagy in Cancer: From Tumor Suppression to Pro-tumor Activity. Front Oncol. 10, 578418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Balcarkova J, Urbankova H, Scudla V, Holzerova M, Bacovsky J, Indrak K. and Jarosova M. (2009) Gain of chromosome arm 1q in patients in relapse and progression of multiple myeloma. Cancer Genet Cytogenet. 192, 68–72 [DOI] [PubMed] [Google Scholar]
- 99.Giunti L, Pantaleo M, Sardi I, Provenzano A, Magi A, Cardellicchio S, Castiglione F, Tattini L, Novara F, Buccoliero AM, de Martino M, Genitori L, Zuffardi O. and Giglio S. (2014) Genome-wide copy number analysis in pediatric glioblastoma multiforme. Am J Cancer Res. 4, 293–303 [PMC free article] [PubMed] [Google Scholar]
- 100.Lo KC, Ma C, Bundy BN, Pomeroy SL, Eberhart CG and Cowell JK (2007) Gain of 1q is a potential univariate negative prognostic marker for survival in medulloblastoma. Clin Cancer Res. 13, 7022–7028 [DOI] [PubMed] [Google Scholar]
- 101.Sargent LM, Ensell MX, Ostvold AC, Baldwin KT, Kashon ML, Lowry DT, Senft JR, Jefferson AM, Johnson RC, Li Z, Tyson FL and Reynolds SH (2008) Chromosomal changes in high- and low-invasive mouse lung adenocarcinoma cell strains derived from early passage mouse lung adenocarcinoma cell strains. Toxicol Appl Pharmacol. 233, 81–91 [DOI] [PubMed] [Google Scholar]
- 102.Winkel D. and Bernstein L. (2022) Movement Disorders. Med Clin North Am. 106, 519–525 [DOI] [PubMed] [Google Scholar]
- 103.Singh S. and Seth PK (2019) Functional association between NUCKS1 gene and Parkinson disease: A potential susceptibility biomarker. Bioinformation. 15, 548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y. and Toda T. (2009) Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 41, 1303–1307 [DOI] [PubMed] [Google Scholar]
- 105.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefansson H, Liu X, Pliner H, Lee JH, Cheng R, International Parkinson’s Disease Genomics, C., Parkinson’s Study Group Parkinson’s Research: The Organized, G. I., and Me, GenePd, NeuroGenetics Research, C., Hussman Institute of Human, G., Ashkenazi Jewish Dataset, I., Cohorts for, H., Aging Research in Genetic, E., North American Brain Expression, C., United Kingdom Brain Expression, C., Greek Parkinson’s Disease, C., Alzheimer Genetic Analysis, G., Ikram MA, Ioannidis JP, Hadjigeorgiou GM, Bis JC, Martinez M, Perlmutter JS, Goate A, Marder K, Fiske B, Sutherland M, Xiromerisiou G, Myers RH, Clark LN, Stefansson K, Hardy JA, Heutink P, Chen H, Wood NW, Houlden H, Payami H, Brice A, Scott WK, Gasser T, Bertram L, Eriksson N, Foroud T. and Singleton AB (2014) Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 46, 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pihlstrom L, Rengmark A, Bjornara KA, Dizdar N, Fardell C, Forsgren L, Holmberg B, Larsen JP, Linder J, Nissbrandt H, Tysnes OB, Dietrichs E. and Toft M. (2015) Fine mapping and resequencing of the PARK16 locus in Parkinson’s disease. J Hum Genet. 60, 357–362 [DOI] [PubMed] [Google Scholar]
- 107.Wang L, Cheng L, Lu ZJ, Sun XY, Li JY and Peng R. (2016) Association of three candidate genetic variants in RAB7L1/NUCKS1, MCCC1 and STK39 with sporadic Parkinson’s disease in Han Chinese. J Neural Transm (Vienna). 123, 425–430 [DOI] [PubMed] [Google Scholar]
- 108.Shimizu F, Katagiri T, Suzuki M, Watanabe TK, Okuno S, Kuga Y, Nagata M, Fujiwara T, Nakamura Y. and Takahashi E. (1997) Cloning and chromosome assignment to 1q32 of a human cDNA (RAB7L1) encoding a small GTP-binding protein, a member of the RAS superfamily. Cytogenet Cell Genet. 77, 261–263 [DOI] [PubMed] [Google Scholar]
- 109.Freeze B, Acosta D, Pandya S, Zhao Y. and Raj A. (2018) Regional expression of genes mediating trans-synaptic alpha-synuclein transfer predicts regional atrophy in Parkinson disease. Neuroimage Clin. 18, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yoon YS, You JS, Kim TK, Ahn WJ, Kim MJ, Son KH, Ricarte D, Ortiz D, Lee SJ and Lee HJ (2022) Senescence and impaired DNA damage responses in alpha-synucleinopathy models. Exp Mol Med. 54, 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.De Angelis PM, Schjolberg AR, Hughes JB, Huitfeldt HS, Norheim Andersen S. and Ostvold AC (2018) Nondysplastic Ulcerative Colitis Has High Levels of the Homologous Recombination Repair Protein NUCKS1 and Low Levels of the DNA Damage Marker Gamma-H2AX. Inflamm Bowel Dis. 24, 593–600 [DOI] [PubMed] [Google Scholar]
- 112.Kalinin S, Marangoni N, Kowal K, Dey A, Lis K, Brodsky S, van Breemen R, Hauck Z, Ripper R, Rubinstein I, Weinberg G. and Feinstein DL (2017) The Long-Lasting Rodenticide Brodifacoum Induces Neuropathology in Adult Male Rats. Toxicol Sci. 159, 224–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, Albrecht E, Ang WQ, Corre T, Cousminer DL, Feenstra B, Franceschini N, Ganna A, Johnson AD, Kjellqvist S, Lunetta KL, McMahon G, Nolte IM, Paternoster L, Porcu E, Smith AV, Stolk L, Teumer A, Tsernikova N, Tikkanen E, Ulivi S, Wagner EK, Amin N, Bierut LJ, Byrne EM, Hottenga JJ, Koller DL, Mangino M, Pers TH, Yerges-Armstrong LM, Zhao JH, Andrulis IL, Anton-Culver H, Atsma F, Bandinelli S, Beckmann MW, Benitez J, Blomqvist C, Bojesen SE, Bolla MK, Bonanni B, Brauch H, Brenner H, Buring JE, Chang-Claude J, Chanock S, Chen J, Chenevix-Trench G, Collee JM, Couch FJ, Couper D, Coveillo AD, Cox A, Czene K, D’Adamo A P, Smith GD, De Vivo I, Demerath EW, Dennis J, Devilee P, Dieffenbach AK, Dunning AM, Eiriksdottir G, Eriksson JG, Fasching PA, Ferrucci L, Flesch-Janys D, Flyger H, Foroud T, Franke L, Garcia ME, Garcia-Closas M, Geller F, de Geus EE, Giles GG, Gudbjartsson DF, Gudnason V, Guenel P, Guo S, Hall P, Hamann U, Haring R, Hartman CA, Heath AC, Hofman A, Hooning MJ, Hopper JL, Hu FB, Hunter DJ, Karasik D, Kiel DP, Knight JA, Kosma VM, Kutalik Z, Lai S, Lambrechts D, Lindblom A, Magi R, Magnusson PK, Mannermaa A, Martin NG, Masson G, McArdle PF, McArdle WL, Melbye M, Michailidou K, Mihailov E, Milani L, Milne RL, Nevanlinna H, Neven P, Nohr EA, Oldehinkel AJ, Oostra BA, Palotie A, Peacock M, Pedersen NL, Peterlongo P, Peto J, Pharoah PD, Postma DS, Pouta A, Pylkas K, Radice P, Ring S, Rivadeneira F, Robino A, Rose LM, Rudolph A, Salomaa V, Sanna S, Schlessinger D, Schmidt MK, Southey MC, Sovio U, Stampfer MJ, Stockl D, Storniolo AM, Timpson NJ, Tyrer J, Visser JA, Vollenweider P, Volzke H, Waeber G, Waldenberger M, Wallaschofski H, Wang Q, Willemsen G, Winqvist R, Wolffenbuttel BH, Wright MJ, Australian Ovarian Cancer, S., Network, G., kConFab, LifeLines Cohort S, InterAct C, Early Growth Genetics C, Boomsma DI, Econs MJ, Khaw KT, Loos RJ, McCarthy MI, Montgomery GW, Rice JP, Streeten EA, Thorsteinsdottir U, van Duijn CM, Alizadeh BZ, Bergmann S, Boerwinkle E, Boyd HA, Crisponi L, Gasparini P, Gieger C, Harris TB, Ingelsson E, Jarvelin MR, Kraft P, Lawlor D, Metspalu A, Pennell CE, Ridker PM, Snieder H, Sorensen TI, Spector TD, Strachan DP, Uitterlinden AG, Wareham NJ, Widen E, Zygmunt M, Murray A, Easton DF, Stefansson K, Murabito JM and Ong KK (2014) Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 514, 92–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cousminer DL, Berry DJ, Timpson NJ, Ang W, Thiering E, Byrne EM, Taal HR, Huikari V, Bradfield JP, Kerkhof M, Groen-Blokhuis MM, Kreiner-Moller E, Marinelli M, Holst C, Leinonen JT, Perry JR, Surakka I, Pietilainen O, Kettunen J, Anttila V, Kaakinen M, Sovio U, Pouta A, Das S, Lagou V, Power C, Prokopenko I, Evans DM, Kemp JP, St Pourcain B, Ring S, Palotie A, Kajantie E, Osmond C, Lehtimaki T, Viikari JS, Kahonen M, Warrington NM, Lye SJ, Palmer LJ, Tiesler CM, Flexeder C, Montgomery GW, Medland SE, Hofman A, Hakonarson H, Guxens M, Bartels M, Salomaa V, ReproGen C, Murabito JM, Kaprio J, Sorensen TI, Ballester F, Bisgaard H, Boomsma DI, Koppelman GH, Grant SF, Jaddoe VW, Martin NG, Heinrich J, Pennell CE, Raitakari OT, Eriksson JG, Smith GD, Hypponen E, Jarvelin MR, McCarthy MI, Ripatti S, Widen E. and Early Growth Genetics C. (2013) Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum Mol Genet. 22, 2735–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu L, Xia C, Sun W, Qin X, Qiu Y. and Zhu Z. (2017) Genetic Polymorphism of NUCKS1 Is Associated With the Susceptibility of Adolescent Idiopathic Scoliosis. Spine (Phila Pa 1976). 42, 1629–1634 [DOI] [PubMed] [Google Scholar]
- 116.Yin Y, Jiang L, Fang D, Jiang L. and Zhou J. (2013) Differentially expressed genes of human microvascular endothelial cells in response to anti-dengue virus NS1 antibodies by suppression subtractive hybridization. Viral Immunol. 26, 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ouyang J, Yadav T, Zhang JM, Yang H, Rheinbay E, Guo H, Haber DA, Lan L. and Zou L. (2021) RNA transcripts stimulate homologous recombination by forming DR-loops. Nature. 594, 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chhabra RR,A; Dutta P. (2019) Phosphorylation of Nucks1 on Serine 79: A Prognostic Marker of Recurrent Nonfunctioning Pituitary Adenoma. Journal of Neurological Surgery. 80 [Google Scholar]
- 119.Liu X, Chen H, Xu X, Ye M, Cao H, Xu L, Hou Y, Tang J, Zhou D, Bai Y. and Ma X. (2018) Insulin-like growth factor-1 receptor knockdown enhances radiosensitivity via the HIF-1alpha pathway and attenuates ATM/H2AX/53BP1 DNA repair activation in human lung squamous carcinoma cells. Oncol Lett. 16, 1332–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J. and Bartek J. (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 434, 864–870 [DOI] [PubMed] [Google Scholar]