Abstract
Coronavirus has emerged as a global health threat due to its accelerated geographic spread over the last two decades. This article reviews the current state of knowledge concerning the origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19). Historically, it has caused two pandemics: severe acute respiratory syndrome and Middle East respiratory syndrome followed by the present COVID-19 that emerged from China. The virus is believed to be acquired from zoonotic source and spreads through direct and contact transmission. The symptomatic phase manifests with fever, cough and myalgia to severe respiratory failure. The diagnosis is confirmed using reverse transcriptase PCR. Management of COVID-19 is mainly by supportive therapy along with mechanical ventilation in severe cases. Preventive strategies form the major role in reducing the public spread of virus along with successful disease isolation and community containment. Development of a vaccine to eliminate the virus from the host still remains an ongoing challenge.
Keywords: Pathology, Histopathology, Education and training (see medical education & training), Medical education and training, Surgery, Transplant surgery, Hepatobiliary surgery, Basic sciences, Pathology
Introduction
Coronavirus (CoV) is derived from the word ‘corona’ meaning ‘crown’ in Latin.1 It causes a range of human respiratory tract infections varying from mild cold to severe respiratory distress syndrome.2 The present novel CoV disease also called as severe acute respiratory syndrome (SARS)-CoV-2 and coronavirus disease 2019 (COVID-19) is an emerging global health threat.3 The COVID-19 epidemic started from Wuhan city of China towards the end of December 2019 and since then spread rapidly to Thailand, Japan, South Korea, Singapore and Iran in the initial months.4–6 This was followed by wide viral dissemination around the world including Spain, Italy, USA, UAE and the UK.7 The WHO declared the COVID-19 outbreak as a pandemic.8 As of 6 May 2020, outbreaks and sporadic human infections have resulted in 3 732 046 confirmed cases and 261 517 deaths.7
The CoV has posed frequent challenges during its course ranging from virus isolation, detection, prevention to vaccine development.9 CoV belongs to the order Nidovirales and has the largest RNA genome.10 It is known to be acquired from a zoonotic source and typically spreads through contact and droplet transmission. The infected person presents with non-specific clinical features requiring virological detection and confirmation by molecular techniques.11–13 This article aims to give a detailed insight into the evolution, transmission and diagnosis of COVID-19. We further discuss the challenges encountered in the management of patients with COVID-19 and the current limitations in the investigational vaccine. Due to the rapidly evolving nature of COVID-19, the readers are requested to update themselves with the nature of change with this particular type of CoV.
ORIGIN
Historic perspective
CoV was discovered during the 1960s. The Coronavirus Study Group under the International Committee on Taxonomy of Viruses used the principle of comparative genomics to further assess and partition the replicative proteins in open reading frames to identify the factors that differentiate CoV at different cluster ranks.14 15 CoV is associated with illness of varied intensity. The most severe type resulting in large-scale pandemics in the past are the SARS (in 2002–2003) and Middle East respiratory syndrome (MERS) (in 2012).16 17
Aetiology
CoV are RNA viruses of the subfamily Coronavirinae. They belong to the family Coronaviridae and the order Nidovirales (nido Latin for ‘nest’). The order Nidovirales is composed of Coronaviridae, Arteriviridae, Mesovirididae and Roniviridae families.10 18 The characteristic features of Nidovirales are as follows: they (1) contain very large genomes for RNA viruses, (2) are highly replicative due to conserved genomic organisation, (3) exhibit several unique enzymatic activities and (4) have extensive ribosomal frameshifting due to the expression of numerous non-structural genes. The Coronaviridae family have two subfamilies: Coronavirinae and Torovirinae. The subfamily Coronavirinae consist of alpha CoV, beta CoV, gamma CoV and delta CoV based on genomic structure.19
Viral structure
The CoV are enveloped positive single-stranded RNA viruses having the largest known viral RNA genomes of 8.4–12 kDa in size.20 The viral genomes are made up of 5ʹ and 3ʹ terminal. The 5ʹ terminal constitutes a major part of the genome and contains open reading frames, which encodes proteins responsible for viral replication. The 3ʹ terminal contains the five structural proteins, namely the spike protein (S), membrane protein (M), nucleocapsid protein (N), envelope protein (E) and the haemagglutinin-esterase (HE) protein.21 22 The S protein mediates an attachment and fusion between the virus and host cell membrane and also between the infected and adjacent uninfected cells. They are the major inducers for neutralising antibodies in a vaccine. The N protein forms RNA complexes that aid in virus transcription and assembly. The M protein is the most abundant structural protein and also defines the viral envelope shape. The E protein is the most enigmatic and the smallest of the major structural protein, which is highly expressed within the infected cell during viral replication cycle. The HE protein is responsible for receptor binding and host specificity.20 23
SARS and MERS
SARS was first recognised in Guangdong province, China, in November 2002. It advanced among 30 countries, infecting 79 000 people by 2003 with a fatality of 9.5%. SARS-CoV was traced and isolated from Himalayan palm civets found in a livestock market in Guangdong, China.16 24 The zoonotic origin of SARS was also discovered in racoon dogs, ferret badgers and in humans working at the same market. These market animals were therefore intermediate hosts that increased the transmission of virus to humans.15 24
Thereon, in 2012, Jeddah, Saudi Arabia, a patient presented with respiratory illness consistent with pneumonia along with features of renal failure.25 The patient’s sputum analysis was done by reverse transciptase (RT-PCR) using pan-CoV primers revealing the viral RNA to be MERS-CoV.26 As of July 2013, 91 patients were infected with MERS-CoV and had a high fatality rate of 34%. Bats and Arabian dromedary camels were identified as potential hosts for MERS-CoV. Intermediate host reservoir species were also seen in goats, sheep and cows.27 28
Novel CoV
In view of taxonomical classification, SARS-CoV-2 (COVID-19) is one among many other viruses in the species, SARS-related CoV. However, SARS-CoV and SARS-CoV-2 vary in terms of disease spectrum, modes of transmission and also diagnostic methods.9 28 The recent report on a cluster cases having respiratory illness in Wuhan, Central China, was followed by a global spread of the disease in a very short duration of time. The samples (oral and anal swabs, blood and broncho-alveolar fluid lavage) from patients admitted to the intensive care unit of Wuhan Jinyintan Hospital were sent to Wuhan Institute of Virology. Pan-CoV PCR primers were used and these samples were positive for CoV.1–3 29 This was followed by metagenomics analysis and genomic sequencing study. The results revealed that this virus was identical (79.6%) to the genetic sequence of SARS-CoVBJ01 leading the WHO to call it novel CoV-2019 (2019-nCoV).30 31
Transmission and Pathogenesis
Zoonosis
CoVs are widespread among birds and mammals with cements bats forming the major evolutionary reservoir and ecological drivers of CoV diversity.32 CoV causes a large variety of diseases in pigs, cows, chicken, dogs and cats. The major diseases caused by CoVs in animals are transmissible gastroenteritis virus, porcine epidemic diarrhoea virus, porcine hemagglutinating encephalomyelitis virus and murine hepatitis virus. In humans, alpha and beta CoV have caused a variety of illness ranging from mild-self-limiting respiratory infections (HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1) to severe acute respiratory distress syndrome (ARDS).16 33–35
Initial cases reported in Wuhan, China, are considered to be an acquired infection from a zoonotic source from Huanan wholesale seafood market which sold poultry, snake, bats and other farm animals.36 37 To isolate the possible virus reservoir, a comprehensive genetic sequence analysis was undertaken among different animal species.9 15 The results suggested that 2019-nCov is a recombinant virus between the bat CoV and an unknown origin CoV. A study revealed, based on relative synonymous codon usage (RSCU) on variety of animal species showed that bats are the most probable wildlife reservoir of 2019-nCov.10 This homologous recombination has proved previously in classical swine fever virus, hepatitis B virus, hepatitis C virus, HIV and dengue virus.38
Modes of spread
Human-to-human transmission occurs through common routes such as direct transmission, contact transmission and airborne transmissions through aerosols and during medical procedures (figure 1). Cough, sneeze, droplet inhalation, contact with oral, nasal and eye mucous membranes are the common modes of spread. Viral shedding occurs from respiratory tract, saliva, faeces and urine resulting in other sources of virus spread.37 39 40 The viral load is higher and of longer duration in patients with severe COVID-19.41 Spread of COVID-19 from patients to health workers and flight attenders who were in close contact with the infected patients are also reported.42
Figure 1.
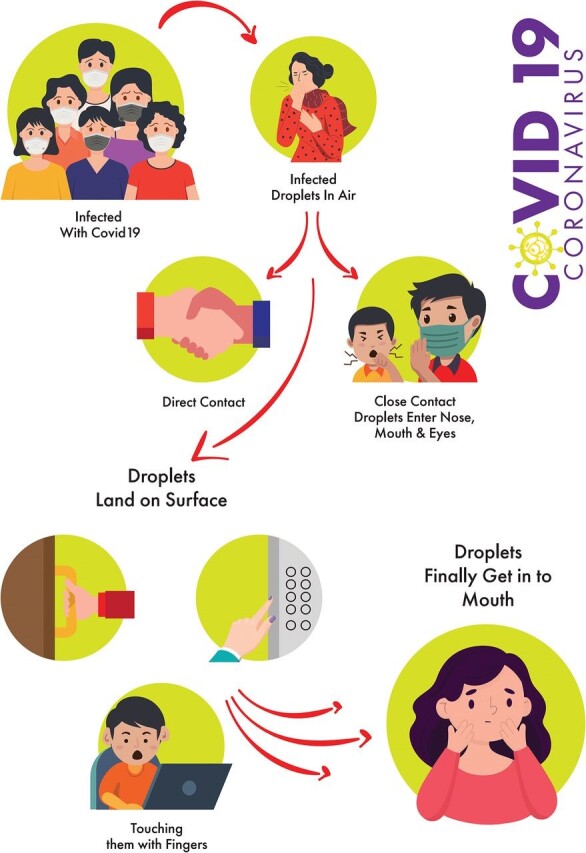
Modes of transmission.
Virus–host interaction
Extensive structural analyses revealed atomic-level interactions between the CoV and the host. Cross-species and human-to-human transmission of COVID-19 is mainly dependent on spike protein receptor-binding domain and its host receptor ACE2.23 43 High expression of ACE2 was identified in lung (type II alveolar cells), oesophagus, ileum, colon, kidney (proximal convoluted tubules), myocardium, bladder (urothelial cells) and also recently the oral mucosa. ACE2 receptors provide entry of the virus into the host cells and also subsequent viral replication. The main factors involved in viral pathogenesis of 2019-nCov are spike 1 subunit protein, priming by transmembrane protease serine-2 (essential for entry and viral replication), ACE2 receptor–2019-nCov interaction and downregulation of ACE2 protein. These factors contribute to atrophy, fibrosis, inflammation and vasoconstriction resulting in host tissue injury.43–45
Clinical Presentation and Diagnosis
Demographics
Based on numerous studies published, the median age was 56 years (range 55–65 years) and males were predominately affected due to high ACE2 concentrations in them. The median onset of illness was 8 days (range 5–13 days).46 47 Due to limited comorbid data availability, it is important to correlate with previously proven susceptible factors to SARS and MERS-CoV infection, which includes smoking, hypertension, diabetes, cardiovascular disease and/or chronic illness.16 24 25 Based on the National Health Institute analysis in Italy, the average mortality age for patients suffering from COVID-19 was 81 years.48 In China, the case fatality rate (CFR) increased with age and showed CFR of 18% for patients above 80 years.49 This striking target to the elderly population is attributed to underlying chronic disorders and declined immune function. Declined immune function has been linked to cytokine storm syndrome (elevated circulating inflammatory cytokines) and hyper-inflammation syndrome. These syndromes are triggered by viral infections and are also predictors of fatality in patients with COVID-19.50 51 Children are less affected due to higher antibodies, lower prior exposure to the virus and relatively low levels of inflammatory cytokines in their systems.
Signs and symptoms
Clinical features varied from mild illness to severe or fatal illness. The most common symptoms of COVID-19 were non-specific and mainly included fever, cough and myalgia. Other minor symptoms were sore throat, headache, chills, nausea or vomiting, diarrhoea, ageusia and conjunctival congestion. The COVID-19 was clinically classified into mild to moderate disease (non-pneumonia and pneumonia), severe disease (dyspnoea, respiratory frequency over 30/min, oxygen saturation less than 93%, PaO2/FiO2 ratio less than 300 and/or lung infiltrates more than 50% of the lung field within 24–48 hours) and critical (respiratory failure, septic shock and/or multi-organ dysfunction/failure).12 52 Many of the elderly patients who had severe illness had evidence of chronic underlying illness such as cardiovascular disease, lung disease, kidney disease or malignant tumours.53
Laboratory evaluation and confirmation
Laboratory findings most consistent with COVID-19 were lymphocytopenia, elevated C reactive protein and elevated erythrocyte sedimentation rate. Lymphocytopenia is due to necrosis or apoptosis of lymphocytes. The severity of lymphocytopenia reflects the severity of COVID-19.54–56 Procalcitonin was commonly elevated and was associated with coinfection in majority of reported paediatric cases.57 58
Detection of COVID-19 is based on virological detection by RT-PCR using swabs (nasopharynx, oropharynx), sputum and faeces, chest radiograph and dynamic monitoring of inflammatory mediators (eg, cytokines).59–61 Faecal specimens detected for COVID-19 nucleic acid was equally accurate as of pharyngeal swab specimens.60 Patients with COVID-19 showed high blood levels of cytokines and chemokines such as interleukin (IL)-7, IL-8, IL-9, IL-10, granulocyte-colony stimulating factor, granulocyte-macrophage colony-stimulating factor ,tumour necrosis factor alpha and VEGFA.50 62 63
Radiological findings
Most standard patterns observed on chest CT were ground-glass opacity, ill-defined margins, smooth or irregular interlobular septal thickening, air bronchogram, crazy-paving pattern and thickening of the adjacent pleura. Chest CT is considered to be a sensitive routine imaging tool for COVID-19.64–66
Management
At the initial presentation of cluster infection, many cases were treated with antiviral therapy, antibacterial therapy and glucocorticoids. Observation forms the mainstay for those who have mild illness. Moderately ill patients with underlying chronic illness, immunocompromised conditions and pregnancy require hospitalisation.67 68
The anti-malarial drugs, hydroxychloroquine and chloroquine, showed promising results in early in vitro study.69 However, the most robust and recent study in patients with COVID-19 have not shown unequivocal evidence of benefits for the treatment with hydroxychloroquine or chloroquine.70–72 In fact, the largest analysis to date of the risks and benefits of treating COVID-19 patients with these anti-malarial drugs was unable to confirm a benefit of hydroxychloroquine or chloroquine, when used alone or with a macrolide, on in-hospital outcomes for COVID-19.70 Besides, this study of 96 000 hospitalised patients on six continents found that those who received the drugs had a significantly higher risk of death and an increased frequency of ventricular arrhythmias compared with those who did not use it.70
Treatment of systemic complications in COVID-19
Extracorporeal membrane oxygenation is an excellent choice for patients with ARDS progressing to respiratory failure. Other modes of treatment include high-flow nasal oxygen and endotracheal intubation. Patients experiencing persistent refractory hypoxemia need prone positioning followed by neuromuscular blockade, inhaled nitric oxide (at 5–20ppm) and also provide optimal end-expiratory pressure by inserting oesophagal balloon.73 74
In the presence of shock with acute renal failure, negative fluid balance needs to be achieved by dialysis. Antimicrobials are used for pre-exposure and post-exposure prophylaxis. This prevents illness from SARS-CoV-2 and also reduces the risk of acquiring secondary infection. Fluid management is important to reduce pulmonary oedema.67 68 75 Glucocorticoids are best avoided due to its harmful effects in viral pneumonia and ARDS.76 Rescue therapy by administration of intravenous infusion of vitamin C has been suggested to attenuate vascular injury and systemic inflammation in sepsis and ARDS.77
Role of vaccines
Vaccine development is underway for COVID-19, but there are various limitations. This includes (1) the place for phase 3 vaccine trials are to be conducted in the locality of the ongoing transmission of disease, (2) vaccine manufactures need to work closely with biotechnology companies to develop effective vaccines which probably takes a minimum of 12–18 months and (3) regulators should evaluate safety with a range of virus strains in more than one animal model.78–80
The investigational vaccine has been currently developed using mRNA as its genetic platform using prior studies related to SARS and MERS.16 24 The basis of effective vaccine is immune targeted and involves identifying of B cell and T cell epitopes derived from the spike (S) and nucleocapsid (N) proteins among 120 available SARS-CoV-2 genetic sequences.24 Effective vaccination would play a vital role in reducing the viral spread and eliminate the virus from the host.81
Conclusion
COVID-19 has presented itself as a global pandemic in a short time period resulting in rapid curve shift of infected patients, increasing death rates, huge global economic burden and widespread mobilisation of medical resource across the globe. Being a novel disease, COVID-19 has presented itself as a mystery infection to the medical field, also requiring tremendous research and insights about the nature of the virus, and posing frequent challenges for a successful vaccine outcome. The approach to this disease requires active loco-regional to international collaboration with regards to disease containment, preventive strategies and treatment approach.
Main messages.
This article reviews the current state of knowledge concerning the origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19).
It traces the origin of coronavirus as it emerged and differentiates the COVID-19 with specific features from SARS and MERS.
We give a detailed insight into the modes of transmission, clinical manifestations, diagnosis and management. Also, it highlights the recent trends on vaccine development.
Key references.
Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9.
Raoult D, Zumla A, Locatelli F, et al. Coronavirus infections: epidemiological, clinical and immunological features and hypotheses. Cell Stress 2020.
Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507–13.
Dong Y, Mo X, Hu Y, et al. Epidemiological characteristics of 2143 pediatric patients with 2019 coronavirus disease in China. Pediatrics 2020; e20200702.
Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181–92.
Self-assessment questions.
Dromedary camel was involved in zoonotic transmission in MERS form of CoV?
Based on COVID-19 genomic structure, the protein that mediates an attachment and fusion between the virus and the host cell membrane is membrane (M) protein.
Cross-species and human-to-human transmission of COVID-19 is mainly dependent on the host receptor ACE2 protein.
Most consistent laboratory finding in COVID-19 patient is lymphopenia
COVID-19 is confirmed by RT-PCR.
Current research questions.
How does COVID-19 differ from SARS and MERS?
Mention the factors responsible for virus–host interaction.
List the specific laboratory findings in suspected cases of COVID-19.
Elaborate the scheme of management in COVID-19.
Self-assessment answers.
True
False
True
True
True
Supplementary Material
Footnotes
Correction notice: This article has been corrected since it appeared Online First. Minor typographical errors have been corrected.
Contributors: SU contributed to the conception and design of the work, data collection, drafting, revision and final version. PS contributed to the data analysis, interpretation, drafting the article and critical revision. AVR contributed to the data collection, analysis, revision and image concept. MMB contributed to the data collection and drafting. JSR contributed to the data analysis and interpretation. LFA-M, SD and HK contributed to the data collection and analysis DKV contributed to the data collection. All the contributors were involved in revising the final version and granting final approval for the article to be submitted for publication.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; internally peer reviewed.
Contributor Information
Srikanth Umakanthan, Paraclinical Sciences, The University of the West Indies at Saint Augustine Faculty of Medical Sciences, Saint Augustine, Trinidad and Tobago.
Pradeep Sahu, Centre for Medical Sciences Education, The University of the West Indies at Saint Augustine, Saint Augustine, Trinidad and Tobago.
Anu V Ranade, Basic Medical Sciences, University of Sharjah, Sharjah, United Arab Emirates.
Maryann M Bukelo, Department of Anatomical Pathology, North Central Regional Health Authority, Eric Williams Medical Sciences Complex, Mount Hope, Trinidad and Tobago.
Joseph Sushil Rao, Department of Surgery, University of Minnesota, Minneapolis, Minnesota, USA.
Lucas Faria Abrahao-Machado, Department of Pathology, Bacchi Laboratory, Sao Paulo, Brazil.
Samarika Dahal, Department of Oral Pathology, Institute of Medicine, Kathmandu, Nepal.
Hari Kumar, Department of International Public Health and Disease Surveillance, Midwest, Australia.
Dhananjaya KV, Department of Radiology, Indiragandhi Institute of Child Health, Bangalore, India.
References
- 1. Weiss SR, Navas-Martin S Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 2005;69:635–64. 10.1128/MMBR.69.4.635-664.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heymann DL, Shindo N, WHO Scientific and Technical Advisory Group for Infectious Hazards. COVID-19: what is next for public health? Lancet 2020;395:542–5. 10.1016/S0140-6736(20)30374-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fisher D, Heymann D. Q&A: the novel coronavirus outbreak causing COVID-19. BMC Med 2020;18:57. 10.1186/s12916-020-01533-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El Zowalaty ME, Jarhult JD. From SARS to COVID-19: a previously unknown SARS-related coronavirus (SARS-CoV-2) of pandemic potential infecting humans - call for a one health approach. One Health 2020;9:100124 10.1016/j.onehlt.2020.100124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sahu P. Closure of universities due to coronavirus disease 2019 (COVID-19): impact on education and mental health of students and academic staff. Cureus 2020;12:4 e7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coronavirus COVID-19 global cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) [May;2020]; Available https://coronavirus.jhu.edu/map.html 2020
- 8. WHO . Coronavirus disease (COVID-2019) situation reports. Situation report-51. 2020. (Available https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10)
- 9. Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science 2003;302:276–8. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- 10. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peckham R. COVID-19 and the anti-lessons of history. Lancet 2020;395:850–2. 10.1016/S0140-6736(20)30468-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raoult D, Zumla A, Locatelli F, et al. Coronavirus infections: epidemiological, clinical and immunological features and hypotheses. Cell Stress 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu J, Zhao S, Teng T, et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 2020;12:E244. 10.3390/v12020244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gorbalenya AE, Baker SC, Baric RS, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020;5:536–44. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woo PC, Lau SK, Chu CM, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 2005;79:884–95 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng VC, Lau SK, Woo PC, et al. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev 2007;20:660–94. 10.1128/CMR.00023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cong Y, Verlhac P, Reggiori F. The interaction between nidovirales and autophagy components. Viruses 2017;9:182. 10.3390/v9070182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med 2004;10:368–73 10.1038/nm1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beniac DR, Andonov A, Grudeski E, et al. Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol 2006;13:751–2. 10.1038/nsmb1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol 1990;64:5367–75. 10.1128/JVI.64.11.5367-5375.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Armstrong J, Niemann H, Smeekens S, et al. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature 1984;308:751–2. 10.1038/308751a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Doremalen N, Miazgowicz KL, Milne-Price S, et al. Host species restriction of middle east respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J Virol 2014;88:9220–32. 10.1128/JVI.00676-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zaki AM, van Boheeman S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 26. De Groot RJ, Baker SC, Baric RS, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 2013;87:7790–2. 10.1128/JVI.01244-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan JF, Lau SK, To KK, et al. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 2015;28:465–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gorbalenya AE, Baker SC, Baric R, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020;5:536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meo SA, Alhowikan AM, Al-Khlaiwi T, et al. Novel coronavirus 2019-nCoV: prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. Eur Rev Med Pharmacol Sci 2020;24:2012–9. [DOI] [PubMed] [Google Scholar]
- 30. Malik YS, Sircar S, Bhat S, et al. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q 2020;40:68–76. 10.1080/01652176.2020.1727993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005;310:676–9. 10.1126/science.1118391 [DOI] [PubMed] [Google Scholar]
- 33. Godet M, L’Haridon R, Vautherot JF, et al. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology 1992;188:666–75. 10.1016/0042-6822(92)90521-P [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McIntosh K, Becker WB, Chanock RM. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci U S A 1967;58:2268–73. 10.1073/pnas.58.6.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bárcena M, Oostergetel GT, Bartelink W, et al. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc Natl Acad Sci U S A 2009;106:582–7. 10.1073/pnas.0805270106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MMalta M, Rimoin AW, Strathdee SA. The coronavirus 2019-nCoV epidemic: is hindsight 20/20? EClinicalMedicine 2020;20:100289. 10.1016/j.eclinm.2020.100289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020;382:1199–207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia L, Hu F, Li H, et al. Characterization of small genomic regions of the hepatitis B virus should be performed with more caution. Virol J 2018;15:188. 10.1186/s12985-018-1100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang B, Bragazzi NL, Li Q, et al. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCoV). Infect Dis Model 2020;5:248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 2020; S1473–3099:30232–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haines A, de Barros EF, Berlin A, et al. National UK programme of community health workers for COVID-19 response. Lancet 2020;395:1173‐5. 10.1016/S0140-6736(20)30735-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci 2020;12:8. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wan Y, Shang J, Graham R, et al. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol 2020;94:e00127–20 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baig AM, Khaleeq A, Ali U, et al. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci 2020;11:995‐998. 10.1021/acschemneuro.0c00122 [DOI] [PubMed] [Google Scholar]
- 46. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708‐20. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497‐506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Porcheddu R, Serra C, Kelvin D, et al. Similarity in case fatality rates (CFR) of COVID-19/SARS-COV-2 in Italy and China. J Infect Dev Ctries 2020;14:125–8. 10.3855/jidc.12600 [DOI] [PubMed] [Google Scholar]
- 49. Wilson N, Kvalsvig A, Barnard LT, et al. Case-fatality risk estimates for COVID-19 calculated by using a lag time for fatality. Emerg Infect Dis 2020;26. 10.3201/eid2606.200320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mehta P, McAuley DF, Brown M, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033‐1034. 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Karakike E, Giamarellos-Bourboulis EJ Macrophage activation-like syndrome: a distinct entity leading to early death in sepsis. Front Immunol 2019;10:55. 10.3389/fimmu.2019.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Singhal TA Review of coronavirus disease-2019 (COVID-19). Indian J Pediatr 2020;87:281‐6. 10.1007/s12098-020-03263-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang T, Du Z, Zhu F, et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395:e52. 10.1016/S0140-6736(20)30558-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Special Expert Group for Control of the Epidemic of Novel Coronavirus Pneumonia of the Chinese Preventive Medicine Association. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41:139‐144. [DOI] [PubMed] [Google Scholar]
- 55. Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis 2020;34:101623. 10.1016/j.tmaid.2020.101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chu H, Zhou J, Wong BH, et al. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 2016;213:904–14 10.1093/infdis/jiv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xia W, Shao J, Guo Y, et al. Clinical and CT features in pediatric patients with COVID-19 infection: different points from adults. Pediatr Pulmonol 2020;55:1169‐74. 10.1002/ppul.24718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dong Y, Mo X, Hu Y, et al. Epidemiological characteristics of 2143 pediatric patients with 2019 coronavirus disease in China. Pediatrics. 2020; e20200702.32179660 [Google Scholar]
- 59. Lan L, Xu D, Ye G, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA 2020;e202783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang J, Wang S, Xue Y Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 2020;S1473–3099:30086–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Conti P, Ronconi G, Caraffa A, et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies [published online ahead of print, 2020]. J Biol Regul Homeost Agents 2020;34:1. [DOI] [PubMed] [Google Scholar]
- 63. McNicholl JM, Smith DK, Qari SH, et al. Host genes and HIV: the role of the chemokine receptor gene CCR5 and its allele [published correction appears in emerg infect dis 1997(4):584]. Emerg Infect Dis 1997;3:261–71. 10.3201/eid0303.970302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang YXJ, Liu WH, Yang M, et al. The role of CT for COVID-19 patient’s management remains poorly defined. Ann Transl Med 2020;8:145. 10.21037/atm.2020.02.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chung M, Bernheim A, Mei X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 2020;295:202–7 10.1148/radiol.2020200230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lin C, Ding Y, Xie B, et al. Asymptomatic novel coronavirus pneumonia patient outside Wuhan: the value of CT images in the course of the disease. Clin Imaging 2020;63:7–9. 10.1016/j.clinimag.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu K, Cai H, Shen Y et al. Management of corona virus disease-19 (COVID-19): the Zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban 2020;49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mitja O, Clotet B. Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob Health 2020;8:e639‐e640. 10.1016/S2214-109X(20)30114-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yao X, Ye F, Zhang M, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome conronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020ciaa237. 10.1093/cid/ciaa237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mehra MR, Desai SS, Ruschitzka F, et al. Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet. 2020. 10.1016/S0140-6736(20)31180-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71. Geleris J, Sun Y, Platt J, et al. Observational study of hydroxychloroquine in hospitalized patients with COVID-19. N Engl J Med. 2020. 10.1056/NEJMoa2012410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mahevas M, Tran V, Roumier M, et al. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalised for COVID-19 infection with oxygen requirement: results of a study using routinely collected data to emulate a target trial. medRxiv. 2020. [Google Scholar]
- 73. Matthay MA, Aldrich JM, Gotts JE Treatment for severe acute respiratory distress syndrome from COVID-19. Lancet Respir Med 2020;8:433‐4. 10.1016/S2213-2600(20)30127-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Goh KJ, Choong MC, Cheong EH, et al. Rapid progression to acute respiratory distress syndrome: review of current understanding of critical illness from COVID-19 infection. Ann Acad Med Singapore 2020;49:1–9. [PubMed] [Google Scholar]
- 75. Wujtewicz M, Dylczyk-Sommer A, Aszkiełowicz A, et al. COVID-19 - what should anaesthesiologists and intensivists know about it? Anaesthesiol Intensive Ther 2020;52:34–41. 10.5114/ait.2020.93756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arabi YM, Mandourah Y, Hameed FA, et al. Corticosteroid therapy for critically ill patients with middle east respiratory syndrome. Am J Respir Crit Care Med 2018;197:757–67 10.1164/rccm.201706-1172OC [DOI] [PubMed] [Google Scholar]
- 77. Fowler AA, Truwit JD, Hite RD, et al. Effect of vitamin C infusion on organ failure and biomarkers of inflammation and vascular injury in patients with sepsis and severe acute respiratory failure: the CITRIS-ALI randomized clinical trial. JAMA 2019;322:1261–70. 10.1001/jama.2019.11825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV: a target for vaccine and therapeutic development. Nat Rev Microbiol 2009;7:226–36. 10.1038/nrmicro2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet 2020;395:470‐473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lin Y, Shen X, Yang RF, et al. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res 2003;13:141–5 10.1038/sj.cr.7290158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ahmed SF, Quadeer AA, McKay MR Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020;12:254. 10.3390/v12030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


