Abstract
The COVID-19 pandemic has resulted in a significant increase in the number of pneumonia cases, including those caused by the Coronavirus. To detect COVID pneumonia, RT-PCR is used as the primary detection tool for COVID-19 pneumonia but chest imaging, including CT scans and X-Ray imagery, can also be used as a secondary important tool for the diagnosis of pneumonia, including COVID pneumonia. However, the interpretation of chest imaging in COVID-19 pneumonia can be challenging, as the signs of the disease on imaging may be subtle and may overlap with normal pneumonia. In this paper, we propose a hybrid model with the name COVINet which uses ResNet-101 as the feature extractor and classical K-Nearest Neighbors as the classifier that led us to give automated results for detecting COVID pneumonia in X-Rays and CT imagery. The proposed hybrid model achieved a classification accuracy of 98.6%. The model's precision, recall, and F1-Score values were also impressive, ranging from 98-99%. To back and support the proposed model, several CNN-based feature extractors and classical machine learning classifiers have been exploited. The outcome with exploited combinations suggests that our model can significantly enhance the accuracy and precision of detecting COVID-19 pneumonia on chest imaging, and this holds the potential of being a valuable resource for early identification and diagnosis of the illness by radiologists and medical practitioners.
Keywords: COVID-19, COVID pneumonia, ResNet-101, Machine learning, Deep learning
1. Introduction
The COVID-19 pandemic caused by the SARS-CoV-2 virus has resulted in a significant increase in illness and fatalities around the world. The first COVID-19-infected case was reported in December 2019 and initially affected the human nose and throat. However, with frequent mutations, new variants emerged in 2021 and 2022 that affected the human lungs, leading to pneumonia-like symptoms that posed diagnostic challenges for medical professionals. COVID-19 pneumonia presents with symptoms such as fever, cough, shortness of breath, and fatigue, and can result in severe respiratory distress leading to hospitalization or death. The pandemic has caused widespread concern and has been a leading cause of morbidity and mortality worldwide, resulting in the implementation of strict public health measures such as lockdowns, travel restrictions, and social distancing guidelines. The RT-PCR test is the primary diagnosis for COVID-19 pneumonia. Chest imaging, including CT scans and X-Ray imagery, is also a crucial diagnostic tool for detecting and managing pneumonia, including COVID-19 pneumonia. These scans can provide valuable information about the extent of lung involvement and help with the diagnosis and assessment of the disease's severity. However, the interpretation of chest imaging in COVID-19 pneumonia can be difficult, as the signs of the disease on imaging may be subtle and overlap with other respiratory infections. Nonetheless, this method can be more effective than the primary diagnosis and help in stopping the spread. To address these challenges, artificial intelligence (AI) is playing an increasingly important role in classifying COVID-19 pneumonia through X-Rays and CT scans. AI algorithms are designed to analyze large amounts of imaging data and identify patterns that may be missed by human radiologists. For instance, studies have shown that AI systems can accurately classify COVID pneumonia in CT scans with an accuracy of 91%, outperforming human radiologists who are not familiar with the specific patterns of COVID pneumonia (Hasija et al., 2022). Additionally, another study found that an AI algorithm was able to correctly identify COVID pneumonia on X-Rays with an accuracy of 97%, significantly higher than the accuracy of human radiologists who were not trained in the specific patterns of COVID pneumonia (Akter et al., 2021). These findings demonstrate the potential for AI to be a valuable tool in classifying COVID-19 pneumonia through chest imaging. Additionally, the AVNC model proposed by Wang, Fernandes, Zhu, and Zhang (2021) achieved a micro-averaged F1-Score of 96.87%, which is higher than 11 state-of-the-art approaches in the detection of COVID-19 in radio images. These findings demonstrate the potential for deep CNNs to be a valuable tool in accurately classifying COVID-19 pneumonia through chest imaging.
In recent years, deep neural networks have also been used in other fields, such as in the classification of malware threats using the Internet of Things (IoT) (Naeem et al., 2022), prediction of Industrial Internet of Things (IIoT) traffic (Ikram et al., 2022), and arrhythmia classification (Jangra et al., 2021). Moreover, a few studies exploited deep neural networks for audio-visual speech recognition (Kumar et al., 2022), medical image fusion (Li, Zhao, Lv, & Li, 2021), segmentation and classification of the brain tumour (Agrawal, Katal, & Hooda, 2022), and prediction of diabetes mellitus (Kumar, Perumal, Nadesh, & Arivuselvan, 2020). These studies have shown that deep neural networks can outperform traditional machine learning algorithms in classification tasks, highlighting the potential for deep learning in a variety of applications. Overall, these studies demonstrate the potential of deep neural networks to be a valuable tool in classifying COVID-19 pneumonia through chest imaging. Furthermore, these studies also show that deep neural networks outperform other traditional methods in this area. In regards to traditional methods, a study by Zhang et al. (2020) demonstrates that the SVM classifier can prove to be the best machine learning classifier for the detection of seam carving in grey-scale images by adopting local contextual features. As such, the development of AI algorithms for COVID-19 diagnosis through chest imaging holds great promise for improving the accuracy and efficiency of COVID-19 diagnosis, which is critical for effective patient management and control of the pandemic. To leverage the potential of the deep neural network as shown in various classification tasks, in this study a hybrid model named COVINet is developed to enhance the detection accuracy of COVID-19 on chest imaging. To achieve this, an imagery dataset of 8718 chest CT scans and X-Ray images for both COVID and Non-COVID pneumonia was collected from the work (Kumar, 2022). To propose the COVINet model several experiments have been carried out by exploiting deep learning feature extractors such as ResNet-101, InceptionResNetV2, DenseNet201, ResNet152, and EfficientNetV2L to generate the feature vector for COVID and Non-COVID pneumonia. The feature vectors are then passed through various machine learning classifiers such as Decision Trees, Multi-Layer Perceptron, Support Vector Machines (SVM), Random Forest, K-Nearest Neighbors (KNN), Extra Trees, and Gaussian Naive Bayes, to classify images into COVID pneumonia and Non-COVID pneumonia. The results of the experiments showed that the combination of ResNet-101 and K-Nearest Neighbors (KNN) yielded the best performance, with an accuracy of 98.6%, a precision of 98-99%, a recall of 99%, and an F1-score of 99% therefore, we proposed this combination as COVINet. The high values for accuracy and precision with ResNet-101 and K-Nearest Neighbors are due to the stable architecture of ResNet-101 which uses residual connections with its convolutional layers and a selective, and balanced number of filters with each convolution layer and K-Nearest Neighbors (KNN) being the best classifier in the cases where appropriate information in the form of the feature vector is available, which makes it easier to classify based on a distance measure. On the other hand, the worst combination was EfficientNetV2L and Decision Trees with an accuracy of 70.2% and precision, recall, and F1-score ranging between 70-71%.
Major contributions of this work are:
-
1.
The proposed hybrid model, combining ResNet-101 and multiple machine learning classifiers, was developed to accurately classify COVID-19 and Non-COVID pneumonia in CT scans and X-Rays.
-
2.
The integration of ResNet-101 and K-Nearest Neighbors (KNN) yielded an accuracy of 98.6% and precision of 99% which was the highest among all the tested combinations.
-
3.
The model's performance was rigorously tested and validated through experimentation with 35 different combinations of CNNs and machine learning classifiers.
-
4.
The proposed hybrid model outperforms existing solutions and effectively classifies COVID-19 and Non-COVID pneumonia in CT scans and X-Ray imagery.
The rest of this paper is as follows: Section 2 presents related research; Section 3 details the dataset and the proposed model; Section 4 outlines the performed experiments, results, and observations, and; Section 5 concludes and outlines potential future directions.
2. Related work
Numerous studies have focused on classifying COVID-19 by analyzing CT scans and X-Ray image datasets. These studies have found that various deep learning models such as MobileNetV3Large, SeNet154, MCRA network, U-Net hybrid with ResNet50, deep classification network, AB-ResNet neural network, modified MobileNet and ResNet, and a deep feature fusion module can effectively classify COVID-19 from other diseases such as common pneumonia and healthy individuals. Additionally, some studies have utilized image processing and data augmentation techniques to further improve the diagnosis accuracy.
Baghdadi et al. (2022) analyzed two datasets, one with two classes and the other with three classes. They found that MobileNetV3Large and SeNet154 were the best models available for each dataset, respectively. Liu et al. (2022) presented a unique solution for chest X-Ray image classification by proposing a multiscale class residual attention network. This network takes advantage of augmented images and addresses data shortages while reducing noise. Their approach demonstrated remarkable results on the COVIDx dataset. Zhang et al. (2022) conducted research in the area of CT image classification for COVID-19. They successfully segmented lesions from the images and were able to differentiate COVID-19 patients from healthy individuals and patients with common pneumonia, achieving a performance that was at the cutting-edge of the field. Kumar (2022) took a different approach, creating a well-annotated dataset of CT and X-Ray images and then improving performance by incorporating a residual network into the YOLO v4-tiny detector. This integration resulted in an improvement of 10-14% over the original YOLO model. Salama and Aly (2022) used a comprehensive framework to segment and classify CT images, aiming to determine COVID-19 status. The best performance was achieved using the U-Net hybrid with ResNet50, with metrics values ranging from 97%-99%.
Fang and Wang (2021) proposed a deep classification network for COVID pneumonia and verified the classification precision for different convolution layers and learning rates. The algorithm achieved good performance, with metrics values ranging from 96% to 98%. introduced a cutting-edge approach called DCCNet for the rapid diagnosis of COVID-19 through chest X-Ray imagery. The DCCNet model outperformed the previously established state-of-the-art models by a margin of 6.7%. Ahila and Subhajini (2022) utilized deep learning and image processing to classify X-Ray, Ultrasound, and CT scans for COVID-19 diagnosis with an accuracy of 99%. Jia, Lam, and Xu (2021) proposed modified MobileNet and ResNet architectures for classifying COVID-19 CXR and CT images, respectively, outperforming comparative models and specific COVID-19 detection models in all metrics on both datasets. Liu et al. (2022) proposed an innovative approach for COVID-19 diagnosis using X-Ray images. They combined a deep feature fusion module and a multi-disease classification module with data augmentation techniques to achieve an accuracy of 99.89% in classifying COVID-19, pneumonia, and healthy images. This result was significantly higher than what had been previously achieved in the field. Wang et al. (2021) presented an explainable diagnosis approach for COVID-19 using a PatchShuffle stochastic pooling neural network (PSSPNN) combined with multiple-way data augmentation techniques. Their proposed model achieved a high F1-Score of 95.79% in distinguishing COVID-19, bacterial pneumonia, and healthy cases on chest X-Ray images. Additionally, they provided visual explanations for the model's classification decisions using Grad-CAM, a popular technique for generating class activation maps from deep neural networks. These findings suggest that PSSPNN can be a useful tool for improving COVID-19 diagnosis accuracy and providing interpretable results. Zhang et al. (2023) proposed a model called SNELM for COVID-19 recognition using X-Ray images. Their model utilizes a combination of SqueezeNet and Extreme Learning Machine (ELM) algorithms to achieve high accuracy in COVID-19 recognition. The proposed model was trained and tested on two datasets of a total of 936 chest CT scan images. The experimental results showed that SNELM achieved an accuracy of 97.1%, a sensitivity of 97.5%, a specificity of 98.3%, and an F1-Score of 97% for COVID-19 recognition. These results demonstrate the effectiveness of combining SqueezeNet and ELM algorithms for accurate COVID-19 recognition in X-Ray images. Wang, Satapathy, Xie, and Zhang (2023) developed an Explainable Localized Unsupervised Convolutional Neural Network (ELUCNN) for COVID-19 diagnosis. ELUCNN utilizes localized unsupervised convolutional layers to extract features from chest X-Ray images and generates an explainable diagnosis by highlighting the significant features. The proposed method had a sensitivity of 94.41%, a specificity of 94.84%, an accuracy of 94.62%, and an F1-Score of 94.61% in classifying COVID-19, pneumonia, and healthy images, which outperformed some of the existing state-of-the-art methods in the field. Wang, Fernandes, Zhu, and Zhang (2021) proposed an attention-based VGG-style network for COVID-19 diagnosis called AVNC, which utilizes the Convolutional Block Attention Module (CBAM) to improve the performance of the model. The dataset used for training and testing consisted of 1164 chest CT scan images. The proposed model achieved a high level of accuracy with sensitivity, precision, and F1-Score for each class all above 95%. Notably, AVNC outperformed 11 state-of-the-art approaches, achieving a micro-averaged F1-Score of 96.87%. A detailed summary of the related work is presented in Table 1 .
Table 1.
Summary of related work.
| Work | Technique | Result (Accuracy/F1-Score) |
|---|---|---|
| Baghdadi et al. (2022) | SeresNext50, SeresNext101, SeNet154, MobileNet, MobileNetV2, MobileNetV3Small, MobileNetV3Large | SeresNext50: 95.25%, SeresNext101: 97.61%, SeNet154: 98%, MobileNet: 94.8%, MobileNetV2: 76.15%, MobileNetV3Small: 93.7%, MobileNetV3Large: 93.73% |
| Liu et al. (2022) | CNN model with multiscale class residual attention and class residual attention | 98.1% |
| Zhang et al. (2022) | Dynamic Transfer- learningClassification Network (DTCN) | 77% |
| Kumar (2022) | RYOLO v4-tiny | 88.18% |
| Salama et al. (2022) | ResNet50 | 97-99% |
| Fang et al. (2021) | Convolution and Deconvolution local enhancement | 97% |
| Ayalew et al. (2022) | DCCNet and HOG | 99.67% |
| Jia et al. (2021) | MobileNet and ResNet. | 99.45% |
| Jiang et al. (2022) | DFFCNet | 99.89% |
| Wang et al. (2021) | PSSPNN | F1-Score: 95.79% |
| Zhang et al. (2023) | SqueezeNet and Extreme Learning Machine (ELM) | 97.1% |
| Wang et al. (2023) | Explainable localized unsupervised convolutional neural network | 94.62% |
| Wang et al. (2021) | Convolutional block attention module (CBAM) | F1-Score: 96.87% |
3. Materials and methods
3.1. Dataset characteristics
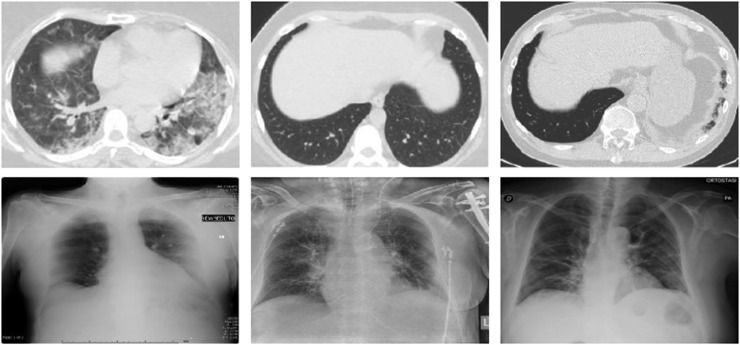
The custom image dataset used for accurately identifying COVID and Non-COVID pneumonia cases included a total of 8718 CT scans and X-Ray images of both classes extracted from (Kumar, 2022). Among these, CT COVID pneumonia had 3006 images, CT Non-COVID pneumonia had 2952 images, X-Ray COVID pneumonia had 1380 images, and X-Ray Non-COVID pneumonia had 1380 images. The images were preprocessed by applying resizing as per the input size of the deep learning feature extractors and further, split in the ratio of 70:30 respectively for training and testing. A combination of CNN feature extractors and machine learning classifiers was used to evaluate the classification accuracy of images belonging to COVID and Non-COVID pneumonia. Figs. 1 and 2 show the visual representation of the two image types (CT scans and X-Rays) and the corresponding classes.
Fig. 1.
Dataset sample of class with with_COVID.
Fig. 2.
Dataset sample of class with without_COVID.
3.2. The proposed model
In this study, CT scans and X-Ray images of COVID and Non-COVID pneumonia patients were pre-processed and labeled to their respective classes. These labeled images were then processed through a deep convolutional neural network, ResNet-101, which was pre-trained on over a million images from the ImageNet database and has learned to recognize intricate features in a diverse range of images. The ResNet-101 acted as a feature extractor, where the final feature vector obtained after advanced pooling was utilized as input to several machine learning classifiers. The fully connected and softmax layer of ResNet-101 was discarded to produce only the feature vector. The network is designed to accept an input of 224×224 image size. Further, the feature vector obtained from ResNet-101 was passed to several machine learning classifiers, including Decision Trees, Multi-Layer Perceptron, Support Vector Machines (SVM), Random Forest, K-Nearest Neighbors (KNN), Extra Trees, and Gaussian Naive Bayes to classify the images as infected with COVID or Non-COVID pneumonia. The ResNet-101 uses fully connected and softmax layers to classify the input fed to it, however, in this work classification has been performed by the machine learning classifiers therefore, we have removed the fully connected and softmax layers and passed the feature vector generated by the ResNet-101 to the machine learning classifiers. In conclusion, the proposed model is a hybrid approach that leverages the feature extraction capabilities of ResNet-101 and the classification capabilities of multiple machine learning algorithms to attain high accuracy and precision in the classification of COVID and Non-COVID pneumonia in CT scans and X-Ray images. The other advantage of the proposed hybridization is that it utilizes lesser computation resources and can be trained and tested on small computation devices such as Jetson Nano, etc. A detailed description of the proposed model is presented in Fig. 3 . Further, the details of ResNet-101 and machine learning classifiers are provided below.
Fig. 3.
Proposed hybrid classifier.
3.2.1. ResNet-101 as a feature extractor
The ResNet-101 model, which is a deep CNN architecture with identity shortcut connections was used for feature extraction. To reduce computational complexity and avoid training the ResNet-101 model from scratch, transfer learning was employed. Specifically, the feature representation produced by the ResNet-101 model was selected using the average pool layer applied after the 101st layer of the network, resulting in a feature representation of size 1×1×1000. A dense layer of size 1024 was applied after the average pool layer to convert the feature representation from 1×1×1000 to a 2-D feature vector. The transfer learning mechanism was used to extract features, and the softmax layer used by ResNet-101 for classification was removed. The obtained feature vector was then passed to various machine learning classifiers which can classify unknown patterns with similar distinguishing features, including noisy and incomplete inputs. This approach was applied to both CT scans and X-Ray images for COVID and Non-COVID pneumonia classification.
The proposed technique involves using a CNN-based ResNet-101 feature extractor to process input images from COVID-19 and Non-COVID pneumonia dataset consisting of CT scans and X-Ray imagery. Each layer of the feature extractor takes input: where, R: RGB image having w: width, h: height and c: channel, and a set of parameters W, and outputs a new image Y = f(X, W). ResNet-101 uses residual connections and identity shortcut connections, enabling it to learn features from the input image while skipping one or more layers and the output image as shown in Eq. (1).
| (1) |
The resulting image in Eq. (1) is a combination of the input image where, f(X) and X have different dimensions and the output of the ResNet-101 feature extractor, which includes as 1×1 convolutions added as identity shortcuts. This technique uses ResNet-101 as a feature extractor generating a 2-D feature vector in the form of numbers. Further, the obtained 2-D feature vector is then utilized to classify COVID and Non-COVID pneumonia samples using various machine learning classifiers. These classifiers distinguish between the obtained features by performing probability-based predictions based on the two classes, i.e., COVID pneumonia and Non-COVID pneumonia. This feature extraction process is crucial for accurately classifying COVID and Non-COVID as both the sets share similar features.
3.2.2. Machine learning classifiers
The feature vector generated by the ResNet-101 was fed to machine learning classifiers to classify the COVID and Non-COVID pneumonia with high accuracy. In order to achieve this, several machine learning classifiers have been used and exploited with the ResNet-101 feature extractor. The details about the machine learning classifiers are as follows:
3.2.2.1. Decision trees
The Decision Tree is a well-known algorithm in the field of machine learning, often used for solving classification problems. It splits data into smaller and smaller subgroups in a tree-like structure based on entropy function and information gain to determine which features are the most relevant for prediction or categorization. This model visually represents the decision-making process by creating branches and leaves, each representing a class label or predicted outcome.
The entropy function is used to measure the impurity or randomness of a set of data, and the goal of the decision tree is to split the data in such a way that the entropy is reduced as much as possible. The information gain is represented by Eq. (2).
| (2) |
In Eq. (2), IG(S,F) calculates the reduction in entropy achieved by splitting the data on a particular feature F. Here, S is the set of data, t are the subgroups created by the split, and p(t) is the proportion of data in subgroup t.
3.2.2.2. Support vector machine
Support Vector Machines (SVM) is a type of machine learning algorithm that is widely used for classification and regression problems. It is particularly effective in categorization tasks where the data points are separable by a clear boundary or hyperplane. The SVM algorithm works by identifying the optimal hyperplane that maximizes the margin between the two classes, separating the data points with the highest accuracy. The size of the hyperplane is determined by the number of features in the data, making it a highly customizable tool for various data sets.
3.2.2.3. Random forest
The Random Forest Method is a popular supervised machine learning algorithm for classification and regression tasks. It is based on the concept of a "forest" of decision trees, where the more trees in the forest, the more robust and accurate the algorithm is. In Random Forest, multiple decision trees are trained on different subsets of the data, and their predictions are combined to give a final output, improving the overall performance and accuracy of the model. The process of combining multiple models to solve a complex issue is known as ensemble learning.
3.2.2.4. Extra trees
The Extremely Randomized Trees Classifier (Extra Trees Classifier) is a form of ensemble learning that combines the results of several randomized decision trees to make a final prediction. This approach is similar to Random Forest Classifier, with the main difference being that the decision trees in the forest are constructed using random sampling of features from the feature set at each node. The classifier selects the best feature to split the data using mathematical criteria such as the Gini Index. The resulting de-correlated decision trees enhance the performance of the model and increase its accuracy.
3.2.2.5. K-Nearest neighbors
K-Nearest Neighbors (KNN) is a widely used and simple classification algorithm in Machine Learning. It is a non-parametric method that operates based on the idea of similarity between instances. In KNN, the classification of a new instance is determined by the class label of its nearest neighbors in the training data. The nearest neighbors are determined based on a distance metric such as Euclidean distance. Given a test instance, the algorithm finds the k-nearest neighbors in the training set and assigns the majority class label among them to the test instance. KNN is highly applicable in real-world problems as it doesn't make any underlying assumptions about the data distribution.
3.2.2.6. Multi-layer perceptron
The Multi-layer Perceptron (MLP) is a type of deep learning network used for classification tasks. It consists of dense, fully connected layers that can transform an input of any dimension into the desired output dimension. By connecting neurons in such a way that some outputs serve as inputs, an MLP can be built to form a multi-layer perception network. It consists of several layers, including an input layer that receives the input features, a number of hidden layers that allow the network to identify complex relationships within the data, and an output layer that produces the final prediction. The number of hidden layers and the number of neurons within each layer can be adjusted to optimize the network's performance.
3.2.2.7. Gaussian naive bayes
Gaussian Naive Bayes is a popular machine learning algorithm used for classification tasks. It is a probabilistic method that assumes strong independence between features and applies Bayes' theorem to make predictions. The algorithm assumes that the features in a dataset follow a normal or Gaussian distribution and uses this information to compute the probabilities of each class. In order to make accurate predictions, Gaussian Naive Bayes requires the mean and variance of the features in the dataset, which are used in the calculation of the probabilities. The "naive" aspect of the algorithm refers to the assumption that features are independent, meaning the existence of one feature has no impact on the existence of another. Despite its limitations, Gaussian Naive Bayes is a fast and simple algorithm that can provide good results in certain classification tasks. The Gaussian Naive Bayes is represented by Eq. (3).
| (3) |
where C is the class, X is the data point, P(X|C) is the probability of the features X given class C, P(C) is the prior probability of the class, and P(X) is the probability of the data point X. The final result is the posterior probability of class C given the data point X. The class with the highest posterior probability is selected as the prediction for that data point.
4. Experiments and results
The experimentation was followed in 4 ways: Finding the relevant dataset and pre-processing it before training and testing, Training the dataset with a combination of CNN feature extractors with machine learning classifiers, Evaluating the performance metrics, and using them to find out the best combination among the exploited hybrid techniques and plotting the comparison plots of the best combination trained on Google Colab with python libraries using various parameters and deep learning neural networks with machine learning classifiers. In this work, we have exploited five deep learning-based feature extractors. Table 2 presents a description of the CNN-based feature extractors used in this study, including their input size, number of CNN layers, and trainable parameters. During the training, batch-normalization and ReLU activation were applied across the CNN network, and a learning rate of 1.1e-05, and momentum of 0.9 has been set for each exploited deep learning feature extractor.
Table 2.
CNN feature extractor parameters.
| CNN feature extractor | Input size in pixels | CNN layers | Trainable Parameters |
|---|---|---|---|
| ResNet-101 | 299×299 | 101 Conv layers | 4,25,52,832 |
| ResNet-152 | 224×224 | 152 Conv layers | 6,01,92,808 |
| InceptionResNetV2 | 299×299 | 164 Conv layers | 2,17,68,352 |
| DenseNet201 | 224×224 | 201 Conv layers | 2,00,13,928 |
| EfficientNetV2L | 384×384 | 57 Conv layers | 11,85,15,272 |
4.1. Evaluation criteria
In this study, it is crucial to evaluate the effectiveness of the classifiers by examining performance metrics. Four commonly used evaluation metrics, namely accuracy, precision, recall, and F1-Score, are calculated and represented in Eqs. (4-7).
| (4) |
| (5) |
| (6) |
| (7) |
These measurements are derived from the confusion matrix of the tested image samples, where the number of false positive samples (FP), false negative samples (FN), true positive samples (TP), and true negative samples (TN) are considered.
4.2. Results
To embrace the proposed model based on ResNet-101 and K-Nearest Neighbors (KNN), we evaluated different deep learning-based CNN feature extractors such as ResNet152, EfficientNetV2L, DenseNet201, and InceptionResNetV2 and trained them with multiple machine learning classifiers like Support Vector Machine, Random Forest, Extra-Trees, K-Nearest Neighbors, Multiple Layer Perceptron, Decision Trees, and Gaussian Naïve Bayes. Our results showed that the ResNet-101 neural network performed the best among all feature extractors and has proven its effectiveness in various problems (Jain, Mittal, Thakur, & Mittal, 2020). As a result, we chose to use ResNet-101 as the feature extractor in our proposed COVINet model for classifying COVID and Non-COVID pneumonia in CT scans and X-Ray imagery. The results of the combinations between feature extractors and machine learning classifiers are summarized in Table 3, Table 4, Table 5, Table 6, Table 7 .
Table 3.
ResNet-101 performance with different ML classifiers.
| ResNet-101 | |||||
|---|---|---|---|---|---|
| ML Classifier | Classes | Accuracy | Precision | Recall | F1-Score |
| SVM | COVID | 92.80% | 0.93 | 0.93 | 0.93 |
| Non-COVID | 0.92 | 0.93 | 0.93 | ||
| Extra Trees | COVID | 92.50% | 0.94 | 0.91 | 0.92 |
| Non-COVID | 0.91 | 0.94 | 0.92 | ||
| Random Forest | COVID | 91.20% | 0.92 | 0.9 | 0.91 |
| Non-COVID | 0.9 | 0.92 | 0.91 | ||
| Multi-layer Perceptron | COVID | 96.10% | 0.96 | 0.96 | 0.96 |
| Non-COVID | 0.96 | 0.96 | 0.96 | ||
| Gaussian Naive Bayes | COVID | 85.90% | 0.87 | 0.86 | 0.86 |
| Non-COVID | 0.85 | 0.86 | 0.86 | ||
| Decision Tree | COVID | 73.10% | 0.73 | 0.74 | 0.74 |
| Non-COVID | 0.73 | 0.72 | 0.72 | ||
| K-Nearest Neighbors | COVID | 98.60% | 0.99 | 0.99 | 0.99 |
| Non-COVID | 0.98 | 0.99 | 0.99 | ||
Table 4.
ResNet152 performance with different ML classifiers.
| ResNet152 | |||||
|---|---|---|---|---|---|
| ML Classifiers | Classes | Accuracy | Precision | Recall | F1-Score |
| SVM | COVID | 91.20% | 0.93 | 0.9 | 0.91 |
| Non-COVID | 0.89 | 0.93 | 0.91 | ||
| Extra Trees | COVID | 92.70% | 0.94 | 0.91 | 0.93 |
| Non-COVID | 0.91 | 0.94 | 0.93 | ||
| Random Forest | COVID | 90.80% | 0.92 | 0.9 | 0.91 |
| Non-COVID | 0.9 | 0.92 | 0.91 | ||
| Multi-layer Perceptron | COVID | 95.50% | 0.96 | 0.95 | 0.96 |
| Non-COVID | 0.95 | 0.96 | 0.95 | ||
| Gaussian Naive Bayes | COVID | 83.00% | 0.83 | 0.84 | 0.84 |
| Non-COVID | 0.83 | 0.82 | 0.83 | ||
| Decision Tree | COVID | 72.90% | 0.73 | 0.73 | 0.73 |
| Non-COVID | 0.72 | 0.73 | 0.72 | ||
| K-Nearest Neighbors | COVID | 98.40% | 0.99 | 0.98 | 0.98 |
| Non-COVID | 0.98 | 0.99 | 0.98 | ||
Table 5.
InceptionResNetV2 performance with different ML classifiers.
| InceptionResNetV2 | |||||
|---|---|---|---|---|---|
| ML Classifiers | Classes | Accuracy | Precision | Recall | F1-Score |
| SVM | COVID | 77.00% | 0.86 | 0.66 | 0.74 |
| Non-COVID | 0.71 | 0.88 | 0.79 | ||
| Extra Trees | COVID | 87.40% | 0.86 | 0.9 | 0.88 |
| Non-COVID | 0.89 | 0.85 | 0.87 | ||
| Random Forest | COVID | 88.50% | 0.88 | 0.9 | 0.89 |
| Non-COVID | 0.89 | 0.87 | 0.88 | ||
| Multi-layer Perceptron | COVID | 86.70% | 0.87 | 0.87 | 0.87 |
| Non-COVID | 0.87 | 0.86 | 0.86 | ||
| Gaussian Naive Bayes | COVID | 55.60% | 0.87 | 0.15 | 0.26 |
| Non-COVID | 0.53 | 0.98 | 0.68 | ||
| Decision Tree | COVID | 76.60% | 0.77 | 0.77 | 0.77 |
| Non-COVID | 0.76 | 0.76 | 0.76 | ||
| K-Nearest Neighbors | COVID | 92.10% | 0.92 | 0.93 | 0.92 |
| Non-COVID | 0.92 | 0.92 | 0.92 | ||
Table 6.
DenseNet201 performance with different ML classifiers.
| DenseNet201 | |||||
|---|---|---|---|---|---|
| ML Classifiers | Classes | Accuracy | Precision | Recall | F1-Score |
| SVM | COVID | 87.90% | 0.88 | 0.88 | 0.88 |
| Non-COVID | 0.88 | 0.87 | 0.88 | ||
| Extra Trees | COVID | 91.00% | 0.94 | 0.88 | 0.91 |
| Non-COVID | 0.88 | 0.94 | 0.91 | ||
| Random Forest | COVID | 90.70% | 0.93 | 0.89 | 0.91 |
| Non-COVID | 0.89 | 0.93 | 0.91 | ||
| Multi-layer Perceptron | COVID | 94.30% | 0.95 | 0.94 | 0.94 |
| Non-COVID | 0.94 | 0.95 | 0.94 | ||
| Gaussian Naive Bayes | COVID | 74.00% | 0.8 | 0.65 | 0.72 |
| Non-COVID | 0.7 | 0.83 | 0.76 | ||
| Decision Tree | COVID | 77.50% | 0.79 | 0.77 | 0.78 |
| Non-COVID | 0.76 | 0.79 | 0.77 | ||
| K-Nearest Neighbors | COVID | 96.50% | 0.97 | 0.97 | 0.97 |
| Non-COVID | 0.96 | 0.96 | 0.96 | ||
Table 7.
EfficientNetV2L performance with different MLclassifiers.
| EfficientNetV2L | |||||
|---|---|---|---|---|---|
| ML Classifiers | Classes | Accuracy | Precision | Recall | F1-Score |
| SVM | COVID | 91.80% | 0.93 | 0.91 | 0.92 |
| Non-COVID | 0.91 | 0.93 | 0.92 | ||
| Extra Trees | COVID | 88.60% | 0.91 | 0.86 | 0.89 |
| Non-COVID | 0.87 | 0.91 | 0.89 | ||
| Random Forest | COVID | 87.50% | 0.89 | 0.86 | 0.88 |
| Non-COVID | 0.86 | 0.89 | 0.88 | ||
| Multi-layer Perceptron | COVID | 93.10% | 0.93 | 0.94 | 0.93 |
| Non-COVID | 0.94 | 0.92 | 0.93 | ||
| Gaussian Naive Bayes | COVID | 80.20% | 0.81 | 0.79 | 0.8 |
| Non-COVID | 0.79 | 0.81 | 0.8 | ||
| Decision Tree | COVID | 70.20% | 0.71 | 0.71 | 0.71 |
| Non-COVID | 0.7 | 0.70 | 0.70 | ||
| K-Nearest Neighbors | COVID | 95.20% | 0.97 | 0.93 | 0.95 |
| Non-COVID | 0.93 | 0.97 | 0.95 | ||
We trained our imagery dataset using ResNet-101 and several machine learning-based classifiers including Multi-Layer Perceptron, Decision Trees, Extra Trees, SVM, K-Nearest Neighbors, Random Forest, and Gaussian Naive Bayes. Among all the models, the K-Nearest Neighbors classifier achieved the highest classification accuracy of 98.6%, while Decision Trees had the lowest accuracy of 73.1%. The K-Nearest Neighbors classifier also showed the best results in terms of precision, recall, and F1-Score, with values ranging from 98% to 99%. On the other hand, Decision Tree showed the least results in these metrics, with values ranging from 72% to 74%. Overall, our results indicate that the combination of ResNet-101 and K-Nearest Neighbors is the most effective solution for classifying COVID and Non-COVID pneumonia in CT and X-Ray imagery. The performance metrics of ResNet-101 compared to all machine learning classifiers are illustrated in Fig. 4, Fig. 5, Fig. 6, Fig. 7 . These figures provide a visual representation of the performance results.
Fig. 4.
ResNet-101 accuracy with different ML classifiers.
Fig. 5.
ResNet-101 precision with different ML classifiers.
Fig. 6.
ResNet-101 recall with different ML classifiers.
Fig. 7.
ResNet-101 F1-Score with different ML classifiers.
The results of the analysis indicate that using the ResNet-101 feature extractor in combination with the K-Nearest Neighbors machine learning algorithm provided the strongest evaluation metrics, including accuracy, precision, recall, and F1-Score in detecting COVID and Non-COVID pneumonia. This particular combination showed the best performance compared to all other feature extractor-classifier combinations.
4.3. Summarization of findings
The results of the experiments carried out support the validity of the hybrid COVID and Non-COVID pneumonia classification method, COVINet, using a combination of ResNet-101 and K-Nearest Neighbors. A summary of the findings is presented, accompanied by key observations and explanations.
-
(1)
The experiments have demonstrated that using a combination of ResNet-101 for feature extraction and K-Nearest Neighbors classifier yields the highest accuracy, reaching 98.6%. This result confirms the effectiveness of the proposed technique.
-
(2)
The results show that among all the machine learning classifiers, the K-Nearest Neighbors classifier demonstrated the best performance when combined with multiple feature extractors. The K-Nearest Neighbors classifier produced impressive accuracy levels when paired with various feature extractors including ResNet152 (98.4%), InceptionResNetV2 (92.1%), DenseNet201 (96.5%), and EfficientNetV2L (95.2%). KNN outperforms other classifiers due to its simplicity and ability to handle complex, non-linear data. The KNN classifier is robust to noise and can handle missing or irrelevant features, making it well-suited for medical imaging classification tasks where data quality can be an issue. Furthermore, the KNN algorithm doesn't require training, which makes it computationally efficient, particularly when working with large datasets.
-
(3)
Additionally, the results suggest that ResNet-101, with its smaller feature extraction network and fewer trainable parameters, performed better in terms of accuracy, precision, recall, and F1-Score compared to ResNet-152. Despite having additional layers, ResNet-152 did not provide a significant improvement in the feature vector for machine learning classification.
-
(4)
The ResNet-101 produced high accuracy due to a higher number of trainable parameters as compared to other tested feature extractors and further, due to the usage of residual connections and appropriated convolution filters that aided in generating a suitable feature vector that can be classified accurately.
-
(5)
In the present work, the dataset consisting of COVID and Non-COVID pneumonia has similar features in images. In the cases, where the images have similar features, the K Nearest Neighbors (KNN) classifier performs better due to its ability to perform classification by searching the distance between similar neighbors that is, the data points having the same feature. However, other machine learning classifiers such as Multi-Layer Perceptron, Support Vector Machine, etc. are good but only in the cases where previous outputs are known to learn the parameters for the next output.
-
(6)
The ResNet-101 takes an input size of size therefore, its complexity can be stated as . However, the best results are obtained with the combination of ResNet-101 and K-NN, therefore the time complexity of K-NN can be stated as where n is the number of data points and d is the distance between the datapoints. The overall time complexity of the proposed ResNet-101 and K-NN-based model can be stated as the time consumed to find relevant data points and the distance between them in a image.
-
(7)
The results of the experiments showed that the decision trees classifier did not perform well. This can be attributed to the instability of decision trees, where small changes in the data can result in significant alterations in the optimal decision tree. Additionally, when classifying categorical variables with different levels, the information gain in decision trees tends to favor attributes with a higher number of levels.
4.4. Comparison with related work
Recently, there has been a surge in studies aimed at classifying COVID and Non-COVID pneumonia in CT scans and X-Ray imagery over the past three years. Many of these works have leveraged Convolutional Neural Network (CNN) architectures for classification purposes. However, in this research, we present a hybrid approach that is computationally more efficient by using CNN as a feature extractor and machine learning algorithms as classifiers. A comparison matrix of our proposed approach with previous studies in the field is provided in Table 8 .
Table 8.
Comparison with related work.
| Author | Technique | Result (Accuracy) |
|---|---|---|
| Baghdadi et al. (2022) | SeresNext50, SeresNext101, SeNet154, MobileNet, MobileNetV2, MobileNetV3Small, MobileNetV3Large | SeresNext50: 95.25%, SeresNext101: 97.61%, SeNet154: 98%, MobileNet: 94.8%, MobileNetV2: 76.15%, MobileNetV3Small: 93.7%, MobileNetV3Large: 93.73% |
| Liu et al. (2022) | CNN model, Multiscale class residual attention, Class Residual Attention | 98.1% |
| Kumar (2022) | RYOLO v4-tiny | 88.18% |
| Ayalew et al. (2022) | DCCNet and HOG | 99.67% |
| Ahuja et al. (2021) | ResNet18, ResNet50, ResNet-101, SqueezeNet | ResNet18: 99.4%, ResNet50: 98.8%, ResNet-101: 97%, SqueezeNet: 95.2% |
| Jain et al. (2020) | ResNet18, VGG16, DenseNet121, ResNet-101 | ResNet18: 97.33%, VGG16: 96.66%, DenseNet121: 97.6%, ResNet-101: 98.98% |
| Gupta et al. (2023) | DLM, DarkNet19, MobileNetV2 | DLM: 95.93%, DarkNet19: 98.91%, MobileNetV2: 97.61% |
| Wang, Lin, & Wong (2020) | COVID-Net, ResNet50, VGG19 | COVID-Net: 93.33%, ResNet50: 90.6%, VGG19: 83% |
| Ours | ResNet-101 and KNN | 98.6% |
The proposed model in this work achieved higher accuracy values compared to the related work as shown in Table 6. Specifically, our model achieved an accuracy of 98.6%, whereas the compared work achieved lower and comparable accuracy values than our proposed model. This indicates that our model, COVINet, is overall more effective in correctly identifying and classifying COVID-19 cases, resulting in fewer false positives and false negatives. Additionally, our model's performance also showed greater robustness and generalization capability. The findings of the study indicate that the proposed model demonstrates a higher level of reliability and accuracy in detecting and classifying COVID-19 cases.
4.5. Ablation study
To gauge the general performance and to validate the proposed ResNet-101 and K-Nearest Neighbors (K-NN) based model, we trained and tested it on the COVID Pneumonia datasets utilized by Hasija et al. (2022) and Baghdadi et al. (2022). The dataset employed by Hasija et al. (2022) consists of 6,244 images of COVID pneumonia and 7,400 images of Non-COVID pneumonia, whereas, the dataset employed by Baghdadi et al. (2022) consists of 14,486 image samples for both COVID and Non-COVID pneumonia. The test results on the datasets (Hasija et al., 2022) and (Baghdadi et al., 2022) are presented in Table 9 .
Table 9.
Ablation study results.
| Model | Dataset | Accuracy |
|---|---|---|
| ResNet-101 and KNN (Ours) | COVID and Non-COVID CT (Hasija et al., 2022) | 99.10% |
| ResNet-101 and KNN (Ours) | COVID and Non-COVID CT (Baghdadi et al., 2022) | 98.9% |
| ResNet-101 and KNN (Ours) | Ours | 98.6% |
The results as shown in Table 9 indicate that when the proposed model is trained and tested on a bigger dataset such as the dataset (Hasija et al., 2022) and (Baghdadi et al., 2022) having almost double the images of the dataset used in this work, the accuracy improves significantly that shows the ability of the proposed model to generalize and achieve better results on a bigger dataset. The better results indicate that the proposed ResNet-101 and K-Nearest neighbors (KNN) model is capable of achieving benchmark results on different datasets in this domain and other related tasks.
5. Conclusion
This work proposed a hybrid classifier, COVINet, for the classification of COVID and Non-COVID pneumonia in CT scans and X-Ray imagery by combining the ResNet-101 feature extractor with K-Nearest Neighbors machine learning classifier. This model achieved an accuracy of 98.6% on the self-preprocessed dataset. These results were achieved after training and testing the dataset with multiple CNNs (ResNet-101, ResNet152, InceptionResNetV2, DenseNet201, and EfficientNetV2L) and machine learning classifiers (Multi-layer Perceptron, K-Nearest Neighbors, Decision Trees, Extra Trees, Support Vector Machines, Random Forest, and Gaussian Naïve Bayes) where ResNet-101 outperformed in classifying COVID and Non-COVID pneumonia in images when combined with the best machine learning classifier: K-Nearest Neighbors. In order to use ResNet-101 as a feature extractor for classification, we removed the soft-max and sigmoid layers to use the only feature vector after advanced pooling. Furthermore, the dataset consisted of CT scans and X-Ray imagery of normal pneumonia and COVID pneumonia enabling the dataset to be robust in nature. The highest accuracy of 98.6% was achieved by using ResNet-101 and K-Nearest Neighbors, while Decision Tree achieved the least accuracy of 73.1%. The proposed model has the potential to efficiently aid in the rapid and accurate detection of COVID-19 from CT scans and X-Ray images and may be developed into an application in the future.
Funding
No funding has been received to carry out this work.
Declaration of Competing Interest
The authors declare that they have no competing interest.
Availability of Data and Material
The dataset and code of the developed model is available on a reasonable request to the corresponding author.
CRediT authorship contribution statement
Vasu Mittal: Conceptualization, Formal analysis, Investigation, Software, Visualization, Writing – original draft. Akhil Kumar: Conceptualization, Data curation, Methodology, Resources, Software, Supervision, Validation, Writing – review & editing.
Acknowledgements
Not applicable.
References
- Agrawal P., Katal N., Hooda N. Segmentation and classification of brain tumor using 3D-UNet deep neural networks. International Journal of Cognitive Computing in Engineering. 2022;3:199–210. [Google Scholar]
- Ahila T., Subhajini A.C. E-GCS: Detection of COVID-19 through classification by attention bottleneck residual network. Engineering Applications of Artificial Intelligence. 2022;116 doi: 10.1016/j.engappai.2022.105398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja S., Panigrahi B.K., Dey N., Rajinikanth V., Gandhi T.K. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Applied Intelligence. 2021;51(1):571–585. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Shamrat FMJM, Chakraborty S, Karim A, Azam S. COVID-19 detection using deep learning algorithm on Chest X-Ray images. Biology (Basel) 2021 Nov 13;10(11):1174. doi: 10.3390/biology10111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew A.M., Salau A.O., Abeje B.T., Enyew B. Detection and classification of COVID-19 disease from X-Ray images using convolutional neural networks and histogram of oriented gradients. Biomedical Signal Processing and Control. 2022;74 doi: 10.1016/j.bspc.2022.103530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Wang X. COVID-19 deep classification network based on convolution and deconvolution local enhancement. Computers in Biology and Medicine. 2021;135 doi: 10.1016/j.compbiomed.2021.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K., Bajaj V. Deep learning models-based CT-scan image classification for automated screening of COVID-19. Biomedical Signal Processing and Control. 2023;80 doi: 10.1016/j.bspc.2022.104268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasija S, Akash P, Bhargav Hemanth M, Kumar A, Sharma S. A novel approach for detection of COVID-19 and Pneumonia using only binary classification from chest CT-scans. Neuroscience Informatics. 2022 Dec;2(4) doi: 10.1016/j.neuri.2022.100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram S.T., Priya V., Anbarasu B., Cheng X., Ghalib M.R., Shankar A. Prediction of IIoT traffic using a modified whale optimization approach integrated with random forest classifier. The Journal of Supercomputing. 2022;78(8):10725–10756. [Google Scholar]
- Jain G., Mittal D., Thakur D., Mittal M.K. A deep learning approach to detect COVID-19 coronavirus with X-Ray images. Biocybernetics and biomedical engineering. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra M., Dhull S.K., Singh K.K., Singh A., Cheng X. O-WCNN: an optimized integration of spatial and spectral feature map for arrhythmia classification. Complex & Intelligent Systems. 2021:1–14. doi: 10.1007/s40747-021-00371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Lam H.K., Xu Y. Classification of COVID-19 chest X-Ray and CT images using a type of dynamic CNN modification method. Computers in Biology and Medicine. 2021;134 doi: 10.1016/j.compbiomed.2021.104425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. RYOLO v4-tiny: A deep learning based detector for detection of COVID and Non-COVID Pneumonia in CT scans and X-RAY images. Optik. 2022;268 [Google Scholar]
- Kumar L.A., Renuka D.K., Rose S.L., Shunmuga Priya M.C., Wartana I.M. Deep learning based assistive technology on audio visual speech recognition for hearing impaired. International Journal of Cognitive Computing in Engineering. 2022;3:24–30. [Google Scholar]
- Kumar P.B.M., Perumal R.S., Nadesh R.K., Arivuselvan K. Type 2: Diabetes mellitus prediction using Deep Neural Networks classifier. International Journal of Cognitive Computing in Engineering. 2020;1:55–61. [Google Scholar]
- Li Y., Zhao J., Lv Z., Li J. Medical image fusion method by deep learning. International Journal of Cognitive Computing in Engineering. 2021;2:21–29. [Google Scholar]
- Liu J., Sun W., Zhao X., Zhao J., Jiang Z. Deep feature fusion classification network (DFFCNet): Towards accurate diagnosis of COVID-19 using chest X-Rays images. Biomedical Signal Processing and Control. 2022;76 doi: 10.1016/j.bspc.2022.103677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai T., Tang X., Zhang Y., Wang C. COVID-19 diagnosis via chest X-Ray image classification based on multiscale class residual attention. Computers in Biology and Medicine. 2022;149 doi: 10.1016/j.compbiomed.2022.106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghdadi N.A., Malki A., Abdelaliem S.F., Balaha H.M., Badawy M., Elhosseini M. An automated diagnosis and classification of COVID-19 from chest CT images using a transfer learning-based convolutional neural network. Computers in Biology and Medicine. 2022;144 doi: 10.1016/j.compbiomed.2022.105383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H., Cheng X., Ullah F., Jabbar S., Dong S. A deep convolutional neural network stacked ensemble for malware threat classification in internet of things. Journal of Circuits, Systems and Computers. 2022 [Google Scholar]
- Salama W.M., Aly M.H. Framework for COVID-19 segmentation and classification based on deep learning of computed tomography lung images. Journal of Electronic Science and Technology. 2022;20(3) [Google Scholar]
- Tran K.A., Kondrashova O., Bradley A., Williams E.D., Pearson J.V., Waddell N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Medicine. 2021;13(1):1–17. doi: 10.1186/s13073-021-00968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lin Z.Q., Wong A. COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. Scientific Reports. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.H., Fernandes S.L., Zhu Z., Zhang Y.D. AVNC: attention-based VGG-style network for COVID-19 diagnosis by CBAM. IEEE Sensors Journal. 2021;22(18):17431–17438. doi: 10.1109/JSEN.2021.3062442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.H., Satapathy S.C., Xie M.X., Zhang Y.D. ELUCNN for explainable COVID-19 diagnosis. Soft Computing. 2023:1–17. doi: 10.1007/s00500-023-07813-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.H., Zhang Y., Cheng X., Zhang X., Zhang Y.D. PSSPNN: PatchShuffle stochastic pooling neural network for an explainable diagnosis of COVID-19 with multiple-way data augmentation. Computational and Mathematical Methods in Medicine. 2021;2021:1–18. doi: 10.1155/2021/6633755. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang D., Yang G., Li F., Wang J., Sangaiah A.K. Detecting seam carved images using uniform local binary patterns. Multimedia Tools and Applications. 2020;79:8415–8430. [Google Scholar]
- Zhang X., Jiang R., Huang P., Wang T., Hu M., Scarsbrook A.F., Frangi A.F. Dynamic feature learning for COVID-19 segmentation and classification. Computers in biology and medicine. 2022;150 doi: 10.1016/j.compbiomed.2022.106136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Khan, M. A., Zhu, Z., & Wang, S. (2023). SNELM: SqueezeNet-guided ELM for COVID-19 recognition. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset and code of the developed model is available on a reasonable request to the corresponding author.