Abstract
Liquid biopsy is a diagnostic repeatable test, which in last years has emerged as a powerful tool for profiling cancer genomes in real-time with minimal invasiveness and tailoring oncological decision-making. It analyzes different blood-circulating biomarkers and circulating tumor DNA (ctDNA) is the preferred one. Nevertheless, tissue biopsy remains the gold standard for molecular evaluation of solid tumors whereas liquid biopsy is a complementary tool in many different clinical settings, such as treatment selection, monitoring treatment response, cancer clonal evolution, prognostic evaluation, as well as the detection of early disease and minimal residual disease (MRD). A wide number of technologies have been developed with the aim of increasing their sensitivity and specificity with acceptable costs. Moreover, several preclinical and clinical studies have been conducted to better understand liquid biopsy clinical utility. Anyway, several issues are still a limitation of its use such as false positive and negative results, results interpretation, and standardization of the panel tests. Although there has been rapid development of the research in these fields and recent advances in the clinical setting, many clinical trials and studies are still needed to make liquid biopsy an instrument of clinical routine. This review provides an overview of the current and future clinical applications and opening questions of liquid biopsy in different oncological settings, with particular attention to ctDNA liquid biopsy.
Keywords: Liquid biopsy, circulating tumor DNA, precision medicine, clinical trials, minimal residual disease
Introduction
In the last decade, the concept of precision medicine in oncology has emerged as an innovative approach based on the ability to sequence each patient’s tumor. The precision medicine aims to optimize treatments and understand the dynamic evolution of cancer under therapeutic pressure [1–3].
Tissue biopsy is currently the standard method for diagnosis and molecular characterization of the tumors. However, it has many limitations due to its invasiveness and potential complications, such as bleeding, injury, infection, pain, tissue accessibility, and sample adequacy (such as insufficient amount and inadequate quality of the sample) [4].
In this context, the liquid biopsy has rapidly demonstrated its utility of being a non-invasive repeatable diagnostic test, able to profile the molecular asset of every single tumor and to guide the clinical management of cancer patients.
The U.S. National Cancer Institute (NCI) defines liquid biopsy as “a test done on a sample of blood to look for cancer cells from a tumor that are circulating in the blood or for pieces of DNA from tumor cells that are in the blood; a liquid biopsy may be used to help find cancer at an early stage which may also be used to help plan treatment or to find out how well treatment is working or if cancer has come back. Being able to take multiple samples of blood over time may also help doctors understand what kind of molecular changes are taking place in a tumor.” [5].
The wide and rapid diffusion of the liquid biopsy in different oncological clinical settings is due to the development of several different comprehensive genomic profile assays and a wide number of preclinical and clinical studies to better define the clinical utility of liquid biopsy [2, 6].
In the last years, many technologies and assays have been developed, many clinical applications have been investigated and different biomarkers have been discovered for the liquid biopsy test. Among these, the circulating tumor DNA (ctDNA) has emerged as a promising biomarker in the liquid biopsy test for clinical needs.
In this review, the current and future clinical applications of liquid biopsy for different solid tumors will be discussed, exploring the diagnostic value and the limitations of this test. This work aims to stimulate discussion and encourage further studies.
Liquid biopsy
History of liquid biopsy
For a long time, the effort of researchers and clinicians in the oncology field has been addressed to find sensible and specific cancer biomarkers or tests, allowing early cancer detection and better cancer management. The current biomarkers of liquid biopsy have been discovered many decades ago but only recently have been used in the management of cancer patients, maybe due to the limitations of available technologies.
As early as 1869, Ashworth [7] identified for the first time the circulating tumor cells (CTCs) in the metastatic cancer patient’s blood. In 1948, Mandel and Métais [8] recognized cell-free DNA (cfDNA) for the first time, but only in the 1970s, cfDNA was identified in cancer patients’ blood. In the 1960s, extracellular vesicles (EVs) were observed for the first time, but only in 2017, they were recognized as cancer biomarkers [9].
Even if they were discovered many decades ago, only in the last years they have captured the attention of scientific community as useful diagnostic tools, due to the diffusion of commercial and homemade liquid biopsy tests [10]. In fact, the development of sequencing technologies and the improvement of cancer genomic knowledge [11] lead to the diffusion of several comprehensive genomic profiling assays to personalize cancer patients’ management, promoting the diffusion of liquid biopsy.
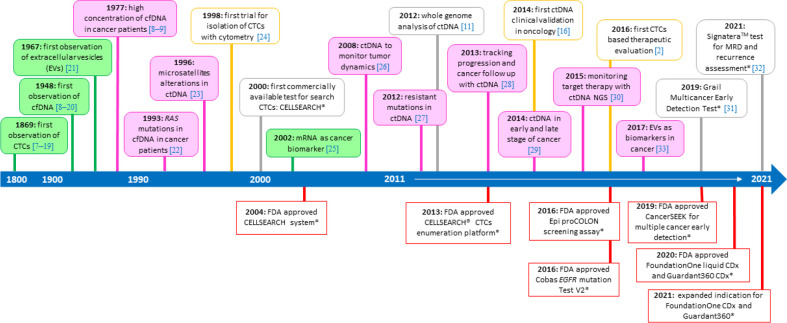
The concept of the liquid biopsy appears in the 1970s on Pubmed platform referred to the analysis of CTCs from the blood of patients affected by different solid tumors, but only in the last decade the number of articles about liquid biopsy has increased progressively, up to the present days when the literature about liquid biopsy is extremely wide (Figure 1) [12–33].
Figure 1.
The timeline of liquid biopsy development. The figure illustrates some of the significant events in the development and spread of the liquid biopsy in clinical practice. * Some of the Food and Drug Administration (FDA) approved tests, addressed in the following text of the manuscript (“ctDNA liquid biopsy assays” and “Clinical applications of ctDNA liquid biopsy”); NGS: next generation sequencing; MRD: minimal residual disease
Biomarkers of liquid biopsy
The term liquid biopsy is usually applied to blood samples mainly for the analysis of ctDNA and in part of CTCs, but many other biomarkers and types of samples are emerging [34].
The tumor microenvironment includes a mix of cellular and non-cellular components, many of which are released from cancer cells, which contribute to cancer survival, invasion, metastatic process, and resistance to treatments. Among these components, CTCs, ctDNA, cfDNA, EVs, or exosomes stood out for their characteristics as useful biomarkers circulating in the peripheral blood of cancer patients.
In blood samples, both in the plasma or serum, and in the cellular fraction, different circulating biomarkers can be detected and characterized [6].
In the plasma or serum, tumor-derived EVs or exosomes [35], proteins, circulating noncoding and messenger RNA [circulating free RNA (cfRNA)] [36], ctDNA [37], and tumor-educated platelets (TEP) [38, 39] can be detected. In the cellular fraction, single CTC and CTC clusters (constituted with CTCs exclusively or with CTCs escorted by immune cells) [40], circulating endothelial cells (CEC) [41], and cancer-associated (CA) fibroblasts can be identified [6]. Leukocytes and T-cell receptor profiling may also be potential biomarkers for metastatic patients treated with immunotherapy [42, 43].
The liquid biopsy biomarkers are present also in different body fluids, expanding the concept of liquid biopsy not only to the peripheral cancer patients’ blood but also to other fluids like prostatic fluid, urine (particularly for renal, bladder, and prostate cancers) [44], cerebrospinal fluid (CSF, particularly for central nervous system metastasis and primary brain cancers) [45], saliva or sputum, pleural effusion (particularly for head and heck and lung cancers) [46], tears, bile, peritoneal lavage, and bone marrow (particularly for breast, prostate, and lung cancers), stool, breast milk, and ascitic fluid [47–49].
CTCs
The CTCs are cancer cells with a median half-life of 1–2.5 h, released in the blood from the primary tumor or metastatic sites at any stage of tumorigenesis [50]. They are present in the circulation as individual cells or in clusters (in this case, they have an increased metastatic ability). The CTCs have a high heterogeneity, according to their genomic, transcriptomic, proteomic, and metabolomic characteristics [51] and their phenotypic aspect depends on cancer type and stage of disease (usually associated with the worst prognosis) [52]. Their interactions with blood and tissue components are crucial for cancer survival and promotion of metastatic disease. The CTC blood concentration is very low and metastatic patients may have 1–10 CTCs per mL of blood [53].
Due to the very low concentration of CTCs, their analysis is difficult, expensive, and technically challenging. Complex technologies are needed to isolate and analyze CTCs and many platforms and assays have been developed for detecting CTCs through recognition of cell size, or expression of cell-surface antigens. In fact, CTC isolation requires different steps such as enrichment, detection, and cell characterization [54–57]. The enrichment step is based on the identification of their physical (size, elasticity, and density) and biological properties to increase CTC concentration. The detection step is based on immune-cytologic, molecular, or functional assays [54, 57, 58]. In the end, the CTC characterization is possible and CTC cultures, CTC lines [59–61] and CTC xenografts can be established [62–64]. There are several commercial assays for the enrichment, detection, and characterization of the CTCs. The CellSearch® system is the most used and it is the only FDA-approved assay for searching CTCs [65]. The EPithelial ImmunoSPOT (EPISPOT) assay is also a clinical validated assay for many different cancers, including breast, prostate, head and neck cancers, and melanoma [66–68]. Other examples of CTC commercial assays for the liquid biopsy are Oncotype diagnosis (DX) androgen receptor splice variant 7 (AR-V7), oncotype sequencing (SEQ), Adnatest®, Epic Sciences, RareCyte, and EPithelial ImmunoSPOT in a DROP (EPIDROP). The CellSearch® system was approved by FDA in 2004 to predict outcomes in metastatic breast cancers. In 2013, FDA expanded the indication of CellSearch® CTC enumeration platform, to monitor patients with colon and prostate cancers [69–71]. In fact, the CTCs have been detected and studied in several different types of cancer, but their role as useful biomarker to predict outcomes, identify MRD, guide therapeutic choice, and monitor cancer progression is more evident in breast, prostate, colon, and lung cancer patients [40, 72–75].
The identification of CTCs allows the preservation of cellular contents and the expression of gene information, consenting functional analysis of CTC cultures, molecular characterization of genome, and protein analysis [76]. For example, the expression of AR-V7 splice variant on the CTCs of metastatic prostate cancer patients strongly predicts the resistance to abiraterone and enzalutamide [77]. Different studies have demonstrated that CTC’s functional and molecular characterization may have a prognostic value in cancer patients and may indicate the tumor’s capacity to become metastatic [53]. They are also associated with shorter progression-free survival (PFS) and overall survival (OS; metastatic breast, colorectal, and prostate cancers), as well as they may be used to monitor cancer evolution and response to therapy [78, 79]. In fact, the CTCs appeared to be generally more expressed in aggressive diseases [80, 81]. In addition to the prognostic value, their molecular characterization can provide additional information about the site of CTC origin (different organ microenvironments can select different types of CTCs), the ability to disseminate, the drug susceptibility or resistance in cell culture or xenograft, and the transcriptional plasticity [80–82].
Furthermore, the whole genome analysis (WGA) of a single CTC has shown great variability, confirming the tumor heterogeneity [83]. Based on these considerations, a blood sample may contain multiple cellular subpopulations and the analysis of a single CTC may not be representative of whole tumor biology [84].
However, CTCs are very difficult to detect due to the very complex and expensive method and the small number of cells in the blood at the early stage of diagnosis, during follow up and in the metastatic disease (often < 1 CTC per mL of blood). For these reasons, CTCs have limited clinical utility [85–89]. Further studies are needed to better understand the role of CTCs in the oncological precision medicine and detection technologies needed to be improved.
Tumor-associated EVs
The tumor-derived EVs or exosomes are small round vesicles, generally 30–120 nm in diameter, delimited by a lipid bilayer that carries out proteins, DNA, RNA, small non-coding RNAs [microRNA (miRNA)], and lipids [53, 90, 91]. They are released by the tumor cells and used for communication between the tumor and other different cells [92, 93]. Each cancer cell type secretes a specific EV, allowing it to determinate the presence of the cancer and the cancer type [94, 95]. They are also found in abundant quantities in biological fluids, increasing the detection sensitivity [39, 93, 94]. The lack of unified adequate isolation methods and standardized analysis is a great limitation of EV implementation in the clinical setting [96]. The most diffused EV isolation technique is ultracentrifugation (UC), which separates EVs based on their size and density, and it requires dedicated equipment, excessive time, and costs [97]. The combination of different procedures, such as ultrafiltration and density gradient centrifugation, can increase the quality of EV isolated but can also reduce the amount of EVs [98–101]. In the last years, many commercial isolation kits have been developed trying to save time but with high costs [102].
Despite these limitations, EVs have been used as a novel biomarker in liquid biopsy for cancer monitoring, staging, and as a prognostic and dynamic tool for cancer therapy management [10, 93–95, 103–107]. However, the advantage of EV application to clinical settings is related to their various cargoes, allowing the identification of different and more specific molecular targets necessary for a personalized treatment [93–95]. The most common EV marker is the RNA and, in particular, the miRNA signature as predictive biomarker of prognosis and survival in different tumors, such as lung [108–110], liver [111, 112], colorectal [113], prostate [114], hepatocellular [115], breast [116], and pancreatic [117] cancers [10] and glioma [118]. For their nature and role, EVs can also be used to target tumor cells and to deliver and transfer drugs such as chemotherapies or immunotherapies in other cancer cells [10, 119–121]. For these reasons, recently, EVs are recognized as a promising potential liquid biopsy resource in oncology, but standardized and convenient methods for isolation are needed [91, 122].
The attention of the scientific community to the role of EVs as biomarkers for liquid biopsy in cancer is relatively recent, so further studies are needed to better understand their clinical utility in the oncological field.
ctDNA
The DNA is released in fragments (cfDNA) through apoptosis, necrosis, and active secretion by normal (particularly lymphoid and myeloid cells, due to the frequent turnover of hematopoietic lineage cells of blood) [123], and cancer (ctDNA) cells [34, 37, 124, 125]. The concentration of ctDNA might only be < 0.01% of total cfDNA [58, 126]. The ctDNA is constituted of small DNA fragments (180–200 bp) released in the bloodstream of cancer patients. It has a short half-life (from 15 min to about 2.5 h) and a concentration range from 0 ng/mL to 1,000 ng/mL in blood (an average of 180 ng/mL in cancer patients) [125, 127, 128]. The fragment size of the ctDNA is variable, according to the different release mechanisms in the blood and the incomplete and random digestion of the ctDNA. The concentration and length of the ctDNA fragment can be a signature for prediction, diagnosis, and prognosis in cancer [129–133]. The ctDNA concentrations are higher in patients with metastatic cancers than those with localized cancers [134]. It is cleared through nucleosomes in the liver (liver macrophages), by circulating nucleases and in the kidney [124, 125, 129, 133, 134]. However, ctDNA shedding depends on several factors, like the tissue cell turnover (proliferation and apoptosis rate), the burden of disease, and the tumor site [124, 125, 129, 133, 134].
For its nature, the ctDNA provides information about point mutations, copy number variations (CNVs), structural rearrangements, loss of heterozygosity, gene fusions, methylation changes, integrated viral sequences associated with the tumor, and other genomic signatures [135–139].
The variant allele frequency (VAF) describes the proportion of ctDNA molecules containing a mutation over the total number of molecules containing the same allele [140–142].
The ctDNA recapitulates with accuracy the tumor characteristics and appears to be a diagnostic tool for many different solid tumors, for example, colorectal [143, 144], endometrial [145, 146], ovarian [78], breast [147], non-small cell lung cancer (NSCLC) [148–150], oropharyngeal [151], pancreatic [152], prostate [153, 154] cancers, and melanoma [155].
Currently, the ctDNA is the preferred biomarker for liquid biopsy analyses of druggable mutations. In fact, it can overcome costs, limitations and the difficulties in identification, extraction, and characterization of CTCs and EVs. Recently, a great number of technologies and commercial assays have been rapidly developed to detect ctDNA, and numerous studies have spread to investigate the potential role of this biomarker.
ctDNA liquid biopsy assays
In the contest of ctDNA liquid biopsy, many different technologies and panel tests have been developed to analyze the molecular alterations for different purposes: polymerase chain reaction (PCR)-based sequencing can be used for single-locus/multiplexed assays and targeted sequencing for point mutations analysis, NGS-based sequencing can be used for single-locus/multiplexed assays, targeted sequencing, genome-wide analysis for point mutations, detection of rearrangements, chromosomal copy-number changes, and WGA [156–158].
The PCR methods use specific DNA probes to target specific known genes and they provide a quantitative measurement of the number of targets in the sample. They are highly sensitive and can detect a low tumor fraction DNA in plasma [159]. The NGS methods also use probes to capture specific DNA fragments and to target comprehensive known and unknown genes, but the data provide a ratio measurement and the sequences of the captured DNA. Moreover, a higher tumor DNA fraction than PCR is needed [142, 159].
Many authors have already examined different PCR and NGS techniques, so this review will make only a quick overview.
The real-time quantitative PCR (qPCR) is a fast and inexpensive tool [160] and it can detect mutant allele fraction (MAF) > 10% [161]. It is efficient when it analyzes a small number of variants, but it can assess only specified variant types, thus offering little discovery value [162].
The digital PCR (dPCR) is like qPCR in principle, but it is superior to qPCR in accuracy, although there are additional costs. It divides the sample into thousands of parallel PCR reactions, so the background noise is reduced. It is fast, inexpensive, and can detect MAF < 0.1% [163]. Multiplexed patient-specific panels can be used [164].
The droplet dPCR (ddPCR) is a type of dPCR. It genotypes a small number of known genes (also extremely infrequent mutations), interrogating multiple genes and at least 5–10 mutations simultaneously with a single ddPCR reaction. It is a sensitive, cost-effective, and fast technique. The ddPCR identifies and quantifies target DNA and provides results as absolute copies per mL. The ddPCR also offers little discovery value, because the number and types of targets are limited by assay design [162].
The dPCR has other different variants as beads, emulsion, amplification, and magnetics (BEAMing) [165]. It is extremely sensitive and has a detection rate of 0.02%, but it is technically complicated and relatively expensive for routine use [166].
The PCR-based assays are sensitive and convenient, and they are the most diffused assays in clinical practice, but they can only search for a limited number of known molecular alterations [157].
The NGS technology is based on massive parallel ultra-deep sequencing of millions of different DNA fragments in parallel, followed by computational analysis of reads, ensuring a high degree of sensitivity [16, 167]. The NGS is characterized by different steps: generation of a fragment DNA library, single fragment clonal amplification, massively parallel sequencing, and data analysis [168]. It has high throughput and it can search for many known and unknown multiple molecular alterations in multiple genes, involving single-nucleotide resolution of DNA sequences. Furthermore, it can discover new molecular alterations without prior knowledge at the end of the test, offering a higher discovery value [157]. Although it is more expensive than PCR, the comprehensive genomic profile nature of NGS can provide clinical knowledge (for its ability to cover more molecular targets) and cost when it is used for many patients at the same time [162]. The NGS detects MAF < 1% and many technical strategies are developed to increase sensitivity and reduce false negatives [169, 170].
Many commercial NGS tests are used to assess somatic alterations (including mutations, fusions, and copy number alterations), targeting simultaneously different genes. In fact, to improve sensitivity and detection power, the NGS methods have been applied to target panels, such as tagged-amplicon deep sequencing (TAm-Seq), safe-Seq system (Safe-SeqS), and cancer personalized profiling by deep-Seq (CAPP-Seq). The TAm-Seq identifies MAF of about 2% with a sensitivity > 97% [171]. The enhanced TAmSeq (eTAmSeq) can detect MAF of < 0.25% and it can identify single-nucleotide variants (SNVs), short insertions/deletions (indels), and CNVs [172]. The Safe-SeqS can reduce the sequencing errors about 70-fold and has a sensitivity of > 98% for detecting tumor mutations [173], while the CAPP-Seq detects MAF of < 0.02% and has a sensitivity of nearly 100% [174].
The whole-genome-Seq (WGS) is a non-targeted NGS technology, it searches for the whole genomic profile of the tumor DNA [159, 160, 175], providing a great amount of information. However, it is expensive and less sensitive and it requires a long time for the analysis. The whole-exome-Seq (WES), sequencing only the exons, is less expensive than WGS [176]. These two methodologies require high concentration of ctDNA. However, the WGA may be not convenient for cost, sensitivity, ctDNA amount, and necessary technologies. For these reasons, they have not yet found an application in clinical practice [157, 175, 176].
The PCR assays appear to be a good option on a large scale. They are the most diffuse technologies for high sensitivity and low cost. They are able to identify very low MAF of ctDNA, but for their nature of detecting only known point mutations, insertions and deletions, the knowledge about the tumor DNA is limited.
In the last years the NGS assays have had a huge spread, thanks to the increased sensitivity, diffusion of commercial companion diagnostic and agnostic panels (which can detect also low MAF of ctDNA), and the accessible costs [37, 142, 159]. The NGS is also applied to the untargeted panels, which do not need prior knowledge of molecular alterations, allowing to find genome-wide DNA variation [177, 178].
The possibility to search for a large number of actionable biomarkers and different known and unknown molecular alterations with a single test is essential for the development of precision medicine. The identification of gene alterations can be searched in both preclinical and clinical settings, allowing the development of new targeted therapies. The absence of standardization of several methods makes it difficult to compare different assays, therefore, it is not possible to define which is the better test to use in clinical practice. In fact, each test is useful for different cancer types and stage of disease and different purposes. In particular, the ctDNA concentration in plasma correlates with tumor size [133] and stage [179], therefore, patients with early cancer stages can have < 10 copies per 5 mL of tumor mutations versus patients with late cancer stages, in which the copies can be increased from 10 times to 100 times [180]. For this reason, in early cancer stages, the ctDNA assays need to be usually highly sensitive, while in late stages the sensitivity and the costs of ctDNA assays can be moderate. Therefore, the PCR assays may be useful for cancers with well-known genetic profiles, for molecular markers routinely searched and for diagnosis or monitoring the therapeutic response. The NGS panels may be useful for poorly defined or unknown origin cancers with an unexpected molecular profile with the aim to identify potential rare targeted alterations.
In the last years, many commercial tests have been developed, some examples are Archer® Reveal ctDNA 28 (28 genes); OncoDNA OncoSTRAT&GO (27 genes); Guardant360® companion diagnostic (CDx) [73 genes, microsatellite instability (MSI)]; Memorial Sloan Kettering-Analysis of Circulating cfDNA to Examine Somatic Status (MSK-ACCESS) (129 genes); FoundationOne® Liquid CDx Assay [> 300 genes, blood TMB (bTMB), MSI, and tumor fraction]; and multiple other assays (Inivata InVision, AVENIO ctDNA panels, Oncomine Pan-Cancer cell-free Assay, Tempus xF Liquid Biopsy Assay, SignateraTM, UW-OncoPlex CT, elio Plasma Resolve, Therascreen®) [181]. In this context, the FDA approved some of them as companion diagnostic tests [182]. Examples of FDA-approved commercial tests for clinical application of ctDNA liquid biopsy are in Table 1 [182].
Table 1.
Examples of FDA-approved commercial tests for clinical application of ctDNA liquid biopsy
| Test | Manufacturer | Cancer type indications | Biomarker (single cancer indication, pan-cancer/multi-cancer indications) | Technology |
|---|---|---|---|---|
| Therascreen®, PIK3CA RGQ PCR Kit | Qiagen Manchester, Ltd. |
Companion diagnostic Breast cancer: piqray (alpelisib) |
PIK3CA | PCR |
| Cobas EGFR mutation Test v2 | Roche Molecular Systems, Inc. |
Companion diagnostic NSCLC: tarceva (erlotinib), tagrisso (osimertinib), iressa (gefitinib) |
EGFR | PCR |
| Epi ProColon® | Epigenomics AG | Ancillary screening: colorectal cancer | SEPT9 methylation | Bisulfite converted DNA and PCR |
| Guardant360® CDx | Guardant Health, Inc. |
Companion diagnostic NSCLC: tagrisso (osimertinib), rybrevant (amivantamab-vmjw), lumakras (sotorasib) |
EGFR | NGS |
| FoundationOne® Liquid CDx | Guardant Health, Inc. | Tumor mutation profiling: any solid tumor | 73 genes | |
| FoundationOne® Liquid CDx | Foundation Medicine, Inc. |
Companion diagnostic NSCLC: iressa (gefitinib), tagrisso (osimertinib), tarceva (erlotinib), alecensa (alectinib), tabrecta (capmatinib) Metastatic castrate resistant prostate cancer (mCRPC): rubraca (rucaparib), lynparza (olaparib) Ovarian cancer: rubraca (rucaparib) Breast cancer: piqray (alpelisib) |
EGFR, ALK BRCA1/2, ATM BRCA1/2, PIK3CA | NGS |
| Tumor mutation profiling: any solid tumor | 324 genes | |||
| BRAC Analysis CDx | Myriad Genetic Laboratories, Inc. |
Companion diagnostic Breast cancer: lynparza (olaparib), talzenna (talazoparib) Ovarian cancer: lynparza (olaparib), rubraca (rucaparib) Pancreatic cancer: lynparza (olaparib) mCRPC: lynparza (olaparib) |
BRCA | PCR |
The table illustrates the characteristics of some FDA-approved commercial tests as companion diagnostic tests and as ancillary screening tests for the ctDNA liquid biopsy. PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; RGQ: rotor-gene Q; EGFR: epidermal growth factor receptor; SEPT9: septin 9; ALK: anaplastic lymphoma kinase; BRCA: breast cancer susceptibility genes; ATM: ataxia- telangiectasia mutated
Strengths and weaknesses of ctDNA liquid biopsy
The molecular analysis of solid cancers alterations on formalin-fixed paraffin-embedded (FFPE) tissue has technical and sample availability challenges, such as longtime of processing, blindness of tumor heterogeneity, and limited amount or poor quality of samples, although sensitivity and specificity are excellent [183, 184].
The liquid biopsy assay is a minimally invasive option, with better sample quality and quick execution time, repeatable several times during the oncological history of patients, capturing spatial and temporal heterogeneity and clonal evolution of the cancer. Moreover, it has a high concordance rate with the tissue NGS [4, 183, 185]. A unique characteristic of the liquid biopsy is the possibility to perform the blood draw necessary for the analysis at the patient’s home [186]. It usually is technically faster than the tissue biopsy analysis and it can help to detect and genetically profile an occult malignancy in patients without tissue cancer available [187]. In addition, the ctDNA is more sensitive than the serum protein biomarkers [carcinoembryonic antigen (CEA), for example] [72].
However, the liquid biopsy increases costs, particularly when it is used concurrently with the tissue tests (it is often necessary when the analysis is negative). It is not able to analyze non-DNA biomarkers and it does not give histopathological and phenotypical information [such as programmed death-ligand 1 (PDL1) status] [4, 184]. Anyway, the liquid biopsy has also several limitations. First, the tumor signatures and actionable molecular alterations are not as frequent as it is expected. Furthermore, the ctDNA analysis requires high sensitivity and specificity to avoid false negative and false positive results. For these purposes, many technologies have rapidly developed, from commercial to homemade panels with different features, costs, and genomic information, underlying the need for standardized assays [7, 188–190].
Interestingly, the cancer and patient characteristics and the natural history of the tumor are important for the effectiveness of the liquid biopsy in the therapeutic choices. To reach these crucial needs, recently some expert recommendations for clinical use of ctDNA testing have been published, such as the European Society of Medical Oncology (ESMO) Scale for Actionability of Molecular Targets (ESCAT) and the OncoKB [191–193].
The false-positive and false-negative results are actually the major challenges that need to be addressed.
The false negative results are associated with a low percentage of mutated fragments below the limit of detection of the assay used. For example, a low tumor burden of disease, low or no ctDNA shedding due to the tumor site, vascularization or histology of the cancer lesions (such as the isolated brain metastasis), and rare allelic frequencies and variants limit the assay’s ability to detect mutations. The false negative results can also be given by a low sensitivity or by the absence of specific molecular alterations in the panel used (particularly in the small panels) [124, 125, 129, 133, 134, 159, 162, 172, 173, 194].
The false-positive results are associated with the sequencing errors, heterogeneity of tumor (for example, the difficulty to identify which clone dominates in which site), and background noises, like cfDNA shedding from cellular sources other than the tumor (for example, in elderly patient or patients with sepsis or inflammatory diseases) [195]. The molecular alterations from the cfDNA may originate from ctDNA, germline alterations, or non-tumor somatic alterations from white blood cells, such as clonal hematopoiesis of indeterminate potential (CHIP), that could not be identified in tumor biopsy [196–198]. In fact, the NGS liquid biopsy may be able to detect sequential mutational events in high turnover compartments, such as the bone marrow, which may not always reflect tumor genotype. These findings arise doubts regarding mutations detected in the cfDNA, especially in specific genes [for example, DNA methyltransferase 3 alpha (DNMT3A), additional sex combs like 1 (ASXL1), and ten-eleven translocation methylcytosine dioxygenase 2 (TET2), as well as tumour suppressor protein p53 (TP53), janus kinase 2 (JAK2), splicing factor 3B subunit 1 (SF3B1), guanine nucleotide-binding protein subunit beta-1 (GNB1), protein phosphatase magnesium-dependent 1 delta (PPM1D), guanine nucleotide-binding protein alpha stimulating (GNAS), and B-cell lymphoma (BCL) 6 corepressor-like 1 (BCORL1)] [196–198]. Determining the source of the cfDNA (tumor versus healthy tissue) remains a limitation of the cfDNA analysis. Finally, it must be considered that the identification of tissue from which cancer has raised, particularly for occult malignancy, is not always possible [187, 194].
The rapid growth of knowledge about genomic profile of different cancer types, the diffusion of many commercial panels and target therapies, and the big variability of information available have led to the rise of the multidisciplinary molecular tumor boards (MTBs) for overcoming the issues. The MTBs are groups of different specialists with expertise in different fields, with the purpose of interpreting the results obtained by the analysis of the ctDNA with liquid biopsy or the tissue NGS to support clinical application of these data [199]. Anyhow, further studies are needed to better understand how to integrate and interpret the liquid biopsy information in clinical practice.
Clinical applications of ctDNA liquid biopsy
Although the tissue biopsy is actually the gold standard technique for cancer diagnosis and biomarkers evaluation, the use of the liquid biopsy has spread predominantly and rapidly in the management of patients with solid cancers at different stages of disease, integrating and improving the standard clinical care at different moments of patients’ cancer history. The advantage of profiling cancer genotypes without invasive procedures, particularly when the tumor tissue is insufficient or not available, makes the liquid biopsy an important tool in therapeutic decisions [200, 201]. The liquid biopsy is a minimally invasive and repeatable assay that can be used during longitudinal monitoring, making this test feasible for different aims, including screening. The ctDNA is the most investigated and used biomarker of the liquid biopsy in clinical practice applications [202–204]. It offers a snapshot of the genomic cancer profile, giving a real, instantaneous, and complete picture of the inter and intra-tumor heterogeneity [43, 202–205].
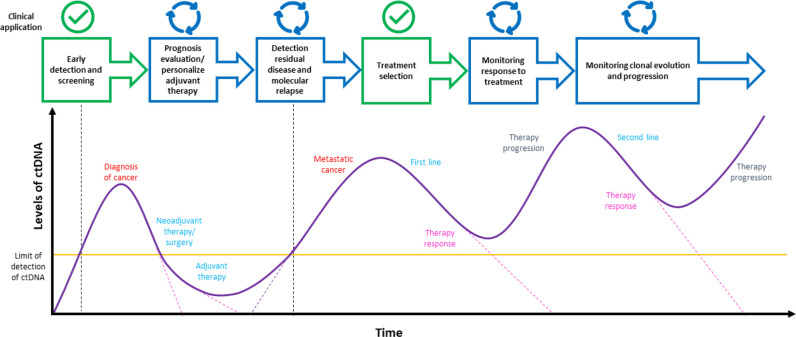
There are many clinical applications of ctDNA liquid biopsy, some of which are widely applied and approved in clinical practice, others are under investigation and for all of them many works and clinical trials have been conducted and others are still in progress [202–204, 206–210]. Some clinical applications of the liquid biopsy are depicted in Figure 2, like the identification of cancer biomarkers and the detection of therapeutic targets for treatment selection [211], the real-time monitoring of therapeutic efficacy and response [212], the disease progression assessment and the identification of resistance mechanisms [213, 214], the early detection of cancer and the MRD assessment [58, 215], and the cancer screening [90, 216, 217].
Figure 2.
A schematic view of clinical applications of ctDNA liquid biopsy
Treatment selection and resistant mechanisms
In the metastatic setting, the therapy decision-making process needs the genomic tumor analysis at the diagnosis and at the restaging of the disease [6]. In the targeted therapy era, the detection of druggable molecular alterations and the respective tailored treatment options can improve patient outcomes [218].
Despite the tissue biopsy is a gold standard technique for cancer diagnosis and molecular characterization, the liquid biopsy can be a complementary and even an alternative tool, considering a good concordance between the two methods and the high specificity and the moderate sensitivity of the latest one [174, 219–225]. For these reasons, the companion diagnostic tests have spread in the routinely clinical practice, particularly when the tissue is unavailable or insufficient (to search clonal alterations) and to detect acquired resistance mechanisms to therapy (to search clonal and subclonal alterations) [199, 226].
The NCI defines the companion diagnostic test as “a test used to help match a patient to a specific drug or therapy. For example, a companion diagnostic test may identify whether a patient’s tumor has a specific gene change or biomarker that is targeted by the drug. This helps determine if the patient should receive the drug or not. Companion diagnostic tests can also be used to find out whether serious side effects may occur from treatment or how well the treatment is working. Most drugs with a companion diagnostic test are cancer drugs that target specific tumor mutations.” [227]. Many guidelines have been developed to help clinicians in the use of the liquid biopsy, mainly for the interpretation of its results, such as ESMO recommendations and OncoKB recommendations [including FDA labeling, National Comprehensive Cancer Network (NCCN) guidelines, conference proceedings, disease-focused expert group recommendations, and the scientific literature] [181, 191, 193, 228].
The advanced NSCLC adenocarcinoma is the best example for the liquid biopsy application in molecular cancer profiling at the time of diagnosis [such as EGFR, ALK, c-ros oncogene 1 (ROS1), v-raf murine sarcoma viral oncogene homolog B1 (BRAF), MET, rearranged during transfection (RET) analysis)], when the tissue is insufficient for the molecular testing (about 50% of the cases) [181, 229] and a new tissue biopsy cannot be performed, or at the time of progressive disease after a targeted therapy with tyrosine kinase inhibitors (TKIs) [200, 201, 230]. For both purposes, the liquid biopsy has been integrated into the recommended guidelines and many companion diagnostic tests are available (such as Guardant360®, FoundationOne® Liquid CDx, and Cobas). It has been shown that ctDNA assays can detect guideline-recommended biomarkers with a high concordance with the tissue analysis, and the power of detection increases when tissue and liquid biopsy are used complementary [163, 231]. The ctDNA can identify the acquired molecular resistances developed after treatment with EGFR TKIs (first or second generation), such as EGFR T790M mutation, and can guide the subsequent therapeutic choice (osimertinib) [232]. When osimertinib is prescribed in the first line therapy, EGFR C797S, BRAF V600E mutation, MET amplification or amplification or human epidermal growth factor receptor 2 (HER2) amplification can be detected in liquid biopsy as acquired resistance alterations [233–237]. In the metastatic EGFR-mutant patients, when the EGFR-positive ctDNA was detected after six weeks of osimertinib and bevacizumab treatment, it identified patients with high risk for early progression and lower OS [238]. The ctDNA analysis can also identify ALK fusion and related acquired resistance alterations, guiding the target therapies with TKIs [239], such as for ROS1 [240] and MET molecular alterations [241]. Also, for ALK TKI therapies, the liquid biopsy may be a surrogate of outcomes, predicting the therapy response, according to the clearance of the blood ctDNA [242]. The emergence of HER2 and Kirsten rat sarcoma (KRAS)G12C alterations powered the use of the liquid biopsy in the choice of the targeted therapy [241–245].
Anyway, if the liquid biopsy is negative (EGFR T790M, exon 19 deletion, and/or exon 21 L858R mutations and/or ALK fusion are not detected in ctDNA analysis, for example), a tissue testing is recommended [229, 246].
Furthermore, the NGS panels can also detect a plethora of unexpected molecular alterations of known or unknown meaning, that could give information about cancer evolution. Unfortunately, the NSCLC can switch under target therapeutic pressure to SCLC. In this scenario, the liquid biopsy is useless and the tissue biopsy is necessary [247].
In the HER2-negative and hormone receptor-positive (HER2–/HR+) metastatic breast cancer, the detection of PIK3CA mutations with the liquid biopsy can guide therapeutic choice with cyclin-dependent kinases 4/6 (CDK4/6) inhibitor alpelisib in combination with fulvestrant after endocrine therapy progression. Even in this case, when the ctDNA PIK3CA is negative, the tissue analysis is always necessary [248, 249]. The FDA approved as companion diagnostic test the Therascreen®, PIK3CA RGQ PCR Kit for the detection of the PIK3CA mutations with the liquid biopsy in breast cancer patients with this setting, based on the results of SOLAR-1 trial [249]. Furthermore, in HER2 mutant non-amplified metastatic breast cancer patients treated with neratinib, the liquid biopsy in NGS demonstrated 100% specificity [250]. In the clinical trial plasma-based molecular profiling of advanced breast cancer to inform therapeutic choices (plasmaMATCH), the ctDNA was used for randomized patients in 5 cohorts based on mutation profiles [251]. In estrogen receptor-positive (ER+) metastatic breast cancer patients, the identification of estrogen receptor 1 (ESR1) and phosphatase and tensin homolog (PTEN) alterations by liquid biopsy is associated with the intrinsic resistance to aromatase inhibitors and alpelisib treatment [252].
For the metastatic colorectal cancer (mCRC) the NCCN guidelines do not directly address ctDNA tests and ESCAT guidelines consider the NGS only an alternative option to the PCR if it does not result in additional costs [228]. Despite this, the ctDNA was used for studying resistance mechanisms of cancer for the first time and showed high concordance with the tissue analysis [179, 253–256]. In the mCRC, the ctDNA liquid biopsy has shown its clinical utility in guiding first-line therapeutic choice by identifying rat sarcoma (RAS) mutations, which are negative biomarkers of response to anti-EGFR monoclonal antibodies therapy. The KRAS and neuroblastoma RAS viral oncogene homolog (NRAS) are also often acquired resistance alterations to target therapy and their identification is important for the re-challenge option with the anti-EGFR therapy in advanced lines [185, 255–259]. The BRAF V600E mutation detected by the ctDNA plays an important role in the choice of combined BRAF and EGFR inhibitors targeted therapy. This genomic profiling detection is important for the evaluation of acquired resistances to target therapy, too [260–262]. The mCRC is the best example to show the clinical utility of the ctDNA to identify and assess spatial and temporal heterogeneity in cancer [263]. In contrast to the tissue analysis, the ctDNA liquid biopsy gives an instantaneous collective snapshot of the cancer heterogeneity from all sites of cancer and at different time points in the history of a cancer patient [264–266]. An example of the cancer spatial heterogeneity of different metastatic sites is shown by the study of Parikh et al. [205] about mCRC patients, in which the post-progression ctDNA liquid biopsy captured all the acquired resistance alterations, that were later found separately in the distinct metastasis lesions by tissue biopsies. Siravegna et al. [267] showed how the ctDNA liquid biopsy can identify the different genomic evolution of each metastasis in mCRC, treated with HER2 blockade therapy [trastuzumab and lapatinib on the HER2 Amplification for Colo-rectaL cancer Enhanced Stratification (HERACLES) study]. They also showed how the ctDNA may help to understand the mixed radiological responses, highlighting the heterogeneity of anatomical response to the targeted therapy [261, 267]. An example of the cancer temporal heterogeneity [268, 269] is given by Parseghian et al. [270] study in which in the mCRC RAS wild type patients, after discontinuation of anti-EGFR antibodies, the ctDNA liquid biopsy predicts the decline of KRAS mutant-clones and provides evidence for re-challenge therapy with the anti-EGFR antibodies. Anyway, further studies are needed to validate the results of liquid biopsy as a tool to identify actionable molecular alterations to detect and manage the cancer’s spatial and temporal heterogeneity.
In 2020 the FDA approved the FondationOne liquid CDx as a companion diagnostic test for the use of rucaparib in BRCA-mutated mCRPC patients, demonstrating the feasibility and utility of the ctDNA in clinical practice also for this cancer type [182, 271].
In advanced melanoma, the liquid biopsy can improve the identification of BRAF and receptor tyrosine kinase (KIT) mutations for targeted therapy choice [272]. The BRAF V600 mutations were detected with the liquid biopsy in patients treated with BRAF and mitogen-activated protein kinase kinase (MEK) inhibitors before the radiological progression, with the lead time reduction of 110 days [273].
The PDL1 expression as a biomarker to select patients for immunotherapy remains limited for few cancer types and its determination requires tissue biopsy. The FDA approved pembrolizumab for genome signatures microsatellite instability-high (MSI-H) and high tumor mutational burden (TMB, ≥ 10 mut/Mb) in different tumors [274–276]. Some ctDNA assays can explore TMB and MSI status, overcoming the sampling bias and allowing pembrolizumab therapy with liquid biopsy analysis [206, 277, 278]. Particularly, the study that used ctDNA Guardant360® assay to identify the MSI-H status showed clinical benefit from anti-PD(L)1 antibody therapy [277]. Wang et al. [279] have demonstrated that it is necessary a high-quality DNA to avoid underestimation of TMB for a correct clinical evaluation. Chauhan et al. [280] have detected and calculated the TMB from the ctDNA in urine rather than in blood [281]. Although many different guidelines (such as NCCN and ESMO guidelines) do not address plasma-based testing TMB or other tumor agnostic biomarkers like neurotrophic tyrosine receptor kinase (NTRK), the ctDNA analysis can help to choose the best treatment in selected cases [194].
Cancer resistance evolves dynamically under targeted therapy selective pressure and the ctDNA liquid biopsy can be a useful tool to detect these mechanisms of resistance [82].
Treatment response monitoring and prognostic value
The usefulness of the ctDNA has been explored in many studies in different settings, including prognosis and treatment response monitoring [282–286]. The response monitoring using the ctDNA was born to track actionable oncogenic alterations during the targeted therapy. The feasibility of the ctDNA for this purpose became evident in different tumor types and in several settings [287, 288]. The monitoring of treatment response is also important to avoid ineffective therapies and unnecessary side effects. Moreover, many studies have analyzed the ctDNA value as a prognostic marker for clinical outcomes and as a tool for monitoring treatment response [289–291].
In the routine practice, the imaging is used to monitor treatment response sometimes in association with the tumor markers, unfortunately, their use is limited in sensitivity and specificity [58]. The ctDNA could be used as a personalized tumor-specific marker for patient surveillance and treatment efficacy monitoring [170, 270, 292]. Although the ctDNA has many issues, such as limited tumor shedding and burden of disease, the dynamic liquid biopsy can allow to monitor not only the tumor response but also the tumor evolution. Furthermore, in many scientific articles, the ctDNA decreasing or increasing clearance is associated with cancer response or progression to therapy respectively, with a shorter leading time than imaging [170, 270, 292, 293].
For example, in advanced EGFR-mutated NSCLC, early changes in the ctDNA value under targeted treatment with osimertinib providing efficacy information, particularly early clearance or decreasing levels of the ctDNA EGFR mutation (also known as molecular response) were associated with subsequent imaging response and with an improvement in response rates (RRs), PFS, and OS [294–296]. Conversely, higher ctDNA levels were associated with poor prognosis [297]. Several groups have demonstrated that early decreasing or clearance of the ctDNA in advanced NSCLC treated with immunotherapy or immunochemotherapy was associated with longer OS and a high RR, conversely increasing ctDNA, was correlated with the disease progression [170, 206, 298–300]. The ctDNA dynamics may also help to identify immunotherapy pseudoprogression, detecting a decrease of ctDNA levels versus increased tumor radiological volume, and conversely detecting an increase of ctDNA in true tumor progression [301]. Despite the diffusion of many works with similar results, further studies are needed to better investigate the role of the ctDNA as a biomarker for therapy response in different solid tumors.
In the metastatic breast cancer, the early ctDNA detection can predict response to palbociclib and fulvestrant therapy [302]. It can also predict therapy response and prognosis in the early breast cancer patients receiving neoadjuvant chemotherapy. In fact, the patients with positive ctDNA before neoadjuvant chemotherapy had significantly shorter disease-free survival (DFS) and OS than those with negative ctDNA [303].
In the advanced pancreatic cancer, a decrease of KRAS mutant ctDNA identified an earlier tumor response to the therapy, and it is strongly associated with clinical outcomes than blood levels of tumor markers [304, 305].
In a study by Lim et al. [306] conducted in RAS wild-type mCRC patients treated with chemotherapy and anti-EGFR antibody, the changes of the ctDNA VAF had linear correlation with tumor size and clinical outcome [307]. Moreover, in the KRAS mutant mCRC patients, the increasing ctDNA KRAS mutation allows to detect progression of disease earlier than imaging [308].
The dynamic ctDNA can correlate with outcomes and with radiological responses also in melanoma patients. Already in 2015, Gray et al. [309] evaluated the ctDNA to monitor treatment response in metastatic melanoma patients treated with targeted therapy or with immunotherapy. The patients who had persistently elevated ctDNA on both therapies had a poor prognosis; conversely, the decreasing or clearance of ctDNA at the first imaging follows up correlated with a better prognosis.
Anyway, the ctDNA has yet several limitations that must be overcome and many interventional clinical trials are needed to investigate its real clinical value, so the ctDNA is not actually used for monitoring treatment response or for prognostication in clinical practice and this remains an active challenge [310].
MRD and risk assessment
The National Institutes of Health (NIH) defined the MRD as “a very small number of cancer cells that remain in the body during or after treatment. MRD can be found only by highly sensitive laboratory methods that are able to find one cancer cell among one million normal cells. Checking to see if there is MRD may help to plan treatment, find out how well treatment is working or if cancer has come back, or make a prognosis. MRD testing is used mostly for blood cancers such as lymphoma and leukemia, also called MRD.” [311].
Therefore, the MRD represents the detection of residual cancer cells (for example, ctDNA) without evidence of cancer disease at the conventional tests (for example, imaging, protein cancer biomarkers) during routine surveillance, after complete resection, neoadjuvant and/or adjuvant therapies [312]. If the cancer cells are still present after treatment, they may induce a relapse of disease. The MRD can help to classify patients with high or low risk of relapse and to predict short- or long-term relapse [313, 314]. The utility of the liquid biopsy to identify MRD is evident, considering the absence of other tools that can detect microscopic disease and its capacity to identify residual disease with a lead time of several months earlier than imaging. Moreover, the liquid biopsy can guide treatment adjustment and the surveillance during or after a curative treatment (for example, surgery and adjuvant chemotherapy) [315].
Although many different studies have investigated the clinical utility of the ctDNA to search the MRD, many limitations have been highlighted. For example, the MRD cannot be assessed by standard clinical and imaging technologies, the CHIP may be a confounding factor [316, 317], the timing of the ctDNA sampling between surgery and starting adjuvant therapy is often a crucial challenging point because it is difficult to choose the best moment (not too soon to detect the ctDNA set free from surgery, but not too late to start therapy) [318]. It is also very difficult to detect the ctDNA at a very low concentration with standard ctDNA non-personalized and uninformed assays, due to low sensitivity and lack of standardization of methodologies and technologies. Recently, many personalized informed technologies are developed to detect the MRD accurately, sequencing single tumor genotypes and designing a PCR or a NGS assay tailored to each patient. Tumor-informed ctDNA analysis is more sensitive than tumor naive analysis, but it is expensive and less feasible for clinical practice [310, 319, 320]. Examples of commercial panels to detect the MRD are SignateraTM [Natera/Foundation Medicine (FMI)], Archer®Dx/Invitae-personalized cancer monitoring (PCM) assay, Guardant Reveal (LUNAR 1), Roche Avenio ctDNA surveillance kit, Inivata RaDaR/InVision MRD kit. Anyway, in a few years studies and clinical trials are increasingly trying to overcome these limitations and to select the best clinical and treatment approach in this new field [320–324].
Researchers have investigated the role of the ctDNA in the MRD not only for prognosis, but mainly for evaluated escalation or de-escalation treatments [325]. In patients with the MRD detected by the liquid biopsy after a curative treatment, an intensification or escalation treatment may be an optional strategy to improve DFS and OS. Conversely, in patients with the absence of the MRD after a curative treatment, a de-intensification or de-escalation therapy (such as omitting or depowering the adjuvant therapy schedule) can reduce side effects or toxicities [314, 326, 327].
In the NSCLC, adjuvant treatment has a modest survival benefit, thus the MRD can help to select the patients that really benefit from therapy [328, 329]. The TRACERx study has shown that the ctDNA MRD-positive predicted relapse before conventional imaging, with a median time of 164 days versus 362 days of imaging, with a significant lead time [330].
The DYNAMIC study evaluated the ctDNA dynamics after radical surgery, highlighting that the ctDNA MRD detected 3 days after surgery (R0) may be used as a baseline for lung cancer monitoring [331, 332]. The ctDNA can be used also in the unresectable locoregionally advanced NSCLC, to guide the escalation or de- escalation treatments. Moding et al. [333] have found that the detection of the ctDNA after chemoradiotherapy (CRT) or early on consolidation with the durvalumab was a strong predictor of risks of progression of disease. In contrast, the patients with the ctDNA not detected after CRT had better outcomes with or without consolidation immunotherapy, identifying patients in which therapy can be de-escalated [333]. A great number of studies are ongoing to better understanding where patients can benefit from adjuvant therapy and are cured by surgery alone, investigating the role of the ctDNA MRD in the management of the early-stage NSCLC.
The role of the ctDNA liquid biopsy has been evaluated for further different tumors. Garcia-Murillas et al. [334, 335] showed that, in the early breast cancer patients, the detection of the ctDNA after a curative therapy was associated with the metastatic recurrence, with a median lead time of 7.9 months over imaging and clinical recurrence.
Another study showed that in the ctDNA-positive muscle-invasive urothelial cancer patients, the adjuvant immunotherapy with atezolizumab can improve survival [336].
In the early CRC, a great number of studies have shown the utility of the ctDNA to detect the MRD and its role as a prognostic factor [337–341]. Reinert et al. [342] have shown that stage I–III CRC patients, with ctDNA- positive after a definitive therapy, had an increased risk of recurrence (about 40 times) than ctDNA negative patients during the follow-up. Notably, the lead time of the disease recurrence was 16.5 months (with respect to the imaging). Tie et al. [343] showed that, in locally advanced rectal cancer, the post-surgery ctDNA detection was strongly predictive of recurrence [344]. Tie et al. [345] also showed that in stage III CRC, the post-surgery ctDNA positive patients have a poor outcome, despite adjuvant therapy, with a 3-year-recurrence-free interval of 47% versus 76% in the ctDNA negative patients. Loupakis et al. [346] have evaluated the mCRC patients who had undergone metastatic resection with curative intent (PREDATOR clinical trial). The postsurgical MRD status was a strong prognostic biomarker associated with DFS. In fact, the MRD-positive patients had an inferior OS than the MRD-negative ones, also, the MRD-negative patients who did not receive systemic therapy had an OS of 100%.
The ctDNA has also been evaluated as a prognostic biomarker for the risk of the assessment in many different cancer types, as described above.
In another study, in the I–III stage NSCLC patients, the pre-surgery ctDNA detection identified patients at a higher risk of recurrence, which may reflect the micrometastatic disease [347].
Moreover, in the metastatic triple-negative breast cancer, the tumor fraction ≥ 10% was associated with a significantly worse survival [348].
In the metastatic BRAF V600 mutant melanoma, the BRAF ctDNA negative-patients at baseline therapy had a high RR to target therapy, while the BRAF-positive patients had a shorter PFS and OS [349].
Several studies have investigated the role of the ctDNA as prognostic biomarker, and many studies identified poor prognosis associated with high ctDNA levels.
Even if the ctDNA MRD may become a real-time biomarker for relapse of disease, many clinical questions are still open [210, 310, 350].
A great number of studies are ongoing in this field trying to delineate clinical utility of the liquid biopsy in the MRD.
Early detection and cancer screening
The use of the ctDNA liquid biopsy in cancer early detection and screening is further investigated [351–355]. The liquid biopsy is a minimally invasive test, which can be repeated multiple times, and the blood draw is well accepted by people. However, in this setting further limitations arise due to the type of the population studied.
First, in the early detection and screening setting, a positive liquid biopsy identifies an “occult” cancer, with unknown origin, not detected by clinical and imaging standard. The detection of the CHIP mutations can give false positive results, limiting the specificity of the test in a setting in which high specificity is required high specificity. Furthermore, in the early cancer stage, the tumor burden is very low and consequently, the ctDNA levels are very low, giving false negative results due to the limited sensitivity of assays [330]. Additionally, some sensitive and specific tests are very expensive [356]. So, the liquid biopsy can over-diagnose or under-diagnose, or can just give an unclear clinical result, limiting its utility [310].
Multiple assays have been developed to implement the liquid biopsy as a good cancer screening test, such as the new ultrasensitive assays or using methylation markers.
Cohen et al. [357] have demonstrated the value of the liquid biopsy CancerSEEK platform for early detection of 8 different cancers, and its specificity was > 99% and sensitivity was 69–98% (all patients knew to have cancer).
The combination of different diagnostic approaches was investigated by Lennon et al. [216] in the Detecting Cancers Earlier Through Elective Mutation-Based Blood Collection and Testing (DETECT_A) study, using a multicancer test coupled with positron emission tomography/computed tomography (PET/CT) imaging to detect early cancer (no patient knew to have cancer), confirming the potential utility of this approach.
The ctDNA levels in the early-stage patients are lower than those in metastatic patients [330], and the sensitivity of a test is also given by both the blood volume sampled and by the number of mutations analyzed [358], so many commercial panels for early cancer screening probes multiple mutations [359].
Despite many limitations, the liquid biopsy can be a helpful tool for the cancer screening when other methods are not available, or patients refuse or are not fit for them.
The FDA has approved some liquid biopsy tests for the cancer screening, such as the Epi proColon® for CRC, and have designed other liquid biopsy tests breakthrough device, such as the Grail’s Galleri, Natera’s SignateraTM, and CancerSEEK multicancer test, to support the current standard screening tests. The Epi proColon® (Epigenomics) is a blood-based screening test, available for patients with an average risk of CRC, who have not completed the conventional guideline-recommended screening (unwilling or unable). It detects the tumor-associated epigenetic changes [360–362]. The CancerSEEK is a blood-based screening test; it identifies 8 different early-stage cancer types (breast, colon, esophagus, lung, liver, ovary, pancreas, and stomach), complementing other screening standard tools. It detects 8 biomarkers and mutations on 16 genes [363].
Furthermore, the liquid biopsy on urine may be useful for screening, and tests such as the UroSEEK or UroVysion have developed [364]. The UroSEEK detects mutations in 11 genes and aneuploidy for the identification of bladder and upper urothelial cancers. It has a sensitivity of 83% [365].
The early diagnosis remains a challenge, with sensitivity, specificity, and costs as critical factors. The liquid biopsy in this setting needs to overcome technical and clinical challenges. Further interventional and prospective, large-scale clinical trials are needed to demonstrate the clinical utility and the positive predictive value for cancer screening.
Conclusions
In the era of precision medicine, it is well established that the cancer genomic profile is crucial to guide clinical decision-making. For this reason, the liquid biopsy assay has emerged as an innovative tool to guide the personalized approach for cancer patients, especially when the tissue samples are insufficient or limited in quality and quantity [366, 367]. The ctDNA analysis provides a real-time, non-invasive, and repeatable snapshot of the cancer molecular profile, offering several advantages in the diagnostic, prognostic field, and treatment selection. The wide diffusion of the ctDNA liquid biopsy is also due to the rapid development of commercial cost-effectiveness panel tests, available in the routinely clinical practice. Some evidence has demonstrated the attractive role of the ctDNA liquid biopsy in different clinical settings, such as early cancer detection, detection of MRD and early relapse, treatment selection, serial treatment response monitoring, prediction of outcome, risk assessment, and detection of resistance mechanisms. Actually, the ctDNA liquid biopsy is approved only for the treatment selection (companion diagnostic tests) and as an ancillary screening tool, many additional trials are needed to validate the clinical utility of this test in other settings. The future applications will be the therapy response assessment, the resistance monitoring, and MRD detection.
There is still to understand how to interpret the liquid biopsy results, as well as how to overcome the liquid biopsy clinical and technical limitations and the lack of technology standardization. The diffusion of new different fluids and biomarkers (EVs, miRNA), other than ctDNA and CTCs, and the development of patient personalized technologies for liquid biopsy could expand its applications. In fact, using them as complementary tools, each biomarker’s limitations could be partially overcome, moving from a genomic, transcriptomic, or proteomic evaluation to a multi-omic analysis of the cancer for better management of the patients. Furthermore, innovative clinical trials are ongoing, and many others will be designed to investigate the real clinical utility of the liquid biopsy and to integrate complementary information for the best clinical decision-making at any crucial moment of the cancer disease. Anyway, it is undoubted that the liquid biopsy is transforming the precision medicine in oncology as a promising revolutionary weapon for real-life oncology in the near future.
Abbreviations
- ALK:
anaplastic lymphoma kinase
- BRCA:
breast cancer susceptibility genes
- CDx:
companion diagnostic
- cfDNA:
cell-free DNA
- CHIP:
clonal hematopoiesis of indeterminate potential
- CTCs:
circulating tumor cells
- ctDNA:
circulating tumor DNA
- ddPCR:
droplet digital polymerase chain reaction
- DFS:
disease-free survival
- dPCR:
digital polymerase chain reaction
- EGFR:
epidermal growth factor receptor
- ESMO:
European Society of Medical Oncology
- EVs:
extracellular vesicles
- FDA:
Food and Drug Administration
- HER2:
human epidermal growth factor receptor 2
- KRAS:
Kirsten rat sarcoma
- MAF:
mutant allele fraction
- mCRPC:
metastatic castrate-resistant prostate cancer
- miRNA:
microRNA
- MRD:
minimal residual disease
- MSI:
microsatellite instability
- NCCN:
National Comprehensive Cancer Network
- NGS:
next generation sequencing
- NSCLC:
non-small cell lung cancer
- OS:
overall survival
- PCR:
polymerase chain reaction
- PFS:
progression-free survival
- PIK3CA:
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- RAS:
rat sarcoma
- RRs:
response rates
- TAM-Seq:
tagged-amplicon deep sequencing
- TKIs:
tyrosine kinase inhibitors
- TMB:
tumor mutational burden
- WGA:
whole genome analysis
Declarations
Author contributions
CMDC, GM: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing—review & editing. VC, FC, TT, SN: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Software, Project administration, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
Not applicable.
Copyright
© The Author(s) 2023.
References
- 1.Watanabe K, Nakamura Y, Low SK. Clinical implementation and current advancement of blood liquid biopsy in cancer. J Hum Genet. 2021;66:909–26. 10.1038/s10038-021-00939-5 [DOI] [PubMed] [Google Scholar]
- 2.Borad MJ, LoRusso PM. Twenty-first century precision medicine in oncology: genomic profiling in patients with cancer. Mayo Clin Proc. 2017;92:1583–91. 10.1016/j.mayocp.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 3.Savonarola A, Palmirotta R, Guadagni F, Silvestris F. Pharmacogenetics and pharmacogenomics: role of mutational analysis in anti-cancer targeted therapy. Pharmacogenomics J. 2012;12:277–86. 10.1038/tpj.2012.28 [DOI] [PubMed] [Google Scholar]
- 4.Rodríguez J, Avila J, Rolfo C, Ruíz-Patiño A, Russo A, Ricaurte L, et al. When tissue is an issue the liquid biopsy is nonissue: a review. Oncol Ther. 2021;9:89–110. 10.1007/s40487-021-00144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liquid biopsy [Internet]. Bethesda: National Cancer Institute; [cited 2022 Mar 16]. Available from: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/liquid-biopsy [Google Scholar]
- 6.Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021;11:858–73. 10.1158/2159-8290.CD-20-1311 [DOI] [PubMed] [Google Scholar]
- 7.Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med J Aust. 1869;14:146–9. [Google Scholar]
- 8.Mandel P, Métais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil. 1948;142:241–3. French. [PubMed] [Google Scholar]
- 9.Wu K, Xing F, Wu SY, Watabe K. Extracellular vesicles as emerging targets in cancer: recent development from bench to bedside. Biochim Biophys Acta Rev Cancer. 2017;1868:538–63. 10.1016/j.bbcan.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abouali H, Hosseini SA, Purcell E, Nagrath S, Poudineh M. Recent advances in device engineering and computational analysis for characterization of cell-released cancer biomarkers. Cancers (Basel). 2022;14:288. 10.3390/cancers14020288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The cancer genome atlas [Internet]. Bethesda: National Human Genome Research Institute; [cited 2022 Apr 10]. Available from: https://www.genome.gov/Funded-Programs-Projects/Cancer-Genome-Atlas [Google Scholar]
- 12.Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398–406. 10.1016/j.molmed.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 13.Chimonidou M, Strati A, Tzitzira A, Sotiropoulou G, Malamos N, Georgoulias V, et al. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem. 2011;57:1169–77. 10.1373/clinchem.2011.165902 [DOI] [PubMed] [Google Scholar]
- 14.Lianidou ES, Markou A. Circulating tumor cells in breast cancer: detection systems, molecular characterization, and future challenges. Clin Chem. 2011;57:1242–55. 10.1373/clinchem.2011.165068 [DOI] [PubMed] [Google Scholar]
- 15.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110–8. 10.1373/clinchem.2012.194258 [DOI] [PubMed] [Google Scholar]
- 16.Couraud S Vaca-Paniagua F Villar S Oliver J Schuster T Blanché Het al.; BioCAST/IFCT-1002 investigators . Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res. 2014;20:4613–24. 10.1158/1078-0432.CCR-13-3063 [DOI] [PubMed] [Google Scholar]
- 17.Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5:3591. 10.1038/ncomms4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu S, Ichikawa D, Takeshita H, Morimura R, Hirajima S, Tsujiura M, et al. Circulating miR-18a: a sensitive cancer screening biomarker in human cancer. In Vivo. 2014;28:293–7. [PubMed] [Google Scholar]
- 19.Zhou H, Zhu L, Song J, Wang G, Li P, Li W, et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol Cancer. 2022;21:86. 10.1186/s12943-022-01556-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–50. [PubMed] [Google Scholar]
- 21.Prince AM, Adams WR. Virus-like particles in human plasma and serum: role of platelet lysosomes. J Natl Cancer Inst. 1966;37:153–66. 10.1093/jnci/37.2.153 [DOI] [PubMed] [Google Scholar]
- 22.Tada M, Omata M, Kawai S, Saisho H, Ohto M, Saiki RK, et al. Detection of RAS gene mutations in pancreatic juice and peripheral blood of patients with pancreatic adenocarcinoma. Cancer Res. 1993;53:2472–4. [PubMed] [Google Scholar]
- 23.Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–7. 10.1038/nm0996-1035 [DOI] [PubMed] [Google Scholar]
- 24.Pachmann K, Heiss P, Demel U, Tilz G. Detection and quantification of small numbers of circulating tumour cells in peripheral blood using laser scanning cytometer (LSC®). Clin Chem Lab Med. 2001;39:811–7. 10.1515/CCLM.2001.134 [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18:1177–85. 10.1158/1078-0432.CCR-11-0564 [DOI] [PubMed] [Google Scholar]
- 27.Isobe K, Hata Y, Kobayashi K, Hirota N, Sato K, Sano G, et al. Clinical significance of circulating tumor cells and free DNA in non-small cell lung cancer. Anticancer Res. 2012;32:3339–44. [PubMed] [Google Scholar]
- 28.Figg WD 2nd, Reid J. Monitor tumor burden with circulating tumor DNA. Cancer Biol Ther. 2013;14:697–8. 10.4161/cbt.25361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bidard FC, Madic J, Mariani P, Piperno-Neumann S, Rampanou A, Servois V, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int J Cancer. 2014;134:1207–13. 10.1002/ijc.28436 [DOI] [PubMed] [Google Scholar]
- 30.Olsson E, Winter C, George A, Chen Y, Howlin J, Tang MH, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015;7:1034–47. 10.15252/emmm.201404913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.New technologies offer the possibility of identifying cancer from a single blood draw [Internet]. Alexandria: American Society of Clinical Oncology and Association for Clinical Oncology; [cited 2019 Oct 10]. Available from: https://www.asco.org/about-asco/press-center/news-releases/new-technologies-offer-possibility-identifying-cancer-single [Google Scholar]
- 32.FDA grants two new breakthrough device designations for Natera’s SignateraTM MRD test [Internet]. Austin: Natera; [cited 2021 Mar 24]. Available from: https://www.natera.com/company/news/fda-grants-two-new-breakthrough-device-designations-for-nateras-signatera-mrd-test-2/ [Google Scholar]
- 33.Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–88. 10.1016/j.tcb.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Y, Li W, Wang K, Xu C, Hao M, Ding L. Perspectives of the application of liquid biopsy in colorectal cancer. Biomed Res Int. 2020;2020:6843180. 10.1155/2020/6843180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs — an update. Nat Rev Clin Oncol. 2018;15:541–63. 10.1038/s41571-018-0035-x [DOI] [PubMed] [Google Scholar]
- 37.Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Best MG, Sol N, In’ t Veld SGJG, Vancura A, Muller M, Niemeijer AN, et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell. 2017;32:238–52.E9. 10.1016/j.ccell.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joosse SA, Pantel K. Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell. 2015;28:552–4. 10.1016/j.ccell.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 40.Lianidou ES, Strati A, Markou A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit Rev Clin Lab Sci. 2014;51:160–71. 10.3109/10408363.2014.896316 [DOI] [PubMed] [Google Scholar]
- 41.Danova M, Comolli G, Manzoni M, Torchio M, Mazzini G. Flow cytometric analysis of circulating endothelial cells and endothelial progenitors for clinical purposes in oncology: a critical evaluation (review). Mol Clin Oncol. 2016;4:909–17. 10.3892/mco.2016.823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofman P, Heeke S, Alix-Panabières C, Pantel K. Liquid biopsy in the era of immuno-oncology: is it ready for prime-time use for cancer patients? Ann Oncol. 2019;30:1448–59. 10.1093/annonc/mdz196 [DOI] [PubMed] [Google Scholar]
- 43.Russano M, Napolitano A, Ribelli G, Iuliani M, Simonetti S, Citarella F, et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: the potentiality of blood samples. J Exp Clin Cancer Res. 2020;39:95. Erratum in: J Exp Clin Cancer Res. 2020;39:120. 10.1186/s13046-020-01601-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batista R, Vinagre N, Meireles S, Vinagre J, Prazeres H, Leão R, et al. Biomarkers for bladder cancer diagnosis and surveillance: a comprehensive review. Diagnostics (Basel). 2020;10:39. 10.3390/diagnostics10010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. 10.1038/ncomms9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JS, Hur JY, Kim IA, Kim HJ, Choi CM, Lee JC, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer. 2018;18:1236. 10.1186/s12885-018-5138-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson DJ, Imperiale TF. Stool testing for colorectal cancer screening. Gastroenterology. 2015;149:1286–93. 10.1053/j.gastro.2015.05.045 [DOI] [PubMed] [Google Scholar]
- 48.Halvaei S, Daryani S, Eslami-S Z, Samadi T, Jafarbeik-Iravani N, Bakhshayesh TO, et al. Exosomes in cancer liquid biopsy: a focus on breast cancer. Mol Ther Nucleic Acids. 2018;10:131–41. 10.1016/j.omtn.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villatoro S, Mayo-de-Las-Casas C, Jordana-Ariza N, Viteri-Ramírez S, Garzón-Ibañez M, Moya-Horno I, et al. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol Oncol. 2019;13:2633–45. 10.1002/1878-0261.12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–62. 10.1158/1078-0432.CCR-04-1110 [DOI] [PubMed] [Google Scholar]
- 51.Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827–40. 10.1101/gad.305805.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parkinson DR, Dracopoli N, Petty BG, Compton C, Cristofanilli M, Deisseroth A, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med. 2012;10:138. 10.1186/1479-5876-10-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domínguez-Vigil IG, Moreno-Martínez AK, Wang JY, Roehrl MHA, Barrera-Saldaña HA. The dawn of the liquid biopsy in the fight against cancer. Oncotarget. 2017;9:2912–22. 10.18632/oncotarget.23131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–31. 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 55.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–91. 10.1158/2159-8290.CD-15-1483 [DOI] [PubMed] [Google Scholar]
- 56.Li N, Zuo H, Chen L, Liu H, Zhou J, Yao Y, et al. Circulating tumor cell detection in epithelial ovarian cancer using dual-component antibodies targeting EpCAM and FRα. Cancer Manag Res. 2019;11:10939–48. 10.2147/CMAR.S211455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu X, Zang X, Lv Y. Detection of circulating tumor cells: advances and critical concerns (review). Oncol Lett. 2021;21:422. 10.3892/ol.2021.12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease — latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16:409–24. 10.1038/s41571-019-0187-3 [DOI] [PubMed] [Google Scholar]
- 59.Cayrefourcq L, Mazard T, Joosse S, Solassol J, Ramos J, Assenat E, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015;75:892–901. 10.1158/0008-5472.CAN-14-2613 [DOI] [PubMed] [Google Scholar]
- 60.Koch C, Kuske A, Joosse SA, Yigit G, Sflomos G, Thaler S, et al. Characterization of circulating breast cancer cells with tumorigenic and metastatic capacity. EMBO Mol Med. 2020;12:e11908. 10.15252/emmm.201911908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–20. 10.1126/science.1253533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–44. 10.1038/nbt.2576 [DOI] [PubMed] [Google Scholar]
- 63.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903. 10.1038/nm.3600 [DOI] [PubMed] [Google Scholar]
- 64.Faugeroux V, Pailler E, Oulhen M, Deas O, Brulle-Soumare L, Hervieu C, et al. Genetic characterization of a unique neuroendocrine transdifferentiation prostate circulating tumor cell-derived eXplant model. Nat Commun. 2020;11:1884. 10.1038/s41467-020-15426-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cristofanilli M, Broglio KR, Guarneri V, Jackson S, Fritsche HA, Islam R, et al. Circulating tumor cells in metastatic breast cancer: biologic staging beyond tumor burden. Clin Breast Cancer. 2007;7:471–9. 10.3816/CBC.2007.n.004 [DOI] [PubMed] [Google Scholar]
- 66.Ramirez JM, Fehm T, Orsini M, Cayrefourcq L, Maudelonde T, Pantel K, et al. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin Chem. 2014;60:214–21. 10.1373/clinchem.2013.215079 [DOI] [PubMed] [Google Scholar]
- 67.Garrel R, Mazel M, Perriard F, Vinches M, Cayrefourcq L, Guigay J, et al. Circulating tumor cells as a prognostic factor in recurrent or metastatic head and neck squamous cell carcinoma: the CIRCUTEC prospective study. Clin Chem. 2019;65:1267–75. 10.1373/clinchem.2019.305904 [DOI] [PubMed] [Google Scholar]
- 68.Cayrefourcq L, De Roeck A, Garcia C, Stoebner PE, Fichel F, Garima F, et al. S100-EPISPOT: a new tool to detect viable circulating melanoma cells. Cells. 2019;8:755. 10.3390/cells8070755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 70.Negin BP, Cohen SJ. Circulating tumor cells in colorectal cancer: past, present, and future challenges. Curr Treat Options Oncol. 2010;11:1–13. 10.1007/s11864-010-0115-3 [DOI] [PubMed] [Google Scholar]
- 71.Resel Folkersma L, Olivier Gómez C, San José Manso L, Veganzones de Castro S, Galante Romo I, Vidaurreta Lázaro M, et al. Immunomagnetic quantification of circulating tumoral cells in patients with prostate cancer: clinical and pathological correlation. Arch Esp Urol. 2010;63:23–31. 10.4321/S0004-06142010000100004 [DOI] [PubMed] [Google Scholar]
- 72.Morgan TM. Liquid biopsy: where did it come from, what is it, and where is it going? Investig Clin Urol. 2019;60:139–41. 10.4111/icu.2019.60.3.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leroy S Benzaquen J Mazzetta A Marchand-Adam S Padovani B Israel-Biet Det al.; AIR Project Study Group . Circulating tumour cells as a potential screening tool for lung cancer (the AIR study): protocol of a prospective multicentre cohort study in France. BMJ Open. 2017;7:e018884. 10.1136/bmjopen-2017-018884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sumanasuriya S, Omlin A, Armstrong A, Attard G, Chi KN, Bevan CL, et al. Consensus statement on circulating biomarkers for advanced prostate cancer. Eur Urol Oncol. 2018;1:151–9. 10.1016/j.euo.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 75.Obermayr E, Bednarz-Knoll N, Orsetti B, Weier HU, Lambrechts S, Castillo-Tong DC, et al. Circulating tumor cells: potential markers of minimal residual disease in ovarian cancer? A study of the OVCAD consortium. Oncotarget. 2017;8:106415–28. 10.18632/oncotarget.22468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene. 2016;35:1216–24. 10.1038/onc.2015.192 [DOI] [PubMed] [Google Scholar]
- 77.Maillet D, Allioli N, Peron J, Plesa A, Decaussin-Petrucci M, Tartas S, et al. Improved androgen receptor splice variant 7 detection using a highly sensitive assay to predict resistance to abiraterone or enzalutamide in metastatic prostate cancer patients. Eur Urol Oncol. 2021;4:609–17. 10.1016/j.euo.2019.08.010 [DOI] [PubMed] [Google Scholar]
- 78.Asante DB, Calapre L, Ziman M, Meniawy TM, Gray ES. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: ready for prime time? Cancer Lett. 2020;468:59–71. 10.1016/j.canlet.2019.10.014 [DOI] [PubMed] [Google Scholar]
- 79.Banys-Paluchowski M, Fehm T, Neubauer H, Paluchowski P, Krawczyk N, Meier-Stiegen F, et al. Clinical relevance of circulating tumor cells in ovarian, fallopian tube and peritoneal cancer. Arch Gynecol Obstet. 2020;301:1027–35. 10.1007/s00404-020-05477-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vanharanta S, Massagué J. Origins of metastatic traits. Cancer Cell. 2013;24:410–21. 10.1016/j.ccr.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17:302–17. 10.1038/nrc.2017.6 [DOI] [PubMed] [Google Scholar]
- 82.Senft D, Leiserson MDM, Ruppin E, Ronai ZA. Precision oncology: the road ahead. Trends Mol Med. 2017;23:874–98. 10.1016/j.molmed.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019;19:553–67. 10.1038/s41568-019-0180-2 [DOI] [PubMed] [Google Scholar]
- 84.Barriere G, Fici P, Gallerani G, Fabbri F, Zoli W, Rigaud M. Circulating tumor cells and epithelial, mesenchymal and stemness markers: characterization of cell subpopulations. Ann Transl Med. 2014;2:109. 10.3978/j.issn.2305-5839.2014.10.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alvarez Cubero MJ, Lorente JA, Robles-Fernandez I, Rodriguez-Martinez A, Puche JL, Serrano MJ. Circulating tumor cells: markers and methodologies for enrichment and detection. Methods Mol Biol. 2017;1634:283–303. 10.1007/978-1-4939-7144-2_24 [DOI] [PubMed] [Google Scholar]
- 86.Pantel K, Alix-Panabières C. Cell lines from circulating tumor cells. Oncoscience. 2015;2:815–6. 10.18632/oncoscience.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kolostova K, Spicka J, Matkowski R, Bobek V. Isolation, primary culture, morphological and molecular characterization of circulating tumor cells in gynecological cancers. Am J Transl Res. 2015;7:1203–13. [PMC free article] [PubMed] [Google Scholar]
- 88.Kolostova K, Zhang Y, Hoffman RM, Bobek V. In vitro culture and characterization of human lung cancer circulating tumor cells isolated by size exclusion from an orthotopic nude-mouse model expressing fluorescent protein. J Fluoresc. 2014;24:1531–6. 10.1007/s10895-014-1439-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maheswaran S, Haber DA. Ex vivo culture of CTCs: an emerging resource to guide cancer therapy. Cancer Res. 2015;75:2411–5. 10.1158/0008-5472.CAN-15-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res. 2015;4:280–90. 10.3978/j.issn.2218-676X.2015.06.05 [DOI] [Google Scholar]
- 91.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 93.Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal. 2015;2015:657086. 10.1155/2015/657086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–8. 10.1016/j.bbagen.2012.03.017 [DOI] [PubMed] [Google Scholar]
- 95.Tamura T, Yoshioka Y, Sakamoto S, Ichikawa T, Ochiya T. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracell Vesicles Circ Nucleic Acids. 2021;2:148–74. 10.20517/evcna.2021.06 [DOI] [Google Scholar]
- 96.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. 10.7150/thno.18133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. 2020;5:144. 10.1038/s41392-020-00258-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nordin JZ, Lee Y, Vader P, Mäger I, Johansson HJ, Heusermann W, et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine. 2015;11:879–83. 10.1016/j.nano.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 99.Gupta S, Rawat S, Arora V, Kottarath SK, Dinda AK, Vaishnav PK, et al. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res Ther. 2018;9:180. 10.1186/s13287-018-0923-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3:24858. 10.3402/jev.v3.24858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120:1632–48. 10.1161/CIRCRESAHA.117.309417 [DOI] [PubMed] [Google Scholar]
- 102.Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One. 2017;12:e0170628. 10.1371/journal.pone.0170628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu J, Chen Y, Pei F, Zeng C, Yao Y, Liao W, et al. Extracellular vesicles in liquid biopsies: potential for disease diagnosis. Biomed Res Int. 2021;2021:6611244. 10.1155/2021/6611244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Soekmadji C, Corcoran NM, Oleinikova I, Jovanovic L, Australian Prostate Cancer Collaboration BioResource. Ramm GA, Nelson CC, Jenster G, Russell PJ. Extracellular vesicles for personalized therapy decision support in advanced metastatic cancers and its potential impact for prostate cancer. Prostate. 2017;77:1416–23. 10.1002/pros.23403 [DOI] [PubMed] [Google Scholar]
- 105.Zhou E, Li Y, Wu F, Guo M, Xu J, Wang S, et al. Circulating extracellular vesicles are effective biomarkers for predicting response to cancer therapy. EBioMedicine. 2021;67:103365. 10.1016/j.ebiom.2021.103365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466–77. 10.1016/j.annonc.2021.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao Z, Fan J, Hsu YS, Lyon CJ, Ning B, Hu TY. Extracellular vesicles as cancer liquid biopsies: from discovery, validation, to clinical application. Lab Chip. 2019;19:1114–40. 10.1039/C8LC01123K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017;8:13048–58. 10.18632/oncotarget.14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sandfeld-Paulsen B, Aggerholm-Pedersen N, Bæk R, Jakobsen KR, Meldgaard P, Folkersen BH, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol. 2016;10:1595–602. 10.1016/j.molonc.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sandfeld-Paulsen B, Jakobsen KR, Bæk R, Folkersen BH, Rasmussen TR, Meldgaard P, et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. 2016;11:1701–10. 10.1016/j.jtho.2016.05.034 [DOI] [PubMed] [Google Scholar]
- 111.Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:135–42. 10.1002/jcb.27165 [DOI] [PubMed] [Google Scholar]
- 112.Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444–52. 10.1002/ijc.31931 [DOI] [PubMed] [Google Scholar]
- 113.Peng ZY, Gu RH, Yan B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J Cell Biochem. 2018;120:1457–63. 10.1002/jcb.27291 [DOI] [PubMed] [Google Scholar]
- 114.Foj L, Ferrer F, Serra M, Arévalo A, Gavagnach M, Giménez N, et al. Exosomal and non-exosomal urinary miRNAs in prostate cancer detection and prognosis. Prostate. 2017;77:573–83. 10.1002/pros.23295 [DOI] [PubMed] [Google Scholar]
- 115.Hou Y, Yu Z, Tam NL, Huang S, Sun C, Wang R, et al. Exosome-related lncRNAs as predictors of HCC patient survival: a prognostic model. Am J Transl Res. 2018;10:1648–62. [PMC free article] [PubMed] [Google Scholar]
- 116.Wang X, Zhong W, Bu J, Li Y, Li R, Nie R, et al. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol Carcinog. 2019;58:674–85. 10.1002/mc.22960 [DOI] [PubMed] [Google Scholar]
- 117.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–82. Erratum in: Nature. 2022;610:E15–7. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lan F, Yue X, Xia T. Exosomal microRNA-210 is a potentially non-invasive biomarker for the diagnosis and prognosis of glioma. Oncol Lett. 2020;19:1967–74. 10.3892/ol.2020.11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nam GH, Choi Y, Kim GB, Kim S, Kim SA, Kim IS. Emerging prospects of exosomes for cancer treatment: from conventional therapy to immunotherapy. Adv Mater. 2020;32:e2002440. 10.1002/adma.202002440 [DOI] [PubMed] [Google Scholar]
- 120.Bellavia D, Raimondo S, Calabrese G, Forte S, Cristaldi M, Patinella A, et al. Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo chronic myelogenous leukemia cell growth. Theranostics. 2017;7:1333–45. 10.7150/thno.17092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Srivastava A, Amreddy N, Babu A, Panneerselvam J, Mehta M, Muralidharan R, et al. Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Sci Rep. 2016;6:38541. 10.1038/srep38541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Geeurickx E, Lippens L, Rappu P, De Geest BG, De Wever O, Hendrix A. Recombinant extracellular vesicles as biological reference material for method development, data normalization and assessment of (pre-)analytical variables. Nat Protoc. 2021;16:603–33. 10.1038/s41596-020-00446-5 [DOI] [PubMed] [Google Scholar]
- 123.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000;906:161–8. 10.1111/j.1749-6632.2000.tb06608.x [DOI] [PubMed] [Google Scholar]
- 125.Sterzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 126.García-Silva S, Gallardo M, Peinado H. DNA-loaded extracellular vesicles in liquid biopsy: tiny players with big potential? Front Cell Dev Biol. 2021;8:622579. 10.3389/fcell.2020.622579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Siravegna G, Bardelli A. Genotyping cell-free tumor DNA in the blood to detect residual disease and drug resistance. Genome Biol. 2014;15:449. 10.1186/s13059-014-0449-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van der Pol Y, Mouliere F. Toward the early detection of cancer by decoding the epigenetic and environmental fingerprints of cell-free DNA. Cancer Cell. 2019;36:350–68. 10.1016/j.ccell.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 129.Jiang P, Lo YMD. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 2016;32:360–71. 10.1016/j.tig.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 130.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. 10.1097/00006676-199807000-00012 [DOI] [PubMed] [Google Scholar]
- 131.Wang BG, Huang HY, Chen YC, Bristow RE, Kassauei K, Cheng CC, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–8. [PubMed] [Google Scholar]
- 132.Krishnamurthy N, Spencer E, Torkamani A, Nicholson L. Liquid biopsies for cancer: coming to a patient near you. J Clin Med. 2017;6:3. 10.3390/jcm6010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38:6159–75. 10.1093/nar/gkq421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011;6:e23418. 10.1371/journal.pone.0023418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7:48832–41. 10.18632/oncotarget.9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li J, Dittmar RL, Xia S, Zhang H, Du M, Huang CC, et al. Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol Oncol. 2017;11:1099–111. 10.1002/1878-0261.12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Soave A, Chun FK, Hillebrand T, Rink M, Weisbach L, Steinbach B, et al. Copy number variations of circulating, cell-free DNA in urothelial carcinoma of the bladder patients treated with radical cystectomy: a prospective study. Oncotarget. 2017;8:56398–407. 10.18632/oncotarget.17657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Husain H, Nykin D, Bui N, Quan D, Gomez G, Woodward B, et al. Cell-free DNA from ascites and pleural effusions: molecular insights into genomic aberrations and disease biology. Mol Cancer Ther. 2017;16:948–55. 10.1158/1535-7163.MCT-16-0436 [DOI] [PubMed] [Google Scholar]
- 139.Li Z, Guo X, Tang L, Peng L, Chen M, Luo X, et al. Methylation analysis of plasma cell-free DNA for breast cancer early detection using bisulfite next-generation sequencing. Tumour Biol. 2016;37:13111–9. 10.1007/s13277-016-5190-z [DOI] [PubMed] [Google Scholar]
- 140.Stewart CM, Kothari PD, Mouliere F, Mair R, Somnay S, Benayed R, et al. The value of cell-free DNA for molecular pathology. J Pathol. 2018;244:616–27. 10.1002/path.5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bos MK, Nasserinejad K, Jansen MPHM, Angus L, Atmodimedjo PN, de Jonge E, et al. Comparison of variant allele frequency and number of mutant molecules as units of measurement for circulating tumor DNA. Mol Oncol. 2021;15:57–66. Erratum in: Mol Oncol. 2021;15:2818–9. 10.1002/1878-0261.12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lin C, Liu X, Zheng B, Ke R, Tzeng CM. Liquid biopsy, ctDNA diagnosis through NGS. Life (Basel). 2021;11:890. 10.3390/life11090890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hamfjord J, Guren TK, Dajani O, Johansen JS, Glimelius B, Sorbye H, et al. Total circulating cell-free DNA as a prognostic biomarker in metastatic colorectal cancer before first-line oxaliplatin-based chemotherapy. Ann Oncol. 2019;30:1088–95. 10.1093/annonc/mdz139 [DOI] [PubMed] [Google Scholar]
- 144.Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y, et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer. 2014;111:1482–9. 10.1038/bjc.2014.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cicchillitti L, Corrado G, De Angeli M, Mancini E, Baiocco E, Patrizi L, et al. Circulating cell-free DNA content as blood based biomarker in endometrial cancer. Oncotarget. 2017;8:115230–43. 10.18632/oncotarget.23247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vizza E, Corrado G, De Angeli M, Carosi M, Mancini E, Baiocco E, et al. Serum DNA integrity index as a potential molecular biomarker in endometrial cancer. J Exp Clin Cancer Res. 2018;37:16. Erratum in: J Exp Clin Cancer Res. 2018;37:35. 10.1186/s13046-018-0688-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fernandez-Garcia D, Hills A, Page K, Hastings RK, Toghill B, Goddard KS, et al. Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Res. 2019;21:149. 10.1186/s13058-019-1235-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Soliman SE, Alhanafy AM, Habib MSE, Hagag M, Ibrahem RAL. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. Biochem Biophys Rep. 2018;15:45–51. 10.1016/j.bbrep.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leng S, Zheng J, Jin Y, Zhang H, Zhu Y, Wu J, et al. Plasma cell-free DNA level and its integrity as biomarkers to distinguish non-small cell lung cancer from tuberculosis. Clin Chim Acta. 2018;477:160–5. 10.1016/j.cca.2017.11.003 [DOI] [PubMed] [Google Scholar]
- 150.Ai B, Liu H, Huang Y, Peng P. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget. 2016;7:44583–95. 10.18632/oncotarget.10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hanna GJ, Supplee JG, Kuang Y, Mahmood U, Lau CJ, Haddad RI, et al. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann Oncol. 2018;29:1980–6. 10.1093/annonc/mdy251 [DOI] [PubMed] [Google Scholar]
- 152.Wei T, Zhang Q, Li X, Su W, Li G, Ma T, et al. Monitoring tumor burden in response to FOLFIRINOX chemotherapy via profiling circulating cell-free DNA in pancreatic cancer. Mol Cancer Ther. 2019;18:196–203. 10.1158/1535-7163.MCT-17-1298 [DOI] [PubMed] [Google Scholar]
- 153.Jung K, Stephan C, Lewandowski M, Klotzek S, Jung M, Kristiansen G, et al. Increased cell-free DNA in plasma of patients with metastatic spread in prostate cancer. Cancer Lett. 2004;205:173–80. 10.1016/j.canlet.2003.11.023 [DOI] [PubMed] [Google Scholar]
- 154.Patsch K, Matasci N, Soundararajan A, Diaz P, Agus DB, Ruderman D, et al. Monitoring dynamic cytotoxic chemotherapy response in castration-resistant prostate cancer using plasma cell-free DNA (cfDNA). BMC Res Notes. 2019;12:275. 10.1186/s13104-019-4312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Valpione S, Gremel G, Mundra P, Middlehurst P, Galvani E, Girotti MR, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer. 2018;88:1–9. 10.1016/j.ejca.2017.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–61. 10.1158/2159-8290.CD-13-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34. 10.1186/s40246-019-0220-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112–23. 10.1373/clinchem.2014.222679 [DOI] [PubMed] [Google Scholar]
- 159.Ye P, Cai P, Xie J, Wei Y. The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: a systematic review and meta-analysis. PLoS One. 2021;16:e0248775. 10.1371/journal.pone.0248775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Köhn L, Johansson M, Grankvist K, Nilsson J. Liquid biopsies in lung cancer-time to implement research technologies in routine care? Ann Transl Med. 2017;5:278. 10.21037/atm.2017.04.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sorber L, Zwaenepoel K, Deschoolmeester V, Van Schil PE, Van Meerbeeck J, Lardon F, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer. 2017;107:100–7. 10.1016/j.lungcan.2016.04.026 [DOI] [PubMed] [Google Scholar]
- 162.Olmedillas-López S, Olivera-Salazar R, García-Arranz M, García-Olmo D. Current and emerging applications of droplet digital PCR in oncology: an updated review. Mol Diagn Ther. 2022;26:61–87. 10.1007/s40291-021-00562-2 [DOI] [PubMed] [Google Scholar]
- 163.Pérez-Callejo D, Romero A, Provencio M, Torrente M. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res. 2016;5:45–65. 10.21037/tlcr.2016.10.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Narayan A, Carriero NJ, Gettinger SN, Kluytenaar J, Kozak KR, Yock TI, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012;72:3492–8. 10.1158/0008-5472.CAN-11-4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. 10.1038/s41576-018-0071-5 [DOI] [PubMed] [Google Scholar]
- 166.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003;100:8817–22. 10.1073/pnas.1133470100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Paweletz CP, Sacher AG, Raymond CK, Alden RS, O’Connell A, Mach SL, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res. 2016;22:915–22. 10.1158/1078-0432.CCR-15-1627-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Malapelle U, Pisapia P, Rocco D, Smeraglio R, di Spirito M, Bellevicine C, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res. 2016;5:505–10. 10.21037/tlcr.2016.10.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baer C, Kern W, Koch S, Nadarajah N, Schindela S, Meggendorfer M, et al. Ultra-deep sequencing leads to earlier and more sensitive detection of the tyrosine kinase inhibitor resistance mutation T315I in chronic myeloid leukemia. Haematologica. 2016;101:830–8. 10.3324/haematol.2016.145888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Goldberg SB, Narayan A, Kole AJ, Decker RH, Teysir J, Carriero NJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res. 2018;24:1872–80. 10.1158/1078-0432.CCR-17-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. 10.1126/scitranslmed.3003726 [DOI] [PubMed] [Google Scholar]
- 172.Gale D, Lawson ARJ, Howarth K, Madi M, Durham B, Smalley S, et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13:e0194630. 10.1371/journal.pone.0194630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–5. 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Napoli GC, Chau CH, Figg WD. Single whole genome sequencing analysis blazes the trail for precision medicine. Cancer Biol Ther. 2022;23:134–5. 10.1080/15384047.2022.2033058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tailor TD, Rao X, Campa MJ, Wang J, Gregory SG, Patz EF Jr. Whole exome sequencing of cell-free DNA for early lung cancer: a pilot study to differentiate benign from malignant CT-detected pulmonary lesions. Front Oncol. 2019;9:317. 10.3389/fonc.2019.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Belic J, Koch M, Ulz P, Auer M, Gerhalter T, Mohan S, et al. Rapid identification of plasma DNA samples with increased ctDNA levels by a modified FAST-SeqS approach. Clin Chem. 2015;61:838–49. 10.1373/clinchem.2014.234286 [DOI] [PubMed] [Google Scholar]
- 178.Belic J, Koch M, Ulz P, Auer M, Gerhalter T, Mohan S, et al. mFast-SeqS as a monitoring and pre-screening tool for tumor-specific aneuploidy in plasma DNA. Adv Exp Med Biol. 2016;924:147–55. 10.1007/978-3-319-42044-8_28 [DOI] [PubMed] [Google Scholar]
- 179.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Haque IS, Elemento O. Challenges in using ctDNA to achieve early detection of cancer. BioRxiv 237578 [Preprint]. 2017. [cited 2017 Dec 21]. Available from: https://www.biorxiv.org/content/10.1101/237578v1 10.1101/237578 [DOI] [Google Scholar]
- 181.Hasenleithner SO, Speicher MR. A clinician’s handbook for using ctDNA throughout the patient journey. Mol Cancer. 2022;21:81. 10.1186/s12943-022-01551-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.List of cleared or approved companion diagnostic devices (in vitro and imaging tools) [Internet]. Credit to the U.S. Food and Drug Administration; [cited 2022 Apr 18]. Available from: https://www.fda.gov/medical-devices/in-vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-in-vitro-and-imaging-tools [Google Scholar]
- 183.Reck M, Hermes A, Tan EH, Felip E, Klughammer B, Baselga J. Tissue sampling in lung cancer: a review in light of the MERIT experience. Lung Cancer. 2011;74:1–6. 10.1016/j.lungcan.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 184.Al-Kateb H, Nguyen TT, Steger-May K, Pfeifer JD. Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (NGS). Mol Oncol. 2015;9:1737–43. 10.1016/j.molonc.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Vitiello PP, De Falco V, Giunta EF, Ciardiello D, Cardone C, Vitale P, et al. Clinical practice use of liquid biopsy to identify RAS/BRAF mutations in patients with metastatic colorectal cancer (mCRC): a single institution experience. Cancers (Basel). 2019;11:1504. 10.3390/cancers11101504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Napolitano S, Caputo V, Ventriglia A, Martini G, Della Corte CM, De Falco V, et al. Liquid biopsy at home: delivering precision medicine for patients with cancer during the COVID-19 pandemic. Oncologist. 2022;27:e633–41. 10.1093/oncolo/oyac071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Aldea M, Cerbone L, Bayle A, Parisi C, Sarkozy C, Vasseur D, et al. Detection of additional occult malignancy through profiling of ctDNA in late-stage cancer patients. Ann Oncol. 2021;32:1642–5. 10.1016/j.annonc.2021.09.002 [DOI] [PubMed] [Google Scholar]
- 188.Campos CDM, Jackson JM, Witek MA, Soper SA. Molecular profiling of liquid biopsy samples for precision medicine. Cancer J. 2018;24:93–103. 10.1097/PPO.0000000000000311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Finkle JD, Boulos H, Driessen TM, Lo C, Blidner RA, Hafez A, et al. Validation of a liquid biopsy assay with molecular and clinical profiling of circulating tumor DNA. NPJ Precis Oncol. 2021;5:63. 10.1038/s41698-021-00202-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Russo A, Incorvaia L, Del Re M, Malapelle U, Capoluongo E, Gristina V, et al. The molecular profiling of solid tumors by liquid biopsy: a position paper of the AIOM-SIAPEC-IAP-SIBioC-SIC-SIF Italian Scientific Societies. ESMO Open. 2021;6:100164. 10.1016/j.esmoop.2021.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1:1–16. 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Deans ZC, Butler R, Cheetham M, Dequeker EMC, Fairley JA, Fenizia F, et al. IQN path ASBL report from the first European cfDNA consensus meeting: expert opinion on the minimal requirements for clinical ctDNA testing. Virchows Arch. 2019;474:681–9. 10.1007/s00428-019-02571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mateo J, Chakravarty D, Dienstmann R, Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets (ESCAT). Ann Oncol. 2018;29:1895–902. 10.1093/annonc/mdy263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cheng ML, Pectasides E, Hanna GJ, Parsons HA, Choudhury AD, Oxnard GR. Circulating tumor DNA in advanced solid tumors: clinical relevance and future directions. CA Cancer J Clin. 2021;71:176–90. 10.3322/caac.21650 [DOI] [PubMed] [Google Scholar]
- 195.Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021;124:345–58. 10.1038/s41416-020-01047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24:4437–43. 10.1158/1078-0432.CCR-18-0143 [DOI] [PubMed] [Google Scholar]
- 197.Pisapia P, Pepe F, Iaccarino A, Sgariglia R, Nacchio M, Russo G, et al. Liquid biopsy analysis in clinical practice: focus on lung cancer. J Mol Pathol. 2021;2:241–54. 10.3390/jmp2030021 [DOI] [Google Scholar]
- 198.Chan HT, Chin YM, Nakamura Y, Low SK. Clonal hematopoiesis in liquid biopsy: from biological noise to valuable clinical implications. Cancers (Basel). 2020;12:2277. 10.3390/cancers12082277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.van de Haar J, Hoes L, Voest E. Advancing molecular tumour boards: highly needed to maximise the impact of precision medicine. ESMO Open. 2019;4:e000516. 10.1136/esmoopen-2019-000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Leighl NB, Page RD, Raymond VM, Daniel DB, Divers SG, Reckamp KL, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res. 2019;25:4691–700. 10.1158/1078-0432.CCR-19-0624 [DOI] [PubMed] [Google Scholar]
- 201.Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 2019;5:173–80. 10.1001/jamaoncol.2018.4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Guerrera LP, Suarato G, Napolitano R, Perrone A, Caputo V, Ventriglia A, et al. Mixed neuroendocrine non-neuroendocrine neoplasms of the gastrointestinal tract: a case series. Healthcare (Basel). 2022;10:708. 10.3390/healthcare10040708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Caputo V, De Falco V, Ventriglia A, Famiglietti V, Martinelli E, Morgillo F, et al. Comprehensive genome profiling by next generation sequencing of circulating tumor DNA in solid tumors: a single academic institution experience. Ther Adv Med Oncol. 2022;14:17588359221096878. 10.1177/17588359221096878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sardo E, Napolitano S, Della Corte CM, Ciardiello D, Raucci A, Arrichiello G, et al. Multi-omic approaches in colorectal cancer beyond genomic data. J Pers Med. 2022;12:128. 10.3390/jpm12020128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019;25:1415–21. Erratum in: Nat Med. 2019;25:1949. 10.1038/s41591-019-0561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lu C, Zhang YC, Chen ZH, Zhou Q, Wu YL. Applications of circulating tumor DNA in immune checkpoint inhibition: emerging roles and future perspectives. Front Oncol. 2022;12:836891. 10.3389/fonc.2022.836891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Markou A, Tzanikou E, Lianidou E. The potential of liquid biopsy in the management of cancer patients. Semin Cancer Biol. 2022;84:69–79. 10.1016/j.semcancer.2022.03.013 [DOI] [PubMed] [Google Scholar]
- 208.Chakraborty S, Hosen MI, Ahmed M, Shekhar HU. Onco-multi-OMICS approach: a new frontier in cancer research. Biomed Res Int. 2018;2018:9836256. 10.1155/2018/9836256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ding SC, Lo YMD. Cell-free DNA fragmentomics in liquid biopsy. Diagnostics (Basel). 2022;12:978. 10.3390/diagnostics12040978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Peng Y, Mei W, Ma K, Zeng C. Circulating tumor DNA and minimal residual disease (MRD) in solid tumors: current horizons and future perspectives. Front Oncol. 2021;11:763790. 10.3389/fonc.2021.763790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40:172–86. 10.1016/j.tips.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 212.Ulz P, Heitzer E, Geigl JB, Speicher MR. Patient monitoring through liquid biopsies using circulating tumor DNA. Int J Cancer. 2017;141:887–96. 10.1002/ijc.30759 [DOI] [PubMed] [Google Scholar]
- 213.Pasini L, Ulivi P. Liquid biopsy for the detection of resistance mechanisms in NSCLC: comparison of different blood biomarkers. J Clin Med. 2019;8:998. 10.3390/jcm8070998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Venesio T, Siravegna G, Bardelli A, Sapino A. Liquid biopsies for monitoring temporal genomic heterogeneity in breast and colon cancers. Pathobiology. 2018;85:146–54. 10.1159/000473882 [DOI] [PubMed] [Google Scholar]
- 215.Angeles AK, Janke F, Bauer S, Christopoulos P, Riediger AL, Sültmann H. Liquid biopsies beyond mutation calling: genomic and epigenomic features of cell-free DNA in cancer. Cancers (Basel). 2021;13:5615. 10.3390/cancers13225615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lennon AM, Buchanan AH, Kinde I, Warren A, Honushefsky A, Cohain AT, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369:eabb9601. 10.1126/science.abb9601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vacante M, Ciuni R, Basile F, Biondi A. The liquid biopsy in the management of colorectal cancer: an overview. Biomedicines. 2020;8:308. 10.3390/biomedicines8090308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Remon J, García-Campelo R, de Álava E, Vera R, Rodríguez-Peralto JL, Rodríguez-Lescure Á, et al. Liquid biopsy in oncology: a consensus statement of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin Transl Oncol. 2020;22:823–34. Erratum in: Clin Transl Oncol. 2020;22:961–2. 10.1007/s12094-019-02211-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20:430–5. 10.1038/nm.3511 [DOI] [PubMed] [Google Scholar]
- 220.Malapelle U, Mayo de-Las-Casas C, Rocco D, Garzon M, Pisapia P, Jordana-Ariza N, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer. 2017;116:802–10. 10.1038/bjc.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang F, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2015;24:206–12. 10.1158/1055-9965.EPI-14-0895 [DOI] [PubMed] [Google Scholar]
- 222.Hao YX, Fu Q, Guo YY, Ye M, Zhao HX, Wang Q, et al. Effectiveness of circulating tumor DNA for detection of KRAS gene mutations in colorectal cancer patients: a meta-analysis. Onco Targets Ther. 2017;10:945–53. 10.2147/OTT.S123954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Xie W, Xie L, Song X. The diagnostic accuracy of circulating free DNA for the detection of KRAS mutation status in colorectal cancer: a meta-analysis. Cancer Med. 2019;8:1218–31. 10.1002/cam4.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4:6269. 10.1038/srep06269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tang M, Deng Z, Li B, Peng Y, Song M, Liu J. Circulating tumor DNA is effective for detection of KRAS mutation in colorectal cancer: a meta-analysis. Int J Biol Markers. 2017;32:e421–7. 10.5301/ijbm.5000295 [DOI] [PubMed] [Google Scholar]
- 226.Hedtke M, Pessoa Rejas R, Froelich MF, Ast V, Duda A, Mirbach L, et al. Liquid profiling of circulating tumor DNA in colorectal cancer: steps needed to achieve its full clinical value as standard care. Mol Oncol. 2021;16:2042–56. 10.1002/1878-0261.13156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Companion diagnostic test [Internet]. Bethesda: National Cancer Institute; [cited 2022 Apr 26]. Available from: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/companion-diagnostic-test [Google Scholar]
- 228.Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–505. 10.1016/j.annonc.2020.07.014 [DOI] [PubMed] [Google Scholar]
- 229.Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. NCCN guidelines insights: non-small cell lung cancer, version 2.2021. J Natl Compr Canc Netw. 2021;19:254–66. 10.6004/jnccn.2021.0013 [DOI] [PubMed] [Google Scholar]
- 230.Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13:1248–68. 10.1016/j.jtho.2018.05.030 [DOI] [PubMed] [Google Scholar]
- 231.Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142:321–46. 10.5858/arpa.2017-0388-CP [DOI] [PubMed] [Google Scholar]
- 232.Mok TS Wu YL Ahn MJ Garassino MC Kim HR Ramalingam SSet al.; AURA3 Investigators . Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sequist LV, Han JY, Ahn MJ, Cho BC, Yu H, Kim SW, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol. 2020;21:373–86. 10.1016/S1470-2045(19)30785-5 [DOI] [PubMed] [Google Scholar]
- 234.Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24:3097–107. 10.1158/1078-0432.CCR-17-2310 [DOI] [PubMed] [Google Scholar]
- 235.Papadimitrakopoulou VA, Wu YL, Han JY, Ahn MJ, Ramalingam SS, John T, et al. Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Ann Oncol. 2018;29:viii741. 10.1093/annonc/mdy424.064 [DOI] [Google Scholar]
- 236.Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121:725–37. 10.1038/s41416-019-0573-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–34. 10.1001/jamaoncol.2018.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yu HA, Schoenfeld AJ, Makhnin A, Kim R, Rizvi H, Tsui D, et al. Effect of osimertinib and bevacizumab on progression-free survival for patients with metastatic EGFR-mutant lung cancers: a phase 1/2 single-group open-label trial. JAMA Oncol. 2020;6:1048–54. 10.1001/jamaoncol.2020.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dagogo-Jack I, Brannon AR, Ferris LA, Campbell CD, Lin JJ, Schultz KR, et al. Tracking the evolution of resistance to ALK tyrosine kinase inhibitors through longitudinal analysis of circulating tumor DNA. JCO Precis Oncol. 2018;2018:1–14. 10.1200/PO.17.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dagogo-Jack I, Rooney M, Nagy RJ, Lin JJ, Chin E, Ferris LA, et al. Molecular analysis of plasma from patients with ROS1-positive NSCLC. J Thorac Oncol. 2019;14:816–24. 10.1016/j.jtho.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Recondo G, Bahcall M, Spurr LF, Che J, Ricciuti B, Leonardi GC, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14-mutant NSCLC. Clin Cancer Res. 2020;26:2615–25. 10.1158/1078-0432.CCR-19-3608 [DOI] [PubMed] [Google Scholar]
- 242.Madsen AT, Winther-Larsen A, McCulloch T, Meldgaard P, Sorensen BS. Genomic profiling of circulating tumor DNA predicts outcome and demonstrates tumor evolution in ALK-positive non-small cell lung cancer patients. Cancers (Basel). 2020;12:947. 10.3390/cancers12040947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tsurutani J, Iwata H, Krop I, Jänne PA, Doi T, Takahashi S, et al. Targeting HER2 with trastuzumab deruxtecan: a dose-expansion, phase I study in multiple advanced solid tumors. Cancer Discov. 2020;10:688–701. Erratum in: Cancer Discov. 2020;10:1078. 10.1158/2159-8290.CD-19-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. 10.1158/2159-8290.CD-19-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Dueling KRASG12C inhibitors achieve responses. Cancer Discov. 2020;10:10. 10.1158/2159-8290.CD-ND2019-012 [DOI] [PubMed] [Google Scholar]
- 246.Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36:1631–41. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 247.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165–72. 10.1016/S1470-2045(14)71180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.André F Ciruelos E Rubovszky G Campone M Loibl S Rugo HSet al.; SOLAR-1 Study Group . Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–40. 10.1056/NEJMoa1813904 [DOI] [PubMed] [Google Scholar]
- 249.Ciruelos EM, Loibl S, Mayer IA, Campone M, Rugo HS, Arnedos M, et al. Abstract PD2-06: clinical outcomes of alpelisib plus fulvestrant in hormone receptorpositive, human epidermal growth factor receptor-2-negative advanced breast cancer with PIK3CA alterations detected in plasma ctDNA by next-generation sequencing: biomarker analysis from the SOLAR-1 study. Cancer Res. 2021;81:PD2–06. 10.1158/1538-7445.SABCS20-PD2-06 [DOI] [Google Scholar]
- 250.Ma CX, Bose R, Gao F, Freedman RA, Telli ML, Kimmick G, Winer E, et al. Neratinib efficacy and circulating tumor DNA detection of HER2 mutations in HER2 nonamplified metastatic breast cancer. Clin Cancer Res. 2017;23:5687–95. 10.1158/1078-0432.CCR-17-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Turner NC, Kingston B, Kilburn LS, Kernaghan S, Wardley AM, Macpherson IR, et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020;21:1296–308. 10.1016/S1470-2045(20)30444-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Razavi P, Dickler MN, Shah PD, Toy W, Brown DN, Won HH, et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat Cancer. 2020;1:382–93. 10.1038/s43018-020-0047-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:795–801. Erratum in: Nat Med. 2015;21:827. 10.1038/nm.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Siravegna G, Sartore-Bianchi A, Nagy RJ, Raghav K, Odegaard JI, Lanman RB, et al. Plasma HER2 (ERBB2) copy number predicts response to HER2-targeted therapy in metastatic colorectal cancer. Clin Cancer Res. 2019;25:3046–53. 10.1158/1078-0432.CCR-18-3389 [DOI] [PubMed] [Google Scholar]
- 255.Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. 10.1038/nature11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. 10.1038/nature11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28:1325–32. 10.1093/annonc/mdx125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Schmiegel W, Scott RJ, Dooley S, Lewis W, Meldrum CJ, Pockney P, et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol. 2017;11:208–19. 10.1002/1878-0261.12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Fenocchio E, et al. Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: the CHRONOS trial. JCO. 2021;39:3506. 10.1200/JCO.2021.39.15_suppl.3506 35915157 [DOI] [Google Scholar]
- 260.Tabernero J, Grothey A, Van Cutsem E, Yaeger R, Wasan H, Yoshino T, et al. Encorafenib plus cetuximab as a new standard of care for previously treated BRAF V600E-mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON study. J Clin Oncol. 2021;39:273–84. 10.1200/JCO.20.02088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–46. Erratum in: Lancet Oncol. 2016;17:e420. 10.1016/S1470-2045(16)00150-9 [DOI] [PubMed] [Google Scholar]
- 262.Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, et al. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: a randomized clinical trial. JAMA. 2017;317:2392–401. 10.1001/jama.2017.7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 264.Walens A, Lin J, Damrauer JS, McKinney B, Lupo R, Newcomb R, et al. Adaptation and selection shape clonal evolution of tumors during residual disease and recurrence. Nat Commun. 2020;11:5017. 10.1038/s41467-020-18730-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ciardiello F, Ciardiello D, Martini G, Napolitano S, Tabernero J, Cervantes A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin. 2022;72:372–401. 10.3322/caac.21728 [DOI] [PubMed] [Google Scholar]
- 266.Ciardiello D, Martini G, Famiglietti V, Napolitano S, De Falco V, Troiani T, et al. Biomarker-guided anti-EGFR rechallenge therapy in metastatic colorectal cancer. Cancers (Basel). 2021;13:1941. Erratum in: Cancers (Basel). 2022;14:3900. 10.3390/cancers13081941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Siravegna G, Lazzari L, Crisafulli G, Sartore-Bianchi A, Mussolin B, Morris VK, et al. Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell. 2018;34:148–62. 10.1016/j.ccell.2018.06.004 [DOI] [PubMed] [Google Scholar]
- 268.Morelli MP, Overman MJ, Dasari A, Kazmi SMA, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26:731–6. 10.1093/annonc/mdv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Khan KH, Cunningham D, Werner B, Vlachogiannis G, Spiteri I, Heide T, et al. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C phase II colorectal cancer clinical trial. Cancer Discov. 2018;8:1270–85. 10.1158/2159-8290.CD-17-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Parseghian CM, Loree JM, Morris VK, Liu X, Clifton KK, Napolitano S, et al. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol. 2019;30:243–9. 10.1093/annonc/mdy509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. 10.1038/s41467-017-00965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, De Mattos-Arruda L, Muñoz-Couselo E, Manzano JL, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res. 2015;25:486–95. 10.1097/CMR.0000000000000187 [DOI] [PubMed] [Google Scholar]
- 273.Haselmann V, Gebhardt C, Brechtel I, Duda A, Czerwinski C, Sucker A, et al. Liquid profiling of circulating tumor DNA in plasma of melanoma patients for companion diagnostics and monitoring of BRAF inhibitor therapy. Clin Chem. 2018;64:830–42. 10.1373/clinchem.2017.281543 [DOI] [PubMed] [Google Scholar]
- 274.FDA approves first-line immunotherapy for patients with MSI-H/dMMR metastatic colorectal cancer [Internet]. Credit to the U.S. Food and Drug Administration; [cited 2022 Apr 26]. Available from: https://www.fda.gov/news-events/press-announcements/fda-approves-first-line-immunotherapy-patients-msi-hdmmr-metastatic-colorectal-cancer [Google Scholar]
- 275.FDA approves first cancer treatment for any solid tumor with a specific genetic feature [Internet]. Credit to the U.S. Food and Drug Administration; [cited 2022 Jun 2]. Available from: https://www.fda.gov/news-events/press-announcements/fda-approves-first-cancer-treatment-any-solid-tumor-specific-genetic-feature [Google Scholar]
- 276.FDA approves pembrolizumab for adults and children with TMB-H solid tumors [Internet]. Credit to the U.S. Food and Drug Administration; [cited 2022 June 2]. Available from: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors [Google Scholar]
- 277.Kasi PM, Klempner SJ, Starr JS, Shergill A, Bucheit LA, Weipert C, et al. Clinical utility of microsatellite instability (MSI-H) identified on liquid biopsy in advanced gastrointestinal cancers (aGI). JCO. 2022;40:56. 10.1200/JCO.2022.40.4_suppl.056 [DOI] [Google Scholar]
- 278.Spencer KR, Wang J, Silk AW, Ganesan S, Kaufman HL, Mehnert JM. Biomarkers for immunotherapy: current developments and challenges. Am Soc Clin Oncol Educ Book. 2016;35:e493–503. 10.1200/EDBK_160766 [DOI] [PubMed] [Google Scholar]
- 279.Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 2019;5:696–702. 10.1001/jamaoncol.2018.7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Chauhan PS, Chen K, Babbra RK, Feng W, Pejovic N, Nallicheri A, et al. Urine tumor DNA detection of minimal residual disease in muscle-invasive bladder cancer treated with curative-intent radical cystectomy: a cohort study. PLoS Med. 2021;18:e1003732. Erratum in: PLoS Med. 2021;18:e1003876. 10.1371/journal.pmed.1003732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hita-Millan J, Carracedo A, Fernandez-Rozadilla C. Liquid Biopsy biomarkers for immunotherapy in non-small cell lung carcinoma: lessons learned and the road ahead. J Pers Med. 2021;11:971. 10.3390/jpm11100971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kilgour E, Rothwell DG, Brady G, Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. 2020;37:485–95. 10.1016/j.ccell.2020.03.012 [DOI] [PubMed] [Google Scholar]
- 283.Mauri G, Vitiello PP, Sogari A, Crisafulli G, Sartore-Bianchi A, Marsoni S, et al. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br J Cancer. 2022;127:394–407. 10.1038/s41416-022-01769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:79. 10.1186/s12943-022-01543-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Cecchini MJ, Yi ES. Liquid biopsy is a valuable tool in the diagnosis and management of lung cancer. J Thorac Dis. 2020;12:7048–56. 10.21037/jtd.2020.04.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Bracht JWP, Mayo-de-Las-Casas C, Berenguer J, Karachaliou N, Rosell R. The present and future of liquid biopsies in non-small cell lung cancer: combining four biosources for diagnosis, prognosis, prediction, and disease monitoring. Curr Oncol Rep. 2018;20:70. Erratum in: Curr Oncol Rep. 2020;22:52. 10.1007/s11912-018-0720-z [DOI] [PubMed] [Google Scholar]
- 287.Barták BK, Fodor T, Kalmár A, Nagy ZB, Zsigrai S, Szigeti KA, et al. A liquid biopsy-based approach for monitoring treatment response in post-operative colorectal cancer patients. Int J Mol Sci. 2022;23:3774. 10.3390/ijms23073774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Nagasaka M, Uddin MH, Al-Hallak MN, Rahman S, Balasubramanian S, Sukari A, et al. Liquid biopsy for therapy monitoring in early-stage non-small cell lung cancer. Mol Cancer. 2021;20:82. 10.1186/s12943-021-01371-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Fattore L, Ruggiero CF, Liguoro D, Castaldo V, Catizone A, Ciliberto G, et al. The promise of liquid biopsy to predict response to immunotherapy in metastatic melanoma. Front Oncol. 2021;11:645069. 10.3389/fonc.2021.645069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Osei-Bordom DC, Sachdeva G, Christou N. Liquid biopsy as a prognostic and theranostic tool for the management of pancreatic ductal adenocarcinoma. Front Med (Lausanne). 2022;8:788869. 10.3389/fmed.2021.788869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Patelli G, Vaghi C, Tosi F, Mauri G, Amatu A, Massihnia D, et al. Liquid biopsy for prognosis and treatment in metastatic colorectal cancer: circulating tumor cells vs. circulating tumor DNA. Target Oncol. 2021;16:309–24. Erratum in: Target Oncol. 2021;16:413. 10.1007/s11523-021-00795-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AF, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;8:364ra155. 10.1126/scitranslmed.aai8545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Cheng ML, Lau CJ, Milan MSD, Supplee JG, Riess JW, Bradbury PA, et al. Plasma ctDNA response is an early marker of treatment effect in advanced NSCLC. JCO Precis Oncol. 2021;5:393–402. 10.1200/PO.20.00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ricciuti B, Jones G, Severgnini M, Alessi JV, Recondo G, Lawrence M, et al. Early plasma circulating tumor DNA (ctDNA) changes predict response to first-line pembrolizumab-based therapy in non-small cell lung cancer (NSCLC). J Immunother Cancer. 2021;9:e001504. 10.1136/jitc-2020-001504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Gray JE, Okamoto I, Sriuranpong V, Vansteenkiste J, Imamura F, Lee JS, et al. Tissue and plasma EGFR mutation analysis in the FLAURA trial: osimertinib versus comparator EGFR tyrosine kinase inhibitor as first-line treatment in patients with EGFR-mutated advanced non-small cell lung cancer. Clin Cancer Res. 2019;25:6644–52. 10.1158/1078-0432.CCR-19-1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Shepherd FA, Papadimitrakopoulou V, Mok T, Wu YL, Han JY, Ahn MJ, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib in the AURA3 trial. JCO. 2018;36:9027. 10.1200/JCO.2018.36.15_suppl.9027 [DOI] [Google Scholar]
- 297.Song Y Hu C Xie Z Wu L Zhu Z Rao Cet al.; written on behalf of AME Lung Cancer Collaborative Group . Circulating tumor DNA clearance predicts prognosis across treatment regimen in a large real-world longitudinally monitored advanced non-small cell lung cancer cohort. Transl Lung Cancer Res. 2020;9:269–79. 10.21037/tlcr.2020.03.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Wei J, Feng J, Weng Y, Xu Z, Jin Y, Wang P, et al. The prognostic value of ctDNA and bTMB on immune checkpoint inhibitors in human cancer. Front Oncol. 2021;11:706910. 10.3389/fonc.2021.706910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1:873–81. 10.1038/s43018-020-0096-5 [DOI] [PubMed] [Google Scholar]
- 300.Zhang Q, Luo J, Wu S, Si H, Gao C, Xu W, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 2020;10:1842–53. 10.1158/2159-8290.CD-20-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Guibert N, Mazieres J, Delaunay M, Casanova A, Farella M, Keller L, et al. Monitoring of KRAS-mutated ctDNA to discriminate pseudo-progression from true progression during anti-PD-1 treatment of lung adenocarcinoma. Oncotarget. 2017;8:38056–60. 10.18632/oncotarget.16935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Darrigues L, Pierga JY, Bernard-Tessier A, Bièche I, Silveira AB, Michel M, et al. Circulating tumor DNA as a dynamic biomarker of response to palbociclib and fulvestrant in metastatic breast cancer patients. Breast Cancer Res. 2021;23:31. 10.1186/s13058-021-01411-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Li S, Lai H, Liu J, Liu Y, Jin L, Li Y, et al. Circulating tumor DNA predicts the response and prognosis in patients with early breast cancer receiving neoadjuvant chemotherapy. JCO Precis Oncol. 2020;4:244–57. 10.1200/PO.19.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Del Re M, Vivaldi C, Rofi E, Vasile E, Miccoli M, Caparello C, et al. Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci Rep. 2017;7:7931. 10.1038/s41598-017-08297-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kruger S, Heinemann V, Ross C, Diehl F, Nagel D, Ormanns S, et al. Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann Oncol. 2018;29:2348–55. 10.1093/annonc/mdy417 [DOI] [PubMed] [Google Scholar]
- 306.Lim Y, Kim S, Kang JK, Kim HP, Jang H, Han H, et al. Circulating tumor DNA sequencing in colorectal cancer patients treated with first-line chemotherapy with anti-EGFR. Sci Rep. 2021;11:16333. 10.1038/s41598-021-95345-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Garlan F, Laurent-Puig P, Sefrioui D, Siauve N, Didelot A, Sarafan-Vasseur N, et al. Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL Study). Clin Cancer Res. 2017;23:5416–25. 10.1158/1078-0432.CCR-16-3155 [DOI] [PubMed] [Google Scholar]
- 308.Salvianti F, Gelmini S, Mancini I, Pazzagli M, Pillozzi S, Giommoni E, et al. Circulating tumour cells and cell-free DNA as a prognostic factor in metastatic colorectal cancer: the OMITERC prospective study. Br J Cancer. 2021;125:94–100. 10.1038/s41416-021-01399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Gray E, Rizos H, Reid A, Suzanah B, Michelle P, Johnny L, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6:42008–18. 10.18632/oncotarget.5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312. 10.1038/s41571-020-00457-x [DOI] [PubMed] [Google Scholar]
- 311.Minimal residual disease [Internet]. Credit to the U.S. Food and Drug Administration; [cited 2022 May 25]. Available from: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/minimal-residual-disease [Google Scholar]
- 312.Szczepański T, Orfão A, van der Velden VH, San Miguel JF, van Dongen JJ. Minimal residual disease in leukaemia patients. Lancet Oncol. 2001;2:409–17. 10.1016/S1470-2045(00)00418-6 [DOI] [PubMed] [Google Scholar]
- 313.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 314.Moding EJ, Nabet BY, Alizadeh AA, Diehn M. Detecting liquid remnants of solid tumors: circulating tumor DNA minimal residual disease. Cancer Discov. 2021;11:2968–86. 10.1158/2159-8290.CD-21-0634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.DiNardo CD, Korde LA, Yurgelun MB. A case-based approach to understanding complex genetic information in an evolving landscape. Am Soc Clin Oncol Educ Book. 2021;41:1–11. 10.1200/EDBK_321041 [DOI] [PubMed] [Google Scholar]
- 316.Huang F, Yang Y, Chen X, Jiang H, Wang H, Shen M, et al. Chemotherapy-associated clonal hematopoiesis mutations should be taken seriously in plasma cell-free DNA KRAS/NRAS/BRAF genotyping for metastatic colorectal cancer. Clin Biochem. 2021;92:46–53. 10.1016/j.clinbiochem.2021.03.005 [DOI] [PubMed] [Google Scholar]
- 317.Chera BS, Kumar S, Beaty BT, Marron D, Jefferys S, Green R, et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res. 2019;25:4682–90. 10.1158/1078-0432.CCR-19-0211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Rolfo C, Russo A. Liquid biopsy for early stage lung cancer moves ever closer. Nat Rev Clin Oncol. 2020;17:523–4. 10.1038/s41571-020-0393-z [DOI] [PubMed] [Google Scholar]
- 319.Ulrich B, Pradines A, Mazières J, Guibert N. Detection of tumor recurrence via circulating tumor DNA profiling in patients with localized lung cancer: clinical considerations and challenges. Cancers (Basel). 2021;13:3759. 10.3390/cancers13153759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Siravegna G, Mussolin B, Venesio T, Marsoni S, Seoane J, Dive C, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol. 2019;30:1580–90. 10.1093/annonc/mdz227 [DOI] [PubMed] [Google Scholar]
- 321.Naidoo M, Gibbs P, Tie J. ctDNA and adjuvant therapy for colorectal cancer: time to re-invent our treatment paradigm. Cancers (Basel). 2021;13:346. 10.3390/cancers13020346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Coakley M, Garcia-Murillas I, Turner NC. Molecular residual disease and adjuvant trial design in solid tumors. Clin Cancer Res. 2019;25:6026–34. 10.1158/1078-0432.CCR-19-0152 [DOI] [PubMed] [Google Scholar]
- 323.Wills B, Gorse E, Lee V. Role of liquid biopsies in colorectal cancer. Curr Probl Cancer. 2018;42:593–600. 10.1016/j.currproblcancer.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 324.Chin RI, Chen K, Usmani A, Chua C, Harris PK, Binkley MS, et al. Detection of solid tumor molecular residual disease (MRD) using circulating tumor DNA (ctDNA). Mol Diagn Ther. 2019;23:311–31. 10.1007/s40291-019-00390-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Honoré N, Galot R, van Marcke C, Limaye N, Machiels JP. Liquid biopsy to detect minimal residual disease: methodology and impact. Cancers (Basel). 2021;13:5364. 10.3390/cancers13215364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Larribère L, Martens UM. Advantages and challenges of using ctDNA NGS to assess the presence of minimal residual disease (MRD) in solid tumors. Cancers (Basel). 2021;13:5698. 10.3390/cancers13225698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Kasi PM, Fehringer G, Taniguchi H, Starling N, Nakamura Y, Kotani D, et al. Impact of circulating tumor DNA-based detection of molecular residual disease on the conduct and design of clinical trials for solid tumors. JCO Precis Oncol. 2022;6:e2100181. Erratum in: JCO Precis Oncol. 2022;6:e2200193. 10.1200/PO.21.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Pignon JP Tribodet H Scagliotti GV Douillard JY Shepherd FA Stephens RJet al.; LACE Collaborative Group . Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–9. 10.1200/JCO.2007.13.9030 [DOI] [PubMed] [Google Scholar]
- 329.Wu YL Tsuboi M He J John T Grohe C Majem Met al.; ADAURA Investigators . Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2020;383:1711–23. 10.1056/NEJMoa2027071 [DOI] [PubMed] [Google Scholar]
- 330.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. TRACERx consortium. PEACE consortium. Swanton C. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–51. Erratum in: Nature. 2018;554:264. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Killock D. DYNAMIC insights into MRD responses early after resection of NSCLC. Nat Rev Clin Oncol. 2019;16:661. 10.1038/s41571-019-0274-5 [DOI] [PubMed] [Google Scholar]
- 332.Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–403. 10.1158/2159-8290.CD-17-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Moding EJ, Liu Y, Nabet BY, Chabon JJ, Chaudhuri AA, Hui AB, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small cell lung cancer. Nat Cancer. 2020;1:176–83. 10.1038/s43018-019-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. 10.1126/scitranslmed.aab0021 [DOI] [PubMed] [Google Scholar]
- 335.Garcia-Murillas I, Chopra N, Comino-Méndez I, Beaney M, Tovey H, Cutts RJ, et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 2019;5:1473–8. Erratum in: JAMA Oncol. 2020;6:162. 10.1001/jamaoncol.2019.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–7. 10.1038/s41586-021-03642-9 [DOI] [PubMed] [Google Scholar]
- 337.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Diehn M, Alizadeh AA, Adams HP, Lee JJ, Klassen S, Palma JF, et al. Early prediction of clinical outcomes in resected stage II and III colorectal cancer (CRC) through deep sequencing of circulating tumor DNA (ctDNA). JCO. 2017;35:3591. 10.1200/JCO.2017.35.15_suppl.3591 [DOI] [Google Scholar]
- 340.Schøler LV, Reinert T, Ørntoft MW, Kassentoft CG, Árnadóttir SS, Vang S, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23:5437–45. 10.1158/1078-0432.CCR-17-0510 [DOI] [PubMed] [Google Scholar]
- 341.Anandappa G, Starling N, Begum R, Bryant A, Sharma S, Renner D, et al. Minimal residual disease (MRD) detection with circulating tumor DNA (ctDNA) from personalized assays in stage II–III colorectal cancer patients in a U.K. multicenter prospective study (TRACC). JCO. 2021;39:102. 10.1200/JCO.2021.39.3_suppl.102 [DOI] [Google Scholar]
- 342.Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell- free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124–31. Erratum in: JAMA Oncol. 2019;5:1232. 10.1001/jamaoncol.2019.0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Tie J, Cohen J, Wang Y, Li L, Kinde I, Elsaleh H, et al. The potential of circulating tumor DNA (ctDNA) to guide adjuvant chemotherapy decision making in locally advanced rectal cancer (LARC). J Clin Oncol. 2017;35:3521. 10.1200/JCO.2017.35.15_suppl.3521 [DOI] [Google Scholar]
- 344.Hirahata T, Ul Quraish R, Quraish AU, Ul Quraish S, Naz M, Razzaq MA. Liquid biopsy: a distinctive approach to the diagnosis and prognosis of cancer. Cancer Inform. 2022;21:11769351221076062. 10.1177/11769351221076062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5:1710–7. Erratum in: JAMA Oncol. 2019;5:1811. 10.1001/jamaoncol.2019.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Loupakis F, Sharma S, Derouazi M, Murgioni S, Biason P, Rizzato MD, et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol. 2021;5:1166–77. 10.1200/PO.21.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Moding EJ, Stehr H, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580:245–51. 10.1038/s41586-020-2140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Stover DG, Parsons HA, Ha G, Freeman SS, Barry WT, Guo H, et al. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative breast cancer. J Clin Oncol. 2018;36:543–53. 10.1200/JCO.2017.76.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Santiago-Walker A, Gagnon R, Mazumdar J, Casey M, Long GV, Schadendorf D, et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res. 2016;22:567–74. 10.1158/1078-0432.CCR-15-0321 [DOI] [PubMed] [Google Scholar]
- 350.Tarazona N, Gimeno-Valiente F, Gambardella V, Zuñiga S, Rentero-Garrido P, Huerta M, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30:1804–12. 10.1093/annonc/mdz390 [DOI] [PubMed] [Google Scholar]
- 351.Crosby D. Delivering on the promise of early detection with liquid biopsies. Br J Cancer. 2022;126:313–5. 10.1038/s41416-021-01646-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Freitas C, Sousa C, Machado F, Serino M, Santos V, Cruz-Martins N, et al. The role of liquid biopsy in early diagnosis of lung cancer. Front Oncol. 2021;11:634316. 10.3389/fonc.2021.634316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Santarpia M, Liguori A, D’Aveni A, Karachaliou N, Gonzalez-Cao M, Daffinà MG, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis. 2018;10:S882–97. 10.21037/jtd.2018.03.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Cowling T, Loshak H. An overview of liquid biopsy for screening and early detection of cancer. In: CADTH issues in emerging health technologies. Ottawa (ON): Canadian Agency for Drugs and Technologies in Health. 2019;179:1–13. [PubMed] [Google Scholar]
- 355.Zheng H, Wu X, Yin J, Wang S, Li Z, You C. Clinical applications of liquid biopsies for early lung cancer detection. Am J Cancer Res. 2019;9:2567–79. [PMC free article] [PubMed] [Google Scholar]
- 356.Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer. 2020;1:276–90. 10.1038/s43018-020-0043-5 [DOI] [PubMed] [Google Scholar]
- 357.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–30. 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.McDonald BR, Contente-Cuomo T, Sammut SJ, Odenheimer-Bergman A, Ernst B, Perdigones N, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11:eaax7392. 10.1126/scitranslmed.aax7392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res. 2019;25:4255–63. 10.1158/1078-0432.CCR-18-3663 [DOI] [PubMed] [Google Scholar]
- 360.Jones RM, Devers KJ, Kuzel AJ, Woolf SH. Patient-reported barriers to colorectal cancer screening: a mixed-methods analysis. Am J Prev Med. 2010;38:508–16. 10.1016/j.amepre.2010.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Steele RJ, McDonald PJ, Digby J, Brownlee L, Strachan JA, Libby G, et al. Clinical outcomes using a faecal immunochemical test for haemoglobin as a first-line test in a national programme constrained by colonoscopy capacity. United European Gastroenterol J. 2013;1:198–205. 10.1177/2050640613489281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Lamb YN, Dhillon S. Epi proColon® 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther. 2017;21:225–32. 10.1007/s40291-017-0259-y [DOI] [PubMed] [Google Scholar]
- 363.Duffy MJ, Diamandis EP, Crown J. Circulating tumor DNA (ctDNA) as a pan-cancer screening test: is it finally on the horizon? Clin Chem Lab Med. 2021;59:1353–61. 10.1515/cclm-2021-0171 [DOI] [PubMed] [Google Scholar]
- 364.Sugeeta SS, Sharma A, Ng K, Nayak A, Vasdev N. Biomarkers in bladder cancer surveillance. Front Surg. 2021;8:735868. 10.3389/fsurg.2021.735868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Springer SU, Chen CH, Rodriguez Pena MDC, Li L, Douville C, Wang Y, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife 2018;7:e32143. Erratum in: eLife. 2018;7:e43237. 10.7554/eLife.32143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Haselmann V, Hedtke M, Neumaier M. Liquid profiling for cancer patient stratification in precision medicine-current status and challenges for successful implementation in standard care. Diagnostics (Basel). 2022;12:748. 10.3390/diagnostics12030748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.García-Pardo M, Makarem M, Li JJN, Kelly D, Leighl NB. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: opportunities and challenges. Br J Cancer. 2022;127:592–602. 10.1038/s41416-022-01776-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.