Summary
Error-free chromosome segregation in mitosis and meiosis relies on the assembly of a microtubule-based spindle that interacts with kinetochores to guide chromosomes to the cell equator before segregation in anaphase. Microtubules sprout from nucleation sites such as centrosomes, but kinetochores can also promote microtubule formation. It is unclear, however, how kinetochore-derived microtubules are generated and what their role is in chromosome segregation. Here, we show that the transient outer-kinetochore meshwork known as the fibrous corona serves as an autonomous microtubule nucleation platform. The fibrous corona is essential for the nucleation of kinetochore-derived microtubules, and when dissociated from the core kinetochore, it retains microtubule nucleation capacity. Nucleation relies on a fibrous-corona-bound pool of the LIC1 subunit of the dynein motor complex, which interacts with the γ-tubulin-tethering protein pericentrin (PCNT). PCNT is essential for microtubule nucleation from fibrous coronas, and in centrosome-depleted cells, where nearly all mitotic nucleation occurs at fibrous coronas, chromosome congression is fully dependent on PCNT. We further show that chromosomes in bovine oocytes, which naturally lack centrosomes, have highly expanded fibrous coronas that drive chromosome-derived microtubule nucleation. Preventing fibrous corona expansion in these cells impairs chromosome congression and causes spindle assembly defects. Our results show that fibrous coronas are autonomous microtubule-organizing centers that are important for spindle assembly, which may be especially relevant in acentrosomal cells such as oocytes.
Keywords: cell division, mitosis, meiosis, chromosome, kinetochore, fibrous corona, microtubule, pericentrin, microtubule nucleation, oocyte
Graphical abstract
Highlights
-
•
The fibrous corona, a transient outer-kinetochore meshwork, nucleates microtubules
-
•
Microtubule nucleation is driven by PCNT, which binds fibrous corona protein LIC1
-
•
Fibrous-corona-derived microtubules contribute to chromosome congression
-
•
Extreme fibrous coronas in oocytes drive chromatin-mediated microtubule nucleation
Wu et al. show that microtubule nucleation from kinetochores in mitosis is driven by fibrous coronas, the transient expansions of outer kinetochores, and that this is mediated by the centrosomal protein pericentrin. Microtubule nucleation from fibrous coronas promotes chromosome congression, which is especially important in acentrosomal oocytes.
Introduction
Error-free chromosome segregation is critical for tissue homeostasis and early human development. Segregation occurs by spindle microtubules, which connect to chromosomes via kinetochores. Prior to segregation, these kinetochore-spindle interactions position the chromosomes at the spindle equator, a process called chromosome congression.1,2,3,4 Centrosomes serve as an important source of microtubules in mitosis and, as such, are a major microtubule-organizing center (MTOC) during cell division.5,6,7,8 However, many studies, including pioneering work from the 1970s and 1980s, showed that chromatin and kinetochores can also promote microtubule growth.9,10,11,12,13,14,15,16,17 Centromeric chromatin and the associated kinetochores appear to be the dominant site of chromosome-driven microtubule nucleation.18 Moreover, most microtubule fibers connected to kinetochores (k-fibers) seem to be generated from non-centrosomal microtubules nucleated near kinetochores.19
The origins of the kinetochore-derived microtubules are unclear, but there is an important role for the small GTPase Ran. Chromatin-localized RCC1 is a guanine nucleotide exchange factor that generates GTP-bound Ran. RanGTP in turn releases the microtubule nucleation factor TPX2 from importin-α and further promotes microtubule nucleation by the Nup107-160 complex (including ELYS/MEL-28), which binds the γ-tubulin ring complex (γ-TuRC).16,20,21,22,23,24,25 As a result, RanGTP promotes spindle assembly around chromatin and is essential for kinetochore-driven microtubule growth.26 Recent studies also indicate a role for augmin, a protein complex that triggers branching microtubule nucleation in conjunction with TPX2.27 Augmin is recruited to kinetochores by the Ndc80 complex in Drosophila S2 cells28 and promotes k-fiber maturation in Indian muntjac cells.29
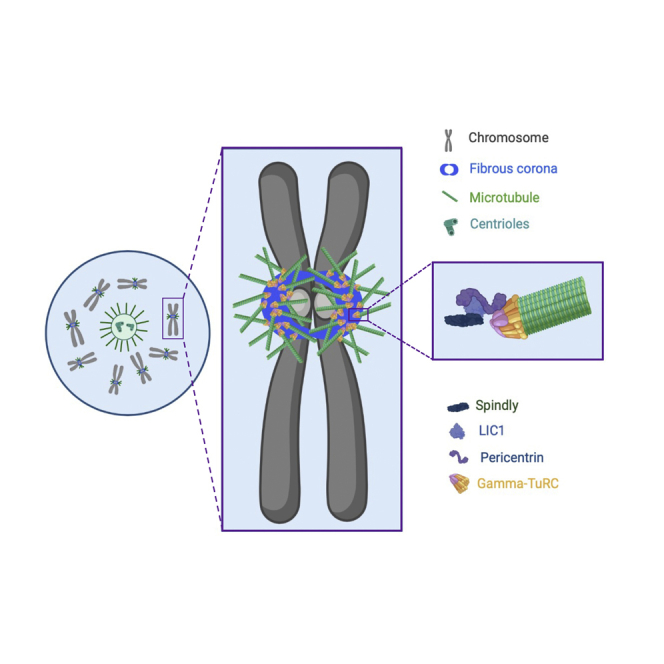
Early electron microscopy studies on cells and isolated chromosomes allowed to recover from microtubule poisons showed that nascent short microtubules were attached to the external surface of outer kinetochores known as the fibrous corona,14,30 suggesting that this is the main nucleation site for chromosome-derived microtubule assembly. The fibrous corona is a transient fibrous meshwork that expands from the outer kinetochore from late prophase onward and is removed by end-on kinetochore-microtubule interactions.31,32 The fibrous corona is generated by the oligomerization of ROD-ZW10-ZWILCH-Spindly (RZZS) complexes that form the core corona scaffold.33,34,35,36,37 This scaffold enables the recruitment of other proteins and protein complexes involved in, for example, the spindle assembly checkpoint (MAD1-MAD2-p31comet), microtubule motor activity (CENP-E, dynein/dynactin), and microtubule dynamics regulation (CLASPs).31 As such, the fibrous corona has important functions in initial microtubule capture, lateral-to-end-on conversion and subsequent stability of kinetochore-microtubule attachments, spindle assembly checkpoint strength, and eventual chromosome biorientation.32,35,38,39,40,41,42,43,44,45,46,47,48
In our quest to understand the contributions of the fibrous corona to chromosome segregation, we report here that we found it to be both required and sufficient for kinetochore-driven microtubule nucleation. We provide new insights into the molecular mechanisms of microtubule nucleation from the fibrous corona and show that it is required for efficient chromosome congression. We show that the fibrous corona is particularly prominent in oocytes, which rely heavily on chromatin-mediated spindle assembly.15,49,50
Results
Kinetochore-derived microtubules form at fibrous coronas
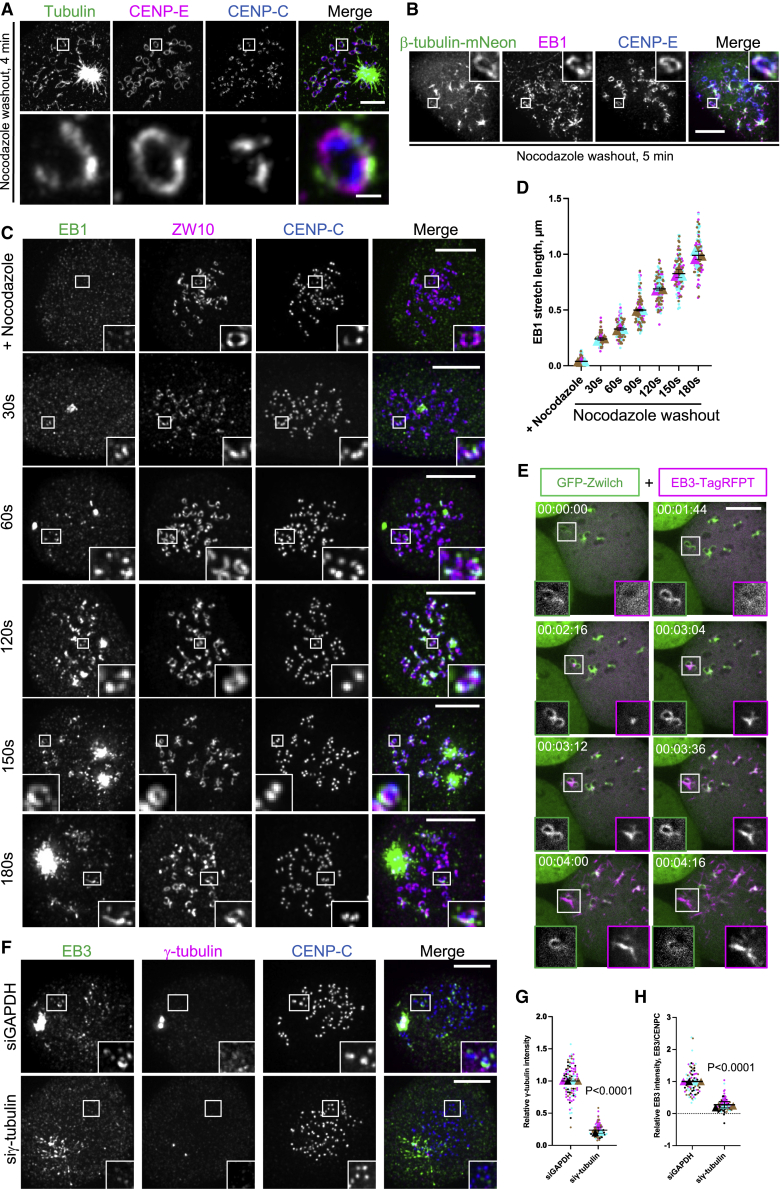
To examine the contribution of the fibrous corona to spindle assembly, chromosome-microtubule interactions, and chromosome segregation, we used a nocodazole-washout assay in which mitotic human RPE1 cells were examined for microtubule outgrowth after washout of the microtubule depolymerizing drug nocodazole (Figure S1A). Super resolution imaging using expansion microscopy (ExM)51 4 min after nocodazole washout showed that newly nucleated microtubules localized to the fibrous corona, marked by CENP-E35,52 (Figure 1A). To enable visualization of microtubules at earlier time points, we imaged end-binding protein 1 (EB1), a more sensitive marker for microtubule growth, which co-localized with tubulin and the fibrous corona (Figures 1B and S1B). EB1 was absent from fibrous coronas in nocodazole-arrested cells (Figures 1C and S1C). As soon as 30 s after washout of nocodazole, EB1 appeared in discrete dots that co-localized with centrosomes and several fibrous coronas, marked by ZW10 (Figure 1C). EB1 patches proximal to fibrous coronas had an average length of ∼0.2 μm (Figure 1D). After 1 min of recovery from nocodazole, short EB1-marked microtubules (∼0.3 μm in length) were prominent at both ends of the fibrous corona crescents (Figures 1C and 1D). With longer recovery time (1.5, 2, and 2.5 min), we observed that EB1-positive structures increased in length and expanded along the fibrous coronas. After 3 min of recovery, longer EB1 structures (∼1 μm) protruded out from the fibrous coronas (Figures 1C and 1D). Similar microtubule outgrowth dynamics were seen by live imaging of RPE1 cells co-expressing GFP-Zwilch and EB3-TagRFPT (Figure 1E; Video S1). Microtubule outgrowth from fibrous coronas was also observed in other cell types, such as human foreskin BJ fibroblasts and primary human umbilical vein endothelial cells (HUVECs) (Figure S1D). As expected,5,6,7,53 microtubule nucleation at the fibrous corona was dependent on γ-tubulin (Figures 1F–1H) and Plk1 (but not Mps1 or Aurora B) kinase activity (Figures S1E and S1F). Furthermore, and in agreement with earlier findings,20,26,54 microtubule outgrowth from fibrous coronas required an active Ran-TPX2 pathway (Figures S1G–S1L). We conclude that kinetochore-derived microtubules form at fibrous coronas (Figure S1M).
Figure 1.
Fibrous coronas promote microtubule growth
(A) Super resolution (ExM) images of RPE1 cells immunostained with tubulin (green), CENP-E (magenta), and CENP-C (blue) after 4 min recovery from nocodazole washout. Scale bars, 1 μm.
(B) Immunostaining of EB1 (magenta), CENP-E (blue), and mNeon (green) in RPE1βtub-mNeon cells 5 min after recovery from nocodazole washout. Scale bars, 5 μm.
(C) Immunostaining of EB1 (green), ZW10 (magenta), and CENP-C (blue) in RPE1 cells after recovery from nocodazole washout for indicated times. Scale bars, 5 μm.
(D) Quantification of the length of EB1 stretches on fibrous coronas in RPE1 cells from experiments, as depicted in (C). The error bars are standard deviations.
(E) Stills from live imaging of RPE1 cells co-expressing GFP-Zwilch and EB3-TagRFPT after nocodazole washout. Scale bars, 5 μm.
(F) Immunostaining of EB3 (green), γ-tubulin (magenta), and CENP-C (blue) in control and γ-tubulin-knockdown RPE1 cells 3 min after recovery from nocodazole washout. Scale bars, 5 μm.
(G) Relative γ-tubulin intensity at the centrosome in control and γ-tubulin-knockdown RPE1 cells after recovery of 3 min and 30 s from nocodazole washout. Wilcoxon signed-rank test.
(H) Relative EB3 intensity at kinetochores in control and γ-tubulin-knockdown RPE1 cells 3.5 min after nocodazole washout. p values were obtained from Wilcoxon signed-rank test.
Live imaging of RPE1 cells co-expressing GFP-Zwilch and EB3-TagRFPT before, during and after nocodazole washout.
Fibrous coronas are necessary and sufficient for kinetochore-dependent microtubule nucleation
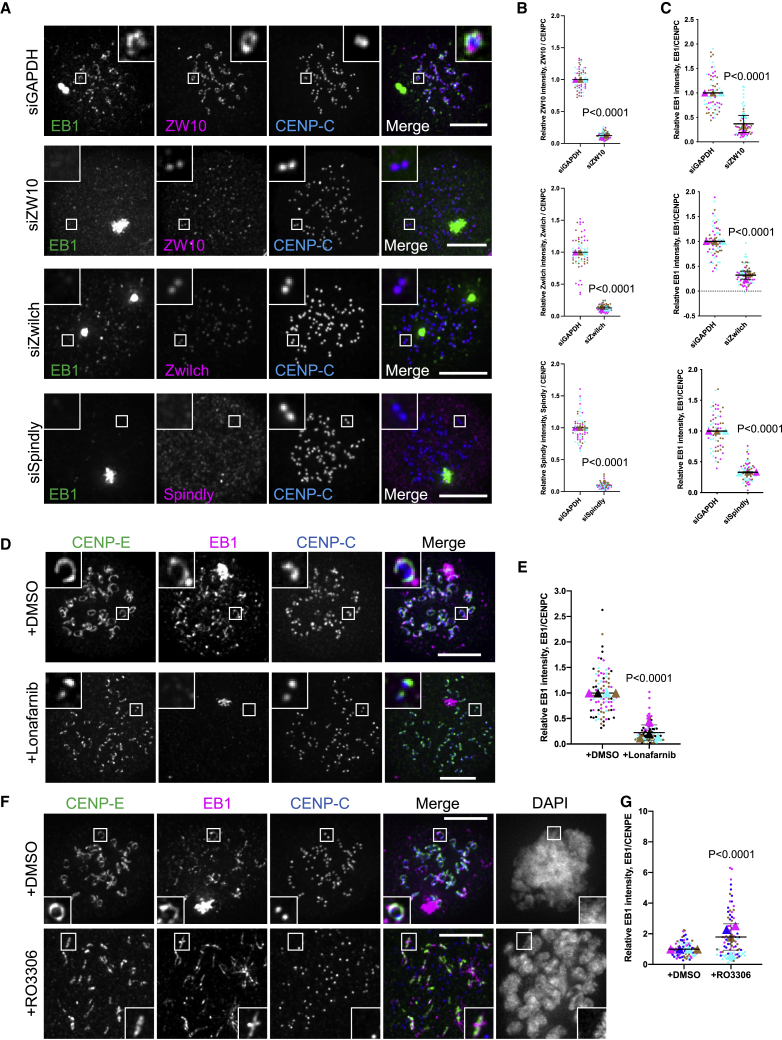
Having observed microtubule nucleation at or close to fibrous coronas, we next asked whether the fibrous coronas were required for this. Preventing fibrous corona assembly by depletion of Spindly or members of the ROD-ZWILCH-ZW10 (RZZ) complex, or by treatment with the farnesyl transferase inhibitor lonafarnib,34,35,40 indeed abolished microtubule nucleation near kinetochores but not centrosomes (Figures 2A–2E). To examine whether fibrous coronas can act as autonomous microtubule nucleation platforms, we performed the nocodazole-washout assay in cells treated briefly with the CDK1 inhibitor RO3306, which causes detachment of fibrous coronas from the core kinetochores.34,35 Strikingly, the crescent or rod-like fibrous corona structures that were no longer connected to the underlying kinetochores maintained a strong capacity to nucleate microtubules (Figures 2F, 2G, S2A, and S2B). Together, these findings show that fibrous coronas provide the predominant microtubule nucleation activity at mitotic kinetochores and can do so independent of connections to the core kinetochore.
Figure 2.
Fibrous coronas are necessary and sufficient for microtubule nucleation
(A) Immunostaining of EB1 (green), ZW10 (magenta)/Zwilch (magenta)/Spindly (magenta), and CENP-C (blue) in control/ZW10-depleted/Zwilch-depleted and Spindly depleted RPE1 cells after recovery of 3 min from nocodazole washout. Scale bars, 5 μm.
(B) Relative ZW10/Zwilch/Spindly intensity at the kinetochore compared with CENP-C in control (siGAPDH) and ZW10-depleted (siZW10), Zwilch-depleted (si-Zwilch), or Spindly depleted (siSpindly) RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
(C) Relative EB1 intensity at the kinetochore compared with CENP-C in control (siGAPDH) and ZW10-depleted (siZW10), Zwilch-depleted (si-Zwilch), or Spindly depleted (siSpindly) RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
(D) Immunostaining of CENP-E (green), EB1 (magenta), and CENP-C (blue) in DMSO- and lonafarnib-treated RPE1 cells after recovery of 3 min from nocodazole washout. Scale bars, 5 μm.
(E) Relative EB1 intensity at the kinetochore compared with CENP-C in DMSO- and lonafarnib-treated RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
(F) Immunostaining of CENP-E (green), EB1 (magenta), and CENP-C (blue) in DMSO- and RO3306-treated RPE1 cells after recovery of 3 min from nocodazole washout. Scale bars, 5 μm.
(G) Relative EB1 intensity at the fibrous corona compared with CENP-E in DMSO- and RO3306-treated RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
Related to Figure S2.
Kinetochore-derived microtubule growth depends on corona-localized dynein LIC1
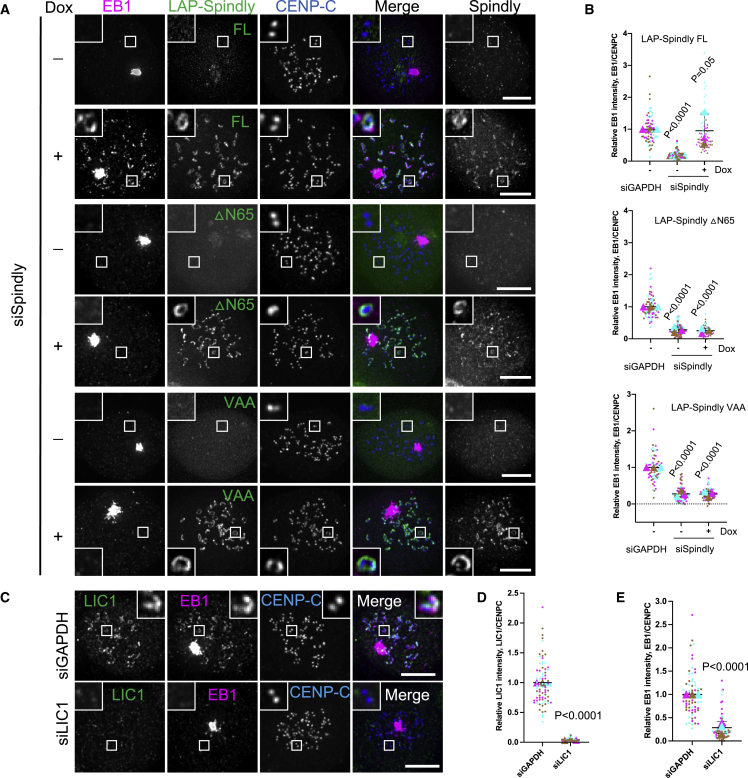
After having established a critical role for fibrous coronas in nucleating kinetochore-derived microtubules, we next wished to examine the mechanistic basis for this. Although RNAi was not maximally penetrant, substantial depletion of members of the Nup107-160 complex, previously proposed to drive kinetochore-derived microtubule nucleation in HeLa cells,20 did not affect fibrous-corona-proximal microtubule nucleation in RPE1 cells (Figures S2C–S2E). Consistently, the robust microtubule nucleation seen on detached fibrous coronas (after CDK1 inhibitor treatment) occurred in absence of detectable Nup133 (Figures S2F–S2J). In search of the mechanism by which fibrous coronas promote microtubule nucleation, we noticed that the expression of a SpindlyΔN mutant prevented fibrous-corona-dependent microtubule nucleation (Figures 3A and 3B). SpindlyΔN bypasses regulatory mechanisms of fibrous corona assembly35 and is deficient in binding the dynein/dynactin complex.35,47 Strikingly, point mutations that specifically perturb the interaction of Spindly with dynein/dynactin (A24V/Y60A/F258A [VAA]),35 while allowing full fibrous corona expansion (Figures S3A and S3B), abolished microtubule nucleation at the fibrous corona (Figures 3A and 3B). Recent work showed that the N-terminal CC1 box motif in Spindly, absent in both SpindlyΔN and SpindlyVAA, interacts directly with the light intermediate chain 1 (LIC1) subunit of dynein.47 In agreement with this, Spindly depleted cells expressing SpindlyΔN or SpindlyVAA failed to localize LIC1 to fibrous coronas (Figures S3C and S3D). Importantly, fibrous coronas could not nucleate microtubules in cells depleted of LIC1 (Figures 3C–3E). LIC1 depletion did not affect centrosomal microtubule nucleation, fibrous corona assembly, or localization of the dynein-dynactin complex to fibrous coronas (Figures S3E–S3K). Spindly bound LIC1 is therefore likely to have a more direct role in driving microtubule nucleation from fibrous coronas.
Figure 3.
Fibrous-corona-derived microtubule nucleation depends on dynein LIC1
(A) Immunostaining of EB1 (magenta), Spindly (gray), and CENP-C (blue) in Spindly knockdown RPE1 cells that were non-induced (−Dox) and induced (+Dox) to overexpress the indicated Spindly mutant after recovery of 3 min from nocodazole washout. Scale bars, 5 μm.
(B) Relative EB1 intensity at the kinetochore compared with CENP-C in control and Spindly depleted RPE1 cells that were non-induced (−Dox) and induced (+Dox) to overexpress the indicated Spindly mutant after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
(C) Immunostaining of LIC1 (green), EB1 (magenta), and CENP-C (blue) in control and LIC1-depleted RPE1 cells after recovery of 3 min from nocodazole washout. Scale bars, 5 μm.
(D) Relative LIC1 intensity at the kinetochore compared with CENP-C in control and LIC1-depleted RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
(E) Relative EB1 intensity at the kinetochore compared with CENP-C in control and LIC1-depleted RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
Related to Figure S3.
Fibrous coronas promote microtubule nucleation through a LIC1-PCNT interaction
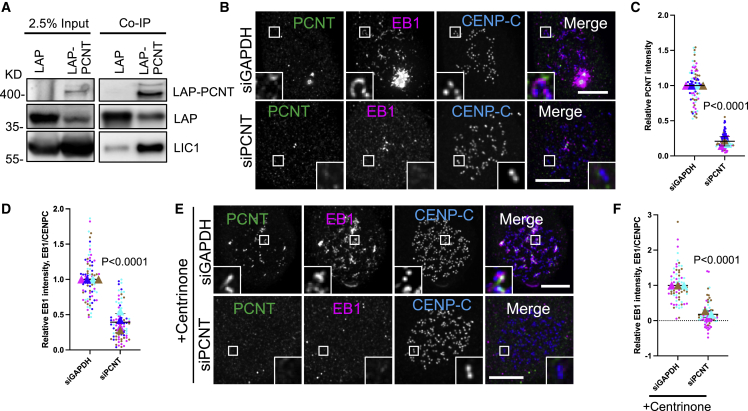
Through its role in adaptor protein engagement, LIC1 has an important contribution to microtubule binding and the motor activity of dynein.55 DHC1 depletion, however, had no significant impact on LIC1 localization to, or microtubule nucleation from, fibrous coronas (Figures S3L–S3P). Whether LIC1 performs its role in nucleation in the context of the dynein complex or not remains to be examined, but this shows that LIC1’s role is unrelated to its contributions to dynein motor function. Prior work had reported that LIC1 interacts with the γ-tubulin-tethering protein pericentrin (PCNT) and that dynein can be displaced from kinetochores by PCNT overexpression.56,57 Co-immunoprecipitation experiments indeed verified that LIC1 and PCNT interact in mitotic HeLa cells (Figure 4A). Because PCNT binds to γ-TuRC and is essential for microtubule nucleation from centrosomes in mitosis,58,59,60 we next tested whether it also mediates nucleation from fibrous coronas. Strikingly, depletion of PCNT abolished microtubule nucleation from fibrous coronas (Figures 4B–4D). Depletion of CDK5RAP2, which binds PCNT in the pericentriolar material and also binds γ-TuRC,61,62 had no effect (Figures S4A–S4C). To examine whether PCNT’s impact could be indirect through its role at centrosomes, we generated centrosome-less RPE1 cells by depleting centrioles with the PLK4 inhibitor centrinone63 (Figures S4D and S4E). Interestingly, fibrous-corona-based microtubule nucleation was substantially enhanced in the absence of centrosomes (Figures S4F and S4G), possibly due to the increased availability of nucleation factors at fibrous coronas. The size of fibrous coronas was not affected by the absence of centrosomes (Figure S4H). In these acentrosomal cells, all microtubule nucleation from fibrous coronas was fully dependent on TPX2 (Figures S4I and S4J) and, more importantly, on PCNT (Figures 4E and 4F).
Figure 4.
Fibrous coronas promote microtubule nucleation through a LIC1-PCNT interaction
(A) GFP-trap co-immunoprecipitation assays performed with the extracts of HeLa cells co-expressing LAP-PCNT or LAP, together with LIC1, and analyzed by western blotting with anti-GFP (upper panels) or anti-LIC1 antibodies (lower panel).
(B) Immunostaining of PCNT (green), EB1 (magenta), and CENP-C (blue) in control and PCNT-depleted RPE1 cells after recovery of 3 min from nocodazole washout. Scale bars, 5 μm.
(C) Relative PCNT intensity at the centrosome in control and PCNT-knockdown RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
(D) Relative EB1 intensity at the kinetochore compared with CENP-C in control and PCNT-knockdown RPE1 cells after recovery of 3 min from nocodazole washout. Wilcoxon signed-rank test.
(E) Immunostaining of PCNT (green), EB1 (magenta), and CENP-C (blue) in control and PCNT-depleted p53-null RPE1 cells treated with centrinone after recovery of 2.5 min from nocodazole washout. Scale bars, 5 μm.
(F) Relative EB1 intensity at the kinetochore compared with CENP-C in control and PCNT-depleted p53-null RPE1 cells treated with centrinone after recovery of 2.5 min from nocodazole washout. Wilcoxon signed-rank test.
Related to Figure S4.
Although we identified a LIC1-PCNT interaction and showed a key, centrosome-independent role for PCNT in fibrous-corona-derived microtubule nucleation, we were unable to visualize PCNT on fibrous coronas. Notably, however, we observed that upon depletion of centrioles, PCNT and γ-TuRC formed large clusters that tended to associate with the fibrous coronas (Figures S4K and S4L) in a manner dependent on PCNT and LIC1 (Figures S4L–S4N). Although the origins and relevance of such clusters are unclear, these data suggest that PCNT can bind LIC1 at fibrous coronas in cells but may normally be present there in relatively low abundance. Together, our findings support the hypothesis that PCNT directly promotes microtubule nucleation from the fibrous corona through its interaction with LIC1.
Fibrous-corona-derived microtubule nucleation promotes efficient chromosome congression
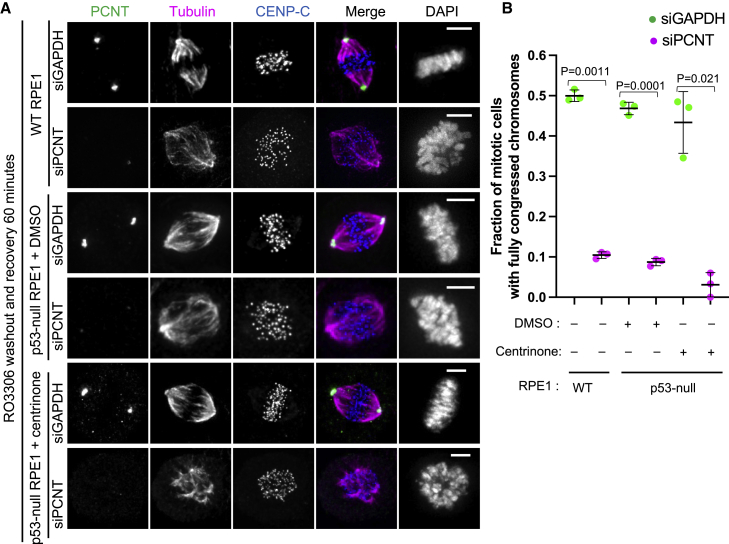
Kinetochores have long been known to drive the formation of microtubules, but their exact contribution to spindle assembly and chromosome segregation has thus far remained enigmatic. Our finding that LIC1-PCNT at fibrous coronas is primarily responsible for microtubule outgrowth from kinetochores enabled us to examine the role of kinetochore-derived microtubules in spindle assembly. Cells subjected to longer recovery time after nocodazole washout displayed various EB1-positive asters that co-localized with fibrous coronas and organized kinetochores into multiple clusters (Figure S5A). Kinetochore clustering depended on the fibrous corona and, in acentrosomal cells, on PCNT (Figures S5A and S5B), suggesting that it resulted from fibrous-corona-dependent microtubule nucleation, likely complemented by dynein activity. To further examine how microtubule nucleation impacts chromosome congression, we first assessed the congression dynamics in RPE1 cells treated with lonafarnib, which largely prevents microtubule nucleation from poorly expanded fibrous coronas (Figures 2D and 2E). Compared with DMSO controls, lonafarnib treatment doubled the time needed for chromosomes to align on the metaphase plate after nuclear envelope breakdown (NEBD) (Figures S5C and S5D; Videos S2 and S3). To perturb nucleation more specifically without affecting the fibrous corona structure and its associated proteins, we next depleted PCNT and assessed chromosome congression following release from a RO3306-induced G2 block or from a nocodazole-induced prometaphase block. These block-and-release approaches allowed us to assess the timing of congression by immuno-imaging of cells verified to be depleted of PCNT. In both approaches, PCNT depletion strongly affected chromosome congression (Figures 5A, 5B, S5E, and S5F). More importantly, PCNT depletion almost completely prevented chromosome congression in centrinone-treated (acentrosomal) RPE1 cells (Figures 5A, 5B, S5E, and S5F) in which the vast majority of nucleation takes place at kinetochores (Figures 4E and 4F). The expression of inactive Ran likewise inhibited congression64 (Figures S5G and S5H). PCNT’s role was unrelated to potential PCNT pools at the Golgi apparatus or at acentriolar PCM (Figures S5I–S5N). We conclude that our data are consistent with a model in which abolishing microtubule nucleation from fibrous coronas compromises chromosome congression.
Figure 5.
Disruption of microtubule nucleation from fibrous coronas delays chromosome congression
(A) Immunostaining of PCNT (green), tubulin (magenta), and CENP-C (blue) in RPE1 cells with indicated conditions after RO3306 washout and recovery for 60 min in medium containing MG132. Scale bars, 5 μm.
(B) Fraction of mitotic cells with fully congressed chromosomes of RPE1 cells with indicated conditions after RO3306 washout and recovery for 60 min in medium containing MG132. Student’s t test. A centrosomal RPE1 cell without misaligned chromosomes and with a length-to-width ratio of chromosome region (DAPI) > 3 is regarded as having fully congressed chromosomes. An acentrosomal RPE1 cell without misaligned chromosomes and with a length-to-width ratio of chromosome region (DAPI) > 2 is regarded as having fully congressed chromosomes.
Live imaging of MAD1-mNeon KI RPE1 cells over-expressing CENPA-mCherry in the presence of DMSO.
Live imaging of MAD1-mNeon KI RPE1 cells over-expressing CENPA-mCherry in the presence of 5uM lonafarnib.
Bovine oocytes have large fibrous coronas that are critical for spindle assembly
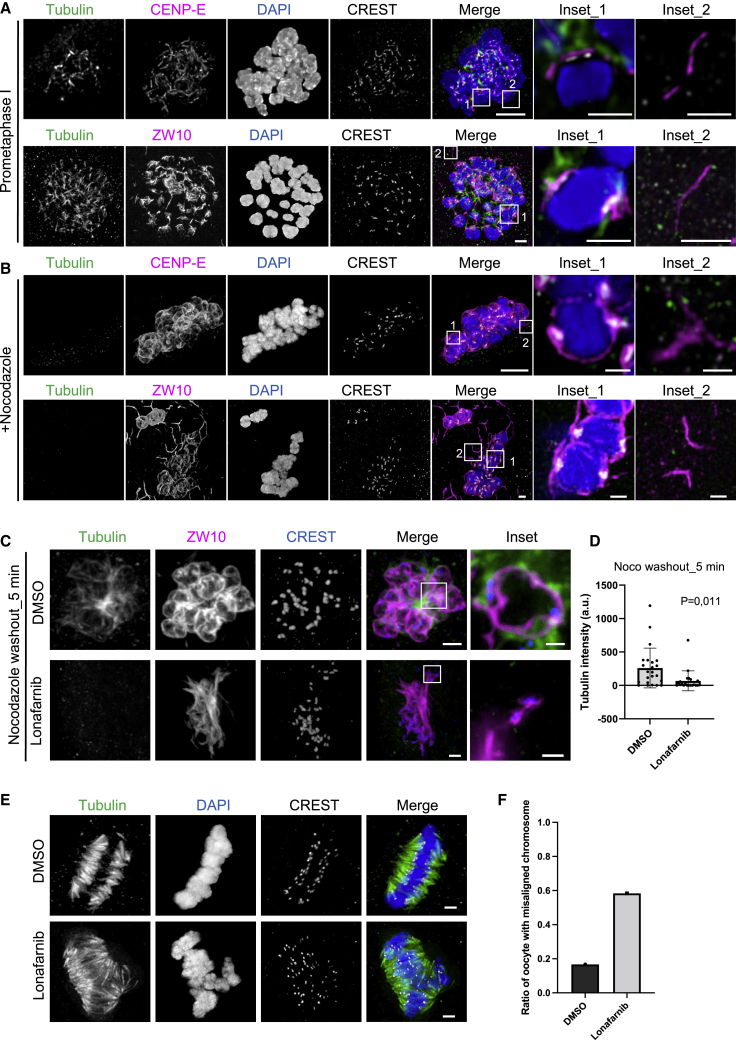
As suggested by our findings in centrinone-treated RPE1 cells, fibrous-corona-derived microtubule formation might be especially relevant in acentrosomal cells. Meiotic divisions in oocytes, which lack centrosomes, rely strongly on microtubule formation from chromosomes.49 Bovine oocytes show mechanisms of spindle assembly that are very similar to such mechanisms in humans,65 and the rate of aneuploidy in bovine oocytes closely matches that of human counterparts.66,67,68 Immunostaining of CENP-E and ZW10 in bovine prometaphase I oocytes showed large, expanded fibrous coronas that covered approximately two-thirds of the bivalent chromosomes (Figure 6A). Fibrous corona expansion was even more pronounced after the addition of nocodazole, when it completely enveloped the bivalent chromosomes (Figure 6B). Of note, in both prometaphase I and nocodazole-treated oocytes, filamentous structures positive for ZW10 and CENP-E could be observed in the cytoplasm (Figures 6A and 6B). Similar to RPE1 cells, washout of nocodazole resulted in prominent microtubule growth in chromosome-proximal locations that co-localized with fibrous coronas but not with the cytoplasmic filament, and this growth was abolished when fibrous corona expansion was prevented by treatment with lonafarnib (Figures 6C, 6D, and S6A–S6D). Oocytes, matured in the presence of lonafarnib, showed severe chromosome misalignment and spindle multipolarity (Figures 6E and 6F). Together, these data show that fibrous coronas in mammalian oocytes substantially contribute to chromatin-derived microtubule nucleation and proper chromosome alignment on the meiotic metaphase plate.
Figure 6.
Extremely expanded fibrous coronas in bovine oocytes promote chromosomal stability
(A) Immunofluorescence staining of bovine oocytes fixed at 13–14 h after IVM. Insets show maximum projection of a single bivalent (inset_1) and corona component filaments independent from chromatin localization (inset_2), where no microtubule nucleation is observed from these filaments. Max intensity projection of confocal sections are shown for tubulin (green), CENP-E/ZW10 (magenta), chromatin (blue), and CREST (gray). Scale bars, 2 μm; inset scale bars, 1 μm.
(B) Immunofluorescence staining of bovine oocytes fixed after incubation with 5 μM nocodazole for 4 h (starting at 13 h of IVM). Insets show maximum projection of a single bivalent (inset_1) and corona component filaments independent from chromatin localization (inset_2). Max intensity projection of confocal sections are shown for tubulin (green), CENP-E/ZW10 (magenta), chromatin (blue), and CREST (gray). Scale bars, 2 μm; inset scale bars, 1 μm.
(C) Immunofluorescence staining of bovine oocytes fixed 5 min after washout from incubation at 5 μM nocodazole for 4 h (starting at 13 h of IVM) in the absence or presence of farnesylation inhibitor lonafarnib. Insets show maximum projection of a single bivalent. Confocal sections are shown for tubulin (green), CENP-E/ZW10 (magenta), and CREST (blue). Scale bars, 2 μm; inset scale bars, 1 μm.
(D) Tubulin intensity at the chromosomes in control and lonafarnib-treated bovine oocytes after recovery of 5 min from nocodazole washout. Student’s t test.
(E) Meiotic spindle immunofluorescent staining of bovine oocytes in the presence and absence of lonafarnib. Max intensity projection of confocal sections are shown for tubulin (green), chromatin (blue), and CREST (gray). Scale bars, 2 μm.
(F) Ratio of bovine oocyte with misaligned chromosome in the presence and absence of lonafarnib. Data from 6 experiments.
Related to Figure S6.
Discussion
Our data show that microtubule nucleation at or close to the kinetochores relies on the fibrous corona. Nucleation from the fibrous corona is driven by LIC1, which binds PCNT and thereby promotes microtubule nucleation in a γ-TuRC-dependent manner. Fibrous-corona-derived microtubules promote efficient chromosome congression, which we show is particularly important in cells devoid of centrioles, such as oocytes.
Recent studies have shown that kinetochores are responsible for the generation of the majority of microtubules that derive from mitotic chromatin and that these constitute a major part of the microtubules for k-fiber formation during spindle assembly.18,19 They can be seen in early prometaphase as short microtubule stubs with lateral and end-on interactions with the kinetochore. Our findings show that these microtubules are generated by the fibrous corona. This is consistent with the early electron microscopy observations of mouse and CHO cells undergoing microtubule growth after washout of spindle poisons, and of isolated chromosomes incubated with tubulin.14,30 Nucleation from the fibrous corona rather than the core kinetochore may help restrict it to the early phases of mitosis, when interactions between kinetochores and the spindle are formed. Fibrous corona stripping by end-on attached microtubules then couples the presence of such attachments with the halting of kinetochore-derived microtubule formation at a time when this is no longer required.
Whether kinetochores can themselves drive microtubule nucleation or recruit microtubules pre-formed in their vicinity has remained largely unresolved. Our findings suggest that nucleation can occur directly at the fibrous corona: LIC1 is a bona fide fibrous corona component required for the formation of kinetochore-derived microtubules and interacts with the known γ-TuRC recruiter PCNT (this study; Purohit et al.56; Tynan et al.57). Consistently, we show that PCNT is essential for microtubule nucleation from fibrous coronas. This process further requires the RanGTP-TPX2 pathway, as expected from prior studies,21,26 possibly to stabilize nascent microtubules25 generated by LIC1-PCNT-γ-TuRC. RanGTP has been observed on unattached kinetochores in U2OS cells,26 raising the possibility that it too is a fibrous corona component. Nevertheless, we were unable to detect fibrous-corona-localized pools of γ-TuRC or PCNT, indicating that low levels of the LIC1-PCNT-γ-TuRC module at fibrous coronas may be sufficient to drive substantial local microtubule growth. We cannot exclude, however, that the entire process, including LIC1’s contribution, occurs in the cytoplasm and that the fibrous corona rapidly binds and concentrates nascent microtubule stubs, for example, via CENP-E, dynein, or other MAPs.19,31 Future studies on this will need to focus on the mechanistic details of the LIC1-PCNT interaction and the regulation thereof, and the application of sensitive detection methods to probe the presence and activity of nucleation factors on fibrous coronas. Kinetochores may additionally promote microtubule growth and k-fiber formation beyond initial nucleation. Augmin interacts with the Ndc80 complex of the core kinetochore in Drosophila S2 cells28 and was shown to promote microtubule amplification for k-fiber self-organization and maturation in Indian muntjac cells.29 Kinetochores can also generate microtubules in budding yeast, which do not make fibrous coronas for their lack of RZZS.69 They do so in a manner dependent on Stu2/XMAP215/chTOG,70 and it will be interesting to examine whether this pathway also contributes to k-fiber formation in animal cells.
Despite many reports of kinetochore-derived microtubule nucleation in cells of various species, their contribution to spindle assembly and chromosome segregation fidelity has remained unclear. Previous studies showed that kinetochore-derived microtubules are incorporated into the spindle by interaction with the lattice of astral microtubules, after which end-on interactions between kinetochores and spindle microtubules can be obtained.17,70 In animal cells, the kinetochore-derived microtubules were shown to contribute the majority of microtubules in k-fibers, suggesting they are crucial for chromosome biorientation and correct segregation. Although we were unable to specifically perturb the nucleation process from kinetochores in normal human somatic cells, we could do so in centriole-depleted cells by virtue of PCNT depletion. In the absence of centrosomes, PCNT’s contribution to spindle formation was almost exclusively restricted to fibrous coronas. PCNT depletion in such cells, although allowing the formation of a spindle-like structure, severely affected chromosome congression, verifying the substantial contribution of fibrous-corona-derived microtubules predicted. In-depth analysis of k-fiber formation in centriole-less cells now enables the probing of the mechanisms by which fibrous-corona-derived microtubules ensure chromosome congression.
The contribution of microtubule growth from fibrous coronas may be especially relevant in cells naturally devoid of centrosomes. Our observation that fibrous coronas in bovine oocytes are extremely expanded and can nucleate microtubules supports this hypothesis. It will be of great interest to understand the assembly and regulation of fibrous coronas in oocytes and probe, by specific perturbations, their significance for correct spindle assembly and chromosome congression in these cells. Interestingly, we observed robust fibrous-corona-derived microtubules in RPE1 cells, BJ fibroblasts, and primary HUVECs (Figure S1D), while they were quite limited in multiple cancer cell lines (data not shown). It will be of interest to examine the cause for this, as it may impact the fidelity of chromosome segregation in these cancer lines and thus contribute to their chromosomal instability phenotype.71
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea pig polyclonal anti-CENP-C | MBL International | Cat# PD030; RRID: AB_10693556 |
| Rabbit polyclonal anti-CENP-E | Brown et al.85 | N/A |
| Rabbit polyclonal anti-ZW10 | Abcam | Cat# ab21582; RRID: AB_779030 |
| Rabbit polyclonal anti-TPX2 | Novus | Cat# NB500-179; RRID: AB_10002747 |
| Rabbit polyclonal anti-Spindly | Bethyl | Cat# A301-354A; RRID: AB_937753 |
| Rabbit polyclonal anti-LIC1 | GeneTex | Cat# GTX120114; RRID: AB_10720534 |
| Rabbit polyclonal anti-pericentrin | Abcam | Cat# ab4448; RRID: AB_304461 |
| Rabbit polyclonal anti-CDK5RAP2 | Bethyl | Cat# A300-554A; RRID: AB_477974 |
| Rabbit polyclonal anti-centrin 1 | Abcam | Cat# ab101332; RRID: AB_10676086 |
| Rabbit polyclonal anti-NUP133 | Abcam | Cat# ab155990 |
| Mouse monoclonal anti-α-tubulin | Sigma-Aldrich | Cat# T9026; RRID: AB_477593 |
| Mouse monoclonal anti-EB1 | BD Biosciences | Cat# 610534; RRID: AB_397891 |
| Mouse monoclonal anti-γ-tubulin | Santa Cruz Biotechnology | Cat# sc-17787; RRID: AB_628417 |
| Mouse monoclonal anti-CENP-E | Abcam | Cat# ab5093; RRID: AB_304747 |
| Mouse monoclonal anti-AKAP450 | BD Biosciences | Cat# 611518; RRID: AB_398978 |
| Mouse monoclonal anti-DHC1 | Santa Cruz | Cat# sc-514579 |
| Mouse monoclonal anti- p150Glued | BD Biosciences | Cat# 612708; RRID: AB_399947 |
| Human polyclonal anti-centromere (CREST) | Antibodies Incorporated | Cat# 15-234-0001; RRID: AB_2687472 |
| Goat anti-guinea pig Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21450; RRID: AB_2735091 |
| Goat anti-rabbit Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11034; RRID: AB_2576217 |
| Goat anti-rabbit Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11036; RRID: AB_10563566 |
| Donkey anti-rabbit Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206, RRID: AB_2535792 |
| Goat anti-mouse Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11031, RRID: AB_144696 |
| Goat anti-rabbit Alexa Fluor 405 | Thermo Fisher Scientific | Cat# A-31556; RRID: AB_221605 |
| Goat anti-mouse Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21236; RRID: AB_2535805 |
| GFP-Booster Atto 488 | ChromoTek | Cat# gba488-100; RRID: AB_2631386 |
| Chemicals, peptides, and recombinant proteins | ||
| Hygromycin | Roche | Cat# 10843555001 |
| Fugene HD | Promega | Cat# E2311 |
| Puromycin | Sigma | Cat# P7255 |
| Nutlin-3a | Sigma | Cat# SML0580-5MG |
| Blasticidine | Gibco | Cat# A1113903 |
| HiPerFect | Qiagen | Cat# 301704 |
| Nocodazole | Sigma-Aldrich | Cat# M1404 |
| Doxycycline | Sigma-Aldrich | Cat# D9891 |
| MG132 | Sigma-Aldrich | Cat# C2211 |
| BI2536 | Advanced ChemBlocks | Cat# 10293 |
| ZM447439 | Tocris Bioscience | Cat# 2458 |
| Cpd5 | R. H. Medema lab, Netherlands Cancer Institute | N/A |
| Lonafarnib | Selleckchem | Cat# S2797 |
| RO3306 | Tocris Bioscience | Cat# 4181 |
| Centrinone | MedChemExpress | Cat# HY-18682 |
| Thymidine | Sigma-Aldrich | Cat# T1895 |
| GFP-trap beads | ChromoTek | Cat# M-270 |
| Experimental models: Cell lines | ||
| RPE1 FlpIn | J. Pines lab, The Institute of Cancer Research, UK | N/A |
| HeLa FlpIn | S. Taylor lab, University of Manchester | N/A |
| HEK 293T | Hubrecht Institute, the Netherlands | N/A |
| BJ fibroblasts | Hubrecht Institute, the Netherlands | N/A |
| HUVEC | A. Akhmanova lab, Utrecht University, the Netherlands | N/A |
| Bovine oocyte | This study | N/A |
| Oligonucleotides | ||
| siRNA targeting sequence: GAPDH | Dharmacon | D-001830-01-05 |
| siRNA targeting sequence: γ-tubulin (5’-GGAGGACAUGUUCAAGGA -3’)80 | Dharmacon | N/A |
| siRNA targeting sequence: TPX2 (5’-GAAUGGAACUGGAGGGCUU-3’)81 | Dharmacon | N/A |
| siRNA targeting sequence: ZW10 (5’-UGAUCAAUGUGCUGUUCAA-3’)35 | Dharmacon | N/A |
| siRNA targeting sequence: Zwilch (5’-UCUACAACGUGGUGAUAUA-3’)35 | Dharmacon | N/A |
| siRNA targeting sequence: Spindly (5’-GAAAGGGUCUCAAACUGAA-3’)35 | Dharmacon | N/A |
| siRNA targeting sequence: LIC1 (5’-AGAUGACAGUGUAGUUGUA-3’)82 | Dharmacon | N/A |
| siRNA targeting sequence: PCNT (5’-AAAAGCUCUGAUUUAUCAAAAGAAG-3’)83 | Dharmacon | N/A |
| siRNA targeting sequence: NUP133 (5’-GCCUAUCUGUAUAACGAAA-3’) | Dharmacon | N/A |
| siRNA targeting sequence: NUP160 (5’-GCCUUAACUUCCACGGAUA-3’) | Dharmacon | N/A |
| siRNA targeting sequence: DHC1 (5’-CGUACUCCCGUGAUUGAUG-3’)84 | Dharmacon | N/A |
| Recombinant DNA | ||
| pOG44 Flp-Recombinase expression vector | Invitrogen | Cat# V600520 |
| pcDNA5-LAP-Spindly FL | Sacristan et al.35 | N/A |
| pcDNA5-LAP-Spindly △N65 | Sacristan et al.35 | N/A |
| pcDNA5-LAP-Spindly A24V/Y60A/F258A | Sacristan et al.35 | N/A |
| pcDNA5-LAP-Ran T24N | This study | N/A |
| pcDNA5-LAP-Ran WT | This study | N/A |
| pcDNA5-LAP-PCNT | This study | N/A |
| pcDNA5-LAP-CENP-E | This study | N/A |
| EB3-TagRFPT-ires-GFP-Zwilch in pLVX-IRES-Puro vector | This study | N/A |
| LIC1 in pLVX-IRES-Puro vector | This study | N/A |
| pSpCas9(BB)-2A-Puro (PX459)- CDK5RAP2 (sgRNA 5’-AAGTCTGAAGCGGGAACTCC-3’) | This study | N/A |
| lentiCRISPR-TP53 (sgRNA 5’-CCGGTTCATGCCGCCCATGC-3’)75 | This study | N/A |
| pLenti6-CENP-A-mCherry | Addgene | Cat# 89767 |
| pSPgRNA | Addgene | Cat# 47108 |
| pSpCas9(BB)-2A-Puro (PX459)-GCP3 (5’-AGGACCGCGAGCTTCACGTG-3’)77 | This study | N/A |
| pSPgRNA- β-tubulin (5’-GAGGCCGAAGAGGAGGCCTAAGG-3’)77 | This study | N/A |
| pSPgRNA- MAD1L1 (5’-CGAGCCTGCAGGCTACGCCACGG-3’)77 | This study | N/A |
| pCAS9-mCherry-Frame +0 | Addgene | Cat# 66939 |
| pCAS9-mCherry-Frame +1 | Addgene | Cat# 66940 |
| pCAS9-mCherry-Frame +2 | Addgene | Cat# 66941 |
| mNeon-targeting plasmid79 | H. Clevers lab, Hubrecht Institute, the Netherlands | N/A |
| Software and algorithms | ||
| Fiji | ImageJ | https://imagej.net/software/fiji/downloads |
| Protein levels near the kinetochores analysis macro | Saurin et al.87/ This study. | https://github.com/Kopsgroup/Wu_etal/tree/main/KT-immuno_Intensity_Quanti |
| Kinetochore protein projection area analysis macro | This study | https://github.com/Kopsgroup/Wu_etal/tree/main/KT-immuno_projectionarea_Quanti |
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Huygens Professional 20.10 | Scientific Volume Imaging | https://svi.nl/Huygens-Professional |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Geert Kops (g.kops@hubrecht.eu).
Materials availability
Plasmids and cell lines generated in this study can be obtained through the lead contact.
Experimental model and subject details
Human cell lines
RPE1 FlpIn cells, HeLa FlpIn cells, HEK 293T cells and BJ fibroblasts were grown in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12 GlutaMAX; Gibco 10565018) supplemented with 9% tetracycline-free fetal bovine serum and 100 μg /ml penicillin-streptomycin (Sigma P0781). Human Umbilical Vein Endothelial Cells (HUVECs) were were a kind gift of Prof. A. Akhmanova (Utrecht University, the Netherlands) grown in endothelial basal medium (EGM-2) supplemented with growth supplements (SingleQuots, Lonza): 2% Fetal Bovine Serum (FBS), human Epidermal Growth Factor (hEGF), Vascular Endothelial Growth Factor (VEGF), R3- Insulin-like Growth Factor-1 (R3-IGF-1), Ascorbic Acid, Hydrocortisone human Fibroblast Growth Factor-Beta (hFGF-β), Heparin, Gentamicin/Amphotericin-B (GA). All cell lines were cultured at 37°C with 5% CO2.
Bovine oocyte
Bovine oocytes were collected from slaughterhouse ovaries as previously described transported to the laboratory within two hours at 37°C. Ovaries were washed upon arrival with physiological saline containing 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C. Cumulus-oocyte complexes (COCs) were aspirated by a 19-gauge needle connected to a vacuum pump from follicles with a diameter of 2-8 mm and collected in 50ml falcon tubes. The COCs were washed in Hepes-buffered M199 (22340-020; Gibco-BRL) and those containing a minimum of three layers of compact cumulus cells were selected and held in synthetic oviduct fluid (SOF) (H-SOF; Avantea) for 8 hours at room temperature. Subsequently, oocytes were washed and cultured in maturation medium (31100-027, NaHCO3-buffered M199 [11150059; Gibco-BRL] supplemented with 1% (vol/vol) penicillin-streptomycin [15140122; Gibco-BRL], 0. 02 IU/ml Follicle Stimulating Hormone [FSH; Sioux Biochemical], 0.02 IU/ml Luteinizing Hormone [LH; Sioux Biochemical), 7.7 μg/ml cysteamine [30070; Sigma-Aldrich), and 10 ng/ml epidermal growth factor [E4127; Sigma-Aldrich]) at 38.5°C and 5% CO2. To study the chromosome alignment during metaphase I, COCs were incubated in maturation medium for 15-16 hours. Before fixation or microtubule re-growth assay, bovine oocytes were vortexed for 3 minutes to remove cumulus cells.
Method details
Plasmids and cloning
LAP-Spindly FL, △N65, and A24V/Y60A/F258A were used previously.35 The LAP-Ran T24N was cloned based on the cDNA of Ran with T24N mutation (addgene 37396),72 and the LAP-Ran WT was cloned by PCR-based reversing the Ran T24N to wildtype. LAP-PCNT was cloned based on the pericentrin cDNA, which was a kind gift from Prof. A. Akhmanova (Utrecht University, the Netherlands). EB3-TagRFPT-ires-GFP-Zwilch cassette and LIC1 cDNA (addgene 74601)73 were cloned into the pLVX-IRES-Puro vector (Clontech). Cloning was performed by PCR-based Gibson assembly and restriction-ligation strategies.
The pSpCas9(BB)-2A-Puro (PX459) vector that was used for CDK5RAP2 knockout was purchased from Addgene (62988).74 The sgRNA of CDK5RAP2 (5’-AAGTCTGAAGCGGGAACTCC-3’) was cloned into pSpCas9(BB)-2A-Puro (PX459) to construct pSpCas9(BB)-2A-Puro (PX459)- CDK5RAP2. The sgRNA of TP53 (5’- CCGGTTCATGCCGCCCATGC-3’)75 was cloned in the lentiCRISPR v2 vector (addgene 52961)75 to construct lentiCRISPR-TP53 for making lentiviruses. The plasmid of pLenti6-CENP-A-mCherry was from addgene (89767).76
The sgRNAs used for making endogenously mNeon-tagged proteins are as follows:
GCP3 (5’-AGGACCGCGAGCTTCACGTG-3’),77 which was cloned into the pSpCas9(BB)-2A-Puro (Addgene, 62988)74;
β-tubulin (5’- GAGGCCGAAGAGGAGGCCTAAGG-3’)77;
MAD1L1 (5’-CGAGCCTGCAGGCTACGCCACGG),77 which were cloned into the pSPgRNA (Addgene, 47108).78 The frame-selector plasmids with a sgRNA that targets and linearizes the mNeon-targeting plasmid, the Cas9 and mCherry cassettes were from Addgene (66939, 66940 and 66941).77 The mNeon-targeting plasmid was a gift from Prof. Hans Clevers.79 RPE1 FlpIn cells were co-transfected with pSPgRNA containing β-tubulin sgRNA, mNeon-targeting plasmid and frame-selector #1 plasmid and sorted by mNeon signal.
Virus production
Viruses were produced by co-transfection of HEK 293T cells with the lentiviral vector containing gene of interest (EB3-TagRFPT-ires-GFP-Zwilch / LIC1/ CENPA-mCherry/ lentiCRISPR-TP53) and separate plasmids that express VSV-G, Tat, Rev and Gag-Pol. After 72 hours transfection, cell supernatant was harvested and filtered.
Cell lines generation
To generate stably integrated RPE1 FlpIn cell line with LAP-tagged gene of interest integrated in the FRT site and TetR inducible, RPE1 FlpIn were co-transfected with pCDNA5-constructs and construct of pOG44 recombinase in a 1:2 ratio with protocol U-017 of the electroporation machine (Amaxa Nucleofector II), and RPE1 FlpIn were selected with 100 μM hygromycin (Roche, 10843555001) for about 3 weeks. To generate stably integrated HeLa FlpIn cell line with LAP-tagged gene of interest integrated in the FRT site and TetR inducible, HeLa FlpIn were co-transfected with pCDNA5 constructs and construct of pOG44 recombinase in a 1:9 ratio with Fugene HD (Promega, E2311) following the manufacturer’s instructions. HeLa FlpIn were selected with 200 μM hygromycin (Roche, 10843555001) for about 3 weeks.
To generate RPE1 CDK5RAP2 KO cell lines, RPE1 FlpIn cells were transfected with pSpCas9(BB)-2A-Puro (PX459)- CDK5RAP2 and selected with puromycin. After selection, single cell clones were isolated by dilution into 96-well plates.
To generate RPE1 GCP3-mNeon, β-tubulin-mNeon and MAD1-mNeon knockin cells, RPE1 FlpIn cells were co-transfected with pSpCas9(BB)-2A-Puro (PX459)-GCP3 or pSPgRNA containing β-tubulin/MAD1L1 sgRNA, together with mNeon-targeting plasmid and frame-selector plasmid and sorted by mNeon signal. To generate RPE1 MAD1-mNeon stabling expressing CENP-A-mCherry cells, RPE1 MAD1-mNeon cells were infected with pLenti6-CENP-A-mCherry lentiviruses and selected with 5 μg/mL blasticidine (Gibco, A1113903) for 1 week.
To generate RPE1 β-tubulin-mNeon knockin cells, RPE1 FlpIn cells were co-transfected with pSPgRNA containing β-tubulin sgRNA, mNeon-targeting plasmid and frame-selector plasmid and sorted by mNeon signal.
To generate RPE1 FlpIn cell stably expressing EB3-TagRFPT-ires-GFP-Zwilch, cells were infected with lentiviruses and selected with 1 μg/mL puromycin (Sigma, P7255) for 1 week. To generate HeLa FlpIn cell stably expressing LIC1, cells were infected with lentiviruses and selected with 1 μg/ml puromycin for 1 week. To generate RPE1 p53 knockout cells, cells were infected with lentiviruses produced by lentiCRISPR-TP53 and selected with 10 μg/mL Nutlin-3a (Sigma, SML0580-5MG) for 2 weeks.
siRNAs transfection
The siRNAs targeting γ-tubulin (5’-GGAGGACAUGUUCAAGGA -3’),80 GAPDH (Dharmacon, D-001830-01-05), TPX2 (5’-GAAUGGAACUGGAGGGCUU-3’),81 ZW10 (5’-UGAUCAAUGUGCUGUUCAA-3’),35 Zwilch (5’-UCUACAACGUGGUGAUAUA-3’),35 Spindly (5’-GAAAGGGUCUCAAACUGAA-3’),35 LIC1 (5’-AGAUGACAGUGUAGUUGUA-3’),82 PCNT (5’- AAAAGCUCUGAUUUAUCAAAAGAAG-3’),83 Nup133 (5’-GCCUAUCUGUAUAACGAAA-3’), Nup160 (5’-GCCUUAACUUCCACGGAUA-3’), DHC1 (5’-CGUACUCCCGUGAUUGAUG-3’)84 were purchased from Dharmacon. RPE1 cells were transfected with siRNAs at 100 nM using the HiPerFect (Qiagen, 301704), according to manufacturer’s instructions.
Cell treatments
For nocodazole treatment and washout assay, 6.6 μM nocodazole (Sigma-Aldrich, M1404) was added to the culture medium for 15hrs before the fixation and washout. For cells under siRNA treatment, nocodazole was added 48hrs after siRNA transfection. For the rescue experiments, 1 μg/ml doxycycline (Sigma-Aldrich, D9891) was added 40hrs after siRNA transfection. For the nocodazole washout assay in the presence of indicated drugs: 5 μM MG132 (Sigma-Aldrich, C2211); 100 nM BI2536 (Advanced ChemBlocks, 10293); 250 nM Cpd5 (a kind gift from R. H. Medema, Netherlands Cancer Institute); 2 μM ZM447439 (Tocris Bioscience, 2458), cells were treated with indicated drugs for 4hrs before the nocodazole washout assay. In the experiments in which farnesyl transferase was inhibited, 5 μM lonafarnib (Selleckchem, S2797) was added together with nocodazole. For the nocodazole treatment or washout assay in which the CDK1 was inhibited, 10 μM RO3306 (Tocris Bioscience, 4181) was added to the cells 30mins before the fixation or washout assay. In the experiments in which centrosome duplication was inhibited, cells were treated with 250 nM centrinone (MedChemExpress, HY-18682) for 10 days. In the co-immunoprecipitation experiments, HeLa cells overexpressing LIC1 were treated with 1 μg/mL doxycycline and 2mM thymidine (Sigma-Aldrich, T1895) for 24hrs to induce the expression of LAP-PCNT or LAP, and then were released into 6.6 μM nocodazole for 8hrs before harvest. Harvest the cells by shaking off the mitotic cells. Wash the cells once with ice-cold PBS, and lysate the cells with lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, protease inhibitor, phosphatase inhibitor, 0.5 % Triton X-100) on ice for 10 min. After lysis, the samples were centrifuged at 14000rpm for 20 min at 4°C. The supernatants of the centrifuged samples were incubated with the GFP-trap beads (ChromoTek, M-270) for 1hr in cold room. Wash the beads with wash buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 0.1 % Triton X-100) 3 times. The samples were analysed by western blot.
Antibodies
We used Guinea pig polyclonal antibody against CENP-C (MBL, PD030), and rabbit polyclonal antibodies against CENP-E,85 ZW10 (Abcam, ab21582), EB3 (a gift from Prof. A. Akhmanova, Utrecht University, the Netherlands), TPX2 (Novus Biologicals, NB500-179), Zwilch (a gift from Prof. Andrea Musacchio, Max Planck Institute of Molecular Physiology), Spindly (Bethyl, A301-354A-1), LIC1 (Genetex, GTX120114), GFP ( custom, NT001), PCNT (Abcam, ab4448), NUP133 (Abcam, ab155990), CDK5RAP2 (Bethyl, A300-554A) and centrin1 (Abcam, ab101332). We used mouse monoclonal antibodies against α-tubulin (Sigma-Aldrich, T9026), EB1 (BD Biosciences, 610534), γ-tubulin (Santa Cruz, sc-17787), CENP-E (Abcam, ab5093), AKAP450 (BD Biosciences, 611518), Zwilch (a gift from Prof. Andrea Musacchio, Max Planck Institute of Molecular Physiology), DHC1 (Santa Cruz, sc-514579), p150Glued (BD Biosciences, 612708), and human antibody against Centromere (Antibodies Incorporated, 15-234-0001). We used the following secondary antibodies: Goat anti-guinea pig Alexa Fluor 647 (Thermo Fisher Scientific, A21450), Goat anti-rabbit Alexa Fluor 488 (Thermo Fisher Scientific, A11034), Goat anti-rabbit Alexa Fluor 568 (Thermo Fisher Scientific, A11036), Donkey anti-rabbit Alexa Fluor 488 (Thermo Fisher Scientific, A21206), Goat anti-mouse Alexa Fluor 568 (Thermo Fisher Scientific, A11031), Goat anti-mouse Alexa Fluor 647 (Thermo Fisher Scientific, A21236), Goat anti-rabbit Alexa Fluor 405 (Thermo Fisher Scientific, A-31556) and GFP-Booster Atto 488 (Chromotek, gba-488).
Immunofluorescence staining
Cells grown on 12 mm coverslip were fixed for 5min with -20 °C methanol. After fixation, coverslips were washed 3 times with PBS, followed by permeabilization with 0.1% triton X-100 in PBS for 2mins. After permeabilization, cells were washed 3 times with 0.05% Tween20 in PBS (PBST), and were blocked with 2% BSA in PBST for 30 mins. After blocking, cells were incubated with primary antibodies diluted in 2% BSA in PBST for 1hr at room temperature in humid condition. Subsequently, cells were washed three times with PBST and incubated with secondary antibodies and DAPI diluted in PBST for 1hour at room temperature, and followed by three times wash with PBST. At the end, coverslips were rinsed in 70% and 100% ethanol, air-dried and mounted on glass slides with Prolong Gold antifade.
Expansion Microscopy (ExM) used the protocol described previously.86 Cells are seeded and fixed and permeabilized with the same protocol described above. For the antibody incubation, a 4-fold excess of primary antibodies overnight at 4°C and 4-fold excess secondary antibodies 1hr at room temperature were used. After 3 times wash with PBST, samples were then fixed with 0.25% glutaraldehyde in PBS for 30 min, and washed 3 times with PBS. Cells were then incubated for 1 h in monomer solution (1× PBS, 2 M NaCl, 2.5% (wt/wt) acrylamide 0.15% (wt/wt) N, N′-methylenebisacrylamide 8.625% (wt/wt) sodium acrylate). Coverslips were placed on top of a drop of 97 μl of freshly prepared gelation solution (monomer solution supplemented with 0.2% (wt/wt) TEMED and 0.2% (wt/wt) APS) and incubated for 70 min at room temperature. Gels were then incubated in digestion solution (8 units/mL proteinase K, 1× TAE, 0.5% TX-100, 0.8 M guanidine HCl) for 2 hours at 37 °C, and expanded in Milli-Q water in petri dish overnight.
Image acquisition by microscopy
All images of non-expanded samples were acquired on a deconvolution system (DeltaVision Elite Applied Precision / GE Healthcare) with a 100X /1.40 NA or 40X /0.95 NA UPlanSApo objective (Olympus) using SoftWorx 6.0 software (Applied Precision/GE Healthcare). Images were acquired as z-stacks at 0.2 μm intervals for 32 stacks and deconvolved using SoftWoRx. The images are maximum intensity projections of deconvoluted stacks. Expanded samples were imaged on Zeiss LSM900 confocal microscope with Airyscan 2 module, using a HC PL APO CS2 63x/1.20 Water objective.
Drug treatments and microtubule re-growth assay in bovine oocytes
When microtubule depolymerization was needed, Nocodazole (1228, Tocris Bioscience, UK), at the final concentration of 13.3μM, was added to the maturation medium at 13 hours of in vitro maturation and the COCs were further incubated for 4 hours. To inhibit fibrous corona expansion, Lonafarnib (Selleckchem, S2797) at the final concentration of 5 μM was added to the maturation medium before COCs incubation.
For the microtubule regrowth assay bovine oocytes incubated in Nocodazole were washed three times for 1 minute and three times for 5 minutes in ice-cold maturation medium (with or without addition of Lonafarnib 5 μM). Immediately after, oocytes were transferred into warm maturation medium medium (38.5°C, 5% CO2, with or without addition of Lonafarnib 5 μM) for 2, 5 and 10 minutes to allow microtubule growth. Bovine oocytes were subsequently fixed and immunofluorescently stained.
Bovine oocyte fixation, immunofluorescent staining and confocal imaging
Before fixation bovine oocytes were pre-permeabilized for 5 seconds in 0.25% Triton X-100 (Sigma-Aldrich, Missouri, USA) and then fixed in 500μl of fixation medium (0.1M PIPES (Sigma-Aldrich, Missouri, USA), 0.1mM CaCl2 (Sigma-Aldrich, Missouri, USA), 1.0mM MgCl2 (Sigma-Aldrich, Missouri, USA) 0.1% Triton X-100 (Sigma-Aldrich, Missouri, USA), 1.0% paraformaldehyde (Electron Microscopy Sciences, Pennsylvania, USA) in MQ) for 30 minutes at room temperature. Bovine oocytes were then washed four times 5 minutes in PBS-T (PBS (B. Braun, Germany) supplemented with 0.1% Triton X-100 (Sigma-Aldrich, Missouri, USA)) and stored in PBS-T at 4°C overnight. All the following treatments were done within wells of ibidi μ-Slides (81501, μ-Slide Angiogenesis; ibidi) filled with 40 μl of solution per well. Oocytes were blocked in 5% normal goat serum and 3% BSA in PBS-T and then incubated with the primary antibodies in blocking solution overnight at 4°C. Thereafter, oocytes were washed in 3% BSA in PBS-T three times for 5 minutes and incubated with the secondary antibodies and fluorescent DNA stain for 3 hours in blocking solution. Finally, oocytes were washed three times for 10 minutes with 3% BSA in PBS-T and two times for 10 minutes with PBS and mounted on glass slides using anti-fade mounting medium (Vectashield; Vector Laboratories). Microtubules were stained with chicken anti-tubulin (ab89984, Abcam, Cambridge, UK) and goat anti-chicken Alexa Fluor 647 antibody (A21449, Molecular Probes, Oregon, USA), inner kinetochore with human anti-CREST (15-235, Antibodies Incorporated, California, USA) and goat anti-human Alexa Fluor 488 antibody (A11013, Life Technologies, California, USA), fibrous corona components were labelled with rabbit anti-ZW10 (ab21582, Abcam, Cambridge, UK), rabbit anti-CENP-E85 and goat anti-rabbit Alexa Fluor 568 antibody (A11036, Invitrogen, Massachusetts, USA). DNA was stained with DAPI (D1306, Thermo Fisher, UK).
Fixed bovine oocytes were imaged at 20-22°C using a Leica SPE-II- DMI4000, 1X PMT spectral detector equipped with a 63× oil-immersion objective (1.3 numerical aperture). Z-stacks of ∼80 μm were acquired at 42.7 nm in xy and 170 nm in z. Images were deconvolved using Huygens Professional (20.10). The images are maximum intensity projections of deconvoluted stacks.
Live-cell imaging
Imaging of EB3-TagRFPT and GFP-Zwilch of RPE1 cultured in 24-well glass bottom plate (Cellvis, P24-0-N) was performed by acquiring images every 4 seconds at 1 × 1 binning on an Andor CSU-W1 spinning disk (50 μm disk) with Nikon 100× 1.45 NA oil objective. 488 nm and 561 nm lasers were used for sample excitation and images were acquired using an Andor iXon-888 EMCCD camera. Nocodazole washout was done during the imaging with warm culture medium. Imaging of MAD1-mNeon and CENPA-mCherry of RPE1 cultured in 8-well plates (μ-Slide 8 well, Ibidi) was performed by acquiring images every 3 mins with z-stacks at 2 μm intervals for 8 stacks at 1 × 1 binning on an Andor CSU-W1 spinning disk (50 μm disk) with Nikon 30× silicon objective. 488 nm and 561 nm lasers were used for sample excitation and images were acquired using an Andor iXon-888 EMCCD camera. Cells were kept at 37 °C and 5% CO2 using a cage incubator and Boldline temperature/CO2 controller (OKO-Lab).
Quantification and statistical analysis
All images collected from DeltaVision Elite Applied Precision / GE Healthcare and used for quantification were deconvolved by SoftWoRx and analysed using Fiji (https://fiji.sc). For quantification of proteins levels, all images of immunostaining experiments were acquired with identical illumination settings. Protein levels near the kinetochores were determined on maximum projections of z-stacks images using an ImageJ macro87 which thresholds the CENP-C signal within the DAPI area or within the cytoplasm area in the images without DAPI staining. For quantification of EB1 and TPX2 levels on the kinetochores, the centrosome area was excluded. The length of EB1 stretches on the fibrous coronas was quantified by measuring the top two longest axis of EB1 stretches per cell by Fiji. For fibrous corona volume measurement, we determined Zwilch or CENP-E maximum projection area by an ImageJ macro thresholds intensity of the Zwilch or CENP-E. The proteins co-localizing ratio was performed on 3D images using Huygens Professional.
All statistical analysis was performed using Excel (Microsoft) and Prism (9, GraphPad). The intensity of proteins level was normalized with Excel, and plot were generated with Prism. Wilcoxon signed-rank test or Student’s t-test were performed for statistical analysis, and the figure legend indicate which statistical analysis method was used for the specific quantification.
Acknowledgments
We thank A. Akhmanova (Utrecht University), H. Clevers (Hubrecht Institute), and A. Musacchio (Max Planck Institute of Molecular Physiology) for sharing reagents. We thank members of the Kops lab for discussions. The graphical abstract and other cartoons were created with biorender.com. The Kops lab is a member of the Oncode Institute, which is partly financed by the Dutch Cancer Society. This study was financially supported by grants from the European Research Council (ERC-SyG 855158) and the Dutch Cancer Society (11080).
Author contributions
J.W. and G.J.P.L.K. conceived the project. J.W. and G.J.P.L.K. designed all experiments, except those performed with bovine oocytes, which were designed by J.W., G.J.P.L.K., A.L.-A., and M.d.R.-V. J.W., A.L.-A., and E.T. performed and analyzed experiments. M.L.A.L. generated the endogenously tagged β-tubulin-mNeon and MAD1-mNeon RPE1 cells. S.J.K. generated the p53-null RPE1 cells. D.L.B. wrote the Fiji macro for kinetochore protein projection area measurement and modified the Fiji macro for kinetochore protein intensity measurement. J.W. and G.J.P.L.K. wrote the manuscript, with input from M.d.R.-V.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 30, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2023.01.010.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
References
- 1.Maiato H., Gomes A.M., Sousa F., Barisic M. Mechanisms of chromosome congression during mitosis. Biology (Basel) 2017;6:13. doi: 10.3390/biology6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maiato H., Sunkel C.E. Kinetochore-microtubule interactions during cell division. Chromosome Res. 2004;12:585–597. doi: 10.1023/B:CHRO.0000036587.26566.81. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka K. Dynamic regulation of kinetochore-microtubule interaction during mitosis. J. Biochem. 2012;152:415–424. doi: 10.1093/jb/mvs109. [DOI] [PubMed] [Google Scholar]
- 4.Foley E.A., Kapoor T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moritz M., Braunfeld M.B., Sedat J.W., Alberts B., Agard D.A. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 6.Petry S., Vale R.D. Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 2015;17:1089–1093. doi: 10.1038/ncb3220. [DOI] [PubMed] [Google Scholar]
- 7.Wu J., Akhmanova A. Microtubule-organizing centers. Annu. Rev. Cell Dev. Biol. 2017;33:51–75. doi: 10.1146/annurev-cellbio-100616-060615. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann I. Centrosomes in mitotic spindle assembly and orientation. Curr. Opin. Struct. Biol. 2021;66:193–198. doi: 10.1016/j.sbi.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 9.McGill M., Brinkley B.R. Human chromosomes and centrioles as nucleating sites for the in vitro assembly of microtubules from bovine brain tubulin. J. Cell Biol. 1975;67:189–199. doi: 10.1083/jcb.67.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Telzer B.R., Moses M.J., Rosenbaum J.L. Assembly of microtubules onto kinetochores of isolated mitotic chromosomes of HeLa cells. Proc. Natl. Acad. Sci. USA. 1975;72:4023–4027. doi: 10.1073/pnas.72.10.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pepper D.A., Brinkley B.R. Microtubule initiation at kinetochores and centrosomes in lysed mitotic cells. Inhibition of site-specific nucleation by tubulin antibody. J. Cell Biol. 1979;82:585–591. doi: 10.1083/jcb.82.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergen L.G., Kuriyama R., Borisy G.G. Polarity of microtubules nucleated by centrosomes and chromosomes of Chinese hamster ovary cells in vitro. J. Cell Biol. 1980;84:151–159. doi: 10.1083/jcb.84.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witt P.L., Ris H., Borisy G.G. Origin of kinetochore microtubules in Chinese hamster ovary cells. Chromosoma. 1980;81:483–505. doi: 10.1007/BF00368158. [DOI] [PubMed] [Google Scholar]
- 14.Mitchison T.J., Kirschner M.W. Properties of the kinetochore in vitro. I. Microtubule nucleation and tubulin binding. J. Cell Biol. 1985;101:755–765. doi: 10.1083/jcb.101.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 16.Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., Mattaj I.W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 17.Maiato H., Rieder C.L., Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connell C.B., Loncarek J., Kaláb P., Khodjakov A. Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J. Cell Biol. 2009;187:43–51. doi: 10.1083/jcb.200903076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sikirzhytski V., Renda F., Tikhonenko I., Magidson V., McEwen B.F., Khodjakov A. Microtubules assemble near most kinetochores during early prometaphase in human cells. J. Cell Biol. 2018;217:2647–2659. doi: 10.1083/jcb.201710094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra R.K., Chakraborty P., Arnaoutov A., Fontoura B.M., Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 2010;12:164–169. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoyama H., Koch B., Walczak R., Ciray-Duygu F., González-Sánchez J.C., Devos D.P., Mattaj I.W., Gruss O.J. The nucleoporin MEL-28 promotes RanGTP-dependent gamma-tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat. Commun. 2014;5:3270. doi: 10.1038/ncomms4270. [DOI] [PubMed] [Google Scholar]
- 22.Ohba T., Nakamura M., Nishitani H., Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- 23.Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I.W. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 24.Brunet S., Sardon T., Zimmerman T., Wittmann T., Pepperkok R., Karsenti E., Vernos I. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell. 2004;15:5318–5328. doi: 10.1091/mbc.e04-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roostalu J., Cade N.I., Surrey T. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol. 2015;17:1422–1434. doi: 10.1038/ncb3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torosantucci L., De Luca M., Guarguaglini G., Lavia P., Degrassi F. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol. Biol. Cell. 2008;19:1873–1882. doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petry S., Groen A.C., Ishihara K., Mitchison T.J., Vale R.D. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bucciarelli E., Pellacani C., Naim V., Palena A., Gatti M., Somma M.P. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr. Biol. 2009;19:1839–1845. doi: 10.1016/j.cub.2009.09.043. [DOI] [PubMed] [Google Scholar]
- 29.Almeida A.C., Soares-de-Oliveira J., Drpic D., Cheeseman L.P., Damas J., Lewin H.A., Larkin D.M., Aguiar P., Pereira A.J., Maiato H. Augmin-dependent microtubule self-organization drives kinetochore fiber maturation in mammals. Cell Rep. 2022;39:110610. doi: 10.1016/j.celrep.2022.110610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ris H., Witt P.L. Structure of the mammalian kinetochore. Chromosoma. 1981;82:153–170. doi: 10.1007/BF00286101. [DOI] [PubMed] [Google Scholar]
- 31.Kops G.J.P.L., Gassmann R. Crowning the kinetochore: the fibrous corona in chromosome segregation. Trends Cell Biol. 2020;30:653–667. doi: 10.1016/j.tcb.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Magidson V., Paul R., Yang N., Ault J.G., O'Connell C.B., Tikhonenko I., McEwen B.F., Mogilner A., Khodjakov A. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 2015;17:1134–1144. doi: 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosalaganti S., Keller J., Altenfeld A., Winzker M., Rombaut P., Saur M., Petrovic A., Wehenkel A., Wohlgemuth S., Müller F., et al. Structure of the RZZ complex and molecular basis of its interaction with Spindly. J. Cell Biol. 2017;216:961–981. doi: 10.1083/jcb.201611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira C., Reis R.M., Gama J.B., Celestino R., Cheerambathur D.K., Carvalho A.X., Gassmann R. Self-assembly of the RZZ complex into filaments drives kinetochore expansion in the absence of microtubule attachment. Curr. Biol. 2018;28:3408–3421.e8. doi: 10.1016/j.cub.2018.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacristan C., Ahmad M.U.D., Keller J., Fermie J., Groenewold V., Tromer E., Fish A., Melero R., Carazo J.M., Klumperman J., et al. Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat. Cell Biol. 2018;20:800–810. doi: 10.1038/s41556-018-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raisch T., Ciossani G., D'Amico E., Cmentowski V., Carmignani S., Maffini S., Merino F., Wohlgemuth S., Vetter I.R., Raunser S., Musacchio A. Structure of the RZZ complex and molecular basis of Spindly-driven corona assembly at human kinetochores. EMBO J. 2022;41:e110411. doi: 10.15252/embj.2021110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altenfeld A., Wohlgemuth S., Wehenkel A., Vetter I.R., Musacchio A. Complex assembly, crystallization and preliminary X-ray crystallographic analysis of the human Rod-Zwilch-ZW10 (RZZ) complex. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:438–442. doi: 10.1107/S2053230X15004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffis E.R., Stuurman N., Vale R.D. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang G., Kruse T., Guasch Boldú C., Garvanska D.H., Coscia F., Mann M., Barisic M., Nilsson J. Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J. 2019;38:e100977. doi: 10.15252/embj.2018100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Rodriguez J.A., Lewis C., McKinley K.L., Sikirzhytski V., Corona J., Maciejowski J., Khodjakov A., Cheeseman I.M., Jallepalli P.V. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol. 2018;28:3422–3429.e5. doi: 10.1016/j.cub.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maiato H., Fairley E.A.L., Rieder C.L., Swedlow J.R., Sunkel C.E., Earnshaw W.C. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- 42.Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magidson V., O'Connell C.B., Lončarek J., Paul R., Mogilner A., Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Famulski J.K., Vos L.J., Rattner J.B., Chan G.K. Dynein/dynactin-mediated transport of kinetochore components off kinetochores and onto spindle poles induced by nordihydroguaiaretic acid. PLoS One. 2011;6:e16494. doi: 10.1371/journal.pone.0016494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrestha R.L., Draviam V.M. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 2013;23:1514–1526. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gama J.B., Pereira C., Simões P.A., Celestino R., Reis R.M., Barbosa D.J., Pires H.R., Carvalho C., Amorim J., Carvalho A.X., et al. Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J. Cell Biol. 2017;216:943–960. doi: 10.1083/jcb.201610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auckland P., Roscioli E., Coker H.L.E., McAinsh A.D. CENP-F stabilizes kinetochore-microtubule attachments and limits dynein stripping of corona cargoes. J. Cell Biol. 2020;219:e201905018. doi: 10.1083/jcb.201905018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holubcová Z., Blayney M., Elder K., Schuh M. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science. 2015;348:1143–1147. doi: 10.1126/science.aaa9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullen T.J., Davis-Roca A.C., Wignall S.M. Spindle assembly and chromosome dynamics during oocyte meiosis. Curr. Opin. Cell Biol. 2019;60:53–59. doi: 10.1016/j.ceb.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen F., Tillberg P.W., Boyden E.S. Optical imaging. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cooke C.A., Schaar B., Yen T.J., Earnshaw W.C. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X.Y., Chen Q., Feng J., Hou J.J., Yang F.Q., Liu J.J., Jiang Q., Zhang C.M. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gamma TuRC to the centrosome. J. Cell Sci. 2009;122:2240–2251. doi: 10.1242/jcs.042747. [DOI] [PubMed] [Google Scholar]
- 54.Tulu U.S., Fagerstrom C., Ferenz N.P., Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumari A., Kumar C., Wasnik N., Mylavarapu S.V.S. Dynein light intermediate chains as pivotal determinants of dynein multifunctionality. J. Cell Sci. 2021;134:jcs254870. doi: 10.1242/jcs.254870. [DOI] [PubMed] [Google Scholar]
- 56.Purohit A., Tynan S.H., Vallee R., Doxsey S.J. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 1999;147:481–492. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tynan S.H., Purohit A., Doxsey S.J., Vallee R.B. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 2000;275:32763–32768. doi: 10.1074/jbc.M001536200. [DOI] [PubMed] [Google Scholar]
- 58.Alonso A., Fabritius A., Ozzello C., Andreas M., Klenchin D., Rayment I., Winey M. Yeast pericentrin/Spc110 contains multiple domains required for tethering the gamma-tubulin complex to the centrosome. Mol. Biol. Cell. 2020;31:1437–1452. doi: 10.1091/mbc.E20-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dictenberg J.B., Zimmerman W., Sparks C.A., Young A., Vidair C., Zheng Y., Carrington W., Fay F.S., Doxsey S.J. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doxsey S.J., Stein P., Evans L., Calarco P.D., Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 61.Choi Y.K., Liu P., Sze S.K., Dai C., Qi R.Z. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fong K.W., Choi Y.K., Rattner J.B., Qi R.Z. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore W.J., Zhang C.M., Clarke P.R. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 2002;12:1442–1447. doi: 10.1016/S0960-9822(02)01076-X. [DOI] [PubMed] [Google Scholar]
- 65.So C., Menelaou K., Uraji J., Harasimov K., Steyer A.M., Seres K.B., Bucevičius J., Lukinavičius G., Möbius W., Sibold C., et al. Mechanism of spindle pole organization and instability in human oocytes. Science. 2022;375:eabj3944. doi: 10.1126/science.abj3944. [DOI] [PubMed] [Google Scholar]
- 66.Gruhn J.R., Zielinska A.P., Shukla V., Blanshard R., Capalbo A., Cimadomo D., Nikiforov D., Chan A.C., Newnham L.J., Vogel I., et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science. 2019;365:1466–1469. doi: 10.1126/science.aav7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luciano A.M., Franciosi F., Lodde V., Tessaro I., Corbani D., Modina S.C., Peluso J.J. Oocytes isolated from dairy cows with reduced ovarian reserve have a high frequency of aneuploidy and alterations in the localization of progesterone receptor membrane component 1 and Aurora kinase B. Biol. Reprod. 2013;88:58. doi: 10.1095/biolreprod.112.106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ménézo Y.J., Hérubel F. Mouse and bovine models for human IVF. Reprod. Biomed. Online. 2002;4:170–175. doi: 10.1016/s1472-6483(10)61936-0. [DOI] [PubMed] [Google Scholar]
- 69.van Hooff J.J., Tromer E., van Wijk L.M., Snel B., Kops G.J. Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 2017;18:1559–1571. doi: 10.15252/embr.201744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kitamura E., Tanaka K., Komoto S., Kitamura Y., Antony C., Tanaka T.U. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev. Cell. 2010;18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vasudevan A., Schukken K.M., Sausville E.L., Girish V., Adebambo O.A., Sheltzer J.M. Aneuploidy as a promoter and suppressor of malignant growth. Nat. Rev. Cancer. 2021;21:89–103. doi: 10.1038/s41568-020-00321-1. [DOI] [PubMed] [Google Scholar]
- 72.Kiyomitsu T., Cheeseman I.M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schroeder C.M., Ostrem J.M., Hertz N.T., Vale R.D. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. eLife. 2014;3:e03351. doi: 10.7554/eLife.03351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pemble H., Kumar P., van Haren J., Wittmann T. GSK3-mediated CLASP2 phosphorylation modulates kinetochore dynamics. J. Cell Sci. 2017;130:1404–1412. doi: 10.1242/jcs.194662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmid-Burgk J.L., Höning K., Ebert T.S., Hornung V. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat. Commun. 2016;7:12338. doi: 10.1038/ncomms12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W., et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Artegiani B., Hendriks D., Beumer J., Kok R., Zheng X., Joore I., Chuva de Sousa Lopes S.C.D., van Zon J., Tans S., Clevers H. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat. Cell Biol. 2020;22:321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 80.Lüders J., Patel U.K., Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 81.Gruss O.J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Silljé H., Karsenti E., Mattaj I.W., Vernos I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- 82.Jones L.A., Villemant C., Starborg T., Salter A., Goddard G., Ruane P., Woodman P.G., Papalopulu N., Woolner S., Allan V.J. Dynein light intermediate chains maintain spindle bipolarity by functioning in centriole cohesion. J. Cell Biol. 2014;207:499–516. doi: 10.1083/jcb.201408025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gavilan M.P., Gandolfo P., Balestra F.R., Arias F., Bornens M., Rios R.M. The dual role of the centrosome in organizing the microtubule network in interphase. EMBO Rep. 2018;19:e45942. doi: 10.15252/embr.201845942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Splinter D., Tanenbaum M.E., Lindqvist A., Jaarsma D., Flotho A., Yu K.L., Grigoriev I., Engelsma D., Haasdijk E.D., Keijzer N., et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown K.D., Wood K.W., Cleveland D.W. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J. Cell Sci. 1996;109:961–969. doi: 10.1242/jcs.109.5.961. [DOI] [PubMed] [Google Scholar]
- 86.Chozinski T.J., Halpern A.R., Okawa H., Kim H.J., Tremel G.J., Wong R.O., Vaughan J.C. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M.A., Kops G.J.P.L. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2011;2:316. doi: 10.1038/ncomms1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live imaging of RPE1 cells co-expressing GFP-Zwilch and EB3-TagRFPT before, during and after nocodazole washout.
Live imaging of MAD1-mNeon KI RPE1 cells over-expressing CENPA-mCherry in the presence of DMSO.
Live imaging of MAD1-mNeon KI RPE1 cells over-expressing CENPA-mCherry in the presence of 5uM lonafarnib.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.