Abstract
The idea of mRNA therapy had been conceived for decades before it came into reality during the Covid-19 pandemic. The mRNA vaccine emerges as a powerful and general tool against new viral infections, largely due to its versatility and rapid development. In addition to prophylactic vaccines, mRNA technology also offers great promise for new applications as a versatile drug modality. However, realizing the conceptual potential faces considerable challenges, such as minimal immune stimulation, high and long-term expression, and efficient delivery to target cells and tissues. Here we review the applications of mRNA-based therapeutics, with emphasis on the innovative design and future challenges/solutions. In addition, we also discuss the next generation of mRNA therapy, including circular mRNA and self-amplifying RNAs. We aim to provide a conceptual overview and outlook on mRNA therapeutics beyond prophylactic vaccines.
Keywords: mRNA therapeutics, mRNA medicine, mRNA vaccine, Protein-replacement therapy, Circular RNA, RNA delivery
1. Introduction
In the battle against diseases, people tend to learn from life itself and take advantage of what nature offers. From secondary metabolites to macromolecules, from cells to organs, these life components can be engineered and applied as new medical treatments. Being one of the key macromolecules, the messenger RNA (mRNA) also has the potential to be applied as new therapeutics. The concept of mRNA therapeutics is quite simple, which uses in vitro transcribed mRNA to direct the translation of proteins with pharmacological activity in vivo. Since mRNA carries the message of any gene, it can theoretically produce almost any functional protein/peptide for vaccination or protein replacement therapy. However, it has taken researchers several decades and great efforts to successfully produce a final commercial product since the concept of mRNA- encoded drugs was conceived, which was marked by the discovery that in vitro synthesized mRNAs can serve as templates to drive protein expression when they were directly transfected into various eukaryotic cells [1] or injected in mice [2].
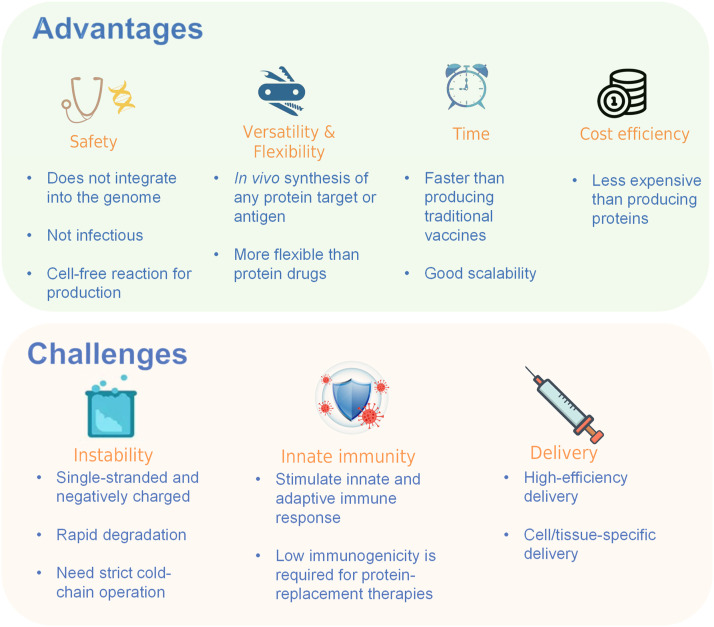
The emergency use authorizations of the mRNA vaccines during the COVID-19 pandemic (BNT162b2 and mRNA-1273) highlight the potential of mRNA as a new type of vaccine and lead to a new era of mRNA therapeutics. Currently, mRNA-based therapy has been considered a distinctive niche between gene therapy and protein therapy. It exhibits several extraordinary advantages as compared to the other two macromolecule medicines. Since mRNA is the template for protein translation, mRNA-based drugs overwhelm protein drugs in many aspects. To be specific, mRNA overcomes the obstacles of post-translational modification (PTM), folding, assembly, and location of exogenously expressed proteins. Meanwhile, it can encode multiple proteins for immunogens composed of multiple subunits at one time, which solves the challenge of reconstituting the protein subunits with the correct stoichiometry [3]. Furthermore, the production and manufacture of mRNA are faster, more flexible, more convenient, and less expensive than those of proteins, especially as tools for rapid screening and developing vaccine products during a pandemic [4]. On the other hand, when compared to DNA therapy, mRNAs do not need to enter the nucleus to become functional, thus having negligible risks of insertional mutagenesis [5]. In addition, mRNA is transiently active compared to DNA, reducing the possibility as a burden to the host homeostasis [6]. These impressive advantages make mRNA therapy the latest yet the most promising medical strategy, gaining tremendous attention from the research community, funding agencies, and the biomedical industry (Fig. 1).
Fig. 1.
The advantages and challenges of mRNA therapy.
Although mRNA therapy has achieved safe, programable, flexible, and cost-effective design and production, the inherent properties of mRNA and other technology limitations pose considerable challenges. The three main challenges are the stability and duration of expression, the immune modulation, and the in vivo targeted delivery (Fig. 1). These challenges evoke researchers’ great interest, with extraordinary efforts being made to find out the corresponding solutions. In this review, we provide a detailed overview of the progress and challenges in mRNA-based therapeutics, emphasizing innovations and solutions that contribute to extending mRNA therapeutics beyond vaccines Readers are also referred to other comprehensive or categorial reviews in association with this field [3,[5], [6], [7], [8], [9], [10], [11], [12].
2. mRNA therapeutics as vaccines and protein-replacement medicines
Based on the application areas, the current mRNA therapeutics can roughly be divided into three basic categories, the prophylactic vaccines against infectious diseases, the therapeutic vaccines against cancers, and the protein-replacement medicines either as direct immune therapies or as other protein drugs. Several recent commentaries have focused on the clinical advances of different categories of mRNA-based therapeutics [3,8,11], which are not discussed in this review. Readers are referred to the research papers, company press releases, and government websites of clinical trials for up-to-date information on clinical advances. In this review, we mainly focus on the challenges and difficulties in the research and development of each category of mRNA therapy.
2.1. mRNA as prophylactic vaccines for infectious diseases
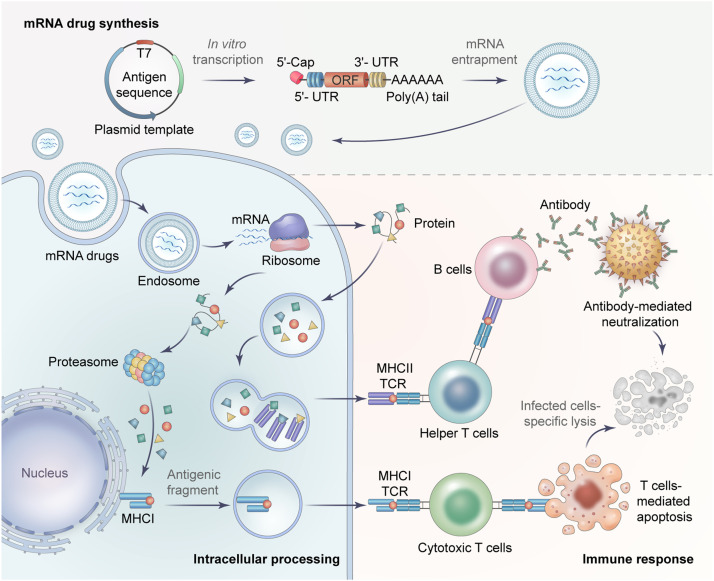
The principle of mRNA preventative vaccines is delivering the in vitro transcribed (IVT) mRNA into the human body to translate antigenic proteins, thereby stimulating the immune system to defend against the pathogen. The antigenic proteins generated from mRNAs can be recognized by the immune system either through major histocompatibility complexes I (MHC I) to activate cytotoxic T cells (CD8+ T cells) that “kill” infected cells or through MHC II and helper T cells (CD4+ T cells) that stimulate B cells to produce neutralizing antibodies (Fig. 2).
Fig. 2.
Strategies of the antigen-coding mRNA therapy. The principle of antigen-encoded mRNA therapy is utilizing the in vitro transcribed mRNA to produce targeted antigens in vivo which can stimulate the host's immune response to defend against diseases. This therapy mainly involves three processes, including mRNA synthesis, intracellular processing, and immune response.
Currently, most clinical applications of mRNA therapy have been focused on prophylactic vaccines, which can partially be attributed to the inherent immunostimulatory properties of mRNA. Proteins are generally unable to activate pattern recognition receptors (PRRs) by themselves, but instead require adjuvants to initiate adaptive immune responses [6]. However, exogenous mRNA is inherently immunostimulatory and has a self-adjuvating effect, providing a huge advantage as vaccines.
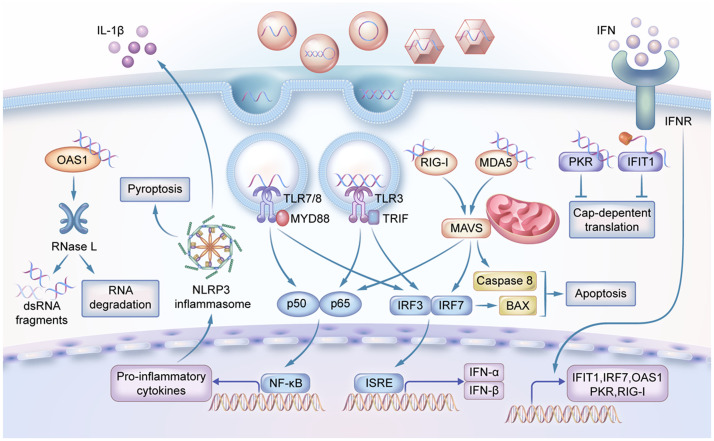
Foreign RNAs can be first detected by the host's innate immune system [13] and subsequently induce both innate immunity and adaptive immunity. In this context, PRRs exert crucial roles in detecting pathogen-associated molecular patterns (PAMPs), including exogenous RNAs within the endosome or cytoplasm (Fig. 3). The in vitro produced mRNA, like viral RNA, can also be recognized by various PRR sensors, including the Toll-like receptor (TLR) and the retinoic acid-inducible gene I (RIG-I)-like receptor (RLR). The TLR3 recognizes double-stranded RNA (dsRNA), whereas TLR7 and TLR8 recognize single-stranded RNA (ssRNA) [14,15]. The TLRs are mainly located in the plasma membrane, endosomes, and lysosomes of the immune cells [16], where the antigen-presenting cells (APCs) can present mRNA-encoded antigen to activate TLRs that induce secretion of interferons (IFNs). The RLRs are located in the cytosol of most non-immune cells [17], [18], [19]. Some RLRs, like RIG-I, can be activated by 5′-triphosphorolyated ssRNA or short dsRNA (18–19 bp), whereas the melanoma differentiation-associated protein 5 (MDA5) is activated by long (>1000 bp) dsRNA [3,18]. The different length preference of RIG-I and MDA5 is mainly determined by their Hel-CTD motifs (Helicase and C-terminal Domain) that adopt different orientations relative to the dsRNA. The RIG-I Hel-CTD is tilted to short dsRNAs, whereas the MDA5 Hel-CTD runs parallel to the dsRNA where they can form filaments to bind very long dsRNA [20]. These two PRR systems together orchestrate an adaptive immune response by producing type I INFs and inflammatory cytokines (Fig. 3), facilitating powerful and prolonged immune protection of mRNA vaccines [21]. For example, the immune response elicited by two 25-µg doses of mRNA-1273 vaccine was found to be comparable to that induced by natural infection with SARS-CoV-2 throughout the 7-month study duration [22].
Fig. 3.
The underlying mechanisms for cells to identify foreign RNAs. In the endosome, dsRNA and ssRNA are detected by TLRs. In the cytoplasm, dsRNA or 5′-triphosphorolyated ssRNA can be recognized by RLRs. The foreign RNAs can also be detected by RNA receptor 2′-5′-oligoadenylate synthetase 1 (OAS1) which produces 2′-5′-oligoadenylates and activate ribonuclease L (RNase L) to directly degrade RNA. Other RNA receptors like dsRNA-activated protein kinase R (PKR) and IFN-induced protein with tetratricopeptide repeats 1 (IFIT1) can recognize viral RNA and inhibit cap-dependent translation.
In addition to the inherent immunogenicity of RNA, some advantages in mRNA drug generation, including rapid development, low-cost production, and safe administration, facilitate mRNAs as successful vaccines during the global COVID-19 pandemic [7]. In fact, many efforts had been made before COVID-19 in developing mRNA vaccines to combat different contagious viruses, including influenza virus, Ebola virus, Zika virus, rabies virus, and HIV. Some of these R&D efforts have proceeded to clinical trials [3,10,11,23]. Nevertheless, the field of applying mRNA as a prophylactic vaccine still awaits further validation of mRNA products against pathogens other than SARS-CoV-2.
Although mRNA therapy exhibited extraordinary advantages in vaccine development, the rational design and optimization of the epitope or the antigen-specific sequence still need careful consideration. For instance, the two mRNA vaccine candidates against SARS-CoV-2 developed by BioNTech adopted different antigen sequences. Specifically, BNT162b1 utilized the S protein receptor-binding domain (RBD) as an antigen with the addition of a T4 fibritin-derived foldon trimerization domain for enhancing its immunogenicity [24], while the clinically approved BNT162b2 encodes full-length S protein modified by two proline substitutions in order to lock it in the prefusion conformation [25]. To take another example, the mRNA-1893 was finally chosen instead of mRNA-1325 in the development of the Zika virus vaccines, because the mRNA-1893 has a different sequence and exhibited 20-fold potency in primate studies [26]. Additional discussion on the selection and design of antigen sequences will be included later in this review.
2.2. mRNA as therapeutic vaccines for cancer immunotherapy
With the constant improvement of vaccines, a paradigm shift has recently taken place in using vaccines not only for disease prevention but also for disease treatment [26]. Studies regarding therapeutic vaccines mainly focus on cancers, not only due to the inherent characteristics of cancer, but also because of the recent explosion and successes in the field of cancer immunotherapy [27], including immune checkpoint blockade (ICB), chimeric antigen receptor T cells (CAR-T) mediated therapy, and cancer-specific antigens. The key idea for cancer therapeutic vaccines is to use cancer-specific antigens to train the human immune system, which in turn kills cancer cells with high specificity. As mRNA has increased immunogenicity compared to DNA vaccines [7], the capability to encode multiple antigens or whole antigens with many epitopes, and the ability of rapid manufacturing, it seems to have great potential as therapeutic vaccines, especially as cancer vaccines to initiate and amplify the anticancer immune response.
It has been reported in early 2000 that intranodal injection of naked mRNA encoding tumor antigen can inhibit cancer growth in a mouse model [28]. In 2008, patient-derived autologous mRNA was first applied as a vaccine to treat patients bearing melanoma [29]. The naked self-adjuvanted mRNA has later been applied as the mRNA vaccine for the first time in phase I/II clinical trials by intradermal injection in prostate cancer patients [30]. However, the success of the mRNA cancer vaccine has yet to be achieved because of some key challenges in cancer complexity and mRNA technology.
Unlike preventative vaccines for infectious diseases, therapeutic cancer vaccines must also induce a strong cytotoxic cellular response to eradicate cancerous cells. Therefore, the selection of proper antigens capable of inducing tumor-specific immune responses becomes an essential task in the design of therapeutic mRNA vaccines. Tumor-associated antigens (TAA) and tumor-specific antigens (TSA) are considered to be reasonable candidates for cancer vaccine development. Typical TAAs are overexpressed antigens (like HER-2 in 30% of breast cancer) and differentiation-specific antigens (like PSA in Prostate cancer). However, as TAAs are also expressed in normal tissues, sometimes they have central tolerance due to the clonal deletion of autoreactive lymph cells during ontogenesis [31], leading to the consequence that the self-antigens cannot activate an appropriate immune response. Therefore, cancer cells may evade immunosurveillance and resist immunotherapy. A combination of multiple therapeutic mRNAs may help to overcome this obstacle. At present, there are several mRNA vaccines in clinical trials using multiple TAAs as immunologic stimulants (NCT03480152, NCT03313778, NCT02410733, etc.). Compared with TAAs, TSAs (especially neoantigens) can bypass central tolerance with high immunogenicity, because they generate the accumulation of aberrant proteins that are absent in normal tissues. However, tumor neoantigens are highly heterogeneous, suggesting that cancer immunotherapy using mRNA vaccines may be highly personalized. Several clinical trials have been launched recently to further evaluate the antitumor efficacy of personal mRNA vaccines (NCT03908671, NCT03468244, etc.).
A rational design of epitope/antigen is expected to elicit both humoral and cellular immune responses. Therefore, a major challenge in antigen design is the lack of knowledge regarding T cell receptor (TCR) epitopes compared with B cell receptor (BCR) epitopes. Since TCRs only recognize the primary structure of the antigen, we need to learn additional features/motifs for further prediction of rational antigens/epitopes [26]. Therefore, the selection and design of antigens and epitopes require a more careful design in therapeutic vaccines than in preventative vaccines.
Because cancer vaccines require antigen presentation to elicit the immune response, another challenge in mRNA cancer vaccines is to increase its targeting to APCs, particularly to dendritic cells (DCs). DCs initiate the adaptive immune response by internalizing and proteolytically processing antigens, thereby presenting them to CD8+ T cells on MHC I and CD4+ T cells on MHC II, respectively (Fig. 2). In addition, DCs are highly amenable to mRNA transfection [7], and thus are an attractive APC target for mRNA vaccines both in vivo and in vitro. Previous studies usually used the in vitro method to introduce mRNA into DCs and load the DCs to the cancer-bearing host. However, a lot of DC-based mRNA vaccines failed to provide clinical benefit, partially due to the low immune-stimulation efficacy. To address this issue, people have to use immunostimulatory adjuvants to improve the efficacy of DCs-base mRNA therapy [21,32]. Furthermore, mRNA therapy had demonstrated various results in different cancer types, including melanoma, glioblastoma, acute myeloid leukemia (AML), and renal cell carcinoma [11], indicating that additional complexity remains to be explored.
For cancer vaccination, even if an antigen can induce cellular immune responses, the suppressive tumor microenvironment could prevent T cell infiltration into tumors, leading to T cell exhaustion [3]. Therefore, therapeutic cancer vaccines may need to be combined with other treatments for better efficacy. For example, the BNT111 that encodes four cancer-specific antigens had to be used in combination with immune checkpoint inhibitors (e.g. PD-1 and LAG-3) to improve the vaccine efficacy in the prostate cancer model [33]. The mRNA-2416, the mRNA encoding OX40 ligand (OX40L) that lacks efficient effects when used as monotherapy, was taken forward to a phase II expansion cohort in the combined treatment with durvalumab for ovarian cancer (NCT03323398, Moderna, Inc. United States Securities and Exchange Commission Form 10-K (2020)). Collectively, due to the complexity of cancer immunity, the successful development of therapeutic cancer vaccines is pretty challenging and has to be further optimized in both antigen selection and the design of clinical trials.
2.3. mRNA as protein-replacement therapy
In addition to mRNA vaccines, mRNA also has great potential in various protein-replacement therapy, either as immune therapy (e.g. antibodies and cytokines) or other protein drugs. Unlike mRNA vaccines that only require a low dose, short-term local expression of antigens, most protein-replacement treatments require a long-term expression of therapeutic proteins in specific tissues. The distinct desirable features of different mRNA medicines were summarized in Table 1.
Table 1.
Property comparison across different categories of mRNA medicine.
| Preventive vaccine | Therapeutic vaccine | Protein-replacement medicine | |
|---|---|---|---|
| Sequence design | medium | difficult | relatively easy |
| Expression time | short | medium | long |
| Dosage | low | medium | high |
| Immunogenicity | medium | high | low |
| Cell-specific expression | not necessarily required | local expression may work | fully required (except when encoding antibodies, growth factors, hormones, etc.) |
2.3.1. mRNA as passive immunotherapy
Passive immunotherapy can be divided into two types: the in vivo delivery of immune reagents (such as antibodies or cytokines), and the ex vivo generation of genetically modified immune cells for adoptive cellular immunotherapies. The mRNA technology provides a general platform for in vivo production of antibodies, cytokines, and ICB regulators, thus becoming a new modality for passive immunotherapy. For example, an mRNA encoded potently neutralizing human monoclonal antibody was reported to protect against chikungunya infections [34]. In addition, a replicon RNA encoding ZIKV-117 mAb exhibited high-level mAb expression and zika virus protection in mice [35]. For anti-cancer application, researchers observed an improved pharmacokinetic profile through a comparison between trastuzumab (an anti-HER2 antibody) mRNA and directly injected trastuzumab protein [36]. Bispecific T cell-engaging antibody (bsAb) has also shown great potential in cancer therapy but has been impeded by manufacturing obstacles. However, an mRNA-based bsAb can efficiently inhibit tumor growth in mice [37].
Unlike mRNA-encoded antibodies that are administrated systematically, mRNA-encoded cytokines and ICB regulators are mainly administrated locally to stimulate the immune response in the cancer microenvironment. For example, mRNA-encoded immune checkpoint inhibitors (like PD-1 and LAG-3 inhibitors) or regulators (like OX40L) have shown antitumor activity by intratumor administrated. In addition, mRNA-2752 which encodes cytokines IL-23 and IL-36γ in addition to OX40L is being evaluated in solid tumors and lymphoma (NCT03739931).
Another promising application for mRNA-based therapy is the ex vivo generation of CAR with mRNA, rather than retroviral vectors, in lymphocytes (e.g. T cells or natural killer (NK) cells). Early in 2017, Tchou et al. observed that CAR-T generated by mRNA targeting c-Met induced inflammatory responses within breast cancer tissues [38]. Interestingly, this ex vivo method can be extended for engineering graft CAR-T cells with TCR deficiency, which avoided graft-versus-host reactions by using mRNA to translate CRISPR/Cas or TALEN [39]. As long as the delivery to lymphocytes is safe and effective, mRNAs exhibit advantages in loading capacity and redosing over viral delivery.
Besides ex vivo CAR immunocyte generation, mRNA can generate CAR-T cells or high-affinity TCR-T cells in vivo. Several studies have demonstrated the possibility of in vivo CAR-T cell generation by mRNA [40], [41], [42], which was achieved with the help of innovative LNP formulation. Interestingly, the mRNA-based CAR-T technology can not only be applied to defend against cancers but also be used to treat other diseases such as cardiac injury. Recently, mRNA-encoded FAP (fibroblast activation protein) CAR was formulated with CD5-targeted LNPs, and specifically delivered into a mouse model of heart failure. This approach successfully produced CD5+ CAR T cells in vivo, reducing fibrosis and restoring cardiac function in the mouse model [43]. Altogether, these impressive outcomes suggest the potential application of mRNA in adoptive immune cell therapy.
2.3.2. mRNA as protein-coding medicines
In addition to passive immunotherapy, mRNA-based therapeutics have an extensive application as protein-replacement medicine. Using mRNA medicine has several advantages over direct protein delivery, including robust production of proteins from a single mRNA molecule and easy manufacture of mRNAs compared to proteins. Great efforts have already been made in using mRNAs to treat cardiac diseases, lung diseases, metabolic diseases, neurogenic disorders, as well as some genetic rare diseases [11,12]. Nevertheless, the majority of these treatments are still in the preclinical stage. Recently good progress has been made in cardiac diseases, with a primary focus on heart failure and myocardial infarction. The mRNA-encoded VEGF (NCT03370887) and CFTR (NCT03375047) were shown to protect mice from heart failure and reduce apoptosis of myocardial cells, and have later been advanced to phase II clinical trials. In addition, mRNA-encoded transcription factors Stemin and YAP5SA can regenerate adult cardiomyocytes and repair infarcted mouse hearts [44], indicating the particular potential of mRNA technology in cardiac diseases. mRNA therapeutics have another encouraging application in the field of genome editing by delivering mRNA that encodes nuclease, including ZFN, TALEN, or CRISPR/Cas. Transient delivery and expressions of the mRNA are preferable for clinical applications with lower mutation risk and reduced off-target effect. mRNA based genome editing has versatile usage. For instance, to advance genome-editing therapies for AIDS, researchers can track edited primary human hematopoietic stem and progenitor cells (HSPCs) in a mutation-specific manner by transfecting macaque-specific CCR5 ZFN mRNA ex vivo into them and then engrafting the modified HSPCs in a large animal model [45]. Liu and colleagues performed an efficient knockdown with BAMEA-O16B formulated Cas9 mRNA and sgRNA, which decreased the proprotein convertase subtilisin/kexin type 9 level in mouse serum down to 20% of nontreatment [46].
2.3.3. The challenges and solutions to protein-replacement therapy
A major challenge in protein-replacement therapy is the dosage and expression duration of the mRNA. In vaccines against pathogens or cancers, nanogram to microgram amounts of highly antigenic proteins are usually sufficient for the efficient induction of immune responses. By contrast, in some cases, such as systemically delivery of growth factors, hormones, or monoclonal antibodies, milligram or even gram amounts of proteins are needed [6], not to mention in certain cases, life-long treatment with repeated dosing may be required. Therefore, large amounts of synthetic mRNAs need to be manufactured and delivered, which could be a challenge for certain mRNAs. Meanwhile, considerable amounts of proteins need to be expressed in vivo, which requires stable, long-term, and high-level expression of mRNA molecules. In order to achieve high-level expression, the idea of using a self-amplifying RNA (saRNA) has been proposed and applied in some mRNA vaccine studies. The saRNA originates from alphaviruses which are positive-sense ssRNA viruses. The self-amplifying ability of alphaviruses is achieved by four nonstructural proteins (nsP1–4) that can combinedly function as a replicase complex (Fig. 4b). The main advantage of saRNA is the dose-sparing effect, which can achieve large-amount production of protein from a single mRNA molecule. However, the main challenge for saRNA is that its long sequence (9–12 kb) is unfavorable in molecular cloning and the following expression. Various efforts have been made to meet such challenges, such as using trans-amplifying sequences to overcome the large-size problem [47] or introducing mutations to promote subgenome expression [48]. An individualized, heterologous chimpanzee adenovirus (ChAd68) and saRNA-based neoantigen vaccine were recently assessed in an ongoing phase 1/2 study in patients with advanced metastatic solid tumors [49]. However, to date, the clinical testing of this technology remains in the early stage, and thus further tuning or the development of alternative methods (such as circular RNA) is required for addressing this issue.
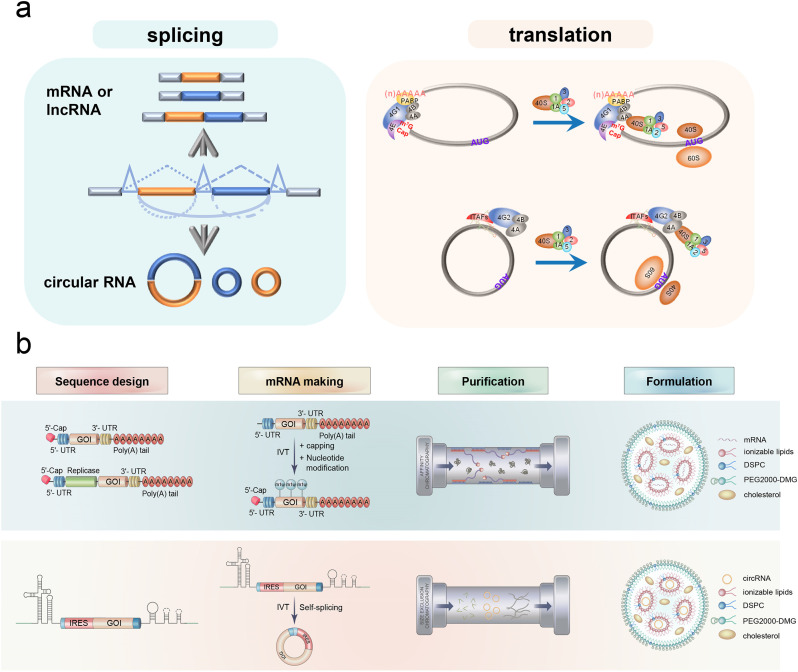
Fig. 4.
Circular mRNA versus linear mRNA. (a) The schematic diagram of the splicing (left) and translation (right) of circular mRNA and linear mRNA. (b) The manufacturing process of mRNA medicine.
A certain degree of RNA immunogenicity is favorable in mRNA vaccines. However, in protein-replacement therapies, the activation of the innate immune system by IVT mRNA is a major disadvantage. This issue can be mostly addressed by nucleotide modification during IVT and stringent purification of IVT products, which will be discussed in detail later in this review.
Another challenge for protein-replacement therapy is the delivery of mRNA to the desired organs/cell types to achieve optimal therapeutic outcomes. When mRNAs are administered systemically in formulation with lipid nanoparticles (LNPs), many tend to accumulate in the liver due to the uptake of apolipoprotein E (ApoE), which will interact with the LNP surface [50]. However, a lot of diseases are induced by the absence or deficiency of certain proteins in specific organs and thus need tissue-specific expression. Moreover, many other situations also need cell/tissue-specific expression. For instance, the cytokines in cancer therapy should best be delivered intratumor to balance the tumor microenvironment. The in vivo CAR-T therapy needs targeted delivery of mRNA to T cells. Moreover, in some cases, certain proteins require further PTMs to become completely functional. However, the PTMs sometimes are tissue-dependent, not simply depending on the mRNA sequence, thus requiring tissue-specific delivery.
In summary, protein-replacement mRNA therapy poses several unique challenges in terms of the efficacy of protein production, immunogenicity reduction, and targeted delivery compared to vaccines. This may explain why the application of mRNA has progressed more slowly than mRNA immunization [3]. Further endeavors are required for its future clinical translation.
3. The common challenges and potential solutions to mRNA-based therapeutics
Although each type of mRNA therapy has its unique challenges, there are some common challenges facing all mRNA therapies. The first challenge is to improve the stability and duration of mRNA expression. mRNA is inherently susceptible to base-catalyzed hydrolysis because the ribose has a highly reactive 2′ hydroxyl group that can attack the adjacent phosphorus. Meanwhile, the rapid degradation of RNA caused by the ubiquitous and hardy ribonucleases (RNase) requires high costs of production, storage, and transportation of RNA products (especially long RNAs). Moreover, the therapeutic vaccine and protein-replacement therapy need long expression time of cellular mRNA medicines, which could be partially achieved by improving the mRNA stability.
The second challenge is the high innate immunogenicity of RNA. Exogenous and formulated mRNA is immunostimulatory when recognized by a variety of innate immune responses (Fig. 3) including TLR [51], RIG-I [52], and dsRNA-activated protein kinase (PKR) [53]. In the application of protein-replacement therapies, the activation of the innate immune system is undesirable due to translation inhibition and cell toxicity. Karikó, Weissman, and colleagues addressed this issue by utilizing modified nucleosides such as pseudouridine to suppress the immunogenicity of RNA and promote protein translation. [54]. This technology has made a crucial contribution to the therapeutic use of mRNA and has been licensed by BioNTech and Moderna to develop COVID-19 vaccines [3] and protein replacement medicines. However, nucleoside modification is relatively expensive and imposes extra constrain in sequence design.
The third challenge is the in vivo delivery of mRNA. mRNA needs to enter the cytoplasm to produce functional proteins. However, mRNA can hardly pass through the anionic phospholipid bilayer of the cell membrane due to its large size and charged properties, limiting the in vivo expression of target protein from mRNA. To obtain the safe and effective intracellular delivery of mRNA, many delivery systems were testified s, among which the LNP exhibited the application potential [55,56]. The LNP-based delivery system was successful in the clinical application of mRNA vaccines by BioNTech/Pfizer [57] and Moderna [58] against SARS-CoV-2. However, the formulation of LNP still has a large space for improvement. In particular, the therapeutic vaccine and protein-replacement application of mRNA need cell or tissue-specific delivery, which urgently requires further development of rational delivery systems.
Substantial efforts have been invested to confront these challenges in all crucial steps of mRNA therapy, including sequence design, mRNA synthesis & production, and targeted delivery. In addition, researchers have broken through the limitation of linear mRNA drugs by developing circular RNA (circRNA) as an alternative modality of mRNA therapy, enabling prolonged-expression and lower immunogenicity. It also provides promising therapeutic potential, especially in protein-replacement medicine beyond mRNA preventative vaccines. Here we will give a more detailed discussion of circRNA as a new generation of mRNA drug modality by introducing the basic concept, strategies and innovations in the sequence design, synthesis, and production of circRNAs. We will also review the new developments in RNA delivery systems, and other findings and technologies contributing to mRNA-based therapeutics.
3.1. Development of circular RNA as a new modality for mRNA therapy
CircRNAs are the covalently closed single-stranded RNA (ssRNA) molecules generated mainly from pre-mRNA back-splicing (Fig. 4a, left). A majority of circRNAs originate from protein-coding genes and contain complete exons [59]. Moreover, the exon-containing circRNAs are primarily localized in the cytoplasm [60]. Collectively, we speculate that circRNAs have the potential to direct protein expression as mRNAs, and further demonstrate that the exogenous circRNA can be translated in vivo [61]. Recently, we and several other groups have collectively proved that endogenous circRNAs can be translated as well [62], [63], [64]. In contrast to linear mRNAs, circRNAs do not have 5′-caps, thus undergo the cap-independent translation. When an internal ribosome entry site (IRES) is introduced in circRNA-expressing vectors, proteins can be effectively translated [63]. The mechanisms of translation are different between mRNA and circRNA in eukaryotes. Briefly, in IRES-dependent translation, most IRESs are regulated by IRES trans-acting factors (ITAFs), which assist in recruiting the eIF4G2-associated complex rather than the eIF4G1-associated complex in mRNA. It can further recruit 43S complex to initiate translation (Fig. 4a, right).
Several features of the circRNA molecule make it an appealing candidate for new-generation mRNA drugs, particularly as protein-replacement therapies. Firstly, they have higher cellular stability than the cognate linear mRNAs as reported by multiple groups [61,65,66], possibly due to their resistance to degradation by exonuclease in vitro and/or by the resistance to the immune-induced linear RNA decay machinery in vivo [67]. The second inherent advantage of circRNAs is that the purified circRNAs have lower immunogenicity compared with linear RNAs [66,68,69], which is desirable for prolonged-expression. It is worth noting that the immunogenicity of circRNAs is a complicated issue. While some early reports show that circRNA may have higher immunogenicity [70], recent reports showed that circRNAs can essentially bypass the innate cellular immune system like RIG-I mediated pathway when carefully purified to eliminate double-stranded RNA (dsRNA) and uncapped precursor RNA [66,68,69], which also contributes to the prolonged-expression in vivo. However, the immunogenicity of circRNAs was also found to be affected by the exact RNA sequence, the method of synthesis, and the type of base modification [71,72].
A major challenge for circRNA application is large-scale in vitro production. The commonly used methods for in vitro synthesis of circRNAs can be classified into two categories: RNA-ligase mediated direct ligation and ribozyme mediated auto-splicing. Using T4 RNA ligase to directly circularize linear precursor RNA is a classic method developed many years ago [73]. This method can generate circRNAs without extraneous fragments, which exhibit minimized immunogenicity [72]. However, a “splint” sequence is usually needed to connect the free ends for ligation. As a result, this method is technically challenging to scale up production. In addition, the use of ligase will add to the complexity in product purification, therefore this strategy is mainly used in research labs.
CircRNAs can also be produced by introducing self-catalyzed group I introns that function as cyclase ribozymes [74,75]. Wesselhoeft and colleagues optimized this strategy to engineer circRNAs for efficient and stable translation in eukaryotes [68,76]. This method, known as PIE (permuted intron-exon), was further adopted to produce circRNA vaccines against SARS-CoV-2 [77]. However, this method leaves a foreign fragment in the circRNA as a “scar” sequence that may distort the structures of circRNAs to provoke innate immunity [72], limiting its application as protein-needed therapies. To address this issue, a Clean-PIE method was developed to produce scarless circRNAs with minimized immunogenicity [78]. Alternatively, we have developed an independent method using self-splicing group II introns to achieve co-transcriptional circularization of circRNAs [66], which can efficiently produce scarless circRNA with low immunogenicity and can be expanded for large-scale production.
The production of circRNA does not require the 5′ capping and the base modification steps, thereby significantly reducing the complexity and costs of circRNA production. Recent studies have already employed this powerful platform in several applications, such as the production of spider silk protein [79], and vaccines against SARS-CoV-2 and the emerging variants [66,77,80]. However, there are still technical hurdles unique to circRNAs as new therapeutics, including sequence design for the efficient circularization and translation, as well as the efficient in vivo delivery.
3.2. Optimization of mRNA translation, stability, and immunogenicity
The sequence design and synthesis of mRNA for improved pharmacological properties are crucial in mRNA therapeutics [11]. All the five functional regions, including the 5′ cap, 5′-UTRs, coding region, 3′-UTRs, and poly(A) tail, have to be carefully tuned to achieve efficient protein translation. The synthetic cap analogs and capping enzymes can be used to add a 5′ cap, which may stabilize mRNA by conferring increased stability and promoting translation via binding to eukaryotic translation initiation factor 4E (eIF4E). As for the poly (A) tail, the masked/unmasked and the length of poly(A) can affect translation [81]. The stem-loop structure in histone mRNA should also be considered in optimization (Patent US 2018/0271964). In addition, the regulatory elements in 5′- and 3′-UTRs can stabilize mRNA and increase protein translation. The sequence design and codon optimization in the open reading frame (ORF) is also critical because it can increase translation efficiency and RNA stability, as well as modulate the stimulation of immune systems. By iterative optimization of these regions, the translational potency and intracellular stability of IVT mRNA can be significantly improved (Fig. 4b, sequence design). There are several sophisticated reviews discussing the strategies for structural modifications of each region [6,11,82], and thus in this review we will mainly discuss the sequence design for circRNAs.
The design of circRNAs should focus on three regions: the self-catalyzed introns, the IRESs, and the ORFs (Fig. 4b, sequence design). In general, the ORF design of circRNA is assumed to be similar to that of linear mRNA, except that the base modification is unnecessary. Notably, IRESs are the focus of the design because they not only affect circRNA translation efficiency but also have the potential to mediate tissue-specific expression by binding tissue-specific ITAFs [63]. The canonical IRESs are mainly derived from viral sequences and are used in the circRNA modality to drive translation. However, recent studies have shown that many additional sequences can function as IRES-like elements or regulatory elements to promote IRES activity and stimulate translation [64,83]. Therefore, screening and optimizing host-friendly and tissue-specific IRESs would be critical for circRNA technology. In addition, the self-catalyzed introns were found to significantly affect circulation efficiency [66], hence screening and optimizing the highly activated self-catalyzing introns may also contribute to circRNA medicine.
Purification of RNA production is the next key step in mRNA manufacturing. During the IVT procedure of mRNA, the capping, nucleotide modification, and poly(A) tailing require additional resources. Therefore, the reduction of cost and labor in these procedures should be taken into consideration in mRNA manufacturing (Fig. 4b). The circRNAs can evade some of these obstacles since they contain neither 5′-cap nor ploy(A) tail. Meanwhile, the superb stability and low immunogenicity of circRNAs seem to make nucleotide modification unnecessary. Moreover, the self-splicing procedure is relatively simple, only requiring GTPs and cationic ions in group I intron strategy [74], [75], [76], and even no GTP in group II intron strategy [66,84] (Fig. 4b, mRNA making). Therefore, the manufacture of circRNAs could circumvent some technical difficulties associated with linear mRNAs. mRNA purity is a key factor for protein production and innate immunity. This is because the removal of contaminants, such as dsRNA generated in IVT by stringent liquid chromatography (LC) purification, can eliminate immune activation and extraordinarily (almost 1000-fold) improve the translation of linear nucleoside-modified mRNA [85] (Fig. 4b, purification). For circRNAs, despite several studies have discussed the roles of RIG-I and PKR in detecting foreign circRNA molecules [70,76,84], the cellular mechanisms to detect foreign circRNAs remain largely unknown. Breuer and colleagues reported that circRNAs can bypass cellular antiviral responses [69], which is consistent with our unpublished results. However, the IVT and back-splicing contaminants in circRNA generation also affected the cellular immune response and the duration of circRNA expression [66,68]. Unlike the mRNA platform that uses affinity LC methods to purify the mRNA, circRNAs need to be purified from RNA precursor and intron RNA via size-exclusion chromatography (SEC) (Fig. 4b, purification). Certainly, the SEC is not necessarily the only method to purify circRNAs, and new strategies can also be developed in circRNA purification.
3.3. Optimization of mRNA delivery
The targeted delivery of large-size RNA in vivo has been a major challenge for decades in mRNA therapy. In such case, many delivery systems have been considered, of which LNP is the only approved delivery systemin SARS-CoV-2 vaccines for clinic. Currently, it has been the most widely used method for mRNA delivery, while many other methods are in the trail of laboratory and clinical research.
3.3.1. LNP delivery system
Tipically, LNP is consisted by four components, including ionizable lipids, cholesterol, helper phospholipid, and polyethylene glycol (PEG)-lipids [86]. These four components can form stable nanoparticles to encapsulate and protect vulnerable mRNA. The nanoparticles enter the cells through endocytosis, and become positively charged in the acidic environment of endosomes, facilitating the subsequent release of the RNAs into the cytoplasm. However, LNP-formulated mRNAs tend to accumulate in the liver, possibly due to the binding of apolipoprotein E (ApoE) onto the LNP surface followed by the ApoE receptor-mediated uptake to hepatocytes [50]. Most of the LNP-mRNA drugs are concentrated in the liver and spleen after tail vein administration, while a small amount preserved in adipose tissues [87]. In the case of the intromuscular injection, the expression of mRNA drugs is mainly found in the liver, muscle, and lymph nodes [88,89].
In addition, organ delivery of LNP other than liver may be achieved by modifying lipid compositions, which includes the adjustment of the lipid identities and/or lipid ratios. The LNP compositions can substantially affect the efficientcy of intracellular delivery , determine cell specificity in delivery, and modulate immunogenicity. For instance, the addition of SORT lipids to LNP changed the organ-targeting properties of LNP in vivo, achieving mRNA delivery to other organs and tissues instead of live only [90]. Moreover, SORT is compatible with various gene-editing technologies, such as Cas9 mRNA/sgRNA and Cas9 ribonucleoprotein (RNP) complex. The chemical changes in PEG-lipid structures of LNP surface were found to assist LNP targeting to bone marrow endothelial cells in the hematopoietic stem cell niche [91], which may also be applied in mRNA delivery. Interestingly, LNP technologies can achieve targeted delivery by adjusting the stoichiometry of LNP components, which was exemplified by the success in lymph node-targeting mRNA delivery in mice [92].
Other strategies, such as additional non-lipid components in LNP formulation, also contribute to the targeted delivery of mRNA. For example, the introduction of CD31 (PECAM) antibody to LNP can target lung endothelial cells [93]. A similar approach using VCAM ligands can successfully direct LNPs to inflammatory sites in the brain and attenuated the TNF-α-triggered brain edema [94]. In another study, a novel lung-targeted mRNA delivery system was developed through in vivo screening of a library of synthetic lipid nanoparticles. They demonstrated that by simply changing the head structure of LNPs, mRNA can be directed to different lung subcellular types [95].
3.3.2. Additional delivery methods
In addition to LNP, several other technologies have also been applied in mRNA delivery, including the use of polyethyleneimine (PEI) polymers. Despite its excellent delivery efficacy, the application of PEI polymers is limited by the toxicity resulting from its high surface charge density. Several strategieshave been studied to reduce the toxicity, such as the use of a low molecular weight PEI [96], incorporation of PEG [97], and binding to cyclodextrin [98]. In addition, new biodegradable polymers with less toxicity have been developed as aalternatives. For example, poly(β-amino esters) are promising in mRNA delivery, especially to the lungs [99]. Since they can be easily synthesized by the Michael reaction, scientists have constructed large libraries of poly(β-amino esters) to facilitate their structure-function studies [100].
Similar to poly(β-amino esters), polyamides are also biodegradable biopolymers that can be synthesized by the Michael reaction, with their backbone and periphery groups can be easily modified [101]. Polyamide-based amines can form hyperbranched dendritic spherical dendrimers, which can effectively form mRNA complexes due to the high amine density at their periphery. However, even though the charge density contributes positively to mRNA complexation, an excessive charge density can lead to cell toxicity and serum aggregation. Introducing disulfide bonds or adding PEG to their cores may resolve these problems [102].
Another type of polymers, pH-responsive polymer, is currently utilized for mRNA delivery. They have attracted wide attention due to their unique mRNA delivery mechanisms. These charge-altering releasable transporters are not protonated within the endosome. Instead, they can self-degrade into neutral, nontoxic byproducts at the pH of the cytoplasm, resulting in the rapid release of mRNA into the cytoplasm in the mouse model [103].
In addition to lipid-based and polymer-based carriers, amphipathic peptides can also help to deliver mRNA into cells owing to the cationic or amphipathic amine groups (e.g., arginine) that can bind to mRNA electrostatically to form nano complexes. The fusion cell-penetrating peptides with repetitive arginine-alanine-leucine-alanine (RALA) motifs can change the conformation at pH level in endosomes. As a result, it can promote the pore formation between membranes and endosomes, thereby mediating mRNA delivery into cells [104]. Moreover, RALA can deliver mRNA to dendritic cells and trigger T cell mediated immunity [105]. Recently, the amphipathic CPP/mRNA complexes with a size less than 200 nm showed high cellular uptake and protein expression in mice colonic cancer cells [106].
3.4. Other progresses contribute to the development of mRNA medicines
3.4.1. Bioinformatics as a driving force in the design of mRNA medicine
When modifying mRNA sequences for better druggability, various bioinformatic approaches are necessary in the mRNA drug design [107,108]. With the aid of artificial intelligence (AI) and machine learning (ML) methods, researchers can optimize the UTRs and ORFs for higher translation efficiency and suitable immunogenicity in a systematic fashion. For example, the current mRNA therapies mostly adopt UTRs from highly expressed genes like β-globin. However, the position of the globin UTR sequence can affect protein expression [109]. Moreover, the regulatory function of UTRs seems to be cell-type and tissue-specific [110], which may account for the differential expression of regulators such as RNA-binding proteins (RBPs). Taken together, there is still space for improvement of the UTRs.
To address this issue, a neural network model was developed in training the high-throughput sequencing data of polysome profiling, aiming to quantify ribosome loading for 5′-UTRs to develop predictive models [111,112]. They also engineered de novo sequences that can increase protein expression for mRNA. Similar ideas can also be applied in the optimization of 3′-UTR and coding regions, and in the screening and optimization of self-catalyzed introns and IRESs in the circRNA platform as well.
In addition, AI can be used to facilitate epitope prediction for the design of mRNA vaccines against infectious diseases [113,114], including epitope prediction to trigger a better immune response. For example, AI was applied in the retrieval of the SARS-CoV-2 spike glycoprotein sequence and the prediction of epitopes for cytotoxic T cells, helper T cells, and B cells [115]. Moreover, a supervised ML model was developed to predict the conserved regions of SARS-CoV-2 and calculate the protective antigenicity score. According to these results, S protein and nsp3 (containing MHC-I and MHC-II T-cell epitopes) were identified as the most promising vaccine candidates against diseases [116].
Predicting antigen/epitope also presents a great challenge in mRNA cancer vaccines, which can be addressed computationally in determining the peptides presented by MHC from the immunopeptidome. In particular, AI/ML can facilitate the development of personalized vaccines by predicting the neoantigen and optimization of the epitopes [117], [118], [119], [120]. For example, AI-based algorithms predict the MHC presentation of neoantigens discovered through whole exome sequencing of tumors [121]. However, AL/ML has a long way to keep up with the accumulation of big data, and further endeavors are needed in meeting the challenges in this field. These endeavors may include the optimization of B cell epitope and MHC-I allele-specific predictor (IEDB server, http://tools.iedb.org/bcell/ & BepiPred); prediction of T cell epitope and MHC-II allele-specific response (NetMHCII and PropPred); prediction of BCR and TCR epitopes. In addition to epitope prediction and optimization, Gonzalez-Dias et al. have developed an ML framework, DAMIP, to predict vaccine efficacy [122], while the ML method was also used in predicting tumor antigen immunogenicity to facilitate clinical translation of the selected antigens [123].
3.4.2. Adjuvants development
Adjuvants play crucial roles in vaccine development to maximize the effectiveness of primary antigens. Interestingly, both BNT162b2 and mRNA-1273 do not require additional adjuvants, probably attributed to the self-adjuvant properties of exogenous RNA combined with the immuno-stimulating effect of the LNP delivery system itself [124]. Nevertheless, additional adjuvants can sometimes help to achieve superior performance when high immunogenicity is warranted, such as in cancer vaccines. Numerous studies have already made efforts to develop various types of adjuvants to improve efficacy and potency. For instance, CureVac has generated the RNActive platform, where protamine is utilized to form a complex with the antigen-coding mRNA, thus activating TLR7 and TLR8 signaling to induce innate immunity [125]. In other cases, single-stranded RNA can be applied as an adjuvant to stimulate innate immunity [126,127].
3.4.3. Route of administration
In drug delivery, the route of administration is also an important consideration, especially for targeting mucosa and inducing the right systemic and local immune response. Currently, most RNA preventative vaccines are administrated by intramuscular injection. However, intranasal administration is under development due to its easy, painless, and safe use [128]. Intranasal administration also reduces the risk of transmitting blood-borne infections through needles and avoids the hurdles of the gastrointestinal tract through the oral route. For cancer vaccines, the intratumoral administration is a popular route, because the systemic injection cannot bypass the microenvironment of tumors. In some cases, the route of administration can be a determining element. For example, in the trial of CureVac rabies vaccine candidate CV7021, volunteers were administrated intradermally or intramuscularly by needle-syringe or needle-free devices. It was demonstrated that, regardless of the safety of CV7201 in all groups, CV7201 exhibited immunogenicity and induced functional antibodies only when it was administrated by needle-free devices [129], proving that the route of administration is a consideration in the development of mRNA medicines.
Aside from the advances in the above-mentioned aspects of mRNA pharmacology, other considerations have also contributed to the R&D of mRNA medicine, such as drug combination, dosing regimens, stock, and transportation, which are not discussed in this review due to space constraints. On all accounts, with all efforts from academia and industry, we will see success in new mRNA-based medicines in the near future.
4. Conclusion
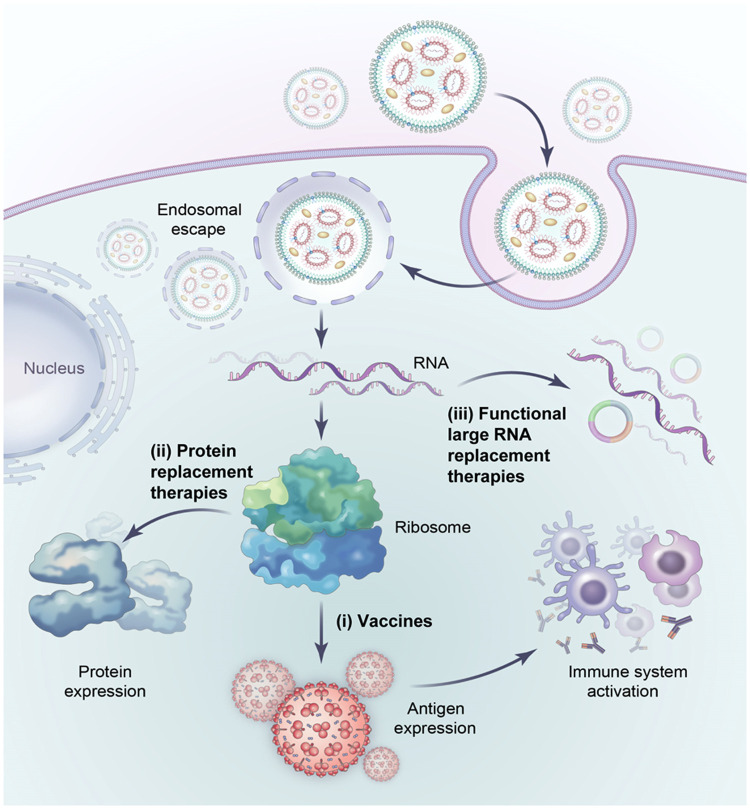
Since the proof-of-concept in the 1990s, mRNA therapy has undergone a long route of development to overcome several major technical difficulties, including poor stability, innate immunity, and delivery issues. The pressure of the Covid pandemic has played a critical role in finally bringing this new technology into reality, which in turn paved the way for additional applications of this new drug modality. Despite early success, there are still many technical challenges in this field, which are currently under intensive investigation. The development of new RNA platform technology, including circRNA and saRNAs, as well as new delivery methods, will likely expand the mRNA therapy into new frontiers like in vivo programming of cells. In addition, the approaches developed for mRNA therapy will likely be useful for future RNA drugs including long non-coding RNA (lncRNA) therapy (Fig. 5).
Fig. 5.
Future applications of RNA-based therapy. Large RNAs have the potential for functional RNA replacement therapies (iii) as well as vaccines (i) or protein-replacement therapies (ii).
Declaration of competing interest
Zefeng Wang is the scientific founder of CirCode Biomedicine in Shanghai. The rest authors declare that they have no conflicts of interest in this work.
Acknowledgment
This work is partially supported by the National Natural Science Foundation of China (NSFC) to Z.W. (91940303 and 31730110) and H-H W. (32171294); the Science and Technology Commission of Shanghai Municipality (STCSM) to H-H W. (20ZR1467300); and the National Key Research and Development Program of China (2021YFA1300503) to Z.W. Z.W. is also sponsored by the type A CAS Pioneer 100-Talent program, and the Starry Night Science Fund at Shanghai Institute for Advanced Study of Zhejiang University (SN-ZJU-SIAS-009).
Biographies

Huan-Huan Wei is an associate professor at the CAS Shanghai Institute of Nutrition and Health (SINH). She obtained her bachelor's degree in Life Science from SiChuan University and her Ph.D. degree in Natural Medicine Chemistry from CAS South China Institute of Botany. After graduation, she worked as a youth backbone teacher at FuDan University before joining Zefeng Wang's group at SINH in 2016. Her current research focuses on the mechanism and regulation of aberrant alternative splicing in cancers, as well as the research and development of RNA medicines.

Zefeng Wang is a Professor at the CAS Shanghai Institute of Nutrition and Health, and the scientific founder of both CirCode BioMed and Enzerna Bioscience. He obtained a double bachelor's degree in Biological Science and Computer Technology at Tsinghua University, and a Ph.D. at Johns Hopkins University. Following his post-doctoral training at MIT, he became a tenured faculty at UNC at Chapel Hill. In 2015, he moved his lab to Shanghai and became the director of PICB. Zefeng has made significant contributions to the field of RNA biology by developing a series of genomic approaches to study RNA splicing and dysregulation systematically. His research group was the first to discover that circular RNAs can be translated within cells, and they continues to investigate the regulatory mechanisms underlying circRNA translation.
Contributor Information
Huan-Huan Wei, Email: weihuanhuan@sinh.ac.cn.
Zefeng Wang, Email: wangzefeng@sinh.ac.cn.
References
- 1.Malone R.W., Felgner P.L., Verma I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. U.S.A. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolff J.A., Malone R.W., Williams P., et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 3.Barbier A.J., Jiang A.Y., Zhang P., et al. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 2022;40:840–854. doi: 10.1038/s41587-022-01294-2. [DOI] [PubMed] [Google Scholar]
- 4.Corbett K.S., Edwards D.K., Leist S.R., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maruggi G., Zhang C., Li J., et al. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27:757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin U., Kariko K., Tureci O. mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discovery. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 7.Pardi N., Hogan M.J., Porter F.W., et al. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discovery. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhary N., Weissman D., Whitehead K.A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discovery. 2021;20:817–838. doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng R., Patil S., Zhao X., et al. RNA therapeutics - research and clinical advancements. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.710738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To K.K.W., Cho W.C.S. An overview of rational design of mRNA-based therapeutics and vaccines. Expert Opin. Drug Discov. 2021;16:1307–1317. doi: 10.1080/17460441.2021.1935859. [DOI] [PubMed] [Google Scholar]
- 11.Qin S., Tang X., Chen Y., et al. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct. Target Ther. 2022;7:166. doi: 10.1038/s41392-022-01007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semple S.C., Leone R., Barbosa C.J., et al. Lipid nanoparticle delivery systems to enable mRNA-based therapeutics. Pharmaceutics. 2022;14 doi: 10.3390/pharmaceutics14020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diebold S.S., Massacrier C., Akira S., et al. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur. J. Immunol. 2006;36:3256–3267. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 15.Lee I.A., Park Y.J., Joh E.H., et al. Soyasaponin Ab ameliorates colitis by inhibiting the binding of lipopolysaccharide (LPS) to Toll-like receptor (TLR)4 on macrophages. J. Agric. Food Chem. 2011;59:13165–13172. doi: 10.1021/jf2033818. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C., Maruggi G., Shan H., et al. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wies E., Wang M.K., Maharaj N.P., et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatematsu M., Funami K., Seya T., et al. Extracellular RNA sensing by pattern recognition receptors. J. Innate Immun. 2018;10:398–406. doi: 10.1159/000494034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneyama M., Kikuchi M., Matsumoto K., et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 20.Brisse M., Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front. Immunol. 2019;10:1586. doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards D.K., Jasny E., Yoon H., et al. Adjuvant effects of a sequence-engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J. Transl. Med. 2017;15:1. doi: 10.1186/s12967-016-1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mateus J., Dan J.M., Zhang Z., et al. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knezevic I., Liu M.A., Peden K., et al. Development of mRNA vaccines: scientific and regulatory issues. Vaccines. 2021;9:81. doi: 10.3390/vaccines9020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulligan M.J., Lyke K.E., Kitchin N., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 25.Walsh E.E., Frenck R.W., Jr., Falsey A.R., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brisse M., Vrba S.M., Kirk N., et al. Emerging concepts and technologies in vaccine development. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.583077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortezaee K. Immune escape: a critical hallmark in solid tumors. Life Sci. 2020;258 doi: 10.1016/j.lfs.2020.118110. [DOI] [PubMed] [Google Scholar]
- 28.Granstein R.D., Ding W., Ozawa H. Induction of anti-tumor immunity with epidermal cells pulsed with tumor-derived RNA or intradermal administration of RNA. J. Invest. Dermatol. 2000;114:632–636. doi: 10.1046/j.1523-1747.2000.00929.x. [DOI] [PubMed] [Google Scholar]
- 29.Weide B., Carralot J.P., Reese A., et al. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 30.Kubler H., Scheel B., Gnad-Vogt U., et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J. Immunother. Cancer. 2015;3:26. doi: 10.1186/s40425-015-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein L., Hinterberger M., Wirnsberger G., et al. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 32.Jansen Y., Kruse V., Corthals J., et al. A randomized controlled phase II clinical trial on mRNA electroporated autologous monocyte-derived dendritic cells (TriMixDC-MEL) as adjuvant treatment for stage III/IV melanoma patients who are disease-free following the resection of macrometastases. Cancer Immunol. Immunother. 2020;69:2589–2598. doi: 10.1007/s00262-020-02618-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zahm C.D., Moseman J.E., Delmastro L.E., et al. PD-1 and LAG-3 blockade improve anti-tumor vaccine efficacy. Oncoimmunology. 2021;10 doi: 10.1080/2162402X.2021.1912892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kose N., Fox J.M., Sapparapu G., et al. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erasmus J.H., Archer J., Fuerte-Stone J., et al. Intramuscular delivery of replicon RNA encoding ZIKV-117 human monoclonal antibody protects against zika virus infection. Mol. Ther. Methods Clin. Dev. 2020;18:402–414. doi: 10.1016/j.omtm.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rybakova Y., Kowalski P.S., Huang Y., et al. mRNA delivery for therapeutic anti-HER2 antibody expression in vivo. Mol. Ther. 2019;27:1415–1423. doi: 10.1016/j.ymthe.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stadler C.R., Bahr-Mahmud H., Celik L., et al. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 38.Tchou J., Zhao Y., Levine B.L., et al. Safety and efficacy of intratumoral injections of chimeric antigen receptor (CAR) T cells in metastatic breast cancer. Cancer Immunol. Res. 2017;5:1152–1161. doi: 10.1158/2326-6066.CIR-17-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knipping F., Osborn M.J., Petri K., et al. Genome-wide specificity of highly efficient TALENs and CRISPR/Cas9 for T cell receptor modification. Mol. Ther. Methods Clin. Dev. 2017;4:213–224. doi: 10.1016/j.omtm.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billingsley M.M., Singh N., Ravikumar P., et al. Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X., Chen J., Qiu M., et al. Imidazole-based synthetic lipidoids for in vivo mRNA delivery into primary T lymphocytes. Angew. Chem. Int. Ed Engl. 2020;59:20083–20089. doi: 10.1002/anie.202008082. [DOI] [PubMed] [Google Scholar]
- 42.Tombácz I., Laczkó D., Shahnawaz H., et al. Highly efficient CD4+ T cell targeting and genetic recombination using engineered CD4+ cell-homing mRNA-LNPs. Mol. Ther. 2021;29:3293–3304. doi: 10.1016/j.ymthe.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rurik J.G., Tombacz I., Yadegari A., et al. Epstein J.A. CAR T cells produced in vivo to treat cardiac injury. Science. 2022;375:91–96. doi: 10.1126/science.abm0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao S., Liang R., Lucero E., et al. STEMIN and YAP5SA synthetic modified mRNAs regenerate and repair infarcted mouse hearts. J. Cardiovasc. Aging. 2022;2 doi: 10.20517/jca.2022.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson C.W., Wang J., Norman K.K., et al. Long-term multilineage engraftment of autologous genome-edited hematopoietic stem cells in nonhuman primates. Blood. 2016;127:2416–2426. doi: 10.1182/blood-2015-09-672337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Chang J., Jiang Y., et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019;31 doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beissert T., Koste L., Perkovic M., et al. Improvement of in vivo expression of genes delivered by self-amplifying RNA using vaccinia virus immune evasion proteins. Hum. Gene Ther. 2017;28:1138–1146. doi: 10.1089/hum.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y., Teague B., Zhang Y., et al. In vitro evolution of enhanced RNA replicons for immunotherapy. Sci. Rep. 2019;9:6932. doi: 10.1038/s41598-019-43422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer C.D., Rappaport A.R., Davis M.J., et al. Individualized, heterologous chimpanzee adenovirus and self-amplifying mRNA neoantigen vaccine for advanced metastatic solid tumors: phase 1 trial interim results. Nat. Med. 2022;28:1619–1629. doi: 10.1038/s41591-022-01937-6. [DOI] [PubMed] [Google Scholar]
- 50.Akinc A., Querbes W., De S., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kariko K., Buckstein M., Ni H., et al. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Gil L., Goff P.H., Hai R., et al. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J. Virol. 2013;87:1290–1300. doi: 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lihoradova O., Kalveram B., Indran S.V., et al. The dominant-negative inhibition of double-stranded RNA-dependent protein kinase PKR increases the efficacy of Rift Valley fever virus MP-12 vaccine. J. Virol. 2012;86:7650–7661. doi: 10.1128/JVI.00778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kariko K., Muramatsu H., Welsh F.A., et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flemming A. Vaccines: self-amplifying RNA in lipid nanoparticles: a next-generation vaccine? Nat. Rev. Drug Discovery. 2012;11:748–749. doi: 10.1038/nrd3854. [DOI] [PubMed] [Google Scholar]
- 56.Lutz J., Lazzaro S., Habbeddine M., et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polack F.P., Thomas S.J., Kitchin N., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baden L.R., El Sahly H.M., Essink B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeck W.R., Sorrentino J.A., Wang K., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo J.U., Agarwal V., Guo H., et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y., Fan X., Mao M., et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Y., Wang Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019;11:911–919. doi: 10.1093/jmcb/mjz091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C.K., Cheng R., Demeter J., et al. Structured elements drive extensive circular RNA translation. Mol. Cell. 2021;81:4300–4318. doi: 10.1016/j.molcel.2021.07.042. e4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Enuka Y., Lauriola M., Feldman M.E., et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.C. Chen, H. Wei, K. Zhang, et al., A flexible, efficient, and scalable platform to produce circular RNAs as new therapeutics, bioRxiv, (2022) 2022.2005.2031.494115.
- 67.Li Y., Zheng Q., Bao C., et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell. 2019;74:508–520. doi: 10.1016/j.molcel.2019.02.015. e504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Breuer J., Barth P., Noe Y., et al. What goes around comes around: artificial circular RNAs bypass cellular antiviral responses. Mol. Ther. Nucleic Acids. 2022;28:623–635. doi: 10.1016/j.omtn.2022.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Y.G., Kim M.V., Chen X., et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67:228–238. doi: 10.1016/j.molcel.2017.05.022. e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y.G., Chen R., Ahmad S., et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109. doi: 10.1016/j.molcel.2019.07.016. e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu C.X., Guo S.K., Nan F., et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell. 2022;82:420–434. doi: 10.1016/j.molcel.2021.11.019. e426. [DOI] [PubMed] [Google Scholar]
- 73.Beaudry D., Perreault J.P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 1995;23:3064–3066. doi: 10.1093/nar/23.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Puttaraju M., Been M.D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992;20:5357–5364. doi: 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ford E., Ares M., Jr. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc. Natl. Acad. Sci. U.S.A, 1994;91:3117–3121. doi: 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qu L., Yi Z., Shen Y., et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–1744. doi: 10.1016/j.cell.2022.03.044. e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Z. Qiu, Q. Hou, Y. Zhao, et al., Clean-PIE: a novel strategy for efficiently constructing precise circRNA with thoroughly minimized immunogenicity to direct potent and durable protein expression, bioRxiv, (2022) 2022.2006.2020.496777.
- 79.Liu L., Wang P., Zhao D., et al. Engineering circularized mRNAs for the production of spider silk proteins. Appl. Environ. Microbiol. 2022;88 doi: 10.1128/aem.00028-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.K. Huang, N. Li, Y. Li, et al., Delivery of circular mRNA via degradable lipid nanoparticles against SARS-CoV-2 delta variant, bioRxiv, (2022) 2022.2005.2012.491597.
- 81.Mockey M., Goncalves C., Dupuy F.P., et al. mRNA transfection of dendritic cells: synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem. Biophys. Res. Commun. 2006;340:1062–1068. doi: 10.1016/j.bbrc.2005.12.105. [DOI] [PubMed] [Google Scholar]
- 82.Weissman D. mRNA transcript therapy. Expert Rev. Vaccines. 2015;14:265–281. doi: 10.1586/14760584.2015.973859. [DOI] [PubMed] [Google Scholar]
- 83.Fan X., Yang Y., Chen C., et al. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 2022;13:3751. doi: 10.1038/s41467-022-31327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jarrell K.A. Inverse splicing of a group II intron. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8624–8627. doi: 10.1073/pnas.90.18.8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kariko K., Muramatsu H., Ludwig J., et al. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H., Han X., Alameh M.G., et al. Rational design of anti-inflammatory lipid nanoparticles for mRNA delivery. J. Biomed. Mater. Res. A. 2022;110:1101–1108. doi: 10.1002/jbm.a.37356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cui L., Hunter M.R., Sonzini S., et al. Mechanistic studies of an automated lipid nanoparticle reveal critical pharmaceutical properties associated with enhanced mRNA functional delivery in vitro and in vivo. Small. 2022;18 doi: 10.1002/smll.202105832. [DOI] [PubMed] [Google Scholar]
- 88.Bahl K., Senn J.J., Yuzhakov O., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2022;30:2874. doi: 10.1016/j.ymthe.2022.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pardi N., Tuyishime S., Muramatsu H., et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng Q., Wei T., Farbiak L., et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krohn-Grimberghe M., Mitchell M.J., Schloss M.J., et al. Nanoparticle-encapsulated siRNAs for gene silencing in the haematopoietic stem-cell niche. Nat. Biomed. Eng. 2020;4:1076–1089. doi: 10.1038/s41551-020-00623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen J., Ye Z., Huang C., et al. Lipid nanoparticle-mediated lymph node-targeting delivery of mRNA cancer vaccine elicits robust CD8(+) T cell response. Proc. Natl. Acad. Sci. U.S.A. 2022;119 doi: 10.1073/pnas.2207841119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parhiz H., Shuvaev V.V., Pardi N., et al. PECAM-1 directed re-targeting of exogenous mRNA providing two orders of magnitude enhancement of vascular delivery and expression in lungs independent of apolipoprotein E-mediated uptake. J. Control. Release. 2018;291:106–115. doi: 10.1016/j.jconrel.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marcos-Contreras O.A., Greineder C.F., Kiseleva R.Y., et al. Selective targeting of nanomedicine to inflamed cerebral vasculature to enhance the blood-brain barrier. Proc. Natl. Acad. Sci. U.S.A. 2020;117:3405–3414. doi: 10.1073/pnas.1912012117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qiu M., Tang Y., Chen J., et al. Lung-selective mRNA delivery of synthetic lipid nanoparticles for the treatment of pulmonary lymphangioleiomyomatosis. Proc. Natl. Acad. Sci. U.S.A. 2022;119 doi: 10.1073/pnas.2116271119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kircheis R., Wightman L., Schreiber A., et al. Polyethylenimine/DNA complexes shielded by transferrin target gene expression to tumors after systemic application. Gene Ther. 2001;8:28–40. doi: 10.1038/sj.gt.3301351. [DOI] [PubMed] [Google Scholar]
- 97.Ke X., Shelton L., Hu Y., et al. Surface-functionalized PEGylated nanoparticles deliver messenger RNA to pulmonary immune cells. ACS Appl. Mater. Interfaces. 2020;12:35835–35844. doi: 10.1021/acsami.0c08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tan L., Zheng T., Li M., et al. Optimization of an mRNA vaccine assisted with cyclodextrin-polyethyleneimine conjugates. Drug Deliv. Transl. Res. 2020;10:678–689. doi: 10.1007/s13346-020-00725-4. [DOI] [PubMed] [Google Scholar]
- 99.Kaczmarek J.C., Kauffman K.J., Fenton O.S., et al. Optimization of a degradable polymer-lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett. 2018;18:6449–6454. doi: 10.1021/acs.nanolett.8b02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chintakunta R., Buaron N., Kahn N., et al. Synthesis, characterization, and self-assembly with plasmid DNA of a quaternary ammonium derivative of pectic galactan and its fluorescent labeling for bioimaging applications. Carbohydr. Polym. 2016;150:308–318. doi: 10.1016/j.carbpol.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 101.Zhou K., Nguyen L.H., Miller J.B., et al. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl. Acad. Sci. U.S.A. 2016;113:520–525. doi: 10.1073/pnas.1520756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ulkoski D., Bak A., Wilson J.T., et al. Recent advances in polymeric materials for the delivery of RNA therapeutics. Expert Opin. Drug Deliv. 2019;16:1149–1167. doi: 10.1080/17425247.2019.1663822. [DOI] [PubMed] [Google Scholar]
- 103.Haabeth O.A.W., Blake T.R., McKinlay C.J., et al. mRNA vaccination with charge-altering releasable transporters elicits human T cell responses and cures established tumors in mice. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E9153–E9161. doi: 10.1073/pnas.1810002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McCarthy H.O., McCaffrey J., McCrudden C.M., et al. Development and characterization of self-assembling nanoparticles using a bio-inspired amphipathic peptide for gene delivery. J. Control. Release. 2014;189:141–149. doi: 10.1016/j.jconrel.2014.06.048. [DOI] [PubMed] [Google Scholar]
- 105.Udhayakumar V.K., De Beuckelaer A., McCaffrey J., et al. Arginine-rich peptide-based mRNA nanocomplexes efficiently instigate cytotoxic T cell immunity dependent on the amphipathic organization of the peptide. Adv. Healthc. Mater. 2017;6 doi: 10.1002/adhm.201601412. [DOI] [PubMed] [Google Scholar]
- 106.Kim Y., Kim H., Kim E.H., et al. The potential of cell-penetrating peptides for mRNA delivery to cancer cells. Pharmaceutics. 2022;14 doi: 10.3390/pharmaceutics14061271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McCaffrey P. Artificial intelligence for vaccine design. Methods Mol. Biol. 2022;2412:3–13. doi: 10.1007/978-1-0716-1892-9_1. [DOI] [PubMed] [Google Scholar]
- 108.T.E.K. Kramps, RNA Vaccines: Methods and Protocols, in: T.E.K. Kramps (Ed.), Humana Press, 2017.
- 109.Sultana N., Hadas Y., Sharkar M.T.K., et al. Optimization of 5′ untranslated region of modified mRNA for use in cardiac or hepatic ischemic injury. Mol. Ther. Methods Clin. Dev. 2020;17:622–633. doi: 10.1016/j.omtm.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Holtkamp S., Kreiter S., Selmi A., et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 111.Sample P.J., Wang B., Reid D.W., et al. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019;37:803–809. doi: 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Linder J., Seelig G. Fast activation maximization for molecular sequence design. BMC Bioinformatics. 2021;22:510. doi: 10.1186/s12859-021-04437-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mohanty E., Mohanty A. Role of artificial intelligence in peptide vaccine design against RNA viruses. Inform. Med. Unlocked. 2021;26 doi: 10.1016/j.imu.2021.100768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lv H., Shi L., Berkenpas J.W., et al. Application of artificial intelligence and machine learning for COVID-19 drug discovery and vaccine design. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbab320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ahammad I., Lira S.S. Designing a novel mRNA vaccine against SARS-CoV-2: an immunoinformatics approach. Int. J. Biol. Macromol. 2020;162:820–837. doi: 10.1016/j.ijbiomac.2020.06.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ong E., Wong M.U., Huffman A., et al. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front. Immunol. 2020;11:1581. doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jabbari P., Rezaei N. Artificial intelligence and immunotherapy. Expert Rev. Clin. Immunol. 2019;15:689–691. doi: 10.1080/1744666X.2019.1623670. [DOI] [PubMed] [Google Scholar]
- 118.Xu Z., Wang X., Zeng S., et al. Applying artificial intelligence for cancer immunotherapy. Acta Pharm. Sin. B. 2021;11:3393–3405. doi: 10.1016/j.apsb.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou X., Qu M., Tebon P., et al. Screening cancer immunotherapy: when engineering approaches meet artificial intelligence. Adv. Sci. 2020;7 doi: 10.1002/advs.202001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thomas S., Abraham A., Baldwin J., et al. Artificial intelligence in vaccine and drug design. Methods Mol. Biol. 2022;2410:131–146. doi: 10.1007/978-1-0716-1884-4_6. [DOI] [PubMed] [Google Scholar]
- 121.Shao X.M., Bhattacharya R., Huang J., et al. High-throughput prediction of MHC class I and II neoantigens with MHCnuggets. Cancer Immunol. Res. 2020;8:396–408. doi: 10.1158/2326-6066.CIR-19-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gonzalez-Dias P., Lee E.K., Sorgi S., et al. Methods for predicting vaccine immunogenicity and reactogenicity. Hum. Vaccin. Immunother. 2020;16:269–276. doi: 10.1080/21645515.2019.1697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smith C.C., Chai S., Washington A.R., et al. Machine-learning prediction of tumor antigen immunogenicity in the selection of therapeutic epitopes. Cancer Immunol. Res. 2019;7:1591–1604. doi: 10.1158/2326-6066.CIR-19-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pati R., Shevtsov M., Sonawane A. Nanoparticle vaccines against infectious diseases. Front. Immunol. 2018;9:2224. doi: 10.3389/fimmu.2018.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rauch S., Lutz J., Kowalczyk A., et al. RNActive(R) Technology: generation and Testing of Stable and Immunogenic mRNA Vaccines. Methods Mol. Biol. 2017;1499:89–107. doi: 10.1007/978-1-4939-6481-9_5. [DOI] [PubMed] [Google Scholar]
- 126.Kim H.J., Kwak H.W., Kang K.W., et al. MERS-CoV spike protein vaccine and inactivated influenza vaccine formulated with single strand RNA adjuvant induce T-cell activation through intranasal immunization in mice. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12050441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim Y.H., Bang Y.J., Park H.J., et al. Inactivated influenza vaccine formulated with single-stranded RNA-based adjuvant confers mucosal immunity and cross-protection against influenza virus infection. Vaccine. 2020;38:6141–6152. doi: 10.1016/j.vaccine.2020.07.022. [DOI] [PubMed] [Google Scholar]
- 128.Dhaliwal H.K., Fan Y., Kim J., et al. Intranasal delivery and transfection of mRNA therapeutics in the brain using cationic liposomes. Mol. Pharm. 2020;17:1996–2005. doi: 10.1021/acs.molpharmaceut.0c00170. [DOI] [PubMed] [Google Scholar]
- 129.Alberer M., Gnad-Vogt U., Hong H.S., et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]