Abstract
T cells are a critical component of the response to SARS-CoV-2, but their kinetics after infection and vaccination are insufficiently understood. Using “spheromer” peptide-MHC multimer reagents, we analyzed healthy subjects receiving two doses of the Pfizer/BioNTech BNT162b2 vaccine. Vaccination resulted in robust spike-specific T cell responses for the dominant CD4+ (HLA-DRB1∗15:01/S191) and CD8+ (HLA-A∗02/S691) T cell epitopes. Antigen-specific CD4+ and CD8+ T cell responses were asynchronous, with the peak CD4+ T cell responses occurring 1 week post the second vaccination (boost), whereas CD8+ T cells peaked 2 weeks later. These peripheral T cell responses were elevated compared with COVID-19 patients. We also found that previous SARS-CoV-2 infection resulted in decreased CD8+ T cell activation and expansion, suggesting that previous infection can influence the T cell response to vaccination.
Keywords: SARS-CoV-2, COVID-19, spheromer, peptide-MHC multimer, Pfizer/BioNTech mRNA vaccine, antigen-specific T cell responses, peripheral CD8+ T cell responses, peripheral CD4+ T cell responses, SARS-CoV-2 variants of concern
Graphical abstract
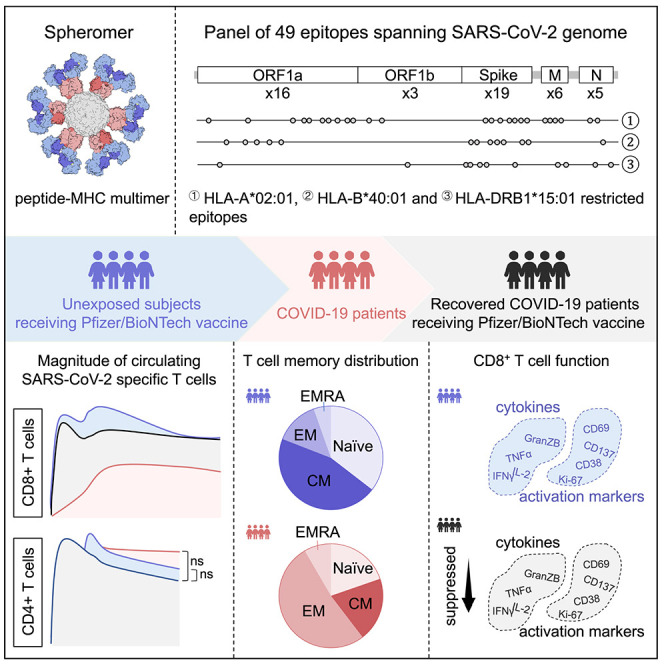
Our understanding of T cell responses in COVID-19 and vaccination is incomplete. Gao et al. examine SARS-CoV-2-specific T cell responses to infection and vaccination, revealing disparate kinetics between CD4+ and CD8+ T cells. Furthermore, compared with vaccination alone, circulating CD8+ T cells are attenuated during infection and in subsequent vaccination.
Introduction
The COVID-19 pandemic has resulted in the rapid development of several novel vaccine platforms, including the mRNA-based Pfizer/BioNTech BNT162b2 vaccine.1 , 2 The mRNA vaccine formulations show high levels of protection and stimulate robust innate and adaptive immune responses.3 , 4 , 5 , 6 They induce neutralizing antibodies, although circulating titers decrease after just months.5 , 7 By contrast, analyses of the magnitude and durability of SARS-CoV-2-specific T cell responses are limited, with most studies relying on bulk measurements after in vitro peptide stimulation.4 , 8 Although rapid and useful, these studies underestimate the frequency of epitope-specific T cells6 and may not be able to identify specific immunodominant epitopes efficiently. Peptide-major histocompatibility complex (pMHC) multimers address these limitations and provide a more quantitative and epitope-specific picture of the T cell response.9 , 10 , 11 , 12
T cell responses play a critical role in controlling disease after SARS-CoV-2 infection. Breakthrough virus in the nasal swabs is seen in all convalescent rhesus macaques with waning or suboptimal neutralizing antibody titers on rechallenge with SARS-CoV-2 after CD8+ T cell depletion.13 Recovery from COVID-19 in patients undergoing B cell depleting therapies further highlights the importance of T cells in SARS-CoV-2 viral clearance.14 CD8+ T cell responses to conserved coronavirus epitopes correlate with mild COVID-19 disease symptoms.15 Rapid expansion of cross-reactive T cells is also seen in individuals with abortive SARS-CoV-2 infection, suggesting their protective role.16 Thus, it is important to understand the kinetics of T cell priming and how these events compare across SARS-CoV-2 naive vaccinees versus COVID-19 patients.
In this study, we used the spheromer technology to identify dominant T cell epitopes after BNT162b2 vaccination. This platform is based on an engineered form of maxiferritin, where 12 pMHCs carried by each nanoparticle are able to detect ∼3- to 5-fold more antigen-specific T cells compared with other multimers.15 Here, we designed a panel of forty-nine predicted epitopes, spanning both spike and non-spike proteins from the original Wuhan-Hu-1 SARS-CoV-2 strain. We probed a total of 351 blood samples collected from vaccinated volunteers with time points ranging from pre-vaccination up to 4 months after the first dose. Overall, BNT162b2 vaccination resulted in polyfunctional CD8+ and CD4+ T cell responses across all volunteers, likely contributing to its remarkable efficacy. We observed distinct CD8+ and CD4+ T cell kinetics after mRNA vaccination. This disparity between the two major T cell responses is unusual, since in other vaccination studies both CD4+ and CD8+ peak in circulation approximately 1 week after stimulating a recall response.17 , 18 , 19 This coordination of T cell subsets was also seen in a Celiac challenge study.20 We speculate that this may be a unique feature of mRNA vaccines. To assess the differences in T cell responses elicited by vaccination versus natural infection, we determined the response in two independent local patient cohorts.15 , 21 , 22 We observed lower frequencies of spike-specific T cells in circulation after infection compared with mRNA vaccination, especially in the CD8+ T cell compartment with a skewing of the response hierarchy among the tested epitopes. We also noticed qualitative differences in the virus-specific T cells. Vaccination led to the rapid induction of effector T cells that contracted by day 90, concomitant with an increase in the frequency of memory T cells. By contrast, only low levels of virus-specific memory CD8+ T cells could be detected in COVID-19 patients, even at 5 months post-symptom onset.
We also evaluated the impact of BNT162b2 vaccination on T cell responses after SARS-CoV-2 infection. Although previous infection had almost no effect on the CD4+ T cell response induced on vaccination, we observed a decrease (3.6- to 54.1-fold at peak) in the frequency of circulating spike-specific CD8+ T cells, and these had attenuated functionality compared with naive vaccinees. This suggests that SARS-CoV-2 virus infection may cause long-term damage to the patients’ immune system well after viral clearance.
Results
The BNT162b2 vaccine encodes a stabilized spike protein from SARS-CoV-2 (Wuhan-Hu-1 strain).1 To analyze the T cell responses, we selected nineteen epitopes across multiple HLA alleles spanning the entire spike protein of 1,273 amino acids. In addition to five HLA-A∗02:01 epitopes used previously for characterizing the response in a COVID-19 patient cohort,15 we included two more HLA-A∗02:01 epitopes and seven HLA-B∗40:01 epitopes to measure CD8+ T cell responses (Table S1). For the CD4+ T cell response, we selected five HLA-DRB1∗15:01 epitopes (Table S1). In addition, we analyzed thirty non-spike epitopes from three different SARS-CoV-2 genes (for CD8+ T cells restricted to HLA-A∗02:01—ORF1ab = 12, M = 4, N = 2; and HLA-B∗40:01—ORF1ab = 5, N = 1; for CD4+ T cells restricted to HLA-DRB1∗15:01—ORF1ab = 2, M = 2, N = 2) in infected individuals (Table S1). Briefly, these peptides were selected based on a combination of the following criteria: literature search,6 , 9 , 10 , 11 , 12 , 15 , 23 , 24 , 25 , 26 bioinformatic analysis,27 , 28 , 29 and an MHC stabilization assay.15
Previously, we described spheromers, an improved 12 pMHC T cell staining platform that has superior sensitivity versus other pMHC multimers.15 We used our SARS-CoV-2 specific spheromers to characterize the T cell response kinetics in three independent cohorts: (1) SARS-CoV-2 naive individuals who received the BNT162b2 vaccine, (2) COVID-19 patients with SARS-CoV-2 infections, and (3) Individuals who received the BNT162b2 vaccine after recovery from a SARS-CoV-2 infection. Blood was collected at the indicated time points. Combinatorial staining was performed as described previously to probe for multiple specificities.15 , 30
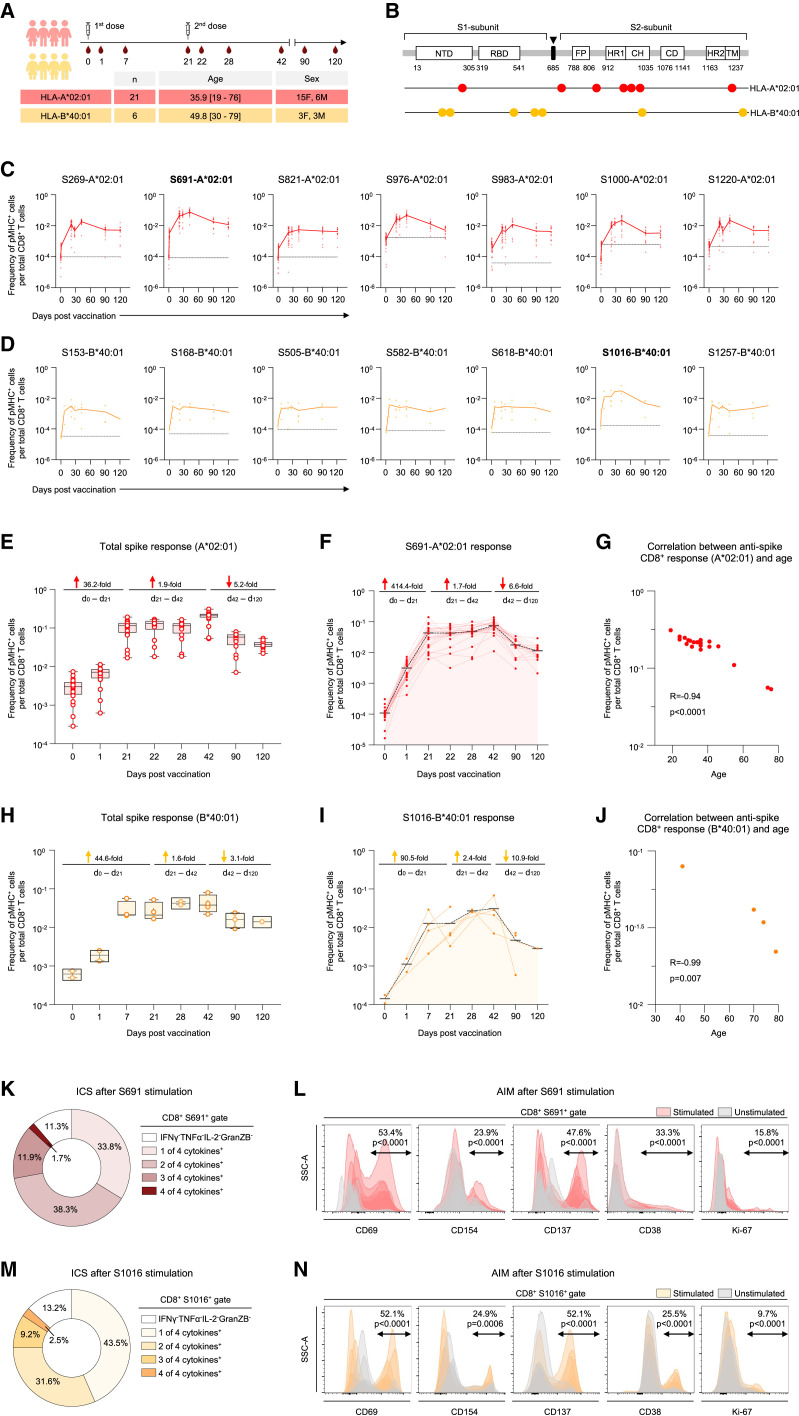
We first measured the spike-specific CD8+ T cell response in SARS-CoV-2 naive vaccinees to estimate the response kinetics to the vaccine. The samples were collected from individuals on day 0 (within 12 h of the first dose) and subsequently followed up to 4 months with blood draws (Figure 1 A). PBMCs from unvaccinated individuals collected at least 1 year before the pandemic were used to ascertain the baseline frequency of SARS-CoV-2 epitopes. We tested fourteen epitopes across HLA-A∗02:01 and HLA-B∗40:01 alleles spanning the entire spike sequence (Figures 1B–1D; Table S1). On day 0, SARS-CoV-2 specific CD8+ T cells were detectable with total HLA-A∗02:01 anti-spike responses ranging between ∼0.007% and 0.1% (Figure 1C), similar to that observed in pre-pandemic samples. We observed an extremely rapid mobilization of antigen-specific CD8+ T cells (Figure 1E). The efficient induction of the immune response after mRNA vaccination resulted in a 36.2-fold increase in spike-specific CD8+ T cells after first dose, consistent with a previous report11 (Figure 1E). The frequency of total spike-specific CD8+ T cells increased from 0.31% at baseline to 10.5% before the second dose (Figure 1E). High frequencies of HLA-A∗02:01 spike-specific CD8+ T cells persisted for several weeks after the second dose with the nominal peak at day 42 (Figure 1E). At day 42, 19.9% CD8+ T cells were specific for the HLA-A∗02:01 epitopes tested. A 5.2-fold contraction was observed by days 42 and 120, but the frequencies remained high in comparison versus day 0 (Figure 1E). We also measured the response to seven distinct HLA-B∗40:01 spike epitopes (Figure 1D) and observed similar kinetics, with a 44.6-fold increase in the frequency of HLA-B∗40:01-restricted spike-specific CD8+ T cells after the first dose (Figure 1H). The frequencies went up further following the second dose of vaccination (Figure 1H). However, the magnitude of spike-specific response to the HLA-B∗40:01 epitopes was lower than that observed for HLA-A∗02:01 (Figures 1E and 1H), showing that some alleles may be much better at stimulating T cell responses than others. The spike-specific CD8+ T cell response was inversely correlated with age but did not show an association with sex (Figures 1G, 1J, S1A, and S1B).
Figure 1.
Vaccine elicited spike-specific CD8+ T cell responses
(A) The experimental design to evaluate the CD8+ T cell response to BNT162b2 vaccination. Timeline showing sequential blood draws post vaccination (first dose [day 0] and second dose [day 21]) in HLA-A∗02:01 and HLA-B∗40:01 donors. The number of donors (n), age, and sex are indicated.
(B–F) (B) Fourteen CD8+ T cell epitopes from SARS-CoV-2 spike protein were evaluated. The magnitude of CD8+ T cell responses to distinct SARS-CoV-2 spike epitopes in (C) HLA-A∗02:01 and (D) HLA-B∗40:01 vaccinees. Baseline for each epitope is shown by a dotted line, determined using pre-pandemic samples (n = 5). Each donor is represented by a dot. Fold-change in the CD8+ T cell response to (E) the spike protein and to (F) the dominant epitope (S691) in HLA-A∗02:01 restricted vaccinees.
(G–I) (G) Correlation between spike-specific CD8+ T cell response at day 42 and age in HLA-A∗02:01 donors. The CD8+ T cell response dynamics to (H) the spike protein and to (I) the dominant epitope (S1016) in HLA-B∗40:01 restrictecinees.
(J) Correlation between spike-specific CD8+ T cell response (day 42) and age in HLA-B∗40:01 donors.
(K and M) Fraction of cytokine producing CD8+ T cells within (K) S691/A∗02:01 and (M) S1016/B∗40:01 specific CD8+ T cells at peak after peptide stimulation.
(L and N) Fraction of cells expressing activation-induced markers (AIM) within (L) S691/A∗02:01 and (N) S1016/B∗40:01 specific CD8+ T cells at peak after peptide stimulation. Data are presented as mean ± range. The Pearson correlation coefficient and statistical significance are noted in (G) and (J). See also Figures S1 and S2.
The CD8+ T cell response to different epitopes varied considerably (Figures 1C and 1D). Nevertheless, we observed very similar kinetics for all the tested epitopes (Figures 1C and 1D). S691 was the most prominent among the seven HLA-A∗02:01 epitopes, with a peak median frequency of 7.5% of the CD8+ T cells (Figures 1C, 1F, S2A, and S2B). The epitope S976, well-conserved across human coronaviruses (hCoVs), also contributes prominently to the overall response with a peak median frequency of 4.6% (Figure 1C). The rest of the HLA-A∗02:01 epitopes had lower frequencies at peak, from 0.5% to 2.2% (Figure 1C). Among the seven HLA-B∗40:01 epitopes, S1016 was the most dominant, peaking at 3.1%, whereas other epitopes ranged from 0.15% to 0.28% (Figures 1D, 1I, and S2B). The baseline epitope-specific CD8+ T cell response is strongly correlated with the epitope conservation across seasonal hCoVs, whereas the peak epitope-specific CD8+ T cell frequencies demonstrated a moderate correlation with epitope conservation across seasonal hCoVs (Figures S1D and S1E). Our results suggest that mRNA vaccination can induce robust responses to novel spike epitopes and is not limited to cross-reactive specificities imprinted from past seasonal hCoV exposures.
Next, we evaluated the functional capacity of the antigen-specific CD8+ T cells following peptide stimulation. PBMC samples collected at day 42 were stimulated with peptides corresponding to the dominant epitopes identified in this study, HLA-A∗02:01/S691 and HLA-B∗40:01/S1016. After stimulation, we performed cytokine profiling by intracellular staining (ICS) of pMHC-spheromer+ CD8+ T cells (Figures 1K and 1M). Most antigen-specific cells made IFNγ and were also able to produce TNF-α and IL-2. A minor subset also produced Granzyme B. We also measured activation-induced markers (AIMs) (Figures 1L and 1N). As shown, the dominant epitopes induced the expression of multiple activation markers; CD69, CD154, CD137, CD38, and a marker of proliferation, Ki-67. This durable and stable induction of polyfunctional CD8+ T cells might contribute to the high efficacy of mRNA vaccines.
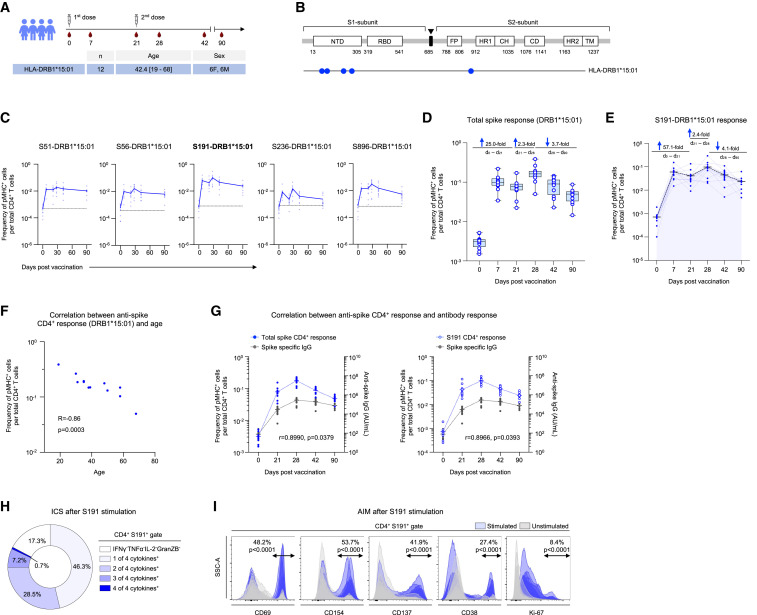
We also surveyed the spike-specific CD4+ T cell response after vaccination (Figures 2 A and 2B; Table S1). At day 0, the frequency of epitope-specific CD4+ T cells ranged from 0.05% to 0.07%, which was comparable with the levels in pre-pandemic samples (Figure 2C). We observed a rapid increase in the frequencies of spike-specific CD4+ T cells within a week after the first dose (Figure 2D). The second dose led to a smaller increase (2.3-fold) in the overall anti-spike CD4+ T cell response (Figure 2D). However, in contrast to the CD8+ T cells, a decrease in the circulating anti-spike CD4+ T cells was observed by day 42 (Figure 2D). This discordance in the kinetics of the major T cell subsets may relate to the distinct functions they execute. Even so, spike-specific CD4+ T cells were detectable at higher frequencies in circulation in comparison to day 0, even 3 months after vaccination (Figure 2D). Among the tested epitopes, the most dominant response was observed against S191, with a median frequency of 9.7% on day 28 (Figures 2C, 2E, and S2B). The other epitopes varied between 1.5% and 2.9% (Figure 2C). The kinetics of CD4+ T cells specific to the dominant epitope, S191, followed the total spike response (Figure 2E). As with the CD8+ T cells, the CD4+ T cell response was decreased in older individuals but showed no sex association (Figures 2F and S1C). The total spike-specific and dominant S191 epitope-specific CD4+ T cell response kinetics further correlated with SARS-CoV-2 spike-specific IgG levels (Figure 2G).
Figure 2.
Vaccine elicited spike-specific CD4+ T cell response
(A) The experimental design to evaluate the epitope-specific CD4+ T cell response to BNT162b2 vaccine in longitudinal samples. The number of donors (n), age and sex are indicated.
(B–E) (B) Five CD4+ T cell epitopes from SARS-CoV-2 spike protein were evaluated. The magnitude of CD4+ T cell responses to SARS-CoV-2 epitopes in (C) HLA-DRB∗15:01 vaccinees. Baseline for each epitope is shown (dotted line), determined using pre-pandemic samples (n = 5). Each donor is represented by a dot. Fold-change in the CD4+ T cell response to (D) the spike protein and to (E) the dominant epitope (S191).
(F) Correlation between spike-specific CD4+ T cell response (day 28) and age. The Pearson correlation coefficient and statistical significance are given.
(G) Pearson correlation between the kinetics of vaccine elicited spike-specific IgG response, total spike-specific CD4+ T cell response (left) and DRB∗15:01/S191 specific CD4+ T cell response (right).
(H) Fraction of cytokine producing cells within S191/DRB∗15:01 specific CD4+ T cells (day 28) after peptide stimulation.
(I) Fraction of AIM+ CD4+ T cells within S191/DRB∗15:01 specific CD4+ T cells (day 28) after peptide (S191) stimulation. Data are presented as mean ± range. See also Figures S1 and S2.
We next evaluated the cytokine profile of spike-specific CD4+ T cells after stimulating day 28 PBMCs with the dominant peptide, S191. The pMHC-spheromer+ CD4+ T cells produced IFNγ, TNF-α, IL-2, and Granzyme B, indicating a Th1-skewing as reported previously31 (Figure 2H). These cells also expressed multiple activation markers after stimulation, further validating the functional capacity of vaccine-induced CD4+ T cells (Figure 2I). In contrast to the CD8+ T cell response, the epitope conservation across seasonal hCoVs did not correlate with the baseline or peak CD4+ T cell frequencies, which suggests that the vaccine-induced responses to novel SARS-CoV-2 epitopes (Figures S1F and S1G). Taken together, these robust T cell responses induced by the BNT162N2 mRNA vaccine likely contribute to its remarkable efficacy.
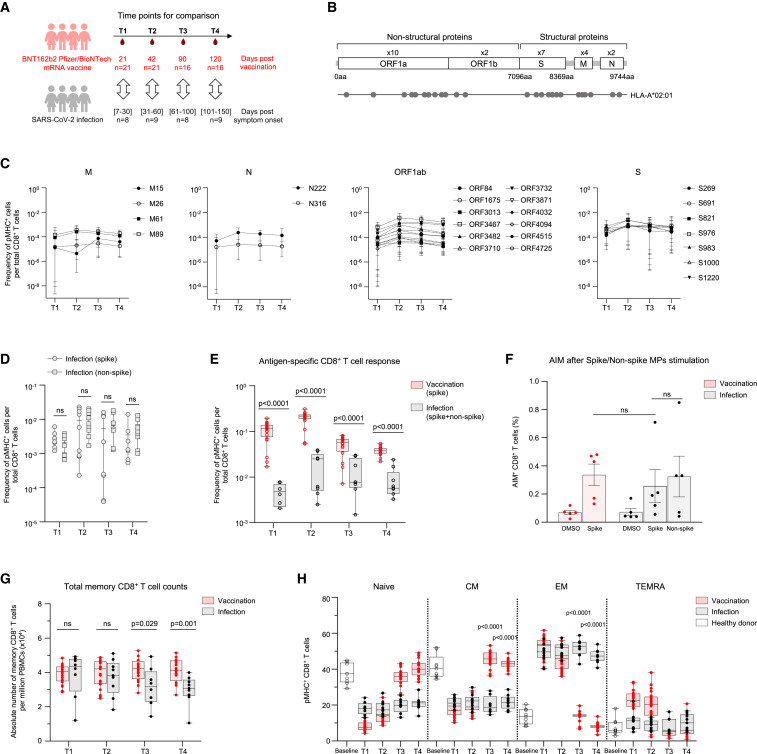
To study the development of anti-SARS-CoV-2 CD8+ T cell immunity mediated by vaccination versus natural infection, we compared the responses of SARS-CoV-2 naive vaccinees and COVID-19 patients. The patient samples were grouped by days since symptom onset and matched with samples from BNT162b2 vaccinees as indicated (Figure 3 A). The patient cohort was established during the first wave of the pandemic (June–December 2020) and was most likely infected by the Wuhan-Hu-1 SARS-CoV-2 strain that matches the vaccine formulation. To perform an integrated analysis, we compiled 12 features of spike-specific CD8+ T cell response derived from flow assays (Figure S3A). Overall, BNT162b2 vaccination and SARS-CoV-2 infection resulted in distinct spike-specific CD8+ T cell profiles indicated by non-overlapping clusters in UMAP space. We observed divergent spike-specific CD8+ T cell response after vaccination and infection in terms of the preferred epitopes (Figure S3B). Although the dominant epitope within spike protein in vaccinees is S691, the main response after infection was against S976 and S983, with median peak frequencies of 0.25% and 0.24%, respectively (Figure S3B). The total spike-specific CD8+ T cell response in circulation elicited by infection was lower in magnitude in comparison to vaccination (Figure S3C). After a single vaccine dose (T1), the spike-specific CD8+ T cell response in circulation was 40.6-fold higher than natural infection (Figure S3C). This difference in median frequency after the second dose of vaccination (T2) ranged from 9.5- to 21.6-fold (Figure S3C). The response to S691 in the COVID-19 patient cohort, the dominant epitope in vaccinated individuals, was 25.1- to 143.4-fold lower across the sampled time points (Figure S3D). As for durability, anti-spike CD8+ T cells were detectable at higher frequencies in circulation in comparison to COVID-19 patients even during the contraction phase (T3 and T4) of the immune response (Figures S3C and S3D). BNT162b2 vaccination induces a T cell response exclusively to spike peptides since the vaccine encodes only that protein. By contrast, SARS-CoV-2 infection generates a response against the whole virus.8 Therefore, to capture that response, we tested eighteen additional epitopes derived from three different genes (ORF1ab = 12, M = 4, and N = 2) (Figure 3B). The magnitude of T cell response to both spike and non-spike epitopes in COVID-19 patients was comparable (Figures 3C and 3D). At the nominal peak after vaccination (T2), the CD8+ T cell response (spike only) in naive vaccinees was 10.6-fold higher than that in COVID-19 patients (spike and non-spike epitopes) (Figure 3E). We also performed peptide mega pool (MP) stimulation assay since it enables profiling a much broader landscape of T cell responses. We did not observe any difference in the response to spike and non-spike peptide pools among COVID-19 patients (Figures 3F and S4A). In contrast to pMHC-spheromer staining (Figures 3F and S4A), we observed a slight but not significant 1.3-fold decrease in the CD8+ T cell response to spike peptide pool stimulation in COVID-19 patients in comparison to vaccinees by AIM assay (Figures 3F and S4A). This discrepancy between pMHC-spheromer staining and AIM assay could in part be due to the limitation of the peptide stimulation assay to capture all relevant T cells due to the relative lack of sensitivity. We recently observed that Mycobacterium tuberculosis (Mtb) MP captures only a fraction (33.6%) of the total T cell response defined by TCR specificity groups identified from the analysis of 19,044 unique TCRβ sequences derived from individuals with latent Mtb infection using GLIPH2 algorithm.32 To investigate this further, we performed stimulation with the dominant CD8+ spike peptide (A2/S691) and evaluated the T cell response using both pMHC-spheromer and AIM markers in 16 vaccine donors (Figures S4C and S4D). This allowed us to directly compare pMHC-spheromer+ and AIM+ CD8+ T cell responses. We found that pMHC spheromers captured most (94.6% ± 9.5%) AIM+ CD8+ T cells ((Figure S4C). By contrast, only a fraction (18.1% ± 10.1%) of all pMHC-spheromer+ cells were positive for both CD69 and CD137 (Figure S4C). For the dominant spike peptide, pMHC spheromers detect 9.5-fold more epitope-specific CD8+ T cells compared with the AIM assay (Figure S4D). Thus, we speculate that stimulation assays are able to capture only a fraction of the total responses compared with pMHC spheromers.
Figure 3.
BNT162b2 vaccination and SARS-CoV-2 infection induce distinct CD8+ T cell response
(A) The experimental design to compare the epitope-specific CD8+ T cell response to BNT162b2 vaccine and SARS-CoV-2 infection. Samples were matched by time points for comparison as shown. The number of subjects (n) is indicated.
(B) The twenty-five evaluated CD8+ T cell epitopes mapped onto the SARS-CoV-2 genome.
(C) The magnitude of CD8+ T cell responses to SARS-CoV-2 epitopes in HLA-A∗02:01 restricted COVID-19 patients.
(D) The comparison of spike and non-spike-specific CD8+ T cell response in COVID-19 patients.
(E) The comparison of antigen-specific CD8+ T cell response to BNT162b2 vaccine and SARS-CoV-2 infection. Data in (C)–(E) represented as mean ± range.
(F) Fraction of AIM+ CD8+ T cells in day 42 samples after spike peptide mega pool (spike MP), non-spike peptide mega pool (non-spike MP) or DMSO stimulation. Data presented as mean ± SD.
(G) Total memory CD8+ T cell counts in vaccinees and patients. Data presented as mean ± range.
(H) Antigen-specific memory CD8+ T cell distribution in vaccinees and patients. (CM, central memory; EM, effector memory; EMRA effector memory T cells expressing CD45RA). Data presented as mean ± range. p values were determined by Mann-Whitney test with Holm-Šídák method. See also Figures S3 and S4.
Next, we characterized the memory T cell compartment in these cohorts. The absolute number of the total memory CD8+ T cells at early time points (T1 and T2) was similar between the two cohorts (Figure 3G). The total memory CD8+ T cell counts during late convalescence in COVID-19 patients were 1.3- and 1.4-fold lower compared with vaccinated individuals at T3 and T4, respectively (Figure 3G). We next measured the spike-specific T cell memory subset distribution (Figure 3H). Antigen-mediated activation of spike-specific CD8+ T cells after vaccination led to an effector phenotype (CD45RA+/−CCR7−). The progressive contraction of effector cells after vaccination was coupled with the establishment of robust central memory (CD45RA+CCR7−) (Figure 3H). By contrast, infection resulted in chronic activation of spike-specific CD8+ T cells, with effector cells (CD45RA−CCR7−) dominating the early to late convalescent phases (Figure 3H).
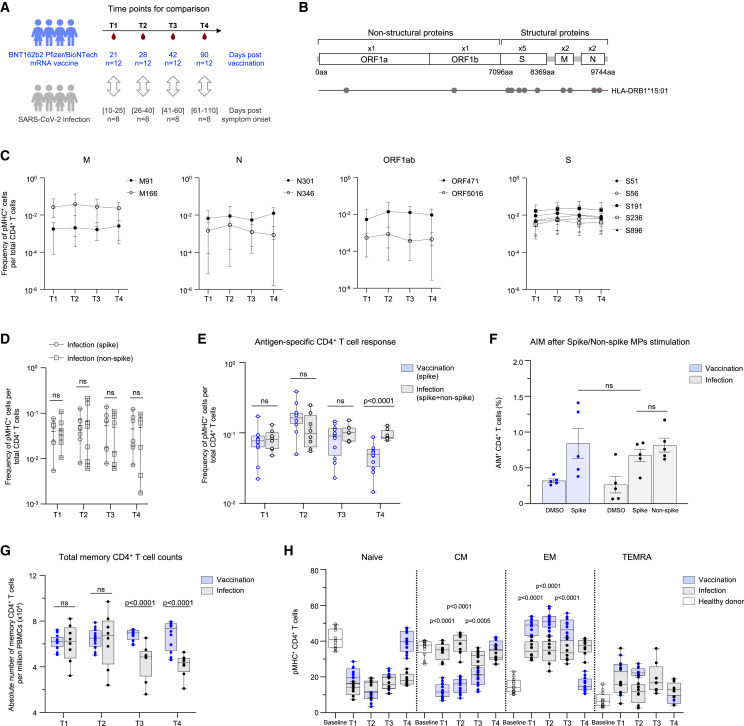
We also measured the effect of BNT162b2 vaccination or SARS-CoV-2 infection on CD4+ T cells (Figure 4 A). The distinct route of exposure to viral antigens, which is vaccination or infection, resulted in non-overlapping spike-specific CD4+ T cell clusters, again suggesting a divergent T cell response (Figure S3E). However, we did not observe any shift in the favored spike epitope between vaccinees and COVID-19 patients, with both cohorts focused on S191 (Figure S3F). The magnitude of spike-specific peripheral CD4+ T cells induced by vaccination demonstrated a higher flux than that in COVID-19 patients (Figure S3G). A single dose of the vaccine (T1) resulted in similar frequencies of spike-specific CD4+ T cells as SARS-CoV-2 infection (Figure S3G), but the second dose of the vaccine resulted in a 3.3-fold higher response in naive vaccinees versus COVID-19 patients (Figure S3G). At later time points (T3 and T4), the spike-specific CD4+ T cells in vaccinees declined to be comparable with COVID-19 patients (Figure S3G). The response to the dominant epitope (S191) followed the same kinetics as the total CD4+ T cell response (Figure S3H). We also measured the CD4+ T cell response to non-spike epitopes (ORF1ab = 2, M = 2, and N = 2) in COVID-19 patients (Figures 4B and 4C) and found that they were comparable with that of the spike epitopes (Figure 4D). The CD4+ T cell response between COVID-19 patients (spike and non-spike) and naive vaccinees (spike only) was comparable at all time points except at T4 (Figure 4E). We did not observe any difference in the CD4+ T cell activation between COVID-19 patients and naive vaccinees by AIM assay at the nominal peak (T2) after vaccination (Figure 4F).
Figure 4.
BNT162b2 vaccination and SARS-CoV-2 infection elicited CD4+ T cell response
(A) The experimental design to compare the epitope-specific CD4+ T cell response with BNT162b2 vaccine and SARS-CoV-2 infection. Samples matched for comparison as shown. The number of subjects (n) is indicated.
(B) The eleven evaluated CD4+ T cell epitopes are mapped onto the SARS-CoV-2 genome.
(C) The magnitude of CD4+ T cell responses to SARS-CoV-2 epitopes in COVID-19 patients.
(D) The comparison of spike and non-spike-specific CD4+ T cell response in COVID-19 patients.
(E) The comparison of antigen-specific CD4+ T cell response to BNT162b2 vaccine and SARS-CoV-2 infection. Data in (C)–(E) are represented as mean ± range.
(F) Fraction of AIM+ CD4+ T cells in day 28 samples after spike MP, non-spike MP or DMSO stimulation. Data represented as mean ± SD.
(G) Total memory CD4+ T cell counts in vaccinees and patients. Data represented as mean ± range.
(H) Antigen-specific memory CD4+ T cell distribution in vaccinees and patients. Data represented as mean ± range. p values determined by Mann-Whitney test with Holm-Šídák method. See also Figures S3 and S4.
However, we saw a marked difference in memory CD4+ T cells between the two cohorts. Although we saw higher frequencies of antigen-specific CD4+ T cells in COVID-19 patients during late convalescence (T4), there was a reduction in the total memory CD4+ T cells at these time points (T3 and T4) compared with naive vaccinees (Figure 4G). Analogous to the CD8+ T cell response, mRNA vaccination resulted in the rapid recruitment of spike-specific effector CD4+ T cells (CD45RA+/−CCR7−) (Figure 4H). The contraction of effector cells was concomitant with central memory (CD45RA+CCR7−) spike-specific CD4+ T cells (Figure 4H). By contrast, natural infection resulted in a more even distribution of spike-specific CD4+ T cells across the effector (CD45RA−CCR7−) and central memory (CD45RA+CCR7−) subsets throughout convalescence (Figure 4H). Taken together, these results suggest differences in how CD8+ and CD4+ T cell responses are triggered by SARS-CoV-2 infection versus BNT162b2 vaccination. Although we cannot exclude the possibility of virus-specific T cell localization in the lung during the course of infection for the noticeably lower circulating spike-specific CD8+ T cells,33 this difference could also be a consequence of the virus’s ability to dampen protective host immune responses via the inhibition of MHC-I expression.34 , 35 , 36
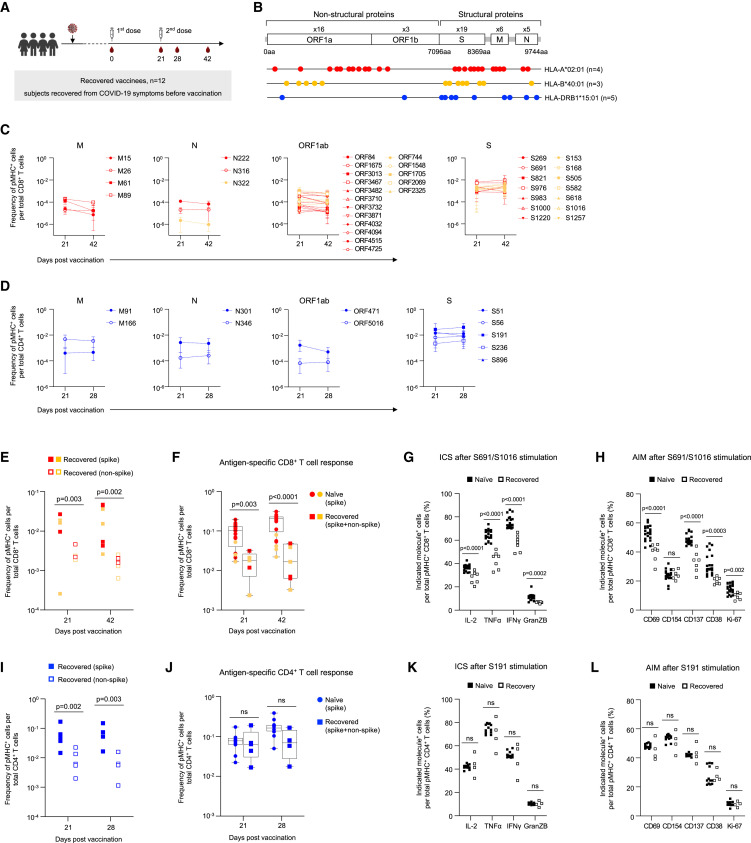
We also investigated the effect of mRNA vaccination in subjects who had previously recovered from SARS-CoV-2 infection (Figures 5 A and 5B). Not surprisingly, the major response in these individuals was to the spike epitopes (Figures 5C and 5D), which were 12.5-fold (at day 42) and 11.3-fold (at day 28) higher than non-spike epitopes for CD8+ and CD4+ T cells, respectively (Figures 5E and 5I).
Figure 5.
Reduced peripheral vaccine-induced CD8+ T cell response in recovered COVID-19 patients
(A) The experimental design to study the CD8+ and CD4+ T cell responses to BNT162b2 vaccine in individuals recovered from previous COVID-19 infection. Timeline indicating the collection of sequential blood samples from HLA-A∗02:01, HLA-B∗40:01 (days 21 and 42) and HLA-DRB1∗15:01 (days 21 and 28) recovered vaccinees. The number of donors (n) is indicated.
(B) Thirty-eight CD8+ T and eleven CD4+ T cell epitopes evaluated in this study are mapped onto the SARS-CoV-2 genome. The number of donors (n) is indicated.
(C) The magnitude of CD8+ T cell responses to SARS-CoV-2 epitopes in HLA-A∗02:01 (red) and HLA-B∗40:01 (yellow) donors.
(D) The magnitude of CD4+ T cell responses to SARS-CoV-2 epitopes in HLA-DRB1∗15:01 donors. Data in (C) and (D) are represented as mean ± range.
(E) The comparison of spike and non-spike-specific HLA-A∗02:01 (red) and HLA-B∗40:01 (yellow) CD8+ T cell responses.
(F) The comparison of HLA-A∗02:01 (red) and HLA-B∗40:01 (yellow) CD8+ T cell responses to BNT162b2 vaccine in naive and recovered vaccinees. Data represented as mean ± range.
(G and H) Fraction of (G) cytokine producing and (H) AIM expressing T cells within S691/A∗02:01 and S1016/B∗40:01 specific CD8+ T cells (day 42 samples) after peptide stimulation (S691 and S1016, respectively).
(I) The comparison of spike and non-spike-specific CD4+ T cell response in recovered vaccinees.
(J) The comparison of antigen-specific CD4+ T cell response to BNT162b2 vaccine in naive and recovered vaccinees.
(K and L) Fraction of (K) cytokine producing and (L) AIM expressing T cells within S191/DRB∗15:01 specific CD4+ T cells (day 28) after peptide stimulation (S191). p values were determined by Mann-Whitney test with Holm-Šídák method. See also Figure S5.
As with SARS-CoV-2 naive individuals, the dominant CD8+ T cell response was against HLA-A∗02:01/S691 and HLA-B∗40:01/S1016 (Figure 5C). However, the total peripheral CD8+ T cell response in convalescent individuals after vaccination was 5.5-fold lower than naive vaccinees after the first dose (day 21) (Figure 5F). Furthermore, we observed minimal boosting of the CD8+ T cell response after the second dose of vaccination, resulting in 7.3-fold lower CD8+ T cell levels in the circulation in comparison to naive vaccinees at day 42 (Figure 5F). By contrast, there was no dampening of specific CD4+ T cell responses between the SARS-CoV-2 naive and pre-exposed individuals (Figure 5J). We also performed a detailed characterization of the spike-specific CD8+ and CD4+ T cell response kinetics in a subset of these individuals (Figure S5A). We noticed that the previous infection did not affect the early spike-specific CD8+ T cell response (days 0–7) (Figure S5B). However, attenuation of the circulating CD8+ response was apparent by day 21. The boost in the CD8+ T cell response after the second dose was minimal and could be due to faster response kinetics in convalescent individuals in comparison to naive vaccinees as previously reported.37 This could also contribute to the difference in the total CD8+ T cell response (spike and non-spike) that was maximum at day 42 (Figure 5F). This difference in the spike-specific CD8+ T cell response was no longer significant 3 months after the first vaccination (Figure S5B). This suggests that BNT162b2 vaccination can partially rescue the lower circulating CD8+ T cell responses observed after SARS-CoV-2 infection. The decrease in the magnitude of circulating spike-specific CD8+ T cells after vaccination in recovered COVID-19 patients was also associated with reduced functionality. PBMCs (day 42) stimulated with spike peptides (S691 or S1016) had a reduced capacity to produce cytokines such as IFNγ, -α, and IL-2 and dampened cytotoxic potential (Granzyme B) (Figure 5G). They were also refractory to activation as seen by the lower expression of multiple activation markers such as CD69, CD137, CD38, and Ki-67, but not CD154 (Figure 5H). However, we did not observe any impaired functionality of spike-specific CD4+ T cell responses after vaccination (Figures 5K, 5L, and S5C). Overall, our results show that SARS-CoV-2 infection impairs CD8+ T cell responses to the BTN162b2 vaccine but not CD4+ T cell responses.
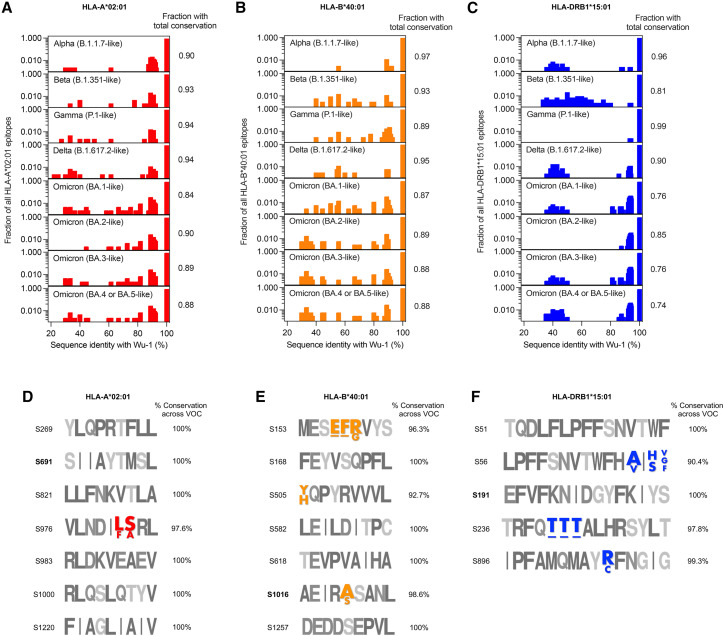
Finally, the emergence of several new SARS-CoV-2 variants raises the question of immune evasion. A high degree of functional preservation is seen in memory T cell responses against early SARS-CoV-2 variants by the AIM assay.38 In total, 84% (CD4+) and 85% (CD8+) of the memory T cell response induced on vaccination with the Wu-1 strain is preserved against the Omicron variant (B.1.1.529).38 However, multiple lineages of the Omicron (B.1.1.529) variant have since emerged that escape from vaccine or infection-induced neutralizing antibodies.39 Therefore, we analyzed the conservation of predicted spike-derived T cell epitopes from the Wu-1 strain across the SARS-CoV-2 variants, including the subvariants BA.4 and BA.5 (Figures 6 A–6C). Overall, the T cell epitopes are fairly conserved across all the analyzed variants, with an average total conservation score of 90.3% and 90.8% for HLA-A∗02:01 and HLA-B∗40:01, respectively (Figures 6A and 6B). The average total conservation score for HLA-DRB1∗15:01 restricted T cell epitopes was marginally lower (84.6%) (Figure 6C). The Omicron subvariant BA.4 and BA.5 had the least conservation of both CD8+ and CD4+ T cell epitopes compared with the Wu-1 strain (Figures 6A–6C). A total conservation of 88% for both HLA-A∗02:01 and HLA-B∗40:01 T cell epitopes was observed between Wu-1 and Omicron subvariants BA.4 and BA.5 (Figures 6A and 6B), as opposed to only 74% for HLA-DRB1∗15:01 (Figure 6C). These results indicate that continued virus evolution could attenuate T cell responses. However, the epitopes we tested in this study are fairly conserved across all variants (Figures 6D–6F). The dominant epitopes, HLA-A∗02:01/S691 and HLA-DRB1∗15:01/S191, are completely conserved across all analyzed variants including BA.4 and BA.5 (Figures 6D and 6E). HLA-B∗40:01/S1016 is 97.6% conserved across all variants (Figure 6F). Presently, the BQ and XBB subvariants of SARS-CoV-2 Omicron are spreading rapidly across the globe, and their neutralization by sera from vaccinees and infected individuals is low.40 Even so, the dominant epitopes for HLA-A∗02:01/S691, HLA-B∗40:01/S1016, and HLA-DRB1∗15:01/S191 as described here are completely conserved in these variants. In this context, Poon et al. monitored the viral diversity in individuals after vaccination and observed that T cell responses do not appear to have a substantial impact on the emergence of these recent viral variants.41
Figure 6.
T cell epitope conservation across SARS-CoV-2 variants
The fractional conservation of all predicted spike-derived T cell epitopes from the SARS-CoV-2 reference Wuhan-1 (Wu-1) strain against the indicated SARS-CoV-2 variant for (A) HLA-A∗02:01 (B) HLA-B∗40:01 and (C) HLA-DRB1∗15:01 are shown. The Pango lineage for each SARS-CoV-2 variant is also mentioned. The fraction of spike epitopes from Wu-1 strain that are fully conserved in each SARS-CoV-2 variant is listed. The logograms show the conservation of all spike-derived T cell epitopes tested in this study for (D) HLA-A∗02:01, (E) HLA-B∗40:01, and (F) HLA-DRB1∗15:01. The mutated residues are colored and labeled accordingly. An amino acid deletion is marked as “-.”
Discussion
The SARS-CoV-2 pandemic has had an enormous health and economic impact worldwide, and thus, a detailed investigation of the mechanisms mediating the high efficacy of the novel RNA vaccines3 , 4 , 5 , 8 , 9 , 10 , 11 , 12 is warranted and should help in the design of vaccines against other pathogens. Using the spheromer technology,15 we probed the kinetics and durability of epitope-specific CD8+ and CD4+ T cell responses after mRNA vaccination in naive and COVID-19 patients. Spheromers can detect ∼3- to 5-fold more specific T cells than tetramers.15 Here, we analyzed the response to the BNT162b2 vaccine and observed a rapid induction of CD8+ and CD4+ T cells, with an increase in the total HLA-A∗02:01 spike-specific response as early as day 1 after vaccination. Here, extending previous results with CD8 T cells,11 we surveyed multiple epitopes and also CD4+ T cell specificities. Previously we found that the frequency of SARS-CoV-2 specific CD8+ T cells in unexposed individuals correlates with epitope conservation across seasonal hCoVs.15 We saw a similar correlation in spike-specific CD8+ T cells and sequence conservation prior to vaccination here, but by day 42 post vaccination, there was only a weak correlation with epitope conservation. Specifically, the dominant CD8+ T cell response at the nominal peak (day 42) was against HLA-A∗02:01/S691 and HLA-B∗40:01/S1016 with frequencies of 7.5% and 3.1%, respectively. These results suggest that mRNA vaccination can efficiently induce a response to novel spike epitopes. Antonio et al. found a high degree of structural convergence of physico-chemical properties of A2/S691 peptide with the immunodominant influenza virus matrix epitope (A2/M1) despite poor sequence conservation.42 TCRs that are specific to both influenza-M1 and SARS-CoV-2 antigens have also been reported.43 This cross-reactivity may explain the higher response we observed against A2/S691 in comparison to A2/S269. Regarding the CD4+ T cell response, the dominant HLA-DRB1∗15/S191 epitope constituted 9.7% of all CD4+ T cells at the nominal peak (day 28). This observation of a higher spike-specific CD4+ T cell response compared with CD8+ T cells is consistent with previous studies.3 , 4 However, in contrast to the results from peptide pool stimulation,3 with pMHC spheromers, we found that the CD4+ and CD8+ responses did not follow the same kinetics. The CD4+ T cell kinetics were synchronous with the spike-specific antibody response, with the peak at day 28 (1 week after the second dose), followed by a contraction. By contrast, we observed a steady increase in the antigen-specific CD8+ T cell response all the way up to day 42 (3 weeks after the second dose). This discordance is unusual compared with other studies where both CD4+ and CD8+ responses peak in the blood about 6–8 days after stimulation in a memory response.17 , 18 , 19 , 20 This may be due to the distinct features of the mRNA vaccine platform. This prolonged induction of CD8+ T cells after vaccination may also relate to the striking increase in IFNγ levels observed after the second dose of BNT162b2 vaccine3 , 44 as opposed to an earlier cytokine surge observed with other vaccines. Although the frequency of spike-specific CD8+ and CD4+ T cells in circulation decreased with time in comparison to the peak levels, they were still detectable 3–4 months after vaccination, indicating a durable T cell response. An elegant study by Mudd et al.10 shows the persistence of spike-specific T follicular helper cells (DP4/S167) in the lymph nodes at a relatively higher frequency in comparison to peripheral circulation at matched time points. The considerable longitudinal sampling of vaccinees further allowed us to study the development of T cell memory. Although we observed differences in the magnitude of response to distinct spike epitopes, the formation of CD8+ and CD4+ T cell memory after vaccination was quite similar across different epitopes. Overall, there was an increase in antigen-specific effector T cells (CCR7-CD45RA+/−) by day 21 that contracted to nearly pre-vaccination levels by day 90. Concomitantly, the spike-specific T cells in circulation after 3–4 months after vaccination exhibited a central memory phenotype (CCR7+CD45RA−). This is important since a stable memory pool could effectively protect against future SARS-CoV-2 infections by their rapid recruitment in the immune response.
We also compared T cell responses after vaccination with natural infection. We found that the circulating antigen-specific CD8+ T cell response was much lower in SARS-CoV-2 infection versus vaccination. Specifically, the nominal peak after vaccination was 10.6-fold higher than that in infected individuals and decreased to 4.3-fold at 4 months after vaccination for spike-specific responses. We also observed a skewing in the preferred CD8+ T cell epitopes targeted after infection in comparison to vaccination, with the maximal spike response against HLA-A∗02:01/S976 with a median frequency of 0.25% at peak. The difference in preferred spike specificities between the two cohorts is likely due to the differences in antigen localization, processing, and presentation after infection versus vaccination. The infection-induced spike-specific CD4+ T cell response in circulation was marginally reduced (3.3-fold) at the peak in comparison to vaccination, but no difference was observed in the total (spike and non-spike) CD4+ T cell frequencies. This marginal reduction in the spike-specific peripheral CD4+ T cells could explain the lower antibody titers observed in individuals experiencing mild symptoms after SARS-CoV-2 infection in comparison to the post-vaccination antibody titers observed in SARS-CoV-2 naive vaccinees.45 The migration of virus-specific T cells after infection to the respiratory tract33 or lymphopenia after SARS-CoV-2 infection46 can also cause lower spike-specific T cells in the periphery. We suggest that this may also be a consequence of the virus’s strategy to escape host defense by specifically inhibiting the MHC-I expression, as reported recently.34 , 35 , 36 Here, we were only able to analyze peripheral T cell responses, as is a typical limitation of human studies. A recent study using pMHC multimers did not observe any difference in the frequency of SARS-CoV-2-specific CD8+ T cells between infected and vaccinated individuals.9 We speculate that this could be a combined effect of the different specificities and time points used, both crucial factors as shown here. We also observed that spike-specific CD8+ T cells induced after infection exhibited an effector phenotype even 5 months after symptom onset. This could be a consequence of viral persistence.47 We suggest that chronic activation probably leads to reduced virus-specific memory CD8+ T cells in comparison to BNT162b2 vaccination. This may contribute to the increased prevalence of breakthrough SARS-CoV-2 infection in COVID-19 patients compared with vaccinees seen in some studies. Eggink et al., observed an increased risk of Omicron infection in previously infected individuals (odds ratio [OR]: 4.2; 95% confidence interval [CI]: 3.8–4.7) compared with naive vaccinated individuals. The OR of Omicron infection among vaccinated individuals was 3.6 (95% CI: 3.4–3.7). This is in contrast to susceptibility to infection by other SARS-CoV-2 variants.48 In another study evaluating protection conferred by mRNA vaccines and previous infection against Omicron in a prison cohort (a high-risk population), the authors observed higher levels of effectiveness from vaccination among staff in comparison to previous infection. However, no difference was observed in the inmates.49 However, it is important to note that these results are contrary to that observed by Altarawneh et al.50 They observed a higher effectiveness of previous infection (alone) against symptomatic BA.2 infection in comparison to two doses of BNT162b2 mRNA vaccine (>6 months before infection) in naive individuals.
We also analyzed the impact of previous SARS-CoV-2 infections on BNT162b2 vaccine-induced T cell responses. Previous studies found no deficit in neutralizing antibody titers to the ancestral Wuhan-Hu-1 SARS-CoV-2 strain after vaccination in pre-exposed individuals.5 Accordingly, we observed no effect on the CD4+ T cell response. However, we did observe a major reduction in both the magnitude and functionality of peak CD8+ T cell responses in previously infected individuals after vaccination. This could be a result of the disproportionate effect of infection on the CD8+ T cell compartment in comparison to CD4+ T cells, as discussed previously. The deterioration of CD8+ T cell function is seen in patients with active viral infections that had been either eliminated, in the case of HCV or greatly reduced (HIV).51 This dysfunction persists for a year or more after the active phase of infection, suggesting lasting damage, despite the absence or near absence of the relevant virus. In this context, it may be that these attenuated CD8+ T cell responses contribute to long COVID, perhaps rendering patients unable to respond robustly to subsequent infections by SARS-CoV-2 variants or other pathogens. Another factor that could contribute to the lower circulating spike-specific T cells in convalescent individuals could be due to the reduced immunogenicity of the mRNA vaccine resulting from antigen sequestration mediated by infection-induced antibodies in circulation. Previous studies52 , 53 have reported higher levels of T cell responsiveness after spike peptide pool stimulation in vaccinated individuals undergoing treatment with anti-CD20 antibody monotherapy or anti-CD19 CAR T that result in lower spike-specific antibodies in comparison to healthy individuals.
Finally, we evaluated the conservation of spike-derived T cell epitopes evaluated in this study across SARS-CoV-2 variants. The dominant epitopes identified here are almost completely conserved, including in the BA.4 and BA.5 subvariants. This can be critical since a reduction in the neutralizing antibody titer in comparison to the reference Wu-1 isolate is seen with the Omicron subvariants even after the administration of a booster dose (3rd vaccine dose).39 The neutralizing antibody titer is lower by a factor of 6.4, 7.0, and 14.1 against BA.1, BA.2, and BA.2.12.1 subvariants, respectively. Furthermore, a 21-fold reduction in the neutralizing antibody titer is seen against the BA.4 and BA.5. Considering this continued viral evolution, the identification of conserved, dominant T cell epitopes as reported here may facilitate the much-needed development of pan-coronavirus vaccines.
In summary, our study elucidates the magnitude, diversity, and kinetics of specific CD4+ and CD8+ T cell responses after BNT162b2 vaccination, and the effects of SARS-CoV-2 infection on these responses. It will be interesting to see whether some of the characteristics we see here are common features of RNA vaccines to other pathogens. In addition, the apparent damage of the CD8+ T cell response by viral infection is cause for concern and may leave even vaccinated individuals with a previous infection at risk for subsequent infections or other health issues.
Limitations of the study
Our study has limitations in that we measured only peripheral T cell responses, and differential tissue localization of immune cells after mRNA vaccination and SARS-CoV-2 infection can contribute to the differences observed between the cohorts. We speculate that virus-induced MHC-I suppression drives the specific attenuation of CD8+ T cell responses after infection, but other factors such as differential kinetics and spike antigenicity in pre-exposed individuals can also affect CD8+ T cell responses in convalescent individuals. Future studies are warranted to delineate the relative impact of these factors. Finally, although we used a large panel of forty-nine epitopes to characterize the SARS-CoV-2-specific T cell responses, this is not exhaustive, and other epitopes might conceivably yield different results.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD14 | BioLegend | Cat# 367123; RRID:AB_2716228 |
| anti-CD19 | BioLegend | Cat# 363020; RRID:AB_2564229 |
| anti-CD33 | BioLegend | Cat# 366609; RRID:AB_2566402 |
| anti-γδTCR | BioLegend | Cat# 331220; RRID:AB_2564275 |
| anti-CD3 | BioLegend | Cat# 300406; RRID:AB_314060 |
| anti-CD8 | BD Biosciences | Cat# 563795; RRID:AB_2722501 |
| anti-CD4 | BioLegend | Cat# 300553; RRID:AB_2564381 |
| anti-CCR7 | BioLegend | Cat# 353236; RRID:AB_2563641 |
| anti-CD45RA | BioLegend | Cat# 304138; RRID:AB_2563815 |
| anti-CD28 | BD Biosciences | Cat# 555726; RRID:AB_396069 |
| anti-CD49d | BD Biosciences | Cat# 555501; RRID:AB_2130052 |
| anti-CXCR3 | BD Biosciences | Cat# 557184; RRID:AB_396595 |
| anti-CXCR5 | BD Biosciences | Cat# 552032; RRID:AB_394324 |
| anti-IL2 | Biolegend | Cat# 500332; RRID:AB_2563877 |
| anti-TNFa | Creative Diagnostics | Cat# CABT-WN1553; RRID:AB_2460295 |
| anti-IFNg | BD Biosciences | Cat# 340452; RRID:AB_400428 |
| anti-GranZB | Biolegend | Cat# 372211; RRID:AB_2728378 |
| anti-CD69 | Biolegend | Cat# 310922; RRID:AB_493775 |
| anti-CD154 | Biolegend | Cat# 310823; RRID:AB_10933251 |
| anti-CD137 | Biolegend | Cat# 309833; RRID:AB_2734279 |
| anti-CD38 | Thermo Fisher Scientific | Cat# 15-0389-41; RRID:AB_1834382 |
| anti-Ki-67 | BD Biosciences | Cat# 563756; RRID:AB_2732007 |
| Biological samples | ||
| PBMC samples from BNT162b2 vaccine donors | Stanford Good Clinical Practice | IRB 8629 |
| PBMC samples from COVID-19 patient | Stanford Occupational Health | IRB 55689 and IRB 55619 |
| PBMC samples from COVID-19 recovered BNT162b2 vaccinated donors | Stanford Good Clinical Practice and Stanford Occupational Health | IRB 8629, IRB 55689, and IRB 55619 |
| Chemicals, peptides, and recombinant proteins | ||
| Human TruStain FcX | BioLegend | #422302 |
| Benzonase nuclease | Millipore Sigma | #71206 |
| MHC-I monomer | NIH tetramer facility core | HLA-A∗02:01 and HLA-B∗40:01 |
| MHC-II monomer | NIH tetramer facility core | HLA-DRB1∗15:01 |
| Peptides (synthesized to 95% purity) | Elim Biopharm | Sequences shown in Table S1 |
| Streptavidin PE-Cyanine7 Conjugate | Thermo Fisher Scientific | # 25-4317-82 |
| Streptavidin PE Conjugate | Thermo Fisher Scientific | #12-4317-87 |
| Streptavidin eFluor™ 450 Conjugate | Thermo Fisher Scientific | #48-4317-82 |
| Streptavidin Alexa Fluor™ 647 conjugate | Thermo Fisher Scientific | #S21374 |
| Streptavidin Brilliant Violet 711 conjugate | BioLegend | #405241 |
| Streptavidin Brilliant Violet 785 conjugate | BioLegend | #405249 |
| Streptavidin PE/Dazzle 594 conjugate | BioLegend | #405247 |
| Cytofix/cytoperm buffer | BD Biosciences | #554714 |
| Perm/wash buffer | BD Biosciences | #554723 |
| Brefeldin-A solution | Thermo Fisher Scientific | #00-4506-51 |
| anti-FITC microbeads | Miltenyi Biotec | #130-048-701 |
| Live/dead fixable aqua dead cell stain kit | Invitrogen | #L34957 |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad software | https://www.graphpad.com/scientificsoftware/prism/ |
| UMAP code package | R studio | N/A |
| FlowJo 10 | BD | https://www.flowjo.com/ |
| IEDB | IEDB website | http://tools.iedb.org/ |
| SYFPEITHI | SYFPEITHI website | http://www.syfpeithi.de/ |
| MARIA | MARIA Stanford | https://maria.stanford.edu/about.php |
| Other | ||
| RPMI 1640 media | Thermo Fisher Scientific | #11875085 |
| Fetal Bovine Serum | R&D Systems | S11150 |
| FACS tube with 70-μm mesh cap | ThermoFisher Scientific | #08-771-23 |
| 30K Amicon tubes | Millipore | #UFC903024 |
| 70μm cell strainer | Corning | 07-201-431 |
| anti-FITC microbeads | Miltenyi Biotec | 130-048-701 |
| 96-well plates | Corning | #3916 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mark. M. Davis (mmdavis@stanford.edu).
Materials availability
Upon specific request and execution of a material transfer agreement (MTA) from School of Medicine, Stanford University to the lead contact, the peptide-MHC spheromer reagents will be made available.
Experimental model and subject details
Human blood sample collection
The BNT162b2 vaccine donors were recruited for the study with informed consent. The study was approved by the Stanford University Institutional Review Board (IRB 8629) and was conducted with full compliance of Good Clinical Practice as per the Code of Federal Regulations. Part of the COVID-19 patient peripheral blood mononuclear cells (PBMCs) sample collection for this study was done at the Stanford Occupational Health under an IRB approved protocol (Protocol Director, Kari C. Nadeau). We obtained samples from adults who had a positive test result for the SARS-CoV-2 virus from an analysis of their nasopharyngeal swab specimens obtained at any point from March 2020 - June 2020. Stanford Health Care clinical laboratory developed internal testing capability with a reverse-transcriptase based polymerase-chain-reaction assay (RT-PCR). All participants consented prior to enrolling in the study. The other COVID-19 patient samples used were from 109 participants enrolled in a Phase 2, single-blind, randomized placebo-controlled trial evaluating the efficacy of Peginterferon Lambda-1a in SARS-CoV-2 infected outpatients.21 , 22 The trial was registered at ClinicalTrials.gov (NCT04331899) and was performed as an investigator-initiated clinical trial with the FDA (IND 419217). In brief, symptomatic outpatients aged 18–71 who tested positive for reverse transcription-polymerase chain reaction (RT-PCR) detection of SARS-CoV-2 within 72 h of enrollment were eligible to participate in the study barring any signs of respiratory distress. Asymptomatic patients were eligible if they had not previously had a positive SARS-CoV-2 test. Full eligibility and exclusion criteria are provided in the study protocol and have been published.21 , 22 PBMC samples from healthy donors were obtained from the Stanford Blood Center according to our IRB approved protocol. All healthy donor samples used in the current study were collected between April 2018 to Feb 2019 before the SARS-CoV-2 pandemic.
Method details
Assembly of pMHC-spheromers
A novel multimeric αβ T cell staining reagent, spheromer, that we reported recently was used to analyze the epitope-specific CD8+ and CD4+ T cell responses.15 The MHC protein purification and peptide exchange were conducted as previously described.54 , 55 The list of peptides used in this study are provided in Table S1. The peptides evaluated in our study were chosen based on a combination of the following criteria: literature search,6 , 9 , 10 , 11 , 12 , 15 , 23 , 24 , 25 , 26 bioinformatic analysis, and MHC stabilization assay. A total of 49 peptides across the entire SARS-CoV-2 genome (SARS-CoV-2/USA/WA-CDC-WA1/2020; Wu-1 strain) were profiled in this study. Briefly, a preliminary list was curated using a combination of previous studies6 , 9 , 10 , 11 , 12 , 15 , 23 , 24 , 25 , 26 and predicted binding affinities using the immune epitope database and analysis resource (IEDB) recommendations (http://tools.iedb.org/).28 Peptides identified from a literature search were included for further analysis only if they were predicted as “strong” binders using the IEDB recommended allele-specific affinity cutoff (HLA-A∗02:01 – 255nM and HLA-B∗40:01 – 639nM). For HLA-DRB1∗15:01, peptides were selected based on a consensus percentile rank of ≤10%. Next, we cross-validated these ‘hits’ using the SYFPEITHI29 and MARIA27 algorithms. MARIA is a deep learning-based algorithm that reportedly outperforms existing prediction methods. Furthermore, amino acids at anchor positions were given higher weights. We also used an MHC stabilization assay to experimentally validate the binding of peptides to ectopically expressed MHC molecules in antigen processing (TAP)–deficient T2 cell lines. Accordingly, we built a broad panel of SARS-CoV-2 peptides (CD8 – spike = 14, non-spike = 24; CD4 – spike = 5, non-spike = 6) representing a wide range of sequence conservation across seasonal human coronaviruses. This enabled us to compare the epitope-specific response kinetics between infection and vaccination, and evaluate the contribution of pre-existing, cross-reactive T cells. The Pfizer/BioNTech BNT162b2 vaccine has two proline mutations (K986P and V987P) to stabilize the spike protein. The engineered maxi-ferritin scaffold was also purified as described previously15 and used spheromer assembly. In brief, the assembly was performed in two steps: 1) Generation of a semi-saturated SAv-pMHC2 complex, and 2) Conjugation of SAv-pMHC2 to the functionalized maxi-ferritin scaffold. SAv-pMHC2 was obtained by incubating 1 μM of the pMHC with 0.45 μM of SAv at 25°C for 30 min without agitation. Subsequently, the spheromer complex was assembled by incubating SAv-pMHC2 with the functionalized scaffold for 1h at room temperature with mild rotation. The fluorophore-conjugated SAv was sourced from Invitrogen. For the simultaneous detection of multiple SARS-CoV-2 spike epitopes using the spheromer technology, we adapted a combinatorial staining approach developed previously.30 Briefly, each peptide was assigned a unique fluorophore-barcode that allows the simultaneous detection of 2n-1 specificities in a sample, where n is the number of distinct fluorophore labels. The relative concentrations for pMHC monomers associated with each fluorophore label was experimentally determined.
PBMC staining and flow cytometry
PBMCs were thawed in a water bath set at 37°C and the cells were immediately transferred to warm RPMI media (Thermo Fisher Scientific) supplemented with 10% FBS (R&D Systems) and 100U/ml of penicillin-streptomycin. After washing, the cells were filtered using a 70μm cell strainer and rested for 1h at 37°C. T cells were enriched from PBMCs by negative selection using a FITC-conjugated antibody cocktail including anti-CD14 (Clone HCD14, BioLegend), anti-CD19 (Clone HIB19, BioLegend), anti-CD33 (Clone HIM3-4, BioLegend) and anti-γδ TCR (Clone 5A6.E9, ThermoFisher Scientific) followed by magnetic bead depletion using anti-FITC microbeads (Miltenyi Biotec). The enriched T cells were washed and resuspended in FACS buffer for staining. All spheromer staining was done for 1h after incubating the cells with Human TruStain FcX (BioLegend) for 15 min on ice. The spheromer were used at a monomeric concentration of ∼100nM and ∼500nM for the staining of CD8+ T cells and CD4+ T cells, respectively. The cells were subsequently stained with anti-CD19 (BV510, clone HIB19), anti-γδTCR (BV510, clone B1), anti-CD33 (BV510, clone HIM3-4), anti-CD3 (PE/Cyanine7, clone OKT3), anti-CD8 (BUV396, clone RPA-T8, BD Biosciences), anti-CD4 (BV785, clone RPA-T4), anti-CCR7 (PE/Dazzle 594, clone G043H7), anti-CD45RA (BV711, clone HI100) and an amine-reactive viability stain (Live/dead fixable aqua dead cell stain kit; Invitrogen) for 30 min on ice, washed, resuspended in FACS buffer and acquired on a BD LSRII flow cytometer. The data was analyzed using FlowJo (v10) software.
Peptide mega pool (MP) and single peptide stimulation
Frozen PBMCs were thawed, counted, and resuspended at a density of 15 million live cells per ml in complete RPMI (RPMI with 10% FBS (Gibco) and antibiotics). 100 μl of cell suspension containing 1.5 million cells was added to each well of a 96-well round-bottomed tissue culture plate. The cells were rested overnight at 37 °C in a CO2 incubator. The next morning, each sample was treated with peptide mega pool (1 μg/ml of each peptide) or single peptide (5 μg/ml) or 0.5% v/v DMSO as negative control in the presence of 1 μg/ml of anti-CD28 (clone CD28.2, BD Biosciences), anti-CD49d (clone 9F10, BD Biosciences), anti-CXCR3 (clone 1C6, BD Biosciences) and anti-CXCR5 (clone RF8B2, BD Biosciences). Peptides were synthesized to 95% purity (Elim Biopharm). All wells contained 0.5% v/v DMSO in total volume of 200 μl per well. The samples were incubated at 37 °C in CO2 incubators for 2 h, and then 10 μg/ml brefeldin-A was added. The cells were further incubated for 6-8 h.
Intracellular cytokine staining (ICS) assay
After peptide stimulation, the cells were washed with PBS containing 5% FCS and stained with amine-reactive viability stain (Live/dead fixable aqua dead cell stain kit; Invitrogen) for 30 min at 4°C. After washing, pMHC-spheromers were added to screen the epitope-specific CD8+ and CD4+ T cells. The samples were stained for 30 min at 4°C in 100 μl volume. After spheromer staining, the cells were washed, fixed and permeabilized with cytofix/cytoperm buffer (BD Biosciences) for 20 min. The permeabilized cells were stained with ICS antibodies (anti-IL2 (clone MQ1-17H12, Biolegend), anti-TNFα (clone Mab11, BD Biosciences), anti-IFNγ (clone B27, BD Biosciences) and anti-GranZB (clone QA16A02, Biolegend)) for 20 min at room temperature in 1X perm/wash buffer (BD Biosciences). Cells were then washed twice with perm/wash buffer and once with staining buffer before acquisition using BD LSRII flow cytometer. The data was analyzed using FlowJo (v10) software.
Activation induced marker (AIM) assay
After peptide stimulation, the cells were washed with PBS containing 5% FCS and stained with amine-reactive viability stain (Live/dead fixable aqua dead cell stain kit; Invitrogen) for 30 min at 4°C. After washing, pMHC-spheromers were added to screen the epitope-specific CD8+ and CD4+ T cells. Meanwhile, the antibody cocktail was added for AIM staining (anti-CD69 (clone FN50, Biolegend), anti-CD154 (clone 24-31, Biolegend), anti-CD137 (clone 4B4-1, Biolegend), anti-CD38 (clone HIT2, BD Biosciences) and anti-Ki-67 (clone B56, BD Biosciences)). The cells were stained for 30 min at 4°C in 100μl volume.
Cells were then washed twice with staining buffer before acquisition using BD LSRII flow cytometer. The data was analyzed using FlowJo (v10) software.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism 8, GraphPad Software, San Diego, California, USA. We performed a meta-analysis to combine the p-values from individual hypothesis tests to assess the significance of the overall distribution. Data were considered statistically significant when p < 0.05. Dimensionality reduction analysis were also performed in R. UMAP to visualize multiparametric flow cytometry data was generated using the “umap” package. The statistical details for each experiment are provided in the associated figure legends.
Acknowledgments
We thank all the volunteers and patients for their participation in this study. We thank the Stanford Occupational Health staff and the Sean N. Parker Center for Allergy and Asthma Research staff for enrolling volunteers. We thank members from the laboratories of Mark Davis and Yueh-Hsiu Chien for helpful discussions. Flow cytometry analysis for this project was done on instruments in the Stanford Shared FACS Facility. HLA-typing was performed at the Stanford Functional Genomics Facility. This work was supported by grants from the Howard Hughes Medical Institute, Open Philanthropy, Bill and Melinda Gates Foundation, and NIAID (AI057229) to M.M.D. Additional support was provided by Center for Human Systems Immunology to V.M. (OPP1113682). Further funds were provided by the Sean N Parker Center, the Sunshine Foundation, and U01 AI140498 to M.M.D. and K.C.N.
Author contributions
Project conceptualization and study design was performed by F.G., V.M., and M.M.D. Experiments were performed by F.G. Data analyses was performed by F.G. and V.M. P.S.A., K.v.d.P., M.M., F.Y., O.W., R.H., E.H., J.-Y.L., C.L., A.S.L., M.M.S., S.B.S., and J.G. assisted with PBMC sample collection, processing, and banking. K.R.K. assisted with epitope selection. R.W., V.R., and J.D.A. provided MHC monomers. J.L. assisted with cell staining. K.R. performed antibody measurements with the supervision of S.D.B. T.T.W. and B.P. provided resources for sample processing. P.J. and K.C.N. supervised volunteer recruitment. F.G., V.M., and M.M.D. wrote the manuscript with input from all the authors.
Declaration of interests
V.M. and M.M.D. are inventors on a patent application (PCT/US2021/064378) on the spheromer technology described in this work.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: March 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2023.03.005.
Supplemental information
Data and code availability
The data supporting the findings of this study are available within the published article and summarized in the corresponding tables, figures, and supplemental materials. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Chakraborty S., Mallajosyula V., Tato C.M., Tan G.S., Wang T.T. SARS-CoV-2 vaccines in advanced clinical trials: where do we stand? Adv. Drug Deliv. Rev. 2021;172:314–338. doi: 10.1016/j.addr.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185 doi: 10.1016/j.cell.2022.01.018. 1025–1040.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 7.Sievers B.L., Chakraborty S., Xue Y., Gelbart T., Gonzalez J.C., Cassidy A.G., Golan Y., Prahl M., Gaw S.L., Arunachalam P.S., et al. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 2022;14:eabn7842. doi: 10.1126/scitranslmed.abn7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Galvez R.I., Cortes F.H., Grifoni A., Tarke A., Chang J., et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185 doi: 10.1016/j.cell.2022.05.022. 2434–2451.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minervina A.A., Pogorelyy M.V., Kirk A.M., Crawford J.C., Allen E.K., Chou C.H., Mettelman R.C., Allison K.J., Lin C.Y., Brice D.C., et al. SARS-CoV-2 antigen exposure history shapes phenotypes and specificity of memory CD8(+) T cells. Nat. Immunol. 2022;23:781–790. doi: 10.1038/s41590-022-01184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mudd P.A., Minervina A.A., Pogorelyy M.V., Turner J.S., Kim W., Kalaidina E., Petersen J., Schmitz A.J., Lei T., Haile A., et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185 doi: 10.1016/j.cell.2021.12.026. 603–613.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wragg K.M., Lee W.S., Koutsakos M., Tan H.X., Amarasena T., Reynaldi A., Gare G., Konstandopoulos P., Field K.R., Esterbauer R., et al. Establishment and recall of SARS-CoV-2 spike epitope-specific CD4(+) T cell memory. Nat. Immunol. 2022;23:768–780. doi: 10.1038/s41590-022-01175-5. [DOI] [PubMed] [Google Scholar]
- 13.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger T., Kornek B. T cells step up after SARS-CoV-2 vaccination with B cell depletion. Nat. Rev. Neurol. 2021;17:729–730. doi: 10.1038/s41582-021-00582-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallajosyula V., Ganjavi C., Chakraborty S., McSween A.M., Pavlovitch-Bedzyk A.J., Wilhelmy J., Nau A., Manohar M., Nadeau K.C., Davis M.M. CD8(+) T cells specific for conserved coronavirus epitopes correlate with milder disease in COVID-19 patients. Sci. Immunol. 2021;6:eabg5669. doi: 10.1126/sciimmunol.abg5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2022;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosor Krnic E., Gagro A., Drazenovic V., Kuzman I., Jeren T., Cecuk-Jelicic E., Kerhin-Brkljacic V., Gjenero-Margan I., Kaic B., Rakusic S., et al. Enumeration of haemagglutinin-specific CD8+ T cells after influenza vaccination using MHC class I peptide tetramers. Scand. J. Immunol. 2008;67:86–94. doi: 10.1111/j.1365-3083.2007.02042.x. [DOI] [PubMed] [Google Scholar]
- 18.Su L.F., Kidd B.A., Han A., Kotzin J.J., Davis M.M. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wild K., Smits M., Killmer S., Strohmeier S., Neumann-Haefelin C., Bengsch B., Krammer F., Schwemmle M., Hofmann M., Thimme R., et al. Pre-existing immunity and vaccine history determine hemagglutinin-specific CD4 T cell and IgG response following seasonal influenza vaccination. Nat. Commun. 2021;12:6720. doi: 10.1038/s41467-021-27064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han A., Newell E.W., Glanville J., Fernandez-Becker N., Khosla C., Chien Y.H., Davis M.M. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc. Natl. Acad. Sci. USA. 2013;110:13073–13078. doi: 10.1073/pnas.1311861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagannathan P., Andrews J.R., Bonilla H., Hedlin H., Jacobson K.B., Balasubramanian V., Purington N., Kamble S., de Vries C.R., Quintero O., et al. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat. Commun. 2021;12:1967. doi: 10.1038/s41467-021-22177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Ploeg K., Kirosingh A.S., Mori D.A.M., Chakraborty S., Hu Z., Sievers B.L., Jacobson K.B., Bonilla H., Parsonnet J., Andrews J.R., et al. TNF-alpha(+) CD4(+) T cells dominate the SARS-CoV-2 specific T cell response in COVID-19 outpatients and are associated with durable antibodies. Cell. Rep. Med. 2022;3:100640. doi: 10.1016/j.xcrm.2022.100640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao A., Chen Z., Amitai A., Doelger J., Mallajosyula V., Sundquist E., Pereyra Segal F., Carrington M., Davis M.M., Streeck H., et al. Learning from HIV-1 to predict the immunogenicity of T cell epitopes in SARS-CoV-2. iScience. 2021;24:102311. doi: 10.1016/j.isci.2021.102311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29:1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prachar M., Justesen S., Steen-Jensen D.B., Thorgrimsen S., Jurgons E., Winther O., Bagger F.O. Identification and validation of 174 COVID-19 vaccine candidate epitopes reveals low performance of common epitope prediction tools. Sci. Rep. 2020;10:20465. doi: 10.1038/s41598-020-77466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B., Khodadoust M.S., Olsson N., Wagar L.E., Fast E., Liu C.L., Muftuoglu Y., Sworder B.J., Diehn M., Levy R., et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 2019;37:1332–1343. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P.E., et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rammensee H., Bachmann J., Emmerich N.P., Bachor O.A., Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 30.Newell E.W., Klein L.O., Yu W., Davis M.M. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat. Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bettini E., Locci M. SARS-CoV-2 mRNA vaccines: immunological mechanism and beyond. Vaccines (Basel) 2021;9:147. doi: 10.3390/vaccines9020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H., Wang C., Rubelt F., Scriba T.J., Davis M.M. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat. Biotechnol. 2020;38:1194–1202. doi: 10.1038/s41587-020-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang J., Zeng C., Cox T.M., Li C., Son Y.M., Cheon I.S., Wu Y., Behl S., Taylor J.J., Chakaraborty R., et al. Respiratory mucosal immunity against SARS-CoV-2 after mRNA vaccination. Sci. Immunol. 2022;7:eadd4853. doi: 10.1126/sciimmunol.add4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arshad N., Laurent-Rolle M., Ahmed W.S., Hsu J.C., Mitchell S.M., Pawlak J., SenGupta D., Biswas K.H., Cresswell P. SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to down-regulate MHC-I surface expression. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2208525120. e2208525120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriyama M., Lucas C., Monteiro V.S., Yale SARS-CoV-2 Genomic Surveillance Initiative. Iwasaki A. SARS-CoV-2 Omicron subvariants evolved to promote further escape from MHC-I recognition. Preprint at bioRxiv. 2022 doi: 10.1101/2022.05.04.490614. [DOI] [Google Scholar]
- 36.Yoo J.S., Sasaki M., Cho S.X., Kasuga Y., Zhu B., Ouda R., Orba Y., de Figueiredo P., Sawa H., Kobayashi K.S. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021;12:6602. doi: 10.1038/s41467-021-26910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozano-Ojalvo D., Camara C., Lopez-Granados E., Nozal P., Del Pino-Molina L., Bravo-Gallego L.Y., Paz-Artal E., Pion M., Correa-Rocha R., Ortiz A., et al. Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in naive and COVID-19 recovered individuals. Cell Rep. 2021;36:109570. doi: 10.1016/j.celrep.2021.109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185 doi: 10.1016/j.cell.2022.01.015. 847–859.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hachmann N.P., Miller J., Collier A.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186 doi: 10.1016/j.cell.2022.12.018. 279–286.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poon L., Gu H., Quadeer A.A., Krishnan P., Chang L., Liu G., Ng D., Cheng S., Lam T.T., Peiris M., et al. Within-host diversity of SARS-CoV-2 lineages and effect of vaccination. Preprint at Research Square. 2022 doi: 10.21203/rs.3.rs-1927944/v1. [DOI] [Google Scholar]
- 42.Antonio E.C., Meireles M.R., Bragatte M.A.S., Vieira G.F. Viral immunogenic footprints conferring T cell cross-protection to SARS-CoV-2 and its variants. Front. Immunol. 2022;13:931372. doi: 10.3389/fimmu.2022.931372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidhom J.-W., Baras A.S. Analysis of SARS-CoV-2 specific T-cell receptors in ImmuneCode reveals cross-reactivity to immunodominant Influenza M1 epitope. Preprint at bioRxiv. 2020 doi: 10.1101/2020.06.20.160499. [DOI] [Google Scholar]
- 44.Bergamaschi C., Terpos E., Rosati M., Angel M., Bear J., Stellas D., Karaliota S., Apostolakou F., Bagratuni T., Patseas D., et al. Systemic IL-15, IFN-gamma, and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell Rep. 2021;36:109504. doi: 10.1016/j.celrep.2021.109504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakraborty S., Gonzalez J.C., Sievers B.L., Mallajosyula V., Chakraborty S., Dubey M., Ashraf U., Cheng B.Y., Kathale N., Tran K.Q.T., et al. Early non-neutralizing, afucosylated antibody responses are associated with COVID-19 severity. Sci. Transl. Med. 2022;14:eabm7853. doi: 10.1126/scitranslmed.abm7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma M.J., Qiu S.F., Cui X.M., Ni M., Liu H.J., Ye R.Z., Yao L., Liu H.B., Cao W.C., Song H.B. Persistent SARS-CoV-2 infection in asymptomatic young adults. Signal Transduct. Target. Ther. 2022;7:77. doi: 10.1038/s41392-022-00931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eggink D., Andeweg S.P., Vennema H., van Maarseveen N., Vermaas K., Vlaemynck B., Schepers R., van Gageldonk-Lafeber A.B., van den Hof S., Reusken C.B., Knol M.J. Increased risk of infection with SARS-CoV-2 Omicron BA.1 compared with Delta in vaccinated and previously infected individuals, the Netherlands, 22 November 2021 to 19 January 2022. Euro Surveill. 2022;27:2101196. doi: 10.2807/1560-7917.ES.2022.27.4.2101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chin E.T., Leidner D., Lamson L., Lucas K., Studdert D.M., Goldhaber-Fiebert J.D., Andrews J.R., Salomon J.A. Protection against omicron from vaccination and previous infection in a prison system. N. Engl. J. Med. 2022;387:1770–1782. doi: 10.1056/NEJMoa2207082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altarawneh H.N., Chemaitelly H., Ayoub H.H., Tang P., Hasan M.R., Yassine H.M., Al-Khatib H.A., Smatti M.K., Coyle P., Al-Kanaani Z., et al. Effects of previous infection and vaccination on symptomatic omicron infections. N. Engl. J. Med. 2022;387:21–34. doi: 10.1056/NEJMoa2203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez Angel C.J., Pham E.A., Du H., Vallania F., Fram B.J., Perez K., Nguyen T., Rosenberg-Hasson Y., Ahmed A., Dekker C.L., et al. Signatures of immune dysfunction in HIV and HCV infection share features with chronic inflammation in aging and persist after viral reduction or elimination. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022928118. e2022928118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apostolidis S.A., Kakara M., Painter M.M., Goel R.R., Mathew D., Lenzi K., Rezk A., Patterson K.R., Espinoza D.A., Kadri J.C., et al. Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat. Med. 2021;27:1990–2001. doi: 10.1038/s41591-021-01507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh B.L.Z., Tan N., de Alwis R., Kunasegaran K., Chen Z., Poon M., Chan E., Low J.G.H., Yeoh A.E.J., Bertoletti A., Le Bert N. Enhanced BNT162b2 vaccine-induced cellular immunity in anti-CD19 CAR T cell-treated patients. Blood. 2022;140:156–160. doi: 10.1182/blood.2022016166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toebes M., Rodenko B., Ovaa H., Schumacher T.N.M. Generation of peptide MHC class I monomers and multimers through ligand exchange. Curr. Protoc. Immunol. 2009 doi: 10.1002/0471142735.im1816s87. 18.16.1–18.16.20. [DOI] [PubMed] [Google Scholar]
- 55.Willis R.A., Ramachandiran V., Shires J.C., Bai G., Jeter K., Bell D.L., Han L., Kazarian T., Ugwu K.C., Laur O., et al. Production of Class II MHC proteins in lentiviral vector-transduced HEK-293T cells for tetramer staining reagents. Curr. Protoc. 2021;1:e36. doi: 10.1002/cpz1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the published article and summarized in the corresponding tables, figures, and supplemental materials. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.