Abstract
Background: Acute myeloid leukemia with normal cytogenetics (CN-AML) is the largest group of AML patients with very heterogenous patient outcomes. The revised World Health Organization classification of the hematolymphoid tumours, 2022, has incorporated AML with Nucleophosphmin1 (NPM1) and CCAAT/enhancer binding protein-alpha (CEBPA) mutations as distinct entities. Despite the existing evidence of the prognostic relevance of FMS-like tyrosine kinase-3 internal tandem duplication (FLT3-ITD) in AML, it has not been included in the revised classification. Method: In this prospective study, we determined the prevalence of NPM1, CEBPA, and FLT3 gene mutations in 151 de novo CN-AML adult patients (age ≥18 years) in a tertiary care hospital in north India. Additionally, the prognostic relevance of these mutations was also evaluated. Results: NPM1, FLT3-ITD, and CEBPA mutations were found in 33.11%, 23.84%, and 15.77% of CN-AML patients, respectively. CEBPA mutations were found at 3 domains: transactivation domain 1 (TAD1) in 10 (6.62%), transactivation domain 2 (TAD2) in 5 (3.31%), and basic leucine zipper domain (bZIP) in 11 (7.82%) patients. Patients with NPM1 mutation had better clinical remission rate (CR) (P=0.003), event-free survival (P=0.0014), and overall survival (OS) (P=0.0017). However, FLT3-ITD and CEBPA mutations did not show any association with CR (P=0.404 and 0.92, respectively). Biallelic CEBPA mutations were found in 12 (7.95%) patients and were associated with better OS (P=0.043). Conclusions: These findings indicate that NPM1 and CEBPA mutations can be precisely used for risk stratification in CN-AML patients.
Keywords: NPM1, FLT3, CEBPA, mutations, CN-AML, prognosis
Introduction
Acute myeloid leukemia (AML) is a phenotypically and genetically heterogeneous disease characterised by an abnormal accumulation of blasts in the bone marrow (BM) and peripheral blood. Recent progress in understanding the molecular spectrum and genetic landscape of AML has led to its inclusion in the revised 4th edition of the World Health Organization (WHO) classification of hematolymphoid tumors and the recent 5th edition, 2022 [1-3]. Specifically, this classification now recognizes AML with nucleophosmin1 (NPM1) and CCAAT/enhancer-binding protein alpha (CEBPA) gene mutations as distinct entities. These mutations were considered provisional entities in the WHO 2008 classification as subtypes of AML with recurrent genetic abnormalities [4]. The recent guidelines did not include FMS-like tyrosine kinase 3 - internal tandem duplication (FLT3-ITD) as a distinct entity in the classification, despite it being found in 20-25% of AML patients [5-9]. Recent studies have examined the role of FLT3 mutations in conjunction with NPM1 or CEBPA mutations [10-18].
While multiple genetic mutations often occur in these patients, karyotyping remains the most important factor to predict outcome in AML patients [1,13,14,17,18]. Therefore, cytogenetic evaluation is an essential step in the management of AML patients. As per the 2022 European leukemia network (ELN), cytogenetic evaluation is the first step in the genetic analysis of patients with leukemia, with the guidelines stating that the report should be available within 5-7 days [1]. According to their recommendations, AML patients are categorized into favorable, intermediate, and adverse risk groups depending on cytogenetic abnormalities [1]. Favorable risk group includes those with balanced translocations like t(8;21)(q22;q22.1), inv(16)(p13.1q22) or t(16;16)(p13.1;q22). Adverse risk group includes t(6;9)(p23;q34.1), t(v;11q23.3), t(9;22)(q34.1;q11.2), inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2), -5 or del(5q), -7; -17/abn(17p), complex karyotype (3 or more unrelated chromosome abnormalities in the absence of WHO-designated recurring translocations or inversions) and monosomal karyotype [19]. Intermediate risk category encompasses cytogenetically normal AML (CN-AML) and AML with cytogenetic abnormalities not classified as favorable or adverse.
CN-AML comprises 40 to 50% of adult AML and 25% of pediatric AML cases [19,20]. This group of AML has intermediate risk with a five-year survival rate ranging from 24 to 42% [19,21,22]. There is a wide spectrum of genetic mutations among AML patients regardless of the presence or absence of karyotypic changes. As CN-AML is a heterogeneous group, molecular characterization is imperative for prognostication [6].
NPM1 is the most common mutation in cytogenetically normal and abnormal AML [1,6,10,23,24]. Other mutations found in CN-AML are in genes like CEBPA, KMT2A, FLT3, the neuroblastoma RAS viral oncogene homolog (NRAS), and the Wilms tumor 1 (WT1), and the runt-related transcription factor 1 (RUNX1) [25-27].
NPM1 gene encodes for nucleophosmin protein, also known as nucleolar phosphamatrin or numatrin. It is an abundant ubiquitously expressed protein mainly localized at nucleoli but continuously shuttles between nucleus and cytoplasm [28,29]. It plays a crucial role in the assembly and transport of ribosomal proteins, prevents the aggregation of proteins in the nucleolus, and regulates the stability and transcriptional activity of p53 after stress [30-32]. NPM1-mutated AMLs typically have a secondary mutation targeting the signaling pathway, giving them an advantage over normal cells. This secondary mutation is frequently found in FMS-like tyrosine kinase receptor 3 (FLT3) gene [23]. It also happens to be the most common tyrosine kinase mutation found in approximately 30% of adult AML patients [33].
FLT3 gene encodes a type III receptor tyrosine kinase (RTK) [34]. The function of this protein is to regulate stem cell proliferation and differentiation. It is expressed predominantly on hematopoietic progenitor cells [35,36]. Upon interaction with its ligand, FLT3 dimerizes and autophosphorylates, thereby activating its receptor and inducing various intracellular signaling pathways [37]. This activation enhances the proliferative capacity of AML cells [38]. FLT3 gene is mapped to the chromosome band 13q12 and comprises 24 exons that span a genomic region of approximately 100 kb [34,35]. The structure of FLT3 protein consists of 4 regions: a) a N-terminal extracellular region consisting of five immunoglobulin-like domains, of which the three most distal from the plasma membrane are involved in ligand binding, while the proximal domains are involved in receptor dimerization; b) a transmembrane domain; c) a juxta-membrane (JM) domain; and d) an intracellular C-terminal domain [36]. In AML, two types of FLT3 mutations are found - internal tandem duplication (FLT3-ITD) and point mutation in tyrosine kinase domain (FLT3-TKD). Both FLT3-ITD and FLT3-TKD are constitutively activating, which leads to ligand-independent FLT3 signaling and cellular proliferation.
Another distinct mutation in the AML classification is in the CCAAT/enhancer-binding protein alpha (C/EBPα) gene. The CEBPA gene maps to the chromosome band 19q13.1. It has a GC-rich (more than 70%) coding region within a single exon [37-39]. This gene encodes for a transcription factor involved in the regulation of myelopoiesis [40,41]. Its expression occurs predominantly in myelomonocytic cells and is upregulated during granulocytic differentiation [42,43]. The C/EBPα comprises a homologous C-terminal DNA-binding (basic region), dimerization (leucine zipper) motifs (bZIP), and two less-conserved N-terminal transactivation domains (TAD). The differentiation of granulocytes is further enhanced by Ras-mediated phosphorylation of serine 248 of the C/EBPα transactivation domain [44].
In this prospective study, we evaluated the prevalence and prognostic significance of NPM1, FLT3-ITD, and FLT3-TKD, CEBPA mutations in adult CN-AML patients treated at All India Institute of Medical Sciences, New Delhi, India.
Materials and methods
Patients
This prospective exploratory study included de novo adult (≥18 years) patients with AML presenting to the department of medical oncology between January 2014 and October 2017. The patients were diagnosed as AML based on morphology, cytochemistry, immunophenotyping, and cytogenetics. All patients underwent baseline karyotyping before the initiation of therapy. Only patients with normal cytogenetics were included in the study. Patients with recurrent cytogenetic abnormalities, secondary or relapsed AML, and insufficient samples were excluded. A total of 151 CN-AML patients were recruited in the present study. This study was conducted in accordance with the ethical standards of the World Medical Association’s Declaration of Helsinki after getting approval from the institutional ethics committee. Written informed consent was taken from all patients.
Determination of NPM1, FLT3-ITD, and CEBPA mutations
Baseline BM samples were collected from all patient samples. BM mononuclear samples were isolated by Ficoll-Hypaque density gradient centrifugation. DNA was isolated from BM samples using a DNA extraction kit from Thermo Fisher Scientific, Waltham, Massachusetts, USA. Screening for NPM1, FLT3-ITD, FLT3-TKD and CEBPA gene mutations was carried out using published protocols [26].
Treatment
All patients were treated with a uniform treatment protocol comprising induction therapy with a 3+7 regimen [daunorubicin 60 mg/m2 for three days and cytosine arabinoside (ara-C) 100 mg/m2 as a continuous infusion for seven days] [45]. BM examination was done at the end of induction therapy to assess remission status. Complete remission (CR) was defined as BM blasts <5%, absence of extramedullary blast proliferation, no dependence on blood transfusion, and absolute neutrophil count >1 × 109/L, platelet count >100 × 109/L. The patients were given three cycles of high doses of ara-C at 18 g/m2 after achieving CR. Relapse was defined as the re-emergence of blasts in the peripheral blood, BM blasts >5%, or the development of extramedullary leukemia [1].
Patient follow-up and statistical analysis
The patients were followed up in the Medical Oncology department. The last follow-up was carried out on December 23, 2020. Overall survival (OS) was defined as the duration from the date of diagnosis to last follow-up or, death due to any cause. Event-free survival (EFS) was measured as the time from the date of diagnosis to the date of last follow-up or the first event (relapse or death). The probability of EFS and OS was calculated using the Kaplan-Meier method, with the differences being compared using a two-sided log-rank test.
Descriptive statistics were used to summarize baseline characteristics. Mann-Whitney-U test was used to compare continuous variables, while Fisher’s exact test was used to compare categorical variables. A p-value ≤0.05 (two-sided) was considered significant. The relationship between EFS and OS variables was calculated by constructing multivariate Cox proportional hazard models. All analyses were performed using the SPSS statistical software package, version 20.0/STATA software, version 11.
Results
A total of 151 adult patients ≥18 years were recruited in the study. All of them were cytogenetically normal by conventional karyotyping. The median age of the patients was 39 years (range 18-75 years). There were 115 (63.89%) males and 65 (36.11%) females. The lab parameters were median hemoglobin 7.9 gm/dL (range 2.8-15.4 gm/dL); median total leukocyte count (TLC) 21,300/µL (range 300 to 411,000/µL), median platelet count 52,000/µL (range 1700-283,000). The median blast percentage in peripheral blood was 60% (range 2-98%) while the median blast percentage in bone marrow was 70% (range 24-95%).
Prevalence of genetic mutations
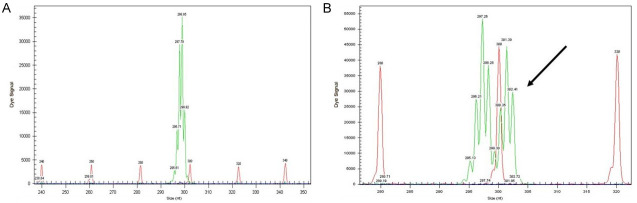
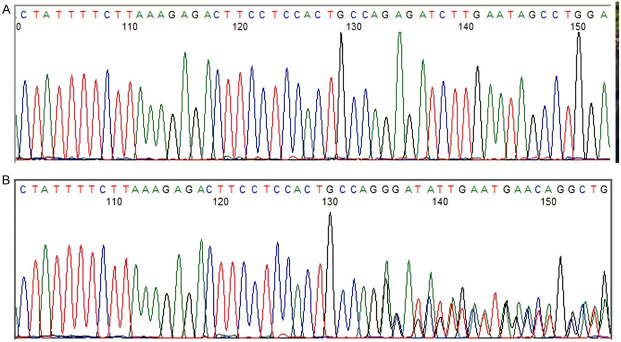
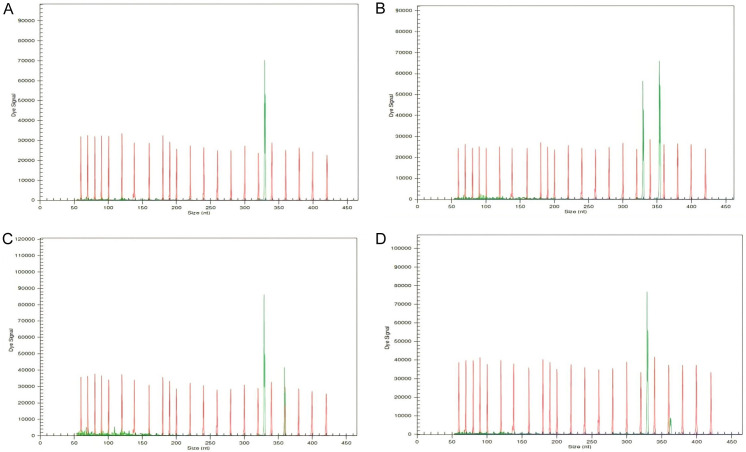
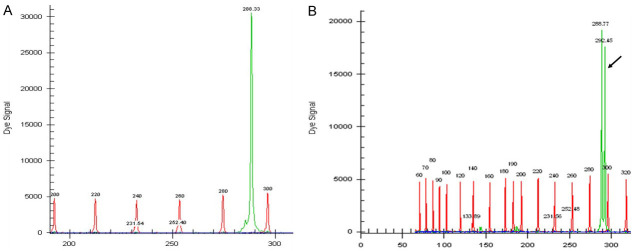
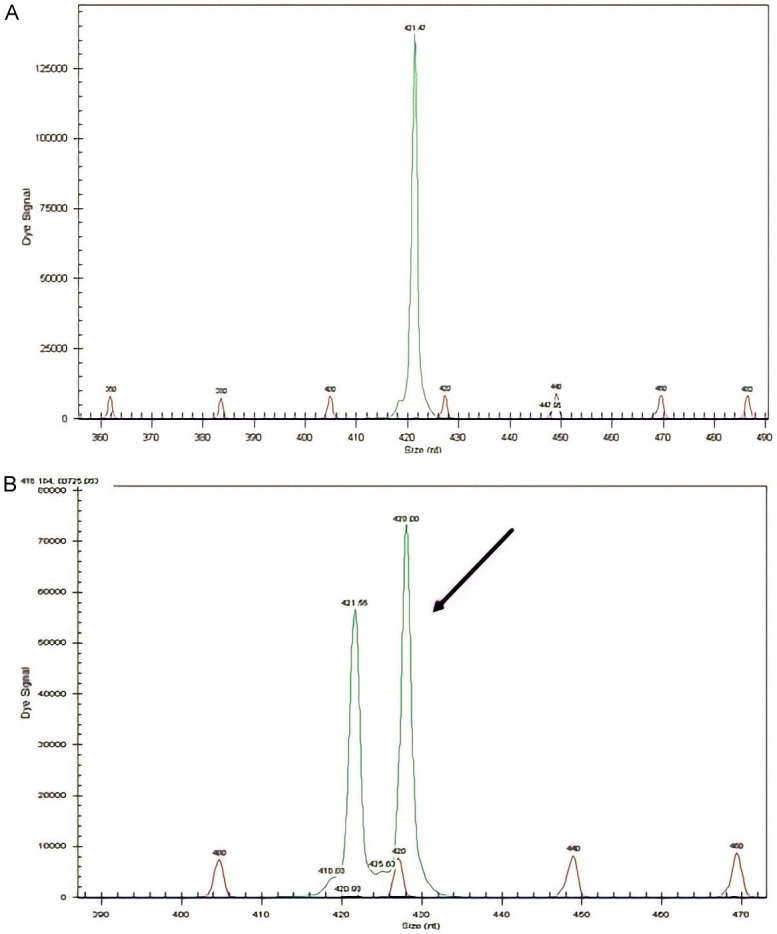
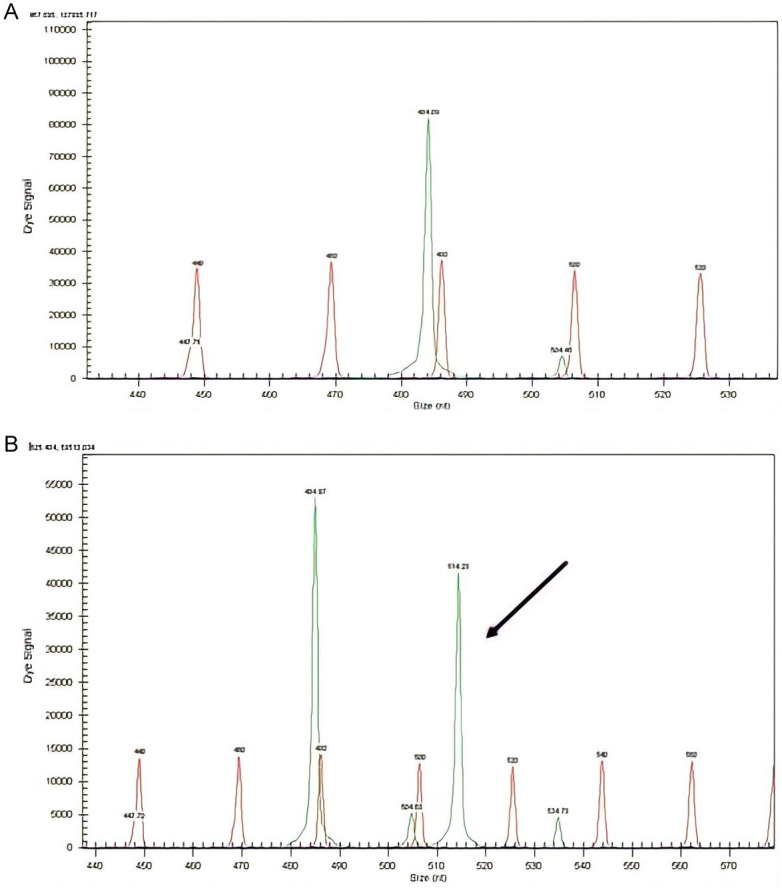
NPM1 mutations were found in 50/151 (33.11%) patients (Figures 1 and 2). The prevalence of different NPM1 mutations found in our patient cohort is shown in Table 1. FLT3-ITD mutation was found in 36 (23.84%) patients (Figure 3). Allelic ratio (mutant/wild allele) was high (>0.5) in 14 (9.27%) patients and low in 22 (14.56%) patients. Fourteen patients had concurrent mutations in the NPM1 gene and FLT3-ITD. FLT3-TKD mutation was positive in 4 (2.64%) cases. CEBPA mutations were found in TAD1 domain in 10 (6.62%) (Figures 4 and 5), TAD2 domain in 5 (3.31%) (Figures 6 and 7) and bZIP domain in 11 (7.82%) patients (Figures 8 and 9). In addition, polymorphism in CEBPA gene was also seen (Figure 10). The overall prevalence of CEBPA mutations in our cohort is shown in Figure 11. Single allele mutations were positive in 11 (7.82%) cases. Biallelic mutations were positive in 12 (7.95%) cases.
Figure 1.
Detection of NPM1 mutation by fragment analysis. (A) NPM1-wild type showing only one peak, (B) Two peaks are seen at 297 bp and the other at 301 bp.
Figure 2.
Detection of NPM1 mutation by sanger sequencing. (A) NPM1-wild type, (B) NPM1-mutated c.861_864insCTGC.
Table 1.
Prevalence of NPM1 mutation in our cohort
| NPM1 GENE | Nucleotide sequence | Protein sequence | Frequency |
|---|---|---|---|
| Wild | GAT CTC TGG CAG TGG AGG AAG TCT CTT TAA GAA AAT AG | DLWQWRKSL stop | 101 |
| c.860-863dupTCTG | GAT CTC TGT CTG GCA GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLCLAVEEVSLRK stop | 40 (80%) |
| c.861_864insCTGC | GAT CTC TGC CTG GCA GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLCLAVEEVSLRK stop | 3 (6%) |
| c.864_867insTATG | GAT CTC TGT ATG GCA GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLCMAVEEVSLRK stop | 2 (4%) |
| c.863_866insGCCG | GAT CTC TGC CGG GCA GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLCRAVEEVSLRK stop | 1 (2%) |
| c.859 TC>CTATGCA | GAT CTA TGC ATG GCA GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLCMAVEEVSLRK stop | 1 (2%) |
| c.861G>ATGCA | GAT CTA TGC ATG GCA GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLCMAVEEVSLRK stop | 1 (2%) |
| c.861_864insATGC | GAT CTA TGC ATG GCA GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLCMAVEEVSLRK stop | 1 (2%) |
| c.867_870insGCGT | GAT CTC TGG CAG CGT GTG GAG GAA GTC TCT TTA AGA AAA TAG | DLWQRVEEVSLRK stop | 1 (2%) |
Figure 3.
Detection of FLT3-ITD mutation by fragment analysis. (A) FLT3-wild type showing only one peak, (B) Two peaks are seen. One at 329 bp and the other at 350 bp signifying internal tandem duplication of 21 bp. The allelic ratio is >0.5, (C) Two peaks are seen. The allelic ratio is <0.5, (D) Two peaks are seen. The allelic ratio is <0.5.
Figure 4.
Detection of CEBPA TAD1 mutation by fragment analysis. (A) CEBPA-wild type showing only one peak-288 bp, (B) Two peaks-269.33 bp and 292 bp are seen in a CEBPA mutated case.
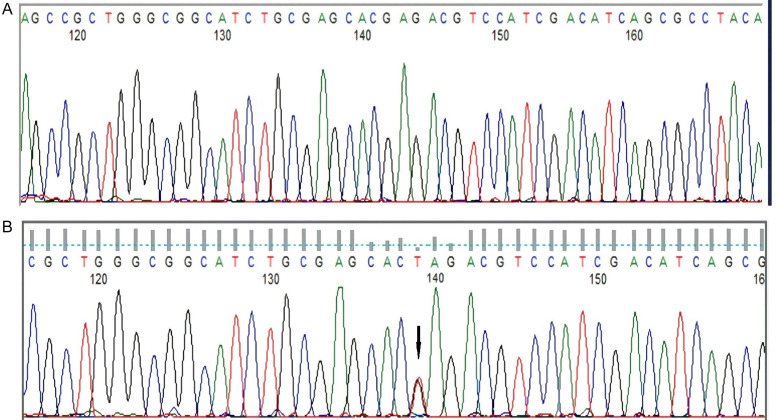
Figure 5.
Detection of CEBPA TAD1 mutation by Sanger sequencing (A) CEBPA-wild type, (B) CEBPA TAD1 mutated c.295G>T p.E89* resulting in premature stop codon: truncated protein.
Figure 6.
Detection of CEBPA TAD2 mutation by fragment analysis. (A) CEBPA-wild type showing only one peak, (B) Two peaks are seen in a CEBPA mutated case.
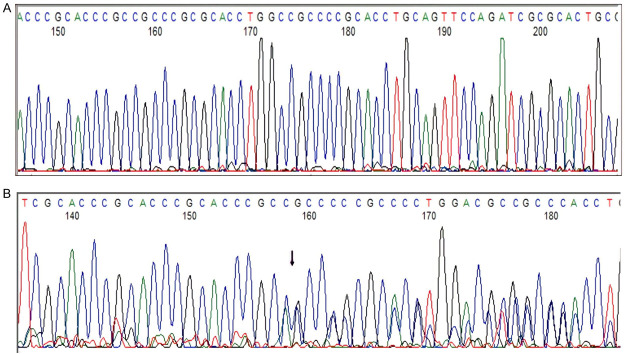
Figure 7.
Detection of CEBPA TAD2 mutation by Sanger sequencing. (A) CEBPA-wild type, (B) CEBPA TAD2 mutated. Out-of-frame insertion.
Figure 8.
Detection of CEBPA bZIP mutation by fragment analysis. (A) CEBPA-wild type showing only one peak, (B) Two peaks are seen in a CEBPA mutated case.
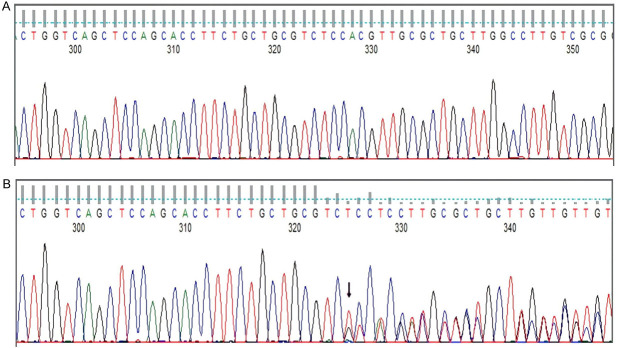
Figure 9.
Detection of CEBPA bZIP mutation by Sanger sequencing. (A) CEBPA-wild type, (B) CEBPA bZIP mutated case.
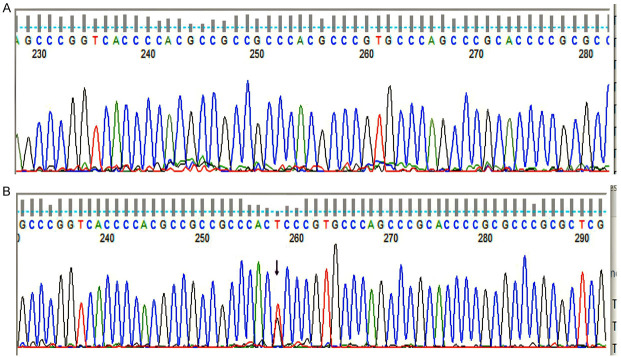
Figure 10.
Detection of CEBPA polymorphisms. (A) CEBPA-wild type, (B) SNP identified: rs34529039. cDNA position 810, protein position 230, amino acids T/T, codons acG/acT, consequences-synonymous variant.
Figure 11.
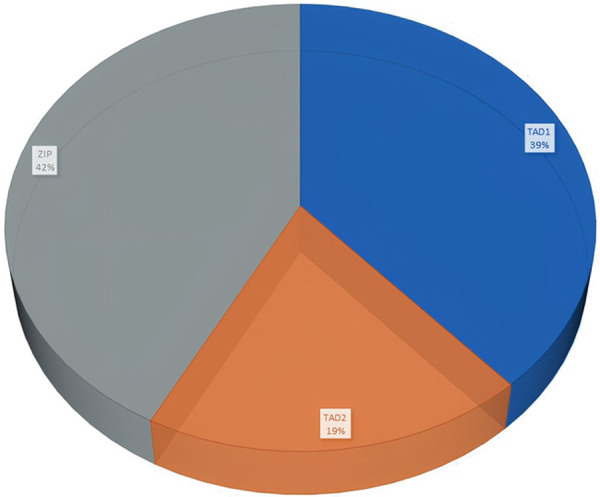
Prevalence of CEBPA mutations in our cohort.
Association of genetic mutations with clinical and lab parameters
We did not find any significant association between the genetic mutations and the clinical and laboratory parameters viz., age, sex, hemoglobin, platelet count, WBC count at diagnosis, and the BM blast percentage. The peripheral blood blast percentage at the time of diagnosis was lower in CN-AML patients with FLT3-ITD than in patients without this mutation (P=0.02). In our study, the presence of the mutations was not associated with CD34 and CD56 expression on leukemic blasts. Table 2 demonstrates the association of genetic mutations with baseline characteristics in CN-AML patients.
Table 2.
Association of genetic mutations with baseline characteristics in CN-AML patients
| Characteristics | NPM1 | FLT3-ITD | CEBPA | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Wild (n=101) | Mutant (n=50) | p-value | Negative (n=115) | Positive (n=36) | p-value | Wild (n=128) | Mutant (n=23) | p-value | |
| Age at diagnosis (years) | 0.48 | 0.68 | 0.86 | ||||||
| Median | 36 | 36.5 | 36 | 36.5 | 37 | 35 | |||
| Range | 18-74 | 18-75 | 18-74 | 18-75 | 18-75 | 18-74 | |||
| Sex, n (%) | 0.066 | 0.16 | 0.12 | ||||||
| Male | 70 (69.3) | 27 (54) | 70 | 27 | 79 | 18 | |||
| Female | 34 (33.66) | 23 (46) | 45 | 9 | 49 | 5 | |||
| Hemoglobin (g/dL) | 0.96 | 0.67 | 0.23 | ||||||
| Median | 8 | 7.8 | 8 | 7.55 | 7.7 | 8.9 | |||
| Range | 2.8-14 | 3.5-15.4 | 2.8-15.4 | 3.8-13.7 | 2.8-17.3 | 6.5-13 | |||
| Platelets (× 109/L) | 0.09 | 0.45 | 0.12 | ||||||
| Median | 52.5 | 69 | 54 | 55 | 57 | 37 | |||
| Range | 1-73 | 15-283 | 1-73 | 6.3-283 | 1-73 | 7-141 | |||
| WBC (× 109/L) | 0.20 | 0.62 | |||||||
| Median | 22.55 | 28.25 | 22.8 | 23.2 | 0.71 | 20.5 | 35.1 | ||
| Range | 0.24-411 | 0.76-282 | 0.24-411 | 4.4-278 | 2.4-411 | 0.68-242 | |||
| Peripheral blood blast, (%) | 0.93 | 0.02 | 0.93 | ||||||
| Median | 61 | 75 | 63 | 55 | 63 | 63 | |||
| Range | 2-98 | 7-96 | 2-98 | 4-96 | 2-98 | 30-95 | |||
| Bone marrow blasts, (%) | 0.36 | 0.22 | 0.87 | ||||||
| Median | 75 | 77.5 | 75 | 77 | 80 | 75 | |||
| Range | 24-95 | 60-95 | 60-95 | 24-95 | 60-95 | 40-95 | |||
Survival analysis
Correlation of genetic mutations with patient outcome
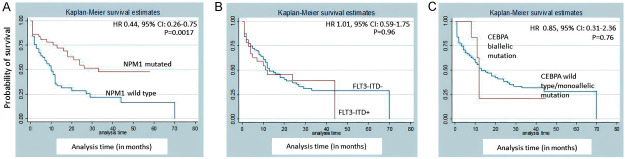
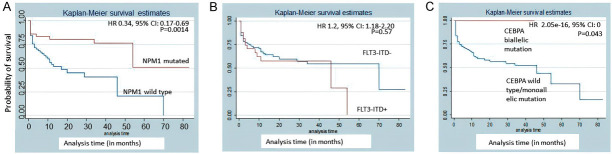
Patients with NPM1 mutation had a better CR rate compared to NPM1 wild-type (P=0.003). However, FLT3-ITD and CEBPA did not correlate with CR (P=0.404 and 0.92, respectively). NPM1 mutation was associated with better EFS [hazard ratio (HR) 0.34, 95% CI: 0.17-0.69; P=0.0014] and OS [HR 0.44, 95% confidence interval (CI): 0.26-0.75; P=0.0017] (Figures 12 and 13). Biallelic mutation of CEBPA was associated with better OS [HR 2.05e-16, 95% CI: 0; P=0.043]. However, it had no effect on EFS [HR 0.85, 95% CI: 0.31-2.36; P=0.76]. FLT3-ITD did not show any correlation with survival. In 22/36 FLT3-ITD mutated patients, the allelic ratio was <0.5. This could explain the non-correlation of FLT3-ITD with survival. Since FLT3-TKD was present only in 4 patients, we did not evaluate it for survival analysis.
Figure 12.
Kaplan-Meier event-free survival (EFS) estimates according to mutations of (A) NPM1, (B) FLT3-ITD, (C) CEBPA.
Figure 13.
Kaplan-Meier overall survival (OS) estimates according to mutations of (A) NPM1, (B) FLT3-ITD, (C) CEBPA.
Discussion
In this study, we determined the prevalence of NPM1, FLT3, and CEBPA mutations in 151 adult CN-AML patients at a tertiary care centre in India. In our study, NPM1 mutations were detected in 33.11% CN-AML patients. NPM1 mutations have been previously reported to have an incidence of 28-35% in adult AML, of which 48-53% are found in CN-AML. It is less frequently mutated in children (2.8%) [10,25,46-49]. NPM1 mutation has been reported to be more frequent in adult female AML patients compared to males [10,20,25,47-50]. Contrary to this, the males were found to have a higher NPM1 mutation rate (54%) in our study. This could be due to presence of more male patients in our cohort. We found a median age of 36.5 years (range 18-75) in our patients, which was lower than those reported by previous studies [10,49-52]. Interestingly, a previous multicentric study has highlighted the biologic differences within immunophenotypically defined subgroups of NPM1-mutated AML that may have prognostic relevance [50]. This aspect has not been investigated in the current study. Akin to previous studies, we also found that NPM1 mutated AML patients had better EFS [HR 0.34, 95% CI: 0.17-0.69; P=0.0014] and OS [HR 0.44, 95% CI: 0.26-0.75; P=0.0017] [10,25,27,47-49,53].
FLT3-ITD was detected in 23.84% CN-AML patients. Although, the prevalence of FLT3-ITD corroborates with previous studies [6-8,16,54], the higher WBC count at diagnosis and poorer prognosis in such patients reported by these researchers were not present in our study. The plausible reason for this may be the presence of a lower allelic ratio (<0.5) in most of the patients. This finding is consistent with ELN criteria that FLT3-ITD low allele ratios predict a better prognosis in AML [1]. A similar prognosis has also been reported by Pratcorona et al., de Jonge et al., Schnittger S et al., Ho A.D. et al. [10,55-57]. Contrary to this, Gale et al., Linch et al., Schlenk et al., Straube et al., and Sakaguchi et al. have reported poor prognoses even in AML patients with FLT3-ITD with low allelic ratios [15,16,54,58,59]. The concurrent FLT3-ITD and NPM1 mutations were found in 14 patients (28%). This incidence is less as compared to 38.1% reported by Schnittger S et al. [56]. de Jonge et al. found simultaneous mutations of NPM1 and FLT3-ITD in 15% AML patients [55]. The low allelic ratio in most of our patients can explain the non-association of FLT3-ITD with survival, even in patients with NPM1 mutations. Straube et al. correlated the NPM1+FLT3-ITD-low allele ratio and NPM1+FLT3-wild type with age and found that in <60 years, the low allele ratio had an inferior outcome compared to the wild type. However, both had similar poor outcomes at age >60 years. In their pediatric cohort, the FLT3-ITD with low allelic ratio without NPM1 mutation was found not to have an adverse prognosis which made them conclude the requirement of a larger-scale study to analyse the biology of the disease [15]. FLT3-ITD mutations have also been associated with a higher percentage of BM blast cells, an increased risk of relapse, and reduced survival by researchers [9,60,61]. In agreement with their findings, the BM blast cells percentage was significantly higher [median 77% (range 24-95%)] in AML patients with FLT3-ITD in our study. A recent report also highlights that NPM1 mutation with high variant allele frequency (VAF) or an abundance of the mutated allele at the time of diagnosis has an independent prognostic implication of poor outcome in de novo AML with or without evidence of co-mutations and clinical variables [51]. FLT3-TKD was positive in only four patients (2.64%) in our study. Hence, the prognostic relevance of FLT3-TKD could not be assessed.
The prevalence of CEBPA mutation was 15.77%. Single allele mutations were positive in 11 (7.82%) cases. Biallelic mutations were positive in 12 (7.95%) cases. This prevalence is slightly higher than the range of 5-14% CEBPA mutations in CN-AML reported by other researchers [33,62-67]. There are, however, variations in the single and double mutations documented. Initially, only biallelic mutations were considered a good prognosis, but recent findings have shown that site-specific mutations are important. The site-wise mutations were further subdivided as TAD1 10 (6.62%), TAD2 5 (3.31%), and bZIP 11 (7.82%). bZIP mutations have gained significance, with two recent studies describing how bZIP site mutations play an independent prognostic role in overall and disease-free survival in single and double mutated allele states [67,68]. Previous studies on gene expression profiling have expressed that in-frame insertion/deletion mutations affecting the bZIP domain in a single or double mutated allele state did not show a unique gene expression profile and were less distinct from the biallelic CEBPA AML [64,65]. The latest finding from Taube et al. showed that 90% of CEBPA bi-mutant cases carry bZIP in-frame mutations explaining the previous analyses [67]. van Doorn-Khosrovani et al. had reported that bZIP mutations were associated with N-terminal mutation in the different alleles in most cases [63].
The survival analysis in our study showed that biallelic mutation of CEBPA was associated with better OS [HR 2.05e-16, 95% CI: 0; P=0.043]. However, it had no effect on EFS [HR 0.85, 95% CI: 0.31-2.36; P=0.76]. The bZIP mutations are the most common mutation site in our study, and a higher occurrence of bi-allelic mutations with them gave the added advantage in the prognosis.
Regarding clinical parameters seen in CEBPA-positive patients, the Hb and WBC counts were higher, and the platelet counts were lower at diagnosis than in NPM1 and FLT3-ITD-positive patients. This finding is in agreement with previous studies [33,62,64,65]. The peripheral blast count was notwithstanding lesser than NPM1 positive AML.
The median age of 35 years (range 18-74), a slightly younger than reported in other studies, also points toward the new inference of bZIP mutation CEBPA being more associated with a younger age group [67,68]. Further hypothesis about presence of increased WBC count at the time of diagnosis regarding bZIP mutation also concurs with our study. Only a few studies have commented on male predisposition in CEBPA positive AML. Van Doorn-Khosrovani S et al. also found a slight male predominance similar to our study.
Limitations of the study
There are certain limitations in the current study. The first limitation of the study is less sample size. The prevalence of genetic mutations in AML should be studied in a larger sample size. Secondly, the bZIP in-frame mutations were not evaluated separately in the present study despite this subset comprising the maximum site of mutations of CEBPA. Recent updates provide critical insight into bZIP mutations, so this mutation should be paramount in managing Indian CN-AML patients. We expect a more extensive study emphasizing the bZIP site mutations of CEBPA to address this issue.
Conclusion
In conclusion, our study has shown the prognostic relevance of NPM1, FLT3, and CEBPA gene mutations in adult CN-AML patients. We found that NPM1 and CEBPA mutations were associated with a good prognosis. We did not find any correlation between FLT3-ITD mutation and patient outcome. However, this should be viewed in light of certain limitations of this study, which primarily relate to a small number of patients. Therefore, there is a need for prospective studies involving a larger number of CN-AML patients. Additionally, the prognostic relevance of in-frame bZIP mutations should also be tested.
Acknowledgements
This work was supported by the Department of Biotechnology (BT/PR5492/MED/30/849/2012) and Wellcome Trust/DBT India Alliance Fellowship (grant number: IA/CPHI/17/1/503333) awarded to AC.
Disclosure of conflict of interest
None.
References
- 1.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz M, Sierra J, Tallman MS, Tien HF, Wei AH, Löwenberg B, Bloomfield CD. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 3.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JKC, Chen W, Chen X, Chng WJ, Choi JK, Colmenero I, Coupland SE, Cross NCP, De Jong D, Elghetany MT, Takahashi E, Emile JF, Ferry J, Fogelstrand L, Fontenay M, Germing U, Gujral S, Haferlach T, Harrison C, Hodge JC, Hu S, Jansen JH, Kanagal-Shamanna R, Kantarjian HM, Kratz CP, Li XQ, Lim MS, Loeb K, Loghavi S, Marcogliese A, Meshinchi S, Michaels P, Naresh KN, Natkunam Y, Nejati R, Ott G, Padron E, Patel KP, Patkar N, Picarsic J, Platzbecker U, Roberts I, Schuh A, Sewell W, Siebert R, Tembhare P, Tyner J, Verstovsek S, Wang W, Wood B, Xiao W, Yeung C, Hochhaus A. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–1719. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 5.Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T, Misawa S. Internal tandem duplication of the FLT3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 6.Fröhling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, Döhner H, Döhner K. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002;100:4372–4380. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 7.Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, Wermke M, Bornhäuser M, Ritter M, Neubauer A, Ehninger G, Illmer T. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 8.Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, Carroll AJ, Mrózek K, Vardiman JW, George SL, Kolitz JE, Larson RA, Bloomfield CD, Caligiuri MA. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 9.Kottaridis PD, Gale RE, Linch DC. FLT3 mutations and leukaemia. Br J Haematol. 2003;122:523–538. doi: 10.1046/j.1365-2141.2003.04500.x. [DOI] [PubMed] [Google Scholar]
- 10.Pratcorona M, Brunet S, Nomdedéu J, Ribera JM, Tormo M, Duarte R, Escoda L, Guàrdia R, Queipo de Llano MP, Salamero O, Bargay J, Pedro C, Martí JM, Torrebadell M, Díaz-Beyá M, Camós M, Colomer D, Hoyos M, Sierra J, Esteve J Grupo Cooperativo Para el Estudio y Tratamiento de las Leucemias Agudas Mieloblásticas. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood. 2013;121:2734–2738. doi: 10.1182/blood-2012-06-431122. [DOI] [PubMed] [Google Scholar]
- 11.Peterlin P, Renneville A, Ben Abdelali R, Nibourel O, Thomas X, Pautas C, de Botton S, Raffoux E, Cayuela JM, Boissel N, Terré C, Celli-Lebras K, Castaigne S, Preudhomme C, Gardin C, Dombret H. Impact of additional genetic alterations on the outcome of patients with NPM1-mutated cytogenetically normal acute myeloid leukemia. Haematologica. 2015;100:e196–199. doi: 10.3324/haematol.2014.115576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakita S, Yamaguchi H, Ueki T, Usuki K, Kurosawa S, Kobayashi Y, Kawata E, Tajika K, Gomi S, Koizumi M, Fujiwara Y, Yui S, Fukunaga K, Ryotokuji T, Hirakawa T, Arai K, Kitano T, Kosaka F, Tamai H, Nakayama K, Fukuda T, Inokuchi K. Complex molecular genetic abnormalities involving three or more genetic mutations are important prognostic factors for acute myeloid leukemia. Leukemia. 2016;30:545–554. doi: 10.1038/leu.2015.288. [DOI] [PubMed] [Google Scholar]
- 13.Kurosawa S, Yamaguchi H, Yamaguchi T, Fukunaga K, Yui S, Wakita S, Kanamori H, Usuki K, Uoshima N, Yanada M, Shono K, Ueki T, Mizuno I, Yano S, Takeuchi J, Kanda J, Okamura H, Inamoto Y, Inokuchi K, Fukuda T. Decision analysis of postremission therapy in cytogenetically intermediate-risk acute myeloid leukemia: the impact of FLT3 internal tandem duplication, nucleophosmin, and CCAAT/enhancer binding protein alpha. Biol Blood Marrow Transplant. 2016;22:1125–1132. doi: 10.1016/j.bbmt.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Versluis J, In’t Hout FE, Devillier R, van Putten WL, Manz MG, Vekemans MC, Legdeur MC, Passweg JR, Maertens J, Kuball J, Biemond BJ, Valk PJ, van der Reijden BA, Meloni G, Schouten HC, Vellenga E, Pabst T, Willemze R, Löwenberg B, Ossenkoppele G, Baron F, Huls G, Cornelissen JJ. Comparative value of post-remission treatment in cytogenetically normal AML subclassified by NPM1 and FLT3-ITD allelic ratio. Leukemia. 2017;31:26–33. doi: 10.1038/leu.2016.183. [DOI] [PubMed] [Google Scholar]
- 15.Straube J, Ling VY, Hill GR, Lane SW. The impact of age, NPM1(mut), and FLT3(ITD) allelic ratio in patients with acute myeloid leukemia. Blood. 2018;131:1148–1153. doi: 10.1182/blood-2017-09-807438. [DOI] [PubMed] [Google Scholar]
- 16.Sakaguchi M, Yamaguchi H, Najima Y, Usuki K, Ueki T, Oh I, Mori S, Kawata E, Uoshima N, Kobayashi Y, Kako S, Tajika K, Gomi S, Shono K, Kayamori K, Hagihara M, Kanda J, Uchiyama H, Kuroda J, Uchida N, Kubota Y, Kimura S, Kurosawa S, Nakajima N, Marumo A, Omori I, Fujiwara Y, Yui S, Wakita S, Arai K, Kitano T, Kakihana K, Kanda Y, Ohashi K, Fukuda T, Inokuchi K. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018;2:2744–2754. doi: 10.1182/bloodadvances.2018020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurosawa S, Yamaguchi H, Yamaguchi T, Fukunaga K, Yui S, Kanamori H, Usuki K, Uoshima N, Yanada M, Takeuchi J, Mizuno I, Kanda J, Okamura H, Yano S, Tashiro H, Shindo T, Chiba S, Tomiyama J, Inokuchi K, Fukuda T. The prognostic impact of FLT3-ITD, NPM1 and CEBPa in cytogenetically intermediate-risk AML after first relapse. Int J Hematol. 2020;112:200–209. doi: 10.1007/s12185-020-02894-x. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Li XQ, Chu TT, Han SY, Qi JQ, Tang YQ, Qiu HY, Fu CC, Tang XW, Ruan CG, Wu DP, Han Y. Clinical significance of FLT3-ITD/CEBPA mutations and minimal residual disease in cytogenetically normal acute myeloid leukemia after hematopoietic stem cell transplantation. J Cancer Res Clin Oncol. 2021;147:2659–2670. doi: 10.1007/s00432-021-03530-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagunas-Rangel FA, Chávez-Valencia V, Gómez-Guijosa M, Cortes-Penagos C. Acute myeloid leukemia-genetic alterations and their clinical prognosis. Int J Hematol Oncol Stem Cell Res. 2017;11:328–339. [PMC free article] [PubMed] [Google Scholar]
- 20.Pourrajab F, Zare-Khormizi MR, Hashemi AS, Hekmatimoghaddam S. Genetic characterization and risk stratification of acute myeloid leukemia. Cancer Manag Res. 2020;12:2231–2253. doi: 10.2147/CMAR.S242479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prada-Arismendy J, Arroyave JC, Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63–76. doi: 10.1016/j.blre.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Zaidi SZ, Owaidah T, Al Sharif F, Ahmed SY, Chaudhri N, Aljurf M. The challenge of risk stratification in acute myeloid leukemia with normal karyotype. Hematol Oncol Stem Cell Ther. 2008;1:141–158. doi: 10.1016/s1658-3876(08)50023-9. [DOI] [PubMed] [Google Scholar]
- 23.Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A, Bullinger L, Fröhling S, Döhner H. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 24.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, Ehninger G. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 25.Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, Habdank M, Späth D, Morgan M, Benner A, Schlegelberger B, Heil G, Ganser A, Döhner H. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 26.Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, Benthaus T, Sauerland MC, Berdel WE, Büchner T, Wörmann B, Braess J, Hiddemann W, Bohlander SK, Spiekermann K. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J. Clin. Oncol. 2010;28:570–577. doi: 10.1200/JCO.2008.21.6010. [DOI] [PubMed] [Google Scholar]
- 27.Verma D, Kumar R, Ali MS, Singh J, Arora M, Singh I, Kumari S, Bakhshi S, Sharma A, Palanichamy JK, Tanwar P, Singh AR, Chopra A. BAALC gene expression tells a serious patient outcome tale in NPM1-wild type/FLT3-ITD negative cytogenetically normal-acute myeloid leukemia in adults. Blood Cells Mol Dis. 2022;95:102662. doi: 10.1016/j.bcmd.2022.102662. [DOI] [PubMed] [Google Scholar]
- 28.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 29.Cordell JL, Pulford KA, Bigerna B, Roncador G, Banham A, Colombo E, Pelicci PG, Mason DY, Falini B. Detection of normal and chimeric nucleophosmin in human cells. Blood. 1999;93:632–642. [PubMed] [Google Scholar]
- 30.Olson MO, Wallace MO, Herrera AH, Marshall-Carlson L, Hunt RC. Preribosomal ribonucleoprotein particles are a major component of a nucleolar matrix fraction. Biochemistry. 1986;25:484–491. doi: 10.1021/bi00350a031. [DOI] [PubMed] [Google Scholar]
- 31.Szebeni A, Olson MO. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 33.Fröhling S, Schlenk RF, Stolze I, Bihlmayr J, Benner A, Kreitmeier S, Tobis K, Döhner H, Döhner K. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J. Clin. Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 34.Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Genomic structure of human FLT3: implications for mutational analysis. Br J Haematol. 2001;113:1076–1077. doi: 10.1046/j.1365-2141.2001.02821.x. [DOI] [PubMed] [Google Scholar]
- 35.Agnès F, Shamoon B, Dina C, Rosnet O, Birnbaum D, Galibert F. Genomic structure of the downstream part of the human FLT3 gene: exon/intron structure conservation among genes encoding receptor tyrosine kinases (RTK) of subclass III. Gene. 1994;145:283–288. doi: 10.1016/0378-1119(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 36.Matthews W, Jordan CT, Wiegand GW, Pardoll D, Lemischka IR. A receptor tyrosine kinase specific to hematopoietic stem and progenitor cell-enriched populations. Cell. 1991;65:1143–1152. doi: 10.1016/0092-8674(91)90010-v. [DOI] [PubMed] [Google Scholar]
- 37.Friedman AD, McKnight SL. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 38.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 39.Antonson P, Xanthopoulos KG. Molecular cloning, sequence, and expression patterns of the human gene encoding CCAAT/enhancer binding protein alpha (C/EBP alpha) Biochem Biophys Res Commun. 1995;215:106–113. doi: 10.1006/bbrc.1995.2439. [DOI] [PubMed] [Google Scholar]
- 40.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 41.Ward AC, Loeb DM, Soede-Bobok AA, Touw IP, Friedman AD. Regulation of granulopoiesis by transcription factors and cytokine signals. Leukemia. 2000;14:973–990. doi: 10.1038/sj.leu.2401808. [DOI] [PubMed] [Google Scholar]
- 42.Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 44.Behre G, Singh SM, Liu H, Bortolin LT, Christopeit M, Radomska HS, Rangatia J, Hiddemann W, Friedman AD, Tenen DG. Ras signaling enhances the activity of C/EBP alpha to induce granulocytic differentiation by phosphorylation of serine 248. J Biol Chem. 2002;277:26293–26299. doi: 10.1074/jbc.M202301200. [DOI] [PubMed] [Google Scholar]
- 45.Tyagi A, Pramanik R, Vishnubhatla S, Bakhshi R, Bakhshi S. Prognostic impact of mitochondrial DNA D-loop variations in pediatric acute myeloid leukemia. Oncotarget. 2019;10:1334–1343. doi: 10.18632/oncotarget.26665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, Liso A, Vignetti M, Fazi P, Meani N, Pettirossi V, Saglio G, Mandelli F, Lo-Coco F, Pelicci PG, Martelli MF GIMEMA Acute Leukemia Working Party. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 47.Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, Suzuki K, Chen F, Asou N, Ohtake S, Miyawaki S, Miyazaki Y, Sakura T, Ozawa Y, Usui N, Kanamori H, Kiguchi T, Imai K, Uike N, Kimura F, Kitamura K, Nakaseko C, Onizuka M, Takeshita A, Ishida F, Suzushima H, Kato Y, Miwa H, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Naoe T. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;28:1586–1595. doi: 10.1038/leu.2014.55. [DOI] [PubMed] [Google Scholar]
- 48.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, Huberman K, Cheng J, Viale A, Socci ND, Heguy A, Cherry A, Vance G, Higgins RR, Ketterling RP, Gallagher RE, Litzow M, van den Brink MR, Lazarus HM, Rowe JM, Luger S, Ferrando A, Paietta E, Tallman MS, Melnick A, Abdel-Wahab O, Levine RL. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pastore F, Dufour A, Benthaus T, Metzeler KH, Maharry KS, Schneider S, Ksienzyk B, Mellert G, Zellmeier E, Kakadia PM, Unterhalt M, Feuring-Buske M, Buske C, Braess J, Sauerland MC, Heinecke A, Krug U, Berdel WE, Buechner T, Woermann B, Hiddemann W, Bohlander SK, Marcucci G, Spiekermann K, Bloomfield CD, Hoster E. Combined molecular and clinical prognostic index for relapse and survival in cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 2014;32:1586–1594. doi: 10.1200/JCO.2013.52.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mason EF, Hasserjian RP, Aggarwal N, Seegmiller AC, Pozdnyakova O. Blast phenotype and comutations in acute myeloid leukemia with mutated NPM1 influence disease biology and outcome. Blood Adv. 2019;3:3322–3332. doi: 10.1182/bloodadvances.2019000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel SS, Kuo FC, Gibson CJ, Steensma DP, Soiffer RJ, Alyea EP 3rd, Chen YA, Fathi AT, Graubert TA, Brunner AM, Wadleigh M, Stone RM, DeAngelo DJ, Nardi V, Hasserjian RP, Weinberg OK. High NPM1-mutant allele burden at diagnosis predicts unfavorable outcomes in de novo AML. Blood. 2018;131:2816–2825. doi: 10.1182/blood-2018-01-828467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki T, Kiyoi H, Ozeki K, Tomita A, Yamaji S, Suzuki R, Kodera Y, Miyawaki S, Asou N, Kuriyama K, Yagasaki F, Shimazaki C, Akiyama H, Nishimura M, Motoji T, Shinagawa K, Takeshita A, Ueda R, Kinoshita T, Emi N, Naoe T. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106:2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 53.Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, Martelli MF, Haferlach T, Hiddemann W, Falini B. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 54.Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, Linch DC. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 55.de Jonge HJ, Valk PJ, de Bont ES, Schuringa JJ, Ossenkoppele G, Vellenga E, Huls G. Prognostic impact of white blood cell count in intermediate risk acute myeloid leukemia: relevance of mutated NPM1 and FLT3-ITD. Haematologica. 2011;96:1310–1317. doi: 10.3324/haematol.2011.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schnittger S, Bacher U, Kern W, Alpermann T, Haferlach C, Haferlach T. Prognostic impact of FLT3-ITD load in NPM1 mutated acute myeloid leukemia. Leukemia. 2011;25:1297–1304. doi: 10.1038/leu.2011.97. [DOI] [PubMed] [Google Scholar]
- 57.Ho AD, Schetelig J, Bochtler T, Schaich M, Schäfer-Eckart K, Hänel M, Rösler W, Einsele H, Kaufmann M, Serve H, Berdel WE, Stelljes M, Mayer J, Reichle A, Baldus CD, Schmitz N, Kramer M, Röllig C, Bornhäuser M, Thiede C, Ehninger G Study Alliance Leukemia. Allogeneic stem cell transplantation improves survival in patients with acute myeloid leukemia characterized by a high allelic ratio of mutant FLT3-ITD. Biol Blood Marrow Transplant. 2016;22:462–469. doi: 10.1016/j.bbmt.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 58.Linch DC, Hills RK, Burnett AK, Khwaja A, Gale RE. Impact of FLT3(ITD) mutant allele level on relapse risk in intermediate-risk acute myeloid leukemia. Blood. 2014;124:273–276. doi: 10.1182/blood-2014-02-554667. [DOI] [PubMed] [Google Scholar]
- 59.Schlenk RF, Kayser S, Bullinger L, Kobbe G, Casper J, Ringhoffer M, Held G, Brossart P, Lübbert M, Salih HR, Kindler T, Horst HA, Wulf G, Nachbaur D, Götze K, Lamparter A, Paschka P, Gaidzik VI, Teleanu V, Späth D, Benner A, Krauter J, Ganser A, Döhner H, Döhner K German-Austrian AML Study Group. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. 2014;124:3441–3449. doi: 10.1182/blood-2014-05-578070. [DOI] [PubMed] [Google Scholar]
- 60.Small D. FLT3 mutations: biology and treatment. Hematology Am Soc Hematol Educ Program. 2006:178–184. doi: 10.1182/asheducation-2006.1.178. [DOI] [PubMed] [Google Scholar]
- 61.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- 62.Preudhomme C, Sagot C, Boissel N, Cayuela JM, Tigaud I, de Botton S, Thomas X, Raffoux E, Lamandin C, Castaigne S, Fenaux P, Dombret H. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA) Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 63.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Meijer J, van Oosterhoud S, van Putten WL, Valk PJ, Berna Beverloo H, Tenen DG, Löwenberg B, Delwel R. Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4:31–40. doi: 10.1038/sj.thj.6200216. [DOI] [PubMed] [Google Scholar]
- 64.Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CA, Wouters BJ, van der Poelvan de Luytgaarde SC, Damm F, Krauter J, Ganser A, Schlenk RF, Löwenberg B, Delwel R, Döhner H, Valk PJ, Döhner K. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117:2469–2475. doi: 10.1182/blood-2010-09-307280. [DOI] [PubMed] [Google Scholar]
- 65.Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pabst T, Eyholzer M, Fos J, Mueller BU. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100:1343–1346. doi: 10.1038/sj.bjc.6604977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taube F, Georgi JA, Kramer M, Stasik S, Middeke JM, Röllig C, Krug U, Krämer A, Scholl S, Hochhaus A, Brümmendorf TH, Naumann R, Petzold A, Mulet-Lazaro R, Valk PJM, Steffen B, Einsele H, Schaich M, Burchert A, Neubauer A, Schäfer-Eckart K, Schliemann C, Krause SW, Hänel M, Noppeney R, Kaiser U, Baldus CD, Kaufmann M, Herold S, Stölzel F, Sockel K, von Bonin M, Müller-Tidow C, Platzbecker U, Berdel WE, Serve H, Ehninger G, Bornhäuser M, Schetelig J, Thiede C Study Alliance Leukemia (SAL) CEBPA mutations in 4708 patients with acute myeloid leukemia: differential impact of bZIP and TAD mutations on outcome. Blood. 2022;139:87–103. doi: 10.1182/blood.2020009680. [DOI] [PubMed] [Google Scholar]
- 68.Bullinger L. CEBPA mutations in AML: site matters. Blood. 2022;139:6–7. doi: 10.1182/blood.2021013557. [DOI] [PubMed] [Google Scholar]