Abstract
Transcriptional Mediator controls diverse gene programs for various developmental and pathological processes. The human Mediator MED23/R617Q mutation was reported in a familial intellectual disability (ID) disorder, although the underlying mechanisms remain poorly understood. Constructed by gene editing, the Med23/R617Q knock-in mutant mice exhibited embryonic lethality due to the largely reduced Med23/R617Q protein level, but the R617Q mutation in HEK293T cells didn’t change its expression and incorporation into Mediator Complex. RNA-seq revealed that MED23/R617Q mutation disturbed gene expression, related to neural development, learning and memory. Specifically, R617Q mutation reduced the MED23-dependent activities of ELK1 and E1A, but in contrast, upregulated the MAPK/ELK1-driven early immediate genes (IEGs) JUN and FOS. ChIP-seq and Hi-C revealed that the MED23 R617Q mutation reprogramed a subset of enhancers and local chromatin interactions, which correlated well with the corresponding gene expression. Importantly, the enhancers and chromatin interactions surrounding IEGs were unchanged by the R617Q mutation, but DACH1, an upstream repressor of IEGs, showed reduced enhancer-promoter interactions and decreased expression in mutant cells, thus relieving its inhibition to the intellectual-related IEGs. Overall, unraveling the MED23-DACH1-IEG axis provides a mechanistic explanation for the effects of the MED23/R617Q mutation on gene dysregulation and inherited ID.
INTRODUCTION
Mammalian genomes are segmented into well-organized domains in the nucleus, organized as compartments, topologically associated domains (TADs) and chromatin loops at different scales (1–4). Transcription is a highly regulated process orchestrated by cis- or trans-DNA elements, transcription factors and cofactors. Precise gene transcription in space and time requires distal enhancers to communicate with target promoters dynamically within the gene regulatory landscape. For this process, enhancers and promoters should be brought physically proximity in the right time and place. Additionally, recent studies have shown the important roles of enhancers in directing lineage- and stage-specific transcription during development (5–9); thus, how enhancers are organized and regulated in the highly dimensioned chromatin environment has become one of the most important questions to be explored.
Mediator is a large, highly conserved and multisubunit complex implicated in nearly all steps of the RNA polymerase II (Pol II) transcription process (10,11). The basic function of Mediator is to help recruit general transcription factors and Pol II for transcription initiation (10). Signaling factors and DNA-binding regulators communicate with the Pol II machinery via Mediator, which transduces developmental and environmental signals to Pol II to generate various mRNA outputs (12). Mediator thus contributes to loop enhancers and promoters in proximity (13), but how exactly Mediator controls enhancer functions and participates in enhancer-promoter communication requires more studies.
The MED23 subunit of Mediator was originally identified as the target of adenoviral oncoprotein E1A in mammalian cells (14,15). In recent years, in vitro and in vivo studies have identified a specific interactome between MED23 and other proteins, including RUNX2, RNA20/40, hnRNPL and CDK9, and revealed versatile roles of MED23 in adipogenesis, osteogenesis, myogenesis, H2B ubiquitination-mediated cell fate choice, mRNA splicing, and Pol II elongation (16–21). Dysfunctions of Mediator subunits are frequently associated with severe human diseases (22), as is MED23. In recently reported cases, several missense mutations of MED23 were found to be associated with familial intellectual disabilities (IDs) (23–25). Patients with MED23 mutations exhibit microcephaly, speech delay, hypotonia, global developmental delay and cognitive defects (25). Among the ID-related mutations of MED23, the R617Q missense mutation was previously reported to lead to mental disorders by disrupting the expression of the immediate early genes (IEGs) FOS and JUN (23). These so-called IEGs, which respond rapidly to stimuli from the external environment, are crucial for neuronal activity (26–29). In the brain, IEGs first respond to various neuronal stimulations and further influence neuronal physiology by regulating the expression of numerous downstream target genes (26). Disruptions in IEG transcriptional regulation are closely related to various neurological disorders (26,27). Here we determined to establish the mouse and the cell line carrying the ID-related MED23 mutation and to elucidate the underlying mechanism through which MED23 R617Q mutation dysregulates IEG expression.
In this study, we utilized CRISPR/Cas9 genomic editing tools to generate ID-related MED23/R617Q mutant mice and cell lines to investigate its effect on transcriptional control. We first determined that the Med23 R617Q mutation in mice results in serious instability of the Med23 protein and leads to early embryonic lethality. In contrast to mice, in the human cell line HEK293T, we found that the R617Q mutation did not alter MED23 expression and Mediator integrity but compromised the transcriptional activity of its binding transcription factors ELK1 and E1A. Even so, the downstream activation targets of ELK1, FOS and JUN were unexpectedly upregulated under the MED23 R617Q mutation. To understand these contradictory results, we further investigated the influence of MED23/R617Q on enhancer activation and chromatin conformation by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) and Hi-C techniques and found that MED23/R617Q remodeled enhancer activity and chromatin compartmentalization and interactions, which corresponded with disrupted Pol II occupancy and gene expression. Through further screening, we found that although the enhancer-promoter interactions or enhancer activities involving FOS and JUN were unchanged, their upstream suppressor DACH1 showed largely decreased enhancer-promoter contacts under MED23 R617Q mutation, along with its enhancer activity and mRNA expression, thus relieving the repression of FOS and JUN expression. Together, our findings provide mechanistic insight into MED23 pathogenic mutations and demonstrate the role of Mediator in the enhancer topology underlying transcriptional control.
MATERIALS AND METHODS
Cell culture and targeted genome editing
Human embryonic kidney 293T cells were cultured in 10% FBS containing high glucose Dulbecco's modified Eagle's medium (DMEM, HyClone, Logan, UT, USA). sgRNAs targeting MED23 exonX were cloned into the CRISPR/Cas9 plasmid vector PX330, which was transfected into 293T cells with a 69 nt single strand DNA template used for homologous directed DNA repair. Cells were sorted with FACS and validated by Sanger sequencing.
Coimmunoprecipitation (Co-IP) and immunoblotting
WT and MED23/R617Q mutant 293T cells were seeded in 10 cm dishes and cultured in an incubator. At nearly 80% confluence, the cells were harvested and lysed in lysis buffer (1% NP-40, 10% glycerol, 135 mM NaCl, 20 mM Tris, pH 8.0) with protease inhibitors. Lysates were subjected to immunoprecipitation with anti-CDK8 antibody (Santa Cruz, California, USA, #sc-1521) or control IgG at 4°C overnight, followed by washing in lysis buffer, SDS–PAGE electrophoresis and immunoblotting with the indicated antibodies (MED6, Santa Cruz, #sc-9434; Med12, Bethyl, TX, USA, # A300-774A; MED23, Abcam, Cambridge, UK, ab200351; MED24, Bethyl, A300-472A; β-actin, Sigma, MA, USA, A5441).
ChIP assay and ChIP-seq
ChIP-seq assays were conducted as previously described (17). Briefly, cells were crosslinked with 0.9% formaldehyde for 9 min at room temperature, and formaldehyde was quenched by glycine at a final concentration of 125 μM for 5 min. Cells were washed twice with ice-cold PBS and collected by a cell scraper. Cell pellets were lysed and sonicated in lysis buffer (1% SDS, 50 mM Tris–HCl (pH 7.4), 10 mM EDTA), and the chromatin was fragmented into 200–500 bp fragments using a COVARIS S220 sonicator. The lysates were then diluted with dilution buffer (0.01% SDS, 1.1% Triton X-100, 16.7 mM Tris–HCl (pH 8.0), 1.2 mM EDTA), and immunoprecipitation was performed overnight at 4°C with 2–4 μg of antibodies against H3K4me1 (Abcam, ab8895), H3K4me3 (Abcam, ab8580), H3K27ac (Abcam, ab4729) or polymerase II (Pol II, Santa Cruz, sc-899) and chromatin corresponding to 1 × 106 cells. The next day, IP samples were incubated with Dynabeads protein G for 2 h at 4°C, and the beads were washed with low salt buffer (0.1% SDS, 1% Triton X-100, 1 mM EDTA, 20 mM HEPES (pH 7.9), 150 mM NaCl, 0.1% deoxycholate), high salt buffer (0.1% SDS, 1% Triton, 1 mM EDTA, 50 mM HEPES (pH 7.9), 500 mM NaCl, 0.1% deoxycholate), LiCl buffer (0.25 M LiCl, 0.5% NP40, 0.5% deoxycholate, 1 mM EDTA, 20 mM Tris–HCl (pH 80)) and TE buffer (10 mM Tris–HCl (pH 8.0), 1 mM EDTA) before elution. Eluates were reverse-crosslinked at 65°C, and DNA was purified. The DNA was subjected to qRT-PCR or library construction (Vazyme, Nanjing, China, ND607) for Illumina sequencing.
qRT–PCR and RNA sequencing (RNA-seq)
Total RNA was extracted from cells lysed in TRIzol. One microgram of total RNA was reverse transcribed (TaKaRa-Bio, Dalian, China, R047A) to cDNA for qRT-PCR. We used a commercial RNA library kit to prepare the RNA-seq library with the manufacturer's specifications. Briefly, mRNA was captured with mRNA Capture Beads before fragmentation. The RNA fragment was reverse transcribed and subjected to library amplification for Illumina sequencing.
ChIP-seq and RNA-seq analysis
ChIP-seq and RNA-seq raw reads were obtained from an Illumina X-10 sequence analyzer. We utilized Fastqc (v0.1.0) for raw data quality summary. Cutadapt was used to remove sequencing adapters and poor-quality base trimming, retaining only reads with a quality score greater than 30 and discarding reads shorter than 50 bp or 75 bp for SE100 or PE150 reads, respectively. For RNA-seq, the filtered clean reads were mapped to the human reference genome hg19 with tophat2, and gene quantification and differential expression analysis were performed by cuffdiff with parameters -b -u. As for ChIP-seq, clean reads were aligned to the human reference genome hg19, and the mapping results were normalized to FPKM (counts per million mapped reads per kilobase) and converted to bigwig files for visualization by bamCoverage. Peak calling was conducted by MACS2 (30) with two or three replicates, and peaks were annotated using the R packages ChIPpeakAnno (31) and ChIPseeker (32). All peaks were concatenated and merged, and peaks within 3 kb of transcription start sites were assigned as promoter peaks. Peaks located out of any annotated genes were assigned as distal peaks. ChIP-seq signal intensity was calculated and normalized by total mapped reads, and P values were calculated using the two-sample Kolmogorov–Smirnov (K–S) test. We defined H3K27ac peaks distributed excluding known gene promoters as enhancers, peaks located within 2 kb of each other were merged together, and super enhancers were identified using the ROSE algorithm (33). The correlation coefficient of gene expression and H3K27ac tag density was calculated with the MA-norm algorithm (34). Finally, all heatmaps and density profiles were generated by DeepTools.
Hi-C library preparation
A total of 3 × 106 cells were collected by centrifugation at 180 × g for 5 min and crosslinked in a final concentration of 1% formaldehyde for 10 min at room temperature. For quenching of crosslink activation, glycine was added to a final concentration of 0.2 M. Crosslinked cell pellets were washed with cold PBS and immediately subjected to lysis and restriction enzyme digestion. Cells were lysed in lysis buffer (10 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.2% IGEPAL CA-630) and digested in 100 U Mbo I restriction enzyme (NEB) at 37°C overnight. The DNA ends were marked with biotin-labeled dATPs, and the 5′ overhang was filled using the Klenow enzyme. The blunt ends were ligated with T4 DNA ligase for 4 h at room temperature. Nuclei pellets were collected and digested with proteinase K. Proteinase-treated nuclei were directly reverse crosslinked at 68°C overnight. Crude DNA was fragmented into ∼400 bp fragments, and biotinylated DNA was pulled down with C1 streptavidin beads. For library preparation, approximately 500 ng of bead DNA was used as the initial material and amplified 5 cycles after adapter ligation.
Hi-C data analysis
Hi-C data analysis was performed with Hic-Pro (35), i.e. sequencing reads were separately aligned to the human reference genome hg19 with a two-step approach using Bowtie2. We filtered out low mapping quality reads, PCR artifacts, multiple hits and singletons. The uniquely mapped reads were used to build the contact maps for a set of resolutions of 5, 10, 25, 50, 100, 250, 500, 1000 and 2500 kb. Contact maps were converted into .hic files by the hicpro2juicebox.sh utility for visualization. We used cooltools (https://github.com/open2c/cooltools) to call diamond insulation scores using a 50 kb bin size and 250 kb bin size with default parameters. Regions between two adjacent boundaries were considered TADs. When calculating the overlap between the TAD boundaries identified from WT and R617Q, we allowed a maximum of 50 kb (size of a single bin) shift. Compartment profiles were calculated for each chromosome in cis as described by Aiden et al. (Lieberman-Aiden, E. et al., 2009, PMID: 19815776) using the cooltool eigs-cis tool (https://github.com/open2c/cooltools). In summary, principal component analysis was applied to the Pearson correlation matrix of the observed/expected matrix under 50- and 500-kb resolution. The PC1 values were further correlated with genome GC content to assign compartment A/B for each chromosome based on GC content. We considered a 50-kb region as the B/A switch region when the sign of the PC1 value of the region changed from negative to positive.
RNA interference
Small interfering RNAs (siRNAs) used for the knockdown of DACH1 and nonspecific siRNA negative controls were designed and synthesized by RiboBio (Guangzhou, China). The cells were seeded in a six-well cell culture plate and cultured in growth medium on the day before transfection. The transfection of siRNAs was carried out using Lipofectamine RNAiMAX Reagent (Invitrogen, CA, USA) according to the manufacturer's instructions. Cells were harvested for qRT−PCR 48 h later.
Statistical analyses
Statistical analyses were carried out using GraphPad Prism software. Data are presented as the mean ± SD, with n indicating the number of independent experiments. The comparison of differences among the groups was carried out by Student's t test. Differences were considered significant with a P value of <0.05. P values are depicted in figures using one to three asterisks (*P < 0.05, ** P < 0.01 and *** P < 0.001).
RESULTS
The human ID-associated MED23 R617Q mutation results in embryonic lethality in mice
To investigate the pathogenic mechanisms of the human MED23 R617Q mutation, we constructed human MED23 R617Q homologous mutant mice by utilizing CRISPR/Cas9 technology. The MED23 R617 amino acid residue is conserved between Homo sapiens and Mus musculus (Supplementary Figure 1A), and successful construction of the Med23/R617Q knock-in mutant mice was confirmed by DNA sequencing (Supplementary Figure 1B). The Med23/R617Q heterozygotes showed no developmental defects and lived normally. However, the homozygous mutant died at embryonic Day 8.5 (E8.5) (Supplementary Figure 1C). By examining the expression of Med23, we found that the Med23 R617Q mutation attenuated the Med23 protein level in mice. The Med23 protein levels of the Med23/R617Q mutant were dramatically decreased (Supplementary Figure 1D), while the RNA level did not change compared with that of the wild-type (WT) (Supplementary Figure 1E). Therefore, the Med23 R617Q mutation in mice leads to more serious consequences than that in humans and can barely mimic the MED23 R617Q mutation in humans. Of note, the possibility of off-target effect of CRISPR/Cas9 here is unlikely, because we have also additionally constructed the Med23/R617Q mice using the traditional gene-targeting strategy based on homologous recombination (Supplementary Figure 1F) and again the generated the R617Q mutant mice died at early embryonic stage, with Med23 protein not being expressed normally (Supplementary Figure 1G-I).
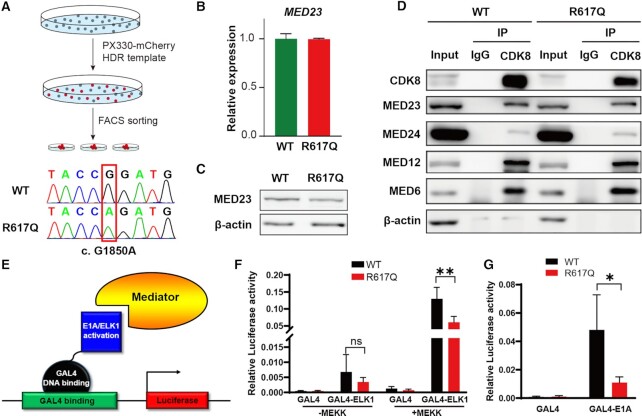
The ID-associated mutation of MED23 does not alter mediator integrity but results in reduced transcriptional activity
To understand the direct impact of ID-associated mutations on transcriptional regulation, we used the CRISPR/Cas9 genome engineering tool to obtain the missense mutation R617Q of MED23 (NM_015979.2, p.R617Q, c.G1850A) in HEK293T cells (Figure 1A). In contrast to the results in the mutant mouse, the G/A base substitution does not affect MED23 transcription; thus, a comparable amount of mRNA was produced (Figure 1B), and the R/Q amino acid residue substitution also did not change the protein level of MED23 (Figure 1C). Since a previous study indicated that MED23 deficiency may impair Mediator subunit components (36), we tested whether this residue change of MED23 may affect its structural association with the Mediator complex. Coimmunoprecipitation using an antibody against CDK8, a subunit located in the kinase module of Mediator, was performed to evaluate Mediator overall integrity. We observed that MED23/R617Q protein can incorporate into whole complex and did not result in a different immunoprecipitated subunit composition compared with the WT MED23, as indicated by the MED6, MED12 and MED24 subunits (Figure 1D). In summary, the R617Q mutation does not alter MED23 expression or MED23 incorporation into the Mediator complex.
Figure 1.
The effects of the MED23 R617Q mutation on Mediator integrity and transcriptional activities of partner transcription factors. (A) The workflow of targeted genome editing using CRISPR/Cas9. (B, C) The expression of MED23 at the RNA level (B, n = 3) and protein level (C) in the WT and MED23/R617Q mutant cells. (D) Co-IP assays to determine the influence of the MED23 R617Q mutation on Mediator organization. (E) Diagram of the luciferase reporter assay. The activation domain of ELK1 and E1A was fused with the GAL4 DNA binding domain, which can bind upstream of the luciferase TSS, thus recruiting Mediator to initiate transcription. (F) The effect of the MED23 R617Q mutation on the transcriptional activity of ELK1 with or without MEKK stimulation. n = 3 for each group. The values are the means ± SDs. **P < 0.01. (G) The effect of the MED23 R617Q mutation on the transcriptional activity of E1A. n = 3 for each group. The values are the means ± SDs. *P < 0.05.
To investigate the regulatory effect of the MED23 mutant in vivo, we performed reporter assays to evaluate the potential changing function of the endogenous WT and R617Q mutant MED23 in activating transcription. The previous reports showed that MED23 can directly interact with the adenoviral oncoprotein E1A to support transcription (14,15). The ternary complex factor (TCF) ELK1 can also interact with MED23 to stimulate transcription when phosphorylated by mitogen-activated protein kinase (MAPK) signaling (20,36). At this point, we employed GAL4-dependent luciferase reporter assays to evaluate the activities of the GAL4 DNA binding domain-fused E1A or ELK1 activation domain (Figure 1E). The chimeric GAL4-ELK1 construct was transfected into WT or MED23/R617Q mutant cells with or without the MAPK kinase kinase (MEKK) plasmid cotransfection. Both WT and mutant MED23 supported reporter gene expression, but the mutant showed significantly reduced transcriptional activity with or without MEKK cotransfection (Figure 1F). Similar results were obtained from GAL4-E1A, WT and R617Q mutant MED23, which were both capable of supporting the reporter gene, while MED23/R617Q exhibited a reduction in transcriptional activity (Figure 1G). Therefore, the ID-associated MED23 R617Q mutation compromised the transcriptional activation capability of the Mediator subunit MED23.
The MED23 R617Q mutant upregulates IEGs involved in developmental process
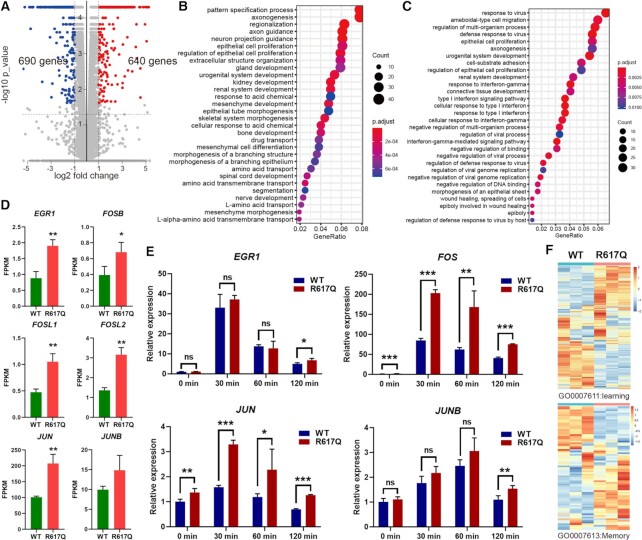
To better understand the changed gene expression at the genome level by the ID-associated mutant of MED23, we performed transcriptome analysis of the WT and MED23/R617Q mutant cells by RNA-seq. A total of 640 upregulated and 690 downregulated genes were identified in the MED23/R617Q mutant cells compared with the WT cells (Figure 2A). To analyze the molecular characteristics of the ID-associated mutant cells, we conducted functional analysis of these differentially expressed genes (DEGs). Gene Ontology (GO) showed that many DEGs involved in nervous system development, pattern specification process, axonogenesis, neuron projection guidance, and spinal cord development process were downregulated (Figure 2B), which is consistent with the phenotypes that patients with ID show developmental abnormalities and intellectual inability. Interestingly, the upregulated genes were also significantly enriched in the development of multiple tissues and organs, including axonogenesis (Figure 2C).
Figure 2.
Analysis of genes dysregulated by the MED23 R617Q mutation. (A) Volcano plots depicting gene expression changes in the WT and R617Q cells (n = 3; fold change ≥ 2, and P value < 0.05, as determined by cuffdiff). (B, C) Gene Ontology Biological Process (GO-BP) analysis of down- (B) and upregulated (C) genes. Terms are ordered by the number of genes hit in each process. (D) FPKM of IEGs affected by the MED23 R617Q mutation in the RNA-seq data. n = 3 for each group. The values are the means ± SDs. *P < 0.05, **P < 0.01. (E) Serum induction assays to determine the influence of the MED23 R617Q mutation on IEG expression. n = 3 for each group. The values are the means ± SDs. *P < 0.05, **P < 0.01, ***P < 0.001. (F) Heatmaps showing expression changes of genes involved in learning (GO: 0007611) and memory (GO: 0007613).
Among these differentially expressed genes, we found that the gene expression levels of the IEGs EGR1, FOS family, JUN and JUNB were all increased in the MED23/R617Q mutant cells compared with the WT cells (Figure 2D). This finding is surprising and unexpected, as we showed that the activation of ELK1 controlled by MAPK is significantly reduced in the MED23/R617Q cells. To better understand the dynamic changes in IEGs induced by the MED23 R617Q mutation, we performed serum stimuli assays, and again, we found significantly higher levels of the IEGs FOS and JUN in the MED23/R617Q mutant cells than in the WT cells in either the steady state or under serum induction (Figure 2E). As we previously found, the ternary complex factor ELK1 transcriptionally activates the expression of many IEGs, especially EGR1, which is dependent on the presence of the Mediator subunit MED23 (36,37). Paradoxically, when ELK1 transcriptional activity was reduced in the MED23/R617Q mutant cells, IEGs were upregulated. These controversial results suggest that MED23/R617Q may influence the expression of IEGs in an alternative mechanism, and we wondered whether the MED23 R617Q mutant exerts its functions beyond transcriptional initiation. Lastly, since IEGs participate in various processes of neuronal development and function, including learning and memory (38–40), and IEGs are dysregulated in MED23 R617Q mutant cells, many learning/memory-related genes were also found to be reprogrammed in the MED23/R617Q mutant cells compared with the WT cells (Figure 2F and Supplementary Table 1). Thus, MED23 R617Q mutation dysregulates IEGs and other genes related to learning and memory.
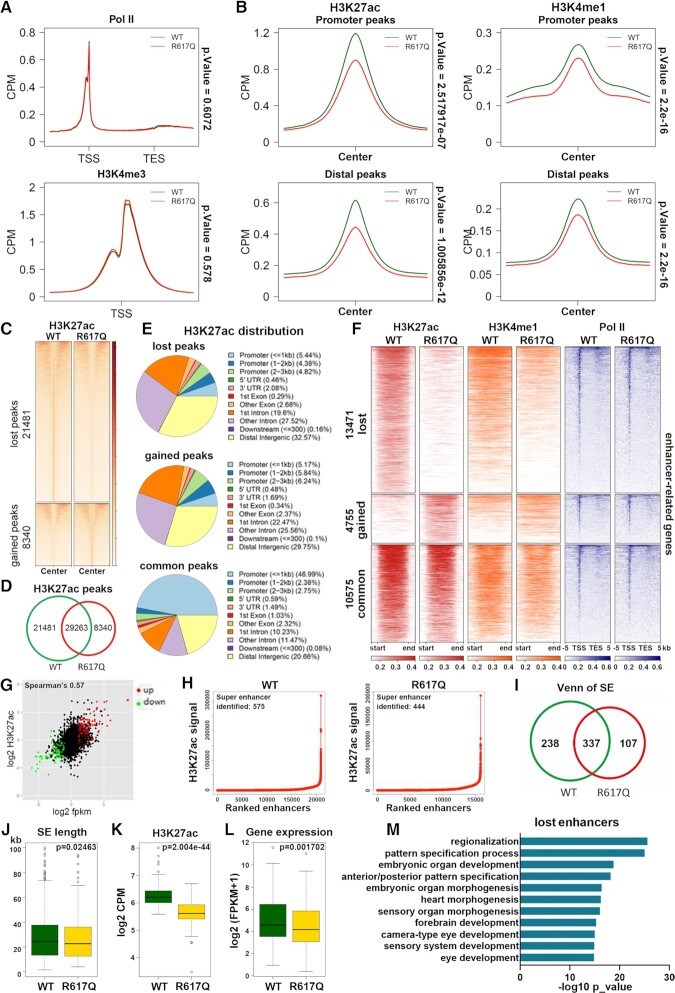
The ID-associated MED23 mutant alters enhancer distribution and activity
Recent studies have revealed that Mediator controls cell identity-related gene expression through looping enhancers and promoters (13,41). Therefore, we wondered whether the MED23 ID-associated mutation may dysregulate specific gene expression by altering enhancer regulation. We investigated whether the MED23 R617Q mutation was involved in epigenetically regulated enhancer functions. Through ChIP-seq, we analyzed the histone modifications of H3K27ac, H3K4me1 and H3K4me3 and RNA Pol II binding in the WT and MED23/R617Q cells. The global Pol II binding profile showed no difference between the WT and MED23/R617Q cells (Figure 3A), supporting our finding that the MED23 mutant specifically changes a subset of gene expression without greatly affecting global transcription (Figure 2A). Consistently, H3K4me3 modification, predominantly occurring at promoters, was almost unchanged between the MED23/R617Q mutant and WT cells (Figure 3A). However, we did observe an overall decrease in the H3K27ac and H3K4me1 signals both at the promoter–proximal and promoter–distal regions (Figure 3B), both of which mark active enhancers, suggesting an overall reduction in enhancer activities. Specifically, there were 21 481 lost and 8340 newly formed H3K27ac peaks in the MED23 R617Q mutant cells compared with the WT cells and 29 263 common peaks in both WT and mutant cells (Figure 3C and D). Genomic distribution analysis on the gained, lost and common H3K27ac peaks revealed that both the lost and gained H3K27ac by MED23 R617Q mutation mainly occurred at noncoding regions, of which >70% of sites are distributed at intron and intergenic regions (Figure 3E), which also conformed with the enhancer locations. By contrast, the common H3K27ac peaks were unaffected by MED23/R671Q, >50% of which were in the promoter region. These results suggest that MED23 R617Q mutation selectively disrupted the H3K27ac modification in the noncoding regions/enhancer locations. Therefore, the MED23 R617Q seems to disturb the functions of a subset of enhancers. We thus further analyzed the role of MED23 R617Q in enhancer regulation.
Figure 3.
Enhancer dysfunction analysis in MED23/R617Q cells. (A) Average tag density plots of H3K4me3 and Pol II ChIP-seq enrichment profiles 3 kb upstream and downstream of the gene locus. Gene bodies are normalized into the same lengths. TSS, transcription start site. TTS, transcription terminal site. n = 2. (B) Average tag density plots of promoter–proximal and promoter–distal H3K27ac and H4K4me1 ChIP-seq enrichment profiles. n = 2. (C) Heatmaps of normalized H3K27ac ChIP-seq reads at differentially acetylated sites in the WT and R617Q cells. The heatmap represents the cpm (counts per million mapped reads) value of two replicates. Sites with decreased and increased signals are plotted separately. (D) Venn diagram of overlapping H3K27ac peaks in the WT and MED23/R617Q cells. (E) Pie chart of H3K27ac signal distribution at genomic loci of gained, lost and common peaks. (F) Heatmaps of normalized H3K27ac and H3K4me1 tag density at enhancers and Pol II density at the enhancer-related genes. Enhancers are hierarchically clustered according to the H3K27ac ChIP-seq signal. The heatmap represents the ChIP-seq signal of two replicates. (G) Scatter plot of fold change in gene expression against fold change in H3K27ac signal at enhancers. Significant genes are indicated with red (upregulated) and green (downregulated) dots. (H) Identification of SEs in the WT and MED23/R617Q cells. Dots representing enhancer clusters are ranked by the H3K27ac signal (as defined by the ROSE algorithm). Peaks located within 12.5 kb of each other are merged. (I) Venn diagram of SEs overlapping in the WT and MED23/R617Q cells. (J) SE length distributions in the WT and MED23/R617Q cells. (K) H3K27ac tag density at lost SEs in the MED23/R617Q cells compared with the WT cells. (L) Analysis of the gene expression of all lost SE-associated genes. (M) Bar plots of GO term enrichment of all lost enhancer-related genes in the MED23/R617Q cells compared with the WT cells. Enrichment was calculated with the R package clusterProfiler.
Conventionally, the H3K27ac modification sites outside the promoter regions were defined as putative enhancers (42). A total of 4755 gained and 13 471 lost putative enhancers were identified in the MED23 R617Q mutant cells compared with the WT cells, as defined by the differential binding signals of H3K27ac and H3K4me1, with the globally unchanged Pol II binding as a control (Figure 3F), in consistence with the aforementioned result (Figure 3A). To determine the correlation between the altered enhancer activity and the corresponding gene expression change, we assigned each enhancer to its nearest promoter and calculated its correlation of H3K27ac reads with RNA transcripts. As a result, H3K27ac signal distributions positively correlated with RNA levels with a Spearman's coefficient of 0.57 (Figure 3G), indicating a significant positive correlation between the enhancer H3K27ac signal intensity and the corresponding gene expression.
Studies have suggested that Mediator binds to cell-specific super enhancers (SEs) to regulate cell fate determination (33). We also analyzed the effect of the MED23 R617Q mutation on SE distribution. Using the rank ordering of super enhancers (ROSE) algorithm (33), by plotting the SE distribution in the WT and MED23/R617Q cells (Figure 3H), we found a total of 238 lost and 107 newly formed SEs in the MED23/R617Q cells compared with the WT cells (Figure 3I). Analysis of SEs in the WT and MED23/R617Q cells showed that the average SE length in MED23/R617Q cells was reduced compared with the WT cells (Figure 3J). Further, we analyzed H3K27ac signal and the gene expression related to the lost and gained SEs in the WT and MED23/R617Q cells (Figure 3K and L, and Supplementary Figure 2A and B). For the lost SEs in MED23/R617Q cells, the H3K27ac signal intensity were reduced (Figure 3K), and the expression of their neighboring genes was downregulated (Figure 3L). For the gained and common SEs, we observed that the H3K27ac of common SEs were significantly lower in MED23/R617Q, in consistent with the globally decreased H3K27ac in the MED23/R617Q cells (Supplementary Figure 2A). The H3K27ac signal of gained SEs in MED23/R617Q cells tended to be increased compared to that in WT cells, although the difference is not significant (Supplementary Figure 2A). As expected, the common SE related gene expression is almost unchanged (Supplementary Figure 2B), and by contrast, the gained SE related gene expression is upregulated in the MED23/R617Q cells (Supplementary Figure 2B). These results suggested that the MED23 R617Q mutation altered the distribution and strength of enhancers, thus disturbing the corresponding gene expression.
To dissect the functional consequence of enhancer malfunctions, we performed GO enrichment of these enhancer-associated genes (Figure 3M, Supplementary Figure 2C). In the genes linked to the lost enhancers, we found multiple enriched biological processes, such as ‘embryonic organ morphogenesis’ and ‘forebrain development’ (Figure 3M). This finding is consistent with the mental development defect of MED23 R617Q mutant patients (23). In summary, the MED23 R617Q mutation may selectively compromise the enhancer enrichment of H3K27ac and H3K4me1, resulting in dysfunction of a set of enhancers that may be key for neural development related to intelligence.
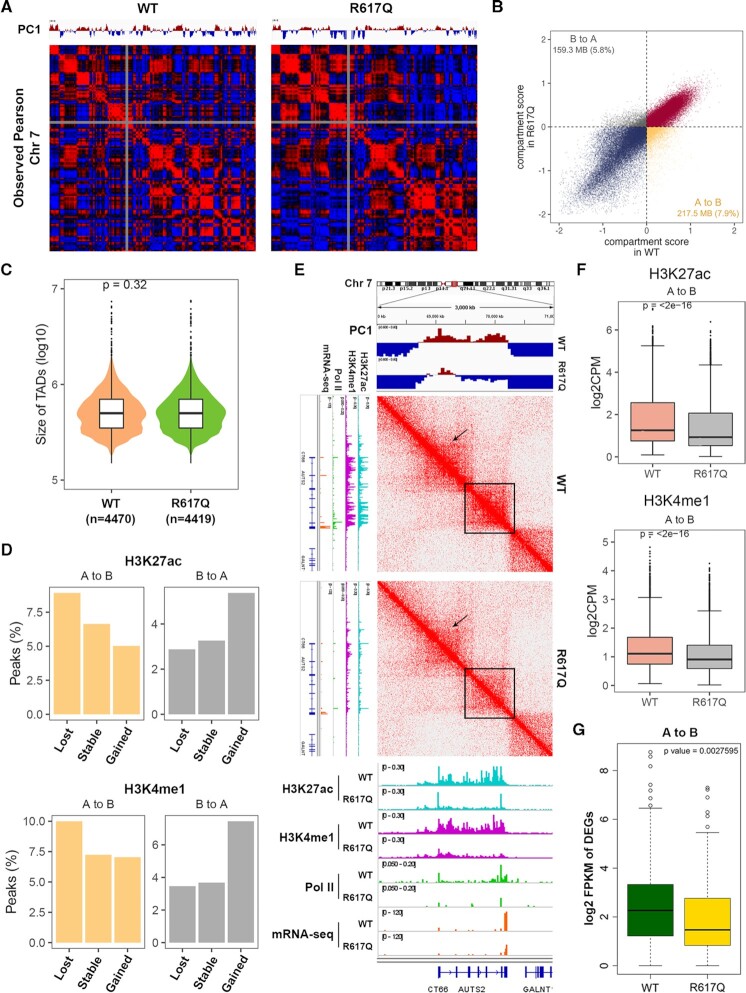
The ID-associated MED23 mutant disrupts a subset of local chromatin interactions
Gene activation by enhancers is mainly achieved by enhancer-promoter looping, and the MED23 R617Q mutant cells alter enhancer figures to control gene expression, which suggests that the MED23 R617Q mutation may change chromatin interactions to a certain extent. We therefore investigated whether the MED23 R617Q mutant impacts 3D chromatin architecture using the Hi-C approach. The mammalian genome is recapitulated with compartments, TADs and loops ranging from megabases to kilobases (2,43), and chromatin is mainly organized into A and B compartments, which represent transcriptionally active euchromatin and transcriptionally inactive heterochromatin, respectively (43,44). To determine whether the MED23 R617Q mutation may influence the A/B compartment organization, we analyzed the compartment types of the genome of the WT and MED23 R617Q mutant cells at 50-kb resolution (Figure 4A). We found that the majority of the compartmentalization was unchanged between the WT and mutant cells (Figure 4A and B). However, 7.9% of the genome regions switched from the A compartment in the WT cells to the B compartment in the MED23 R617Q mutant cells, and 5.8% of the genome regions showed the opposite switch from the B compartment in the WT cells to the A compartment in the mutant cells (Figure 4B). We further investigated whether TADs, which are demarcated by insulators or TAD boundaries (45), were influenced by the MED23 R617Q mutation. We called TADs and identified 4470 and 4419 TADs with the same median length of 500 kb in the WT and MED23 R617Q cells, respectively (Figure 4C), and there were 4036 TAD boundaries overlapping between the WT and MED23 R617Q mutant genomes, suggesting that the TADs were largely unaffected by the MED23 R617Q mutation, which is consistent with previous findings that the key factors in TAD formation are the zinc finger transcription factor CTCF and the multisubunit protein complex cohesin (46,47).
Figure 4.
Chromatin conformation alteration analysis between the WT and MED23/R617Q cells by Hi-C. (A) Comparison of chromosome compartments. An example of the chromosome compartment at chromosome 7 is shown as Pearson correlation matrices. The PC1 of chromosome 7 is shown in each picture. (B) The genomic compartment changes at 50-kb resolution between the WT and MED23/R617Q genomes. ‘A’ and ‘B’ represent the open and closed compartments, respectively. ‘A to B’ denotes compartments that are open in the WT but closed in MED23/R617Q, and ‘B to A’ denotes compartments that are closed in MED23/R617Q and open in the WT. (C) Violin plot showing the size distribution of TADs for the whole genome. The number of identified TADs in the WT (n = 4470) and MED23/R617Q (n = 4419) cells is shown below the violin plots. (D) Bar plots showing the H3K27ac and H3K4me1 peaks residing at regions for different compartmental switch categories (50-kb resolution). (E) A representative snapshot of the Hi-C interaction matrix (middle panel) at 10-kb resolution in one of the ‘A to B’ regions (top panel), and the boxed regions and the arrows within the heatmap indicate the weakened chromatin interaction regions under the MED23 R617Q mutation. The following tracks show normalized H3K27ac (brilliant blue), H3K4me1 (fuchsia) and Pol II (green) ChIP-seq and mRNA-seq (orange) read coverage. (F) Box plots showing the H3K27ac and H3K4me1 signal density in the ‘A to B’ regions in the WT and MED23/R617Q genomes. The horizontal black line within each white box indicates the median value. (G) Box plot showing the log2 FPKM expression levels of DEGs residing in the ‘A to B’ regions. The horizontal black line within each white box indicates the median value.
Therefore, in light of the results above, we further revealed the link between the A/B compartment switch and the distribution alteration of the H3K27ac and H3K4me1 peaks in the genome of the MED23 R617Q mutant cells compared with the WT cells. We analyzed the location of the lost, stable and gained H3K27ac and H3K4me1 peaks in the MED23 R617Q mutant cells. We found that more lost H3K27ac peaks in the MED23 R617Q mutant cells were located within the ‘A to B’ compartment regions compared with the stable and gained peaks, and by contrast, more gained H3K27ac peaks in the MED23 R617Q mutant cells were located within the ‘B to A’ compartment regions compared with the stable and lost peaks (Figure 4D). Additionally, the H3K4me1 peak redistributions behaved in a similar fashion (Figure 4D). These results suggested a positive correlation between genome-wide compartment switching and the changing enhancer epigenetic activities. Specifically, the A/B compartment switch may be caused by the H3K27ac and H3K4me1 modification alterations.
Based on the globally decreased H3K27ac and H3K4me1 signals under the MED23 R617Q mutation (Figure 3B) and the neural development-related GO items that were enriched in the lost enhancer-related genes in the MED23/R617Q cells (Figure 3M), we focused on analyzing the ‘A to B’ compartment regions. Consistent with the results above, we observed that in the ‘A to B’ compartment, the H3K27ac and H3K4me1 signals were both weakened by the MED23 R617Q mutation, and the chromatin interactions were weakened accordingly (Figures 4E and F, Supplementary Figure 3), which further led to weakened Pol II binding and mRNA expression (Figure 4E, Supplementary Figure 3). The expression level of DEGs located in the genomic regions switched from the A to B compartment in the MED23 R617Q mutant cells was significantly lower than that in the WT cells (Figure 4G), suggesting a positive relationship between compartment switching, epigenetic changes of enhancers, and differential gene expression. Collectively, we concluded that the MED23 R617Q mutation modestly changes the global chromatin architecture, affecting the compartment of active and inactive chromatin to some extent, thus disturbing a subset of enhancer activities, which attenuates the related gene expression.
Reduced DACH1 expression relieves the repression of early response genes
After surveying the global chromatin conformation and enhancer epigenetic changes induced by the MED23 R617Q mutation, we wondered whether IEG disruption by the MED23 R617Q mutation could be attributed to chromatin changes. Additionally, we attempted to resolve the paradox that MED23/R617Q attenuated ELK1 activity while upregulating IEG expression, which is thought to be controlled by ELK1. We first analyzed the chromatin interactions surrounding FOS and JUN but found that there were no detectable changes between the WT and MED23 R617Q mutant (Supplementary Figure 4). In addition, the H3K27ac and H3K4me1 modifications around the FOS and JUN loci were generally unchanged in the WT and MED23/R617Q cells (Supplementary Figure 4). Thus, the upregulation of FOS and JUN expression in the MED23/R617Q cells may be not resulted from the changed chromatin conformation and/or enhancer-promoter interactions.
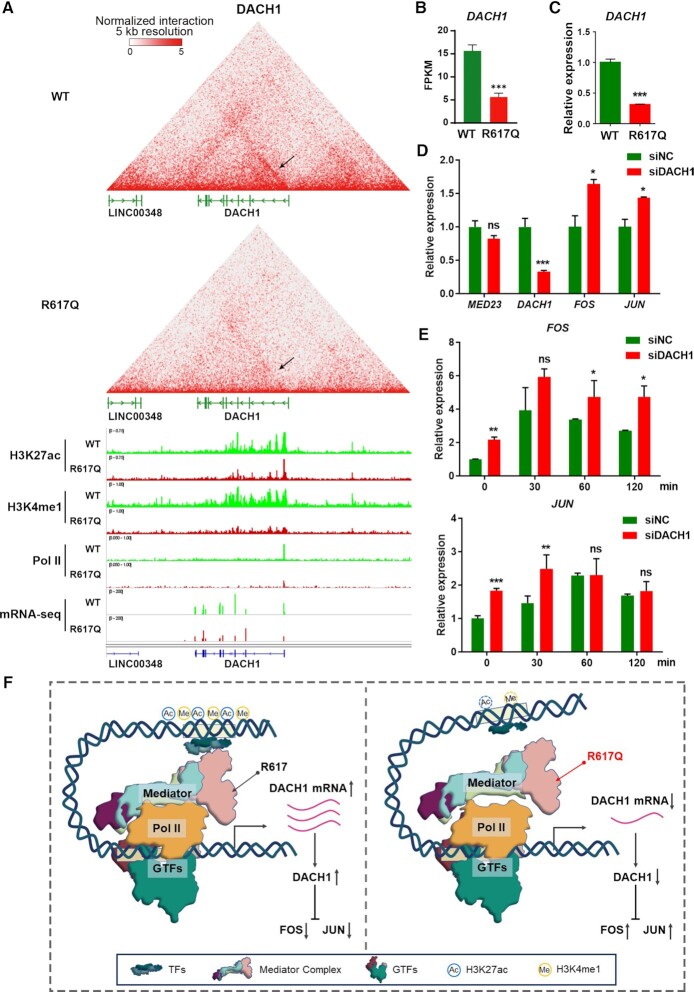
We further examined the upstream regulators of FOS and JUN, and through reanalysis of the Hi-C data, we found greatly decreased enhancer-promoter interactions of Dachshund family transcription factor 1 (DACH1) in the MED23/R617Q mutant cells (Figure 5A). DACH1 was previously shown to repress FOS, JUN and JUNB expression (48). As shown in Figure 5A, the contact frequency between the DACH1 promoter and its putative enhancers, which were modified with H3K27ac and H3K4me1 in DACH1 intron regions, was obviously weakened in the MED23/R617Q mutant cells compared with the WT cells, as the arrows indicated. Similar to the Hi-C data, the expression of DACH1 in the MED23/R617Q cells was also downregulated in the RNA-seq data (Figure 5B), which was also validated by qPCR (Figure 5C). Moreover, the active enhancer marker H3K27ac and H3K4me1 modifications were both decreased at the putative enhancers in DACH1 introns under MED23 R617Q mutation (Figure 5A).
Figure 5.
Mechanistic study of the upregulation of FOS and JUN expression. (A) Representative snapshots of the Hi-C interaction matrix at the DACH1 locus at 5-kb resolution in the WT and MED23/R617Q cells. Heatmaps are shown as a 45° rotated contact map. The following tracks show normalized H3K27ac, H3K4me1, Pol II ChIP-seq and mRNA-seq read coverage in the WT (green) and MED23/R617Q cells (red). (B) FPKMs of DACH1 in the WT and MED23/R617Q cells in the RNA-seq data. n = 3 for each group. The values are the means ± SDs. ***P < 0.001. (C) qPCR assays to determine the expression of DACH1 in the WT and MED23/R617Q cells (n = 3). n = 3 for each group. The values are the means ± SDs. ***P < 0.001. (D) DACH1 knockdown assays to determine its effect on FOS and JUN expression. n = 3 for each group. The values are the means ± SDs. *P < 0.05, ***P < 0.001. (E) Serum induction assays to determine the influence of DACH1 knockdown on FOS and JUN expression in the WT and MED23/R617Q cells. n = 3 for each group. The values are the means ± SDs. *P < 0.05, **P < 0.01, ***P < 0.001. (F) Model of MED23 R617Q mutation-induced transcription changes.
Interestingly, in development, DACH1 is a cell fate determination factor that functions to control multiple progenitor cell proliferation in mammalian organogenesis (49), and Dach1 mutant mice exhibit postnatal lethality (50). Moreover, DACH1 is highly expressed in the proliferating neural progenitors of the developing cortical ventricular and subventricular regions (51), suggesting a fundamental role of DACH1 in neurogenesis and intelligence. These evidences suggested that the downregulation of DACH1 may be the reason for the upregulation of the IEGs FOS and JUN in the MED23/R617Q mutant, which collectively accounts for the intellectual inability related to MED23/R617Q.
We then further tested whether the elevated IEG expression was caused by the inhibitory effect of DACH1. As expected, knocking down DACH1 in HEK293T cells upregulated the expression of both FOS and JUN (Figure 5D). Similarly, the levels of FOS and JUN were also higher under serum induction conditions in the DACH1 knockdown cells than in the control cells (Figure 5E). Thus, the decrease in DACH1 indeed resulted in elevated IEG FOS and JUN expression. Collectively, these results indicated that the disrupted enhancer activity and enhancer-promoter interactions caused by the MED23 R617Q mutation led to the downregulation of DACH1, which subsequently relived its repression and led to the upregulation of the IEGs FOS and JUN (Figure 5F). Overall, MED23-DACH1-IEGs may represent a novel molecular axis controlling the neural development underlying intelligence.
DISCUSSION
Although MED23 is known to be critical for normal intellectual development (23), the mechanistic understanding of MED23 in ID remains to be explored, especially regarding how its inherited mutation dysregulates genes involved in specific processes. In this work, we investigated the impact of the ID-related mutation MED23 R617Q on gene expression and chromatin architecture through the gene-edited cell line and mouse model. We found that the MED23 mutation R617Q impairs the transcriptional activity of its interacting transcription factors. At the genomic level, the MED23 R617Q mutation reprograms gene expression by modulating enhancer landscapes and local chromatin interactions. However, regarding how the MED23 R617Q mutation is associated with intellectual disability, the IEGs FOS and JUN were found to be abnormally elevated in the MED23 mutant cells, which is unexpected due to the weakened transcriptional activity of their upstream activator ELK1 in the MED23/R617Q cells. In addition, the histone modifications and chromatin interactions around FOS and JUN gene loci were found to be unchanged by the MED23 R617Q mutation. Subsequently, by reexamining the histone ChIP-seq and Hi-C data, we found that their upstream regulator DACH1 was downregulated due to the compromised enhancer activity and weakened enhancer-promoter interactions under the MED23 R617Q mutation. DACH1 knockdown indeed led to the upregulation of FOS and JUN expression. Interestingly, previous studies have found that DACH1 is strongly related to intellectual development. Although DACH1 mutation cases have not been reported in humans to date, studies in flies have discovered that dachshund, the DACH1 homolog in Drosophila, plays a pivotal role in controlling mushroom body development, which is critical for memory and learning behavior, and dachshund-deficient Drosophila manifest brain abnormalities (51–53). Therefore, DACH1 may play an important role in human intellectual development as well, which may be partially realized by suppressing FOS and JUN expression. Taken together, in this study we revealed the MED23-DACH1-IEGs axis, which provides a new molecular understanding of the inherited ID with the MED23 mutation.
Although the HEK293 cells are considered the kidney embryonic epithelial cells (54,55), our RNA-seq profiling revealed that the DEGs, related to neuron and intelligence development, were enriched through comparison between the WT and MED23/R617Q cells, which is unexpected and interesting. In fact, increasing evidence suggests that HEK293 cells may have a neuronal linage origin. The neuron-specific genes were observed to be expressing in HEK293 cells, such as neurofilaments, neuroreceptors, neuron-specific ion channel subunits, etc. (56–58). In addition, HEK293 cells exhibited some characteristics of the neuronal lineage cells. For example, HEK293 cells showed neuron-like electrophysiological characteristics and sensitivity to neural-specific agonist, and even formed the functional excitatory synapses when cocultured with neurons (58–64). These observations suggest that HEK293 may share a neural linage origin, therefore the molecular mechanism revealed in HEK293 cells may at least in part reflect what occurs in neuronal linage cells.
Our previous research has established that MED23 deficiency resulted in the defective ELK1 activation in response to MAPK signaling, which further led to the defective serum induction of IEGs EGR1 and FOS (36,37). However, in this study we observed the barely-changed EGR1 and the upregulated FOS expression in MED23/R617Q cells, which is consistent with the unchanged EGR1 and the upregulated FOS in the MED23/R617Q patient's skin fibroblasts (23). The modestly reduced ELK1 transcription activity by MED23 R617Q mutation might be insufficient accounting for the transcription dysregulation of IEGs, simply because that the 50% reduced ELK1-activity by MED23 R617Q mutation here is far more less than the >10-fold decrease by MED23 knockout (36). Therefore, through the genomic-scale analysis, we further attributed the dysregulation of IEGs to the disrupted DACH1 expression.
Regarding the possible mechanisms of how DACH1 regulates gene expression, studies have found that DACH1 can interact with HDAC1 and HDAC3, further suggesting that DACH1 may suppress FOS and JUN expression by promoting their histone deacetylation (48). Herein, we found that FOS and JUN expression was suppressed by DACH1; however, the H3K27ac modification around the FOS and JUN gene loci was unchanged, suggesting that the alternative mechanisms by which DACH1 suppresses FOS and JUN expression may be not through epigenetic regulation. The exact mechanisms await further investigation in the future.
Our findings advance the overall understanding of MED23 in chromatin structure and transcription regulation. As the bridge of enhancers and promoters, Mediator collaborates with cohesion to loop the enhancers and promoters (13). However, less is known about the mechanisms of the Mediator complex in mediating the enhancer-promoter interaction. Herein, we determined that a single amino acid mutation of MED23, R617Q, attenuated enhancer-promoter looping without destroying its incorporation into the Mediator complex, suggesting that MED23 R617 is a critical link through which Mediator functions in looping the enhancers and promoters, at least in part.
In conclusion, this work demonstrated how the MED23 R617Q mutation functions in dysregulating gene expression and further leads to human intellectual conditions; and the dysregulation of the MED23-DACH1-IEGs axis is likely underlying the human intellectual disability.
DATA AVAILABILITY
The sequencing data, including RNA-seq, ChIP-seq and Hi-C were deposited in GEO under the accession number GSE216401.
Supplementary Material
Contributor Information
Yenan Yang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Chonghui Li, State Key Laboratory of Cell Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Ziyin Chen, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Yiyang Zhang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Qing Tian, State Key Laboratory of Cell Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Meiling Sun, State Key Laboratory of Cell Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Shuai Zhang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Miao Yu, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
Gang Wang, State Key Laboratory of Genetic Engineering, School of Life Sciences and Zhongshan Hospital, Fudan University, Shanghai 200438, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [32030028 and 32170735 to G.W.]. Ministry of Science and Technology of China [2022YFA0806200 to M.Y. and G.W.]. Funding for open access charge: National Natural Science Foundation of China [32030028].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O.et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J.et al.. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dixon J.R., Gorkin D.U., Ren B.. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016; 62:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang J., Liu X., Li D., Shao Z., Cao H., Zhang Y., Trompouki E., Bowman T.V., Zon L.I., Yuan G.C.et al.. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell. 2016; 36:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu C.S., Hellmuth J.C., Singh R., Ying H.Y., Skrabanek L., Teater M.R., Doane A.S., Elemento O., Melnick A.M., Roeder R.G.. Unique immune cell coactivators specify locus control region function and cell stage. Mol. Cell. 2020; 80:845–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siersbaek R., Scabia V., Nagarajan S., Chernukhin I., Papachristou E.K., Broome R., Johnston S.J., Joosten S.E.P., Green A.R., Kumar S.et al.. IL6/STAT3 Signaling hijacks estrogen receptor alpha enhancers to drive breast cancer metastasis. Cancer Cell. 2020; 38:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thakurela S., Sahu S.K., Garding A., Tiwari V.K.. Dynamics and function of distal regulatory elements during neurogenesis and neuroplasticity. Genome Res. 2015; 25:1309–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lettice L.A., Williamson I., Devenney P.S., Kilanowski F., Dorin J., Hill R.E.. Development of five digits is controlled by a bipartite long-range cis-regulator. Development. 2014; 141:1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malik S., Roeder R.G.. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010; 11:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richter W.F., Nayak S., Iwasa J., Taatjes D.J.. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2022; 23:732–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allen B.L., Taatjes D.J.. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015; 16:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S.et al.. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010; 467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boyer T.G., Martin M.E.D., Lees E., Ricciardi R.P., Berk A.J.. Mammalian Srb Mediator complex is targeted by adenovirus E1A protein. Nature. 1999; 399:276–279. [DOI] [PubMed] [Google Scholar]
- 15. Wang G., Berk A.J.. In vivo association of adenovirus large E1A protein with the human mediator complex in adenovirus-infected and -transformed cells. J. Virol. 2002; 76:9186–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Z., Yao X., Yan G., Xu Y., Yan J., Zou W., Wang G.. Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat. Commun. 2016; 7:11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao X., Tang Z., Fu X., Yin J., Liang Y., Li C., Li H., Tian Q., Roeder R.G., Wang G.. The Mediator subunit MED23 couples H2B mono-ubiquitination to transcriptional control and cell fate determination. EMBO J. 2015; 34:2885–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Y., Li W., Yao X., Lin Q.J., Yin J.W., Liang Y., Heiner M., Tian B., Hui J., Wang G.. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell. 2012; 45:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang W., Yao X., Huang Y., Hu X., Liu R., Hou D., Chen R., Wang G.. Mediator MED23 regulates basal transcription in vivo via an interaction with P-tefb. Transcription. 2013; 4:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang W., Huang L., Huang Y., Yin J.W., Berk A.J., Friedman J.M., Wang G.. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev. Cell. 2009; 16:764–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yin J.W., Liang Y., Park J.Y., Chen D., Yao X., Xiao Q., Liu Z., Jiang B., Fu Y., Bao M.et al.. Mediator MED23 plays opposing roles in directing smooth muscle cell and adipocyte differentiation. Genes Dev. 2012; 26:2192–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin J.W., Wang G.. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 2014; 141:977–987. [DOI] [PubMed] [Google Scholar]
- 23. Hashimoto S., Boissel S., Zarhrate M., Rio M., Munnich A., Egly J.M., Colleaux L.. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science. 2011; 333:1161–1163. [DOI] [PubMed] [Google Scholar]
- 24. Trehan A., Brady J.M., Maduro V., Bone W.P., Huang Y., Golas G.A., Kane M.S., Lee P.R., Thurm A., Gropman A.L.et al.. MED23-associated intellectual disability in a non-consanguineous family. Am. J. Med. Genet. A. 2015; 167:1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hashemi-Gorji F., Fardaei M., Tabei S.M.B., Miryounesi M.. Novel mutation in the MED23 gene for intellectual disability: a case report and literature review. Clin Case Rep. 2019; 7:331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gallo F.T., Katche C., Morici J.F., Medina J.H., Weisstaub N.V.. Immediate early genes, memory and psychiatric disorders: focus on c-fos, Egr1 and arc. Front. Behav. Neurosci. 2018; 12:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bahrami S., Drabløs F.. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016; 62:37–49. [DOI] [PubMed] [Google Scholar]
- 28. Herdegen T., Leah J.D.. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res. Brain Res. Rev. 1998; 28:370–490. [DOI] [PubMed] [Google Scholar]
- 29. Tischmeyer W., Grimm R.. Activation of immediate early genes and memory formation. Cell. Mol. Life Sci. 1999; 55:564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R.. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinf. 2010; 11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 33. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao Z., Zhang Y., Yuan G.C., Orkin S.H., Waxman D.J.. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012; 13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E.. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015; 16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stevens J.L., Cantin G.T., Wang G., Shevchenko A., Shevchenko A., Berk A.J.. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002; 296:755–758. [DOI] [PubMed] [Google Scholar]
- 37. Wang G., Balamotis M.A., Stevens J.L., Yamaguchi Y., Handa H., Berk A.J.. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell. 2005; 17:683–694. [DOI] [PubMed] [Google Scholar]
- 38. Minatohara K., Akiyoshi M., Okuno H.. Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front. Mol. Neurosci. 2015; 8:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosen J.B., Fanselow M.S., Young S.L., Sitcoske M., Maren S.. Immediate-early gene expression in the amygdala following footshock stress and contextual fear conditioning. Brain Res. 1998; 796:132–142. [DOI] [PubMed] [Google Scholar]
- 40. Guzowski J.F., Lyford G.L., Stevenson G.D., Houston F.P., McGaugh J.L., Worley P.F., Barnes C.A.. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J. Neurosci. 2000; 20:3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andre K.M., Sipos E.H., Soutourina J.. Mediator roles going beyond transcription. Trends Genet. 2021; 37:224–234. [DOI] [PubMed] [Google Scholar]
- 42. Beagan J.A., Pastuzyn E.D., Fernandez L.R., Guo M.H., Feng K., Titus K.R., Chandrashekar H., Shepherd J.D., Phillips-Cremins J.E.. Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression. Nat. Neurosci. 2020; 23:707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dekker J., Mirny L.. The 3D genome as moderator of chromosomal communication. Cell. 2016; 164:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rowley M.J., Corces V.G.. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018; 19:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ibrahim D.M., Mundlos S.. Three-dimensional chromatin in disease: what holds us together and what drives us apart?. Curr. Opin. Cell Biol. 2020; 64:1–9. [DOI] [PubMed] [Google Scholar]
- 47. Kim Y., Shi Z.B., Zhang H.S., Finkelstein I.J., Yu H.T.. Human cohesin compacts DNA by loop extrusion. Science. 2019; 366:1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu K., Liu M., Li A., Donninger H., Rao M., Jiao X., Lisanti M.P., Cvekl A., Birrer M., Pestell R.G.. Cell fate determination factor DACH1 inhibits c-Jun-induced contact-independent growth. Mol. Biol. Cell. 2007; 18:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X., Oghi K.A., Zhang J., Krones A., Bush K.T., Glass C.K., Nigam S.K., Aggarwal A.K., Maas R., Rose D.W.et al.. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003; 426:247–254. [DOI] [PubMed] [Google Scholar]
- 50. Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H.et al.. High-throughput discovery of novel developmental phenotypes. Nature. 2016; 537:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Castiglioni V., Faedo A., Onorati M., Bocchi V.D., Li Z., Iennaco R., Vuono R., Bulfamante G.P., Muzio L., Martino G.et al.. Dynamic and cell-specific DACH1 expression in Human neocortical and striatal development. Cereb. Cortex. 2019; 29:2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kurusu M., Nagao T., Walldorf U., Flister S., Gehring W.J., Furukubo-Tokunaga K.. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:2140–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martini S.R., Roman G., Meuser S., Mardon G., Davis R.L.. The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development. 2000; 127:2663–2672. [DOI] [PubMed] [Google Scholar]
- 54. Ashokkumar B., Vaziri N.D., Said H.M.. Thiamin uptake by the human-derived renal epithelial (HEK-293) cells: cellular and molecular mechanisms. Am. J. Physiol. 2006; 291:F796–F805. [DOI] [PubMed] [Google Scholar]
- 55. Cusick J.K., Mustian A., Goldberg K., Reyland M.E.. RELT induces cellular death in HEK 293 epithelial cells. Cell. Immunol. 2010; 261:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shaw G., Morse S., Ararat M., Graham F.L.. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002; 16:869–871. [DOI] [PubMed] [Google Scholar]
- 57. Thomas P., Smart T.G.. HEK293 cell line: a vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods. 2005; 51:187–200. [DOI] [PubMed] [Google Scholar]
- 58. Stepanenko A.A., Dmitrenko V.V.. HEK293 in cell biology and cancer research: phenotype, karyotype, tumorigenicity, and stress-induced genome-phenotype evolution. Gene. 2015; 569:182–190. [DOI] [PubMed] [Google Scholar]
- 59. He B.J., Soderlund D.M.. Human embryonic kidney (HEK293) cells express endogenous voltage-gated sodium currents and Na(v)1.7 sodium channels. Neurosci. Lett. 2010; 469:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vetter I., Lewis R.J.. Characterization of endogenous calcium responses in neuronal cell lines. Biochem. Pharmacol. 2010; 79:908–920. [DOI] [PubMed] [Google Scholar]
- 61. Fu Z.Y., Washbourne P., Ortinski P., Vicini S.. Functional excitatory synapses in HEK293 cells expressing neuroligin and glutamate receptors. J. Neurophysiol. 2003; 90:3950–3957. [DOI] [PubMed] [Google Scholar]
- 62. Kim S., Burette A., Chung H.S., Kwon S.K., Woo J., Lee H.W., Kim K., Kim H., Weinberg R.J., Kim E.. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat. Neurosci. 2006; 9:1294–1301. [DOI] [PubMed] [Google Scholar]
- 63. Mah W., Ko J., Nam J., Han K., Chung W.S., Kim E.. Selected SALM (Synaptic Adhesion-Like Molecule) Family proteins regulate synapse formation. J. Neurosci. 2010; 30:5559–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Woo J., Kwon S.K., Choi S., Kim S., Lee J.R., Dunah A.W., Sheng M., Kim E.. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat. Neurosci. 2009; 12:428–437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data, including RNA-seq, ChIP-seq and Hi-C were deposited in GEO under the accession number GSE216401.