Abstract
In Caenorhabditis elegans, the N6-methyladenosine (m6A) modification by METT10, at the 3'-splice sites in S-adenosyl-l-methionine (SAM) synthetase (sams) precursor mRNA (pre-mRNA), inhibits sams pre-mRNA splicing, promotes alternative splicing coupled with nonsense-mediated decay of the pre-mRNAs, and thereby maintains the cellular SAM level. Here, we present structural and functional analyses of C. elegans METT10. The structure of the N-terminal methyltransferase domain of METT10 is homologous to that of human METTL16, which installs the m6A modification in the 3'-UTR hairpins of methionine adenosyltransferase (MAT2A) pre-mRNA and regulates the MAT2A pre-mRNA splicing/stability and SAM homeostasis. Our biochemical analysis suggested that C. elegans METT10 recognizes the specific structural features of RNA surrounding the 3'-splice sites of sams pre-mRNAs, and shares a similar substrate RNA recognition mechanism with human METTL16. C. elegans METT10 also possesses a previously unrecognized functional C-terminal RNA-binding domain, kinase associated 1 (KA-1), which corresponds to the vertebrate-conserved region (VCR) of human METTL16. As in human METTL16, the KA-1 domain of C. elegans METT10 facilitates the m6A modification of the 3'-splice sites of sams pre-mRNAs. These results suggest the well-conserved mechanisms for the m6A modification of substrate RNAs between Homo sapiens and C. elegans, despite their different regulation mechanisms for SAM homeostasis.
INTRODUCTION
N 6 -Methyladenosine (m6A) is an abundant modification found in eukaryotic mRNAs (1–4). It regulates mRNA splicing, translation, localization, decay, and stability (5–8), and thereby plays pivotal roles in various cellular processes encompassing cell differentiation, proliferation, development, and cell signaling (9–13).
Two m6A methyltransferase writers that install m6A in mRNAs have been described so far. Most m6A modifications in mRNAs are established by the heteromeric METTL3 and METTL14 complex (METTL3/METTL14). The METTL3/METTL14 complex introduces the m6A in the RRACH motif (modified adenosine is underlined, R = A or G, H = A, C or U), located at positions close to the stop codon or 3′-UTR of mRNAs (2,3,14–16). METTL3 is a catalytic subunit and interacts with S-adenosyl-L-methionine (SAM), while METTL14 contributes to substrate RNA binding (17,18).
Another methyltransferase-like protein, METTL16, was recently identified. Human METTL16 (hMETTL16) installs m6A into distinct target RNAs in a specific structural context (19–23). It methylates the adenosines in the UACAGAGAA (methylated adenosine is underlined) motif within the hairpins of the 3′ UTR of the methionine adenosyltransferase 2A (MAT2A) precursor mRNA (pre-mRNA). The m6A modification regulates the splicing and stability of MAT2A pre-mRNA (19,20). Under conditions with lower SAM concentrations in cells, the longer dwelling time of METTL16 on the 3'-UTR hairpins in the MAT2A pre-mRNA may promote MAT2A pre-mRNA splicing by recruiting splicing factors via the METTL16 C-terminal domain (20). Conversely, in cells with higher concentrations of SAM, the methylation of 3'-UTR hairpins in MAT2A leads to the degradation of MAT2A pre-mRNA. Thereby, METTL16 maintains the intracellular SAM homeostasis (19,20) (Supplementary Figure 1A).
METTL16 also methylates a specific adenosine (A43 in human) of U6 snRNA in the conserved motif sequence, as observed in the 3′-UTR hairpins of MAT2A pre-mRNAs (20–22). A43 lies within the highly conserved ACAGAGA (methylated adenosine is underlined) box of U6 snRNA, which base pairs with the 5′-splice sites of mRNA during pre-mRNA splicing (24–28). In yeast, mutations in the conserved sequence are lethal (29), implying that the m6A modification at this position is essential for pre-mRNA splicing regulation. Recently, the m6A modification of A37 (corresponding to A43 in human U6 snRNA) in the U6 snRNA of the fission yeast, Schizosaccharomyces pombe, was shown to regulate the splicing by facilitating interactions with 5'-splice site adenosines in a subset of S. pombe introns (30).
The homolog of hMETTL16 in Caenorhabditis elegans, METT10 (CeMETT10), reportedly regulates the SAM homeostasis by installing the m6A modification at the distal 3'-splice sites (‘AG’ dinucleotide: methylated adenine is underlined) between intron 2 and exon 3 of SAM synthetase (sams) pre-mRNAs (31,32). Under high nutrient (SAM) conditions, CeMETT10 installs m6A at the 3'-splice sites of sams pre-mRNAs, and this methylation prevents the splicing factor U2AF35 from binding to the 3'-splice site (31–33). As a result, the alternative splicing and nonsense-mediated mRNA decay (AS-NMD) of the pre-mRNAs are induced to degrade the sams pre-mRNAs, and the SAM homeostasis in cells is maintained (Supplementary Figure 1B). Analyses of the nucleotide sequences around the 3'-splice sites of sams pre-mRNAs revealed that the UACAG sequence (methylated adenosine is underlined) is conserved and the region could also form stem–loop hairpin structures similar to the 3'-UTR hairpins in the MAT2A pre-mRNA (22,23). Although the amino acid sequence of the N-terminal methyltransferase domain (MTD) of CeMETT10 is homologous to that of hMETTL16, the amino acid sequence of the C-terminal half of CeMETT10 apparently lacks this homology (20,34,35). In hMETTL16, the C-terminal half region, termed the vertebrate conserved region (VCR), is suggested to recruit splicing factors to promote the MAT2A pre-mRNA splicing (20). A more recent report found that the VCR facilitates the m6A modification of U6 snRNA and MAT2A hairpin RNAs in vitro (35). Thus, the detailed mechanism of the m6A modification of the 3'-splice sites of sams pre-mRNAs by CeMETT10 is not fully understood. In this study, we analyzed the structure and function of CeMETT10 to elucidate the mechanism of m6A modification at the 3'-splice sites of sams pre-mRNAs. Our results suggest that the mechanisms for the methylation of specific RNA substrates are shared between CeMETT10 and hMETTL16, despite their different systems for regulating SAM homeostasis.
MATERIALS AND METHODS
Plasmid constructions
The synthetic DNA encoding the full-length C. elegans METT10 (CeMETT10) gene was purchased from Eurofins Genomics Japan (Supplementary Table 1) and cloned between the NdeI and XhoI sites of the pET15SUMO vector (36), yielding pET15SUMO_CeMETT10-FL. The plasmids for the expression of CeMETT10-FLΔL with the deletion of the putative disordered region (residues 386–430) in the C-terminal half, and CeMETT10-FLΔL(RRR/7E) with mutations in the RRR-motif (arginine-rich-region: residues 360–366) to seven Glu residues in CeMETT10-FLΔL, were prepared by using a KOD-Plus mutagenesis kit (Toyobo, Japan) according to the manufacturer's instructions, yielding pET15SUMO_CeMETT10-FLΔL and pET15SUMO_CeMETT10-FLΔL(RRR/7E), respectively. The expressed proteins have an N-terminal His6-SUMO sequence. The DNA fragments encoding the N-terminal methyltransferase domain (MTD: residues 1–314) of CeMETT10 were PCR-amplified and cloned between the NdeI and XhoI sites of pET22b (Merck Millipore, Japan), yielding pET22_CeMETT10-MTD. The CeMETT10-MTD variants were expressed from the respective plasmids, and mutations were introduced into pET22_CeMETT10-MTD as described above. The expressed proteins have a C-terminal His6-tag sequence. The oligonucleotide sequences used for molecular cloning and mutations are listed in Supplementary Table 2.
Expression and purification of C.elegans METT10 and its variants
For the overexpression of the CeMETT10-FL protein or its variants, Escherichia coli BL21(DE3) was transformed by the corresponding overexpression plasmid and cultured in LB medium containing 50 μg/ml ampicillin at 37°C, until the OD660 reached 0.8. The protein expression was then induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.1 mM, and the culture was continued at 20°C for 20 h. The cells were harvested and lysed in buffer, containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 20 mM imidazole, and 5% (v/v) glycerol. CeMETT10-MTD and its variants were first purified by Ni-NTA agarose (Qiagen, Japan) chromatography, and the proteins were eluted from the column with buffer, containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 300 mM imidazole, and 5% (v/v) glycerol. The proteins were further purified on a HiTrap Heparin column (GE Healthcare, Japan), and finally separated by chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare, Japan), in buffer containing 20 mM Tris–HCl, pH 7.0, 200 mM NaCl, and 10 mM β-mercaptoethanol. For the purification of CeMETT10-FLΔL and its variants, His6-SUMO-tagged proteins were purified with the Ni-NTA agarose column as described above. The His6-SUMO tags were then cleaved by ULP1 (ubiquitin-like-specific protease 1) during dialysis against buffer, containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, and 5% (v/v) glycerol, at 4°C overnight. The ULP1-treated proteins were then passed through the Ni-NTA agarose column again. The proteins were further purified by chromatography on the HiTrap Heparin and HiLoad 16/60 Superdex 200 columns, as described above. The purified proteins were concentrated and stored at –80°C until use.
Crystallization and structural determination of CeMETT10-MTD
CeMETT10-MTD crystals were obtained by the sitting drop vapor diffusion method at 20°C. A 0.2 μl portion of the purified CeMETT10-MTD (5 mg/ml) was mixed with 0.2 μl of reservoir solution, containing 0.2 M ammonium acetate, 0.1 M Tris–HCl, pH 8.5, and 25% (w/v) PEG3350, and incubated at 20°C. Crystals appeared in three days and continued growing for up to two weeks. The crystals were cryo-protected with reservoir solution supplemented with 25% (v/v) ethylene glycol, and flash-cooled in liquid nitrogen. X-ray diffraction data were collected at Beamline 1A at the Photon Factory (KEK, Japan). The diffraction data were indexed, integrated, and scaled with XDS (37). The initial phase of the METT10-MTD structure was determined by molecular replacement with Phaser (38), using the crystal structure of human METTL16-MTD (PDB: 6B91) (34) as the search model. The CeMETT10-MTD structure was further refined by Phenix.refine (39) and manually modified with Coot (40). The final structural model was assessed with the MolProbity program (41).
Preparation of RNAs
Synthetic sams hairpin (sams-hp) RNA and its variants were purchased from Fasmac, Japan. The sams hp-ls RNA and C. elegans U6 snRNA were synthesized by in vitro transcription with T7 RNA polymerase, using plasmids encoding the respective DNA sequence downstream of the T7 promoter as templates (Supplementary Table 1). The synthesized RNAs were purified by 10% (w/v) polyacrylamide gel electrophoresis under denaturing conditions. The nucleotide sequences of the RNAs used in this study are listed in Supplementary Table 3.
In vitro methylation assay
In vitro methylation assays were conducted using the MTase-Glo system (Promega, Japan), as previously described (35) with slight modifications. The reaction mixture (15 μl), containing 0.4 μM CeMETT10-FLΔL (-MTD or its variants), 50 mM HEPES–KOH, pH 8.0, 120 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM SAM (Cayman Chemical, USA), 1× MTase-Glo Reagent, and a series of different concentrations of RNA samples (0.25–8.0 μM), was incubated at 37°C for 4 min. Afterwards, 15 μl of MTase-Glo Detection Solution was added, and the reaction was incubated at room temperature for 30 min. The luminescence was quantified by the GloMax-Multi Detection System (Promega, Japan). The luminescence value of the mixture without the RNA substrate was used as the background and subtracted. One picomole of SAH was estimated to be equivalent to 190 000 LU (light units), according to the standard quantification curve. For the SAM titration experiments, the RNA substrate concentration was fixed at 20 μM, and the SAM concentrations ranged from 0.05 to 1.5 mM. Since the reaction proceeds in a linear range up to 8 min, the luminescence data obtained at the 4 min reaction time point were used to calculate the initial velocities of the reactions.
Gel-shift assays
RNAs (sams-hp, sams-hp-ls, and U6 snRNA) were 5'-32P-labeled by T4 polynucleotide kinase (NEB, Japan) using γ-32P-ATP (3000 Ci/mmol; PerkinElmer, Japan), and the 32P-labeled RNAs were gel-purified. The 32P-labeled RNAs (10 000 cpm) were mixed with various concentrations of CeMETT10-FLΔL, -MTD or their variants in 10 μl of solution, containing 50 mM Tris–Cl, pH 8.0, 120 mM NaCl, 10 mM MgCl2, 5 mM β-mercaptoethanol, 10% (v/v) glycerol and 2.5 μg E. coli tRNA mixture. The solution was incubated at room temperature for 15 min and then placed on ice for 10 min. Loading buffer (2 μl) containing 0.02% (w/v) xylene cyanol and 50% (v/v) glycerol was added, and the RNAs were separated by 6% (w/v) polyacrylamide gel electrophoresis under native conditions at 4°C. The 32P-labeled RNA bands in the gel were visualized and quantified by a BAS-5000 image analyzer (Fujifilm, Japan).
RESULTS
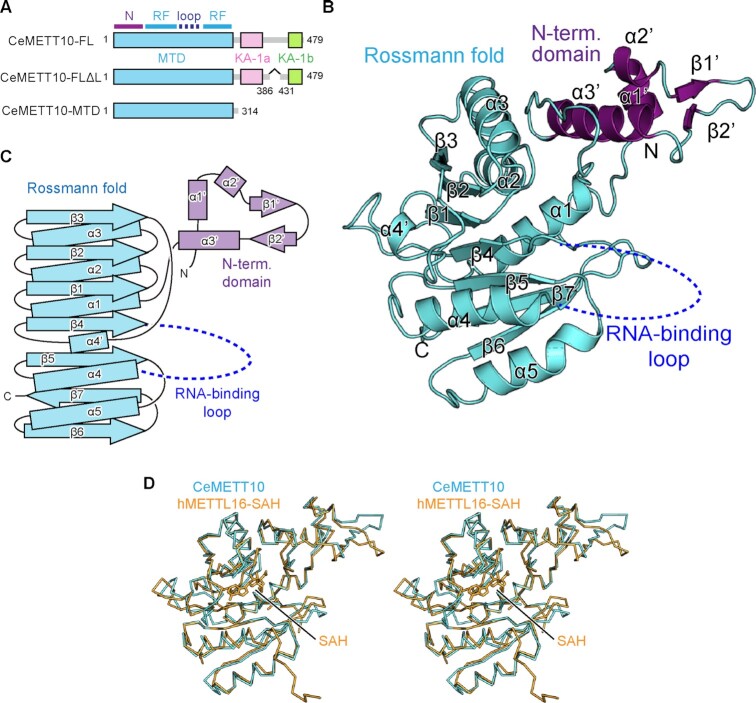
Overall structure of C. elegans METT10-MTD.
To elucidate the mechanism of m6A modification of RNAs by C. elegans METT10 (CeMETT10) (31,32), we tried to obtain crystals of full-length CeMETT10 (CeMETT10-FL) (Figure 1A). However, the expression level of soluble CeMETT10-FL in E. coli was low, and we could not obtain sufficient amounts of highly purified proteins. As described below, we noticed that CeMETT10 has the kinase associated 1 (KA-1) domain in the C-terminal half, and there is a putative disordered loop region in the middle of KA-1, as in human METTL16 (hMETTL16) (35). Thus, we deleted this loop region (residues 386–430 between KA-1a and KA-1b) (Figure 1A), yielding CeMETT10-FLΔL. The CeMETT10-FLΔL protein was expressed well in E. coli and purified. We also expressed the N-terminal methyltransferase domain (MTD: residues 1–314) of CeMETT10 (CeMETT10-MTD) (Figure 1A). As described below, both CeMETT10-FLΔL and CeMETT10-MTD can methylate the hairpin RNA, sams-hp, derived from the nucleotide sequence around the 3'-splice sites of SAM synthetase (sams) pre-mRNAs, and U6 snRNA in vitro.
Figure 1.
Overall structure of C. elegans METT10-MTD. (A) Schematic diagrams of full-length C. elegans METT10 (CeMETT10-FL), its variant CeMETT10-FLΔL lacking the putative disordered region (residues 386–430), and its N-terminal methyltransferase domain (CeMETT10-MTD: residues 1–314). The MTD is colored cyan, and the C-terminal kinase-associated domains, KA-1a and KA-1b, are magenta and green, respectively. The region between KA-1a and KA-1b is predicted to be disordered. The MTD consists of the N-terminal domain (N) and Rossmann fold (RF). (B) Overall structure of CeMETT10-MTD. Residues 6–187 and 236–309 are modeled in the structure. The extended N-terminal domain and the Rossmann fold are colored magenta and cyan, respectively, and the RNA binding loop (dashed line) between β4 and α4 was disordered and not modeled in the structure. (C) Schematic view of the secondary structure of CeMETT10-MTD. The N-terminal domain and the Rossmann fold are colored as in (B). (D) Stereo view of the structural alignment of Cα atoms between CeMETT10-MTD (cyan) and human METTL16-MTD (hMETTL16-MTD) in complex with SAH (orange, PDB: 6B92) (34).
The crystallization trial of CeMETT10-FLΔL was not successful. However, we obtained diffractive crystals for CeMETT10-MTD. The CeMETT10-MTD crystal belongs to the space group P212121, and one molecule of CeMETT10-MTD is contained in the asymmetric unit cell. The initial phase was determined by the molecular replacement method (38), using the structure of the MTD of hMETTL16 (PDB: 6B91) (34) as the search model. Finally, the structure was model-built and refined to an Rwork factor of 24.1% (Rfree of 29.8%) to 3.0 Å resolution (Figure 1B). The final model for CeMETT10-MTD includes residues 6–187 and 236–309. The terminal and internal regions (residues 1–5, 188–235 and 310–314) could not be modeled due to insufficiently clear electron densities. The statistical details for the data collection and refinement are provided in Table 1, and a representative image of the electron density map is shown in Supplementary Figure 2.
Table 1.
Data collection and refinement statistics
| Data collection | METT10-MTD |
|---|---|
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 43.21, 70.08, 94.23 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 1.0000 |
| Resolution (Å)* | 50–3.0 (3.118–3.0) |
| R sym* | 0.113 (1.242) |
| I / σI* | 18 (1.8) |
| CC 1/2* | 0.999 (0.765) |
| Completeness (%)* | 99.9 (99.5) |
| Redundancy* | 12.5 (12.7) |
| Refinement | |
| Resolution (Å) | 50–3.0 |
| No. reflections | 6053 |
| R work/Rfree (%) | 24.11/29.75 |
| No. atoms | |
| Protein | 2039 |
| B-factors (Å2) | |
| Protein | 103.74 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.64 |
*Values in parentheses are for the highest-resolution shell.
The CeMETT10-MTD adopts a classical Rossmann fold with the extended N-terminal domain (Figure 1B, C). The core Rossmann fold consists of seven β-strands (β1–β7) and six α-helices (α1–α5 and α4'), and SAM would interact with the topological switch in the center of the Rossmann fold. The extended N-terminal domain consists of three α-helices (α1'–α3') and two β-strands (β1' and β2'). In the present structure, the region between β4 and α4 (residues 188–235) in the Rossmann fold is disordered.
The structures of the CeMETT10-MTD and the MTD of hMETTL16 (hMETL16-MTD) (PDB: 6B92) (22,34) are homologous, with a root mean square deviation (RMSD) of 1.04 Å for 198 Cα atoms (Figure 1D). This structural homology is expected from the highly homologous amino acid sequences between CeMETT10-MTD and hMETL16-MTD [38% identity between CeMETT10-MTD (residues 1–314) and hMETTL16-MTD (residues 1–306)] (Supplementary Figure 3).
Substrate recognition by CeMETT10-MTD.
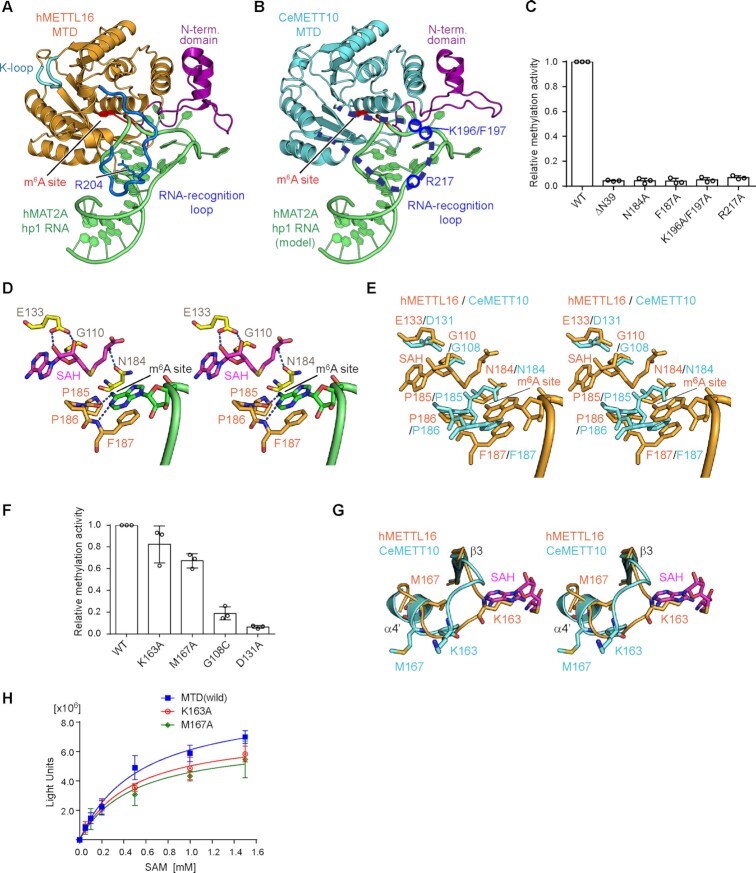
The crystal structure of hMETTL16-MTD in complex with the 3'-UTR hairpin of methionine adenosyltransferase 2A (MAT2A) mRNA (hMAT2A-hp1) was recently reported (PDB: 6DU4) (23). In the structure, the extended N-terminal domain and the loop between β4 and α4 in the Rossmann fold interact with the RNA substrate and clamp the hairpin-structured RNA from various angles (Figure 2A). The structure of CeMETT10-MTD was superimposed on that of hMETTL16-MTD complexed with hMAT2A-hp1 (Figure 2B) (23). In the superimposition, the extended N-terminal domain of CeMETT10-MTD is proximal to the hMAT2A-hp RNA and thus expected to interact with the RNA substrate. The loop between β4 and α4, which was disordered in the apo CeMETT10-MTD structure (Figure 1B), is also expected to interact with the substrate RNA, as observed in the structure of hMETTL16-MTD complexed with hMAT2A-hp1 (23).
Figure 2.
Key residues of CeMETT10- MTD for RNA methylation. (A) The structure of human METTL16-MTD (hMETTL16-MTD) in complex with hMAT2A- hp1 RNA (PDB: 6DU4) (23). hMAT2A-hp1 is shown in green. (B) RNA docking onto the CeMETT10-MTD structure. The structure of hMETTL16-MTD complexed with hMAT2A-hp1 in (A) was superimposed on the structure of CeMETT10-MTD. hMETTL16-MTD was omitted for clarity. hMAT2A-hp1 is colored green. (C) Methylation of sams-hp RNA (see Figure 3B) by METT10-MTD and its variants. The sams-hp RNA (1 μM) was incubated with 0.4 μM METT10-MTD in the presence of 1 mM SAM, at 37°C for 4 min. The methylation of sams-hp RNA by wild-type METT10-MTD was taken as 1.0. The bars in the graphs are the SD of three independent experiments. (D) Model of the hMETTL16 active site with SAH and hMAT2A-hp1. The SAH was modeled into the structure of hMETTL16-MTD in complex with hMAT2A-hp1 by referring to the structure of hMETTL16-MTD in complex with SAH (PDB: 6B92) (34). The methylation site in the RNA (m6A site) is shown in green, and SAH is colored magenta. (E) Superimposition of the active site structures of CeMETT10 (cyan) and hMETTL16 (orange). (F) Methylation of sams-hp RNA by METT10-MTD and its variants, as in (C). The methylation of sams-hp RNA by wild-type METT10-MTD was taken as 1.0. The bars in the graphs are the SD of three independent experiments. (G) Superimposition of the K-loops of CeMETT10-MTD (cyan) and hMETTL16-MTD (orange) in complex with hMAT2A-hp1. SAH was modeled as in (D). (H) Methylation reactions of sams-hp-ls RNA by METT10-MTD and its variants with various concentrations of SAM (0.05, 0.1, 0.2, 0.5, 1.0 and 1.5 mM). The sams-hp RNA (20 μM) was incubated with 0.4 μM METT10-FLΔL or its variants in the presence of various concentrations of SAM, at 37°C for 4 min. The bars in the graphs are the SD of three independent experiments.
The amino acid sequence and the structure of the extended N-terminal region of CeMETT10-MTD are homologous to those of hMETTL16-MTD (Figure 1D, Supplementary Figure 3). The deletion of the N-terminal 39 amino acid residues (ΔN39) from CeMETT10-MTD decreased the methylation of sams-hairpin (sams-hp) RNA, containing the 3'-splice sites of sams pre-mRNAs, in vitro (Figure 2C), as in hMETTL16-MTD (22). The disordered region between β4 and α4 in the CeMETT10-MTD structure has some additional amino acid insertions, and thus is longer than the corresponding loops in hMETTL16 and other vertebrate METTL16 homologs (Supplementary Figure 3). The mutation of Arg217, corresponding to the Arg204 residue in hMETTL16 that hydrogen-bonds with the RNA as described below, to Ala in CeMETT10-MTD decreased the methylation of sams-hp in vitro (Figure 2C). In addition, the mutations of Lys196 and Phe197, conserved only in worm METT10 homologs (Supplementary Figure 4), to Ala (K196A/F197A) also decreased the methylation of sams-hp in vitro. Thus, additional and/or specific interactions may be present between the loop connecting β4 and α4 of CeMETT10-MTD and sams-hp RNA. In hMETTL16, Arg200 in the corresponding loop interacts with MAT2A-hp, and the Arg200Glu mutation increased the methylation of MAT2A-hp (23). The corresponding amino acid in CeMETT10 is Ala213 (Supplementary Figure 3). Thus, as described below, Ala213 in CeMETT10 is expected to allow the sams-hp to adopt the productive structure for methylation.
To further evaluate the involvement of the extended N-terminal region and the loop between β4 and α4 in RNA interactions, gel-shift assays were performed using CeMETT10-MTD and its variants. In these gel shift experiments, distinct shifted bands were not clearly observed, and the shifted bands were smeared in the gels (Supplementary Figure 5A). This would be due to the instability of the CeMETT10-MTD -sams-hp complex under the gel-shift conditions and/or the weaker interactions between CeMETT10MTD and sams-hp. Therefore, the affinities of CeMETT10-MTD and its variants for sams-hp could not be reliably quantified. Nevertheless, the results showed that the extended N-terminal deletion (ΔN39), and the R217A and K196A/F197A mutations in the CeMETT10-MTD, all decreased the affinities for sams-hp RNA (Supplementary Figure 5A). Thus, the loop between β4 and α4 of CeMETT10-MTD and the N-terminal extended domain would interact with RNA substrates.
The structures of hMETTL16-MTD complexed with S-adenosyl-l-homocysteine (SAH) (PDB: 6B92) (34) or hMAT2A-hp1 RNA (PDB: 6DU4) (23) revealed that the adenine base to be methylated (m6A site) in the RNA substrate is captured in the hydrophobic pocket near the Asn-Pro-Pro-Phe motif (residues 184–187, NPPF) (23). The adenine base stacks with Phe187 and forms hydrogen-bonds with the main chain of Pro185-Pro186-Phe187 (Figure 2D). Gly110, Glu133 and Asn184 interact with SAH in hMETTL16 (Figure 2D). The superimposition of the catalytic pockets of CeMETT10-MTD and hMETTL16-MTD showed that these key residues superimposed well (Figure 2E, Supplementary Figure 3). The NPPF motif is conserved in CeMETT10, and Phe187, Gly110, Glu133 and Asn184 in hMETTL16 correspond to Phe187, Gly108, Asp131 and Asn184 in CeMETT10, respectively (Figure 2E). The Phe187Ala, Asn184Ala, Asp131Ala and Gly108Cys mutations in CeMETT10-MTD all decreased the methylation of sams-hp RNA in vitro (Figure 2C, F). Thus, the mechanisms used to recognize SAM and the adenine base of the m6A site in the RNA substrate are shared between CeMETT10-MTD and hMETTL16-MTD.
In hMETTL16, Lys163 and Met167 in the loop between β3 and α4', termed the K-loop (Figure 2A, G), were suggested to regulate the affinity of SAM for the catalytic pocket (23). In the structure of the hMETTL16-MTD complex with hMAT2A-hp1, the side chains of Lys163 and Met167 in the K-loop can occlude the SAM binding site through extensive intramolecular hydrophobic interactions, and the mutations of Lys163Ala and Met167Ala reportedly enhanced the methylation of hMAT2A-hp RNA in vitro and in vivo (23). In contrast, the corresponding region between β3 and α4' in CeMETT10-MTD has an insertion of a few additional amino acid residues, as compared to this region in hMETTL16 (Supplementary Figure 3). The mutations of Lys163Ala and Met167Ala in this region of CeMETT10-MTD did not enhance the methylation of sams-hp RNA in vitro, but rather slightly decreased the methylation of sams-hp RNA (1 μM) in the presence of 1 mM SAM (Figure 2F). To further examine the effects of the mutations on the methylation of sams-hp RNA, a saturating amount of sams-hp RNA (20 μM) was used for the assays over the range of SAM concentrations (50 μM–1.5 mM). The results showed that the mutations did not affect the methylation of sams-hp RNA significantly over the range of tested SAM concentrations (Figure 2H), and the Km values of wild-type CeMETT10-MTD, Lys163Ala and Met167Ala for SAM were estimated to be 551, 480, and 491 μM, respectively. Thus, the mutations did not cause any significant changes in the affinities for SAM. Since the Km value of CeMETT10-MTD is larger than that of hMETTL16-MTD for SAM (∼182 μM) (35), the effects of the Lys163Ala and Met167Ala mutations in CeMETT10 might not be detectable under the assay conditions, and the SAM-dependent response of the methylation of sams pre-mRNA by CeMETT10 might not be as prominent, in comparison with that of hMETTL16 (23). Notably, the Lys163 and Met167 residues of CeMETT10 are not conserved among the METT10 homologs from worms (Supplementary Figure 4); thus, the loop between β3 and α4' in CeMETT10 might tightly occlude the SAM binding site in a different manner from that observed in hMETTL16 complexed with hMAT2A-hp1.
Elements in RNA substrate for methylation by CeMETT10-MTD.
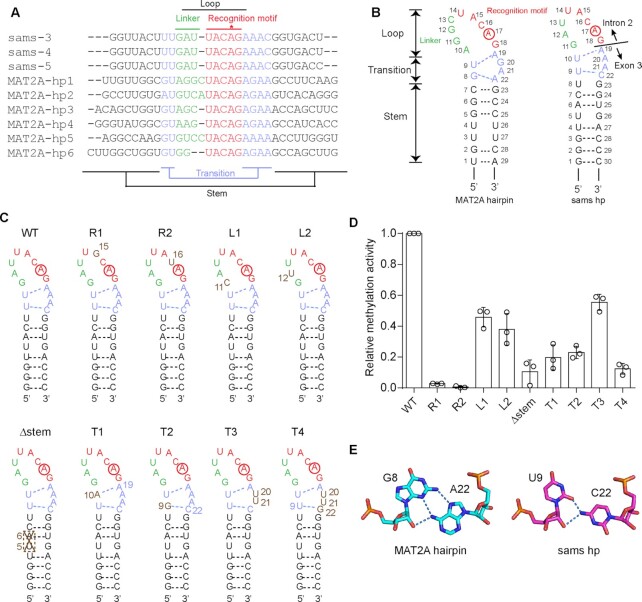
The comparison of the sequences of MAT2A pre-mRNA 3'-UTR hairpins with the sequences around the 3'-splice sites (AG-dinucleotides between intron 2 and exon 3) of sams pre-mRNAs revealed the conservation of the UACAG sequence (methylated adenosine is underlined) (Figure 3A) (31,32). The nucleotide sequences around the 3'-splice sites of sams pre-mRNAs suggested that the region could also form stem–loop hairpin structures similar to the MAT2A 3'-UTR hairpins (22,23) (Figure 3A, B).
Figure 3.
RNA recognition by C. elegans METT10. (A) Alignments of the nucleotide sequences around the junction between intron 2 and exon 3 of C. elegans sams (-3, -4 and -5) pre-mRNAs and the 3'-UTR hairpin regions of human MAT2A (MAT2A-hp1–6) pre-mRNAs (31). (B) Secondary structures of 3'-UTR hairpin RNA (MAT2A-hp1) of human MAT2A pre-mRNA (left) and C. elegans sams hairpin RNA spanning intron 2 and exon 3, including the 3'-splice site (AG-dinucleotides) (right). The sams RNA also adopts a hairpin structure (sams-hp), as in human MAT2A-hp1. The adenine residues targeted for methylation are circled. The hairpin comprises the loop, transition, and stem regions (23). The loop region is composed of the recognition motif (UACAG: red) and linker (green). (C) Secondary structures of C. elegans sams-hp RNA and its variants used for the methylation assays in (D). (D) Methylation of sams-hp RNA and its variants by METT10-FLΔL. The sams-hp RNA (1 μM) and its variants were each incubated with 0.4 μM METT10-FLΔL in the presence of 1 mM SAM, at 37°C for 4 min. The methylation of wild-type sams-hp RNA by METT10-FLΔL was taken as 1.0. The bars in the graph are the SD of three independent experiments. (E) G8–A22 base pairing in the transition region near the stem of hMAT2A-hp (left) and the possible U9–C22 base pairing at the corresponding position in the sams-hp (right). Possible hydrogen bonds between the sugar edge of U9 and the 4-NH2 group of C22 are depicted by dashed lines.
MAT2A-hp bound to hMETTL16-MTD has structural conformation elements consisting of the loop (U14A15C16A17G18 recognition motif and linker), transition, and stem, which are required for productive catalysis (23). Considering the structural similarity between hMETTL16-MTD and CeMETT10-MTD, the nucleotide sequences around the 3'-splice sites of sams pre-mRNAs could also adopt structures analogous to those of MAT2A-hp1 for productive catalysis (Figure 3B).
The structure of hMETTL16-MTD complexed with MAT2A 3'-UTR hairpins (MAT2A-hp1) suggested that the conserved UACAG motif in the MAT2A-hp loop is required to form a productive structure for methylation by hMETTL16 (23), in which the adenine base is captured in the hydrophobic pocket near the catalytic site (Figure 2D, E). In the structure of hMETTL16 complexed with MAT2A-hp, Phe20 and Phe46 in the N-terminal region stack with the uracil (U14) and adenine (A15) bases of the conserved U14A15C16A17G18 motif in the recognition loop of hMAT2A-hp1, respectively (Supplementary Figure 6A, B) (34). These two amino acid residues are conserved as Phe20 and Phe46 in CeMETT10 (Supplementary Figures 3, 6B). In hMETTL16, the 4-NH2 group of C16 forms a hydrogen bond with the phosphate backbone of A15 and stacks with the adenine base of A15 (Supplementary Figure 6A), and G18 stacks with the U9-A19 base-pair (Supplementary Figure 6C, D). As a result, the adenine of A17 (m6A site) is captured in the hydrophobic pocket near the NPPF motif.
To determine whether CeMETT10 methylates RNA containing the UACAG motif in a specific structural context, we prepared sams-hairpin (sams-hp) RNA variants (Figure 3C) and tested their methylation by CeMETT10-FLΔL in vitro (Figure 3D). The mutation of nucleotides in the conserved U14A15C16A17G18 recognition motif in sams-hp (R1: A15G and R2: C16U) decreased the methylation of the RNAs (Figure 3D), suggesting the requirement of the UACAG motif in the loop for methylation by CeMETT10, as with hMETTL16.
The mutations in the linker of the loop (L1: G11C and L2: A12U) moderately decreased the RNA methylation. In the structures of hMETTL16 complexed with MAT2A-hp, the linker region does not interact with hMETTL16-MTD (Supplementary Figure 7A–C) (23). Furthermore, the sequences and numbers of the nucleotides in the linker differ (Figure 3A). Thus, the effects of the mutations would be moderate. The stem-disrupting mutation (Δstem: deletion of U5A6) decreased the methylation of the RNA (Figure 3D). The partial deletion of the stem could disrupt the proper folding of the entire stem–loop structure required for productive catalysis.
The structure of the transition region of MAT2A-hp in the complex with hMETTL16-MTD showed that the U9-A19 pair proximal to the loop is a reverse Watson-Crick base pair, and Arg204 forms hydrogen bonds with the uridine (U9) (Figure 3B). The G8–A22 pair near the stem is formed between the sugar-edge of G8 and the Hoogsteen-edge of A22, and Arg200 forms hydrogen bonds with the Hoogsteen edge of G8 (23). G20 and A21, which are between the two base pairs, interact via a water molecule (Supplementary Figure 7D–F).
Mutations of the corresponding base pair in the transition region of sams-hp also decreased the methylation of the RNAs by CeMETT10-MTD (Figure 3D). The mutation of U10–A19 (corresponding to U9-A19 in MAT2A-hp1) in the transition region to A10-A19 (T1) decreased the methylation of the RNAs, and the mutation of Arg217, corresponding to Arg204 which interacts with U9 in hMETTL16, negatively affected the methylation of sams-hp (Figure 2C). The mutation of U9–C22 to G9–C22 (T2) also decreased the methylation of the RNAs. U9–C22 in the transition region of sams-hp could also form a similar interaction to G8–A22 in MAT2A-hp through the sugar-edge of U9 and the 4-NH2 group of C22 (Figure 3E), and allow the sams-hp to adopt the productive structure for efficient methylation by CeMETT10. Thus, the U9–C22 to G9–C22 (T2) mutation would disrupt the productive structure for the methylation of the substrate RNA. Arg200, which interacts with G8 in the G8–A22 pair near the stem of the hMETTL16-MAT2A-hp structure, is replaced with Ala (Ala213) in CeMETT10 (Supplementary Figure 3). Therefore, Ala213 in CeMETT10 might not directly interact with U9 in the U9–C22 of sams-hp. As described above, in CeMETT10, additional and/or specific interactions may be present between the loop connecting β4 and α4 of CeMETT10-MTD and sams-hp RNA (Figure 2B, C).
The mutation of A20A21 to U20U21 (T3) moderately decreased the methylation of the RNA (Figure 3C, D). The A20 and A21 in the transition region of the sams-hp could adopt a structure similar to the G20A21 of MAT2A-hp, and the two nucleotides between the two base pairs (U10–A19 and U9–C22) in the transition region could impact the methylation of RNA by CeMETT10. In MAT2A-hp1, the substitution of G20 to A20 reportedly did not affect the affinity of the RNA toward hMETTL16, but increased the RNA methylation (23). Thus, the presence of A20 in the sams-hp contributes to the formation of the productive structure for efficient methylation by CeMETT10. Altogether, these results suggest that CeMETT10 and hMETTL16 recognize the same structural features in the RNA substrate for productive catalysis.
The presence of KA-1 in the C-terminal half of CeMETT10.
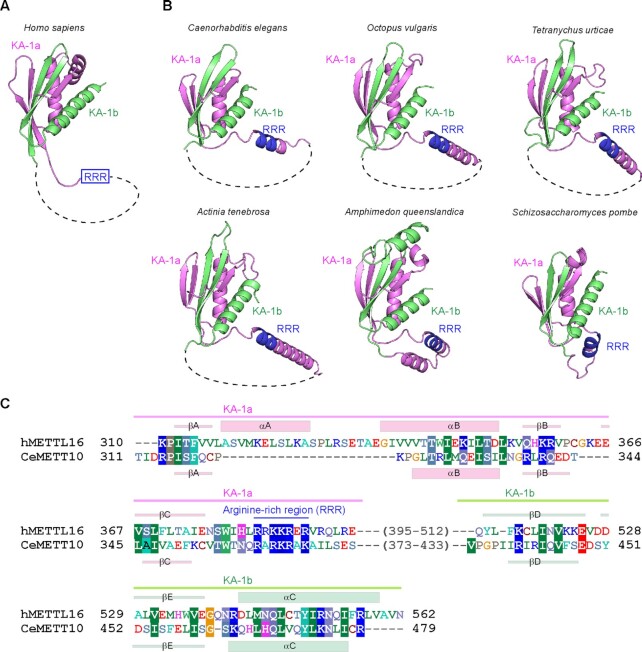
METTL16 proteins from vertebrates possess the distinct C-terminal domain, which was originally termed the vertebrate conserved region (VCR) (19–21). Recent structural and functional studies of the hMETTL16 VCR showed that its structure is homologous to the kinase associated 1 (KA-1) domain structure found in U6-specific terminal uridyltransferase (TUT1) (Figure 4A) (35,42,43). The VCR in hMETTL16 acts as an RNA binding domain to facilitate the methylation of MAT2A-hp RNA and U6 snRNA, through binding to the double-stranded regions of the RNAs (35).
Figure 4.
Predicted structure of the C-terminal region of CeMETT10. (A) Structure of the C-terminal KA-1 domain of human METTL16 (PDB: 6M1U) (35). The disordered loop (dashed line) is inserted between KA-1a (magenta) and KA-1b (green). (B) Model structures of the C-terminal regions of CeMETT10 and hMETTL16 homologs from invertebrates [Caenorhabditis elegans (MET16_CAEEL), Octopus vulgaris (A0A6P7TIF1_OCTVU), Tetranychus urticae (T1JVZ0_TETUR), Actinia tenebrosa (A0A6P8ITS0_ACTTE), and Amphimedon queenslandica (A0A1 × 7U5N0_AMPQE)], and a fission yeast [Schizosaccharomyces pombe (MTL16_SCHPO)] generated by Alphafold2 (44,45). The disordered region between KA-1a (magenta) and KA-1b (green), predicted by Alphafold2 in each structure, is depicted by dashed lines (Supplementary Figures 2, 6). The arginine-rich region, RRR, is colored blue. (C) Alignments of the amino acid sequences of the C-terminal regions of hMETTL16 and CeMETT10. The secondary structural elements (α-helices and β-strands) for hMETTL16 and CeMETT10 are shown in parallel above and below the alignment, respectively.
VCRs were previously considered to be present only in vertebrate METTL16 homologs. The amino acid sequence identity of the C-terminal halves of CeMETT10 and hMETL16 is only 6.9% [CeMETT10 (residues 315–479) and hMETTL16 (307–562)]. However, our careful sequence alignments of the C-terminal regions of invertebrate and fission yeast METTL16 homologs suggested the presence of a domain similar to KA-1 in their C-terminal regions (Supplementary Figures 3 and 4).
Furthermore, the structure prediction by Alphafold2 (44,45) revealed that the C-terminal regions of METTL16 homologs from invertebrates, including C. elegans and S. pombe, adopt similar structures to that of KA-1 in hMETTL16 (PDB: 6M1U) (35) (Figure 4B, C). The flexible loops are inserted in the middle of KA-1, but the N-terminal KA-1a and the C-terminal KA-1b together compose a single domain (Figure 4B, Supplementary Figure 8). Structural alignment of the KA-1 domains of hMETTL16 and CeMETT10 suggested the presence of a basic region, termed RRR (arginine-rich region: RRR), between KA-1a and KA-1b of CeMETT10 (Figure 4C). The RRR in hMETTL16 is involved in RNA binding and facilitates the methylation of MAT2A hp and U6 snRNA (35). Furthermore, the basic regions are also present in other METTL16 homologs from invertebrates and fission yeast, and they align with the RRR of hMETTL16 (Figure 5A). These observations suggest that the C-terminal regions of invertebrate METTL-16 homologs could be functional domains that facilitate the methylation of RNAs.
Figure 5.
The C-terminal KA-1 of CeMETT10 facilitates m6A modification of RNAs. (A) Alignment of amino acid sequences around the arginine-rich region (RRR) in the C-terminal KA-1 domains from various organisms (Supplementary Figures 2 and 3), including invertebrates (C. elegans, O. vulgaris, T. urticae, A. queenslandica and A. tenebosa), a fission yeast (S. pombe), and vertebrates (H. sapiens, G. gallus, C. picta, X. laevis and D. rerio). (B) Schematic diagram of CeMETT10-FL and its variants used in the methylation assays in (D) and (E). MTD, KA-1a and KA-1b are colored cyan, magenta, and green, respectively. RRR (arginine-rich region: RARKRAK) is colored blue, and its mutant is red (RRR/7E). (C) Secondary structures of sams-hp-ls and U6 snRNA. (D, E) Steady-state kinetics of the methylation of sams-hp-ls (D) and U6 snRNA (E) by METT10-FLΔL, -MTD, and -FLΔL_RRR/7E. Various concentrations of RNA [0–8 μM for sams-hp-ls (D) and 1–4 μM for U6 snRNA (E)] were incubated with 0.4 μM METT10- FLΔL, -MTD or -FLΔL _RRR/7E in the presence of 1 mM SAM, at 37°C for 4 min, and the initial reaction velocities were calculated. The bars in the graph are the SD of three independent experiments. (F) Gel-shifts of sams-hp-ls by wild-type METT10-FLΔL (0–1.0 μM), -MTD (0–100 μM) or -FLΔL _RRR/7E (0–20 μM). The graphs on the left in (F) and (G) are magnified views of the graphs on the right at the lower protein concentration ranges. (G) Gel-shifts of U6 snRNA by wild-type METT10-FLΔL (0–1.0 μM), -MTD (0–100 μM) or -FLΔL _RRR/7E (0–20 μM). The bars in the graphs (F, G) are the SD of three independent experiments.
To examine the function of the KA-1 of CeMETT10, the methylations of the sams hairpin with a longer stem (sams-hp-ls) and C. elegans U6 snRNA by CeMETT10 variants were tested (Figure 5B, C). The steady-state kinetics of RNA methylation by CeMETT10 and its variants showed that KA-1 facilitates the methylation (Figure 5D, E) of both the sams-hp-ls and U6 snRNA substrates. The estimated Km value of FLΔL for sams-hp-ls is 1.82 μM, while that of MTD for sams-hp-ls is >>8 μM, since the initial velocity of the reaction did not reach a plateau at 8 μM of substrate RNA (Figure 5D). Thus, the Km value was elevated by at least several-fold by deleting KA-1. The Km value of FLΔL_RRR/7E, in which RRR (residues: RARKRAK) was mutated to seven Glu residues, for sams-hp-ls is also >>8 μM. These results suggest that the KA-1 of CeMETT10 facilitates the methylation of sams-hp-ls by increasing the affinity for the substrate RNA through the RRR. The Km value of FLΔL for U6 snRNA is estimated to be 0.25 μM. However, the Km value of the MTD for U6 snRNA is estimated to >>4 μM, since the initial velocity of the reaction did not reach the plateau at 4 μM of substrate RNA. Thus, the Km value of U6 snRNA increased by at least sixteen-fold by deleting KA-1 (Figure 5E). The Km value of FLΔL_RRR/7E for U6 snRNA also increased and was estimated to be >>4 μM.
To further evaluate the involvement of the C-terminal KA-1 and the RRR region of KA-1 in RNA interactions, gel-shift assays were performed. While the Kd value of FLΔL for sams-hp-ls was estimated to be 0.14 μM, those of MTD and FLΔL_RRR/7E for sams-hp-ls were 22.3 and 8.3 μM, respectively. The estimated Kd value of FLΔL for U6 snRNA was 0.27 μM, and those of MTD and FLΔL_RRR/7E for U6 snRNA were 11.8 and 2.4 μM, respectively. Thus, the deletion of the C-terminal KA-1 domain and the mutation of RRR in KA-1 decreased the affinity of CeMETT10 for RNAs (Figure 5F, G, Supplementary Figure 5B, C).
DISCUSSION
In this study, we performed functional and structural analyses of C. elegans METT10 (CeMETT10), which installs m6A at the 3'-splice sites (‘AG’ dinucleotide) of sams pre-mRNAs and regulates the SAM homeostasis in cells (31,32). Human METTL16 (hMETTL16) installs m6A on specific adenines in the 3'-UTR of the MAT2A pre-mRNA, and regulates the SAM homeostasis (19–21). Despite their distinct mechanisms for the regulation of SAM homeostasis (Supplementary Figure 1), CeMETT10-MTD and hMETTL16-MTD are structurally quite homologous (Figure 1D) (22,23,34) and share the same mechanism of RNA substrate recognition for productive catalysis (Figure 3). These results are consistent with recent data showing that the sams-3 mRNA splicing at the 3'-splice site is inhibited and the alternative splicing is promoted, when the C. elegans sams-3 transgene reporter is introduced into human cells (32). CeMETT10 also possesses a functional domain that is structurally and functionally equivalent to the C-terminal KA-1 domain of hMETTL16. The KA-1 domain facilitates the efficient methylation by increasing the affinity for the RNAs (Figures 4, 5A, Supplementary Figure 8) (35).
The Km values of CeMETT10 and hMETTL16 for SAM are approximately 10- to 100-fold larger than those of other Rossman-fold methyltransferases that use SAM as a methyl donor (46–49). These values explain the biological functions of CeMETT10 and hMETTL16 in SAM biogenesis. CeMETT10 and hMETTL16 negatively regulate the expression of SAM synthetase by only methylating RNAs when the intracellular SAM levels are high. A comparison of the SAM (SAH) binding sites of CeMETT10 (hMETTL16) with those of other Rossmann-fold type methyltransferases (49–53) revealed the diversity of the sequences corresponding to the K-loop of hMETTL16 (Supplementary Figure 9). The corresponding loops in CeMETT10 and hMETTL16 might be tightly anchored to the catalytic core pocket and occlude the SAM binding site, and this may explain the higher Km values of CeMETT10 and hMETTL16 than those of other methyltransferases. Notably, the Lys163Ala and Met167Ala mutations of CeMETT10-MTD do not affect the methylation of sams-hp, in contrast to the corresponding mutations of hMETTL16-MTD, which increase the methylation (23) (Figure 2F, H). The K-loop in CeMETT10, which is longer than that in hMETTL16, might occlude the SAM binding site in a distinct manner from that observed in hMETTL16 complexed with hMAT2A-hp1.
In CeMETT10, KA-1 facilitates the methylation of sams-hp-ls and U6 snRNA through its RRR, as in the KA-1 of hMETTL16, by increasing the affinity of CeMETT10 for RNA substrates (Figure 5D) (35). A previous study suggested that the KA-1 of hMETTL16 interacts with double-stranded regions, and KA-1 binding to these regions of RNA substrates would promote the proper folding of the entire stem–loop structure required for productive catalysis by the MTD. Thus, both hMETTL16 and CeMETT10 methylate the adenosine of the conserved UACAG motif in the specific structural context of RNAs, by cooperatively using the N-terminal MTD and C-terminal KA-1.
KA-1 domains have also been identified in other invertebrate and fission yeast METTL16 homologs (Figure 4B, Supplementary Figures 3, 4, 8). In S. pombe, the METTL16 homolog possesses the KA-1 domain in its C-terminal half (Figures 4B, 5A, Supplementary Figures 3, 8). A recent report showed that S. pombe METTL16 methylates A37 (A43 in human) of U6 snRNA, and regulates the splicing by facilitating an interaction with the 5'-splice site adenosines in a subset of S. pombe introns (30). Although the 3'-UTR hairpins of MAT2A mRNA are conserved in vertebrates, they are not present in other organisms (54). In fact, S. pombe S-adenosylmethionine synthetase 1 (sam1) mRNA, an ortholog of MAT2A, is not affected in the mtl16Δ strain (30). In S. pombe, the SAM homeostasis is partially regulated by the translation of sam1 mRNA, through the interaction of SAM with the 5'-UTR of the sam1 mRNA (55). Thus, the primary target of S. pombe METTL16 would be U6 snRNA, and the KA-1 domain would facilitate its methylation to control the splicing of some pre-mRNAs (30).
Under cellular conditions with a limited concentration of SAM, hMETTL16 dwells on the 3'-UTR hairpins of the pre-MAT2A mRNA, and the KA-1 (VCR) in hMETTL16 may recruit splicing factors for efficient and rapid splicing of the MAT2A pre-mRNA (20). The affinity of hMETTL16 for MAT2A-hp-ls is larger than that of CeMETT10 for sams-hp-ls (Kd values of hMETTL16 and CeMETT10 for RNA are 0.042 μM and 0.14 μM, respectively) (Figure 5F) (35). Under limited SAM concentration conditions, while hMETTL16 dwells on MAT2A pre-mRNA and recruits splicing factors through KA-1 to promote splicing, CeMETT10 might quickly dissociate from the 3'-splice site of sams pre-mRNA and U2AF35 would bind to the 3'-splice site to promote the pre-mRNA splicing. Considering that the KA-1 of CeMETT10 facilitates the efficient methylation of the 3'-splice sites of sams pre-mRNAs (Figure 5D), the KA-1 of CeMETT10 would promote the rapid methylation under conditions with higher SAM concentrations, and the binding of U2AF35 to the 3'-splice site would be blocked; thereby, CeMETT10 inhibits the splicing of sams pre-mRNA and reduces the level of SAM biosynthesis.
The previous biochemical study showed that KA-1 of hMETTL16 interacts with the double-stranded stem region of RNA (35). The KA-1 of CeMETT10 would also interact with double-stranded RNA. However, the detailed mechanism of the interactions between KA-1 and RNA remains elusive (35,42). Full-length hMETTL16 reportedly interacts with the 3'-terminal triple-helix of MALAT lncRNA, while the MTD does not, suggesting that the KA-1 (VCR) and/or the disordered part in the C-terminal region between KA-1a and KA-1b could interact with the 3'-triple helix RNA (34,56). The detailed mechanisms of the RNA recognition by KA-1 and the dynamical cooperative methylation of substrate RNAs by the N-terminal MTD and the C-terminal KA-1 await further structural determinations.
DATA AVAILABILITY
Coordinates and structure factors for the crystal structure of C. elegans METT10-MTD have been deposited in the Protein Data Bank, under the accession code 8GU3.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the beamline staff of BL-1A (KEK, Tsukuba) for technical assistance during data collection.
Contributor Information
Jue Ju, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
Tomohiko Aoyama, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
Yuka Yashiro, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
Seisuke Yamashita, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
Hidehito Kuroyanagi, Department of Biochemistry, Graduate School of Medicine, University of the Ryukyus, Nishihara-cho, Okinawa 903-0125, Japan.
Kozo Tomita, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research (A) [18H03980 to K.T.]; Scientific Research (B) [20H03181 to H.K.] from JSPS; Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [26113002 to K.T., 17H05596, 20H04839 to H.K.]; Kobayashi Foundation; Uehara Memorial Foundation; Terumo Life Science Foundation; Naito Foundation [to K.T.]. Funding for open access charge: Grant-in-Aid for Scientific Research (A) from JSPS [to K.T.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends. Biochem. Sci. 2013; 38:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dominissini D. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 3. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yue Y., Liu J., He C.. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015; 29:1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G.et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., Sun H.-Y., Li A., Ping X.-L., Lai W.-Y.et al.. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 7. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer K.D. 5′ UTR m6A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fustin J.M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M.S., Kakeya H., Manabe I.et al.. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013; 155:793–806. [DOI] [PubMed] [Google Scholar]
- 10. Batista PedroJ., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley DonnaM., Lujan E., Haddad B., Daneshvar K.et al.. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell. Stem. Cell. 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Batista P.J. The RNA modification N6-methyladenosine and its implications in human disease. Genom. Proteom. Bioinform. 2017; 15:154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S.et al.. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015; 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- 13. Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J.et al.. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell. Res. 2017; 27:1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu J. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ke S., Alemu E.A., Mertens C., Gantman E.C., Fak J.J., Mele A., Haripal B., Zucker-Scharff I., Moore M.J., Park C.Y.et al.. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 2015; 29:2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C.et al.. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature. 2016; 534:575–578. [DOI] [PubMed] [Google Scholar]
- 18. Sledz P., Jinek M.. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016; 5:e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., Kumagai S., Ochiai K., Suzuki T., Igarashi K.. S-Adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017; 21:3354–3363. [DOI] [PubMed] [Google Scholar]
- 20. Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K.. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017; 169:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., Sloan K.E., Bohnsack M.T.. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017; 18:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mendel M., Chen K.M., Homolka D., Gos P., Pandey R.R., McCarthy A.A., Pillai R.S.. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 2018; 71:986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doxtader K.A., Wang P., Scarborough A.M., Seo D., Conrad N.K., Nam Y.. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell. 2018; 71:1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sawa H., Shimura Y.. Association of U6 snRNA with the 5'-splice site region of pre-mRNA in the spliceosome. Genes Dev. 1992; 6:244–254. [DOI] [PubMed] [Google Scholar]
- 25. Sawa H., Abelson J.. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5' splice site during the splicing reaction in yeast. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:11269–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wassarman D., Steitz J.. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science. 1992; 257:1918–1925. [DOI] [PubMed] [Google Scholar]
- 27. Gu J., Patton J.R., Shimba S., Reddy R.. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA. 1996; 2 9:909–918. [PMC free article] [PubMed] [Google Scholar]
- 28. Brow D.A., Guthrie C.. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988; 334:213–218. [DOI] [PubMed] [Google Scholar]
- 29. Madhani H.D., Bordonne R., Guthrie C.. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 1990; 4:2264–2277. [DOI] [PubMed] [Google Scholar]
- 30. Ishigami Y., Ohira T., Isokawa Y., Suzuki Y., Suzuki T.. A single m(6)A modification in U6 snRNA diversifies exon sequence at the 5' splice site. Nat. Commun. 2021; 12:3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watabe E., Togo-Ohno M., Ishigami Y., Wani S., Hirota K., Kimura-Asami M., Hasan S., Takei S., Fukamizu A., Suzuki Y.et al.. m(6) A-mediated alternative splicing coupled with nonsense-mediated mRNA decay regulates SAM synthetase homeostasis. EMBO J. 2021; 40:e106434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mendel M., Delaney K., Pandey R.R., Chen K.M., Wenda J.M., Vågbø C.B., Steiner F.A., Homolka D., Pillai R.S.. Splice site m(6)A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell. 2021; 184:3125–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshida H., Park S.Y., Sakashita G., Nariai Y., Kuwasako K., Muto Y., Urano T., Obayashi E.. Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1. Nat. Commun. 2020; 11:4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruszkowska A., Ruszkowski M., Dauter Z., Brown J.A.. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 2018; 8:5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aoyama T., Yamashita S., Tomita K.. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 2020; 48:5157–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J., Yashiro Y., Sakaguchi Y., Suzuki T., Tomita K.. Mechanistic insights into tRNA cleavage by a contact-dependent growth inhibitor protein and translation factors. Nucleic Acids Res. 2022; 50:4713–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kabsch W XDS. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamashita S., Takagi Y., Nagaike T., Tomita K.. Crystal structures of U6 snRNA-specific terminal uridylyltransferase. Nat. Commun. 2017; 8:15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yashiro Y., Tomita K.. Function and regulation of human terminal uridylyltransferases. Front. Genet. 2018; 9:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A.et al.. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021; 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Christian T., Hou Y.-M.. Distinct Determinants of tRNA Recognition by the TrmD and Trm5 Methyl Transferases. J. Mol. Biol. 2007; 373:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu D., Kaur G., Blumenthal R.M., Zhang X., Cheng X.. Enzymatic characterization of three human RNA adenosine methyltransferases reveals diverse substrate affinities and reaction optima. J. Biol. Chem. 2021; 296:100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martinez A., Yamashita S., Nagaike T., Sakaguchi Y., Suzuki T., Tomita K.. Human BCDIN3D monomethylates cytoplasmic histidine transfer RNA. Nucleic Acids Res. 2017; 45:5423–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Y., Martinez A., Yamashita S., Tomita K.. Crystal structure of human cytoplasmic tRNAHis-specific 5′-monomethylphosphate capping enzyme. Nucleic Acids Res. 2020; 48:1572–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song J., Teplova M., Ishibe-Murakami S., Patel D.J.. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012; 335:709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Y., Eichhorn C.D., Wang Y., Cascio D., Feigon J.. Structural basis of 7SK RNA 5'-γ-phosphate methylation and retention by MePCE. Nat. Chem. Biol. 2019; 15:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu R.J., Long T., Li J., Li H., Wang E.D.. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017; 45:6684–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Finer-Moore J., Czudnochowski N., O’Connell J.D. 3rd, Wang A.L., Stroud R.M.. Crystal structure of the human tRNA m(1)A58 methyltransferase-tRNA(3)(Lys) complex: refolding of substrate tRNA allows access to the methylation target. J. Mol. Biol. 2015; 427:3862–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Parker B.J., Moltke I., Roth A., Washietl S., Wen J., Kellis M., Breaker R., Pedersen J.S.. New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Res. 2011; 21:1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang X., Sun W., Chen D., Murchie A.I.H.. Interactions between SAM and the 5' UTR mRNA of the sam1 gene regulate translation in S. pombe. RNA. 2020; 26:150–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brown J.A., Kinzig C.G., DeGregorio S.J., Steitz J.A.. Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:14013–14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the crystal structure of C. elegans METT10-MTD have been deposited in the Protein Data Bank, under the accession code 8GU3.