Abstract
The tumor-suppressor proteins BRCA1 and BARD1 function as an E3 ubiquitin ligase to facilitate transcriptional repression and DNA damage repair. This is mediated in-part through its ability to mono-ubiquitylate histone H2A in nucleosomes. Studies in Caenorhabditis elegans have been used to elucidate numerous functions of BRCA1 and BARD1; however, it has not been established that the C. elegans orthologs, BRC-1 and BRD-1, retain all the functions of their human counterparts. Here we explore the conservation of enzymatic activity toward nucleosomes which leads to repression of estrogen-metabolizing cytochrome P450 (cyp) genes in humans. Biochemical assays establish that BRC-1 and BRD-1 contribute to ubiquitylation of histone H2A in the nucleosome. Mutational analysis shows that while BRC-1 likely binds the nucleosome using a conserved interface, BRD-1 and BARD1 have evolved different modes of binding, resulting in a difference in the placement of ubiquitin on H2A. Gene expression analysis reveals that in spite of this difference, BRC-1 and BRD-1 also contribute to cyp gene repression in C. elegans. Establishing conservation of these functions in C. elegans allows for use of this powerful model organism to address remaining questions regarding regulation of gene expression by BRCA1 and BARD1.
INTRODUCTION
BRCA1 protein protects the genome by signaling for DNA damage repair of double stranded breaks via homologous recombination, halting cell cycle progression in damaged cells, and preventing DNA damage through repression of genes and satellite DNA (reviewed in (1)). BRCA1 performs these functions in many mammalian tissues such as embryonic stem cells, fibroblasts, and neurons (2–4). Yet, heterozygous mutations in BRCA1 are most often associated with increased cancer-risk in estrogen-responsive tissues in women: up to 90% increase in breast cancer risk and 44% increase in ovarian cancer risk (5–7). The role BRCA1, and its partner BARD1, play in repressing the expression of cytochrome P450 (cyp) genes that metabolize estrogen into DNA damaging free radicals has been hypothesized to contribute to the tissue specificity (8,9). The RING domains of BRCA1 and BARD1 form a heterodimer (BCBD) that catalyzes attachment of the small signaling protein, ubiquitin, onto the C-terminal tail of histone H2A that serves as a repressive mark on chromatin. However, many questions remain regarding how ubiquitylated H2A represses genes, how BCBD is recruited to the regulated genes, and which genes are regulated by BCBD.
The model organism Caenorhabditis elegans offers an opportunity to explore the functions of BRCA1 and BARD1. Unlike mammals, C. elegans can grow past the embryonic stage and reproduce without functional BRCA1 or BARD1. Researchers have taken advantage of this to investigate in vivo functions of the C. elegans BRCA1 and BARD1 orthologs, BRC-1 and BRD-1, respectively. Like human BCBD (HsBCBD), the worm orthologs (CeBCBD) also maintain genome stability by acting in pathways such as DNA damage repair, meiosis, satellite DNA repression, and cell cycle checkpoint signaling (10–16). However, it is unknown if CeBCBD plays a conserved role in gene repression through H2A ubiquitylation. Here, we establish that BRC-1 and BRD-1 form a complex that can ubiquitylate histone H2A in nucleosomes in vitro and repress cyp genes in vivo. We show that the interaction of BRC-1 with the nucleosome occurs through a conserved interface, while the BRD-1 nucleosome-binding interface differs from that described for BARD1. The biochemistry of the CeBCBD complex supports recent hypotheses regarding how divergent RING-nucleosome interactions determine the specific lysine target on histones. These findings provide a path to use C. elegans to investigate the many remaining questions regarding BRCA1 function in transcription regulation and the role of BRCA1’s enzymatic activity throughout development.
MATERIALS AND METHODS
Generation of constructs
To investigate the biochemistry of CeBCBD in vitro, a construct coding for BRD-1 residues 1-107 was cloned into the pET28N vector using restriction enzyme cloning. The resulting protein contains an additional glycine as the second residue due to the restriction enzyme site introduced during cloning. A construct coding for BRC-1 residues 1–100 with an N-terminal histidine tag was cloned into the pCOT7 vector. Mutants were synthesized to probe the nucleosome binding interface using the primers described in Supplementary Table S1 and the Agilent QuikChange Site-directed Mutagenesis protocol with modifications described previously (17). The plasmid coding for the H2A chimeric protein and the mutant K125H chimeric H2A were generated from histidine- and VSVG-tagged human H2A on the pHIS vector using New England Biolabs (NEB) Q5 Site-directed mutagenesis protocol. PCR was performed using appropriate forward and reverse primers designed to delete residues 122–130 from human H2A and replace them with residues 123–127 of C. elegans H2A. All constructs generated were confirmed via genetic sequencing analysis using a Hitachi Genetic Analyzer 3130XL.
Protein expression and purification
Both partners of the heterodimer for all CeBCBD and HsBCBD constructs were co-transformed into BL21 (DE3) Escherichia coli according to the New England Biolabs protocol. The transformed cells were grown, and the CeBCBD constructs were synthesized as described for HsBCBD with the following minor modification (18,19). Protein expression was induced for 16 h at 16°C using IPTG at a final concentration of 0.2–0.25 mM. Reducing agent (1 mM DTT or 1 mM TCEP) was present in all steps of purification from lysis to storage of the purified protein and is necessary for production of ligase active protein. Histidine-tag affinity chromatography was performed using a HiTrap Talon Crude cobalt column (GE) on an Äkta Start GE system. Size exclusion chromatography was performed on an NGC Quest 10 Plus Bio-Rad chromatography system in 25 mM HEPES, 150 mM NaCl, 1 mM TCEP–HCl, pH 7.0. The final CeBCBD concentration was determined using absorbance at 280 nm and an extinction coefficient of 15.00 mM cm−1. Construct expression and purity was confirmed with 15% SDS-PAGE (Supplementray Figure S1). All mutant constructs of CeBCBD were purified as described for wild-type with the exception of C21W and C40Y mutants. As reported for HsBCBD containing the equivalent mutations (9), these constructs are more challenging to work with in vitro and size exclusion chromatography could not be performed without loss of the heterodimer to quantities below what is needed for assays. Instead the C21W and C40Y mutant proteins were dialyzed into 25 mM HEPES, 150 mM NaCl, 1 mM TCEP–HCl, pH 7.0 after metal affinity chromatography. Due to impurities that absorb at 280 nm, the concentration of these mutants were adjusted using comparison to wild-type CeBCBD at known concentrations on SDS-PAGE.
Ubiquitin, E1 (UBA1), and E2 (LET-70) were expressed and purified as described previously (20,21). These proteins were stored at –80°C in a buffer containing 25 mM sodium phosphate, 150 mM NaCl at a pH of 7.0. Both chimeric H2A constructs were expressed and assembled into octamers as previously described for human nucleosome reconstitution (22). Reconstitution was verified with 5% polyacrylamide TBE gel electrophoresis. The nucleosomes were stored on ice at 4°C in a buffer solution containing 50 mM NaCl and 20 mM TRIS and used within one week of reconstitution. HsBCBD was co-expressed (His6-BRCA1 residues 1–110 and BARD1 residues 26–140) and purified using the same protocol as for CeBCBD.
H2A ubiquitylation assays and western blotting
All ubiquitylation assays were carried out in reaction mixtures containing a final concentration of 25 mM HEPES, 150 mM NaCl, 20 μM ubiquitin, 8 μM CeBCBD, 4 μM E2 (LET-70), 0.5 μM E1 (UBA1), 0.3 μM nucleosomes, 5 mM ATP, 5 mM MgCl2, 0.1–0.2 mM TCEP–HCl at pH 7.0. All nucleosomes used for assays contain the chimeric H2A with the worm C-terminal tail described in ‘generation of constructs’. The sample for the zero-time point was taken prior to the addition of ATP. Samples of the reaction were taken at 10 and 30 min after the addition of ATP, and the reaction was quenched by adding reducing SDS-PAGE load dye. Samples were subjected to 15% SDS-PAGE and analyzed with western blotting. Blotting was carried out using a 1:10 000 dilution of a rabbit antibody against the VSVG-tag on histone H2A (Millipore Sigma Corp.). A 1:5000 dilution of a goat-anti-rabbit antibody conjugated to alkaline phosphatase (Rockland Immunochemicals Inc.) was used for detection. The secondary antibody was detected using BCIP/NBT Color Development Substrate (Promega Corporation) according to manufacturer specified protocols. Quantification of at least three replicates was carried out by densitometry using ImageJ (NIH) and GraphPad Prism or Microsoft Excel was used to perform the statistical tests described in the figure captions.
C. elegans strains and maintenance
Wild-type (WT) N2 Bristol was gifted by Dr. Phil Hartman (Texas Christian University, Texas, USA). DW103 [brd-1(dw1)] strain was obtained from C. elegans Genetic Center, University of Minnesota, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The VC4248 (gk5332) strain, a CRISPR generated knockout, was created by the Moerman Laboratory and gifted by Dr. Dana Miller (Washington University, Washington, USA) (23). Worms were cultured on nematode growth media (NGM) agar seeded with E. coli OP50 and maintained at 20°C using the standard procedure (24). L4 stage worms were used for all gene expression measurements. Briefly, a synchronized population of L1 worms were obtained using hypochlorite bleaching. The pool of L1 larvae were grown in E. coli seeded NGM plates for 36 hours until the worms reached the L4 stage. L4 larvae were flash-frozen using dry ice and stored at –80°C.
RNA isolation and RT-qPCR
For each biological replicate (four replicates for each worm strain), total RNA was isolated from a pool of 2000 flash frozen L4 worms using the Maxwell 16 LEV simplyRNA Tissue Kit connected with Maxwell 16 AS2000 instrument (both Promega Corporation, Madison, WI, USA) with some modifications. In brief, 210 μl of cold Promega homogenization buffer was added to the sample and sonicated using VCX130 with CL-18 tip at 85% amplification (six pulses with 3 s on time and 3 s off time between pulses) followed by addition of 200 μl Promega lysis buffer. The quantification of RNA was performed using NanoDrop ND-1000 Microvolume UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). All the samples had an absorbance ratio of 260/280 and 260/230 >2 indicating acceptable levels of RNA purity for downstream applications. High starting quantities of worms were necessary to achieve robust quantification of cyp-13A mRNA due to low expression levels. Blinding was not used as it is not standard practice for RT-qPCR experiments with C. elegans.
Quantabio qscript cDNA Supermix was used to convert RNA to cDNA. Briefly, total RNA was diluted to 50 ng/μl and combined with cDNA supermix at a ratio of 4:1 total RNA:supermix. Reactions were carried out using a TC100 thermal cycler (BioRad, Hercules, CA) with a program of 5 min at 25°C followed by 30 min at 42°C and 5 min at 85°C. To quantify gene expression, qPCR reactions containing 0.4 μl of cDNA, 4.3 μl of nuclease free water, 0.3 μl of 10 μM primer mix and 5 μl of PerfeCta SYBR Green FastMix (Quantabio), were performed in triplicate using a BioRad CFX Connect real-time PCR detection system, managed by CFX Manager 3.0 software, with a cycling program consisting of an activation step (95°C, 30 s) and 40 cycles of denaturing (95°C, 10 s) and annealing (primer specific temperature, 15 s). Melt curve analysis was conducted at the end of each qPCR reaction to confirm primer specificity. The primer sequences and annealing temperatures are shown in Table S2 of the SI. A standard curve was generated for each gene to ascertain the efficiency of the primer (80–97%) and estimate the starting quantity (SQ) of target genes. Any replicate deviating more than two standard deviations away from the mean for a particular gene within that strain was considered an outlier and removed before analysis (n = 3–4 biological replicates for analysis). The expression of the reference gene, tba-1, was found to be stable across the strains. The data are presented as the mean SQ normalized to reference gene tba-1 and presented relative to WT. To minimize confounding factors, WT samples prepared and analyzed alongside each strain were used for comparison. A Student's t-test compared the expression of each gene in the transgenic strain to expression levels in WT worms using GraphPad Prism version 9.0.1. Statistical significance was considered P < 0.05. Reporting for experiments using C. elegans follows the recommendations in the ARRIVE guidelines.
RESULTS
Nucleosome ubiquitylation is a conserved function of CeBCBD
Like human BRCA1 and BARD1, C. elegans BRC-1 and BRD-1 contain conserved RING domains, form a heterodimer, and can act as an E3 ubiquitin ligase (10,11). To determine if H2A ubiquitylation is a conserved function in CeBCBD, we expressed recombinant BRC-1 and BRD-1 RING domains in bacteria and purified the heterodimer. The core of histone H2A has 98% sequence similarity in worms and humans, and importantly all of the nucleosome residues that HsBCBD contacts in the available structures are conserved (Supplementary Figure S2a) (22,25). However, the H2A C-terminal tail, which is the target of HsBCBD, is not conserved in worms (Figure 1A). To better mimic the worm histone, we created a chimera of human H2A in which the C-terminal tail is replaced with the worm sequence. Keeping the core consistent allowed for efficient nucleosome assembly in vitro without modification of established protocols. We found when the CeBCBD heterodimer is combined with ubiquitin (Ub), reconstituted mononucleosomes, the worm E2 enzyme (LET-70), the human E1 enzyme (UBA1) and ATP that the RING domain heterodimer catalyzes transfer of Ub onto the chimeric H2A. Ub is attached through a covalent peptide bond allowing for observation of enzyme activity by monitoring the change in molecular weight of H2A in a western blot for the VSVG epitope tag on H2A (Figure 1B). The increasing intensity of higher molecular weight bands corresponding to ubiquitylated H2A and the disappearance of the lower molecular weight band from unmodified H2A both indicate that the enzymatic activity toward H2A in nucleosomes is a conserved property of CeBCBD (lanes 1–3) and HsBCBD (lanes 7–9). CeBCBD also demonstrates preference for modification of H2A over other histone tails similar to HsBCBD (Supplementary Figure S2b) (26).
Figure 1.
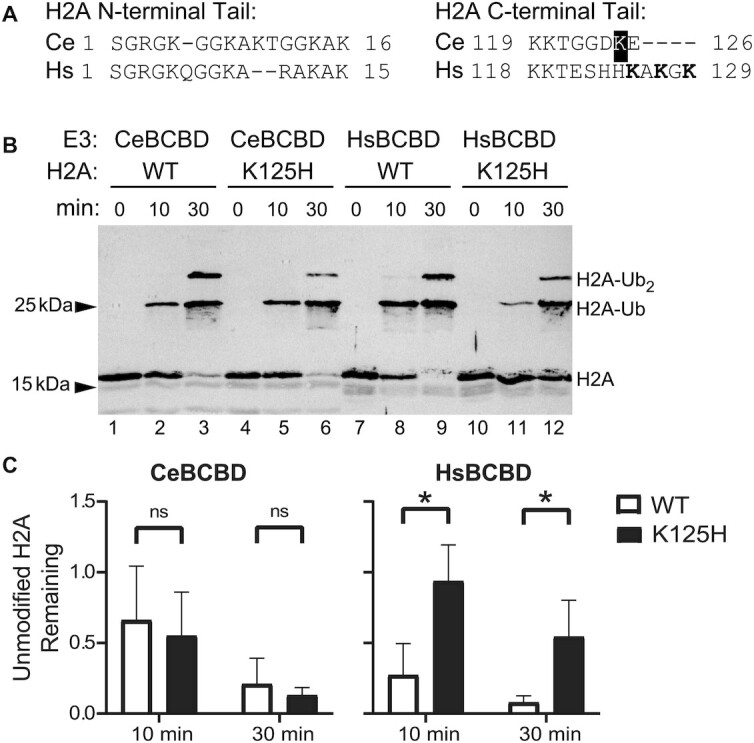
H2A ubiquitylation is a conserved feature of CeBCBD, but does not primarily occur at the predicted location. (A) Amino acid sequence alignment of worm (Ce) and human (Hs) histone H2A tails. The residue highlighted in black, K125, is the lysine closest to the three residues targeted by HsBCBD shown in bold. Dashes indicate gaps in the sequence. (B) In vitro activity of CeBCBD and HsBCBD toward nucleosomes containing H2A with the worm C-terminal tail sequence (WT) or K125H mutation is demonstrated via western blot for H2A. (C) Mean unmodified H2A remaining after 10 and 30 min from two biological and two technical replicates of in vitro H2A ubiquitination assays. Error bars represent the standard deviation. The intensity of unmodified H2A bands in western blots such as (B) were quantified using ImageJ. Means were compared using a Welch's t-test for data with unequal variance (HsBCBD at 30 min) or an unpaired two-sample t-test was used for data with equal variances (all other time points). Statistical significance was determined using a P-value <0.05. An uncropped blot image is available in the supplementary information.
To determine if the site of CeBCBD ubiquitylation is conserved, the lysine in the C-terminal tail closest to those targeted in the human system was eliminated through mutation (K125H) (26). HsBCBD shows a preference for K125 ubiquitylation consistent with the pattern of lysine preference reported in Witus et al. (22). This is most easily observed in Figure 1b by noting the increase in unmodified H2A in lanes 11–12 when compared to lanes 8–9. In contrast, ubiquitylation of the K125H H2A mutant by CeBCBD is not substantially affected as observed by comparing lanes 2–3 to 5–6. A statistical comparison of the average band density for unmodified H2A remaining at 10 and 30 min in Figure 1c, supports these observations. The results suggest that HsBCBD retains its known preference for lysine on the flexible extreme C-terminal tail in the chimeric H2A, but that K125 is not the preferred target of CeBCBD.
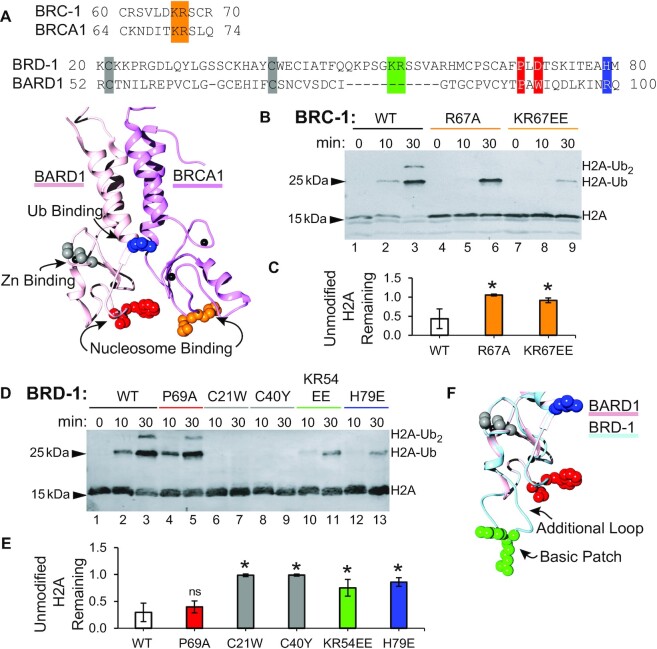
Three human RING E3 ligases ubiquitylate H2A on non-overlapping Lys residues. The placement of ubiquitin onto a specific Lys on the nucleosome is hypothesized to be dictated by the interaction of the RING domains with the histone core on the face of a nucleosome, rather than a specific interaction formed with the tails themselves (22,27,28). To investigate the conservation of the nucleosome binding interface, we mutated the BRC-1 residues that align with the BRCA1 arginine anchor shown to bind the nucleosome in recent structures (Figure 2A) (22,25). Mutation of the arginine anchor to a neutral residue (R67A) or of both basic residues in the loop to the opposite charge (K66E and R67E) resulted in a substantial loss of H2A ubiquitylation activity that can be observed by the intense band of unmodified H2A remaining throughout the assay or the less intense bands corresponding to ubiquitylated H2A after 10 and 30 min (lanes 4–9 in Figure 2B). Quantification of the remaining unmodified H2A after 30 min confirms these BRC-1 mutants have a significant decrease in activity compared to wild-type (Figure 2C). These results are similar to what has been observed when mutating the corresponding residues in BRCA1 consistent with these BRC-1 residues having a conserved nucleosome-targeting function in CeBCBD (22,27).
Figure 2.
BRC-1 utilizes a conserved nucleosome binding interface. (A) Sequence alignments of the relevant regions of the RING domains and solution structure (PDB 1JM7) of HsBCBD highlighting the sidechains of residues from BRCA1 and BARD1 that are important for binding to the nucleosome in orange and red, respectively. The sidechain of BARD1 R99 that non-covalently engages ubiquitin in E2∼Ub is shown in blue. Zinc coordinating residues that were mutated in families with a history of breast cancer are shown in gray. Western blots comparing the ubiquitylation activity of mutant BRC-1 (B) and mutant BRD-1 (D) constructs to wild-type CeBCBD (WT). The substrate observed is H2A incorporated in nucleosomes. Means and standard deviations of unmodified H2A at 30 min are presented for BRC-1 mutants (C) and BRD-1 mutants (E). Asterisk denotes mutants with significant decreases in activity according to Student's t-test with P-value < 0.05, while ‘ns’ denotes activity difference is not significant (P-value > 0.05). (F) BRD-1 homology model generated by SWISS-MODEL (light blue) highlighting the K54 and R55 basic patch in green (44). The additional loop in BRD-1 that contains these residues is hypothesized to be positioned below the nucleosome binding residues for BARD1 (red). Uncropped blot images are available in the supplementary information.
While BRCA1 is vital for binding to the E2 and the nucleosome acidic patch, these roles are also preserved for the other two RING domains that ubiquitylate H2A on alternate lysines, the PRC1 complex and RNF168 (29–31). Additional nucleosome contacts outside of the arginine anchor, such as those formed by BARD1, are hypothesized to dictate lysine specificity (22). To determine if BRD-1 function could be responsible for the different lysine residue targeted by CeBCBD, we aligned the sequences of BRD-1 and BARD1 (Figure 2A). The main residue that forms nucleosome contacts in BARD1, W91, is not present in BRD-1, which has a charged Asp in its place. A nearby proline (P89) that is important for HsBCBD binding to nucleosomes is conserved in CeBCBD (both W91 and P89 are highlighted in red in Figure 2A) (22). To test if the analogous region of the BRD-1 RING interacts with nucleosomes we mutated the proline to alanine (P69A). Unlike in HsBCBD, this mutation has negligible effects on the activity of CeBCBD as observed by decreasing intensity of the unmodified H2A band and formation of ubiquitylated H2A in Figure 2D (lanes 4 and 5). Statistical tests of the quantification of activity do not show significant deviation from wild-type activity levels (Figure 2E). The non-conservative residue at the W91 position coupled with the lack of a functional effect of mutating the P89 analog suggest that the nucleosome binding interface of BARD1 is not conserved in BRD-1.
To determine if BRD-1 is involved in nucleosome binding at an alternative interface, we generated mutations in two conserved zinc-coordinating Cys residues highlighted gray in Figure 2A (C21W and C40Y). The homologous mutations prohibit BARD1 nucleosome binding in HsBCBD and were identified in families with a history of breast cancer (9). These residues do not bind directly to nucleosomes, but through the loss of a zinc coordination site they are predicted to disrupt the RING domain structure and thereby perturb the location of the nucleosome binding residues (32). As the BARD1 RING domain is not directly involved in binding to the E2 but instead interacts with the nucleosome substrate, mutating a zinc-coordinating cysteine residue is an indirect way of determining if BRD-1 contains a nucleosome binding interface (9). As was reported for HsBCBD, mutation of zinc-coordinating cysteine residues in BRD-1 results in a heterodimer that still co-purifies from bacteria, but may be less stable than wild-type heterodimers (Supplementary Figure S1) (9). Nucleosome ubiquitylation assays with C21W and C40Y heterodimers resulted in loss of nucleosome ubiquitylation (lanes 6–9 in Figure 2d–e). These results suggest that BRD-1 contributes to nucleosome interactions.
To predict the alternate interface used by BRD-1 to engage nucleosomes, we built a homology model using the BARD1 structure (Figure 2F). BRD-1 contains an additional loop of eleven residues compared to BARD1 (Figure 2A) that extends past the BARD1 nucleosome binding interface in the predicted structure (Figure 2F and Supplementary Figure S3). Highlighted in green in Figure 2A and F are the only two residues in this loop that are conserved amongst species in the Caenorhabditis genus: two basic residues at position 54 and 55 (see Supplementary Figure S3d for multiple sequence alignment). We hypothesized these residues may interact with acidic residues on the surface of nucleosomes or with the negatively-charged DNA backbone. To test this hypothesis, we mutated these residues to oppositely charged glutamic acid (K54E and R55E, referred to as KR54EE in Figure 2). This mutation significantly decreases nucleosome ubiquitylation consistent with the hypothesis (lanes 10 and 11 in Figure 2D, E). Importantly, CeBCBD containing the KR54EE mutation is still able to bind and activate the E2∼Ub conjugate for transfer when free Lys is used as a proxy substrate (Supplementary Figure S4a, b). This further indicates the mutation disrupts interactions with the nucleosome substrate specifically, rather than affecting the interaction of BRD-1 with BRC-1 or with the E2∼Ub conjugate.
BARD1 plays dual roles in HsBCBD E3 ligase activity toward nucleosomes. In addition to nucleosome binding, BARD1 contains a positively-charged residue (R99) that helps activate the E2∼Ub conjugate for transfer through an ionic interaction between its positively-charged sidechain and a negative charge on Ub (R99 is highlighted in blue in Figure 2). At the analogous position to the Arg, BRD-1 contains a His that could be positively-charged at physiological pH depending on its pKR. We mutated this His to a negatively-charged Glu (H79E). This mutation decreases the activity of CeBCBD as observed by the amount of unmodified H2A remaining and by the decrease in ubiquitylated H2A (lanes 12 and 13 of Figure 2D, E), suggesting BRD-1 may serve an analogous role to BARD1 in activating E2∼Ub conjugate for transfer. We also show that, unlike wild-type CeBCBD, the H79E mutant fails to enhance the reactivity of the E2∼Ub conjugate toward free lysine (Supplementary Figure S4c, d). Together, the results are consistent with the residue playing a role in ubiquitin interactions, as opposed to a being directly involved in nucleosome interactions.
cyp gene repression is a conserved function of CeBCBD
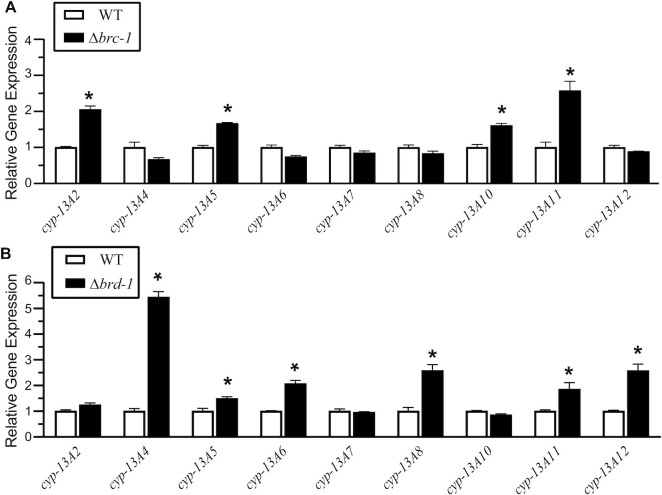
One consequence of H2A ubiquitylation by HsBCBD is the repression of cyp1a1 and cyp3a4, genes involved in estrogen metabolism (8,9). To investigate the conservation of this function in C. elegans, we used the basic local alignment search tool to identify potential cyp homologs. While cyp1a1 returned no close matches in worms, the cyp-13a subfamily of genes in C. elegans contain 27–32% sequence identity to cyp3a4 (Supplementary Figure S5). The regulation of cyp3a4 may also be a conserved feature in the cyp-13a subfamily, as both have expression inducible by the antibiotic rifampicin (33). In human breast epithelial cells (MCF10A), loss of homozygosity for either BRCA1 or BARD1 results in increased expression of cyp3a4 by 2- and 32-fold, respectively (8,9). The cyp-13A gene transcriptome in L4 worms was compared in strains with either brc-1 or brd-1 gene mutations. Of the nine cyp-13A family members with quantifiable expression levels, four genes (cyp-13A2, cyp-13A5, cyp-13A10 and cyp13A11) are upregulated by 1.5- to 2.5-fold compared to wild-type (N2) when brc-1 is knocked out by CRISPR-Cas9 in the strain VC4248 (gk5332) (Δbrc-1 in Figure 3A).
Figure 3.
CeBCBD regulates the expression of cyp-13A genes in vivo. The expression of cyp-13A2, cyp-13A4, cyp-13A5, cyp-13A6, cyp-13A7, cyp-13A8, cyp-13A10, cyp-13A11 and cyp-13A12 in WT (N2) and experimental worm strains Δbrc-1 (gk5332) (A) and Δbrd-1 (dw1) (B) were measured with RT-qPCR. All the data are presented as the mean starting quantity (SQ) normalized to reference gene tba-1 and presented relative to simultaneously measured WT SQ. Each error bar represents ± SEM and * denotes statistically significant deviation from WT as determined by Student's t-test. P-values <0.05 were considered statistically significant.
While characterization of a complete knockout of brd-1 has not been reported in C. elegans, dw1 worms lack the brd-1 exons coding for the ankyrin domain and part of the BRCT domain. These domains are implicated in chromatin localization and increased targeting of HsBCBD to nucleosomes (25,34–36). The dw1 strain shows decreased BRC-1 and BRD-1 protein levels, presumably due to a loss of protein stability (37). The brd-1 (dw1) strain showed significant upregulation of six cyp-13A genes, including cyp-13A4, cyp-13A5, cyp-A6, cyp-13A8, cyp-13A11, and cyp-13A12 (Δbrd-1 in Figure 3B). cyp-13A4 showed the largest change in expression of nearly 5.5-fold while the other genes vary from 1.5- to 2.5-fold increase in expression. Together, these results indicate that CeBCBD is important for gene repression of cyp homologs, a function brought about by BRCA1/BARD1-directed nucleosome ubiquitylation in human cells (9).
DISCUSSION
Here, we show that the role of HsBCBD in transcriptional regulation of cyp genes and H2A ubiquitylation are conserved features in worms. As BARD1 mutants that disrupt nucleosome binding without affecting E3 ligase activity or other known protein interactions have been linked to hereditary breast cancer, nucleosome ubiquitylation is thought to be important for tumor suppression (9). While there is some debate about which roles of BCBD are responsible for tumor suppression, other functions that promote genome stability likely contribute and are also conserved in worms such as heterochromatin-mediated transcriptional silencing, promoting homologous recombination at damaged DNA, and cell cycle checkpoint control (10,11,14,15). Together, these findings suggest that the model organism C. elegans can be exploited to probe the mechanisms and pathways BCBD uses to bring about the myriad functions.
Use of C. elegans to study BCBD function can help overcome current challenges. Human tissue culture is complicated by the lack of a complete BRCA1 or BARD1 knockout in a non-cancer cell line. Mammalian model systems are challenging as loss of either BRCA1 or BARD1 is lethal in the embryonic stage. In contrast, C. elegans without BRCA1 can be stably propagated for generations in an otherwise wild-type background, allowing for investigation of BCBD function throughout development. The protein sequence identity (similarity) of the BRC-1 and BRD-1 RING domains are only 34% (59%) and 20% (72%) conserved respectively. This suggests sequence comparisons may be helpful for predicting which RING domain residues are used for carrying out conserved functions.
While BRD-1 contributes to nucleosome binding, the specific residues used for interacting with the substrate vary from BARD1 (Figure 2). This is reminiscent of several studies that show BCBD functions in axon regeneration and meiosis in both humans and worms, but the mechanism appears to be different for the divergent proteins (16,38). Evolution of differing mechanisms to achieve similar cellular outcomes highlights the importance of these BCBD functions to the fitness of organisms.
The distinction in BARD1 and BRD-1 nucleosome-binding interfaces adds to the growing evidence for the hypothesis that the interaction between the non-E2 binding RING domain partner and the nucleosome dictate the position of ubiquitin placement (22,27). BRD-1, like a non-analogous E3 ligase partner BMI1, contains an additional loop when compared to BARD1. The BMI1 loop contains basic residues that interact with the nucleosome and tilt the complex toward different Lys on the C-terminal tail (27). The BRD-1 loop is located in a different part of the RING domain compared to BMI1, but alignment of the predicted BRD-1 structure with BMI1 or BARD1 when bound to the nucleosome suggests the additional BRD-1 loop will also face the nucleosome (Supplementary Figure S3). Figure 2 suggests that BRD-1 may use basic residues in this loop to interact with the nucleosome. Structure alignments place these basic residues in close proximity to the DNA backbone, rather than the histone residues that BARD1 binds (Supplementary Figure S3). Consistent with the nucleosome interaction of the non-E2 binding partner determining the Lys specificity, we show that CeBCBD does not share the same C-terminal H2A Lys preference as HsBCBD.
The relevance of the location of ubiquitin placement on H2A is unknown, but gene expression data presented here demonstrates that CeBCBD is associated with gene repression much like HsBCBD and RING1/BMI1 (8,9,39,40). H2A is also ubiquitylated in a CeBCBD-dependent manner in silenced satellite DNA repeat sequences in C. elegans (15). This is consistent with H2A-Ub generated by CeBCBD being important for transcriptional repression. The mechanism for repression of genes by H2A-Ub is unknown, but use of C. elegans may help shed light on this and other remaining questions.
Here, we show that CeBCBD is involved in repression of a subset of cyp-13A family members, homologous to cyp3a4 repression by HsBCBD. cyp-13A7 has been observed to increase expression due to rifampicin exposure similar to cyp3a4 in humans, suggesting conserved modes of regulation between these cyp genes (33). BCBD has several recently-identified functions as a histone reader, which may contribute to how it is targeted to select genes (34,35,41). Variation in the expression pattern amongst the different cyp-13A family members may help pinpoint the factors that contribute to BCBD gene selectivity. The primers reported here (Supplementary Table S2) target specific cyp-13A family members and can be used in future studies to parse out the variation in expression conditions of the divergent cyp-13A family members.
Two isoforms, cyp-13A5 and cyp-13A11, showed increased expression upon mutation of either brc-1 or brd-1, but six other isoforms in this gene family showed increased expression in either brc-1 or brd-1 mutant worms. One explanation for this differential regulation would be if BRC-1 and BRD-1 contain some independent functions, which has been noted for BRCA1 and BARD1 (42). Our biochemical assays confirm that functional BRC-1 and BRD-1 are needed for nucleosome ubiquitylation, so if BRC-1 and BRD-1 have independent functions it would suggest they can modulate gene expression through means other than ubiquitylation of H2A. Recent findings using Xenopus have proposed that gene expression can be modulated by BRCA1/BARD1 inhibition of histone acetylation independent of histone ubiquitylation (43). In contrast, expression of a ubiquitin-H2A fusion protein was enough to repress targeted cyp genes in human tissue culture engineered to mimic an inherited BARD1 mutation, suggesting no additional mechanisms are necessary for repression (9). It could be that BCBD participates in redundant mechanisms of gene repression or that use of these mechanisms varies across genes, tissue-type, developmental stage, or species. Establishing the conservation of H2A ubiquitylation and cyp gene repression in the C. elegans model organism is a positive step toward elucidating the epigenetic functions of BCBD throughout development.
DATA AVAILABILITY
Atomic coordinates for the models of BRD-1 generated via alpha fold and homology modeling are available as supplementary information. Supplementary data referenced are available at NAR online.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Phil Hartman for providing the N2 C. elegans strains, C. elegans maintenance training, and for his feedback on presentation of gene expression data. We thank Courtney Skalley, former TCU student, for her contribution in generating the BRC-1 mutants used in this study.
Author contributions: I.T., R.V., S.W., M.J., R.K. and M.S. designed the study. I.T., R.V., S.W., C.L., O.F., M.J. and M.S. collected, analyzed and presented data with I.T., M.J. and M.S. contributing to gene expression studies and R.V., S.W., C.L., O.F. and M.S. contributing to biochemical assays. I.T., R.V., C.L. and M.S. drafted the manuscript. All authors critically reviewed and edited the manuscript.
Contributor Information
Ishor Thapa, Department of Biology, Texas Christian University, Fort Worth, TX 76129, USA.
Russell Vahrenkamp, Department of Biology, Texas Christian University, Fort Worth, TX 76129, USA.
Samuel R Witus, Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.
Caitlin Lightle, Department of Biology, Texas Christian University, Fort Worth, TX 76129, USA.
Owen Falkenberg, Department of Biology, Texas Christian University, Fort Worth, TX 76129, USA.
Marlo K Sellin Jeffries, Department of Biology, Texas Christian University, Fort Worth, TX 76129, USA.
Rachel E Klevit, Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.
Mikaela D Stewart, Department of Biology, Texas Christian University, Fort Worth, TX 76129, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM135900 to M.D.S., CA260834 to R.E.K.]; TCU Biology Department; R.E.K. is the Edmond H. Fisher/Washington Research Foundation Endowed Chair in Biochemistry. Funding for open access charge: TCU Biology Department; NIH [GM135900].
Conflict of interest statement. None declared.
REFERENCES
- 1. Tarsounas M., Sung P.. The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020; 21:284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Snouwaert J.N., Gowen L.C., Latour A.M., Mohn A.R., Xiao A., DiBiase L., Koller B.H.. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 1999; 18:7900–7907. [DOI] [PubMed] [Google Scholar]
- 3. Xu B., Kim S., Kastan M.B.. Involvement of brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 2001; 21:3445–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu Q., Pao G.M., Huynh A.M., Suh H., Tonnu N., Nederlof P.M., Gage F.H., Verma I.M.. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011; 477:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Risch H.A., McLaughlin J.R., Cole D.E.C., Rosen B., Bradley L., Fan I., Tang J., Li S., Zhang S., Shaw P.A.et al.. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin–cohort study in Ontario, Canada. J. Natl. Cancer Inst. 2006; 98:1694–1706. [DOI] [PubMed] [Google Scholar]
- 6. Chen S., Parmigiani G.. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 2007; 25:1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.A., Mooij T.M., Roos-Blom M.J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N.et al.. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. J. Am. Med. Assoc. 2017; 317:2402–2416. [DOI] [PubMed] [Google Scholar]
- 8. Savage K.I., Matchett K.B., Barros E.M., Cooper K.M., Irwin G.W., Gorski J.J., Orr K.S., Vohhodina J., Kavanagh J.N., Madden A.F.et al.. BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 2014; 74:2773–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stewart M.D., Zelin E., Dhall A., Walsh T., Upadhyay E., Corn J.E., Chatterjee C., King M.C., Klevit R.E.. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boulton S.J., Martin J.S., Polanowska J., Hill D.E., Gartner A., Vidal M.. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditiselegans. Curr. Biol. 2004; 14:33–39. [DOI] [PubMed] [Google Scholar]
- 11. Polanowska J., Martin J.S., Garcia-Muse T., Petalcorin M.I., Boulton S.J.. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 2006; 25:2178–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adamo A., Montemauri P., Silva N., Ward J.D., Boulton S.J., La Volpe A.. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008; 9:287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Q., Saito T.T., Martinez-Garcia M., Deshong A.J., Nadarajan S., Lawrence K.S., Checchi P.M., Colaiacovo M.P., Engebrecht J.. The tumor suppressor BRCA1-BARD1 complex localizes to the synaptonemal complex and regulates recombination under meiotic dysfunction in Caenorhabditiselegans. PLoS Genet. 2018; 14:e1007701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Janisiw E., Dello Stritto M.R., Jantsch V., Silva N.. BRCA1-BARD1 associate with the synaptonemal complex and pro-crossover factors and influence RAD-51 dynamics during Caenorhabditiselegans meiosis. PLoS Genet. 2018; 14:e1007653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Padeken J., Zeller P., Towbin B., Katic I., Kalck V., Methot S.P., Gasser S.M.. Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression. Genes Dev. 2019; 33:436–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Q., Hariri S., Engebrecht J.. Meiotic double-strand break processing and crossover patterning are regulated in a sex-specific manner by BRCA1-BARD1 in Caenorhabditiselegans. Genetics. 2020; 216:359–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edelheit O., Hanukoglu A., Hanukoglu I.. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009; 9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brzovic P.S., Meza J., King M.-C., Klevit R.E.. The Cancer-predisposing mutation C61G disrupts homodimer formation in the nH2-terminal BRCA1 RING finger domain. J. Biol. Chem. 1998; 273:7795–7799. [DOI] [PubMed] [Google Scholar]
- 19. Meza J.E., Brzovic P.S., King M.C., Klevit R.E.. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem. 1999; 274:5659–5665. [DOI] [PubMed] [Google Scholar]
- 20. Christensen D.E., Brzovic P.S., Klevit R.E.. E2–BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007; 14:941–948. [DOI] [PubMed] [Google Scholar]
- 21. Pickart C.M., Raasi S.. Methods Enzymol. 2005; 399:Academic Press; 21–36. [DOI] [PubMed] [Google Scholar]
- 22. Witus S.R., Burrell A.L., Farrell D.P., Kang J., Wang M., Hansen J.M., Pravat A., Tuttle L.M., Stewart M.D., Brzovic P.S.et al.. BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1. Nat. Struct. Mol. Biol. 2021; 28:268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Au V., Li-Leger E., Raymant G., Flibotte S., Chen G., Martin K., Fernando L., Doell C., Rosell F.I., Wang S.et al.. CRISPR/Cas9 methodology for the generation of knockout deletions in Caenorhabditiselegans. G3: Genes Genomes. Genet. 2019; 9:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brenner S. The genetics of Caenorhabditiselegans. Genetics. 1974; 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Q., Botuyan M.V., Zhao D., Cui G., Mer E., Mer G.. Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation. Nature. 2021; 596:438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kalb R., Mallery D.L., Larkin C., Huang J.T.J., Hiom K.. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 2014; 8:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McGinty R.K., Henrici R.C., Tan S.. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014; 514:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horn V., Uckelmann M., Zhang H., Eerland J., Aarsman I., le Paige U.B., Davidovich C., Sixma T.K., van Ingen H.. Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nat. Commun. 2019; 10:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taherbhoy A.M., Huang O.W., Cochran A.G.. BMI1–RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun. 2015; 6:7621. [DOI] [PubMed] [Google Scholar]
- 30. Bentley M.L., Corn J.E., Dong K.C., Phung Q., Cheung T.K., Cochran A.G.. Recognition of ubch5c and the nucleosome by the bmi1/ring1b ubiquitin ligase complex. EMBO J. 2011; 30:3285–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mattiroli F., Uckelmann M., Sahtoe D.D., van Dijk W.J., Sixma T.K.. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat. Commun. 2014; 5:3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Witus S.R., Stewart M.D., Klevit R.E.. The BRCA1/BARD1 ubiquitin ligase and its substrates. Biochem. J. 2021; 478:3467–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chakrapani B.P., Kumar S., Subramaniam J.R.. Development and evaluation of an in vivo assay in Caenorhabditiselegans for screening of compounds for their effect on cytochrome P450 expression. J. Biosci. 2008; 33:269–277. [DOI] [PubMed] [Google Scholar]
- 34. Nakamura K., Saredi G., Becker J.R., Foster B.M., Nguyen N.V., Beyer T.E., Cesa L.C., Faull P.A., Lukauskas S., Frimurer T.et al.. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 2019; 21:311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Becker J.R., Clifford G., Bonnet C., Groth A., Wilson M.D., Chapman J.R.. BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination. Nature. 2021; 596:433–437. [DOI] [PubMed] [Google Scholar]
- 36. Dai L., Dai Y., Han J., Huang Y., Wang L., Huang J., Zhou Z.. Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin. Mol. Cell. 2021; 81:2765–2777. [DOI] [PubMed] [Google Scholar]
- 37. Boulton S.J. BRCA1-mediated ubiquitylation. Cell Cycle. 2006; 5:1481–1486. [DOI] [PubMed] [Google Scholar]
- 38. Sakai Y., Hanafusa H., Shimizu T., Pastuhov S.I., Hisamoto N., Matsumoto K.. BRCA1–BARD1 regulates axon regeneration in concert with the Gqα–DAG signaling network. J. Neurosci. 2021; 41:2842–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y.. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004; 431:873–878. [DOI] [PubMed] [Google Scholar]
- 40. Abdouh M., Hanna R., El Hajjar J., Flamier A., Bernier G.. The polycomb repressive complex 1 protein BMI1 is required for constitutive heterochromatin formation and silencing in mammalian somatic cells. J. Biol. Chem. 2016; 291:182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krais J.J., Wang Y., Patel P., Basu J., Bernhardy A.J., Johnson N.. RNF168-mediated localization of BARD1 recruits the BRCA1-PALB2 complex to DNA damage. Nat. Commun. 2021; 12:5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Irminger-Finger I., Ratajska M., Pilyugin M.. New concepts on BARD1: regulator of BRCA pathways and beyond. Int. J. Biochem. Cell Biol. 2016; 72:1–17. [DOI] [PubMed] [Google Scholar]
- 43. Barrows J.K., Fullbright G., Long D.T.. BRCA1-BARD1 regulates transcription through BRD4 in xenopus nucleoplasmic extract. Nucleic Acids Res. 2021; 49:3263–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.AP., Rempfer C., Bordoli L.et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates for the models of BRD-1 generated via alpha fold and homology modeling are available as supplementary information. Supplementary data referenced are available at NAR online.