Abstract
Conjugation of DNA relies on multicomponent protein complexes bridging two bacterial cytoplasmic compartments. Whereas plasmid conjugation systems have been well documented, those of integrative and conjugative elements (ICEs) have remained poorly studied. We characterize here the conjugation system of the ICEclc element in Pseudomonas putida UWC1 that is a model for a widely distributed family of ICEs. By in frame deletion and complementation, we show the importance on ICE transfer of 22 genes in a 20-kb conserved ICE region. Protein comparisons recognized seven homologs to plasmid type IV secretion system components, another six homologs to frequent accessory proteins, and the rest without detectable counterparts. Stationary phase imaging of P. putida ICEclc with in-frame fluorescent protein fusions to predicted type IV components showed transfer-competent cell subpopulations with multiple fluorescent foci, largely overlapping in dual-labeled subcomponents, which is suggestive for multiple conjugation complexes per cell. Cross-dependencies between subcomponents in ICE-type IV secretion system assembly were revealed by quantitative foci image analysis in a variety of ICEclc mutant backgrounds. In conclusion, the ICEclc family presents an evolutionary distinct type IV conjugative system with transfer competent cells specialized in efficient transfer.
INTRODUCTION
Bacterial conjugation is widely appreciated for its importance to horizontal gene distribution and consequent facilitation of adaptation to new selective conditions. Conjugation is mostly attributed to plasmid DNA, but more recently, integrative and conjugative elements (ICEs) have been recognized as additional widespread conjugative vehicles, which, however, are mostly ‘hidden’ (inserted) within the bacterial genome (1–5). ICEs combine both phage-like and plasmid properties. Like temperate phages, they remain mostly integrated in their host genome, and occasionally excise. In contrast to lysogenic phages, excised ICEs spread by conjugation to a new cell, where they re-integrate in the host's chromosome (Figure 1A). Expression of the ICE excision and conjugation functions is silent in the integrated state, being subject to complex, multi-layered regulatory control that varies across different ICEs (6–8). Various physiological and external cues can lead to activation of ICE transfer (6,8,9). Particularly for the model ICEclc of Pseudomonas knackmussii B13 (see below), activation only occurs in a subpopulation of transfer competent (tc) cells, and only these cells are the ones that can excise, replicate and conjugate the ICE (9–11).
Figure 1.
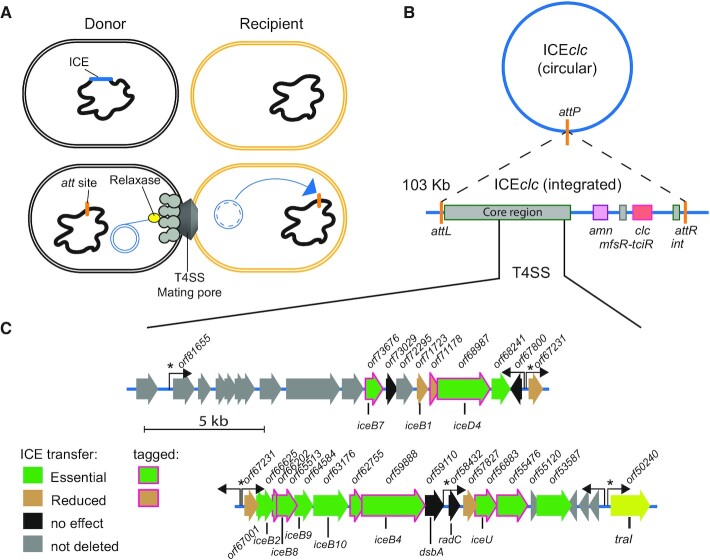
The ICEclc lifestyle. (A) Schematic representation of ICEclc transfer from an integrated state (dark blue) to the excised form (double blue circle) that is conjugated through a type IV secretion system (T4SS) mating pore into a recipient cell (yellow), where integration can occur (blue arrow). (B) Schematic structure of ICEclc with its excised circular form recombined at the attP site and the integrated state between attL and attR. Conserved core region shared with other ICEclc family members is schematically indicated, as well as are key features of ICEclc (int, integrase gene; mfsR-tciR, regulators; amn and clc, metabolic pathway gene clusters). (C) Detailed map of the conjugative gene region of ICEclc. Genes and their orientation are represented by colored arrows, with their original orf designation (as in GenBank AJ617740.2) described above and the deduced relevant gene names below (gene names follow analogous designation as in the vir system). Filled colors correspond to transfer effects as in the colored legend (see Figure 2), whereas magenta arrow outlines point to those components tagged with translational fusions for epifluorescence microscopy. Hooked arrows indicate known promoters; an asterisk pointing to those being active only in transfer competent cells as shown in Ref. (21).
Most known ICEs have a mosaic ‘modular’ genetic structure, being composed of syntenic conserved gene regions coding for core mobility functions, intersparsed with highly variable gene content that can provide adaptive benefits to the host (1,3,5). Different ICE families have been recognized, based on their evolutionary history and relatedness of the different conserved modules (2). Particularly, conserved components of the conjugation machinery, such as the VirB4 and VirD4 ATPases of the Type IV secretion system (T4SS) have been used for the classification (12,13). Based on this, Guglielmini and coworkers proposed eight phylogenetic clades, covering mating pair formation (MPF) systems from both ICEs and plasmids (14). In particular, the MPFG clade is unique for conjugative systems of ICEs, and has so far not been well-characterized (14). The only studied MPFG-class member is ICEHin1056 of Haemophilus influenzae, which non-exhaustively identified a number of T4SS components important for pilus biogenesis (15).
The aim of the current work was to better characterize the MPFG clade conjugative system from the ICEclc element and to uncover features distinct from other (plasmid) MPF systems that might point to specific transfer selection. ICEclc is 103 kb long and contains genes encoding for metabolism of 3-chlorocatechol and 2-aminophenol (Figure 1B). It is present in two identical copies in the P. knackmussii B13 genome at the 3′ end of tRNAGly genes (16,17) and can be conjugated to Pseudomonas putida where it maintains a single integrated copy. The ICEclc core region is highly conserved among a wide range of putative ICEs detected in genomes of Beta- and Gammaproteobacteria (7,17). Transfer only occurs from tc cells, that appear in 3–5% of the stationary phase population, notably after growth on 3-chlorobenzoate (9,18). After its excision, the ICEclc molecule can replicate to up to 7 copies in its host tc cell, which may contribute to more effective and multi-recipient transfer (11). Transfer of ICEclc is dependent on the TraI relaxase (Figure 1C), which nicks the DNA at either one of two origins of transfer (oriT) (19), and is assumed to guide the single-stranded ICE-DNA molecule to the mating pore.
In order to characterize the components of the ICEclc conjugative system, we deployed a combination of genetic dissection, regular and high resolution epifluorescence microscopy and bioinformatics. We focused on a region on ICEclc that was previously suggested to locate the genes for its conjugative system (Figure 1C) (20). Out of the 24 genes present in the locus, we seamlessly deleted 22 by double recombination in a P. putida ICEclc host background, and complemented each deletion individually in trans by a mini-Tn5 delivered single copy wild-type gene, expressed under control of the native ICE promoter upstream of the orf67231 gene (Figure 1C), which is active only in tc cells (21). The effects of gene deletion and complementation on ICE transfer were quantified from isogenic P. putida ICEclc donor-recipient matings. Protein structures were predicted from the genes in this ICE region using AlphaFold2 (22) and Phyre2 (23), and compared to the known protein structure databases to find potential homologs. This analysis was complemented by amino acid similarity searches to archetype T4SS components. Finally, a subset of nine identified components of the ICEclc conjugative machinery was expressed as in-frame fluorescent protein fusions from their native location to study their cellular localization in P. putida ICEclc wild-type or mutant backgrounds, and all fusion proteins were verified for functional ICE transfer. Our results indicate that the ICEclc conjugation machinery is a very distant T4SS, with several functionally analogous components to the F-plasmid system of Escherichia coli (24), the Agrobacterium tumefaciens Vir system (25) or the Dot/Icm system of Legionella pneumophila (26,27), but with a number of unique other components important for its transfer.
MATERIALS AND METHODS
Biological resources, reagents and growth conditions
Escherichia coli DH5α λpir was used for cloning of ICEclc genes and plasmid amplification, and was grown at 37°C on Luria-Bertani (LB) liquid or agar-solidified broth. P. putida UWC1-ICEclc::lacOarray was used as host of the ICE, carrying a single copy ICEclc integrated at the tRNAGly-5 gene (11). The lacOarray modification was introduced in the amnB gene of the ICE with the purpose to follow single-copy ICE-DNA transfer, if needed (11). P. putida UWC1 Tn7 PBADlacI-cfp or UWC1 Tn7 Ptac-mCherry were used as recipient strains for mating experiments. P. putida was cultured at 30°C either on LB, on nutrient agar (Oxoid) or on minimal medium (MM, type 21C (28)). MM was supplemented with either 5 mM succinate or 3 mM 3-chlorobenzoate (3-CBA) as sole carbon source. Both E. coli and P. putida were cultured aerobically in shaking flasks. Where necessary for plasmid maintenance or for selection of genomic constructs, antibiotics were added at the following concentrations: kanamycin (Km, 50 μg ml−1), ampicillin (Amp, 100 μg ml−1 for E. coli, 500 μg ml−1 for P. putida), tetracycline (Tc, 20 μg ml−1 for E. coli, 50 μg ml−1 for P. putida), or gentamycin (Gm, 20 μg ml−1, 10 μg ml−1 in MM). The following supplements were added for screening or promoter induction (see below): 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 20 μg ml−1), isopropyl-β-D-1-thiogalactopyranoside (IPTG, 0.5 mM), or m-toluate (15 mM). All produced strains are listed in Supplementary table S1.
DNA manipulations
DNA manipulations followed standard procedures or as indicated by the reagent supplier. Plasmids were purified from E. coli DH5α λpir using NucleoSpin Plasmid kits (Macherey-Nagel, Duren, Germany). Oligonucleotide primers were purchased from SigmaAldrich. PCR amplicons and digested fragments were purified using NucleoSpin Gel and PCR Clean–up kits (Macherey-Nagel). Clones were initially screened by PCR on resuspended individual colonies in a GoTaq® G2 green master mix (20 μl, Promega, Madison, United States). Plasmid fragments were assembled using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China), digesting the plasmid backbone with appropriate restriction enzymes (New England Biolabs, Ipswich, United States), and mixing with insert-DNA. All of the constructed plasmid inserts were sequenced prior to further use in P. putida, to verify their correctness, using the ICEclc sequence as a reference (29) (GenBank AJ617740.2). Plasmids are listed in Supplementary table S2.
Chromosomal gene knockouts and complementations
Seamless in-frame deletions of ICEclc genes were created using double recombination with marker counterselection (30). Up- and downstream regions (∼800 bp) of the target gene were PCR amplified using Q5 high fidelity polymerase (New England BioLabs) and primers as detailed in Supplementary table S3, and subsequently cloned into the suicide vector pEMG (30). Purified plasmids (300–500 ng) were electroporated into P. putida UWC1-ICEclc::lacOarray as described previously (19). Single recombinants were selected on plates with Km, and colonies were verified by PCR for integration of the suicide vector. Single recombinants were then cultured and transformed with 50 ng of purified plasmid pSW (30). Transformants were cultured and induced by addition of 15 mM m-toluate (Honeywell Fluka™) for 16 h, to express the I-SceI nuclease and force chromosomal repair, after which culture dilutions were spread on LB plates. Candidate colonies were again verified by PCR for the success of the double recombination and the deletion genotype. Successful clones were then cultured in absence of antibiotics to segregate out pSW, and stored at –80°C with 15% v/v glycerol.
Deleted genes on ICEclc were complemented by individual or combinations of the cognate gene(s), placed in trans in single-copy on the P. putida chromosome using mini-Tn5 delivery (31). ICEclc genes for complementation were amplified using Q5 high-fidelity DNA polymerase (New England BioLabs) and primers as in Supplementary table S3, and then brought under control of the native wild-type ICEclc promoter P67231 that is active only in tc cells (21). The 360-bp P67231-promoter region was amplified with primers containing appropriate restriction sites for cloning into the mini-Tn5 delivery vector pBAM (31) and overhangs to allow assembly. Assembled DNA mixtures were transformed by heat-shock into E. coli DH5α λpir for propagation and plasmid DNA purification. Purified plasmids were sequenced to verify the cloned insert, and electroporated into the corresponding P. putida ICEclc gene deletion strain. Integrants were selected for antibiotic resistance from the mini-Tn5 delivered construct and verified by PCR, after which three clones with potentially different integration positions of the complementation insert were frozen at –80°C for further analysis.
Fluorescent protein fusions
The same strategy used to produce gene deletions was employed for translationally fusing fluorescent protein tags (mCherry or sfGFP) at the N- or C-terminal end of target ICEclc conjugative system proteins. In case of N-terminal fusion the open reading frame of the fluorescent protein was inserted after the signal sequence if present (e.g. orf62755), otherwise following the start codon (e.g. iceB8), followed by a short linker peptide as in (32). In case of C-terminal fusion the linker and the fluorescent protein were inserted before the stop codon of the target gene. In the eventuality of overlap between the translation stop and start of two consecutive genes, the insertion was performed in such a way that the ribosome binding site of the following gene remained intact. Primers used for the amplification of the homology regions and fluorescent proteins are listed in Supplementary table S3. Cognate ICE-genes were replaced with the fluorescently-tagged version directly on wild-type integrated ICEclc in P. putida or on ICEclc carrying additional deletions for conjugative components. P. putida without ICEclc was used as control for fluorescent protein auto-aggregation, and transformed with relevant mini-Tn5 constructs and plasmids pME-bisR or pME-bisDC expressing two known transcriptional regulators involved in ICEclc activation to induce the P67231-promoter in all cells (7). P. putida codon-optimized coding sequences (JCat tool (33)) for mCherry and sfGFP were synthesized by ThermoFisher (Waltham, USA).
Protein homology assignments
Homology and functional predictions focused on ICEclc genes in the conserved presumed conjugative gene region (gene designations orf53587–orf73676, as defined in AJ617740.2, Figure 1C). To avoid finding numerous spurious hits to highly homologous ICE genes in other bacterial genomes, we first compared the translated amino acid sequences from individual ICEclc genes to each of the known components of the classical ‘Vir’ T4SS from A. tumefaciens (NZ_CP029048.1) by EMBOSS needle alignment across the full protein length. Next, we identified any conserved domains by BLASTp (34), Phyre2 (23), pfam (35), or Dali (36), and used TMHMM (37) and SignalP (38) to predict the presence of transmembrane domains and signal peptides. Finally, we used AlphaFold2 (22) to predict structures for each of the presumed ICEclc conjugative components (using standard settings on the web server platform https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb), and then compared the highest ranked derived .pdb structure prediction in PDBeFold (39) with the PDB database to find potential structural analogs. PDBeFold analyses were carried out with default parameter settings.
ICEclc transfer experiments
ICEclc transfer was quantified from binary mating experiments. As donors we tested P. putida carrying ICEclc wild-type, or ICEclc with an inserted lacOarray (11), ICEclc gene deletion variants and the various complemented strains, as well as fluorescent protein fusion variants. As recipient we used isogenic but ICEclc-free P. putida UWC1 carrying a chromosomally integrated Gm resistance (Supplementary table S1). Donors and recipient were freshly plated from –80°C stocks on LB agar plates with appropriate antibiotics and grown for 16 h. Three separate clones (donors) and a single colony (recipient) were picked and inoculated each in 10 ml LB with antibiotics. These individual pre-cultures were grown for 24 h, after which a volume of 40 μl was transferred into 8 ml MM with 3 mM 3-CBA (donors) or 5 mM succinate (recipient) as the sole carbon sources, without antibiotics. After 48 h (donor) and 24 h (recipient) of growth the culture turbidities were measured (OD600), and recipient and donor cultures were mixed together in new vials in a 2:1 ratio (OD/OD), respectively, and to a final volume of 1 ml. Donor and recipient cultures alone served as controls for the appearance of background growth on selective plates.
Donor-recipient cell mixtures were centrifuged at 13 000 rpm (Thermo Scientific Fresco™ 21 Microcentrifuge) for 2 min, after which the supernatant was discarded and the cell pellet was resuspended in 1 ml of sterile MM (without carbon source). The suspension was centrifuged as before and cell pellets were now resuspended in 20 μl of MM. The suspension was transferred on top of a 0.2-μm, 25 mm øcellulose acetate filter (Huberlab, Aesch, Switzerland), that had been placed on a MM agar plate with 0.5 mM 3-CBA. Matings were incubated at 30°C for 48 h, after which cells were washed from the filters with 1 ml MM. Cell suspensions were tenfold serially diluted and 5 μl aliquots of every dilution were dropped and dried on MM-agar with 3 mM 3-CBA to select for the donor alone, on MM-agar with 5 mM succinate and Gm to select for the recipient, and on MM-agar with 3 mM 3-CBA and Gm to select for the transconjugants. Plates were incubated at 30°C until colonies appeared. ICEclc transfer rates were then calculated as the ratio of the number of transconjugant colonies per filter and that of the donor. Transfer rates were normalized across different experiments to that of P. putida ICEclc::lacOarray, which was used as positive control in all experiments.
Epifluorescence microscopy
Fluorescently labelled P. putida ICEclc strains were grown for 96 h in liquid MM supplemented with 3 mM 3-CBA to induce the ICEclc transfer competence pathway (7). Cells were then transferred on the surface of 1-mm thick 1% MM-agarose patches (ø1 cm) placed upside down on a round microscope slide and enclosed in an imaging chamber (Perfusion Chamber, H. Saur Laborbedarf, Germany) as described (9). Cells were imaged with a Nikon Eclipse Ti-E inverted microscope with a perfect focus system (PFS), pE-100 CoolLED, a Plan Apo λ 100 × 1.45 oil objective (Nikon) and a Hamamatsu ORCA-Flash4.0 V2 C11440-22CU camera (Hamamatsu, Hamamatsu City, Japan) installed in a controlled temperature room (22°C). sfGFP and mCherry fluorescence were imaged with exposure times of 500 ms and exported as 16-bit .TIF files. Images for display were adjusted to the same level in Adobe Photoshop (v. 2020, Adobe Inc.), downsampled to 8-bit, cropped and saved as 300 dpi.
For improved resolution, samples of the same cell cultures were additionally imaged with a ZEISS LSM 980 Airyscan 2 microscope equipped with ZEN 3.3 software, installed in a controlled temperature room (22°C). Bacterial cells were located on the agarose patches using phase-contrast, after which Z-stacks (Z = 0.17 μm, 14 layers) of individual cells were aquired in the GFP and/or mCherry channels with an exposure time of 314 ms per layer. Images were processed with the Zeiss Deconvolution algorithm with default settings. Images show a single representative deconvoluted 170-nm Z-slice saved as .TIF file, ‘auto-toned’ in Adobe Photoshop (v. 2020), cropped to the final size and saved as 300 dpi.
Image analysis
Individual P. putida cells were segmented on the Nikon 16-bit .TIF phase-contrast images using SuperSegger (40), and the fluorescence intensities, scores, and positions of up to 5 foci in individual cells were extracted. SuperSegger uses 1D-Gaussian fitting on background-normalized and watershed fluorescence cell images to detect foci, as illustrated in Supplementary Figure S1. Subpopulations of tc cells were identified from quantile-quantile plot analysis (41) of the median cell sfGFP or mCherry fluorescent signals. ‘Real’ and spurious fluorescent foci were distinguished by plotting distributions of the normalized foci intensities among tc and non-tc cell populations, and setting a threshold at which the probability for foci detection in non-tc cells was <1%. Custom made MATLAB scripts were used to extract the following parameters: number of foci per cell, median fluorescence intensity per cell, median top 5% pixel intensity per cell (as proxy for foci intensity), distributions of SuperSegger foci scores (quality of the Gaussian fit) and of Gaussian fit sigma values (as proxy for the apparent foci peak-to-size aspect).
Linear fluorescence profiles were derived from the outer 2-pixel (px) perimeter (‘cell envelope’) of each identified tc cell using its phase-contrast logical mask, and a further 1-px dilated mask. Profiles were scaled to a median cell perimeter of 50 px for plotting. The 20th percentile fluorescence value was taken as proxy for the cell ‘membrane’ fluorescence background, from which we calculated the median across all tc cells per replicate (n = 142–1996 cells); which were compared between wild-type fluorescent protein labeled strains and those with gene deletions in ICEclc. The standard deviation of the pixel fluorescence values in the two cell perimeters was calculated as second proxy for potential relocalization of fluorescent protein from foci to the cell envelope.
Statistical comparisons
ICEclc transfer rates were normalized across all experiments to that of P. putida ICEclc::lacOarray, which was taken as a control in all individual mating series. Significance of transfer rate decrease was then calculated by pair-wise two-tailed t-test comparison of independent biological triplicate matings to that of the wild-type. Subpopulations of tc cells were estimated from quantile-quantile plotting of sfGFP or mCherry fluorescence values with a 95% confidence assumption (41). Differences in foci intensities were tested from pair-wise comparison to their wild-type ICE protein fusion of the median top 5% fluorescence pixel value in biological triplicates (as proxy for the foci intensity), and of the proportion of tc cells without detectable foci (above a normalized foci intensity threshold of 6 for –sfGFP, -mCherry fusions, and 3 for Orf55476-mCherry fusions due to lower background). Median 20th percentiles and median standard deviations of cell envelope pixel fluorescence values were compared between wild-type and mutants in a two-sample equal variance t-test.
RESULTS
Function prediction of the genes in the conjugative region of ICEclc
In order to better delineate the genes on ICEclc important for conjugative transfer and to define their potential functional roles, we focused on the region in between orf53587 and orf73676 (Figure 1C) with previously shown low homology to H. influenzae ICEHin1056 (20). Low amino acid similarities (23–30%) were found in full-length alignments of some ICEclc genes and VirB-T4SS components encoded by the Ti plasmid of A. tumefaciens, notably, VirD4, VirB1, VirB2, VirB4, VirB8, VirB9 and VirB10 (Table 1). Further structure prediction using AlphaFold2 and comparison with the PDB database suggested that these may be bona fide T4SS functional homologs (Table 1, Supplementary figure S2). An additional ICE gene product (Orf73676) showed structural homology to DotD of the Dot/Icm system of L. pneumophila and contained a lipoprotein signal peptide (Table 1). DotD is a component of the T4SS outer membrane complex (42) and an analog of VirB7 of the Xanthomonas citri T4SS (43). Another gene (orf56883) showed domain similarity to TraU (Table 1) of the F-plasmid in E. coli (44).
Table 1.
Sequence and structural homology predictions of genes in the ICEclc conjugative transfer region
| ICEclc gene designationa | Length (aa) | Archetype T4SS b | Similarity %/gaps/overlap length | NCBI conserved domains/pfam | E-value | AlphaFold2/ PDBeFOLD prediction | Signal Peptide | Trans-membrane helices | Suggested gene function | Essential for transferd |
|---|---|---|---|---|---|---|---|---|---|---|
| orf53587 (CAE92904.1) | 505 | VirB6 (WP_010974917.1) | 11.9/ 482/641 | TraG_N/07916 | 2.29e-71 | No structural overlapc | – | 5 | Inner membrane protein | + |
| orf55120 (CAE92905.1) | 119 | - | None | - | FrmR formaldehyde transcriptional repressor | – | 3 | Transcription regulator | ND | |
| orf55476 (CAE92906.1) | 465 | VirB11 (WP_010974918.1) | 3.7/ 695/752 | Conjugal TIGR03755 | 0e + 00 | No structural overlap | SPI | 1 | pilT | + |
| orf56883 (CAE92907.1) | 315 | - | - | TraU superfamily | 0e + 00 | No structural overlap | SPI | – | (traU)/iceU | + |
| orf57827 (CAE92908.1) | 148 | - | - | DURF1525/ pfam07511 | 2.07e-55 | Thioredoxin-like protein | SPI | 1 | Disulfide oxidoreductase | +/– |
| orf58432 (CAE92909.1) | 164 | - | - | PRK00024 super family | 1.31e−62 | DNA repair protein RadC | – | – | Putative DNA repair protein | – |
| orf59110 (CAE92910.1) | 254 | - | - | DsbA family/ Thioredoxin-like superfamily | 5.78e−13 | Protein disulfide isomerase | – | 1 | dsbA | – |
| orf59888 (CAE92911.1) | 955 | VirB4 (WP_010974916.1) | 29.4/ 362/1053 | TraC_PFL_4706 | 0e00 | Type VII Secretion ATPase EccC | – | – | Conjugative transfer ATPase (virB4)/iceB4 | + |
| orf62755 (CAE92912.1) | 146 | - | - | conj_TIGR03751 | 5.9e−61 | No structural overlap | SPII | – | Outer membrane protein | + |
| orf63176 (CAE92913.1) | 472 | VirB10 (WP_010891494.1) | 23.2/ 289/569 | conj_TIGR03752 | 0e00 | COMB10 of a COM-type IV secretion system | – | 1 | virB10/iceB10 | + |
| orf64584 (CAE92914.1) | 310 | VirB9 (WP_010891495.1) | 29.1/ 145/374 | TraK superfamily/ pfam11920 | 1.67e−160 | No structural overlap | SPI | 1 | virB9/iceB9 | + |
| orf65513 (CAE92915.1) | 230 | VirB8 (WP_010891496.1) | 30.8/ 111/289 | conj_TIGR03746 | 1.02e−127 | Periplasmic domain of DotI/CagV | – | 1 | virB8/iceB8 | + |
| orf66202 (CAE92916.1) | 136 | VirB3 (WP_010891501.1) | 7.5/ 184/213 | DUF 3487/pfam11990 | 1.35e−48 | No structural overlap | – | 2 | Inner membrane protein | + |
| orf66625 (CAE92917.1) | 119 | VirB2 (WP_010891502.1) | 25.9/76/158 | conj_TIGR03745/ DUF2976 | 4.69e−46 | Conjugative pilus | SPI | 3 | Candidate major pilin/iceB2 | + |
| orf67001 (CAE92918.1) | 77 | - | - | conj_TIGR03758/ DUF3262 | 8.62e−21 | Membrane anchor protein of succinate DH complex | – | 2 | Inner membrane protein | + |
| orf67231 (CAE92919.1) | 127 | - | ICE_RAQPRD | 7.35e−38 | Modular repressor recruitment protein | SPI | – | Unknown | – | |
| orf67800 (CAE92920.1) | 134 | - | - | No match | - | No structural overlap | – | – | Unknown | – |
| orf68241 (CAE92921.1) | 249 | - | - | conj_TIGR03747/ DUF4400 | 3.54e−117 | No structural overlap | – | 4 | Inner membrane protein | + |
| orf68987 (CAE92922.1) | 728 | VirD4 (WP_065698658.1) | 28.3/ 366/881 | conj_TOL_TraD | 0e+00 | Coupling protein TrwB | – | 2 | virD4/iceD4 | + |
| orf71178 (CAE92923.1) | 182 | - | - | DUF 2859/ pfam11072 | 1.67e−72 | Thiol-disulfide oxidoreductase | SPI | – | Thioredoxin-like | +/– |
| orf71723 (CAE92924.1) | 196 | VirB1 (WP_010891503.1) | 31.8/ 107/274 | Lyz-like super family | 2.51e−09 | Lytic transglycosylase | SPI | – | virB1/iceB1 | +/– |
| orf72295 (CAE92925.1) | 239 | - | - | conj_TIGR03759 | 6.73e−112 | Thiol-disulfide oxidoreductase | SPI | – | Thioredoxin-like | ND |
| orf73029 (CAE92926.1) | 216 | - | - | No match | - | Membrane anchoring PilP protein. | – | – | Unknown | – |
| orf73676 (CAE92927.1) | 206 | - | - | conj_PilL | 9.91e−51 | STN superfamily periplasmic signaling domain/DotD | SPII | – | dotD/iceB7 | + |
aDesignation in GenBank AJ617740.2.
bArchetype Vir-T4SS of A. tumefaciens.
cOnly listed when RMSD > 3.
dsee Figure 2; ND, not determined.
In contrast, the products of the ICEclc genes orf53587, orf55476, orf62755, orf66202 and orf68241 displayed conserved domains to other hypothetical proteins but without function prediction, nor structural overlap to other known proteins, although with predicted transmembrane helices and/or translocation signals (Table 1). A lipoprotein signal sequence was further predicted for the orf62755 gene product, suggesting it may be localized in the outer membrane. A further number of genes in this ICEclc region code for proteins with detectable functional domains, but unrelated to the T4SS per se (e.g. a RadC DNA repair domain in Orf58432, Table 1). Several proteins have predicted functional domains related to cysteine modifications (i.e. orf57827 disulfide oxidoreductase, orf59110 protein disulphide isomerase, orf71178 thioredoxin oxidoreductase, orf72295 disulfide oxidoreductase), suggesting their implication in post-translational protein modifications. Some ICE-encoded proteins showed structural similarities with multicomponent ring-like membrane complexes, such as orf67001 and orf67231 (Table 1, Supplementary figure S2), suggesting they may be forming multi-subunit structures in the inner membrane. Finally, the orf73029 gene product showed structural similarity to a protein involved in pilus biogenesis (Table 1). Although none of the software used showed any significant similarities for the orf55476 gene product, previous alignments (20) had pointed to low amino acid similarity (41%) with a proposed PilT homolog of ICEHin1056 (15). The orf55476 protein was further predicted to carry a translocation signal peptide (Table 1). Therefore, it may encode a PilT-like ATPase of the ICEclc conjugative machinery, involved in pilus de-polymerization (45).
The sequence and structural homology searches thus suggested the following protein subassemblies for the ICEclc T4SS (Supplementary figure S2), which, whenever possible, we propose to rename analogous to the Vir-nomenclature (e.g. orf59888 becomes iceB4 in analogy to virB4, Table 1). Two ATPases, IceB4 and IceD4, may associate on the cytoplasmic side of the inner membrane. A third ATPase and PilT analog, encoded by orf55476, may be located in the periplasm. An inner membrane complex (IMC) of the T4SS may be constituted by Orf53587, IceB8, Orf66202, Orf68241 and Orf67001 (Supplementary figure S2). In analogy, IceB10 would contact and span from the inner to the outer membrane complex (OMC), further composed of IceB9 and IceB7, and possibly Orf62755. Finally, structural analysis suggested that the conjugative pilus might be formed from IceB2 (Orf66625) subunits (Supplementary figure S2). In absence of Vir-homolog, we renamed Orf56883 to IceU, following the Tra-nomenclature. A set of periplasmic proteins may complete or associate with the ICE-T4SS complex, including IceU, IceB1 and Orf67231, and the putative post-translational modifier proteins Orf57827, Orf59110, Orf71178, Orf72295 and Orf73029 (Supplementary figure S2).
Gene deletions implicate ICEclc functions in conjugative transfer
To study their potential implication in ICEclc transfer, we individually deleted 22 out of the identified 24 open reading frames in the predicted T4SS-encoding region (Figure 1, Table 1). Deletions were created in-frame by leaving a stretch of 6–10 amino acids at the N- and C-terminal of the protein, to avoid polar effects. Neither single nor double deletions could be recovered of orfs iceB2, 67001 and 67231, perhaps because of their small gene size. This was then replaced by a single deletion removing all the three consecutive genes (i.e. Δorf67231-iceB2).
Deletions of 15 of the targeted genes caused a more than 100-fold decrease of ICEclc transfer rates in isogenic P. putida matings. These comprised both genes analogous to archetype T4SS components (i.e. iceB4, iceB8, iceB9, iceB10, iceD4, and iceB7, Figure 2A; see Figure 1B for original orf nomenclature and location, and Table 1), as well as those without clear T4SS counterparts (i.e. orf53587, orf55476, iceU, orf62755, orf66202, orf67231-iceB2 and orf68241, Figure 2B). Two other deletions significantly affected ICEclc transfer, but less severely than the ones mentioned above. ICE transfer decreased by 50-fold in a ΔiceB1 (orf71723) mutant (Figure 2A), and 10-fold in a Δorf71178 mutant compared to wild type (Figure 2B). Ectopic complementation with a single-copy wild-type gene under control of the ICEclc transfer competence P67231-promoter restored transfer in all cases but one (i.e. orf53587, Figure 2A and B), indicating that deletion of orf53587 might have caused a polar effect. However, for all the other deletions the transfer defect was attributable to the single gene knockout. Several deletions, e.g. of orf55476, orf66202, orf67231-iceB2, orf71178, iceB7 or iceB10, were restored partly by ectopic complementation (Figure 2B). This might indicate that ectopic placement resulted in imbalanced expression levels or timing for functional complementation. Since the deletion of Δorf67231-iceB2 involved three consecutive genes, we further complemented this with each gene individually or in pairs, ectopically placed in single integrated copy under control of the P67231-promoter (Supplementary figure S3). Only the combination of orf67001-iceB2 partly restored transfer, suggesting they are the essential genes, whereas orf67231 is not (Supplementary figure S3). In contrast, a further set of four gene deletions did not cause detectable effects on ICE transfer (orf58432, orf59110, orf67800, and orf73029, Figure 2C), whereas deletion of orf57827 and its complementation reduced transfer by 4-fold (Figure 2C). This suggests them to have minor or no influence on ICE transfer, or to be functionally redundant.
Figure 2.
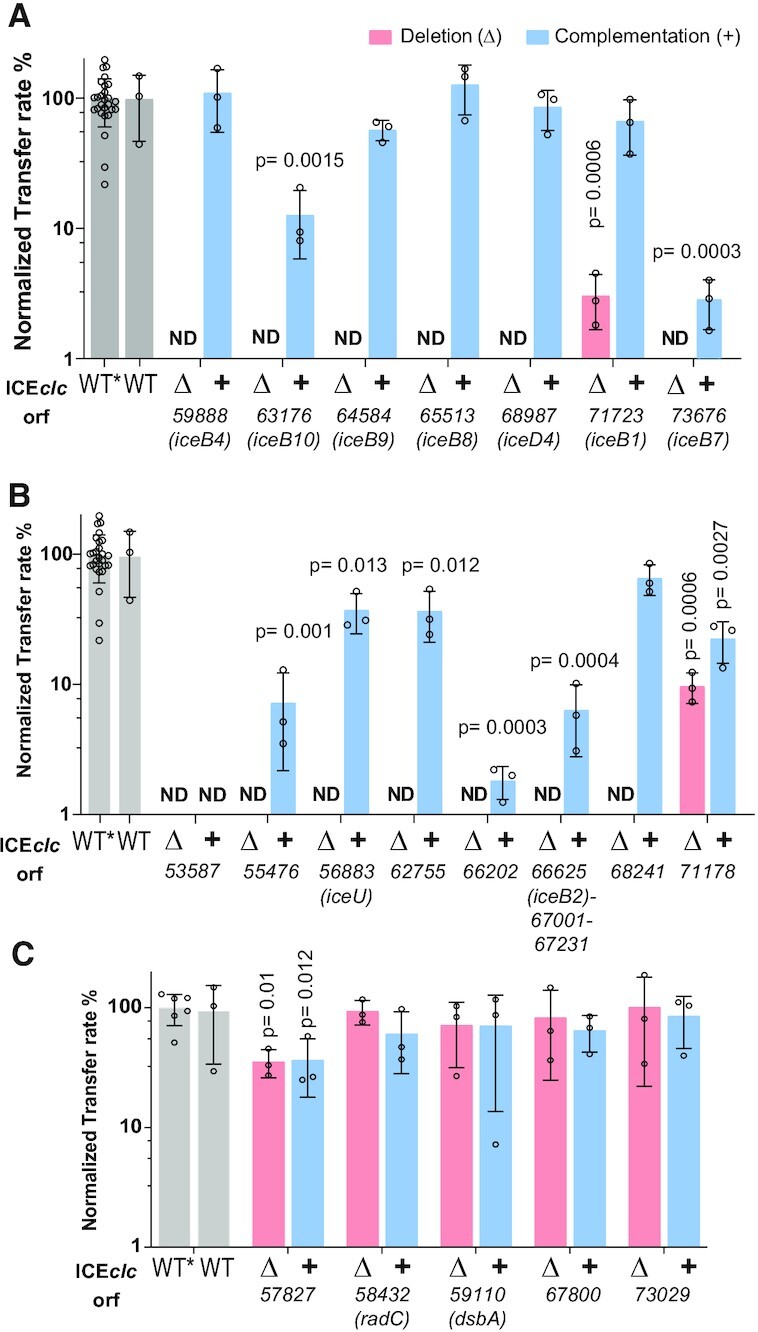
Effects of individual ICEclc gene deletion and complementation on transfer. (A) Deletion effects of ICEclc genes with distinct sequence and/or structural similarity to known T4SS components. (B) ICEclc genes without distinct sequence and structural similarity. (C) ICEclc genes without or with mild effect on transfer. Bars represent the mean normalized transfer rates of ICE with single gene deletions (Δ, salmon), or single copy complementations (+, blue), as percentage of that of wild-type ICEclc (grey) in P. putida UWC1 isogenic matings, each individually quantified as the frequency of transconjugant colony forming units (CFU) on selective medium per donor CFU. Error bars indicate calculated standard deviations on biological replicates, with small open circles showing the individual replicate values. Orf and gene names indicated below. ND, below detection limit (<1%). WT, unmodified ICEclc wild-type. WT*, ICEclc carrying an integrated lacO-array; used for producing the deletions and complementations. Note that the same WT and WT* rates are reproduced in all panels for ease of comparison. P-values were calculated with a two-tailed t-test between wild-type (WT*) and mutant replicate rates (as percentages). Only P-values <0.05 are shown.
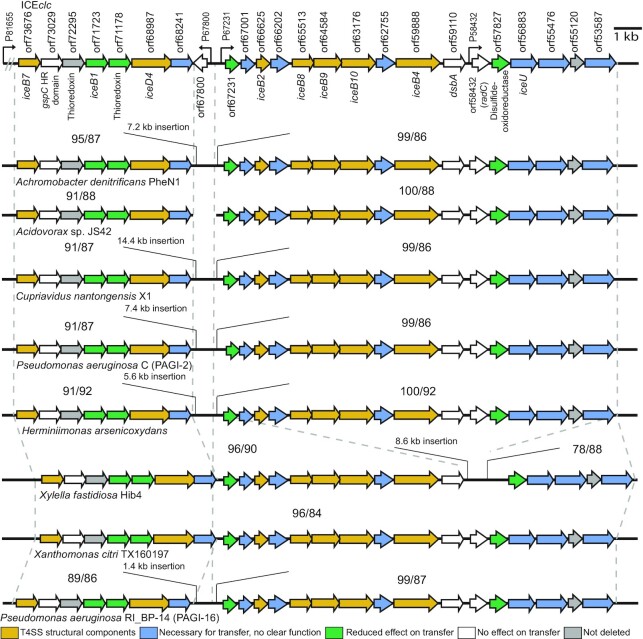
We concluded from these results that the region in between orf53587 and orf73676 on ICEclc is indeed essential for its conjugation. The predicted ICE-T4SS is very dissimilar to archetypal plasmid T4SSs, except for a few distantly structurally related components (Supplementary figure S2). In contrast, the ICE-T4SS loci and gene synteny are highly conserved between ICEclc and genome regions in other β and γ-proteobacteria, suspected to carry similar ICEs (Figure 3). This strongly suggests functional conservation, even for those genes whose deletion on ICEclc did not directly affect isogenic transfer rates. Two sites (nearby orf67800 and orf58432 on ICEclc) were permissive for gene loss in the other compared genomic regions (Figure 3). Nevertheless, the strong conservation of gene order and overall similarity is an indication that these are parts of mobile elements capable of producing T4SS conjugative systems for which ICEclc is a representative model.
Figure 3.
ICEclc transfer region gene and synteny conservation. Genetic organization of the ICEclc conjugative system gene loci (top), aligned with corresponding regions within the genomes of eight other β- and γ-Proteobacteria. The dotted lines delimit the regions of synteny to ICEclc, with numbers representing the sequence coverage (left) and the percentage of nucleotide identity (right), respectively. Arrows indicate coding frames and their orientations, with filled colors representing transfer effects as in the legend on the bottom of the figure.
Subcellular localization of the ICEclc conjugative system proteins
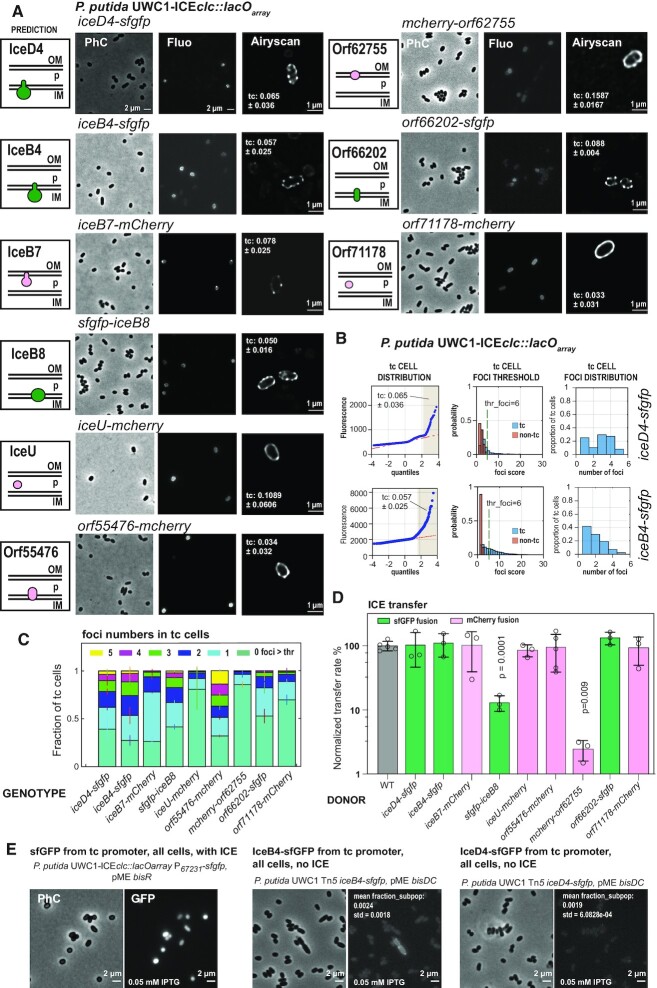
To further corroborate bioinformatic predictions and transfer results, we investigated the subcellular localization of nine of the encoded proteins in the ICEclc conjugative system gene region. This choice was not exhaustive, but contained a number of predicted structurally ‘known’ (e.g. IceB4, IceB7, IceB8) as well as less clear components (e.g. Orf66202, Orf55476, outlined in Figure 1C), and targeting both proteins predicted to be associated to the inner and outer membranes, as well as those with predicted periplasmic localization. In order to visualize protein localization, we translationally fused the target open reading frames within their original ICEclc locus with the reading frame of super-folder green fluorescent protein (sfgfp) or of mcherry, separated by a small linker peptide to avoid folding complications, as in Ref. (32). This approach was expected to best preserve stoichiometry and timing of fusion protein production with respect to the native ICEclc counterpart. Our hypothesis here was that if the targeted protein would be part of a multi-component and multi-subunit T4SS complex, expressing the fluorescent protein fusion might result in a well-defined fluorescent spot, whereas if it would not be part of the T4SS, its fluorescence would be homogenously distributed in the cytoplasm or in the cell envelope. We chose a combination of both sfGFP and mCherry in order to test for colocalization of different components (see below). To test for functionality of the expressed fluorescent fusion protein, we quantified ICE transfer from respective P. putida donors and compared this to wild-type ICE. We assumed similar transfer rates to be the best indication for proper functionality, in which case we concluded that the position and/or distribution of the observed fluorescence was representative for the true (non-labeled) protein localization.
All tested ICE-fusion proteins produced multiple clear fluorescent foci in stationary phase P. putida cells, with in about half of the cases, accentuated fluorescent cell outlines (Figure 4A). As expected from previous studies using transcriptional fusions to key ICEclc transfer competence promoters (7,21), fluorescent fusion protein expression was limited to a subpopulation of cells appearing in stationary phase (Figure 4B). These subpopulations reached similar abundances for the different constructs, as deduced from quantile-quantile plots of observed versus expected normal distribution of fluorescence values (Figure 4B, light brown-shaded regions). A co-expressed cytoplasmic marker for transfer competence (PinR-echerry) labeled the same cells as those displaying the fluorescent fusion protein-signal in stationary phase (Supplementary Figure S4A). This would be in agreement with the hypothesis that only tc cells assemble the ICE conjugative machinery. Foci were situated near the cell edge in high-resolution images (Figure 4A, Airyscan), coinciding with stained cell membranes (Supplementary Figure S4B), as expected for a membrane-spanning protein complex such as the T4SS. Because of the limited throughput in high-resolution microscopy (i.e. Figure 4A, Airyscan) we quantified foci numbers from larger numbers of cells (1000–10 000 per replicate per strain) imaged in regular epifluorescence microscopy, and using a threshold for foci detection based on differences in foci scores among non-tc and tc cells (Figure 4B). This confirmed the high-resolution visual aspects; notably, the occurrence of multiple foci per cell in case of fusions to IceD4, IceB4, IceB7, iceB8, Orf55476 and Orf66202 (Figure 4C). In contrast, fusions to IceU, Orf62755 and Orf71178 showed fewer quantifiable foci, also in agreement with the high resolution aspect of the cell envelope outlines (Figure 4C, Supplementary Figure S5).
Figure 4.
Subcellular localization of predicted ICEclc conjugative system subcomponents. (A) Micrographs of P. putida strains with indicated fluorescent-protein-tagged conjugative system components, imaged in stationary phase cultures after growth on 3-CBA. Panels on the left show the bioinformatic prediction of the protein's localization with respect to inner membrane (IM), periplasmic space (P) or outer membrane (OM); green, sfGFP-tagged protein fusion; magenta, mCherry-fusion. PhC, Phase contrast; Fluo, fluorescent channel. Airyscan, high-resolution deconvoluted 170-nm image slice of the respective strain. Proportion of observed tc cells (mean ± one SD of individual biological triplicates) indicated within the Airyscan panels. (B) Example of tc cell foci quantification on epifluorescence images of P. putida ICEclc hosts expressing either IceB4- or IceD4-sfGFP; quantile-quantile plot of median fluorescence per cell measured on a single replicate set of images for the different strains and showing the derived tc cell subpopulation (highlighted in light brown and its mean proportion ± one SD). Thresholds (thr_foci) used for distinguishing foci for the relevant indicated labeled constructs in tc cells (blue bars) from spurious background ‘foci’ in the majority of non-tc cells (red bars), based on calculated foci intensities from Gaussian fitting. Plots show probability normalized distributions of foci intensities among the two cell groups (differentiated based on quantile-quantile plots). Derived number of foci per cell at the indicated threshold (thr_foci) for a single replicate. (C) Stack plots showing the mean proportions of foci (± one SD from biological triplicates) above threshold in tc cells for all constructs shown in panel (A). (D) Wild-type normalized mean ICEclc transfer rates (± one SD from biological triplicates; circles showing individual data points) from the fluorescent protein tagged donor strains into P. putida recipients. Differences compared to wild-type were assessed with a two-tailed t-test on biological triplicate pairs (if not indicated, p-values were >0.1). (E) Micrographs of control strains P. putida UWC1 ICEclc-lacOarray expressing sfGFP from the P67231-promoter, and of P. putida UWC1 devoid of ICEclc expressing IceB4-sfGFP or IceD4-sfGFP. P67231-promoter activation was induced in all cells by overexpressing the proteins BisR or BisDC in presence of 0.05 mM IPTG, two known regulators of ICEclc activation cascade (7). Numbers indicate the mean proportion of the subpopulation of cells in quantile-quatile plots from biological triplicates (±1 SD).
ICE transfer rates from P. putida donors were unaffected for seven of the nine tested fluorescent protein fusions, suggesting no functional impairments (Figure 4D). In contrast, donors expressing sfGFP-IceB8 or mCherry-Orf62755 reduced ICE transfer by 10- and 50-fold, respectively, compared to wild-type, indicating loss of some aspect of functionality by the fluorescent protein fusion (Figure 4D).
To provide further evidence that the observed fluorescent foci were specific for conjugation complexes in P. putida ICEclc tc cells, and not the result of aberrant or spontaneous fluorescent protein aggregation, we studied control strains of P. putida ICEclc expressing only sfGFP from the same P67231 promoter but artificially induced in all cells by overexpression of the ICE-transcription factor BisR (7). These cells showed no detectable foci but homogenous cytoplasmic fluorescence, indicating that sfGFP does not self-aggregate into fluorescent foci at the cell envelope (Figure 4E and Supplementary figure S4C). Secondly, we expressed the iceB4-sfgfp or iceD4-sfgfp fusions in all cells of P. putida UWC1 devoid of ICEclc, through induction with the ICE-transcription factor complex BisDC (7). These cell populations showed very little fluorescence, with some 0.2% outliers from quantile-plotting (Figure 4E). Sporadic low-intensity foci were observed among this subpopulation of fluorescent cells (Figure 4E, Supplementary figure S4D and E), appearing at polar regions and accompanied by cellular morphological aberrations (Figure 4E). Although there is a tendency of expressed IceB4- or IceD4-sfGFP protein to aggregate into sporadic fluorescent foci, this frequency is 10- to 50-fold lower than appearing clear foci in envelopes of P. putida tc cells carrying ICEclc. It is therefore very unlikely that the multiple bright foci detected with six of the fluorescently tagged ICE conjugative proteins (Figure 4C) consist of autoaggregated misfolded proteins.
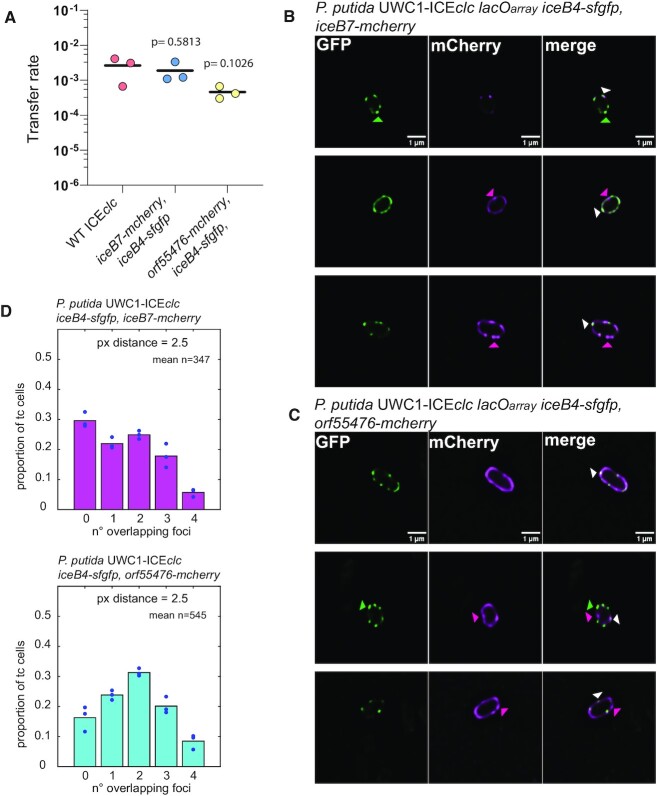
Colocalization of dual-labeled ICEclc conjugative system components
In order to determine whether the multiple foci seen in tc cells for different fluorescently-tagged ICE proteins were likely part of the same macromolecular protein complexes, we quantified the colocalization of sfGFP and mCherry signals in P. putida ICEclc hosts expressing double-labeled components. We chose for this a combination of either iceB4-sfgfp and iceB7-mcherry, or iceB4-sfgfp and orf55476-mCherry. ICE transfer rates from donors with the double label IceB4-IceB7 combination was again very close to wild-type transfer rates (Figure 5A), suggesting retained functionality. Double labeling of IceB4 and Orf55476 resulted in a tenfold decrease in mean transfer rate compared to wild-type – although this was not statistically significant (Figure 5A).
Figure 5.
Colocalization of dual-labeled components of the ICEclc conjugative system in P. putida. (A) Mean transfer rates (lines, as frequency of transconjugant CFU on selective medium per donor CFU) of ICEclc in isogenic P. putida matings from wild-type donors or ICE with dual-labeled fluorescent protein constructs, as indicated. Dots depict individual replicate transfer rates with p-values showing the probability of means being dissimilar in two-tailed t-test comparisons. (B and C) Micrographs of individual tc cells of P. putida ICEclc::lacOarray expressing both IceB4-sfGFP and IceB7-mCherry, or IceB4-sfGFP and Orf55476-mCherry, imaged in stationary phase after growth on 3-CBA in their proper individual fluorescent channel (sfGFP, mCherry), or presented as overlay (merge, GFP is green; mCherry is magenta). Images composed of a single 170–nm deconvoluted slice in Airyscan microscopy. Colored triangles in (B) and (C) point to examples of non-colocalized (green or magenta) or overlapping foci (white). (D) Measured mean proportions (bars) of the number of overlapping GFP and mCherry foci (between 1 and 4) in tc cells of both strains (overlap is within a minimum distance of 2.5 pixel or 165 nm of the fitted 2D-Gaussian foci centres). Circles are individual replicate values. n= mean number of tc cells per replicate.
We assumed foci overlap at geometric distances within 165 nm (2.5 pixels on the images), and imaged potential colocalization in deconvoluted high-resolution 170-nm cell slices. Visual inspection from high-resolution images of P. putida expressing double fusions confirmed that the two proteins are expressed simultaneously in tc cells and suggested mostly overlapping foci, but not exclusively (Figure 5B, C). Quantification of foci distances on larger numbers of tc cells from epifluorescence images, indicated that on average between 1 and 4 foci overlapped in 70% of tc cells with dual IceB4- and IceB7-labeling, and 83% of tc cells in case of strains with both IceB4-sfGFP and Orf55476-mCherry labeling (Figure 5D).
In conclusion, the microscopy and transfer experiments indicated that fluorescent foci observed in stationary phase tc cells of P. putida ICEclc are most likely representative for the formation and positions of multimeric protein complexes such as expected from the T4SS, and not the result of spontaneous aggregation of the fluorescent protein fusions. Obtaining similar foci-per-cell numbers in case of fusions to IceD4, IceB4, IceB7, IceB8 (despite its partial loss of functionality), Orf55476 and Orf66202 is very suggestive for these proteins to be part of the same multiprotein complexes, which was confirmed to some extent by the fluorescent marker overlap in double-labeled strains. The different fluorescence aspect (e.g. stronger cell envelope outlines) of the IceU- and Orf71178- fusions (the Orf62755-fusion being impaired for ICE transfer) may be an indication for them not being integral part of a T4SS and performing a different role in the ICE conjugative process.
Interdependencies among subcomponents of the ICEclc conjugative system
Finally, we studied potential dependencies of the observed foci formation on other predicted components of the ICE conjugative system. The same fluorescent protein fusion constructions were reproduced in their native locus in P. putida that further carried single copy deletions in ICEclc conjugative system genes, and effects of the deletions on fluorescent signals were examined (Figure 6A–C). Apart from direct high-resolution cell images we focused on three aspects of fluorescence measurements that we expected to yield quantifiable differences. First of all, we assumed that observed foci intensities might diminish, for example, if fewer of the tagged protein subunits would assemble into the T4SS structure, or if their turnover would be faster in absence of proper assembly. Secondly, we expected that mutations might lead to fewer fluorescent foci per cell, for example, as a result of T4SS assembly failures. This might then result in enrichment of the fluorescence signal of the tagged protein in, for example, the cell envelope. Finally, we imagined that quantifiable differences in foci ‘aspects’ might occur in mutants (i.e. blurring; visible from fitting scores and Gaussian sigma values, Supplementary Figure S1D and E), due to impaired T4SS assembly or misfolded multimeric structures. We quantified these parameters from epifluorescence imaging of tc cell subpopulations (142–1996 tc cells per replicate) in stationary phase cultures and in comparison to wild-type tagged constructs in biological independent triplicates to calculate statistical significance. We do acknowledge that at the resolution of EFM and the Airyscan, observed differences may have different underlying causes from what we assumed.
Figure 6.
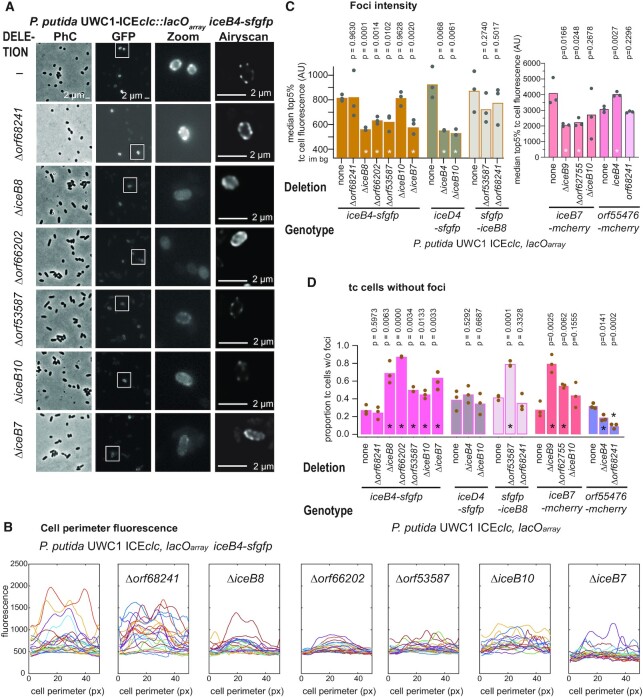
Dependencies among subcomponents of the ICEclc conjugative system. (A) Visible aspect of IceB4-sfGFP foci formation in P. putida in stationary phase after growth on 3-CBA with a single integrated copy of (otherwise) wild-type ICEclc::lacOarray, in comparison to additional deletions of orf68241, iceB8, orf66202, orf53587, iceB10, or iceB7. Micrographs as in Figure 4 with a highlighted cell area (Zoom) from the GFP image and a high-resolution Airyscan image (single 170-nm slice). Images are scaled to the same maximum and minimum intensities. (B) sfGFP fluorescence traces of the perimeter of individual tc cells (1-pixel outer ‘shell’) of the indicated constructs in panel (A) on their absolute fluorescent scales. ‘Peaks’ correspond to foci in the cell micrographs. (C) Mean top 5% median fluorescence among tc cells (bars, indicative for foci intensity) among biological triplicates of the different labeled strains in wild-type or mutant backgrounds (circles showing individual replicate values; im bg, image background fluorescence). P-values result from paired two-tailed t-test comparisons of mutant to wild-type (asterisks indicating significance at 0.05 or below). (D) As C, but for the mean proportion of tc cells without detectable foci (treshold determination as in Figure 4). All other individual strain micrographs and foci analysis are presented in Supplementary figures (see main text).
As an example, in case of IceB4-sfGFP background (Figure 6A), all tested deletions except that of orf68241, affected some aspect of fluorescent foci formation (Supplementary table S4). This was mostly quantifiable as generally weaker and more diffuse foci (Figure 6B, C, Supplementary Figure S6), and more cells without any detectable foci (Figure 6D). There was no increase in the median tc-cell cytoplasmic fluorescence intensity as a result of lowered foci intensities in mutants (Supplementary Table S5), but the estimated sizes of the tc population remained the same (Supplementary Table S5). Altogether, this would indicate that IceB4-sfGFP assembly into fluorescent foci is directly or indirectly dependent on IceB7, IceB8, Orf66202, Orf53587 and IceB10, but not on Orf68241. All of these deletions by themselves completely abolish ICE transfer (Figure 2), and, therefore, loss of IceB4-sfGFP foci formation would be in agreement with an impairment of proper multi-component T4SS assembly.
A number of other deletions were tested in the IceB7-mCherry, sfGFP-IceB8, IceD4-sfGFP and Orf55476-mCherry backgrounds, the effects of which are summarized in Figure 7. Deletions of either iceB9 or orf62755 in IceB7-mCherry background resulted in weaker and fewer foci in tc cells, whereas effects of an iceB10 deletion were inconclusive (Figure 6C and D, Supplementary Figure S7). Similar as in case of IceB4-sfGFP background, the deletion of orf68241 did not measurably change foci numbers of sfGFP-IceB8, whereas deletion of orf53587 caused foci to become weaker and more blurred (Figure 6C and D, Figure 7, Supplementary Figure S8). Both deletions of iceB4 or iceB10 yielded weaker and fewer IceD4-sfGFP foci (Figure 6C and D, Figure 7, Supplementary Figure S9). Finally, deletion of iceB4 in Orf55476-mCherry background produced more foci variability, whereas deletion of orf68241 resulted in weaker foci and clearer cell outlines (Supplementary Figure S10). Since all the deletions individually by themselves abolish ICEclc transfer, these different aspects of foci fluorescence in mutant strains (Figure 7) suggest impairments and/or losses of proper assembly of the multimeric multiprotein conjugative complexes, as will be discussed further below.
Figure 7.
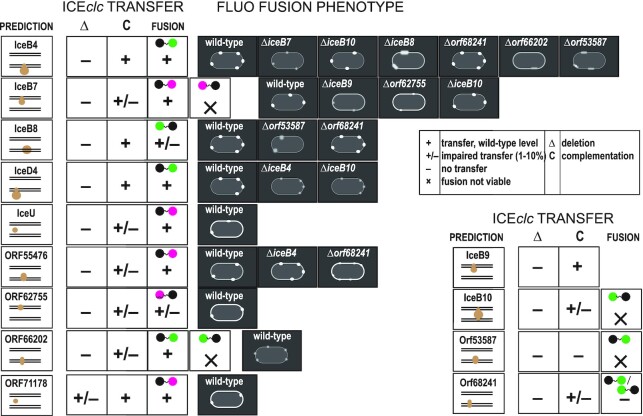
Schematic summary presentation of subcomponent dependencies in the ICEclc conjugative system, derived from transfer, mutant and fluorescent protein fusion studies. Panels show for each of the tested component the bioinformatic prediction of its localization, the ICEclc transfer frequency of its deletion mutant (Δ) and the complementation (C), and of the fusion construct (magenta, mCherry fusion; green, sfGFP; black circle, targeted protein). Fluorescent protein fusion phenotypes schematically indicated for the type (crisp, small, fuzzy) and numbers of foci observed, and cell envelope fluorescence characteristic (thick, more pronounced outline). Qualitative indication of transfer rates as in the legend, corresponding to measured values as in Figure 4D.
DISCUSSION
We performed a systematic analysis of the gene functions involved in ICEclc conjugation as representative of the poorly studied MPFG clade of T4SS conjugation systems (14). Our results indicate that ICEclc encodes a conjugative system that has both a number of structurally analogous components to known T4SS, as well as several crucial unique and unknown components (Table 1). The unique components are common to a wide class of elements related to ICEclc. We further find that formation of the ICEclc-T4SS is restricted to transfer competent (tc) cells, which can carry multiple such systems per cell as judged from microscope imaging. The occurrence of multiple T4SS per P. putida ICEclc tc cell is similar as what was found for labeled Vir-components in case of Agrobacterium tumefaciens (25,46) and from cryo-tomography of Legionella pneumophila Dot/Icm cells (27), and may contribute to the efficiency of the ICE to transfer from specialized cells (11,47).
Despite representing its rightful separate evolutionary lineage, the T4SS of ICEclc has parallel structural and functional analogies to other widespread types such as MpfT and MpfF. Our functionality analysis was based on detailed bioinformatic structural analysis, individual gene deletion and complementation studies, fluorescent fusion protein analysis, and their effects on ICE transfer. Mating-pair formation complexes can comprise between 12 and more than 20 individual proteins (48,49). We found 14–15 essential elements within the ICEclc conjugative gene region (including traI, Figure 1). Weak full length amino acid similarities and predicted structural homologies enabled to classify analogs of archetypal T4SS structural components within the ICEclc system, which we named IceB2 (for VirB2 analog, etc.), IceB4, IceB7, IceB8, IceB9, IceB10, and IceD4, as well as unknown components (e.g. Orf55476, Orf53587; Table 1). Indeed, their deletion abolished ICEclc transfer, whereas ectopic single copy complementation under control of a transfer competence promoter partly or fully restored it, indicating clear gene-dependent effects. Microscopy imaging of strains expressing translational fluorescent protein fusions from their native ICEclc loci showed that IceD4, IceB4, IceB8, and IceB7 fusions form clearly localized fluorescent foci in the cell envelope. Of these, IceD4-, IceB4- and IceB7-fusions retained wild-type transfer rates, confirming their functionality. Mutation studies in backgrounds of fluorescently-tagged ICE proteins, indicated that foci formation of IceB4-sfGFP was impaired by additional deletions of iceB7, iceB8, iceB10, orf53587 and orf66202; of IceD4-sfGFP by deletions of iceB4 and iceB10, and of IceB7-mCherry by iceB9 or orf62755 (Figure 7). Since these deletions in wild-type background completely abolish ICE-transfer, this strongly suggests that foci consist of the ICE-T4SS conjugation machines, which cannot correctly assembly in absence of their core components. This is consistent with a hypothesis of IceB4, IceB7, IceB8, IceB9, IceB10 and IceD4 being part of a multicomponent structure similar to known plasmid-type T4SSs (schematically assigned as in Supplementary figure S2A).
Various analogies can be drawn from other T4SSs and our results on the ICEclc system, which support the hypothesis of their overall structural resemblance. For example, the localization of IceB4 (the predicted cytoplasmic ATPase) to the ICEclc-T4SS required the presence of Orf66202, IceB8, Orf53587, IceB10 and IceB7 proteins (predicted inner and outer membrane components). This would be in agreement with observed dependencies of the VirB4-analog DotO protein of the Dot/Icm T4SS in L. pneumophila (50), and crucial documented protein-protein interactions between VirB4 and either VirB3 or VirB8, and between VirB3 and VirB8 (51–53). Fluorescent protein fusions to Orf66202 also formed multiple visible foci (albeit weaker than to IceB4) in the cell envelope of tc cells (Figure 7), and remained functionally intact. Orf66202 is a predicted inner membrane protein with two transmembrane helices, and, in that sense, may have a similar role as VirB3 (54), although there was no detectable sequence or structural similarity between Orf66202 and VirB3. Although we did not manage to obtain a viable fluorescent protein fusion to Orf53587 (Figure 7), deletion of its gene disturbed formation of both IceB4-sfGFP and sfGFP-IceB8 foci, suggesting it is involved in T4SS assembly. Orf53587 is also predicted to be an inner membrane protein, with five trans-membrane helices, which is analogous to the A. tumefaciens VirB6 protein (55). Therefore, Orf53587 of ICEclc may have a similar role. VirB6 and VirB8 dependencies have also been described (56), which would then be in agreement with the effects observed here for the orf53587 deletion on sfGFP-IceB8 foci. We acknowledge that sfGFP-IceB8 labeling caused a tenfold decrease of ICE transfer, indicative for partial functionality, but its foci were crisp and strongly similar in numbers to those of completely functional fusions, e.g. IceB4-sfGFP. We assume, therefore, that sfGFP-IceB8 foci are still representative for the ICEclc conjugative systems localization in the cell, which in absence of Orf53587 cannot assemble properly (Figure 7).
As far as tested here, IceD4 association to the conjugative systems seemed to be dependent on IceB4 and IceB10, which we concluded from strongly diminished IceD4-sfGFP foci intensity and foci numbers in their absence (Figure 7). IceD4 is the predicted analog of the coupling protein ATPase, which is thought to oligomerize and activate only upon substrate binding (57). IceD4, with its two predicted trans-membrane (TM) helices in the N-terminal region, probably has natural affinity for the cytoplasmic membrane. Weaker IceD4-sfGFP foci intensities in absence of iceB4 may thus be the consequence of IceD4 missing its molecular interaction partner, leading to destabilization of protein contacts, even though the T4SS core complex itself would be present. Cryo-EM studies on the R388 plasmid plasmid system showed that its VirD4 coupling protein indeed locates in close proximity of VirB4 (58). In case of iceB10 deletion, the IceD4-sfGFP foci are hardly distinguishable from spurious background (Supplementary figure S9). Unfortunately, IceB10 fluorescent protein fusions were not viable (Figure 7), and the reciprocal localization experiment could not be tested. Therefore, we have to assume that in absence of IceB10, with its predicted core structural role, the ICE conjugative system is not properly assembled; leaving IceD4 without interaction partners. This idea is supported by previous studies of the A. tumefaciens T4SS, demonstrating conformational changes in VirB10 upon ATP hydrolysis by VirD4 (59).
We further found that IceB7-mCherry foci formation depended on IceB9 (Figure 7), analogs of which are known to form together the outer membrane channel part of the T4SS (43), which suggests similar protein contacts for the ICEclc-system. The dependency of IceB7-mCherry foci formation on Orf62755 has not been previously documented. Orf62755 is a predicted outer membrane protein but without other known structural homology, and its membrane localization was illustrated by the patchy cell envelope fluorescence observed in an mCherry-fusion Orf62755-labeled strain (Figure 7). However, this fusion strain was only partly functional in ICE-transfer, and the localization of its fluorescence may not be completely informative. Orf62755 deletion strongly reduced foci formation of IceB7-mCherry, which would indicate that it is involved in T4SS assembly or a structural component of it.
Finally, one other ICEclc essential transfer protein (Orf55476), may be a further structural part of its T4SS. Orf55476-mCherry fusions were functional in ICE transfer and largely colocalized with IceB4-sfGFP foci in individual cells, but formed without strong dependence on iceB4 (Figure 7). Orf55476 is a distant relative of a proposed PilT-analog in the ICEHin1056 element (15), which are more broadly known as ATPases that energize retraction of the Type IV pilus used for twitching motility and natural transformation (60). We hypothesize, therefore, that Orf55476 is an ICEclc T4SS-associated protein; possibly implicated in its pilus dynamics. Interestingly, Orf55476-mCherry foci formation was dependent on the presence of Orf68241, itself a transfer-essential protein, but for which neither N- nor C-terminal sfGFP-fused protein was functionally active (Figure 7). As mentioned, deletion of orf68241 did not influence IceB4-sfGFP foci formation nor that of sfGFP-IceB8 (Figure 7). These results may thus indicate a specific and transient structural linkage between Orf68241 and Orf55476.
Apart from the structural T4SS analog proteins mentioned above, our analysis included predicted proteins without any sequence homology nor structural similarities to other functionally characterized proteins. One of these (IceU, product of orf56883) has an analog in TraU. The function of TraU remains unclear, although its absence was shown to reduce pilus formation and decrease transfer rates of the F-plasmid in E. coli (44). Our fluorescent reporter fusion to IceU was transfer-functional and localized to the cell envelope, producing occasional foci, in agreement with its predicted periplasmic situation (Figure 7). This suggests its role in transfer to reside in some transient interaction with T4SS complexes.
Deletions of other individual genes in the ICEclc T4SS gene region partly inhibited conjugative transfer or were without any noticeable effect altogether. Some of these have clear predicted functions, such as iceB1 (orf71723, lytic transglycosylase), orf58432 (RadC family), or orf59110 (thiol disulphide oxidoreductase, DsbA). Lytic transglycosylases are frequently present in T4SS conjugative systems (61). Their assumed role is to locally degrade peptidoglycan and facilitate T4SS complex formation. Despite this, they are not essential for conjugation (62). Indeed, iceB1 deletion reduced ICE transfer to ∼3% compared to the wild type, which is similar to what was observed for deletion of the functional analog VirB1 in the A. tumefaciens system (62). Four genes in the ICEclc T4SS locus are predicted to encode thioredoxin homologs (Table 1). Similar numbers of thioredoxins have also been documented for other T4SS, such as from the F- and R27-plasmids of E. coli (63). These proteins are supposed to facilitate folding or stabilization of the periplasmic, pilin and outer membrane subunits of the T4SS. Their mild (10-fold reduction, for orf71178; and 4-fold, for orf57827) to no influence on ICEclc transfer upon individual deletion can be explained by functional redundancies. The Orf71178-mCherry fusion was functional and localized in the cell envelope with diffuse foci (Figure 4 and 7, and Supplementary figure S5), which might be congruent with a presumed chaperone-like role. Attempts to delete orf72295 on ICEclc were not successful; therefore, it may have some other essential role. Finally, the presence of a well-conserved radC-homolog (orf58432) on ICEclc and its family members remains enigmatic. These widespread proteins were initially thought to have a role in DNA repair at the replication fork (64), and some radC are expressed during the competence state in naturally transformable bacteria (65–67). In spite of that, inactivation of radC did not influence the efficiency of DNA uptake (68,69) and also deletion of orf58432 on ICEclc had no measurable effect on conjugation rates.
The combination of mutation analysis and foci quantification gives a relatively coherent picture on the ICEclc conjugative system, but, evidently, the analysis of fluorescent foci in tc cells comes with uncertainty from limited resolution. Controls in isogenic P. putida without ICEclc but with overexpressed fluorescent proteins in all cells clearly indicate that the tc cell foci are not the result of unspecific fluorescent protein aggregation, but must be reflections of T4SS subunit multimerization. Furthermore, the combination of higher resolution Airyscan deconvoluted imaging of 170–nm cell slices with epifluorescence foci imaging across large numbers of tc cells provided quantifiable foci features with statistically sound comparisons among the various tagged protein constructs and additional ICEclc-gene deletions. Finally, there is precedent for observation of multiple fluorescent foci per cell from tagged T4SS proteins in A. tumefaciens (46), which suggested multiple T4SS units to be formed per cell. This was recently confirmed by cryo-tomography of the Dot/Icm system of L. pneumophila showing multiple T4SS complexes in individual cells (27). However, complexes seen in high-resolution microscopy by fluorescence tagging may not be consisting of the same T4SS assembly states, as demonstrated by recent cryo-EM studies of E. coli F-plasmid T4SS complexes in mini-cells, which revealed four distinct structural configurations (24).
In conclusion, we provide a detailed characterization of the ICEclc conjugative system and its subcellular localization, which is congruent to known plasmid-type T4SSs, but with a number of distinct and unknown structural elements as well as hitherto unreported dependencies. The ICEclc gene loci in this region are highly conserved in other Beta- and Gammaproteobacteria, both in sequence and gene synteny. The high conservation of T4SS gene regions in a variety of hosts is supportive for the hypothesis that they are part of other ICEclc–family elements, as proposed previously (7,17). Some of those, particularly in P. aeruginosa isolates, carry antibiotic resistance cassettes (70,71), suggesting that they encode active conjugative systems and are being selected for their efficient transfer. It is thus important to further unravel the details and potential dynamic steps in T4SS assembly and gene transfer.
DATA AVAILABILITY
The data underlying this article are available in the article and in its online supplementary material.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Nicolas Carraro and Stephan Gruber for critical reading of the manuscript, and Justine Collier for advice.
Contributor Information
Andrea Daveri, Department of Fundamental Microbiology, University of Lausanne, 1015 Lausanne, Switzerland.
Valentina Benigno, Department of Fundamental Microbiology, University of Lausanne, 1015 Lausanne, Switzerland.
Jan Roelof van der Meer, Department of Fundamental Microbiology, University of Lausanne, 1015 Lausanne, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [31003A_175638, 310030_204897]. Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Johnson C.M., Grossman A.D.. Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 2015; 49:577–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Juhas M., van der Meer J.R., Gaillard M., Harding R.M., Hood D.W., Crook D.W.. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009; 33:376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delavat F., Miyazaki R., Carraro N., Pradervand N., van der Meer J.R.. The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 2017; 41:512–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wozniak R.A., Waldor M.K.. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010; 8:552–563. [DOI] [PubMed] [Google Scholar]
- 5. Burrus V., Waldor M.K.. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004; 155:376–386. [DOI] [PubMed] [Google Scholar]
- 6. Beaber J.W., Hochhut B., Waldor M.K.. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004; 427:72–74. [DOI] [PubMed] [Google Scholar]
- 7. Carraro N., Richard X., Sulser S., Delavat F., Mazza C., van der Meer J.R.. An analog to digital converter controls bistable transfer competence development of a widespread bacterial integrative and conjugative element. Elife. 2020; 9:e57915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Auchtung J.M., Lee C.A., Monson R.E., Lehman A.P., Grossman A.D.. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:12554–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reinhard F., Miyazaki R., Pradervand N., van der Meer J.R.. Cell differentiation to “mating bodies” induced by an integrating and conjugative element in free-living bacteria. Curr. Biol. 2013; 23:255–259. [DOI] [PubMed] [Google Scholar]
- 10. Minoia M., Gaillard M., Reinhard F., Stojanov M., Sentchilo V., van der Meer J.R.. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20792–20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delavat F., Moritz R., van der Meer J.R.. Transient replication in specialized cells favors transfer of an integrative and conjugative element. Mbio. 2019; 10:e01133-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fronzes R., Christie P.J., Waksman G.. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 2009; 7:703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costa T.R.D., Harb L., Khara P., Zeng L., Hu B., Christie P.J.. Type IV secretion systems: advances in structure, function, and activation. Mol. Microbiol. 2021; 115:436–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guglielmini J., Neron B., Abby S.S., Garcillan-Barcia M.P., de la Cruz F., Rocha E.P.. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014; 42:5715–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Juhas M., Crook D.W., Dimopoulou I.D., Lunter G., Harding R.M., Ferguson D.J., Hood D.W.. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 2007; 189:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sentchilo V., Czechowska K., Pradervand N., Minoia M., Miyazaki R., van der Meer J.R.. Intracellular excision and reintegration dynamics of the ICEclc genomic island of Pseudomonas knackmussii sp. Strain B13. Mol. Microbiol. 2009; 72:1293–1306. [DOI] [PubMed] [Google Scholar]
- 17. Miyazaki R., Bertelli C., Benaglio P., Canton J., De Coi N., Gharib W.H., Gjoksi B., Goesmann A., Greub G., Harshman K.et al.. Comparative genome analysis of Pseudomonas knackmussii B13, the first bacterium known to degrade chloroaromatic compounds. Environ. Microbiol. 2015; 17:91–104. [DOI] [PubMed] [Google Scholar]
- 18. Sentchilo V., Zehnder A.J., van der Meer J.R.. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. Strain B13. Mol. Microbiol. 2003; 49:93–104. [DOI] [PubMed] [Google Scholar]
- 19. Miyazaki R., van der Meer J.R.. A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol. Microbiol. 2011; 79:743–758. [DOI] [PubMed] [Google Scholar]
- 20. Gaillard M., Pradervand N., Minoia M., Sentchilo V., Johnson D.R., van der Meer J.R.. Transcriptome analysis of the mobile genome iceclc in Pseudomonas knackmussii B13. BMC Microbiol. 2010; 10:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sulser S., Vucicevic A., Bellini V., Moritz R., Delavat F., Sentchilo V., Carraro N., van der Meer J.R.. A bistable prokaryotic differentiation system underlying development of conjugative transfer competence. PLoS Genet. 2022; 18:e1010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu B., Khara P., Christie P.J.. Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:14222–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aguilar J., Zupan J., Cameron T.A., Zambryski P.C.. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosal D., Jeong K.C., Chang Y.W., Gyore J., Teng L., Gardner A., Vogel J.P., Jensen G.J.. Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat Microbiol. 2019; 4:1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park D., Chetrit D., Hu B., Roy C.R., Liu J.. Analysis of dot/Icm type IVB secretion system subassemblies by cryoelectron tomography reveals conformational changes induced by DotB binding. Mbio. 2020; 11:e03328-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerhardt P., Murray R.G.E., Costilow R.N., Nester E.W., Wood W.A., Krieg N.R., Briggs Phillips G.. Manual of methods for general bacteriology. 1981; Washington, D.C: American Society for Microbiology. [Google Scholar]
- 29. Gaillard M., Vallaeys T., Vorhölter F.J., Minoia M., Werlen C., Sentchilo V., Pühler A., van der Meer J.R.. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 2006; 188:1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez-Garcia E., de Lorenzo V.. Engineering multiple genomic deletions in gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 2011; 13:2702–2716. [DOI] [PubMed] [Google Scholar]
- 31. Martinez-Garcia E., Calles B., Arevalo-Rodriguez M., de Lorenzo V.. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 2011; 11:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyazaki R., Minoia M., Pradervand N., Sulser S., Reinhard F., van der Meer J.R.. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLos Genet. 2012; 8:e1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grote A., Hiller K., Scheer M., Munch R., Nortemann B., Hempel D.C., Jahn D.. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005; 33:W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 35. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holm L. DALI and the persistence of protein shape. Protein Sci. 2020; 29:128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 38. Nielsen H., Engelbrecht J., Brunak S., von Heijne G.. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 1997; 8:581–599. [DOI] [PubMed] [Google Scholar]
- 39. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta. Crystallogr. D Biol. Crystallogr. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 40. Stylianidou S., Brennan C., Nissen S.B., Kuwada N.J., Wiggins P.A.. SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Mol. Microbiol. 2016; 102:690–700. [DOI] [PubMed] [Google Scholar]
- 41. Reinhard F., van der Meer J.R.. Improved statistical analysis of low abundance phenomena in bimodal bacterial populations. PLoS One. 2013; 8:e78288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakano N., Kubori T., Kinoshita M., Imada K., Nagai H.. Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems. PLoS Pathog. 2010; 6:e1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Durie C.L., Sheedlo M.J., Chung J.M., Byrne B.G., Su M., Knight T., Swanson M., Lacy D.B., Ohi M.D.. Structural analysis of the Legionella pneumophila dot/icm type IV secretion system core complex. Elife. 2020; 9:e59530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore D., Maneewannakul K., Maneewannakul S., Wu J.H., Ippen-Ihler K., Bradley D.E.. Characterization of the F-plasmid conjugative transfer gene traU. J. Bacteriol. 1990; 172:4263–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chlebek J.L., Hughes H.Q., Ratkiewicz A.S., Rayyan R., Wang J.C., Herrin B.E., Dalia T.N., Biais N., Dalia A.B.. PilT and PilU are homohexameric atpases that coordinate to retract type iva pili. PLoS Genet. 2019; 15:e1008448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aguilar J., Cameron T.A., Zupan J., Zambryski P.. Membrane and core periplasmic agrobacterium tumefaciens virulence type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells. Mbio. 2011; 2:e00218-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Delavat F., Mitri S., Pelet S., van der Meer J.R.. Highly variable individual donor cell fates characterize robust horizontal gene transfer of an integrative and conjugative element. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E3375–E3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Christie P.J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E.. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005; 59:451–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kubori T., Nagai H.. The type IVB secretion system: an enigmatic chimera. Curr. Opin. Microbiol. 2016; 29:22–29. [DOI] [PubMed] [Google Scholar]
- 50. Chetrit D., Hu B., Christie P.J., Roy C.R., Liu J.. A unique cytoplasmic atpase complex defines the Legionella pneumophila type IV secretion channel. Nat Microbiol. 2018; 3:678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ward D.V., Draper O., Zupan J.R., Zambryski P.C.. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yuan Q., Carle A., Gao C., Sivanesan D., Aly K.A., Hoppner C., Krall L., Domke N., Baron C.. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 2005; 280:26349–26359. [DOI] [PubMed] [Google Scholar]
- 53. Mossey P., Hudacek A., Das A.. Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J. Bacteriol. 2010; 192:2830–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones A.L., Shirasu K., Kado C.I.. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 1994; 176:5255–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jakubowski S.J., Krishnamoorthy V., Cascales E., Christie P.J.. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 2004; 341:961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Casu B., Mary C., Sverzhinsky A., Fouillen A., Nanci A., Baron C.. VirB8 homolog TraE from plasmid pKM101 forms a hexameric ring structure and interacts with the VirB6 homolog TraD. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:5950–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matilla I., Alfonso C., Rivas G., Bolt E.L., de la Cruz F., Cabezon E.. The conjugative DNA translocase TrwB is a structure-specific DNA-binding protein. J. Biol. Chem. 2010; 285:17537–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Redzej A., Ukleja M., Connery S., Trokter M., Felisberto-Rodrigues C., Cryar A., Thalassinos K., Hayward R.D., Orlova E.V., Waksman G.. Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J. 2017; 36:3080–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cascales E., Christie P.J.. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17228–17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Craig L., Forest K.T., Maier B.. Type IV pili: dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 2019; 17:429–440. [DOI] [PubMed] [Google Scholar]
- 61. Hoppner C., Liu Z., Domke N., Binns A.N., Baron C.. VirB1 orthologs from Brucella suis and pKM101 complement defects of the lytic transglycosylase required for efficient type IV secretion from Agrobacterium tumefaciens. J. Bacteriol. 2004; 186:1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mushegian A.R., Fullner K.J., Koonin E.V., Nester E.W.. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:7321–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Elton T.C., Holland S.J., Frost L.S., Hazes B.. F-like type IV secretion systems encode proteins with thioredoxin folds that are putative DsbC homologues. J. Bacteriol. 2005; 187:8267–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saveson C.J., Lovett S.T.. Tandem repeat recombination induced by replication fork defects in Escherichia coli requires a novel factor, RadC. Genetics. 1999; 152:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Redfield R.J., Cameron A.D., Qian Q., Hinds J., Ali T.R., Kroll J.S., Langford P.R.. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J. Mol. Biol. 2005; 347:735–747. [DOI] [PubMed] [Google Scholar]
- 66. Berka R.M., Hahn J., Albano M., Draskovic I., Persuh M., Cui X., Sloma A., Widner W., Dubnau D.. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 2002; 43:1331–1345. [DOI] [PubMed] [Google Scholar]
- 67. Rimini R., Jansson B., Feger G., Roberts T.C., de Francesco M., Gozzi A., Faggioni F., Domenici E., Wallace D.M., Frandsen N.et al.. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 2000; 36:1279–1292. [DOI] [PubMed] [Google Scholar]
- 68. Ogura M., Yamaguchi H., Kobayashi K., Ogasawara N., Fujita Y., Tanaka T.. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 2002; 184:2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Attaiech L., Granadel C., Claverys J.P., Martin B.. RadC, a misleading name?. J. Bacteriol. 2008; 190:5729–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hong J.S., Yoon E.J., Lee H., Jeong S.H., Lee K.. Clonal dissemination of Pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob. Agents Chemother. 2016; 60:7216–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Roy Chowdhury P., Scott M., Worden P., Huntington P., Hudson B., Karagiannis T., Charles I.G., Djordjevic S.P. Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol. 2016; 6:150175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.