Abstract
Recently, cognitive impairments (CI) and behavioral abnormalities in patients with amyotrophic lateral sclerosis (ALS) have been reported. However, the underlying mechanisms have been poorly understood. In the current study, we explored the role of gut microbiota in CI of ALS patients. We collected fecal samples from 35 ALS patients and 35 healthy controls. The cognitive function of the ALS patients was evaluated using the Edinburgh Cognitive and Behavioral ALS Screen. We analyzed these samples by using 16S rRNA gene sequencing as well as both untargeted and targeted (bile acids) metabolite mapping between patients with CI and patients with normal cognition (CN). We found altered gut microbial communities and a lower ratio of Firmicutes/Bacteroidetes in the CI group, compared with the CN group. In addition, the untargeted metabolite mapping revealed that 26 and 17 metabolites significantly increased and decreased, respectively, in the CI group, compared with the CN group. These metabolites were mapped to the metabolic pathways associated with bile acids. We further found that cholic acid and chenodeoxycholic acid were significantly lower in the CI group than in the CN group. In conclusion, we found that the gut microbiota and its metabolome profile differed between ALS patients with and without CI and that the altered bile acid profile in fecal samples was significantly associated with CI in ALS patients. These results need to be replicated in larger studies in the future.
Keywords: amyotrophic lateral sclerosis, cognitive decline, gut microbiota, fecal metabolites, bile acids
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder characterized by a progressive degeneration of the upper and lower motor neurons[1]. Motor functions of ALS patients would usually deteriorate within three to five years, and they might suffer from respiratory failure in the late stages[1]. ALS is devastating to both patients and caregivers due to an absence of curative treatment. Many neuroscientists and neurologists have observed that cognitive and behavioral abnormalities are common among ALS patients[2]. Approximately 35% of ALS patients have comorbid cognitive impairments (CI)[3]. Cognitive decline severely impairs treatment compliance among patients and causes a burden on caregivers[4], leading to shortened survival spans and dampening the quality of life[5]. Cognitive decline in ALS patients primarily manifests with executive, language, and visual space dysfunction[6]. A subpopulation of ALS patients could develop behavioral impairments, including loss of interest, apathy, absence of insight, irritability, and aggression[7–8]. However, the underlying mechanisms remain unknown.
Among genes related to the pathogenesis of ALS, hexanucleotide GGGGCC repeat expansions in the C9orf72 gene explain the association between ALS and frontotemporal dementia. About 5%–15% of ALS patients satisfy the diagnostic criteria for frontotemporal dementia[9]. However, this C9orf72 mutation barely occurs in Chinese ALS patients[10–11]. Our clinical data suggested that about 38.7% of ALS patients exhibited CI and that 31.5% had behavioral abnormalities[12]. Therefore, we proposed an alternative mechanism other than genetic susceptibility that may contribute to the impaired cognition in ALS patients.
Recent studies have elucidated a link between gut microbiota and CI in neurodegenerative disorders, such as Alzheimer's (AD) and Parkinson's disease[13–14]. The altered gut microbiota play a role in the CI of patients suffering from neurodegenerative diseases[15]. A few microbiomics studies combined with metabolomics have shedded light on cognitive impairment mechanisms in patients with neurodegenerative diseases, including regulating microglial function using secreted metabolites and neurotransmitters[16–17]. Although several global study groups have reported altered gut microbiota in ALS patients compared with healthy controls, the conclusions are inconsistent[18–22]. To date, no studies have uncovered the mechanistic links between CI and gut microbiota in ALS patients.
In the current study, we first conducted 16S rRNA sequencing of fecal samples from both ALS patients and healthy controls. Then we further compared gut microbiota and untargeted fecal metabolites of the ALS patients with and without CI. Biostatistical analyses revealed that the extracted metabolites significantly differed between the two groups and that these metabolites were mapped to metabolic pathways associated with bile acids. We attempted to identify a link between the gut microbiota and CI in ALS patients, providing clues for future mechanistic studies.
Materials and methods
Study design, registrations, and patient consents
We conducted a case-control study of 35 patients with ALS and 35 age- and sex-matched healthy controls and also explored the changes in gut microbiota and fecal metabolites between ALS patients with and without cognitive decline. The current study was approved by the ethics committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (TJ-IRB20201219). All participants provided written informed consent, and the study was conducted following the Declaration of Helsinki.
Subject recruitment and fecal sample collection
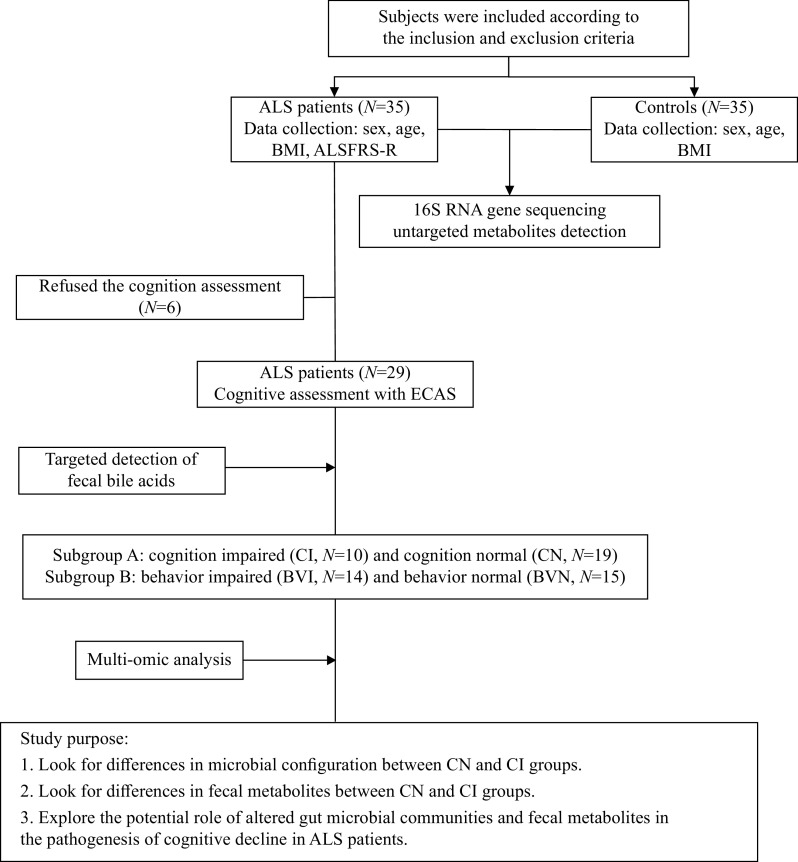
The study design is described in Fig. 1. Briefly, 35 ALS patients diagnosed with the revised El Escorial criteria[23] and 35 age- and sex-matched healthy controls were recruited between September 2020 and December 2021. Exclusion criteria were incorporated to exclude diseases with a clear impact on the gut microbiota (Supplementary Table 1, available online). Specifically, patients with severe dysphagia were excluded to reduce the biases in nutrient intake. Two fecal samples of each participant were collected from the first bowel moment of the day, and the samples were collected in 15 mL falcon tubes and stored at −80 ℃.
Figure 1. The research flow chart.
ALS: amyotrophic lateral sclerosis; BMI: body mass index; ALSFRS-R: revised ALS Functional Rating Scale; ECAS: Edinburgh Cognitive and Behavioral ALS Screen.
Assessment of motor function and cognitive function in ALS patients
The Revised ALS Functional Rating Scale (ALSFRS-R) was performed to assess disease severity of ALS patients[24]. The Edinburgh Cognitive and Behavioral ALS Screen (ECAS) is specifically designed to identify cognitive and behavioral changes in ALS patients[6]. The ECAS evaluates three ALS-specific cognitive domains (language, verbal fluency, and executive function) and two ALS-non-specific cognitive domains (memory and visuospatial function). In the current study, we screened ALS patients with CI and ALS patients with normal cognition (CN), based on the cut-off value of total ECAS scores in Chinese population[6]. Additionally, ECAS includes a brief questionnaire to detect behavioral abnormalities, completed by primary caregivers. Patients with an abnormality in at least one behavioral domain were considered to have behavioral impairments (BVI); otherwise, behavior normal (BVN). Two trained investigators performed the ALSFRS-R and the Chinese version of ECAS.
16S rRNA gene sequencing and microbiome analysis
The amplicon sequencing of the 16S rRNA gene was conducted by the Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (China), as previously described[25]. Briefly, the DNA was extracted (Cat. No. DP712, Tiangen, Beijing, China) and the V3-V4 variable region was amplified with barcode-indexed primers (338F and 806R). Then, amplicons were sequenced using the Illumina MiSeq platform (PE300). The raw sequences were processed as previously described[25]. The sequence data has been deposited in the National Genomics Data Center (https://ngdc.cncb.ac.cn/, PRJCA006335). Sobs, Chao, and Ace index were calculated (Mothur, v1.30.2) to evaluate the richness and evenness (α diversity) of the microbial communities[26–27]. Principal coordinate analysis (PCoA) and permutational multivariate analysis of variance were used for the comparison of β diversity based on the Bray-Curtis distance[28]. The linear discriminant analysis (LDA) of effect size (LEfSe) revealed representative bacterial taxa in each group[29], and the threshold was an LDA score >3.
Fecal metabolite detection and metabolomic analysis
The Shanghai Majorbio Bio-Pharm Technology Co., Ltd. performed the detection of fecal metabolites. The metabolites were separated using an ExionLC AD System (AB Sciex, Framingham, USA). The Ultra Performance Liquid Chromatography (UPLC) system was coupled with a quadrupole time-of-flight mass spectrometer (AB SCIEX-Triple TOF 5600+, Framingham, USA) and electrospray ionization source. The data acquisition was performed in the Data Dependent Acquisition mode. The metabolites were annotated based on the Human Metabolome Database (HMDB) and Metlin mass spectral database. The orthogonal partial least squares discriminate analysis (OPLS-DA) showed the similarity of fecal metabolites among different groups. Representative metabolites from each group were selected based on the Variable Importance in the Projection (VIP) value in OPLS-DA and the P-value within the comparison of concentrations (VIP score >1, P<0.05). The metabolites were mapped to biochemical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) for exploring metabolic functions. In the bile acids-targeted detection, the ion fragments were identified in AB SCIEX quantification software OS and manually checked. The concentration of 46 bile acids was evaluated based on the standard curve plotted using the mass spectral peak area as the ordinate and the analytic concentration.
Statistical analysis
Continuous variables are presented as mean (SD) or median (including 25% quartile and 75% quartile). In comparing the two groups, Student's t-test was used for normally distributed data, Mann-Whitney U-test was used for non-normally distributed data, and Fisher's exact probability test was employed for categorical data. Spearman's correlation coefficient was calculated in the correlation analysis. P<0.05 was considered statistically significant. Statistical analysis was performed using SPSS (version 23.0). Graphs were drawn using R (version 3.4.1) and GraphPad Prism 7.0.
Results
Demographic and clinical characteristics
The same number of ALS patients and healthy controls (35 each) were enrolled. No significant difference was observed among age, body mass index (BMI), and sex ratio between the cases and controls (Table 1). Among the 35 ALS patients, 32 had a spinal onset, and three had a bulbar onset. The median disease duration in these patients with ALS was 10 months, and the median score of ALSFRS-R was 43.
Table 1. Demographic and clinical characteristics.
| Parameters | Participants | P-value1 | P-value2 | |||
| ALS (N=35) | HC (N=35) | CN (N=19) | CI (N=10) | |||
|
Data are presented as mean (SD) or median (25% quantile, 75% quantile). P-values were calculated by Student's t-testa, Mann-Whitney U-testb or Fisher's exact-testc. P-value1: ALS vs. HC; P-value2: CN vs. CI. ALS: amyotrophic lateral sclerosis; HC: healthy control; CN: normal cognition; CI: cognitive impairments; BMI: body mass index; ALSFRS-R: revised ALS Functional Rating Scale; ECAS: the Edinburgh Cognitive and Behavioral ALS Screen; SD: standard deviation. | ||||||
| Demographic information | ||||||
| Age (years) | 54 (8) | 54 (9) | 50 (6) | 59 (10) | 0.967a | 0.030a |
| Female/Male | 14/21 | 14/21 | 8/11 | 4/6 | 1c | 0.615c |
| BMI (kg/m2) | 22.15 (2.69) | 22.77 (2.11) | 22.35 (2.97) | 22.02 (3.01) | 0.291a | 0.776a |
| Clinical characteristics | ||||||
| Duration (months) | 10 (6, 24) | – | 7 (4, 15) | 24 (12, 24) | – | 0.006b |
| ALSFRS-R | 43 (39, 46) | – | 43 (39, 46) | 43 (39, 45) | – | 0.626b |
| ALSFRS-R, Bulb scores | 12 (10, 12) | – | 12 (11, 12) | 11 (9, 12) | – | 0.129b |
| ALSFRS-R, Respiration scores | 12 (12, 12) | – | 12 (12, 12) | 12 (12, 12) | – | 0.490b |
| ECAS | – | – | 97 (11) | 59 (17) | – | - |
Microbial and metabolomic differences between ALS patients and healthy controls
According to the 16S rRNA gene data, there was no difference in the α diversity (P>0.05) between ALS patients and healthy controls (Fig. 2A). Regarding β diversity, PCoA analysis depicted a significant difference between ALS patients and healthy controls (P<0.05) (Fig. 2B). Compared with the healthy control group, the phylum Proteobacteria significantly decreased in the ALS patient group (Fig. 2C). The Firmicutes to Bacteroidetes (F/B) ratio was comparable between ALS patients and healthy controls (Supplementary Table 2, available online). Additionally, the Lefse analysis revealed distinctive bacterial species of each group and the subordination of bacterial species (Fig. 2D and E).
Figure 2. Difference in microbial community and fecal metabolites between ALS patients and healthy controls.
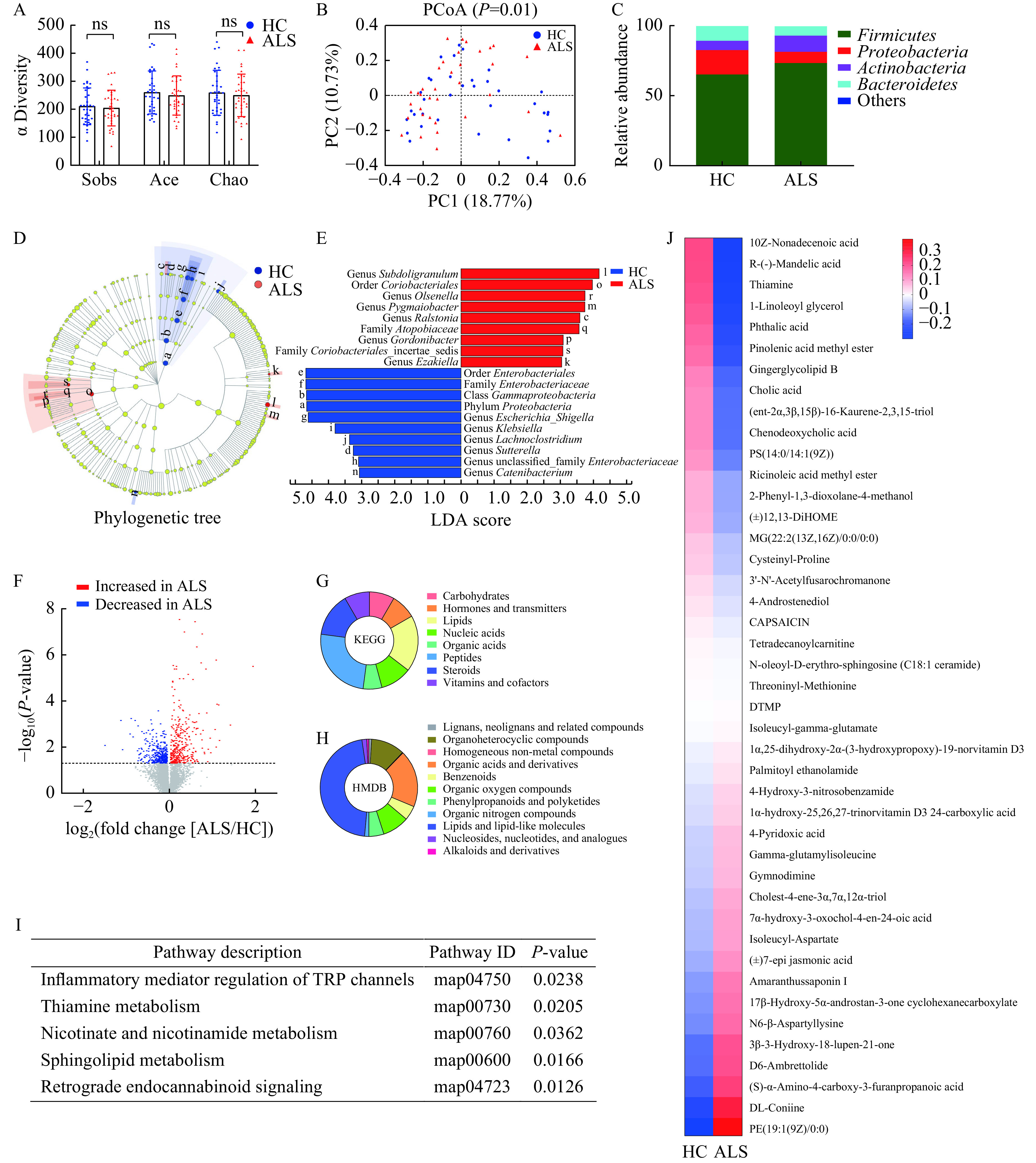
A: Comparison of microbial α diversity (assessed by Sobs, Ace and Chao index). B: Comparison of microbial β diversity (assessed by PCoA). C: Relative abundance of the microbial community at the phylum level. D and E: Distinguishing bacterial taxa identified by LEfSe analysis. Cladogram showing the phylogenetic distribution of the bacterial lineages. Circles indicate phylogenetic levels from domain to genus. The diameter of each circle is proportional to the abundance of the bacterial group (D). The letters show the distinguishing taxa with a LDA score >3 (E). F: Comparison of fecal metabolites. Among 14 108 metabolites detected, 350 increased in ALS (P<0.05), while 339 were reduced in ALS (P<0.05), compared with the HC group. G and H: The classification of 41 distinguishing fecal metabolites (P<0.05, VIP score >1) were identified in database. Compounds with biological roles in KEGG (G). Superclass metabolites in HMDB database (H). I: In KEGG, 41 distinguishing fecal metabolites between ALS and HC were mapped to five metabolic pathways. J: Relative concentrations of 41 distinguishing fecal metabolites (P<0.05, VIP score >1) between ALS and HC. ns: not significant; ALS: amyotrophic lateral sclerosis; HC: healthy controls; PCoA: Principal coordinates analysis; LEfSe: the Linear discriminant analysis of effect size; VIP: Variable Importance in the Projection; KEGG: Kyoto Encyclopedia of Genes and Genomes; HMDB: Human Metabolome Database.
For the untargeted metabolite mapping, 29 ALS patients and 23 healthy controls were included. A total of 14108 metabolites were detected, among which 41 statistically different metabolites were identified from the KEGG and HMDB database (P<0.05) (Fig. 2F–H). Twenty-two metabolites increased, and 19 metabolites decreased, respectively, in the ALS patient group, compared with the healthy control group (Fig. 2J). In the KEGG pathway enrichment analysis, those 41 metabolites were mapped to five metabolic pathways (P<0.05), including the retrograde endocannabinoid signaling, inflammatory mediator regulation of the transient receptor potential channels, sphingolipid, nicotinamide, and thiamine metabolism (Fig. 2I).
Microbial difference between ALS patients with and without cognitive impairment
Among 29 ALS patients who underwent the ECAS, 10 (34.48%) had CI, and 19 patients (65.52%) had normal cognition (Fig. 3A and B), according to the cut-off score of ECAS in Chinese ALS patients[6]. Fourteen patients (48.28%) exhibited at least a single type of BVI (Fig. 3B), and six patients showed CI combined with abnormal behaviors (Fig. 3C). The microbial α diversity was similar between the CN and CI groups (P>0.05) (Fig. 3D). The microbial β diversity was significantly different between the CN and CI groups based on the PCoA (P<0.05) (Fig. 3E). The phylum Bacteroidetes (P<0.05) and Proteobacteria (P>0.05) elevated, while Firmicutes and Actinobacteria reduced in the CI group (P>0.05) compared with the CN group (Fig. 3F and G). The F/B ratio was significantly lower in the CI group than that in the CN group (P<0.05) (Supplementary Table 3, available online). In Lefse analysis, the CI group revealed a higher abundance of bacterial species belonging to Bacteroidia and Verrucomicrobiae classes, while the CN group showed a higher abundance of bacterial species belonging to the class Erysipelotrichia (Fig. 3H and I).
Figure 3. Difference in the microbial community between patients with CN and CI.
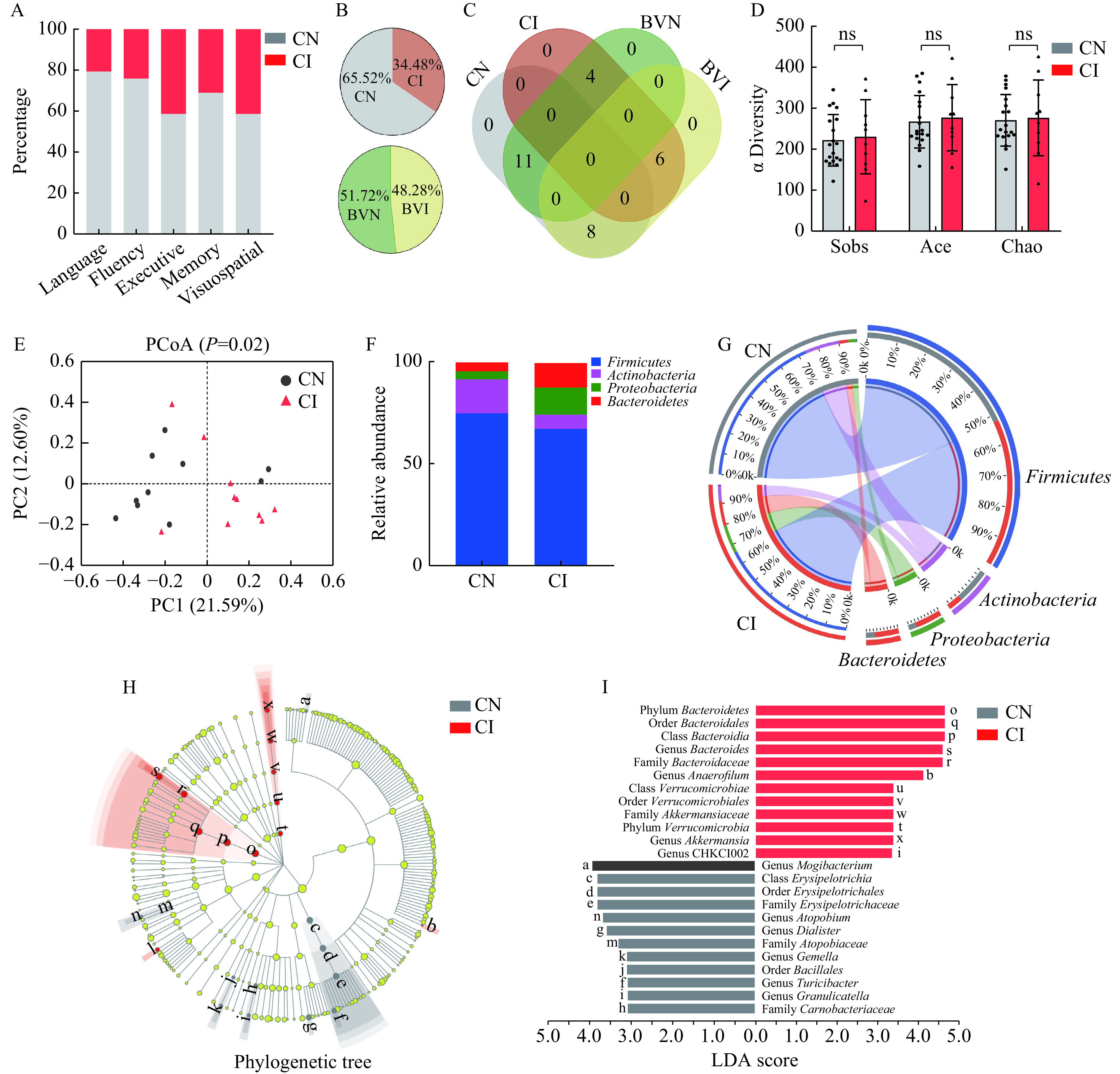
A–C: Cognitive function of patients with ALS in the current study. Frequency of patients with CI in five subdomains of ECAS (A). Frequency of patients with CI and behavioral impairments (B). Coexistence of CI and behavioral impairments in patients with ALS (C). D: Comparison of microbial α diversity between the CN and CI groups (assessed by Sobs, Ace, and Chao index). E: Comparison of microbial β diversity between the CN and CI groups (assessed by PCoA). F and G: Relative abundance of the microbial community at phylum level between CN and CI. H and I: Distinguishing bacterial taxa between the CN and CI groups was identified by LEfSe analysis. Cladogram showing the phylogenetic distribution of the bacterial lineages. Circles indicate phylogenetic levels from domain to genus. The diameter of each circle is proportional to the abundance of the bacterial group (H). The letters show the distinguishing taxa with a LDA score >3 (I). ns: not significant; ECAS: Edinburgh Cognitive and Behavioral ALS Screen; CN: normal cognition; CI: cognitive impairments; BVN: behavior normal; BVI: behavior impaired; PCoA: Principal coordinates analysis; LEfSe: the Linear discriminant analysis of effect size.
Fecal metabolite difference between patients with and without CI
The OPLS-DA analysis separates the CN and CI groups into discrete distributions, indicating distinctive metabolomic characteristics (Fig. 4B). Among 43 metabolites identified in the KEGG and HMDB, 26 metabolites decreased and 17 metabolites increased in the CI group (Fig. 4C–E), respectively, compared with the CN group. Those 43 metabolites were mapped to six metabolic pathways, including the sulfur relay system, antifolate resistance, vitamin B6 metabolism, bile secretion, primary bile acid biosynthesis, and secondary bile acid biosynthesis (Fig. 4F). Patients with CI showed unique metabolic characteristics in fecal samples. Notably, three of six distinctive metabolic pathways were linked to bile acid metabolism.
Figure 4. Difference in fecal metabolites between patients with CN and CI.
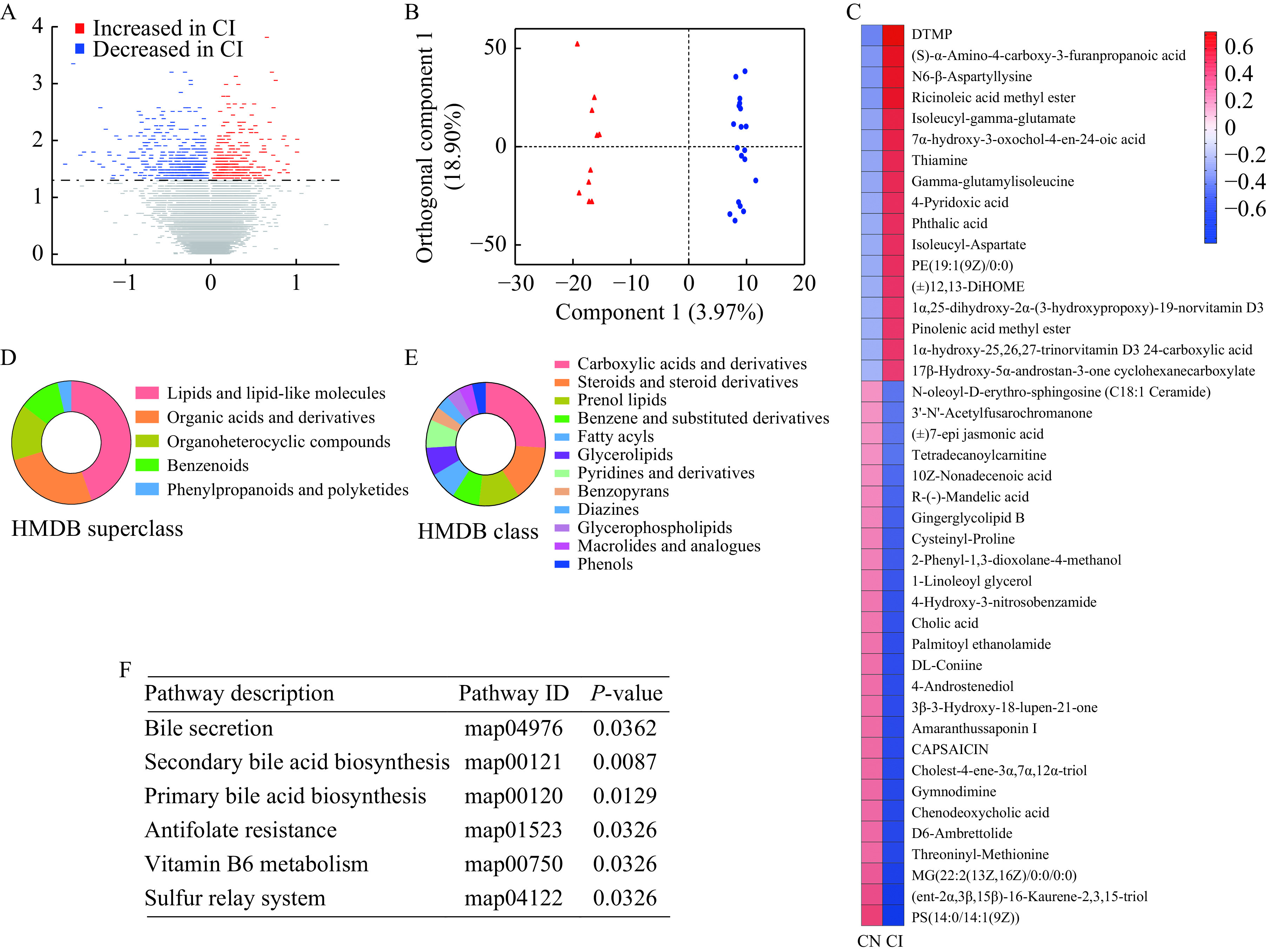
A: Comparison of fecal metabolites. Three hundred and thirty metabolites increased (P<0.05), and 369 metabolites decreased (P<0.05), respectively, in the CI group, compared with the CN group. B: In OPLS-DA, obvious separation indicates a different structure of fecal metabolites between the CN and CI groups. C: Relative concentrations of 43 distinguishing fecal metabolites (P<0.05, VIP score >1) between the CN and CI groups. D and E: The classification of 43 distinguishing fecal metabolites. HMDB Superclass metabolites (D). HMDB Class metabolites (E). F: In KEGG, 43 distinguishing fecal metabolites between the CN and CI groups were mapped to 6 metabolic pathways. CN: normal cognition; CI: cognitive impairments; OPLS-DA: Orthogonal partial least squares discriminate analysis; VIP: Variable Importance in the Projection; HMDB: Human Metabolome Database; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Bile acids profile changed in patients with CI
Based on our findings in the untargeted metabolic mapping (Fig. 4F), we further undertook targeted bile acids quantification between the CN and CI groups. Primary bile acids (PBAs) include cholic acid (CA), chenodeoxycholic acid (CDCA), and their taurine or glycine bound derivatives. Secondary bile acids (SBAs) were the PBA derivates, including deoxycholic acid (DCA, a CA derivative), lithocholic acid (LCA, a CDCA derivative), ursodeoxycholic acid (UDCA), and α-muricholic acid and their conjugated forms[30]. In total, 46 kinds of bile acids were identified. Compared with the CN group, PBAs were significantly lower in the CI group, including CA, CDCA, GlyCA, GlyCDCA, and TaurCDCA (P<0.05) (Table 2 and Supplementary Table 5 [available online]).
Table 2. Statistically different fecal bile acids between CN and CI.
| Bile acids | Classification | Group | P-value | |
| CN (ng/g) | CI (ng/g) | |||
|
Data are expressed as median (25% quantile, 75% quantile). P-values were calculated by Student's t test or Mann-Whitney U-test. CN: normal cognition; CI: cognitive impairments; PBA: primary bile acid; SBA: secondary bile acid. | ||||
| Cholic acid | PBA | 17476.93 (3392.46, 31117.48) | 2298.26 (402.65, 15086.72) | 0.035 |
| Chenodeoxycholic acid | PBA | 13405.17 (4045.43, 23799.13) | 2222.42 (541.38, 13613.2) | 0.017 |
| Glycocholic acid | PBA | 440.68 (163.06, 1255.82) | 99.83 (39.4, 577.28) | 0.022 |
| Glycochenodeoxycholic acid | PBA | 746.89 (398.69, 1124.03) | 218.68 (100.81, 428.34) | 0.004 |
| Taurochenodeoxycholic acid | PBA | 220.9 (120.53, 407.36) | 77.85 (32.10, 223.88) | 0.022 |
| Allocholic Acid | SBA | 1591.32 (212.42, 2767.59) | 152.17 (74.74, 1402.8) | 0.039 |
| Alpha-Muricholic acid | SBA | 36.88 (19.05) | 17.45 (14.45) | 0.009 |
| Beta-Muricholic acid | SBA | 988.52 (398.5, 2676.31) | 265.4 (226.12, 1015.36) | 0.048 |
| Chenodeoxycholic Acid 24-Acyl-β-D-glucuronide | SBA | 3.14 (1.22, 9.08) | 0.39 (0.20, 4.19) | 0.039 |
| Glycohyocholic acid | SBA | 2.4 (1.36, 5.34) | 0.39 (0.34, 1.03) | 0.001 |
| Glycoursodeoxycholic acid | SBA | 268.64 (106.39, 602.2) | 28.84 (23.06, 267.43) | 0.008 |
| Lithocholic acid 3-sulfate | SBA | 2337.19 (941.51, 8830.24) | 461.87 (122.87, 3939.11) | 0.035 |
| Norcholic acid | SBA | 131.32 (84.62, 299.36) | 55.98 (35.71, 157.43) | 0.044 |
| Taurohyodeoxycholic acid | SBA | 25.91 (10.84, 120.13) | 2.44 (0.94, 33.75) | 0.013 |
| Tauroursodeoxycholic acid | SBA | 24.58 (7.75, 95.88) | 2.04 (1.15, 32.88) | 0.019 |
The correlation between the altered bile acids and gut microbial species
We analyzed the correlations between the abundance of the top 10 microbial genus species and the concentration of the top 10 abundant metabolites. CDCA and CA were associated with eight out of 10 top-expressing genera (Fig. 5A). Bacterial species belonging to the family Ruminococcaceae and genus Clostridium are the known bacteria that play a critical role in transforming PBAs to SBAs, converting CA to DCA, and CDCA to LCA[31–32]. Further, we compared the relative abundance of all genera belonging to the family Ruminococcaceae between the CN group and the CI group, 10 of 11 genera belonging to the Ruminococcaceae family increased in the CI group (Fig. 5B).
Figure 5. Difference in bacterial species related to bile acids metabolism between patients with CN and CI.
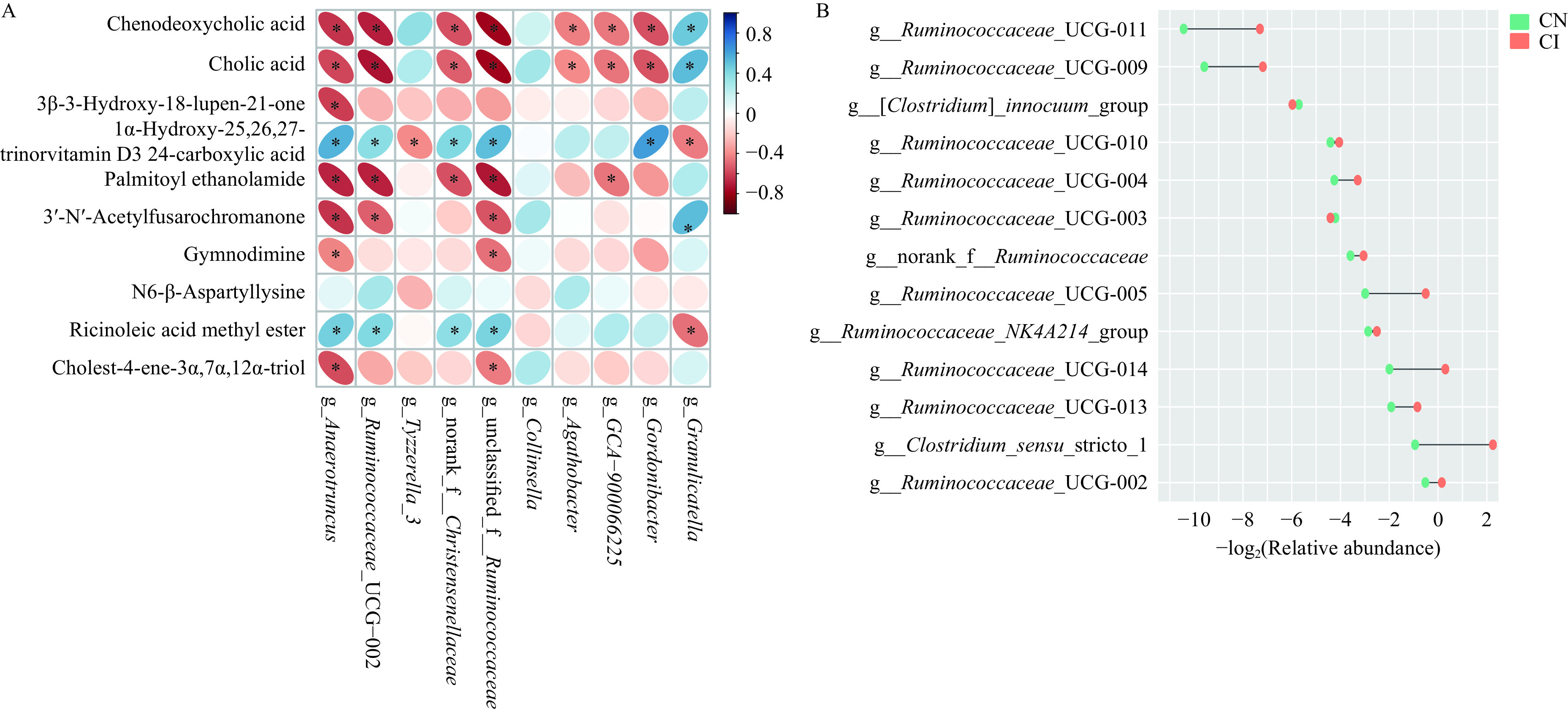
A: The correlation between 10 metabolites with the highest concentrations and 10 bacterial genera with the highest relative abundance (*P<0.05). B: Comparison of genus belonging to family ruminococcaceae between the CN and CI groups. SD: standard deviation; PBA: primary bile acids; SBA: secondary bile acids; CN: normal cognition; CI: cognitive impairments.
Microbial and metabolomic differences between ALS patients with and without behavioral abnormalities
In the current study, 14 ALS patients were divided into patients with behavior impairments (BVI) and 15 ALS patients were divided into patients without behavior impairments (BVN) as described in the methods. There was also no difference in α diversity and β diversity between the BVN and BVI groups (P>0.05) (Supplementary Fig. 1A and B, available online). Both the abundance of four dominant phyla and the ratio of F/B were comparable between the BVN and BVI groups (P>0.05) (Supplementary Fig. 1C and Supplementary Table 4, available online). Additionally, there was no bacterial species with a LDA score >3 in LEfSe analysis. In comparison of metabolites, there were 22 metabolites with significant differences in concentration between the BVI and BVI groups (Supplementary Fig. 1D, available online). However, these metabolites were not significantly mapped to any metabolic pathway. In the quantitative determination of bile acids, only dehydrocholic acid was higher in BVI group compared with the BVN group (Supplementary Table 6, available online).
Discussion
In the current study, we found altered gut microbiota and lowered PBAs levels in fecal samples of ALS patients with CI, compared with patients with normal cognition. Decreased PBAs were associated with increased bacteria that could consume PBAs. The current data suggests that the disruption of gut microbiota-related bile acids metabolism is associated with cognitive decline in ALS.
Gut microbiota and fecal metabolites differed between ALS patients with and without CI
In the current study, we found evident differences in gut microbiota and fecal metabolites between CN and CI patients, but not between BVN and BVI patients. Firmicutes and Bacteroidetes belong to bacterial phyla, lower F/B ratio indicates intestinal homeostasis dysregulation[33]. We found that the F/B ratio was comparable between ALS patients and healthy controls; however, it was significantly lower in the CI group than that in the CN group. Previously, various gut microbiota studies have been conducted in ALS patients (reviewed in Table 3). In the current study, the changes of gut microbiota between ALS patients and healthy controls have some similarities with those found in previous studies, with a similar α diversity but a different β diversity between ALS patients and healthy controls[34–38]. In untargeted metabolic mapping, we found disturbed profiles of lipids and lipid-like molecules in CI patients, including CA CDCA, 1-tetradecanoyl-2-(9Z-tetradecenoyl)-glycero-3-phosphoserine, and 1,3-dihydroxypropan-2-yl (9Z)-tetradec-9-enoate. Particularly, three of six mapped metabolic pathways in KEGG were related to bile acid metabolism, suggesting a potential link between bile acids and cognition in ALS patients. A recent study was conducted with both metagenomics and metabolomics analyses in 10 ALS patients and 10 healthy controls, providing limited conclusive findings[36]. In spite of previous studies about the ALS gut microbiota, to the best of our knowledge, we are the first to reveal the differences in gut microbiota and fecal metabolites between patients with and without cognitive decline using a multi-omics approach.
Table 3. Literature review of clinical studies on gut microbiota in ALS.
| References | Subjects (N) | Study methods | Main results | ||
| ALS | HC | NDC | |||
|
Note: only the results regarding the fecal microbiome and metabolome were summarized in this table. ALS: amyotrophic lateral sclerosis; HC: healthy control; NDC: neurodegenerative control; PCR: polymerase chain reaction; SCFAs: short chain fatty acids; F/B: Firmicutes/Bacteroidetes; IL: interleukin. | |||||
| Fang et al, 2016[34] | 6 | 5 | 0 | High-throughput sequencing (V3–V4 region) | 1. The β-diversity was different between ALS and HC. 2. Dorea genus increased in ALS, and Oscillibacter, Anaerostripes, Lachnospiraceae genus decreased in ALS. |
| Rowin et al, 2017[35] | 5 | 96 | 0 | 1. PCR for specific bacterial species 2. Quantitative determination of short chain fatty acids (SCFAs) in fecal samples using mass spectrometry |
1. The diversity of the microbiome decreased in ALS (not clearly stated whether α-diversity or β-diversity). 2. A low Firmicutes/Bacteroidetes (F/B) ratio was found in 3 ALS patients. 3. The level of SCFAs decreased in 1 ALS patient. |
| Brenner et al, 2017[21] | 25 | 32 | 0 | 454-pyrosequencing (V3–V6 region) | 1. Both α-diversity and β-diversity were comparable between ALS and HC. 2. The total number of operational taxonomic units increased in ALS. 3. The relative abundance of the uncultured Ruminococcaceae genus was different between ALS and HC. |
| Zhai et al, 2019[25] | 8 | 8 | 0 | 1. High-throughput sequencing (V4–V5 region) 2. Determination of endotoxin, SCFAs, NO2-N/NO3-N, and g-aminobutyric acid in fecal samples using enzyme-linked immunosorbent assay (ELISA) kits |
1. The F/B ratio and Methanobrevibacter genus showed an increased tendency in ALS. 2. Faecalibacterium and Bacteroides genus (beneficial micro-organisms) decreased in ALS. 3. No significant difference in levels of endotoxin, SCFAs, NO2-N/NO3-N, and g-aminobutyric acid was found between ALS and HC. |
| Blacher et al, 2019[20] | 37 | 29 | 0 | Shotgun sequencing | 1. The microbiome composition was different between ALS and HC. 2. Only a marginally significant difference in the abundances of specific bacterial species was found between ALS and HC. 3. ALS microbiomes decreased significantly in the global bacterial gene content related to nicotinamide metabolism. |
| Zeng et al, 2020[36] | 20 | 20 | 0 | 1. High-throughput sequencing (V4 region) 2. Shotgun sequencing (10 ALS and 10 HC) 3. Untargeted metabolome using liquid chromatography mass spectrometry (LC-MS) (10 ALS and 10 HC) |
1. The α-diversity (Shannon index) was different between ALS and HC. 2. Bacteroidetes phylum increased in ALS. 3. Firmicutes phylum, Kineothrix, Parabacteroides, Odoribacter, Sporobacter, Eisenbergiella, Mannheimia, Anaerotruncus, and unclassified Porphyromonadaceae genus decreased in ALS. 4. Enterococcus columbae positively correlated with 2-(1-ethoxyethoxy) propanoic acid and 3,7-dihydroxy-12-oxocholanoic acid. |
| Ngo et al, 2020[37] | 49 | 51 | 0 | High-throughput sequencing (V6–V8 region) | 1. The fecal microbiome was not significantly different between ALS and HC. 2. A greater risk for earlier death was reported in ALS patients with increased richness and diversity of the microbiome, and in those with a higher F/B ratio. |
| Nicholson et al, 2020[38] | 66 | 61 | 12 | Shotgun sequencing | 1. Comparable α-diversity and β-diversity between ALS and HC 2. The relative abundance of the dominant butyrate-producing microbial members decreased in ALS. |
| Di Gioia et al, 2021[19] | 43 | 44 | 0 |
High-throughput sequencing (V3–V4 region) |
1. The α-diversity (Chao1 index) and β-diversity were different between ALS and HC. 2. Microbial members of the Cyanobacteria phylum increased in ALS. 3. Microbial members of Clostridiaceae family decreased in ALS. |
| Niccolai, et al, 2021[22] | 19 | 9 | 0 | 1. High-throughput sequencing (V3–V4 region) 2. Determination of 30 kinds of cytokines (test kits) and SCFAs (gas chromatography and mass spectrometry) |
1. The F/B ratio decreased in ALS. 2. The relative abundance of butyrate-producing microbial members decreased in ALS. 3. Interleukin-2 (IL-2) and IL-1β increased in ALS. 4. IL-21 decreased in patients with a fast progression. |
Gut microbiota correlated with cognition by altering fecal bile acids profile in ALS
Based on our findings of altered gut microbiota and in bile acids-related metabolic pathways between CN and CI, we further performed a quantification of fecal bile acids. Decreases in CA, CDCA, and conjugated forms with glycine or taurine and increases in several secondary bile acids were found in the CI group, compared with the CN group. The higher ratios of CA/DCA (CI: 0.63, CN: 0.10) and CDCA/LCA (CI: 0.48, CN: 0.07) demonstrated a higher efficiency of conversion from CA to DCA, CDCA to LCA in the CI group than in the CN group. In humans, PBAs (mainly CA and CDCA) are synthesized in the liver and excreted to the small intestine[30]. As PBAs move from the small intestine to the colon, they are converted to SBAs by the biotransformation of the resident microbial community, and the key step is the 7α-dehydroxylation reaction[32]. In the colon, nearly 100% of CA and CDCA are converted to DCA and LCA, respectively. However, only a few known bacteria, all from the Ruminococcaceae family and Clostridium genus, perform the 7α-dehydroxylation[39]. Interestingly, 11 genera belonging to the family Ruminococcaceae were identified in the current study, and 10 genera increased in the CI group. According to these findings, we propose that the higher efficiency of conversion from PBAs to SBAs in the CI group probably results from alterations in gut microbial communities. However, since 16S sequencing is limited in the annotation of bacteria at the species level[40], we failed to further link the bile acids metabolism to specific bacterial strains. More in-depth basic and clinical studies, such as metagenomic studies and animal intervention studies, need to be further conducted to verify this hypothesis.
In the current study, ALS patients with CI presented with decreased fecal CA and CDCA, leading us to think about whether the impaired cognitive function is potentially linked to altered bile acid metabolic profiles. Bile acids are essential products of cholesterol metabolism, and in addition to playing a key role in lipid metabolism and absorption, recent studies suggest that the bile acid metabolism is associated with cognitive function[41]. In bile duct ligation mice, oral administration of obeticholic acid normalized memory function, prevented hippocampal network deficits, and reversed neuronal senescence by activating the farnesoid X receptor[42]. Interestingly, CDCA is an endogenous activator of the farnesoid X receptor[43], but whether CDCA can protect cognition by activating the farnesoid X receptor needs to be further explored. It was also reported that oral CDCA supplementation ameliorates neurotoxicity and cognitive deterioration via enhancing insulin signaling in AD model rats[44]. In clinical studies, AD patients exhibited significantly lower serum CA and higher DCA levels, compared with healthy controls. The ratio of DCA/CA was significantly correlated with the severity of cognitive decline[45]. Elevated DCA was also found in diabetes patients with cognitive impairment[46]. Both CA and CDCA could diffuse across the blood-brain barrier, generating aligned concentration in the brain and peripheral tissue[47]. Therefore, oral administration of either CA or CDCA could potentially be used as therapeutics for improving cognition. Clinical efficacy of such an approach for slowing cognitive decline in ALS patients could be tested in clinical trials in the future.
Conclusions
In the current study, we found altered gut microbial communities and bile acid metabolism in ALS patients, highlighting a possible role in the pathogenesis of cognitive decline in ALS patients. To our knowledge, we are the first to reveal connections between gut microbiota and CI in ALS patients. Based on the findings of our multi-omics approach, we propose that novel therapeutics could target bile acid metabolites with the aim of reducing CI in ALS patients. The study could be further strengthened by investigating bile acid metabolites in serum and cerebrospinal fluid, and the current findings need to be verified in studies with a larger sample size, particularly in longitudinal studies that may observe the dynamic changes of gut microbiota and metabolomics overtime.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgments
None.
Funding Statement
This study was supported by National Key Research and Development Program of China (Grants No. 2018YFE0118900 and 2021YFC2502200) and the clinical research project of the Bethune Charitable Foundation.
Footnotes
CLC number: R744.8, Ducument code: A
The authors reported no conflict of interests.
Contributor Information
Fengfei Ding, Email: fengfei_ding@fudan.edu.cn.
Min Zhang, Email: zhang_min_3464@126.com.
References
- 1.Kiernan MC, Vucic S, Cheah BC, et al Amyotrophic lateral sclerosis. Lancet. 2011;377(9769):942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 2.Arnoriaga-Rodríguez M, Fernández-Real JM Microbiota impacts on chronic inflammation and metabolic syndrome - related cognitive dysfunction. Rev Endocr Metab Disord. 2019;20(4):473–480. doi: 10.1007/s11154-019-09537-5. [DOI] [PubMed] [Google Scholar]
- 3.Crockford C, Newton J, Lonergan K, et al ALS-specific cognitive and behavior changes associated with advancing disease stage in ALS. Neurology. 2018;91(15):e1370–e1380. doi: 10.1212/WNL.0000000000006317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiò A, Ilardi A, Cammarosano S, et al Neurobehavioral dysfunction in ALS has a negative effect on outcome and use of PEG and NIV. Neurology. 2012;78(14):1085–1089. doi: 10.1212/WNL.0b013e31824e8f53. [DOI] [PubMed] [Google Scholar]
- 5.Elamin M, Phukan J, Bede P, et al Executive dysfunction is a negative prognostic indicator in patients with ALS without dementia. Neurology. 2011;76(14):1263–1269. doi: 10.1212/WNL.0b013e318214359f. [DOI] [PubMed] [Google Scholar]
- 6.Ye S, Ji Y, Li C, et al The edinburgh cognitive and behavioural ALS screen in a Chinese amyotrophic lateral sclerosis population. PLoS One. 2016;11(5):e0155496. doi: 10.1371/journal.pone.0155496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z, Alruwaili ARS, Henderson RD, et al Screening for cognitive and behavioural impairment in amyotrophic lateral sclerosis: frequency of abnormality and effect on survival. J Neurol Sci. 2017;376:16–23. doi: 10.1016/j.jns.2017.02.061. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein LH, Abrahams S Changes in cognition and behaviour in amyotrophic lateral sclerosis: nature of impairment and implications for assessment. Lancet Neurol. 2013;12(4):368–380. doi: 10.1016/S1474-4422(13)70026-7. [DOI] [PubMed] [Google Scholar]
- 9.Balendra R, Isaacs AM C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14(9):544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Z, Zhou Z, Che C, et al Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol, Neurosurg, Psychiatry. 2017;88(7):540–549. doi: 10.1136/jnnp-2016-315018. [DOI] [PubMed] [Google Scholar]
- 11.He J, Tang L, Benyamin B, et al C9orf72 hexanucleotide repeat expansions in Chinese sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2015;36(9):2660.e1–2660.e8. doi: 10.1016/j.neurobiolaging.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Tang J, Yang Y, Gong Z, et al Plasma uric acid helps predict cognitive impairment in patients with amyotrophic lateral sclerosis. Front Neurol. 2021;12:789840. doi: 10.3389/fneur.2021.789840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Sun G, Feng T, et al Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer's disease progression. Cell Res. 2019;29(10):787–803. doi: 10.1038/s41422-019-0216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Xu J, Chen Y Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients. 2021;13(6):2099. doi: 10.3390/nu13062099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matisz CE, Gruber AJ Neuroinflammatory remodeling of the anterior cingulate cortex as a key driver of mood disorders in gastrointestinal disease and disorders. Neurosci Biobehav Rev. 2022;133:104497. doi: 10.1016/j.neubiorev.2021.12.020. [DOI] [PubMed] [Google Scholar]
- 16.Giau VV, Wu S, Jamerlan A, et al Gut microbiota and their neuroinflammatory implications in Alzheimer's disease. Nutrients. 2018;10(11):1765. doi: 10.3390/nu10111765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Białecka-Dębek A, Granda D, Szmidt MK, et al Gut microbiota, probiotic interventions, and cognitive function in the elderly: a review of current knowledge. Nutrients. 2021;13(8):2514. doi: 10.3390/nu13082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mccombe PA, Henderson RD, Lee A, et al Gut microbiota in ALS: possible role in pathogenesis? Expert Rev Neurother. 2019;19(9):785–805. doi: 10.1080/14737175.2019.1623026. [DOI] [PubMed] [Google Scholar]
- 19.Di Gioia D, Bozzi Cionci N, Baffoni L, et al A prospective longitudinal study on the microbiota composition in amyotrophic lateral sclerosis. BMC Med. 2020;18(1):153. doi: 10.1186/s12916-020-01607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blacher E, Bashiardes S, Shapiro H, et al Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572(7770):474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 21.Brenner D, Hiergeist A, Adis C, et al The fecal microbiome of ALS patients. Neurobiol Aging. 2018;61:132–137. doi: 10.1016/j.neurobiolaging.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Niccolai E, Di Pilato V, Nannini G, et al. The Gut Microbiota-Immunity Axis in ALS: A Role in Deciphering Disease Heterogeneity?[J]. Biomedicines, 2021, 9(7).
- 23.Brooks BR, Miller RG, Swash M, et al El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(5):293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 24.Cedarbaum JM, Stambler N, Malta E, et al The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase Ⅲ) J Neurol Sci. 1999;169(1–2):13–21. doi: 10.1016/s0022-510x(99)00210-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhai C, Zheng J, An B, et al Intestinal microbiota composition in patients with amyotrophic lateral sclerosis: establishment of bacterial and archaeal communities analyses. Chin Med J. 2019;132(15):1815–1822. doi: 10.1097/CM9.0000000000000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-García A, Pineda-Quiroga C, Atxaerandio R, et al Comparison of mothur and QIIME for the analysis of rumen microbiota composition based on 16S rRNA amplicon sequences. Front Microbiol. 2018;9:3010. doi: 10.3389/fmicb.2018.03010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugerth LW, Andersson AF Analysing microbial community composition through amplicon sequencing: from sampling to hypothesis testing. Front Microbiol. 2017;8:1561. doi: 10.3389/fmicb.2017.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Wang H, Roberts DJ, et al Persistence of antibiotic resistance genes from river water to tap water in the Yangtze River Delta. Sci Total Environ. 2020;742:140592. doi: 10.1016/j.scitotenv.2020.140592. [DOI] [PubMed] [Google Scholar]
- 29.Segata N, Izard J, Waldron L, et al Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridlon JM, Kang DJ, Hylemon PB Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Duboc H, Rainteau D, Rajca S, et al Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24(6):513–520. doi: 10.1111/j.1365-2982.2012.01893.x. [DOI] [PubMed] [Google Scholar]
- 32.Staley C, Weingarden AR, Khoruts A, et al Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101(1):47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinninella E, Raoul P, Cintoni M, et al What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang X, Wang X, Yang S, et al Evaluation of the microbial diversity in amyotrophic lateral sclerosis using high-throughput sequencing. Front Microbiol. 2016;7:1479. doi: 10.3389/fmicb.2016.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowin J, Xia Y, Jung B, et al Gut inflammation and dysbiosis in human motor neuron disease. Physiol Rep. 2017;5(18):e13443. doi: 10.14814/phy2.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng Q, Shen J, Chen K, et al The alteration of gut microbiome and metabolism in amyotrophic lateral sclerosis patients. Sci Rep. 2020;10(1):12998. doi: 10.1038/s41598-020-69845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ngo ST, Restuadi R, McCrae AF, et al. Progression and survival of patients with motor neuron disease relative to their fecal microbiota. Amyotroph Lateral Scler Frontotemporal Degener. 2020;21(7–8):549–562. doi: 10.1080/21678421.2020.1772825. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson K, Bjornevik K, Abu-Ali G, et al The human gut microbiota in people with amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2021;22(3–4):186–194. doi: 10.1080/21678421.2020.1828475. [DOI] [PubMed] [Google Scholar]
- 39.Sinha SR, Haileselassie Y, Nguyen LP, et al Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27(4):659–670.e5. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson JS, Spakowicz DJ, Hong BY, et al Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10(1):5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahlström A, Sayin SI, Marschall HU, et al Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24(1):41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Gee LMV, Barron-Millar B, Leslie J, et al Anti-cholestatic therapy with obeticholic acid improves short-term memory in bile duct-ligated mice. Am J Pathol. 2022;193(1):11–26. doi: 10.1016/j.ajpath.2022.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Ding L, Yang L, Wang Z, et al Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm Sin B. 2015;5(2):135–144. doi: 10.1016/j.apsb.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bazzari FH, Abdallah DM, El-Abhar HS Chenodeoxycholic acid ameliorates AlCl3-induced Alzheimer's disease neurotoxicity and cognitive deterioration via enhanced insulin signaling in rats. Molecules. 2019;24(10):1992. doi: 10.3390/molecules24101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahmoudiandehkordi S, Arnold M, Nho K, et al Altered bile acid profile associates with cognitive impairment in Alzheimer's disease-An emerging role for gut microbiome. Alzheimers Dement. 2019;15(1):76–92. doi: 10.1016/j.jalz.2018.07.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suhre K, Meisinger C, Döring A, et al Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5(11):e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mertens KL, Kalsbeek A, Soeters MR, et al Bile acid signaling pathways from the enterohepatic circulation to the central nervous system. Front Neurosci. 2017;11:617. doi: 10.3389/fnins.2017.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.