Abstract
National Cancer Institute (NCI) Program for Natural Product Discovery is a new initiative aimed at creating new technologies for natural product-based drug discovery. Here, we present the development of a neural network-based bioinformatics platform for visualization and analysis of natural product high-throughput screening data using the NCI’s 60 human tumor cell anticancer drug screen. We demonstrate how the tool enables visualization of similar patterns of response that can be parsed both chemically and taxonomically, grouping NCI-60 biological profiles in one easy-to-use bioinformatics interface.
Introduction
The National Cancer Institute (NCI) Program for Natural Product Discovery (NPNPD) is a Cancer Moonshot-funded initiative by the US NCI aimed at better integrating natural product samples into high-throughput screening (HTS) programs. Recently, we have presented methods for the automated generation of the NPNPD partially purified fraction library,1 as well as a two-step, rapid procedure for the purification and identification of biologically active natural products from the prefractionated library.2 To date, 500,000 NPNPD fractions have been made available to the public in a 384-well plate format,3 with the goal of enabling this unrivaled supply of chemical diversity to be screened against multiple targets and disease states to find new bioactive compounds for potential therapeutic development. With multiple copies of each fraction set generated, and more than 5,000,000 samples shipped to screening centers so far, the development of methods scalable to a variety of HTS data from many screening campaigns and in various assay systems is another aspect of technology development undertaken by the NPNPD. Thus, we set out to develop a new method of accessing and visualizing the data generated in large screens of test samples derived from source organisms with diverse taxonomic and geographic origins. With the aim of establishing a bioinformatics platform capable of integrating biological activity, taxonomy, and geographic location, as well as chemical structure, we launched a proof-of-concept study using an artificial neural network of the NCI’s 60 human tumor cell anticancer drug screen (NCI-60) data of natural product extracts, fractions, and pure compounds.
The NCI-60 is one of the longest-running biological assays used to identify small molecules with cytotoxic, cytostatic, and antiproliferative activity against cancer cell lines grown in vitro.4,5 Measuring the cell growth of 60 different human tumor cell lines representing leukemia, melanoma, and cancers of the lung, colon, brain, ovary, breast, prostate, and kidney, the assay generates a 60 × 5-point dose–response biological profile. Growth inhibition ranges from no inhibition (100%) to cytostatic (0%) to complete cell death (−100%). To date, the NCI has screened more than 150,000 crude natural product extracts in the 1-dose NCI-60 assay. Of those, 37,054 natural product samples were determined to be sufficiently active to progress to the 5-dose screen. These active extracts comprised 9447 marine, 19,546 plant, and 8061 microbial samples, and the results represent one of the most comprehensive biological assay campaigns involving the use of crude natural product extracts. While there are correlation-based tools available to examine the NCI-60 biological response pattern that has been previously published (such as COMPARE6 and CellMiner7), pairwise comparisons of large datasets generally require secondary or tertiary steps to visualize meaningful relationships. Here, we present the development of a tool that can visualize all the NCI-60 dose–response profiles for the natural product samples in one easy-to-use bioinformatic resource. The utility of this system to help prioritize natural product chemistry efforts on active extracts is also demonstrated.
Results and Discussion
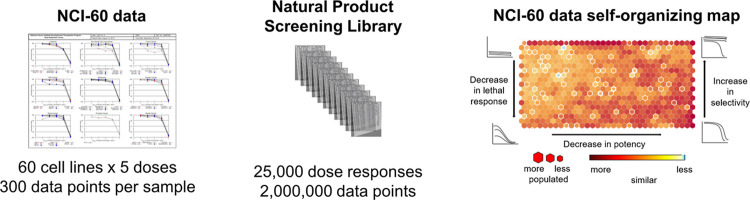
Visualization techniques such as principal component analysis (PCA), or the Kohonen self-organizing map (SOM) can project high-dimensional datasets onto low-dimensional space and have become commonly used tools for the analysis of chemical and biological data.8,9 As we were interested in depicting the similarities between a large number of NCI-60 dose responses, and artificial neural network methods are ideally suited to such data sets, we chose a SOM visualization technique to analyze the data.10 A SOM is an unsupervised artificial neural network data mining tool that has shown extensive use for clustering, organization, and visualization of complex datasets.10,11 The basic concept of the SOM architecture with simplified dose–response curves is shown in Figure 1A. The SOM algorithm arranges data in a topologically ordered fashion, where similar input vectors (e.g., dose–response profiles), are mapped together to form representative nodes. The map in Figure 1A is composed of nine nodes that define the basic map topology shown by different codebook vectors, which are depicted as averages of the underlying data, with new query dose–response or input vectors placed in a representative, closest matched node on the trained SOM.
Figure 1.
(A) Simplified representation of a trained SOM, represented by the 3 × 3 central grid of codebook vectors, with input vectors being organized to the best matching unit on the lattice. (B) Final NCI-60 SOM neighbor distance plot with general organization of input vectors. Prototypic activity profiles are depicted near the region which they represent. Unified distance is presented as a heatmap amongst the nodes. The U-matrix is a visualization of the data distribution, highlighting areas of higher density and any gaps between them. Here, red nodes adjacent to other red nodes will be more similar in biological profile than data assigned to orange or yellow nodes.
For the NCI-60 SOM, all data preparation and SOM construction were done using the Kohonen package for R.12 Dose–response data of natural product extracts and natural product pure compounds tested in the NCI-60 growth inhibition (GI) assay were selected from 1989 until 2015. To reduce the proportion of low information data and focus the SOM on samples with the most differential data points, only extracts with more than 3 cell lines that reached a level of IC25 at 10 μg/mL (inhibition of the growth of at least 3 cancer cell lines by 75%) were included in the training set. This had the effect of omitting marginally active profiles or those that had the response limited to the highest dose and concentrating the SOM on those extracts that had the highest level of actionable cytotoxicity. The resulting “active SOM” dataset reduced the total number of extract experiments plotted from a little over 25,000, to approximately 11,000 (For examples of dose responses that were selected and omitted from the set, see Supporting Information SI Figure S1). A random selection of the resulting 6000 NCI-60 five dose datasets was then mapped to the untrained lattice and the codebook vectors were exported. The final SOM depicted in Figure 1B maps the data set to a hexagonal grid of 33 × 19 nodes with an average occupancy of 10 extracts per node. Node sizes were correlated to the number of input vectors assigned (i.e., smaller hexagons have less data). Map coloring was further correlated with the average calculated distance of a node to its surrounding neighbors with nodes depicted in red having bioactivity patterns most similar and those in yellow and blue the least similar to their nearest neighboring nodes of bioactivity. The map was organized such that broadly cytotoxic extracts are on the upper left corner of the SOM, with a decrease in lethal response from top to bottom. Potency was also used as a descriptor with nodes showing a decrease in activity toward the right side of the map. Representative examples taken from selected map regions are shown in SI Figure S2. In total, the SOM contained NCI-60 five-dose information on 5041 marine invertebrate extracts, 4570 plant extracts, and 2592 microbial extracts, representing 2427 genera. This integrated platform enabled us to visualize and examine taxonomic, geographic, chemical, and NCI-60 dose–response similarity relationships between different extracts, thereby introducing a more comprehensive approach for the prioritization of lead projects from NCI-60 data.
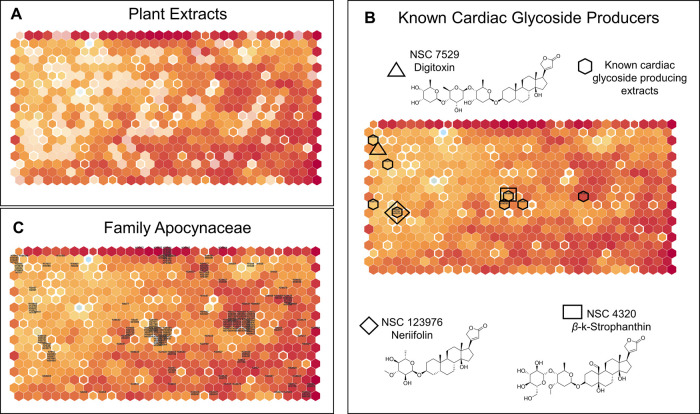
The ability to parse diverse datasets accrued from various source organisms in taxonomic, biological, and even chemical contexts makes the NCI-60 SOM a very powerful tool for oversight of large screening datasets and project prioritization. In Figure 2A, we show the distribution of all plant-sourced extracts on the SOM which covers 80% of the map (i.e., 491 of the 627 total nodes of biological response on the map). Similar visualizations of other source phyla are also possible, for marine invertebrate and microbial-sourced extract coverage on the SOM, see SI Figures SI3 and SI4, respectively. The analysis can be further annotated with the integration of knowledge of chemical structure known to be responsible for a particular NCI-60 cell response. The utility of this approach is shown by the clustering of plant extracts which we have determined in various screening campaigns13,14 possess a biological signature in the NCI-60 panel that is diagnostic of cardiac glycosides (Figure 2B). Also indicated on the SOM are the nodes where three prototypal cardiac glycoside NCI-60 profiles, namely digitoxin (NSC 7529), neriifolin (NSC 123976), and β-k-strophanthin (NSC 44320), are found, with the three compounds positioned separately on the SOM because of differences in potency. Cardiac glycosides are known to be produced across the plant family Apocynaceae,15 and when the map is queried against all samples from this family with NCI-60 data (Figure 2C), many of the extracts cluster in the same regions as shown in 2B. Here, we demonstrate that the main driver for the NCI-60 activity of the Apocynaceae are the cardiac glycosides—a very potent group of compounds that are common natural products found across this plant family.
Figure 2.
Visualization of extracts on the NCI-60 SOM. (A) Distribution of extracts sourced from plant biota on the SOM. Highlighted hexagons represent nodes where at least one plant-sourced extract is present, and the opaque hexagons represent nodes where there are none. (B) Placement of known cardiac glycoside-containing plant extracts on the SOM. Extracts are represented with a six-digit alphanumeric sequence and the nodes they are in are highlighted with a bold hexagon. Bold rectangle, diamond, and triangle represent the positions on the SOM of the NCI-60 dose–response profiles of the three cardiac glycoside compounds. (C) Placement of all NCI-60 dose–response profiles of extracts from the family Apocynaceae on the SOM. Individual extracts are represented with a six-digit alphanumeric sequence on the node and the extract is placed on the map.
When members of the same closely related taxonomic groups do not always produce identical or even similar natural products, and their biological activity profile in the NCI-60 assay is different, the SOM can be used to differentiate them. A clear example is found from two extracts of Taonura sp. sponges (extract NSC numbers C19265 and C17835), which were found separated along the y-axis of the SOM by 18 nodes (Figure 3). Interestingly, the samples were collected in the same geographic region, and photographs taken at the time of collection show a similar appearance and habitat for both organisms (see SI Figures S5 and S6). NCI-60 bioassay-guided isolation determined the active principle of Taonura sp. C19265 extract to be the triterpene heteronemin,16 while the active principle of the C17835 extract was the macrolide latrunculin A.17 The identified compounds displayed very different NCI-60 profiles, heteronemin being a potent cytotoxin and latrunculin A having a largely cytostatic profile across most of the NCI-60 cell lines. Here, although the extracts were from the same genus of sponge, collected in the vicinity of each other, their placement on the SOM indicated a different response profile, and the active principles subsequently isolated and identified were chemically distinct.
Figure 3.
Taonura sp. work summary showing the location of the extracts on the map, NCI-60 data of the crude extract, as well as the structure and NCI-60 activity of the active principles.
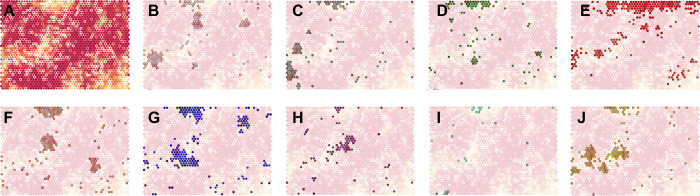
A second version of an NCI-60 SOM based on the calculated GI50 values derived from 5-dose experiments was also developed. Envisioned as a complement to COMPARE6 analysis, the interface provides the ability to refine searches by various parameters such as a cancer panel or a particular cell line within the NCI-60 cell lines. Similar to the COMPARE algorithm, this analysis uses calculated thresholds rather than normalized responses: specifically, the concentration (in μg/mL) at 50% growth inhibition for vector input. An example of the outcome of this process is shown in Figure 4 where we have re-parsed the SOM to highlight data from the nine different NCI-60 panels. By concentrating on a select panel or a cell line, we can find biological activity nodes that coincide with taxa or compounds that have been reported to contain natural products with selective cytotoxic activity against, for example, breast cancer. In SI Figure S7, we show select examples of dose responses that show selectivity against some of the cell lines in the breast cancer panel and their placement on the GI50 mean graph SOM. This type of cell- and panel-specific data parsing can aid the analysis of new NCI-60 data to enable more efficient chemistry efforts on cancer-type-specific natural products.
Figure 4.
GI50 mean graph SOM. (A) General topology of the SOM showing data mapped to a hexagonal grid of 50 × 40 nodes with an average occupancy of 10 extracts per node. Node sizes were correlated to the number of input vectors assigned with smaller nodes containing less data and map coloring was correlated with the average calculated distance of a node to its surrounding neighbors, with nodes depicted in red being the most similar and yellow the least similar to their nearest neighbors. (B–J) GI50 mean graph SOM shows an emphasis on the nodes that show sensitivity to the breast cancer panel in (B), central nervous system cancer panel in (C), colon cancer panel in (D), leukemia cancer panel in (E), melanoma cancer panel in (F), nonsmall cell lung cancer panel in (G), ovarian cancer panel in (H), prostate cancer panel in (I), and renal cancer panel in (J). The sensitivity of cell lines was defined as 1 log dilution less than the median GI50.
In addition to mapping the activity of crude extracts derived from plant, marine invertebrate, and microbial sources, the SOM can also be parsed for pure compounds. In Figure 5, SOM nodes containing one or more pure natural product 5-dose experiments are highlighted, while the opaque nodes represent parts of the map where only natural product extract dose–response profiles are present. Approximately 25% of the NCI-60 dose–response profiles on the SOM are not currently annotated with a chemical structure, indicating that there remains work to do to fully chemically annotate the NCI-60 SOM. While useful for guiding current projects, these opaque nodes represent potential areas of underexplored “biological space” with yet unassigned chemical annotation and potential leads for future projects using NCI-60-guided activity isolation.
Figure 5.

Placement of 1302 dose–response profiles of pure natural products on the SOM. Highlighted nodes contain at least one pure compound dose–response, while the opaque nodes contain only natural product extract dose–response profiles.
Conclusions
Large natural product collections represent an abundance of chemical diversity with drug discovery potential. However, all their biological, analytical, and chemical data are challenging to annotate. Here, we used NCI-60 cell line screen results, a content-rich dataset on one of the largest collections of natural product samples, and developed a neural network-based bioinformatics platform for data analysis, visualization, and project prioritization. Examples shown were focused on relationships of NCI-60 dose–response profiles and the taxonomy and chemistry of the extracts. Using the NCI-60 SOM, we were able to demonstrate taxonomic, chemical, and NCI-60 phenotypic relationships between different extracts, thereby introducing a new approach for the prioritization of lead projects from the NCI-60 data to improve the efficiency of natural products chemistry efforts. When the SOM was further extended to try to deduce, a priori, mechanism of action, it was determined that, except in a few limited examples, a greater amount of annotation on the specific genetic and expression-level features of the cancer cell lines was likely necessary. We have recently used a higher level of cell-line annotation to look at the activity of pure natural products in the NCI-60 assay.18 In the future, we hope to extend this more advanced approach to predict specific genetic signature features that correlate with the activity of natural product mixtures to enable the discovery of new natural products with selectivity for specific cell line mutations or overexpression levels.
Methods
SOM Architecture
All data preparation and SOM construction were done using the R statistical environment with the Kohonen package.10 Dose–response data of natural product extracts and natural product pure compounds tested in the NCI-60 growth inhibition (GI) assay were selected from 1989 until 2015. The data were formatted into a matrix of 15,942 sample experiments versus 300 cell line responses at low to high doses. For preparation, all values above 100% (growth beyond positive control) were set to 100. Raw data were divided by 100 so that all values were between −1 and 1. The ratio of map dimensions were calculated (based initially on MATLAB’s SOM_TOPOL_STRUCT() function): (1) heuristic estimate of the number of nodes in the lattice, (2) calculate covariance matrix of data set, and (3) use the ratio of the two largest eigenvalues to return the estimated width and height of the untrained map. The final map size was adjusted from the initial estimate until the topographic error stabilized to its approximate lowest value of 0.613, with a topological error (average distance of best matching codebook vectors) of 1.74. The R script for the NCI-60 SOM assembly is given in SI.
General Experimental Procedures
General Experimental Procedures. Optical rotations were recorded on a Rudolph Research Analytical Autopol IV polarimeter using a 0.25 dm cell in the solvent indicated. NMR spectra were recorded at 25 °C on a 400 MHz Agilent Inova spectrometer equipped with a 5 mm OneNMR probe. The 1H and 13C NMR chemical shifts were referenced to the solvent peaks for CDCl3 at δH 7.26 and δC 77.0. NMR FID processing and data interpretation was done using MestReNova software, version 14.0. High-resolution mass spectra were recorded on an Agilent 6545 Accurate-Mass Q-TOF LC/MS system (1290 Infinity II) equipped with a dual AJS ESI source. Preparative-scale HPLC purification was performed with a Waters Prep LC system, equipped with a Delta 600 pump and a 996-photodiode array detector using the Phenomenex Kinetex C8 HPLC column [5 μm, 150 × 21.2 mm]. All solvents used for chromatography, UV, and MS were HPLC grade, and the H2O was Millipore Milli-Q PF filtered.
Taonura sp.—Collection, Extraction, and Isolation
The sponge Taonura sp. was collected by SCUBA at a depth of 18 m in Tonga in 1997 under contract through Coral Reef Research Foundation for the National Cancer Institute. The specimen was taxonomically identified by Patricia Bergquist and a voucher specimen (0CDN5587) was deposited at the Smithsonian Institution. The sponge (wet weight 214.18 g) was extracted in water, followed by a MeOH/DCM overnight soak according to the National Cancer Institute’s standard marine extraction procedure detailed in McCloud19 to give the organic solvent extract C19265 (8.3 g). A portion of the organic extract (250 mg) was prefractionated on C8 SPE (2 g) eluting sequentially with: water/methanol (95:5) to yield 16.8 mg of fraction 1, water/methanol (80:20) to yield 10.9 mg of fraction 2, water/methanol (60:40) to yield 4.3 mg of fraction 3, water/methanol (40:60) to yield 2.8 mg of fraction 4, water/methanol (20:80) to yield 9.0 mg of fraction 5, methanol to yield 52.2 mg of fraction 6, and methanol/acetonitrile (50:50) to yield 63.8 mg of fraction 7. A portion of combined fractions 5, 6, and 7 (m = 25.0 mg) were further purified on C8 preparative HPLC eluting with a steep gradient from water/methanol (9:1) to methanol over 48 min. The conditions were isocratic at water/methanol (9:1) for 5 min followed by a gradient from water/methanol (9:1) to methanol over 35 min, followed by isocratic methanol conditions over 8 min. Collections were done in 1-minute increments starting from time = 0 min. Fractions 41 to 44 combined yielded 11.6 mg of heteronemin, (11.6% crude extract yield).
Heteronemin
Clear oil; chirooptical and NMR spectroscopic data in agreement to those previously reported;16 HRESIMS m/z [M + Na]+ 511.30277 (calcd for C29H44NaO6+, 511.30301).
Taonura sp.—Collection, Extraction, and Isolation
The sponge Taonura sp. was collected by SCUBA at a depth of 12 m in Tonga in 1997 under contract through Coral Reef Research Foundation for the National Cancer Institute. The specimen was taxonomically identified by Patricia Bergquist and a voucher specimen (0CDN5502) was deposited at the Smithsonian Institution. The sponge (wet weight 231.55 g) was extracted in water, followed by a MeOH/DCM overnight soak according to the NCI’s standard marine extraction procedure detailed in McCloud19 to give the organic solvent extract C17835 (11.4 g). A portion of the organic extract (250 mg) was prefractionated on C8 SPE (2 g) eluting sequentially with: water/methanol (95:5) to yield 9.6 mg of fraction 1, water/methanol (80:20) to yield 15.0 mg of fraction 2, water/methanol (60:40) to yield 23.3 mg of fraction 3, water/methanol (40:60) to yield 8.5 mg of fraction 4, water/methanol (20:80) to yield 8.6 mg of fraction 5, methanol to yield 50.5 mg of fraction 6, and methanol/acetonitrile (50:50) to yield 83.2 mg of fraction 7. A portion of combined fractions 4, 5, and 6 (m = 50.0 mg) were further purified on C8 preparative HPLC eluting with a steep gradient from water/methanol (9:1) to methanol over 48 min. The conditions were isocratic at water/methanol (9:1) for 5 min followed by a gradient from water/methanol (9:1) to methanol over 35 min, followed by isocratic methanol conditions over 8 min. Collections were done in one-minute increments starting from time = 0 min. Fractions 36 and 37 combined yielded 15.8 mg of latrunculin A (19.2% crude organic extract yield).
Latrunculin A
Clear oil; chirooptical and NMR spectroscopic data in agreement to those previously reported;17 HRESIMS m/z [M + Na]+ 444.18182 (calcd for C22H31NNaO5S+, 444.18151).
Acknowledgments
The authors would like to acknowledge Dr. David Covell of the National Cancer Institute. His guidance and willingness to listen and discuss issues and ideas cleared many hurdles and helped make this project possible. We thank the Molecular Pharmacology Branch, DTP, DCTD, NCI for performing the NCI 60-cell cytotoxicity assays in support of this study. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E and by the National Cancer Institute’s Cancer Moonshot. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c07416.
ExamplesSupporting Information :NCI-60 profiles (Figure S1); representative NCI-60 dose responses (Figure S2); distribution of extracts sourced from marine invertebrate biota (Figure S3); distribution of extracts sourced from microbial biota (Figure S4); collection photograph of the sponge Taonoura sp. that extract C19265 was sourced from (Figure S5); collection photograph of the sponge Taonoura sp. that extract C17835 was sourced from (Figure S6); examples of NCI-60 dose response profiles (Figure S7); 1H NMR spectrum (400 MHz, CDCl3) for heteronemin (Figure S8); 1H NMR spectrum (400 MHz, CDCl3) for latrunculin A (Figure S9); and SOM assembly (List S1) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Thornburg C. C.; Britt J. R.; Evans J. R.; Akee R. K.; Whitt J. A.; Trinh S. K.; Harris M. J.; Thompson J. R.; Ewing T. L.; Shipley S. M.; Grothaus P. G.; Newman D. J.; Schneider J. P.; Grkovic T.; O’Keefe B. R. NCI Program for Natural Product Discovery: A Publicly-Accessible Library of Natural Product Fractions for High-Throughput Screening. ACS Chem. Biol. 2018, 13, 2484–2497. 10.1021/acschembio.8b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic T.; Akee R. K.; Thornburg C. C.; Trinh S. K.; Britt J. R.; Harris M. J.; Evans J. R.; Kang U.; Ensel S.; Henrich C. J.; Gustafson K. R.; Schneider J. P.; O’Keefe B. R. National Cancer Institute (NCI) Program for Natural Products Discovery: Rapid Isolation and Identification of Biologically Active Natural Products from the NCI Prefractionated Library. ACS Chem. Biol. 2020, 15, 1104–1114. 10.1021/acschembio.0c00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Program for Natural Products Discovery Prefractionated Library. https://dtp.cancer.gov/organization/npb/npnpd_prefractionated_library.htm (accessed November 2022).
- Shoemaker R. H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Boyd M. R.; Paull K. D. Some practical considerations and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev. Res. 1995, 34, 91–109. 10.1002/ddr.430340203. [DOI] [Google Scholar]
- Paull K. D.; Shoemaker R. H.; Hodes L.; Monks A.; Scudiero D. A.; Rubinstein L.; Plowman J.; Boyd M. R. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989, 81, 1088–1092. 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- Reinhold W. C.; Varma S.; Sousa F.; Sunshine M.; Abaan O. D.; Davis S. R.; Reinhold S. W.; Kohn K. W.; Morris J.; Meltzer P. S.; Doroshow J. H.; Pommier Y. NCI-60 whole exome sequencing and pharmacological CellMiner analyses. PLoS One 2014, 9, e101670/1–e101670/18. 10.1371/journal.pone.0101670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro R.; Smilde A. K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. 10.1039/C3AY41907J. [DOI] [Google Scholar]
- Brereton R. G. Self organising maps for visualising and modelling. Chem. Cent. J. 2012, 6, S1. 10.1186/1752-153X-6-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T. Self-organizing neural projections. Neural Networks 2006, 19, 723–733. 10.1016/j.neunet.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Kohonen T. Self-organized formation of topologically correct feature maps. Biol. Cybern. 1982, 43, 59–69. 10.1007/BF00337288. [DOI] [Google Scholar]
- Wehrens R.; Buydens L. M. C. Self- and Super-organizing Maps in R: The kohonen Package. J. Stat. Software 2007, 21, 1–19. 10.18637/jss.v021.i05. [DOI] [Google Scholar]
- Grkovic T.; Evans J. R.; Akee R. K.; Guo L.; Davis M.; Jato J.; Grothaus P. G.; Ahalt-Gottholm M.; Hollingshead M.; Collins J. M.; Newman D. J.; O’Keefe B. R. Erythrofordins D and E, two new cassaine-type diterpenes from Erythrophleum suaveolens. Bioorg. Med. Chem. Lett. 2019, 29, 134–137. 10.1016/j.bmcl.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson P. J.; Cai S.; Carver C.; Powell D. R.; Risinger A. L.; Grkovic T.; O’Keefe B. R.; Mooberry S. L.; Cichewicz R. H. Triple-Negative Breast Cancer Cells Exhibit Differential Sensitivity to Cardenolides from Calotropis gigantea. J. Nat. Prod. 2020, 83, 2269–2280. 10.1021/acs.jnatprod.0c00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S.; Chen Y.; Lu Y.; Wang Y.; Ding L.; Jiang M. Cardenolides from the Apocynaceae family and their anticancer activity. Fitoterapia 2016, 112, 74–84. 10.1016/j.fitote.2016.04.023. [DOI] [PubMed] [Google Scholar]
- Kashman Y.; Rudi A. The carbon-13 NMR spectrum and stereochemistry of heteronemin. Tetrahedron 1977, 33, 2997–2998. 10.1016/0040-4020(77)88036-8. [DOI] [Google Scholar]
- Kashman Y.; Groweiss A.; Shmueli U. Latrunculin, a new 2-thiazolidinone macrolide from the marine sponge Latrunculia magnifica. Tetrahedron Lett. 1980, 21, 3629–3632. 10.1016/0040-4039(80)80255-3. [DOI] [Google Scholar]
- Krushkal J.; Negi S.; Yee L. M.; Evans J. R.; Grkovic T.; Palmisano A.; Fang J.; Sankaran H.; McShane L. M.; Zhao Y.; O’Keefe B. R. Molecular genomic features associated with in vitro response of the NCI-60 cancer cell line panel to natural products. Mol. Oncol. 2021, 15, 381–406. 10.1002/1878-0261.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloud T. G. High throughput extraction of plant, marine and fungal specimens for preservation of biologically active molecules. Molecules 2010, 15, 4526–4563. 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.