Abstract
Objective
To investigate the frequencies and bacterial load of three species of periodontal bacteria in samples from oropharyngeal cancer patients versus healthy individuals.
Study design
This is a case-control study based on biopsies collected from tumor tissues obtained from patients with oropharyngeal squamous cell carcinoma between 2016 and 2017 and shed oral mucosal epithelial cells that were collected from controls using the Cepimax® brush, carrying out several brushings towards the posterior third edge of the tongue and the cheek. Porphyromonas gingivalis, Tannerella forsythia and Prevotella intermedia detection and absolute quantification was determined through q-PCR. Statistical analysis included a U- test, X2, Fisher's exact test, odds ratio (OR) and Conditional logistic regression analysis and unconditional regression analysis (p < 0.05).
Results
A total of 48 donors older than 55 years old participated in this study. The population was distributed into 24 patients (cases) and 24 controls. A robust association was established in cases and controls with significance regarding Prevotella intermedia (OR: 15.00) and Porphyromonas gingivalis (OR:11.00). In the comparison between the amount of each bacteria in the groups, P. intermedia showed a higher bacterial load in oropharyngeal cancer patients (p = 0.04). However, multivariate analysis adjusted to the presence of different bacteria and the diverse confounding variables did not reveal significant differences for oropharyngeal cancer association.
Conclusion
P. gingivalis and P. intermedia were detected more frequently in the group of patients with cancer. The bivariate analysis of the bacterial load evidenced significant differences for Prevotella intermedia, suggesting that it could be associated with oropharyngeal cancer.
Keywords: Oropharyngeal cancer, Periodontal bacteria, Dysbiosis, Microbiota
1. Introduction
Head and neck tumors account annually for approximately to 400,000 diagnosed cases, mostly of epithelial type (90%), located with greater frequency in the oral cavity (40%) and 15% in the pharynx [1]. In 2021 Globocan Colombia reported 914 oral cancer cases and 378 deaths, and 530 throat cancer cases and 189 deaths [2]. For both oral and throat cancers the principal risk factors are excessive tobacco and alcohol consumption, recently associated with the processes of oral and pharyngeal microbiota dysbiosis, due to chronic inflammation and production of carcinogenic agents, such as acetaldehyde and toxins, among others [1].
Over 700 bacterial species have been described in the oral cavity, suggesting high microbial diversity [3], to such an extent that oral cavity diseases have been associated with dysbiosis [4], whose main outcomes are dental caries and periodontal disease, which is highly prevalent in adults [5]. In fact, bacteria have been directly associated with this pathology and identified as periodontal pathogens [6].
Identification of bacteria in patients with oral cancer by pyrosequencing of the 16S rRNA gene revealed that the most prevalent genera were Streptococcus, Porphyromonas, Fusobacterium and Lactobacillus, and for the control group: Prevotella, Neisseria, Leptotrichia, Capnocytophaga, Actinobacillus and Oribacterium [7]. In a recent systematic review of the literature we summarized the bacterial genera and species that have been associated with oral cancer, some of them considered periodontal pathogens such as: Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Aggregatibacter actinomycetemcomitans, Prevotella intermedia, Fusobacterium nucleatum, among others [7].
Regarding the oncogenic potential of these periodontal pathogenic bacteria, Porphyromonas gingivalis has been the best described species. In an animal model, Olsen and collaborators demonstrated that chronic infection with P. gingivalis in addition to the administration of 4-nitroquinoline-1-oxide (4NQO), an oral carcinogen, favored squamous cell transformation, augmented IL-6 production and activated the transcription factor signal transducer and activator of transcription 3 (STAT3), resulting in growth and increased aggressiveness of the tumor. Furthermore, Binder and collaborators support these results by describing tumorigenesis stimulation through Toll like receptor activation, Janus kinase survival signaling pathway and glycogen synthase kinase 3 beta (GSK3β). Additionally, the JAK/STAT signaling pathway is capable of activating indiscriminate cell proliferation in the presence of gingipains, lipopolysaccharides (LPS) and fimbria present in bacteria [8,9].
Additionally, Tannerella forsythia is a Gram-negative anaerobic bacterium involved in the red-complex conglomerate in periodontitis. It has been associated with oral squamous cell carcinoma by synergizing with other bacteria such as Porphyromonas gingivalis and Fusobacterium nucleatum. Therefore, it is fundamental to understand how its virulence factors contribute to chronic inflammation and pathogenicity of multiple clinical diseases, such as, proteins rich in leucine that allow adherence to fibronectin and fibrin among others, capsular S proteins that allow co-aggregation with Fusobacterium nucleatum and stimulation of IL-8 production [10].
With respect to Prevotella intermedia, it is a Gram-negative anaerobic bacterium associated with multiple pathogenic processes in the oral cavity, such as periodontal disease, abscesses, cellulitis and squamous cell carcinoma, including systemic diseases such as diabetes, cardiopathies, rheumatoid arthritis, low birth weight, premature birth, etc. Thus, it is vital to understand how virulence factors induce high IL-1, -6, and -8, TNF-alpha and interferon levels, produced by epithelial and endothelial cells, macrophages and fibroblasts. Moreover, it has been shown its capacity to stimulate tyrosine kinase receptors that modulate cell proliferation, migration and differentiation associated with disease progression [11,12].
To date the role that periodontal pathogens play during carcinogenesis has been delineated. However, studies describing the prevalence and quantity of bacteria in patients with oropharyngeal cancer are scarce. Hence, the objective of this study was to determine the prevalence and to quantify the bacterial load of Porphyromonas gingivalis, Tannerella forsythia and Prevotella intermedia in patients with oropharyngeal cancer in comparison with healthy donors.
2. Materials and methods
2.1. Type of study
This was an analytical-observational study approved by the Ethics Committee of the Pontificia Universidad Javeriana School of Dentistry (CIEFOUJ 0193 and May 6, 2016), which complements previous studies carried out at the Centro de Investigaciones Odontológicas (CIO) at the Pontificia Universidad Javeriana, in compliance with the Helsinki Declaration. Each participant in the study signed a detailed informed consent form.
2.2. Sample selection
Biopsies were collected from tumor tissues obtained from patients 18 years old or older with primary oropharyngeal squamous cell carcinoma tumor diagnosed by histopathology, who attended scheduled surgery for tumor resection in hospitals in Bogotá, Colombia, and who had not received previous antineoplastic treatment. Based on age and sex of the patients a group of matched control individuals was selected to whom oral brush cytology was performed. None of the individuals participating in this study had antimicrobial treatment in the previous three months. In addition, pregnancy was an exclusion criterium.
2.3. Sample collection
To collect the biopsies, a surgery was performed using a 6 mm Schumacher biotome to isolate the lesion and facilitate the excision of the tumor tissue fragment, which was then preserved in RNA later®. Shed oral mucosal epithelial cells were collected using the Cepimax® brush, carrying out 5 to 8 brushings towards the posterior third edge of the tongue and the cheek. Subsequently, the brush was placed in a tube with RNA later® for conservation. All samples were maintained at −20 °C until processed.
2.4. DNA extraction
DNA extraction was performed in all collected samples (biopsy and oral mucosal cytology) using the phenol-chloroform method. Briefly, an enzymatic proteinase K (Merck, 20 mg/ml) digestion was carried out overnight, followed by phenol-chloroform-isoamyl alcohol extraction following manufacturer's instructions (Thermo Scientific). The DNA in the aqueous phase was precipitated with isopropanol. Two 70% ethanol washes were performed and last the DNA was allowed to air-dry and re-suspended in TE buffer. Samples DNA concentration was determined in a Nanodrop 1000.
2.5. P. gingivalis, T. forsythia and P. intermedia quantification through qPCR
Real time quantitative PCR (qPCR) was used to specifically detect and quantify Porphyromonas gingivalis, Tannerella forsythia and Prevotella intermedia employing Taqman probes for each organism. Probes were designed based on sequences obtained from Genbank of the 3-deoxy-D-manno-octulosonic-acid gene specific for each microorganism (Table 1). The standardized qPCR contained a final volume of 10 μL, consisting of 1 μl DNA template (Approx. 50 ng), 1X master mix, 300 nM forward and reverse primers, 200 nM probe and 3.2 μl water. Sample were quantified in triplicate.
Table 1.
Primers and probes used to amplificated Porphyromonas gingivalis, Tannerella forsythia, and Prevotella intermedia with qPCR.
| Porphyromonas gingivalis | Tannerella forsythia | Prevotella intermedia | |
|---|---|---|---|
| Primer Forward | ATACGACGGCCTTTCATACG | TTATTTGCCCTTCGATACGC | GACCCGAACGCAAAATACAT |
| Primer Reverse | GCCTCGGGTGTATTGTGAAT | TTATTTGCCCTTCGATACGC | AGGGCGAAAAGAACGTTAGG |
| Probe | Quasar 705-TCCTCCATTTATCGCTACGG –BHQ2 | CALFLUORGOLD540-CAAATGGTACGGAAGCTGGT-BHQ1 | FLUOR RED 610-AAAGAAGGAACACCCCGACT – BHQ2 |
Reactions were performed using the following protocol in a CFX 96 Real Time System BioRad thermocycler: First a 10-min denaturing phase at 95 °C was carried out, followed by 40 cycles at 95 °C for 1 min and 60 °C for 2 min. As a negative control the master mix ran without DNA template. Copy number was established as the quantifying unit.
A paired statistical analysis was employed, where relative and absolute frequencies were used for nominal variables. For the microorganisms of interest quantification data of central tendency and dispersion (median and interquartile range IQR) were calculated. To determine differences between median values obtained for the microorganisms quantified a U Mann–Whitney test was performed.
In addition, a X2 and a Fischer exact test was used to compare clinical, socio-demographic characteristics and presence or absence of microorganisms between cases and controls to determine the association magnitude between variables an odds ratio (OR) was estimated with a 95% confidence interval.
Last, multivariate conditional and unconditional logistic regression models created by stepwise elimination of variables were performed. All variables with a marginal association (p < 0.20) were considered for the model. For all analyzes significance was determined at p < 0.05. For statistical analyses, SPSS Statistics V 22 (IBM SPSS Statistics) and Stata® 14 (StataCorp LLC) software were employed.
3. Results
3.1. Sociodemographic characteristics of the study population
A total of 48 people older than 55 years old participated in this study. The population was distributed in 24 cancer patients (cases) and 24 controls. In each group, 17 subjects were male (70.8%) and 19 female (79.2%). The locations of the tumors were: 11 cases of oropharyngeal (45.8%), followed by the palatine amygdala (4 cases, 16.7%), tongue (2 cases,8.3%) and larynx (2 cases, 8.3%). Regarding their clinical and socio-demographic characteristics in both populations, significant differences were observed in the number of sexual partners (greater than 5) and oral sex practice, greater in the patient group. On the other hand, smoking or passive smoking habits and acid reflux characteristics were grater in the control group. No significant differences were observed for the rest of the analyzed variables (Table 2). For the analysis using risk measures of clinical and sociodemographic variables, none were considered a risk factor, on the contrary they could have been considered protecting factors. The OR for oral sex practice was OR: 0.11, however, it must be taken into account that 3/24 patients and 21/24 in the control group did not respond to this question. The variables, smoking habits, passive smoker and acid reflux presented ORs of OR: 0.18, OR: 0.07 and OR: 0.18, respectively.
Table 2.
Comparison between patients and controls for their clinical and sociodemographic characteristics (X2 and Fischer exact test).
| Patients |
Controls |
p value | OR | 95% CI | |||
|---|---|---|---|---|---|---|---|
| n = 24 | % | n = 24 | % | ||||
| Sex | |||||||
| Female | 7 | 29.2 | 7 | 29.2 | 1.000 | 1 | |
| Male | 17 | 70.8 | 17 | 70.8 | 1.00 | (0.23–4.34) | |
| Age | |||||||
| <55 years old | 5 | 20.8 | 5 | 20.8 | 1.000 | 1 | |
| ≥55 years old | 19 | 79.2 | 19 | 79.2 | 1.00 | (0.07–13.7) | |
| Socio-economic status | |||||||
| Low | 12 | 50.0 | 5 | 20.8 | 0.281 | 1 | |
| Medium | 10 | 41.7 | 16 | 66.7 | 0.29 | (0.05–1.41) | |
| High | 2 | 8.3 | 3 | 12.5 | 0.24 | (0.24–2.35) | |
| Number of sexual partners | |||||||
| 1 to 2 | 9 | 37.5 | 17 | 70.8 | 0.030 | 1 | |
| 3 to 4 | 9 | 37.5 | 6 | 25.0 | 6.99 | (0.86–56.8) | |
| > 5 | 6 | 25.0 | 1 | 4.2 | 2.55 | (0.30 - ND) | |
| Oral sex practice | |||||||
| No | 9 | 37.5 | 3 | 12.5 | 0.0001 | 1 | |
| Yes | 9 | 37.5 | 0 | 0.0 | 0.11 | (0.02–0.80) | |
| DK/NR | 6 | 25.0 | 21 | 87.5 | |||
| Smoking habit | |||||||
| No | 16 | 66.7 | 3 | 12.5 | 0.0001 | 1 | |
| Yes | 8 | 33.3 | 21 | 87.5 | 0.18 | (0.03–0.65) | |
| Passive smoker | |||||||
| No | 20 | 83.3 | 7 | 29.2 | 0.0001 | 1 | |
| Yes | 4 | 16.7 | 17 | 70.8 | 0.07 | (0.02–0.32) | |
| Alcohol consumption | |||||||
| No | 12 | 50.0 | 10 | 41.7 | 0.562 | 1 | |
| Yes | 12 | 50.0 | 14 | 58.3 | 0.77 | (0.24–2.34) | |
| Acid reflux | |||||||
| No | 12 | 50.0 | 8 | 33.3 | 0.0001 | 1 | |
| Yes | 4 | 16.7 | 16 | 66.7 | 0.18 | (0.03–0.50) | |
| DK | 8 | 33.3 | 0 | 0.0 | |||
| Medication in the past 15 days | |||||||
| No | 15 | 62.5 | 9 | 37.5 | 0.099 | ||
| Yes | 9 | 37.5 | 15 | 62.5 | |||
| Tumor location | |||||||
| Amygdala | 4 | 16.7 | – | – | |||
| Hypopharynx | 1 | 4.2 | |||||
| Lower lip | 1 | 4.2 | |||||
| Larynx | 2 | 8.3 | |||||
| Tongue | 2 | 8.3 | |||||
| Oropharynx | 11 | 45.8 | |||||
| Not reported | 3 | 12.5 | |||||
Performed using X2 and Fisher exact test.
DK: does not know.
NR: No response.
3.2. Comparison between the three bacteria species in the oral cavity
We first determined the presence or absence of the P. intermedia, P. gingivalis and T. forsythia in patients and control individuals (Table 3). Our results evidenced significant differences for P. intermedia, which was present in 83.3% (20/24) of the patients in comparison with 25% (6/24) in the control group for a highly significant difference (p = 0.000). Furthermore, P. gingivalis was identified in 66.7% (16/24) of the patients and 25% (6/24) of control individuals, with a p = 0.004. Last, T. forsythia was present in 41.7% (10/24) of patients with lesions and 12.5% (3/24) in controls with a p = 0.023. These results showed that all three periodontal pathogens were more prevalent in patients compared with control individuals. Upon association analysis the presence of P. intermedia and P. gingivalis presented OR: 15 and OR: 11, respectively. Thus, they can be considered risk factors in the analyzed sample.
Table 3.
Presence or absence comparison of Prevotella intermedia, Porphyromonas gingivalis and Tannerella forsythia in patients vs. controls. Significance established using X2 and Fischer exact test.
| Patients |
Controls |
p value | OR | 95% CI | |||
|---|---|---|---|---|---|---|---|
| n = 24 | % | n = 24 | % | ||||
| P. intermedia | |||||||
| Absence | 4 | 16.7% | 18 | 75.0% | 0.000 | 15.00 | 1 |
| Presence | 20 | 83.3% | 6 | 25.0% | (2.3–63.0) | ||
| P. gingivalis | |||||||
| Absence | 8 | 33.3% | 18 | 75.0% | 0.004 | 11.00 | 1 |
| Presence | 16 | 66.7% | 6 | 25.0% | (1.59–47.0) | ||
| T. forsythia | |||||||
| Absence | 14 | 58.3% | 21 | 87.5% | 0.023 | 4.50 | 1 |
| Presence | 10 | 41.7% | 3 | 12.5% | (0.93–42.7) | ||
Performed using X2 and Fischer exact test.
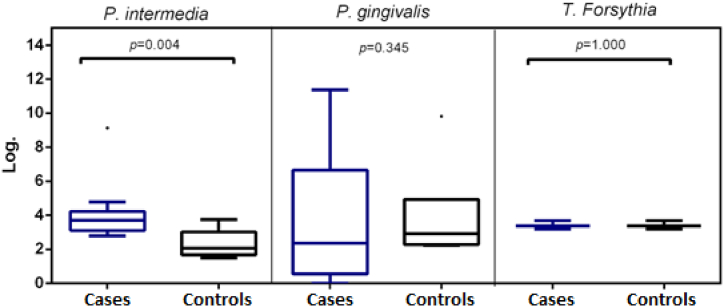
Next, we performed a quantitative analysis of bacterial load for each species using a logarithm transformation for statistical analysis (Table 4 and Fig. 1). For P. intermedia a greater CFU for the patient group with a median of 3.70 CFU (interquartile range (IQR): 3.16 to 4.20) in comparison with controls 2.06 CFU (IQR: 1.75 to 2.79), with a p = 0.04 was evidenced, suggesting that, for Prevotella intermedia, significant changes take place between patients and controls. However, for P. gingivalis, a greater dispersion was evidenced in patients in comparison with controls with no significant differences. Last, for T. forsythia no significant difference was observed between groups.
Table 4.
Colony forming units (CFU) comparison between patients vs. controls.
| Patients |
Controls |
p value | |||||
|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | ||||
| P. intermedia log. | 3.70 | 3.16 | 4.20 | 2.06 | 1.75 | 2.79 | 0.0431 |
| P. gingivalis log. | 2.36 | 0.57 | 6.55 | 2.92 | 2.30 | 3.31 | 0.3454 |
| T. forsythia log. | 4.39 | 3.95 | 5.15 | 3.37 | 3.20 | 3.68 | 1.000 |
Performed by Wilcoxon rank-sum test. Median evidence and interquartile ranges for each bacterium.
IQR: interquartile range.
Fig. 1.
Copy number comparison between patients (cases) and controls for the different evaluated microorganisms.
3.3. Multivariate analysis
The conditional logistic regression model (Table 5) adjusted to the different bacteria species and confounding variables, did not demonstrate significant differences for the presence of oropharyngeal cancer correlating with any of the microorganisms evaluated. In Table 5 a trend of P. intermedia towards an increased risk of oropharyngeal cancer is highlighted. However, no possible association between bacterial presence and oropharyngeal cancer could be established (p > 0.05).
Table 5.
Conditional logistic regression model for the presence or absence of the different bacteria objective of this study with respect to presence of oropharyngeal cancer.
| OR | IC 95% | p value | |
|---|---|---|---|
| P. intermedia | |||
| Absence | 1 | 0.056 | |
| Presence | 8.43 | (0.94–73.6) | |
| P. gingivalis | |||
| Absenc | 1 | 0.333 | |
| Presence | 3.11 | (0.31–31.0) | |
| T. forsythia | |||
| Absence | 1 | 0.749 | |
| Presence | 1.41 | (0.17–11–6) | |
4. Discussion
Most pathogenic bacteria in the oral cavity are Gram negative anaerobes, whose presence is evident in widely known diseases, such as cellulitis, abscesses, pericoronitis, alveolitis, periodontitis (periodontal pathogens), among others. There is evidence that oral microbiota disequilibrium can favor pathogenic bacterial growth, at the same time that eubiotic bacteria, related to healthy processes, become diminished; thus, favoring development of systemic diseases such as diabetes, elevated blood pressure and cancer, associated with virulence factors and subsequent chronic inflammation [13,14].
Socransky et al. (1998) proposed an oral-microbial complexes model associated with periodontal disease, proposing six bacterial complexes: 1. Yellow complex (Streptococcus species) associated with periodontal health. 2. Purple complex (for example Veillonela parvula and Actinomyces odontolyticus), 3. Green complex (for example Eikenella corrodens, Capnocytophaga gingivalis, C. sputigena, C. ochracea) 4. Blue complex (Actinomyces spp), 5. Orange complex (for example Fusobacterium, Prevotella and Campylobacter) and 6. Red complex (for example Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia). Bacteria in complex 5 and 6 are associated with periodontal disease [15].
Such description has been verified by other studies such as Pérez Chaparro et al., 2014, who associated Porphyromonas gingivalis and Tannerella forsythia to the red complex. Moreover, Prevotella intermedia, Parvimonas micra, Fusobacterium nucleatum, Eubacterium nodatum and Aggregatibacter actinomycetemcomitans were associated with the orange complex, related to the etiology of different periodontal conditions [15]. According to Aruni and collaborators (2015), bacteria that initially adhere to the tooth surface are Gram positive, facultative anaerobes, such as Actinomyces spp and oral streptococci. These bacteria interact with other primary colonizers and allow secondary colonizers, such as Prevotella intermedia, P. loescheii, Capnocytophaga spp and Fusobacterium nucleatum, forming the biofilm's matrix. Last, late colonizers are attracted, such as Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia [16].
Concerning oral or oropharyngeal cancer, bacteria in the oral cavity such as Porphyromonas gingivalis, Fusobacterium nucleatum, Tannerella forsythia and Prevotella species have been associated with the initiation, promotion and progression of this neoplasia. It has been described in studies on P. gingivalis that virulence factors such as fimbriae and LPS stimulate pro-inflammatory cytokine production, such as, IL-1, IL-6, IL-8, TNF-alpha, and activation of the JAK2/STAT3, signaling cascade blocking apoptosis and thus promoting cancer development [3,8,9].
With regard to Tannerella forsythia, scientific evidence points out this bacterium possess proteins rich in leucine that allow it to adhere to fibronectin and fibrin in the extracellular matrix of connective tissues. Additionally, the S protein stimulates IL-8 production that allows it to co-aggregate with Fusobacterium nucleatum. In fact, this microorganism can act synergistically with other bacteria, such as Porphyromonas gingivalis and Fusobacterium nucleatum, contributing to chronic inflammation [10].
Furthermore, virulence factors, such as fimbriae that aid in bacterial adhesion and co-aggregation have been described for Prevotella intermedia. Moreover, its capacity to degrade immunoglobins, and its toxic action against fibroblasts and fibrinolytic activity have been described. The pathogenic significance for the rest of the species is not well known; thus, they are related with other bacteria, with a synergistic and polymicrobial characteristic [15]. Results in the present study evidenced that P. gingivalis was detected through qPCR in 16 out of 24 patients with oropharyngeal cancer, corresponding to 66.7% of the patient group.
On the other hand, P. gingivalis was identified in the control group in 6 out of 24 individuals, corresponding to 25% of the population assayed. A significant difference was observed between patients and controls. Kawasaki and collaborators performed a study with 58 cases and 51 controls that aimed to detect periodontal pathogens in esophageal cancer through PCR. Three types of samples were collected: bacterial plaque, saliva and tumor tissue. P. gingivalis detection evidenced a greater presence in patients in comparison with controls, mainly in bacterial plaque, with values of 65.5% (38/58) in patients and 37.5% (18/51) in controls. In saliva samples 78.6% of the patient population was positive for this bacterium and it was detected in 79.03% of the controls. P. gingivalis was detected in 35.13% (13/37) of the samples collected from the patient's tumor, without a control for this last sample group [17]. In addition, Peters et al., 2017, elaborated a study whose objective was to evaluate P. gingivalis in patients with esophageal squamous cell carcinoma. Their results collected from brushing of oral mucosal revealed 32% (9/28) positivity for cancer patients and 20% (10/50) for controls [18]. These results are in agreement with our study, where 66.7% of the patients were positive for this bacterium. Our results are similar and even report a higher prevalence in cancer patients. In is noteworthy that for the Kawasaki study, results between patients and controls were similar, suggesting saliva is not the best sample source to compare cases and controls [17].
Other studies detecting P. gingivalis in samples from cancer patients, contrasting them with a control group were Yuan et al., 2017, who demonstrated presence of this bacterium in 48% (24/50) of the samples collected from esophageal tumors. The authors also performed this analysis in dysplastic tissue with 23.3% (7/30) detections. In contrast, in normal esophageal mucosa only one person was positive 3.3% (1/30) [19]. Gao et al., 2021 detected this bacterium in 71% (71/100) of patients with esophageal cancer and 3.3% (1/30) in controls from healthy esophageal mucosa [20]. Our results are in agreement with Yuan et al. and Gao et al. It should be noted that our patients had a percentage very similar to that reported by Gao et al. with an approximate 5% discrepancy [20]. However, it is clear the trend that P. gingivalis is more prevalent in patients than controls remains. Nevertheless, our controls presented a higher value (25% vs. 3.3%) in comparison with Yuan and Gao studies.
Other authors, such as Kang et al., 2009, described saliva in healthy individuals contained a greater number of periodontal pathogens in comparison with that of cancer patients, due to a variety of factors, such as the patient's health status or cancer therapy [21]. This was corroborated by Na et al., 2013, who found that age, diet and disease had an effect on the presence of different bacterial species [22].
Another hypothesis suggests the higher prevalence of pathogenic bacteria in controls (44%) in comparison with cancer patients (12%), may be due to the diverse P. gingivalis genotypes. Monier et al., 2020, established a relationship between ATCC 33277, W83, W50, ATCC 49417 strains with oral cancer, which presented higher virulence factors. In the present study, P. gingivalis genotype was not determined; hence, in future studies this microorganism must be genotyped. Moreover, Monier et al. noted the importance of evaluating bacteria associated with oral cancer in tumor tissue rather than saliva, since the microbiome composition can be due to particular characteristics of the tumor's microenvironment [23]. Furthermore, Chen et al. described the aforementioned strains associated with other pathologies, such as periodontal disease [24].
These previous studies used different PCR techniques, which without doubt could have influenced the results. Monier and collaborators consider PCR to be the ideal technique for bacterial detection, since it is a specific, sensitive and reproducible technique and widely available, which is not influenced by microorganism growth as observed in culture [23].
In our study Tannerella forsythia was detected in 41.7% (10/24) of the patients and in 12.5% (3/24) of the control individuals. Kawasaki and collaborators (2021) demonstrated its presence in bacterial plaque samples collected from patients 79.3% (46/58) and in controls 25.4% (13/51). In saliva its presence was (57/61) for patients and (57/62) for controls, 93.44% and 91.93%, respectively. Whereas for tumor tissue its presence was 16.21% (6/37), this group did not have a corresponding control [17]. Peters and collaborators (2017) designed a similar study to those previously described, whose objective was to evaluate the presence of the microorganism in patients with esophageal squamous cell carcinoma. They evidenced 52% (15/28) for patients and 58% (29/50) for controls in samples collected by brushing of oral mucosa [18]. Findings from the Peters study are different than ours for both populations collected, this could be attributed to the technique used to detect the microorganism. Peters et al. performed conventional PCRs without significant differences, whereas our study employed qPCR with a significant p value. In regard to the Kawasaki study no control was assayed for the tumor tissue [17]. Thus, it is not entirely comparable with our investigation.
Concerning Prevotella intermedia, its presence was detected in 83.3% (20/24) of the patients in comparison with controls 25% (6/24). According to Kawasaki, this bacterium was detected in bacterial plaque samples in 60% (35/58) of the patients and 39.9% (20/51) of the controls. In saliva it was detected in 68.8% of the cases and 59.9% of the controls. Last, in 35.13% of the tumor tissue evaluated [17]. In our study this microorganism prevails with higher presence in patients in comparison with control individuals.
Due to the lack of case-control studies that associated periodontal pathogens with oropharyngeal carcinoma, other similar studies were taken into account that aimed to establish an association between periodontal pathogens and esophageal carcinoma, because both structures possess a squamous stratified mucosa of the head and neck region. Therefore, more studies investigating oropharyngeal carcinoma are required [25].
From the quantitative point of view, the present study collected brushings of oral mucosa from tumor tissue (patients/cases) and controls. Microorganisms were quantified through qPCR and evidenced for P. intermedia log a median of 3.70 CFU in patients and 2.06 CFU in controls (p = 0.0431), suggesting that an increase in this microorganism could be considered as a poor prognosis marker. For P. gingivalis log medians between cases and control were similar 2.36 and 2.92 CFU, respectively, as well as for T. forsythia log medians of 4.39 CFU in patients and 3.37 CFU in controls.
Kawasaki and collaborators performed qPCR and evidenced low levels (<1 CFU) of P. intermedia, P. gingivalis and T. forsythia in subgingival bacterial plaque for patients as well as controls. For F. nucleatum the quantifications were 100 CFU in patients and 200 CFU in controls. For saliva samples the authors evidenced for patients and controls the following results F. nucleatum 6.5–7 CFU, P. gingivalis 5–5.5 CFU, P. intermedia 3–4 CFU and T. forsythia. 3.2–3.5 CFU, none of these results presented significant differences [17].
The study by Chen and collaborators quantified P. gingivalis through qPCR in patients with esophageal carcinoma and controls finding values of 7.5 CFU for patient samples and less than 2 for controls [26]. Sawant and collaborators performed a case and control study, where P. gingivalis was quantified through qPCR obtaining 4.01 × 105 BGE/ml (bacterial genomic equivalent) in patients with oral cancer in comparison with controls (8.45 × 104 BGE/ml). 120 subjects participated in the study, divided in 40 cases and 40 controls and 40 heavy smokers [27].
In the present study it was evidenced acid reflux (OR = 0.18 95% CI (0.03–0.50) p = 0.001) could be considered a protecting factor, present in 20% patients and 80% in controls. El-Serag et al., 2001, performed a case study with hospitalized and ambulatory laryngeal and pharyngeal cancer patients and controls, including hospitalized and ambulatory patients without cancer. In their multivariate logistic regression analysis controlled by age, sex, ethnicity, smoking habits and alcohol consumption, acid reflux was associated with an adjusted odds ratio OR of 2.40 (95 %CI of: 2.15–2.69, p = 0.0001) for the laryngeal cancer and 2.38 (95 %CI 1.87–3.02, p = 0.0001) for pharyngeal cancer in comparison with controls [28]. In contrast to the present work the results obtained by El-Serag et al. demonstrated acid reflux was considered a risk factor for laryngeal and pharyngeal cancer.
Additionally, smokers and passive smokers obtained ORs of 0.18 with a 95% CI (0.02–0.80) and OR of 0.07 with a 95% CI 95 (0.02–0.32), respectively, a contradictory result for cancer risk when compared with that presently reported in the literature. According to Gowda et al., 2020, smokers at some point demonstrated a 2.5-fold risk of oral cancer (95%CI: 1.6342, 4.026) without adjusting to any probable risk factor. After adjustment for chewing tobacco and alcohol consumption the risk increased by 3.4-fold (95% CI: 1371, 8475) [29]. The contradictory results in the present study could be accounted by the fact that the number of cigarettes smoked per day, the type of cigarette (with and without filter), vaping system, among others were not considered.
In this study the oral sex practice results should be carefully analyzed, since 22% of the case group and 77% of the control group did not respond to this question in the questionnaire. Thus, it is not possible to determine whether it is associated or nor to oropharyngeal cancer.
Concerning our logistic regression analysis, we observed for P. intermedia (OR = 8.43 (95% CI 0.94–73.6), P. gingivalis (OR = 3.11 (95% CI 0.31–31.0), and T. forsythia (OR = 1.41 (95% CI 0.17–11.6) results that did not demonstrate significant differences between patients and controls. This could be accounted by a limited sample size. However, it is noteworthy to highlight for the three bacteria evaluated that our logistic regression model revealed a tendency towards increased oropharyngeal cancer risk.
Studies such as the one performed by Kawasaki et al. demonstrated for the bacteria evaluated in the present study ORs of: OR = 4.46 (95% CI 0.79–25.40) p = 0.09 for P. gingivalis, OR = 79.70 (95% CI 4.34–1460.00) p = 0.003 for T. forsythia and an OR = 0.16 (95% CI 0.02–1.60) p = 0.12 for P. intermedia.17 Therefore, it could be suggested T. forsythia presence could be associated with cancer.
Peters et al. in their regression logistic model were also unable to find an association between P. gingivalis (OR = 1.30 95%CI (0.96–1.77) and T. forsythia (OR = 0.95 95% CI (0.58–1.55) and esophageal cancer (19). They highlighted P. gingivalis is the most relevant periodontal pathogen associated with esophageal cancer, even though no significant differences were determined [18].
Collectively, available evidence is not sufficient to demonstrate an association between oropharyngeal cancer and the presence of bacteria. However, this study does highlight the importance of quantifying mainly P. intermedia, where high levels were observed in the patient group in comparison with controls.
5. Conclusions
In this case-control study a significant difference in the prevalence and bacterial load of Prevotella intermedia was observed in oropharyngeal cancer patients, suggesting that this bacterial strain could be associated with the development of this malignancy.
6. Limitations of the study
This study did not include a comparison of the biopsies taken from patients with tissue obtained from the control group. Moreover, tissue adjacent to the tumor was not included. It was not possible to identify the clinical stage of the disease at the time of sampling.
Statement of clinical relevance
Increase in Prevotella intermedia count is predominantly higher in patients with oropharyngeal cancer compared to the control group, which suggests the importance of oral health care in patients with this diagnosis and as far as possible detect and quantify them.
Declarations
Author contribution statement
Gabriel Jaime Castañeda Corzo; Luís Felipe Infante Rodríguez: Contributed reagents, materials, analysis tools or data; Wrote the paper.Jean Carlos Villamil Poveda: Performed the experiments; Contributed reagents, materials, analysis tools or data.Jairo Bustillo: Conceived and designed the experiments; Wrote the paper.Angel Cid-Arregui: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper Dabeiba-Adriana García-Robayo: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper
Funding statement
Mrs Dabeiba-Adriana García-Robayo was supported by Pontificia Universidad Javeriana . Grant ID 20383 and proposal ID 10377.
Data availability statement
No data was used for the research described in the article.
Declaration of interest's statement
The authors declare no conflict of interest.
Contributor Information
Gabriel-Jaime Castañeda-Corzo, Email: gabrieljcastaneda@javeriana.edu.co.
Luís-Felipe Infante-Rodríguez, Email: pipeinfanterz@gmail.com.
Jean-Carlos Villamil-Poveda, Email: jean.villamil@javeriana.edu.co.
Jairo Bustillo, Email: bustillo@javeriana.edu.co.
Angel Cid-Arregui, Email: cid@Dkfz-Heidelberg.de.
Dabeiba-Adriana García-Robayo, Email: garciad@javeriana.edu.co.
References
- 1.Ferguson B.L., Barber S., Asher I.H., et al. Role of oral microbial infections in oral cancer. Dent. Clin. 2017;61:425–434. doi: 10.1016/j.cden.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 2.The Global Cancer Observatory - All Rights Reserved - March. 2021. https://gco.iarc.fr/today/data/factsheets/populations/170-colombia-fact-sheets.pdf Accessed. [Google Scholar]
- 3.Chattopadhyay I., Verma M., Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol. Cancer Res. Treat. 2019;18 doi: 10.1177/1533033819867354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh P.D., Zaura E. Dental biofilm: ecological interactions in health and disease. J. Clin. Periodontol. 2017;44:S12–S22. doi: 10.1111/jcpe.12679. [DOI] [PubMed] [Google Scholar]
- 5.Wen B.W., Tsai C.S., Lin C.L., et al. Cancer risk among gingivitis and periodontitis patients: a nationwide cohort study. QJM. 2014;107:283–290. doi: 10.1093/qjmed/hct248. [DOI] [PubMed] [Google Scholar]
- 6.Chang C., Geng F., Shi X., et al. The prevalence rate of periodontal pathogens and its association with oral squamous cell carcinoma. Appl. Microbiol. Biotechnol. 2019;103:1393–1404. doi: 10.1007/s00253-018-9475-6. [DOI] [PubMed] [Google Scholar]
- 7.Robayo D.A., Hernandez R.F., Erira A., et al. Oral Microbiota associated with oral and gastroenteric cancer. Open Microbiol. J. 2020;14:1–17. doi: 10.2174/1874285802014010001. [DOI] [Google Scholar]
- 8.Olsen I., Yilmaz Ö. Possible role of Porphyromonas gingivalis in orodigestive cancers. J. Oral Microbiol. 2019;11 doi: 10.1080/20002297.2018.1563410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallimidi A.B., Fischman S., Revach B., et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6:22613–22623. doi: 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma A. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000;54:106–116. doi: 10.1111/j.1600-0757.2009.00332.x. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan S.M., Zhang M., He J.J., et al. Mitogen-activated protein kinases and phosphatidylinositol 3-kinase are involved in Prevotella intermedia-induced proinflammatory cytokines expression in human periodontal ligament cells. Biochem. Biophys. Res. Commun. 2009;386:471–476. doi: 10.1016/j.bbrc.2009.06.058. [DOI] [PubMed] [Google Scholar]
- 12.Guan S.M., Shu L., Fu S.M., et al. Prevotella intermedia induces matrix metalloproteinase-9 expression in human periodontal ligament cells. FEMS Microbiol. Lett. 2008;283:47–53. doi: 10.1111/j.1574-6968.2008.01140.x. [DOI] [PubMed] [Google Scholar]
- 13.Adda G., Aimetti M., Citterio F., et al. Consensus report of the joint workshop of the Italian society of diabetology, Italian society of periodontology and implantology, Italian association of clinical diabetologists (SID-SIdP-AMD) Nutr. Metabol. Cardiovasc. Dis. 2021;31:2515–2525. doi: 10.1016/j.numecd.2021.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Landi L., Grassi G., Sforza N.M., et al. Hypertension and periodontitis: an upcoming joint report by the Italian society of hypertension (SIIA) and the Italian society of periodontology and implantology (SIdP) High Blood Pres. Cardiovasc. Prev. 2021;28:1–3. doi: 10.1007/s40292-020-00430-w. [DOI] [PubMed] [Google Scholar]
- 15.Pérez-Chaparro P.J., Gonçalves C., Figueiredo L.C., et al. Newly identified pathogens associated with periodontitis: a systematic review. J. Dent. Res. 2014;93:846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aruni A.W., Dou Y., Mishra A., et al. The biofilm community-rebels with a cause. Curr Oral Health Rep. 2015;2:48–56. doi: 10.1007/s40496-014-0044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasaki M., Ikeda Y., Ikeda E., et al. Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer. 2021;127:512–519. doi: 10.1002/cncr.33316. [DOI] [PubMed] [Google Scholar]
- 18.Peters B.A., Wu J., Pei Z., et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77:6777–6787. doi: 10.1158/0008-5472.CAN-17-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan X., Liu Y., Kong J., et al. Different frequencies of Porphyromonas gingivalis infection in cancers of the upper digestive tract. Cancer Lett. 2017;404:1–7. doi: 10.1016/j.canlet.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Gao S., Li S., Ma Z., et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect. Agents Cancer. 2016;11:1–9. doi: 10.1186/s13027-016-0049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang M.S., Oh J.S., Kim H.J., et al. Prevalence of oral microbes in the saliva of oncological patients. J. Bacteriol. Virol. 2009;39:277–285. doi: 10.4167/jbv.2009.39.4.277. [DOI] [Google Scholar]
- 22.Na H.S., Kim S., Choi J.Y., et al. Oral microbiota comparison between healthy volunteers, periodontitis patients and oral cancer patients. Int J Oral Biol. 2013;38:181–188. doi: 10.11620/ijob.2013.38.4.181. [DOI] [Google Scholar]
- 23.Monier N.M., Atteya I.M., Askar H., et al. Detection of the periodontal pathogen Porphyromonas gingivalis in oral squamous cell carcinoma. Mansoura J Dent. 2020;7:24–28. doi: 10.21608/mjd.2020.198725. [DOI] [Google Scholar]
- 24.Chen Q., Shao Z., Liu K., et al. Salivary Porphyromonas gingivalis predicts outcome in oral squamous cell carcinomas: a cohort study. BMC Oral Health. 2021;21:228. doi: 10.1186/s12903-021-01580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erira A., García Robayo D.A., Chalá A.I., et al. Bacteriome identified by next-generation sequencing in saliva, dental plaque, and tumor tissue of patients with oral squamous cell carcinoma. Open Microbiol. J. 2021;15:98–110. doi: 10.2174/1874285802115010098. [DOI] [Google Scholar]
- 26.Chen M.F., Lu M.S., Hsieh C.C., et al. Porphyromonas gingivalis promotes tumor progression in esophageal squamous cell carcinoma. Cell. Oncol. 2021;44:373–384. doi: 10.1007/s13402-020-00573-x. [DOI] [PubMed] [Google Scholar]
- 27.Sawant S., Dugad J., Parikh D., et al. Absolute quantitation of oral bacteria involved in oral cancer by real-time PCR. Med Microecol. 2021;7 doi: 10.1016/j.medmic. [DOI] [Google Scholar]
- 28.El-Serag H.B., Hepworth E.J., Lee P., et al. Gastroesophageal reflux disease is a risk factor for laryngeal and pharyngeal cancer. Am. J. Gastroenterol. 2001;96:2013–2018. doi: 10.1111/j.1572-0241.2001.03934.x. [DOI] [PubMed] [Google Scholar]
- 29.Gowda K.L., Vijay C.R., Lokesh V., et al. A hospital based case control study on oral cancer: KMIO (regional cancer center) experience. J Med Sci Clin Res. 2020;8:1022–1031. doi: 10.18535/jmscr/v8i1.17. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.