Abstract
The nucleolus is a multifunctional nuclear domain primarily dedicated to ribosome biogenesis. Certain viruses developed strategies to manipulate host nucleolar proteins to facilitate their replication by modulating ribosomal RNA (rRNA) processing. This association interferes with nucleolar functions resulting in overactivation or arrest of ribosome biogenesis, induction or inhibition of apoptosis, and affecting stress response. The nucleolar protein fibrillarin (FBL) is an important target of some plant and animal viruses. FBL is an essential and highly conserved S-adenosyl methionine (SAM) dependent methyltransferase, capable of rRNA degradation by its intrinsically disordered region (IDR), the glycine/arginine-rich (GAR) domain. It forms a ribonucleoprotein complex that directs 2′-O-methylations in more than 100 sites of pre-rRNAs. It is involved in multiple cellular processes, including initiation of transcription, oncogenesis, and apoptosis, among others. The interaction with animal viruses, including human viruses, triggered its redistribution to the nucleoplasm and cytoplasm, interfering with its role in pre-rRNA processing. Viral-encoded proteins with IDRs as nucleocapsids, matrix, Tat protein, and even a viral snoRNA, can associate with FBL, forcing the nucleolar protein to undergo atypical functions. Here we review the molecular mechanisms employed by animal and human viruses to usurp FBL functions and the effect on cellular processes, particularly in ribosome biogenesis.
Keywords: Fibrillarin, Animal viruses, Human viruses, Nucleolus, Ribosome biogenesis, Intrinsic disordered region
Introduction
The nucleolus is the most extensive membrane-less nuclear body. Although separated from the nuclear space, it is accessible for dynamic exchange. Mammalian cell nucleoli, which are the best studied, contain three phase-separated subcompartments: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). The FC and DFC form functional units distributed in multiple copies throughout a single GC [1]. In addition, the nucleolus is often surrounded by a ring of condensed chromatin, also known as perinucleolar chromatin, enriched in specific genes to react to the environment and other stimuli [2]. The nucleolus´ primary role is the initial steps of ribosomal biogenesis, including RNA polymerase I (Pol I)-driven transcription, processing and modification of ribosomal RNA (rRNA) and the assembly of rRNA containing complexes [3]. Then precursor subunits move from the nucleolus to the nucleoplasm and ultimately to the cytoplasm for further maturation, and resulting in functional ribosomal subunits ready to participate in mRNA translation into protein. These initial steps are proposed to be coordinated by liquid-liquid phase separation (LLPS) (reviewed in [4, 5]), since there is strong evidence that the multilayered architecture of the nucleolus arises from multiphase liquid immiscibility [6]. The nascent transcripts are synthesized in FC and directed to DFC by the interaction of the 5′end of 47 S pre-rRNA and the methyl transferase domain (MD) of fibrillarin (FBL). Then, the FBL-RNA complexes translocate to the DFC by the glycine- and arginine-rich (GAR) domain of FBL, self-association. FBL self-association through its GAR domain, an intrinsically disordered region (IDR), strongly correlates with such pre-rRNA sorting, and is required for pre-rRNA processing. The methylation activity of FBL is not required for rRNA sorting [5].
The presence of different domains along the sequence determines the structure and function of a protein. These domains favor the formation of diverse structures and packaging, which confers the enzymatic properties and interactions with nucleic acids and proteins. The amino acid sequence of a protein motif will define; function and specificity. Arginine is one of the most frequently repeated amino acids in protein motifs, which is known to be regulated during post-transcriptional modifications [7]. Multivalent interactions between repetitive protein-protein or protein-RNA interaction domains/motifs are the key determinants of liquid-liquid phase separation [8, 9]. Multivalent contacts can be achieved with tandem repeats of folded domains [10], by low complexity, intrinsically disordered regions (IDRs) or both [11]. Arginine-serine (RS)-rich repeats have been reported to be associated with proteins in nuclear smears and in splicing processes. These domains are regulated by phosphorylation and play an important role in the condensation of disordered regions, the charge mixture of the domains present in a protein, and the localization in nuclear spots [12, 13].
Diverse studies show that regions rich in RGG/RG are intrinsically disordered sequences, a feature that gives the protein greater conformational plasticity and adaptability, binding more easily to different targets [14]. In some cases, these motifs have been mostly identified in proteins that have RNA-binding domains [15, 16]. These RGG motifs are usually followed by RG repeats and are represented as RGG/RG motifs, which can be divided into tri-RGG, di-RGG, tri-RG, and di-RG [15]. The lack of a rigid structure of intrinsically disordered proteins (IDPs), gives the ability to perform various interactions with several partners [17]. This feature explains the high disorder content of viral proteomes, since viruses need to hijack and manipulate host cellular processes by interacting with multiple host components [18].
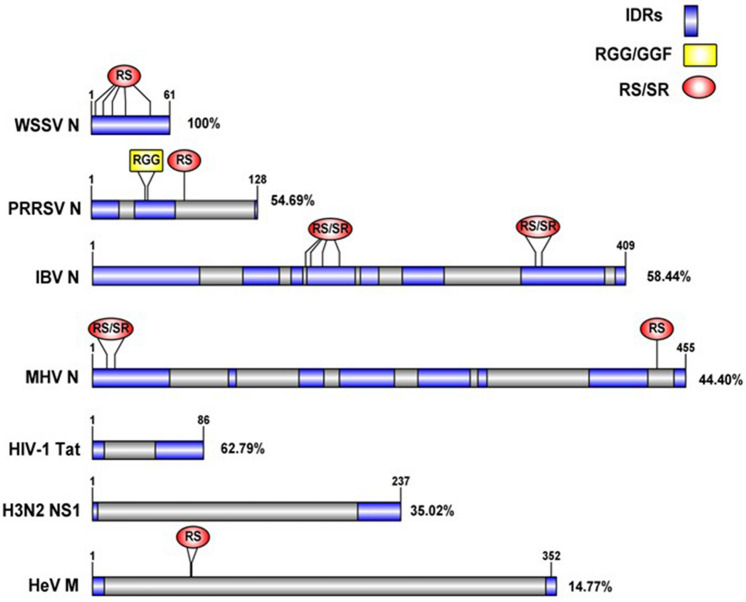
Most nucleolar proteins contain IDRs [19], and many IDRs undergo LLPS in vitro, while IDR containing-proteins have been implicated in numerous condensates [20, 21]. Disordered proteins increase the interaction valency, since they can establish protein-protein and RNA-protein interactions, which are key for phase separation [22]. LLPS are condensates that allow control over the molecular environment and promote rapid exchange between elements. In addition to protein composition (IDRs), concentration plays a critical role to establishing LLPS [23]. For instance, FBL saturation concentration depends on the presence of RNA, so nearly actively transcribing rDNA, FBL preferential condensation arises from the high RNA concentration there [24]. This is particularly important in nucleolar stress, where nucleolar proteins are redistributed, decreasing their concentration and affecting the LLPS in nucleolar processes. An analysis of the sequences of human and animal viruses-encoded proteins known to associate with FBL, described in Table 1, evidence one or more IDRs, an SR, and RGG-rich regions. Can be expected since viral proteomes are characterized by the dominance of short disordered segments [18], as illustrated in Fig. 1. Interestingly, IDPs can bind to globular proteins but also with other IDPs, since the GAR domain (an IDR) tend to be the binding region of viral proteins [17].
Table 1.
Animal and human virus-encoded proteins and snoRNA described to associate with FBL
| Virus name | Genome | Virus-encoded protein | Putative function | References |
|---|---|---|---|---|
| Infectious bronchitis virus | (+)ssRNA | Nucleocapsid | Undetermined | [25] |
| Porcine reproductive and respiratory syndrome virus | (+)ssRNA | Nucleocapsid | Undetermined | [26] |
| Mouse hepatitis virus | (+)ssRNA | Nucleocapsid | Transcription and/or translation of viral mRNAs | [25, 27] |
| White spot syndrome virus | dsDNA | Nucleocapsid (VP15) | Undetermined | [28] |
| Influenza A H3N2 | (-)ssRNA | non-structural protein 1 (NS1) | Impairment of pre-rRNA proccesing | [29] |
| Human immunodeficiency virus | (+)ssRNA | Tat protein | Impairment of pre-rRNA proccesing | [30] |
| Epstein-Barr virus | dsDNA | v-snoRNA1 | Impairment of pre-rRNA proccesing | [31] |
| Hendra virus | (-)ssRNA | Matrix | Viral replication and IRES-dependent translation of viral mRNAs | [32] |
Fig. 1.
IDR prediction, SR boxes, and RGG sequences of animal and human viruses-encoded proteins known to associate with FBL. Disorder percentage is indicated
FBL is an essential protein that has been conserved in sequence and function throughout evolution [33–35]. During the interphase of HeLa cells, it localizes at the transition zone of FC and DFC in the Cajal Bodies (CBs). CBs are small nuclear organelles observed in plant and animal nuclei. CBs can be localized to the nucleolar periphery or within [36, 37]. Primarily, they are involved in processing small nuclear RNAs and small nuclear ribonucleoproteins (RNPs) [36]. CBs disappear from prophase nuclei and reappear in late G1 [38]. Thus, during prophase, FBL is directed to the chromosomal periphery until anaphase [39, 40]. Then, during telophase, FBL is significantly accumulated in prenuclear bodies, which indicates its role in nucleolar assembly [41]. In early G1 phase, FBL localizes in condensed chromatin of nuclei. Depletion of FBL by siRNA (with a reduction of expression levels) in HeLa cells showed nuclear morphology defects. Nucleolin depletion, another abundant nucleolar protein, did not affect nuclear morphology. Also, FBL depletion in HeLa cells showed decreased cell growth. This reduction is thought to arise due to a delay at the G2-M transition of the cell cycle, rather than a direct defect in DNA synthesis [42]. Thus, FBL is involved in nuclear maintenance and cell growth in HeLa cells.
FBL catalyzes the 2′-O-methylation (2′-O-Me) of pre-rRNA[43] and regulates rRNA transcription by methylating histone H2A [44, 45]. In eukaryotes, FBL forms an RNP complex with Nop56, Nop58, and 15.5 ka proteins, which is guided by one of several C/D box small nucleolar RNAs (snoRNA). Depending on the organism, this complex is responsible for more than 100 sites of methylations in pre-rRNA [46].
In eukaryotes, the FBL N-terminal domain consists of the GAR domain, an IDR, and a spacer region. A novel ribonuclease activity for rRNA was described within the GAR domain of human FBL 1 and Arabidopsis thaliana FBL 2 (AtFib2) [47, 48]. Its ribonuclease activity is affected by phosphoinositides and phosphatidic acid. The GAR domain contains a nucleolar localization signal (NoLS) and is responsible for interacting with multiple cellular and viral proteins [35, 49]. The binding of phosphoinositides and FBL may facilitate phase separation necessary for pre-rRNA processing and DFC formation. This ribonuclease activity is blocked when U3 is bound [47]. The most abundant snoRNA, U3, despite to complex with FBL, does not methylates pre-rRNA, but aids in its cleavage [50, 51]. On the other hand, the FBL C-terminal end has an RNA-binding motif[52] and a conserved structure of -sheets and -helical domains, which constitute the AdoMet-dependent MT domain [53]. The alpha-helix domain interacts with Nop56 protein, which is indispensable for the association with the C/D box snoRNPs [54].
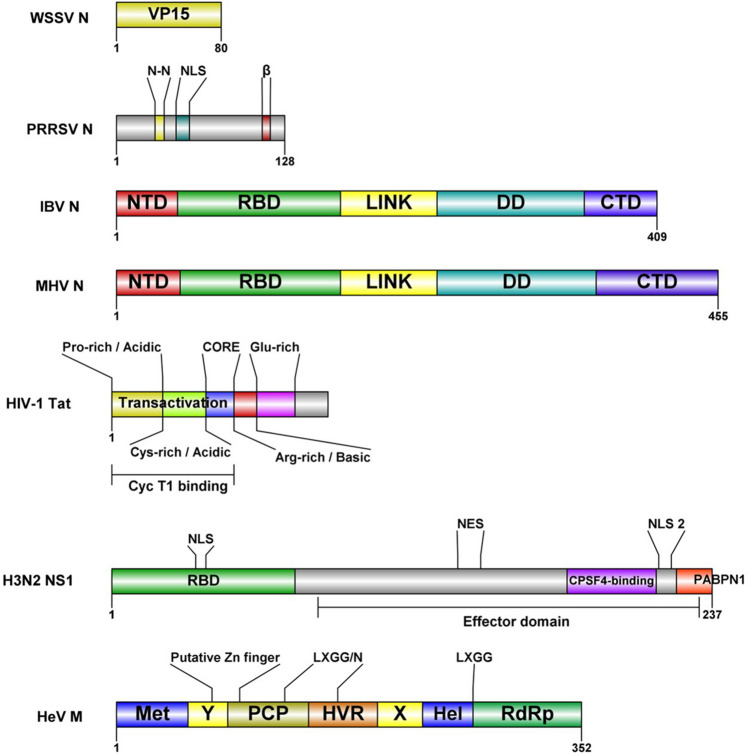
FBL is a target for distinct types of stresses. FBL is implicated in human prostatic intraepithelial neoplasia that can progress to prostate cancer [55] and in human adenocarcinoma [56]. In breast cancer, tumors express high levels of FBL [57]. However, a low level of FBL has also been found [58]. p53 decreases the expression and protein level of FBL by interacting with FBL intron one sequence that contains a p53 regulatory site. Reduced levels of p53 in breast cancer cells result in an increased level of FBL, which promotes abnormal//atypical methylations in rRNA that affects translational fidelity, and an increased internal ribosomal entry sites (IRES) of key cancer genes [57]. Furthermore, FBL has been a target of plant viruses and animal viruses. In certain plant viruses, FBL has been reported to participate in cell-to-cell and long-distance movement and has a putative role in virus‑mediated suppression of RNA silencing (for a review, see reference [49]). The viral proteins known to associate with FBL from animal and human viruses (Table 1) differ in function. However, their coding sequences have common features, for example, IDR, RNA-binding domains (RBDs), nuclear and nucleolar localization signals (NLS/NoLS). Such motifs may be crucial for the association with FBL (Fig. 2). Here, we review the molecular mechanisms of human and animal viruses to manipulate FBL and the implications over the host-cell processes, such as ribosome biogenesis.
Fig. 2.
Domains of viral proteins that are associated with FBL. WSSV (white spot syndrome virus) Nucleocapsid protein VP15 [59]; PRRSV (porcine reproductive and respiratory syndrome virus) Nucleocapsid domains: N-N, N protein-N protein interactive domain [26]; NLS, nuclear localization signal [60]; β-strand conserved among arteriviruses [61]; IBV (infectious bronchitis virus) and MHV (mouse hepatitis virus) Nucleocapsid domains: NTD, predicted intrinsically disordered N-terminal domain; RBD, RNA-binding domain; LINK, predicted disordered central linker; DD, dimerization domain; CTD, predicted disordered C-terminal domain [62]; HIV-1 (human immunodeficiency virus-1) Tat protein: transactivation site ( Proline and Cystein rich acidic site; CORE), arginine rich basic site (functions as a RNA-binding domain (RBD), a protein transduction domain (PTD) and nuclear localization signal (NLS), glutamine-rich region [63]; Influenza A H3N2-NS1 protein domains: RBD, RNA binding domain; NLS, nuclear localization signal; Effector domain: NES, nuclear export signal; NLS2, nuclear localization signal; CPSF4, factor subunit 4; PABPN1, polyadenine binding protein 1 [12]; HeV (Hendra virus) Met, methyl transferase; X and Y domains, PCP, a papain-like cysteine protease; HVR, proline-rich hypervariable region, Hel, RNA helicase; RdRp, RNA-dependent RNA polymerase [64]
Nucleocapsid proteins tend to redistribute FBL
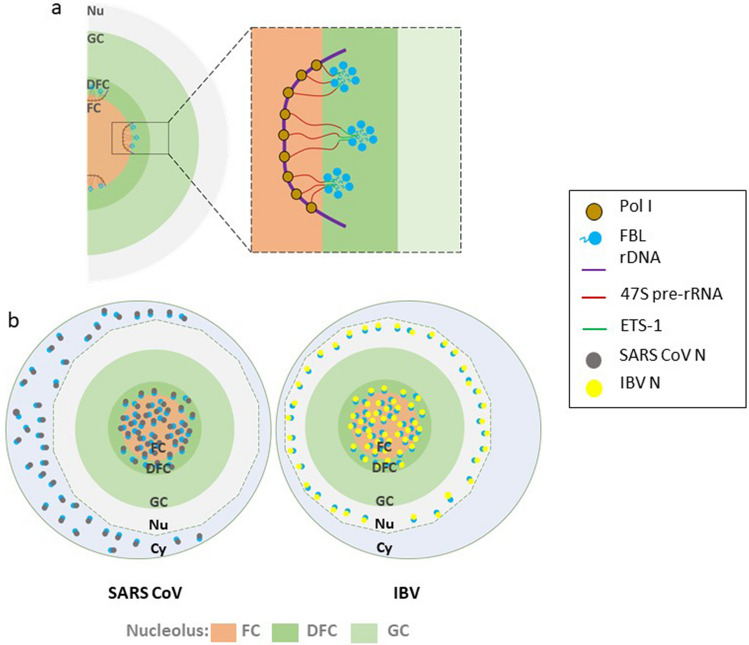
FBL has been shown to co-localize with the nucleocapsid of different mammalian viruses, including white spot syndrome virus (WSSV), infectious bronchitis virus (IBV), mouse hepatitis virus (MHV), porcine reproductive and respiratory syndrome virus (PRRSV) and severe acute respiratory syndrome coronavirus (SARS-CoV) [25, 26, 28, 65, 66]. The infection of these viruses and the expression of the nucleocapsid without its respective virus results in a redistribution of FBL to the perinuclear region, where transcription and RNA metabolism takes place in the host cell. Such redistribution of FBL may affect the early steps of pre-rRNA processing since the nascent transcripts emerge in the FC-DFC interface where they are bound to the RBD of FBL to finally be sorted (as a complex) to the DFC to initiate the pre-rRNA processing (Fig. 3a) [22]. Interestingly, just SARS-Cov N protein has been reported to relocalize a fraction of FBL to the cytoplasm (Fig. 3b) [66]. Compared to adjacent uninfected cells or mock-infected cells, FBL forms its classic “Christmas tree-like” structure [52].Chen et al. (2002) reported that after 20 fields of view in duplicate experiments, the Christmas tree-like structure was inexistent in observed IBV-infected cells. The localization of nucleocapsid proteins was primarily cytoplasmic and nuclear but showed a speckled pattern. In virology, these speckled structures are formed within the nucleus, commonly functioning as a viral assembly matrix or an RNA transcription site [67]. However, it seems that the subcellular localization of nucleocapsid protein depends on the infection stage.Yoo et al. (2003) tracked the subcellular distribution of PRRSV N protein through infection time. From 6 to 20 h p.i., the localization was mainly nuclear and nucleolar, whereas by the time of 30 to 48 h p.i., it turns exclusively cytoplasmic [61]. It is important to highlight that these findings were carried out in different cell lines, such as Vero or HeLa cells. The observations were similar, indicating that the redistribution of FBL is a potential common feature and not unique for a particular cell line.
Fig. 3.
Comparison of FBL redistribution by SARS CoV and IBV N proteins. a FBL nucleolar organization during nascent pre-rRNA sorting. Nascent transcripts synthesized in FC are pulled to the FC/DFC interface by the RBD of FBL. FBL diffuses to the DFC, where self-associates through its GAR domain. The IDR of the GAR domain establishes the DFC phase, where nascent transcripts move for initial pre-rRNA processing. See text for details. b Although SARS CoV and IBV bind FBL with the same viral protein (N), different redistribution of FBL occurs. While SARS CoV N (grey circles) co-localizes with FBL (blue circles) in the nucleolus and cytoplasm, IBV N (yellow circles) redistributes a fraction of FBL to the nucleus. Nu nucleus; Cy cytoplasm
Just the N protein of PRRSV is known, to our knowledge, to form a physical association with FBL [26]. The in vivo interaction of PRRSV N- FBL complex was confirmed by the mammalian two hybrid assay in HeLa cells. This result was corroborated in vitro by the GST-pull down assay. Furthermore, by a series of deletion mutants from both PRRSV N protein and FBL, the authors identified the GAR domain from FBL and eight (30IAQQNQSR37) amino acids of PRRSV N protein, are the interactive domains [26]. Unfortunately, interaction assays between the nucleocapsids mentioned above and FBL are pending to be done. Alternatively, PRRSV N is known to bind not just viral genomic RNA but also with rRNA.Yoo et al. (2003) investigated if the association with rRNA was involved with the FBL interaction. The addition of RNase A to an immobilized GST- FBL fusion protein on glutathione sepharose beads and incubated with radiolabeled PRRSV N protein inhibited the interaction. In contrast, the absence of RNase A enables the physical association. Thus, it is hypothesized that the rRNA binding makes a conformational change in N to establish a stable interaction with FBL. This might be the case since the co-localization between both proteins did not occur in CBs, where traces of FBL exist at this sub-nuclear compartment but not rRNA. On the contrary,Chen et al. (2002) spotted the co-localization in a speckled pattern in the nucleus of Vero cells but expressing (distinct cell lines) the MHV and IBV N proteins, at what appears to be CBs. The co-localization at CBs varies from one virus to another; even so, what remains constant is their distribution between the DFC and the FC within the nucleolus.
According to the findings of each N protein described in this section, the viral protein behaves similarly with FBL. Further studies should be done to elucidate this association’s precise role. Another coronavirus possibly interacting with FBL is the severe acute respiratory syndrome-2 (SARS CoV-2). SARS CoV-2 RNA interacts in the nucleolus with snoRNA U27 and is 2′-O-methylated, while 2′-O-Me levels in host RNAs decrease after SARS CoV-2 infection [68]. snoRNA U27 is responsible for 18 S rRNA methylation and is associated with FBL, NOP56, NOP58 and NHP2L1 [69]. These data may suggest that FBL is hijacked by the strong association between SARS CoV-2 RNA and U27 snoRNA, where U27 guide FBL to perform 2′-O-Me over SARS CoV-2 RNA. This idea corroborates the decrease 2′-O-Me levels in host RNAs since significant amounts of FBL may be away from pre-rRNA processing. Unfortunately, there is no report for an interaction. However, it´s most abundant protein, N, localizes in the cytoplasm and nucleolus [70], just as SARS CoV N [66], suggests it is a possible interactor. Nonetheless, the redistribution of FBL might affect ribosomal biogenesis and, consequently, the host translation machinery. Host cell translation is decreased under MHV infection, while viral mRNAs are unaffected or upregulated [27]. Therefore, N proteins might hijack FBL to disrupt their normal functions and for replication, transcription, and/or translation of viral RNAs.
Human immunodeficiency virus-1 Tat protein hijacks FBL to impair pre-rRNA processing
The human immunodeficiency virus-1 (HIV-1) encodes for two proteins imported to the nucleolus, Rev and Tat protein. NPM1 is reported to import both viral proteins to the nucleus, which are then retained in the nucleolus through interaction with nucleolar components [70, 71]. Unlike the HIV-1 Rev protein, Tat protein localization depends on its expression level and function [72].
The HIV-1Tat protein regulates viral gene transcription and manipulates diverse cellular processes, mainly nuclear, through interaction with nuclear components [73–75]. HIV-1 Tat is classified as an intrinsically disordered protein (IDP) [76], which may explain the ability to associate with several cellular components since IDPs have a greater surface area, giving the ability to bind multiple partners [77]. In fact, through genome-wide occupancy of Tat protein on host cell chromatin in HIV-1-infected T-cells, 568 genes were identified to bind significantly to Tat protein. So, they might be acting as a transcriptional regulator, since Tat protein locates at the promoter of 66% of the reported genes. This is the case for c-Rel, downregulated by the interaction of Tat with kappa B nuclear factor (NFB) binding sites on the promoter [78].
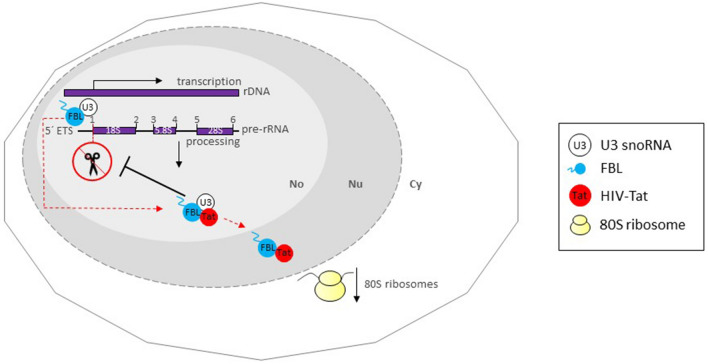
Tat protein localization might depend on its cellular concentration. In HIV-1 infected Jurkat cells, when the expression levels of Tat were high, the localization was nucleolar. Conversely, when the levels were low, it was nucleoplasmic [79]. The same pattern occurred in stable transfected HeLa cells expressing Tat-GFP [80]. Meanwhile, Tat transgenic Drosophila melanogaster cells, localize in both regions, although the concentration difference wasn’t studied [81]. In addition, since Tat protein is secreted by infected cells and internalized by neighboring cells, its intranuclear concentration is lower than in infected cells. Therefore the distribution is mainly nuclear, while in HIV-infected cells is nucleolar. Tat can indirectly modify host gene expression or directly interact with the gene promoters [78, 82, 83]. According to the expression profiles of deregulated genes, a distinct subset of genes are regulated depending on their intranuclear concentration and localization. In Tat stable transfected Jurkat T cells (nucleoplasmic localization), deregulated genes are linked to viral carcinogenesis, stem cell pluripotency, B cell receptor signaling and cancers. Alternatively, nucleolar Tat in HIV-infected T cells, deregulate genes involved mainly with ribosomal biogenesis [72]. This is the case for Drosophila nurse cells expressing Tat protein, where the levels of 80 S ribosome particles are reduced. Ponti et al. (2008), found that Tat affects at least cleavage site 1, impairing pre-rRNA maturation of Drosophila Tat expressing-cells. Tat protein immunoprecipitates with FBL and with U3 snoRNA. Thus, the inhibition of pre-rRNA processing by Tat might be induced through the interaction with these key nucleolar components for the early steps of pre-rRNA processing, preventing them from functioning in ribosomal biogenesis (Fig. 4). Therefore, Tat protein might interfere with FBL binding to the 5´end of nascent 47 S pre-rRNA. In concordance with this, FBL knockdown in HeLa cells showed aberrant accumulation of 47 S and 34 S pre-rRNAs, accompanied by reduced 28 S and 18 S rRNAs [5].Ponti et al. (2008) reported a co-localization of Tat protein and FBL at the nucleoplasm and nucleolus, indicating a redistribution of FBL, since during interphase, FBL localizes in the transition zone between FC and DFC, which is transcriptionally active [5, 35].
Fig. 4.
Association of HIV Tat with FBL and U3 snoRNA impairs pre-rRNA processing. In HIV-infected cells, nucleolar Tat protein inhibits the processing of the pre-rRNA by subtracting (red dotted arrow indicates movement) FBL and U3 snoRNA from the initial cleavage of ETS-18 S pre-rRNA. The Tat-mediated FBL redistribution to the nucleoplasm, as well, affects pre-rRNA processing and a reduced 80 S ribosome particles levels. No nucleolus, Nu nucleus, Cy cytoplasm
Hendra virus orchestrates FBL 2´-O-Me to influence proviral host genes and viral proteins synthesis
Hendra virus (HeV) and Nipa virus (NiV), both highly pathogenic viruses transmitted from bats to animals and humans, belong to the Henipavirus genus classified in the Paramyxoviridae family [84]. Both viruses require an overlapping subset of host genes for infection. A genome-wide analysis of host genes required for henipavirus infection identified multiple genes involved in ribosomal biogenesis, nuclear export/import, and transcriptional regulation. From the 585 proviral genes identified, FBL exhibited the highest impact on henipavirus infection. HeV and NiV infection was inhibited by more than 99.9% when FBL was knockdown. Deffrasnes et al. (2016) tested the dependency on FBL of cytoplasmic replicating viruses belonging to different genera in the same subfamily: measles virus (MeV, genus Morbilivirus), mumps virus (MuV, genus Rubulavirus), respiratory syncytial virus (RSV, genus Pneumovirus), and a nuclear replicating virus: influenza A/WSN/33. Interestingly, the cytoplasmic replicating virus showed a significant reduction in titer cells transfected with siFBL, except for the nuclear replicating virus. In general, FBL is a key factor for paramyxovirus infection.
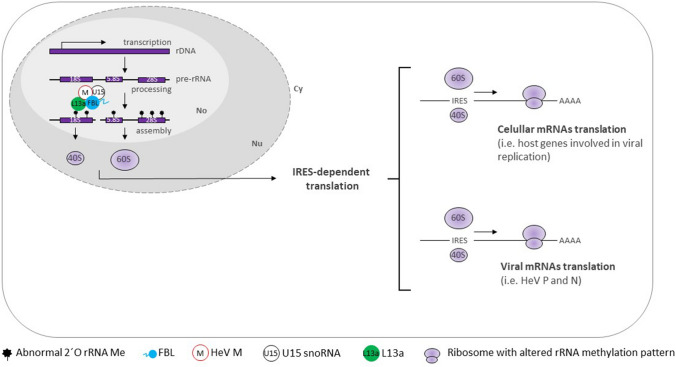
The link between FBL and paramyxoviruses, prompted Deffrascnes et al. (2016) to elucidate the precise role across the viral cycle. HeV-infected HeLa cells transfected with siFBL resulted in significantly reduced levels of viral RNA. The same pattern occurred to HeV gene expression levels since P and N (the most abundant viral proteins) synthesis was barely detected in HeV-infected cells treated with siFBL. (+)ssRNA viruses do not contain 5′ cap, so its viral protein synthesis is initiated through IRESs translation initiation. IRES-dependent translation is influenced over CAP-dependent translation by rRNA 2′-O-Me [85]. In breast cancer cells, changes in FBL expression were correlated with alterations of rRNA 2′-O-Me levels, with efficient translational initiation of mRNAs containing IRES elements [57]. Another host gene required for Henipavirus infection was L13a, a protein necessary for rRNA methylation and IRES-mediated translation [86]. Co-immunoprecipitation and co-localization assays demonstrate that FBL complex with L13A and U15 snoRNA [87]. Therefore, it is conceivable that HeV orchestrates FBL methylation to influence proviral host genes and viral protein synthesis (Fig. 5) since these viruses are known to target host translation factors to shut down translation and/or boost viral translation [88].
Fig. 5.
HeV infections orchestrate FBL 2′-O-Me to influence proviral host genes and viral proteins synthesis by IRES-dependent translation. During HeV infection, viral M proteins (by an unknown mechanism) alter pre-rRNA 2′-O-Me by physically interacting with FBL, U15 snoRNA and protein L13a in the nucleolus. HeV M manipulates FBL 2′-O-Me to induce IRES-dependent translation of proviral host genes (containing IRES elements) and HeV proteins. No nucleolus, Nu nucleus, Cy cytoplasm
The requirement of the methylation activity of FBL for henipavirus infection was studied through a rescue experiment. HeLa cells were transfected with siFBL-2 to knock down endogenous FBL expression, then transfected with a plasmid encoding a yeast FBL (NOP1) E191A mutant. This mutation impaired the methylation activity in vitro, followed by infection with HeV. The E191A mutant was unable to rescue HeV infection, compared to control cells. Thus, suggesting that the methylation activity of FBL is required for HeV infection. Furthermore, to corroborate if the methylation of pre-ribosomes by FBL is precisely the target of henipaviruses, Deffrasnes et al. (2016) reduced the expression of NOP56 and NOP58 by SMART pool siRNAs and tested the impact on HeV infection. Depletion of either NOP56 or NOP58 impaired HeV infection, as measured by TCID50 assays and immunofluorescence. In summary, these findings support the idea that henipavirus infection exploits the role of FBL in pre-rRNA processing.
In addition, by confocal microscopy and reciprocal co-immunoprecipitation studies, the HeV matrix (M) was shown to colocalize with FBL in the nucleolus and, to a lesser extent, in the nucleoplasm, and to associate directly or indirectly, respectively [32]. However, the interaction still needs to be determined. Since FBL is well documented to localize in nucleoli during interphase, HeV infection might redistribute a small fraction of FBL to the nucleoplasm.
Epstain-Barr virus v-snoRNA1 assemble with FBL and NOP56/58
The snoRNAs are the regulatory elements responsible for posttranscriptional rRNA maturation of two different modifications, the C/D box snoRNAs and H/ACA box snoRNAs, which catalyzes 2′-O-Me and pseudouridylations, respectively [89]. Mainly, C/D box snoRNAs are typically 70–90 nt long and serve as the scaffold for the assembly of snoRNP, including FBL [90]. Of note, several known C/D box snoRNAs do not have a predicted rRNA target, so they are classified as “orphan”. Alternatively, some C/D box snoRNAs have been found to associate with proteins other than FBL, NOP56/58, and 15.5. They indicated that C/D box snoRNAs could associate with complexes lacking a methylase.
Evidence indicates that snoRNAs are crucial for viral infectivity [50]. Although most viruses do not encode snoRNAs, and mainly RNA viruses use C/D box snoRNAs, Epstein-Barr virus (EBV) (dsDNA genome) is the exception. EBV belongs to the -subfamily of herpesvirus and infects approximately 95% of the world’s population [70]. EBV encodes a viral-specific v-snoRNA1. v-snoRNA1 is conserved within various EBV strains and among evolution. Therefore, the v-snoRNA1 serves in both the latent and lytic mode of infection, but with different functions [31]. Interestingly, v-snoRNA1 was shown to co-localize and assembled in the nucleolus with NOP56/58 and FBL, while U3 snoRNA, co-localized in the nucleolus with v-snoRNA1 as well. FBL is present in the U3 complex; however, U3 does not methylate pre-rRNA, but aids in its cleavage [51]. In vitro studies determined that human FBL has a ribonuclease activity for rRNA, and this second activity is blocked when U3 is bound [47]. Hence, v-snoRNA1 might compete for FBL and NOP56/58 against U3 snoRNA, affecting pre-rRNA maturation and affecting efficient protein synthesis.Hutzinger et al. (2009) classified v-snoRNA1 as an “orphan” snoRNA, since neither ribosomal or spliceosomal RNA were determined as targets. Also, by comparing a null mutant of v-snoRNA1 against its wild-type version, it was concluded that the v-snoRNA1 mutant remained indistinguishable from its wild-type counterparts in terms of lytic replication, infection and B cell transformation. However, the increment up to 30-fold expression levels upon induction of the lytic replication cycle, undoubtedly assures the relevance of v-snoRNA1, particularly at this stage of infection. Studies may be conducted to establish the precise role of EVB v-snoRNA1.
Influenza a H3N2 NS1 arrests rRNA transcription by the interaction with nucleolin, B23, and FBL
Influenza virus A (IAV) is a (−)ssRNA virus capable of translating viral mRNAs while attenuating host protein synthesis [91]. IAV has evolved a mechanism where the 5′-m7G cap is excised from host mRNAs and snoRNAs, which serves as a primer to synthesize the viral (+)-sense mRNA, and ultimately the synthesis of a 3′ poly(A) tail from a poly(U) stretch. Once the viral mRNAs are exported to the cytoplasm, the translation mechanism remains elusive. As in poliovirus and encephalomyocarditis virus infection, IAV is proposed to induce mTOR pathway and activate the 4E binding proteins (4EBP). 4EBP sequester eIF4E prevents interaction with eIF4G, inhibiting canonical cap-dependent translation initiation therefore the translation of the viral mRNA might be independent of 5´cap recognition by eIF4E.
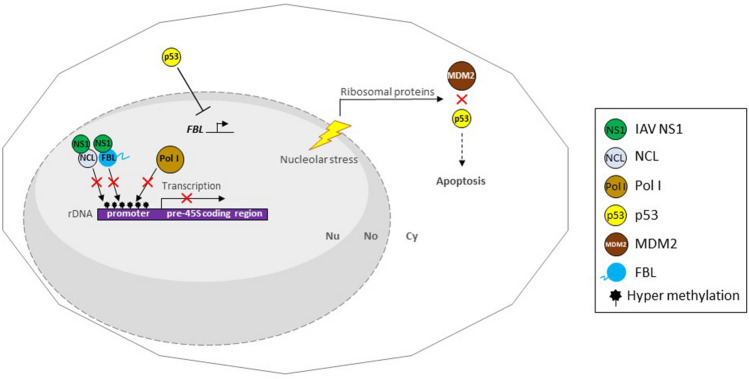
The genome of IAV is segmented into eight RNAs that encode for 12 viral structural and nonstructural proteins [92]. Among those viral proteins, the non-structural protein 1 (NS1), is one of the major virulence factors. IAV NS1 is a multi-functional protein, with a N-terminal dsRNA-binding domain and a C-terminal effector domain. It plays essential roles to counteract immune responses of the infected hosts [93–97]. All types of NS1 have an N-terminal nuclear localization signal 1 (NLS1), but particularly the H3N2 NS1 has an additional NLS2 within the C-terminal domain, which also functions as a NoLS. Thus, only the NS1 protein of IAV H3N2 subtype virus is characterized to distribute inside the nucleolus [29, 98]. Interestingly, the NS1 protein of the human H3N2 virus interacts through the NLS2/NoLS and in a minor extent the NLS1 with the main nucleolar proteins: nucleolin, B23 and FBL. Moreover, confocal laser microscopy revealed that the NS1 protein co-localizes with nucleolin in the nucleolus and nucleoplasm, while with B23 and FBL only in the nucleolus of IAV/Udorn/72 virus-infected A549 cells [29, 99]. B23/nucleophosmin is a nucleolar phosphoprotein involved in 28 S rRNA processing and ribosome assembly that localized in the GC. Knocking down B23 inhibits the processing of pre-rRNA leading to cell death. On the contrary, the overexpression promotes S-phase entry in cells lacking p53 [100]. Nucleolin, a multifactorial protein involved in several cellular processes, is also implicated in the transcription and maturation of rRNA, and ribosome assembly and transport [101]. Nucleolin inactivation induces nucleolar disruption, which leads to apoptosis [102]. H3N2 NS1 protein represses RNA Pol I-dependent transcription through nucleolin interaction, by hyper-methylation in the UCE of rRNA gene promoter. In NS1 expressed cells, an increased association of ribosomal proteins with MDM2, and p53 accumulation, suggests an induced nucleolar stress that would promote apoptosis [103]. These findings imply that H3N2 NS1 mediate depletion of rRNA which in turn triggers nucleolar stress, resulting in p53-dependent apoptosis.
The Pol I repressor, p53, also represses the expression of FBL. FBL expression is inversely associated with p53 activity in human breast cancer cells [57]. Ribosome biogenesis is overactivated in p53-inactivated cancer cells [104]. In the context of IAV H3N2 infection both the accumulation of p53 might regulate FBL expression, and the interaction with NS1 may prevent it to establish its physiological processes. In general, the role of H3N2 NS1 by targeting nucleolin, B23 and FBL strongly suggests the impairment of ribosomal biogenesis and its above-mentioned consequences (Fig. 6).
Fig. 6.
IAV H3N2 NS1 blocks rRNA transcription resulting in nucleolar stress and p53-dependent apoptosis. H3N2 NS1 binds to nucleolin and FBL in the nucleolus. NS1/nucleolin association disrupts the ability of nucleolin to bind rDNA, which promotes hyper-methylation state of UCE region of rDNA promoter. Thus, recruitment of RNA Pol I to hyper-methylated rDNA promoter is compromised, which leads to nucleolar stress. Depletion of rRNA leads to the release of ribosomal proteins and their interaction with MDM2, which blocks p53-degradation. Accumulated p53 leads to apoptosis of IAV-infected cells. In addition, unregulated p53 represses FBL transcription and residual FBL is targeted by NS1, blocking FBL to conduct its role in pre-rRNA processing. No nucleolus, Nu nucleus, Cy cytoplasm
Conclusions and future directions
The nucleolus is the largest nuclear compartment. It contains more than 4500 proteins, which functions are distributed in controlling the cell cycle; DNA replication and repairing; pre-rRNA processing; assembling signal recognition particles; detecting and responding to different kinds of stress stimuli; and more [105, 106]. Thus, the nucleolus as a multifunctional compartment, serves as an interface for the interaction between pathogens and the host cell. Viruses as obligate intracellular parasites, often encode viral proteins that target to the nucleolus to take over host-cell functions, and hijack nucleolar proteins to facilitate an effective infection process [107–109].
In plant viruses, FBL is involved in long-distance movement (LDM) and cell-to-cell movement, and has been involved in virus‑mediated suppression of RNA silencing. The redistribution of FBL is a constant phenomenon in plant and animal viruses. Regardless, cytoplasmic FBL redistribution has been reported only in plants virus infections. This supports the idea that particularly animal viruses targets FBL for its nucleolar functions in rRNA processing [110, 111]. In summary, at least from the described viruses in this review, comply with the following: (1) FBL knockdown impairs viral infection; (2) ribosomal biogenesis is blocked by different mechanisms; and (3) viral proteins with FBL interaction possesses in their sequence IDRs, RBDs and NLS/NoLS.
Most of the viruses described here, reported a co-localization with FBL, but just a few were explored to forma physical interaction. It is important to establish whether there is an interaction or not. Since the FBL GAR domain has been reported to be the binding site for viral proteins from plant[49] and animal viruses, and this domain has a ribonuclease activity[47, 48] and an important role in the early steps of pre-rRNA processing [22], it is possible that these viral interactions may affect these particular roles of FBL. Thus, do these interactions affect the methyl transferase and/or the ribonuclease activity of FBL? Can disrupt the ability of FBL to form RNPs complexes? The viral protein-mediated redistribution of FBL affects the nascent pre-rRNA sorting via phase separation? Answers of these questions may contribute to a more detailed understanding of the precise role of FBL in virology, consequently facilitating the design of novel anti-viral therapies.
Author contributions
SD-C contributed with the writing and research for the review. ALR-P contributed with the writing and research for the review. LCR-Z contributed with the editing and managing. EC defined the subject rewrite and overall management of the review and defined the model.
Funding
This research was funded by CONACYT project FC 1572, and CB-2016-286730-Z-0142.
Data availability
There is no additional data, Not applied.
Code availability
Not applied.
Declarations
Conflict of interest
The authors declare that there is no competing interest and that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thiry M, Lafontaine DLJ. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005;15:194–199. doi: 10.1016/J.TCB.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Németh A, Conesa A, Santoyo-Lopez J, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010 doi: 10.1371/JOURNAL.PGEN.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 4.Lafontaine DLJ. Birth of nucleolar compartments: phase separation-driven ribosomal RNA sorting and processing. Mol Cell. 2019;76:694–696. doi: 10.1016/J.MOLCEL.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Yao RW, Xu G, Wang Y, et al. Nascent Pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human Nucleolus. Mol Cell. 2019;76:767–783e11. doi: 10.1016/J.MOLCEL.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Feric M, Vaidya N, Harmon TS, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/J.CELL.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/J.JMB.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 8.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2014;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung HYJ, Birol M, Rhoades E. IDPs in macromolecular complexes: the roles of multivalent interactions in diverse assemblies. Curr Opin Struct Biol. 2018;49:36–43. doi: 10.1016/J.SBI.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr Opin Struct Biol. 2021;67:41–50. doi: 10.1016/J.SBI.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittag T, Parker R. Multiple modes of protein–protein interactions promote RNP granule assembly. J Mol Biol. 2018;430:4636–4649. doi: 10.1016/J.JMB.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin X. Regulatory network of serine/arginine-rich (SR) proteins: the molecular mechanism and physiological function in plants. Int J Mol Sci. 2022 doi: 10.3390/IJMS231710147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greig JA, Nguyen TA, Lee M, et al. Arginine-enriched mixed-charge domains provide cohesion for Nuclear Speckle Condensation. Mol Cell. 2020;77:1237–1250e4. doi: 10.1016/J.MOLCEL.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Järvelin AI, Noerenberg M, Davis I, Castello A. The new (dis)order in RNA regulation. Cell Commun Signal. 2016;14:1–22. doi: 10.1186/S12964-016-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. doi: 10.1016/J.MOLCEL.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/J.CELL.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 17.Mészáros B, Simon I, Dosztányi Z. The expanding view of protein-protein interactions: complexes involving intrinsically disordered proteins. Phys Biol. 2011 doi: 10.1088/1478-3975/8/3/035003. [DOI] [PubMed] [Google Scholar]
- 18.Xue B, Blocquel D, Habchi J, et al. Structural disorder in viral proteins. Chem Rev. 2014;114:6880–6911. doi: 10.1021/CR4005692. [DOI] [PubMed] [Google Scholar]
- 19.Stenström L, Mahdessian D, Gnann C, et al. Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol Syst Biol. 2020;16:e9469. doi: 10.15252/msb.20209469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–7194. doi: 10.1073/PNAS.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nott TJ, Petsalaki E, Farber P, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/J.MOLCEL.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2020;22(3):165–182. doi: 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- 23.Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in studying liquid-liquid phase separation and Biomolecular Condensates. Cell. 2019;176:419–434. doi: 10.1016/J.CELL.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berry J, Weber SC, Vaidya N, et al. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A. 2015;112:E5237–E5245. doi: 10.1073/PNAS.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Wurm T, Britton P, et al. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J Virol. 2002;76:5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo D, Wootton SK, Li G, et al. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J Virol. 2003;77:12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilton A, Mizzen L, Macintyre G, et al. Translational control in murine hepatitis virus infection. J Gen Virol. 1986;67(Pt 5):923–932. doi: 10.1099/0022-1317-67-5-923. [DOI] [PubMed] [Google Scholar]
- 28.Xing Y, Shi Z. Nucleocapsid protein VP15 of white spot syndrome virus colocalizes with the nucleolar proteins nucleolin and fibrillarin. Can J Microbiol. 2011;57:759–764. doi: 10.1139/W11-061. [DOI] [PubMed] [Google Scholar]
- 29.Melén K, Tynell J, Fagerlund R, et al. Influenza a H3N2 subtype virus NS1 protein targets into the nucleus and binds primarily via its C-terminal NLS2/NoLS to nucleolin and fibrillarin. Virol J. 2012 doi: 10.1186/1743-422X-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponti D, Troiano M, Bellenchi G, et al. The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol. 2008;9:1–10. doi: 10.1186/1471-2121-9-32/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutzinger R, Feederle R, Mrazek J, et al. Expression and processing of a small nucleolar RNA from the Epstein-Barr Virus Genome. PLoS Pathog. 2009;5:e1000547. doi: 10.1371/JOURNAL.PPAT.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deffrasnes C, Marsh GA, Foo CH, et al. Genome-wide siRNA screening at biosafety Level 4 reveals a crucial role for Fibrillarin in Henipavirus infection. PLoS Pathog. 2016;12:e1005478. doi: 10.1371/JOURNAL.PPAT.1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–133. doi: 10.1111/j.1768-322X.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 34.Jansen RP, Hurt EC, Kern H, et al. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol. 1991;113:715–729. doi: 10.1083/JCB.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Corona U, Sobol M, Rodriguez-Zapata LC, et al. Fibrillarin from Archaea to human. Biol Cell. 2015;107:159–174. doi: 10.1111/boc.201400077. [DOI] [PubMed] [Google Scholar]
- 36.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/ANNUREV.CELLBIO.16.1.273. [DOI] [PubMed] [Google Scholar]
- 37.Dundr M, Hebert MD, Karpova TS, et al. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/JCB.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol. 2003;4:975–980. doi: 10.1038/NRM1262. [DOI] [PubMed] [Google Scholar]
- 39.Takata H, Uchiyama S, Nakamura N, et al. A comparative proteome analysis of human metaphase chromosomes isolated from two different cell lines reveals a set of conserved chromosome-associated proteins. Genes Cells. 2007;12:269–284. doi: 10.1111/J.1365-2443.2007.01051.X. [DOI] [PubMed] [Google Scholar]
- 40.Uchiyama S, Kobayashi S, Takata H, et al. Proteome analysis of human metaphase chromosomes. J Biol Chem. 2005;280:16994–17004. doi: 10.1074/JBC.M412774200. [DOI] [PubMed] [Google Scholar]
- 41.Fomproix N, Gébrane-Younès J, Hernandez-Verdun D. Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J Cell Sci. 1998;111(Pt 3):359–372. doi: 10.1242/JCS.111.3.359. [DOI] [PubMed] [Google Scholar]
- 42.Amin MA, Matsunaga S, Ma N, et al. Fibrillarin, a nucleolar protein, is required for normal nuclear morphology and cellular growth in HeLa cells. Biochem Biophys Res Commun. 2007;360:320–326. doi: 10.1016/J.BBRC.2007.06.092. [DOI] [PubMed] [Google Scholar]
- 43.Tollervey D, Lehtonen H, Jansen R, et al. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-F. [DOI] [PubMed] [Google Scholar]
- 44.Tessarz P, Santos-Rosa H, Robson SC, et al. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature. 2014;505:564–568. doi: 10.1038/nature12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loza-Muller L, Rodríguez-Corona U, Sobol M, et al. Fibrillarin methylates H2A in RNA polymerase I trans-active promoters in Brassica oleracea. Front Plant Sci. 2015;6:976. doi: 10.3389/fpls.2015.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunbar DA, Wormsley S, Lowe TM, Baserga SJ. Fibrillarin-associated box C/D small nucleolar RNAs in Trypanosoma brucei. Sequence conservation and implications for 2’-O-ribose methylation of rRNA. J Biol Chem. 2000;275:14767–14776. doi: 10.1074/JBC.M001180200. [DOI] [PubMed] [Google Scholar]
- 47.Guillen-Chable F, Corona UR, Pereira-Santana A, et al. Fibrillarin ribonuclease activity is dependent on the GAR domain and modulated by phospholipids. Cells. 2020;9:1–22. doi: 10.3390/cells9051143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez-Corona U, Pereira-Santana A, Sobol M, et al. Novel ribonuclease activity differs between fibrillarins from arabidopsis thaliana. Front Plant Sci. 2017;8:1–10. doi: 10.3389/fpls.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decle-Carrasco S, Rodríguez-Zapata LC, Castano E. Plant viral proteins and fibrillarin: the link to complete the infective cycle. Mol Biol Rep. 2021;48:4677–4686. doi: 10.1007/S11033-021-06401-1/TABLES/1. [DOI] [PubMed] [Google Scholar]
- 50.Stamm S, Lodmell JS. C/D box snoRNAs in viral infections: RNA viruses use old dogs for new tricks. Noncoding RNA Res. 2019;4:46–53. doi: 10.1016/J.NCRNA.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Granneman S, Vogelzangs J, Lührmann R, et al. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol Cell Biol. 2004;24:8600–8610. doi: 10.1128/MCB.24.19.8600-8610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aris JP, Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc Natl Acad Sci U S A. 1991;88:931–935. doi: 10.1073/PNAS.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng L, Starostina NG, Liu ZJ, et al. Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus. Biochem Biophys Res Commun. 2004;315:726–732. doi: 10.1016/J.BBRC.2004.01.114. [DOI] [PubMed] [Google Scholar]
- 54.Lechertier T, Grob A, Hernandez-Verdun D, Roussel P. Fibrillarin and Nop56 interact before being co-assembled in box C/D snoRNPs. Exp Cell Res. 2009;315:928–942. doi: 10.1016/J.YEXCR.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Koh CM, Iwata T, Zheng Q, et al. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–683. doi: 10.18632/ONCOTARGET.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller DM, Thomas SD, Islam A, et al. c-Myc and Cancer metabolism. Clin Cancer Res. 2012;18:5546. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marcel V, Ghayad SE, Belin S, et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013;24:318–330. doi: 10.1016/J.CCR.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Nguyen F, Lardy-Cleaud A, Carène D, et al. Low level of Fibrillarin, a ribosome biogenesis factor, is a new independent marker of poor outcome in breast cancer. BMC Cancer. 2022 doi: 10.1186/S12885-022-09552-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boonyakida J, Xu J, Satoh J, et al. Identification of antigenic domains and peptides from VP15 of white spot syndrome virus and their antiviral effects in Marsupenaeus japonicus. Sci Rep 2021. 2021;11(1 11):1–12. doi: 10.1038/s41598-021-92002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowland RR, Kervin R, Kuckleburg C, et al. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/S0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 61.Wootton SK, Rowland RRR, Yoo D. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J Virol. 2002;76:10569–10576. doi: 10.1128/JVI.76.20.10569-10576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/J.VIRUSRES.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clark E, Nava B, Caputi M. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget. 2017;8:27569–27581. doi: 10.18632/ONCOTARGET.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pallerla SR, Harms D, Johne R, et al. Hepatitis E Virus infection: circulation, Molecular Epidemiology, and impact on Global Health. Pathogens. 2020;9:1–21. doi: 10.3390/PATHOGENS9100856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You J, Dove BK, Enjuanes L, et al. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol. 2005;86:3303–3310. doi: 10.1099/VIR.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 66.Li FQ, Xiao H, Tam JP, Liu DX. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/J.FEBSLET.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pyper JM, Clements JE, Zink MC. The nucleolus is the site of Borna disease virus RNA transcription and replication. J Virol. 1998;72:7697–7702. doi: 10.1128/JVI.72.9.7697-7702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang SL, DeFalco L, Anderson DE, et al. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat Commun. 2021 doi: 10.1038/S41467-021-25357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falaleeva M, Pages A, Matuszek Z, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci U S A. 2016;113:E1625–E1634. doi: 10.1073/PNAS.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ariumi Y. Host Cellular RNA helicases regulate SARS-CoV-2 infection. J Virol. 2022 doi: 10.1128/JVI.00002-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arizala JAC, Chomchan P, Li H, et al. Identification of nucleolar factors during HIV-1 replication through rev immunoprecipitation and mass spectrometry. J Vis Exp. 2019 doi: 10.1007/S00018-015-2077-X. [DOI] [PubMed] [Google Scholar]
- 72.Musinova YR, Sheval E, Dib C, et al. Functional roles of HIV-1 Tat protein in the nucleus. Cell Mol Life Sci. 2016;73:589–601. doi: 10.1007/S00018-015-2077-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romani B, Engelbrecht S, Glashoff RH. Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J Gen Virol. 2010;91:1–12. doi: 10.1099/VIR.0.016303-0/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- 74.Mbonye U, Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454–455:328–339. doi: 10.1016/J.VIROL.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Lint C, Bouchat S, Marcello A. HIV-1 transcription and latency: an update. Retrovirology. 2013;10:1–38. doi: 10.1186/1742-4690-10-67/FIGURES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shojania S, O’Neil D. Intrinsic disorder and function of the HIV-1 Tat protein. Protein Pept Lett. 2010;17:999–1011. doi: 10.2174/092986610791498993. [DOI] [PubMed] [Google Scholar]
- 77.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/JMR.747. [DOI] [PubMed] [Google Scholar]
- 78.Dhamija N, Choudhary D, Ladha JS, et al. Tat predominantly associates with host promoter elements in HIV-1-infected T-cells – regulatory basis of transcriptional repression of c-Rel. FEBS J. 2015;282:595–610. doi: 10.1111/FEBS.13168. [DOI] [PubMed] [Google Scholar]
- 79.Coiras M, Camafeita E, Ureña T, et al. Modifications in the human T†cell proteome induced by intracellular HIV-1 Tat protein expression. Proteomics. 2006;6:S63–S73. doi: 10.1002/PMIC.200500437. [DOI] [PubMed] [Google Scholar]
- 80.Stauber RH, Pavlakis GN. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology. 1998;252:126–136. doi: 10.1006/VIRO.1998.9400. [DOI] [PubMed] [Google Scholar]
- 81.Ponti D, Troiano M, Bellenchi G, et al. The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol. 2008 doi: 10.1186/1471-2121-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kukkonen S, Martinez-Viedma MDP, Kim N, et al. HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells. Retrovirology. 2014;11:1–16. doi: 10.1186/1742-4690-11-30/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marban C, Su T, Ferrari R, et al. Genome-wide binding map of the HIV-1 Tat protein to the Human Genome. PLoS ONE. 2011;6:e26894. doi: 10.1371/JOURNAL.PONE.0026894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Broder CC, Wong KT. Henipaviruses. Neurotrop Viral Infect. 2016 doi: 10.1007/978-3-319-33133-1_3. [DOI] [Google Scholar]
- 85.Yi Y, Li Y, Meng Q, et al. A PRC2-independent function for EZH2 in regulating rRNA 2’-O methylation and IRES-dependent translation. Nat Cell Biol. 2021;23:341–354. doi: 10.1038/S41556-021-00653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chaudhuri S, Vyas K, Kapasi P, et al. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA. 2007;13:2224–2237. doi: 10.1261/RNA.694007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Basu A, Das P, Chaudhuri S, et al. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol Cell Biol. 2011;31:4482. doi: 10.1128/MCB.05804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mailliot J, Martin F. Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip Rev RNA. 2018;9:e1458. doi: 10.1002/WRNA.1458. [DOI] [PubMed] [Google Scholar]
- 89.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/S0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 90.Henras AK, Dez C, Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol. 2004;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 91.Yángüez E, Nieto A. So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus Res. 2011;156:1–12. doi: 10.1016/J.VIRUSRES.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 92.Jagger BW, Wise HM, Kash JC, et al. An overlapping protein-coding region in Influenza A Virus Segment 3 modulates the host response. Science. 2012;337:199. doi: 10.1126/SCIENCE.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Opitz B, Rejaibi A, Dauber B, et al. IFNβ induction by influenza a virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–938. doi: 10.1111/J.1462-5822.2006.00841.X. [DOI] [PubMed] [Google Scholar]
- 94.Min JY, Li S, Sen GC, Krug RM. A site on the influenza a virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363:236–243. doi: 10.1016/J.VIROL.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 95.Nemeroff ME, Barabino SML, Li Y, et al. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 96.Chen Z, Li Y, Krug RM. Influenza a virus NS1 protein targets poly(A)-binding protein II of the cellular 3’-end processing machinery. EMBO J. 1999;18:2273. doi: 10.1093/EMBOJ/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Min JY, Krug RM. The primary function of RNA binding by the influenza a virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006;103:7100–7105. doi: 10.1073/PNAS.0602184103/ASSET/4FCD07B8-7804-4543-B8C4-2C8583541BD4/ASSETS/GRAPHIC/ZPQ0180620620005.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Melén K, Kinnunen L, Fagerlund R, et al. Nuclear and nucleolar targeting of influenza a virus NS1 protein: striking differences between different virus subtypes. J Virol. 2007;81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murayama R, Harada Y, Shibata T, et al. Influenza a virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem Biophys Res Commun. 2007;362:880–885. doi: 10.1016/J.BBRC.2007.08.091. [DOI] [PubMed] [Google Scholar]
- 100.Itahana K, Bhat KP, Jin A, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/S1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 101.Cong R, Das S, Ugrinova I, et al. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012;40:9441–9454. doi: 10.1093/NAR/GKS720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ugrinova I, Monier K, Ivaldi C, et al. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol. 2007;8:1–16. doi: 10.1186/1471-2199-8-66/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yan Y, Du Y, Wang G, Li K. Non-structural protein 1 of H3N2 influenza a virus induces nucleolar stress via interaction with nucleolin. Sci Rep. 2017 doi: 10.1038/S41598-017-18087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bywater MJ, Poortinga G, Sanij E, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/J.CCR.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andersen JS, Lam YW, Leung AKL, et al. Nucleolar proteome dynamics. Nat 2004. 2005;433:7021. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 106.Bensaddek D, Nicolas A, Lamond AI. Quantitative proteomic analysis of the human nucleolus. Methods Mol Biol. 2016;1455:249–262. doi: 10.1007/978-1-4939-3792-9_20/FIGURES/3. [DOI] [PubMed] [Google Scholar]
- 107.Hiscox JA. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taliansky ME, Brown JWS, Rajamäki ML, et al. Involvement of the plant nucleolus in virus and viroid infections. Parallels with animal pathosystems. Adv Virus Res. 2010;77:119–158. doi: 10.1016/B978-0-12-385034-8.00005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hiscox JA. The nucleolus - a gateway to viral infection? Arch Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tonello F, Massimino ML, Peggion C. Nucleolin: a cell portal for viruses, bacteria, and toxins. Cell Mol Life Sci. 2022 doi: 10.1007/S00018-022-04300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153:169. doi: 10.1083/JCB.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no additional data, Not applied.
Not applied.