Abstract
As an essential horticultural crop, Citrus has carotenoid diversity, which affects its aesthetic and nutritional values. β,β-Xanthophylls are the primary carotenoids accumulated in citrus fruits, and non-heme di-iron carotene hydroxylase (BCH) enzymes are mainly responsible for β,β-xanthophyll synthesis. Previous studies have focused on the hydroxylation of BCH1, but the role of its paralogous gene in citrus, BCH2, remains largely unknown. In this study, we revealed the β-hydroxylation activity of citrus BCH2 (CsBCH2) for the first time through the functional complementation assay using Escherichia coli, although CsBCH2 exhibited a lower activity in hydroxylating β-carotene into β-cryptoxanthin than citrus BCH1 (CsBCH1). Our results showed that overexpression of CsBCH2 in citrus callus increased xanthophyll proportion and plastoglobule size with feedback regulation of carotenogenic gene expression. This study revealed the distinct expression patterns and functional characteristics of two paralogous genes, CsBCH1 and CsBCH2, and illustrated the backup compensatory role of CsBCH2 for CsBCH1 in citrus xanthophyll biosynthesis. The independent function of CsBCH2 and its cooperative function with CsBCH1 in β-cryptoxanthin biosynthesis suggested the potential of CsBCH2 to be employed for expanding the synthetic biology toolkit in carotenoid engineering.
Introduction
Carotenoids are the most widely distributed isoprenoid pigments, and they are synthesized by photosynthetic organisms and multiple non-photosynthetic microorganisms [1]. The prominent role of carotenoids in horticultural crops is responsible for the exterior quality of fruits and vegetables. Carotenoids also serve as vitamin A precursors and potent antioxidants with health benefits for humans [2, 3]. In nature, carotenoids are classified into two categories: carotenes and xanthophylls. In higher plants, the composition of xanthophylls such as lutein, β-cryptoxanthin, and zeaxanthin is remarkably conserved, with oxygen atoms in the structure [4, 5]. Xanthophylls are essential to plant photosynthesis. For example, the xanthophyll cycle influences the efficiency of absorbed light energy transfer to the PSII reaction center [6]. Furthermore, xanthophylls are beneficial to human health. Previous studies have shown that xanthophylls such as lutein and β-cryptoxanthin can effectively prevent eye diseases and inflammation, with high antioxidant activity [4, 7].
One of the main xanthophylls found in nature is β-cryptoxanthin, which serves as the precursor of vitamin A in humans. β-Cryptoxanthin is not commonly accumulated in horticultural crops, and it is only rich in a few fruits, such as citrus, papaya, and persimmon [8]. Citrus, as one of the essential fruits, is a plentiful source of carotenoids. The carotenoid diversity of citrus fruits is higher than that of other carotenoid-rich crops, such as tomato and carrot. Nutritional values and exterior qualities of citrus cultivars vary, with different carotenoid compositions and contents. β,β-Xanthophylls are the primary carotenoids accumulated in citrus fruits and flowers, and they are responsible for the color. According to carotenoid diversity, citrus cultivars are divided into three categories, namely, β-cryptoxanthin-rich cultivars, violaxanthin-rich cultivars, and low-content carotenoid cultivars. The fruit of mandarins exhibits a higher level of β-cryptoxanthin than that of oranges, accounting for >90% of the total carotenoids, and oranges primarily accumulate violaxanthin in the pulp [9–11]. The massive accumulation of β-cryptoxanthin endows citrus fruits with high nutritional value, and citrus fruits act as the major human dietary β-cryptoxanthin source. Therefore, revealing the biosynthesis mechanism of β-cryptoxanthin in citrus fruits is helpful for their quality improvement.
Key genes involved in carotenoid synthesis, such as PSY1 and LCYB, have been isolated, and their functions in citrus fruit have been thoroughly investigated and applied to the engineering of carotenoid synthesis metabolism [9, 12–15]. Carotene hydroxylation is an important part of carotenoid synthesis. Hydroxylation of the carotenoid rings is catalyzed by ring-specific hydroxylase, and it can convert carotenes into xanthophylls such as lutein and zeaxanthin [16]. Two types of carotene hydroxylases have been reported in plants, including non-heme di-iron carotene hydroxylase (also known as BCH, HYD, or HYb) and heme-containing cytochrome P450 (CYP) carotene hydroxylase [7]. BCH can efficiently catalyze the hydroxylation of the β-rings of β-carotene. The first β-carotene hydroxylase (named CrtZ) was detected in the phytopathogenic bacterium Erwinia uredovora [17]. Two BCH genes have been characterized in Arabidopsis thaliana, and they display 30–37% sequence identity with CrtZ. The double-null mutation of BCH1 and BCH2 has been reported to result in a significant decrease in β,β-xanthophylls [5, 18, 19]. Overexpression of BCH1 increases xanthophyll level but decreases β-carotene level in Arabidopsis, tomato, carrot, and kiwifruit [20–23]. Silencing CHY1 and CHY2 in potato tubers can increase β-carotene content and total carotenoid content, with a significantly better effect achieved by silencing CHY1 than silencing CHY2 [24]. A similar observation has been reported in citrus, that BCH1 can hydroxylate carotenes into β-cryptoxanthin and zeaxanthin in Escherichia coli and that silencing BCH1 can increase carotene content, but total carotenoid content is decreased, with β-xanthophylls remaining at the highest proportion in carotenoids [25]. The existing studies of β-carotene hydroxylation have primarily focused on the function of BCH1, and revealed that BCH1 can catalyze β-carotene hydroxylation [7]. However, the functions of BCH2 (paralogous gene of BCH1) in citrus carotenoid metabolism remain largely unknown.
Phylogenetic analyses have indicated that BCH gene duplication events occurred after the monocot–dicot split in higher plants [5]. Gene duplication and subsequent functional divergence induced by mutations in protein-coding regions and/or gene expression pattern change are increasingly recognized as crucial evolutionary mechanisms [5]. Previous studies have demonstrated that two BCH members in some higher plants present similar functions, although they are specifically expressed in different tissues [5, 20, 26]. Examining the functional characteristics of citrus BCH2 (named CsBCH2) is vital for revealing xanthophyll accumulation in citrus fruit and the evolutionary mechanism of paralogous genes. To determine whether CsBCH2 plays the same role as citrus BCH1 (named CsBCH1) in carotenoid synthesis, we analyzed the expression patterns and functional characteristics of CsBCH2 in vivo and in vitro. The functional complementation assay showed that CsBCH2 could hydroxylate β-carotene into β-cryptoxanthin in E. coli. Results of analysis of the expression pattern and functional characteristics of CsBCH2 revealed that it played an essential role in citrus xanthophyll biosynthesis, meanwhile providing backup compensation for CsBCH1.
Results
Isolation and sequence analysis of CsBCH2
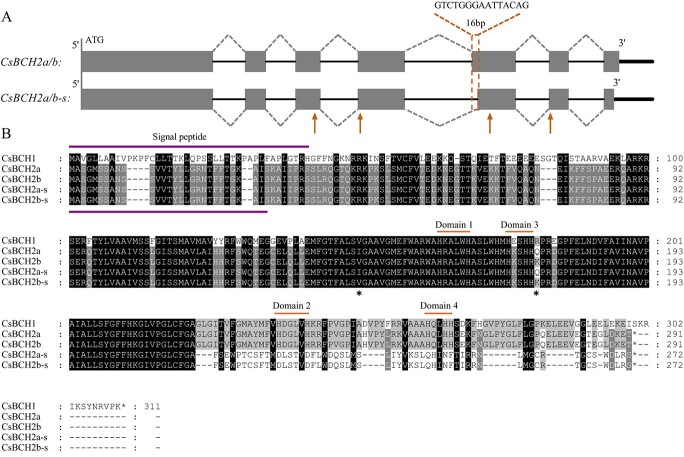
With the continuous release of citrus genomic and transcriptomic data, BCH2 (the paralogous gene of BCH1), has gradually attracted our attention. Given the genetic diversity of citrus species, we isolated BCH2 from the cDNA of three common citrus species fruits, including mandarin (Citrus reticulata Blanco), sweet orange (C. sinensis Osbeck), and pummelo [C. maxima (Burm.) Merr.] and named it CsBCH2, to explore its catalytic activity. Two CsBCH2 alleles (CsBCH2a and CsBCH2b) were obtained from chromosome 5. Both CsBCH2a (from mandarin and sweet orange) and CsBCH2b (from pummelo and sweet orange) contained an 876-bp open reading frame (ORF) and encoded one putative 32.4-kDa protein consisting of 291 amino acids with two non-synonymous sites. We also obtained two different transcripts corresponding to the two alleles (CsBCH2a-s and CsBCH2b-s). These two transcripts encoded a 272-amino acid protein and carried a 16-bp alternative splicing (Fig. 1A). The putative protein encoded by CsBCH2a/b contained highly conserved histidine domains, including two ‘HXXXXH’ domains (Domain 1 ‘HRALWH’ and Domain 2 ‘HDGLVH’) and two ‘HXXHH’ domains (Domain 3 ‘HKSHH’ and Domain 4 ‘HQLHH) (Fig. 1B), which was consistent with the domains of BCH genes in carrot, pepper, and kiwifruit [23, 27, 28]. Alternative splicing of CsBCH2a-s and CsBCH2b-s occurred at 646 bp downstream of the translational start codon of the cDNA sequence, thereby causing the frameshifts of the ‘HDGLVH’ and ‘HQLHH’ conserved domains, which might affect the catalytic activity of the enzyme.
Figure 1.
Gene structures of CsBCH2 alleles and protein sequence alignments of CsBCH1 and CsBCH2. (A) Schematic of CsBCH2 transcript structures. Gray boxes represent exons, black lines indicate introns, bold black lines indicate untranslated regions, orange arrow indicates the position of conserved domain, and the orange dashed box represents the alternative splicing region. (B) Amino acid sequence alignments. Signal peptides and the conserved domains are marked by purple lines and orange lines. Asterisks indicate non-synonymous sites.
To characterize CsBCH2, the coding sequence of CsBCH1 on chromosome 9 was obtained for subsequent comparative analysis. CsBCH1 was 936 bp in length, and it encoded a 34.7-kDa protein with 311 amino acids (Fig. 1B), with 69% amino acid sequence identity with CsBCH2a/b. CsBCH1 contained the same two conserved histidine domains, ‘HDGLVH’ (Domain 2) and ‘HQLHH’ (Domain 4), as CsBCH2. Unlike CsBCH2, the other two conserved domains, ‘HKALWH’ and ‘HESHH’ of CsBCH1 contained two non-synonymous sites. The above results showed similar sequences and conserved domains between CsBCH1 and CsBCH2, implying that the two BCHs might have similar functions in citrus.
Expression patterns of CsBCH genes in citrus
The transcriptome data published on the Citrus Pan-genome to Breeding Database (http://citrus.hzau.edu.cn/) showed that CsBCH1 and CsBCH2 transcript levels differed among different tissues and species (Supplementary Data Fig. S1). Further, we identified the spatial and temporal expression patterns of CsBCH1 and CsBCH2 using quantitative real-time PCR (qRT–PCR) in two carotenoid-rich citrus species, including β-cryptoxanthin-rich cultivar Ponkan mandarin and violaxanthin-rich cultivar Hamlin sweet orange [11].
The qRT–PCR results showed that CsBCH1 was highly expressed in flowers and fruits, especially in fruits, and CsBCH2 was mainly expressed in flowers (Fig. 2A). Both CsBCH1 and CsBCH2 showed tissue-specific expression in xanthophyll-accumulating tissue, and the expression levels of the two genes were higher in sweet orange than in mandarin. Given that citrus fruit is the predominant tissue for carotenoid accumulation, we performed qRT–PCR to compare CsBCH gene expression levels at six stages of fruit development (Fig. 2B and C). The results showed that the two CsBCH genes were expressed differently throughout the developmental stages of the citrus fruit. As the fruit ripened, the relative expression level of CsBCH1 increased gradually and was higher in citrus peel, which was consistent with the β-cryptoxanthin accumulation trend in mandarin [7]. In the peel, the relative expression level of CsBCH2 gene continued to increase in the early stages and then decreased after the color-turning stage in sweet orange. By contrast, in the pulp, it decreased first and then increased in sweet orange with violaxanthin accumulation, but the overall expression level of CsBCH2 was lower than that of CsBCH1. Our results suggested that CsBCH1 and CsBCH2 exhibited distinct expression tissue preferences and patterns as the fruit ripened. The spatial and temporal expression patterns associated with the accumulation of xanthophylls in fruits indicated the distinct roles of these two genes in citrus.
Figure 2.
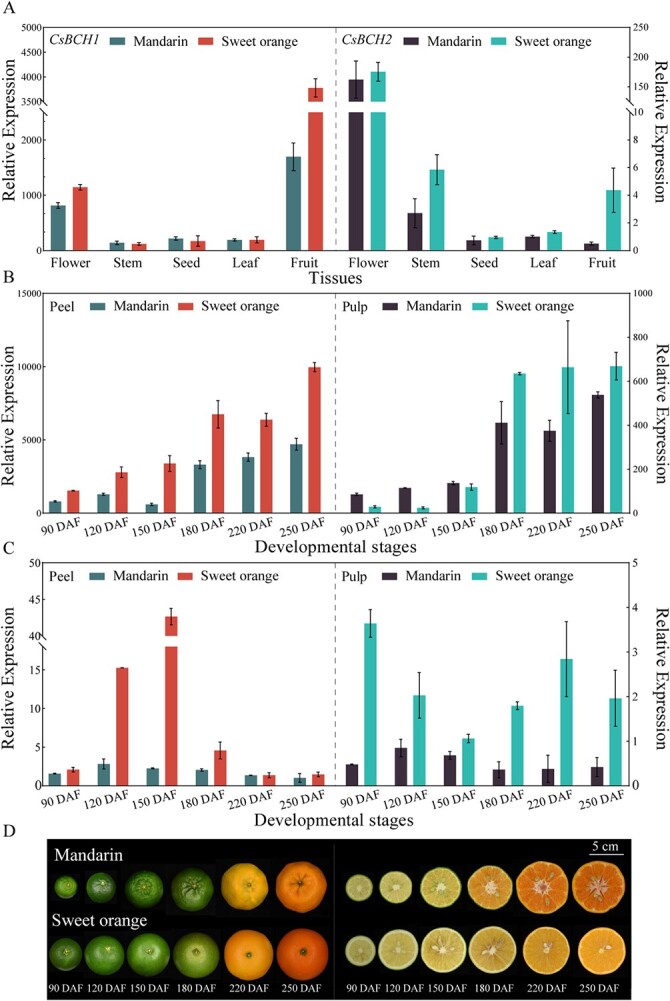
Expression patterns of CsBCH genes in citrus fruit. (A) CsBCH gene expression patterns were compared between mandarin (Ponkan) and sweet orange (Hamlin) in different tissues by qRT–PCR. (B, C) Relative expression levels of CsBCH1 (B) and CsBCH2 (C) at six development stages of citrus peel and pulp. (D) Phenotype of fruit at six development stages. DAF, days after flowering. Data are mean ± standard error of triplicate samples. Scale bar = 5 cm.
Functional complementation assay of CsBCH2 in E. coli
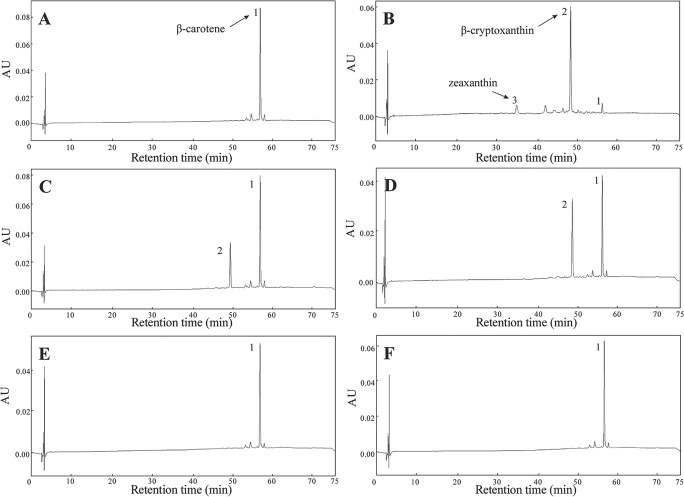
Since the complex and dynamic metabolic network obscures plant enzyme activities, it was not easy to directly investigate substrate specificity. Functional complementation assay of enzymes in vitro is an effective method for exploring enzyme characteristics with E. coli or yeast used as a heterologous host. To characterize the enzyme more directly, we explored the activity and substrate specificity of CsBCH2 through a functional complementation assay in E. coli, in which four transcripts of CsBCH2 (CsBCH2a, CsBCH2b, CsBCH2a-s, and CsBCH2b-s) were expressed in β-carotene-accumulating E. coli, and carotenoid products were extracted from bacteria for subsequent high-performance liquid chromatography (HPLC). The results showed that CsBCH2a- and CsBCH2b-expressing cells accumulated the substrate β-carotene and the mono-hydroxylated product β-cryptoxanthin, but the di-hydroxylated product zeaxanthin was undetectable. By contrast, neither β-cryptoxanthin nor zeaxanthin accumulation was detected in CsBCH2a-s- and CsBCH2b-s-expressing cells, suggesting that the alternative splicing-induced frameshift deprived CsBCH2 of its hydroxylation activity (Fig. 3C–F).
Figure 3.
HPLC analysis of carotenoids in E. coli expressing CsBCH1 or CsBCH2. (A–F) pHIS8 (A), pHIS8-CsBCH1 (B), pHIS8-CsBCH2a (C), pHIS8-CsBCH2b (D), pHIS8-CsBCH2a-s (E), or pHIS8-CsBCH2b-s (F) was co-transformed into E. coli BL21(DE3) cells with pACCAR16∆crtX. Reverse-phase HPLC was used to separate and determine carotenoids with spectra extracted at 450 nm. AU, absorbance unit; peak 1, β-carotene; peak 2, β-cryptoxanthin; peak 3, zeaxanthin.
To better understand the characteristics of CsBCH2, we also expressed CsBCH1 in E. coli for comparative analysis. Consistent with the functions of BCH1 reported in other studies [7, 16], our results showed that CsBCH1 effectively hydroxylated the two β-rings of β-carotene, resulting in the formation of the mono-hydroxylated intermediate β-cryptoxanthin and the di-hydroxylated product zeaxanthin with a higher level of β-cryptoxanthin than zeaxanthin (Fig. 3B). Only a trace amount of β-carotene substrate was detected in the cells. All the above results showed that both CsBCH1 and CsBCH2 could hydroxylate β-carotene, and CsBCH1 exhibited a higher hydroxylation activity.
Overexpression of CsBCH2 in citrus callus Rm
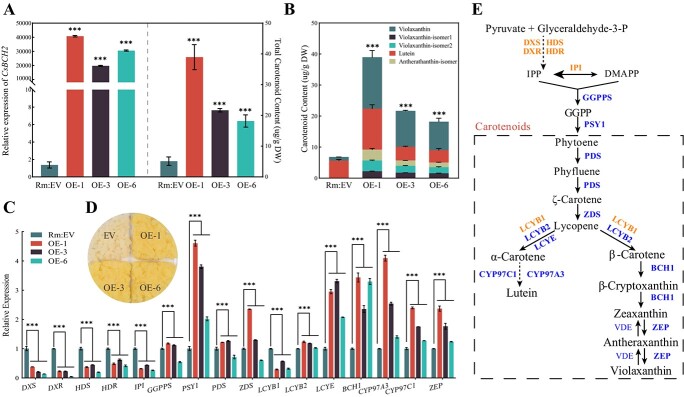
Enzyme activity and regulation in plants may not be accurately reflected in E. coli, which is a limitation of in vitro experiments. Since citrus is a perennial woody tree with a long juvenile period, it will take a long time to obtain the fruit of transgenic lines. Considering this, we explored the function of CsBCH2 in citrus callus, a valuable in vivo system that has been used successfully for evaluating the function of carotenogenic genes [9, 29–31]. The full-length transcript of CsBCH2 from mandarin was overexpressed in citrus callus Rm (white) to investigate the role of CsBCH2 in carotenoid metabolism. The results showed that, compared with empty vector (EV) callus, the CsBCH2-overexpressing transgenic lines all turned light yellow. Subsequently, the carotenoid composition and content in three selected transgenic lines were determined using HPLC (Fig. 4D). The results showed that the total carotenoid content was increased in transgenic lines, and xanthophylls such as violaxanthin and violaxanthin isomers were significantly increased, which explained the callus color change (Fig. 4B, Supplementary Data Table S3).
Figure 4.
Carotenoid accumulation and carotenogenic gene expression levels in CsBCH2-overexpressing transgenic lines of citrus callus Rm. (A) Relative expression levels of CsBCH2 (left) and total content of carotenoid (right) in transgenic calli Rm. (B) Composition and content of carotenoid in transgenic calli Rm. (C) Expression patterns of carotenogenic genes in transgenic lines. (D) Phenotypes of transgenic line callus Rm. (E) Schematic of general pathways of carotenoid biosynthesis in citrus. Upregulated genes in blue; downregulated genes in orange. Data are expressed as mean ± standard error for triplicate samples. Asterisks above the bars indicate statistically significant differences compared with EV (Student’s t-test): ***P < .001.
Since the expression level of endogenous carotenogenic genes is always influenced by manipulating the carotenoid level [24], we analyzed the relative expression levels of major carotenogenic genes to reveal the mechanism of carotenoid variation in transgenic lines (Fig. 4C). The results demonstrated that the expression levels of carotenogenic genes upstream of the geranylgeranyl diphosphate (GGPP) synthesis step were inhibited (Fig. 4E). All the carotenogenic genes downstream of the carotenoid synthesis pathway (except LCYB1) exhibited significant upregulation; especially, the rate-limiting enzyme gene PSY1 was upregulated >4-fold. Overall, the overexpression of CsBCH2 increased the content of xanthophylls and jointly regulated carotenoid synthesis with other carotenogenic genes in citrus callus.
Overexpression of CsBCH2 in β-carotene-accumulating callus Rm33
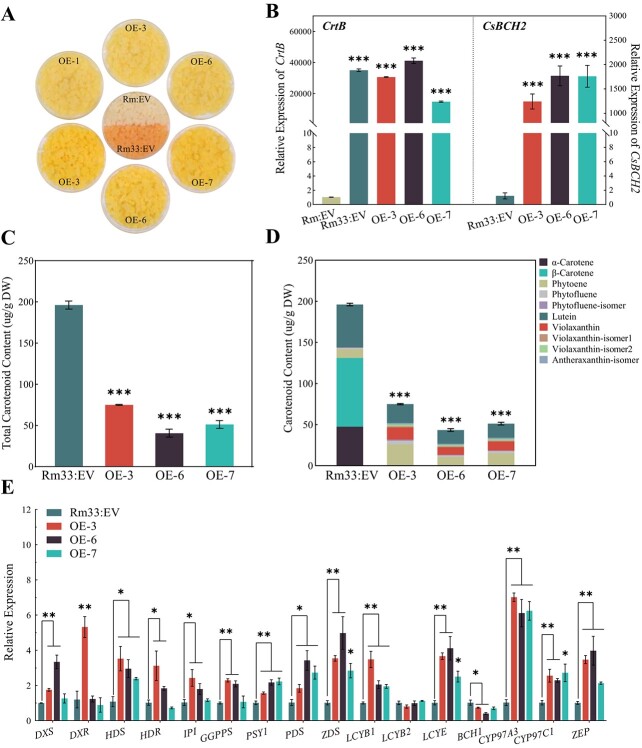
Considering that β-carotene is the hydroxylated substrate of BCH, we overexpressed CsBCH2 in the β-carotene-accumulating callus Rm33 [engineering cell model (ECM), which had already overexpressed a bacterial phytoene synthase gene (CrtB) in callus Rm] [29]. We selected three transgenic lines (OE-3, OE-6, and OE-7) for carotenoid content and carotenogenic gene expression analysis with the premise of not disrupting overexpression of CrtB in callus Rm33 (Fig. 5B). Control lines (callus Rm33 with empty vector) exhibited a high level of β-carotene (42.6%), lutein (26.6%), and α-carotene (24.2%) but only a low level of phytoene (7.4%) and phytofluene (1.5%) (Supplementary Data Table S4). The accumulation of carotenes was not detected in transgenic lines, but the percentages of oxygenated carotenoids in the three transgenic lines were increased to 57.6–69.1%. Of oxygenated carotenoids, the violaxanthin content of the β-branch in three transgenic lines was increased to 26.8–31.5%, and the lutein content of the α-branch was significantly decreased to 16.78–23.13 μg/g dry weight (DW), but as a percentage of total carotenoid content it was increased to 30.8–38.4%. Furthermore, total carotenoid content in transgenic lines (43.71–75.10 μg/g DW) was significantly lower than that in the control lines (196.09 ± 4.92 μg/g DW).
Figure 5.
Carotenoid accumulation and carotenogenic gene expression levels in CsBCH2-overexpressing transgenic lines of callus Rm33. (A) Phenotypes of transgenic line callus Rm (upper part) and Rm33 (lower part). (B) Relative expression levels of CrtB (left) and CsBCH2 (right) in transgenic lines. (C) Total carotenoid content in transgenic lines. (D) Composition and content of carotenoids in transgenic lines. (E) Expression levels of carotenogenic genes in transgenic lines. Data are expressed as mean ± standard error for triplicate samples. Asterisks above the bars indicate statistically significant differences compared with EV (Student’s t-test): *P < .05; **P < .01; ***P < .001.
In addition to the changes in composition and content of carotenoids, our results also showed that the expression levels of all the main carotenogenic genes in transgenic lines were upregulated to varying degrees (Fig. 5E), which was consistent with the trend of the Rm overexpressing CsBCH2 (Fig. 4C). The upregulation of carotenogenic genes upstream of the carotenoid synthesis pathway led to increases in phytoene and phytofluene contents. Notably, LCYE, a cyclase gene involved in α-branch synthesis, was 2- to 4-fold upregulated. CYP97A3 was 6- to 7-fold upregulated, and CYP97C1 was upregulated ~2-fold. By contrast, LCYB2, involved in β-branch synthesis, was not upregulated, and BCH1 expression was inhibited significantly in two transgenic lines (OE-3 and OE-6). Compared with citrus callus Rm, callus Rm33 accumulated more β-branch carotenoids than α-branch carotenoids, which might be attributed to the overexpression of CrtB (PSY1). The increased proportion of lutein in total carotenoids was attributed to the upregulation of genes in α-branch synthesis, and the downregulation of BCH1 expression level might be the regulation mechanism to balance carotenoid synthesis between the α-branch and the β-branch. The above results implied that the overexpression of CsBCH2 in callus Rm33 could simultaneously affect the carotenoid composition and the expression level of other endogenous carotenogenic genes.
Introduction of CsBCH2 into citrus callus affects the plastoglobules of plastids
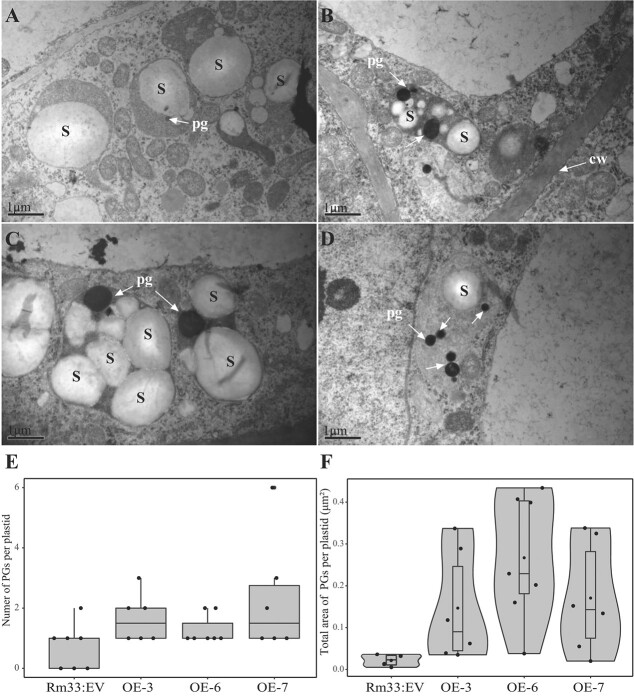
Plastoglobules (PGs) are plastid lipoprotein particles found in chromoplasts, which are unique organelles responsible for synthesizing and storing large amounts of carotenoids [32, 33]. Since carotenoid variation can affect plastid morphology and plastoglobule size, we investigated plastid ultrastructure in Rm33 transgenic lines using transmission electron microscopy (TEM) to explore the effects of CsBCH2 overexpression at the cellular level. The results showed that plastids in callus Rm33 were primarily present in the form of amyloplasts accumulating starch granules and a small number of PGs, which was consistent with the previously reported ultrastructural features of ECMs [29]. With the overexpression of CsBCH2, we observed that the morphology of plastids converted from globular to oval or fusiform. In addition, the CsBCH2-overexpressing transgenic lines exhibited amyloplast-like structures containing starch granules and more visible PGs than callus Rm33 (Fig. 6). Statistical analysis showed that although the number of PGs per plastid did not increase significantly in some cells, the total area of PGs per plastid increased among transgenic lines, which affects the capability of carotenoid storage (Fig. 6E and F). We suggest that the morphology conversion of plastids and the increase in PG size were associated with the change in carotenoid composition and the increased accumulation of xanthophylls in transgenic callus. Taking these results together, the cellular ultrastructural changes in transgenic lines indicated that the overexpression of CsBCH2 in citrus callus could regulate the storage ability of carotenoids by affecting the carotenoid sequestration substructures.
Figure 6.
TEM of plastid ultrastructure from transgenic line callus Rm33. (A) Plastid ultrastructure of transgenic line callus Rm33 carrying the EV. (B–D) Plastid ultrastructure of transgenic callus Rm33 expressing CsBCH2 (OE-3, OE-6, OE-7). (E) Number of PGs per plastid in transgenic lines. (F) Total area of PGs per plastid. S, starch granule; pg, plastoglobule; cw, cell wall. Scale bar = 1 μm.
Discussion
Carotenoids are important secondary metabolites that are beneficial to human health and they act as colorants in most horticultural plants. The oxygenated derivative xanthophylls are the most abundant carotenoids in the light-harvesting complexes, and they serve as substrates for volatile products and precursors of hormone synthesis in plants [5]. As an important horticultural crop, citrus has a large variety of carotenoids, especially abundant β,β-xanthophylls synthesized by BCH [7]. In this study we systematically examined the hydroxylation activity of citrus CsBCH2 and elucidated the vital role of CsBCH2 in the biosynthesis of citrus xanthophyll by comparing CsBCH2 with the well-studied CsBCH1.
CsBCH2 participates in xanthophyll synthesis by hydroxylating β-carotene
Extensive studies have shown that BCH1 can hydroxylate β-carotene [19, 26]. The β,β-xanthophylls (mainly 9-Z-violaxanthin) are the major component in β-CHX (BCH1)-silenced citrus [25], implying that other enzymes are also involved in β-branch hydroxylation to supply substrate for violaxanthin synthesis. BCH2 has been reported to be differentially expressed during fruit development, thus affecting the accumulation of carotenoids [34]. Revealing the role of BCH2 is helpful for understanding xanthophyll biosynthesis in citrus.
Heterologous expression is an efficient approach to exploring enzyme characteristics in vitro. Expressing tomato CrtR-b1 (also named BCH1) and CrtR-b2 (also named BCH2) in β-carotene-accumulating E. coli confirmed that both BCH1 and BCH2 can catalyze β-carotene into β-cryptoxanthin and zeaxanthin [26]. The enzymatic activity analysis results of kiwifruit AcBCH genes in β-carotene-accumulating yeast showed that AcBCH1 could convert β-carotene into β-cryptoxanthin and zeaxanthin, whereas AcBCH2 could not [23]. Our E. coli complementary functional assay results showed that citrus CsBCH1 hydroxylated β-carotene into β-cryptoxanthin and a small amount of zeaxanthin (Fig. 3B). However, CsBCH2a and CsBCH2b hydroxylated β-carotene to generate only β-cryptoxanthin in E. coli, without zeaxanthin. Since no hydroxylation products were detected, we speculated that the transcripts (CsBCH2a-s and CsBCH2b-s) with frameshifts in two conserved histidine domains (Domain 2 and Domain 4) might have lost their hydroxylation activity and the highly conserved histidine domains of CsBCH2 were necessary for the hydroxylation activity (Figs 1B and 3). Similar functions of the two CsBCH genes did not mean that CsBCH2 was completely redundant, since they exhibited different expression patterns in citrus. The spatial and temporal expression patterns showed that the high level of β-cryptoxanthin accumulation in citrus fruits was associated with the expression of CsBCH1 and CsBCH2. The higher expression of CsBCH1 than CsBCH2 in fruits might be attributed to the stronger β-cryptoxanthin synthesis capability of CsBCH1 than CsBCH2 (Fig. 2). Compared with previous functional analyses of BCH genes in other horticultural crops, such as tomato and kiwifruit [23, 26], our results revealed that the specific accumulation of β-cryptoxanthin in citrus fruit might be attributed to the joint action of two CsBCH genes and their strong capability for β-cryptoxanthin synthesis. In addition, the higher concentration of β-cryptoxanthin in mandarin than in sweet orange might be related to the regulation of other factors, such as the participation of unreported enzymes. The differential accumulation of β-cryptoxanthin in different citrus species remains to be further investigated in future studies.
Studies of Arabidopsis have indicated that four enzymes (BCH1, BCH2, CYP97C1, and CYP97A3) are involved in the hydroxylation of carotenes. In spite of the fact that CYP97A can only convert β-carotene into β-cryptoxanthin in E. coli, only dihydroxy β-carotene derivatives (violaxanthin and neoxanthin) rather than β-cryptoxanthin are detected in the Arabidopsis triple mutant bch1 bch2 cyp97c1 [5]. However, our experimental results indicated that CsBCH2 only converted β-carotene into β-cryptoxanthin in E. coli, which did not mean that CsBCH2 could not produce the downstream product zeaxanthin with the assistance of other factors in vivo. Overexpression of CsBCH2 resulted in a higher level of accumulation of β,β-xanthophylls in transgenic callus Rm and Rm33 than in EV. The change in gene expression may be the major factor affecting carotenoid biosynthesis, and thus manipulating endogenous carotenogenic gene expression can change carotenoid content [26]. Our findings in citrus callus showed that the overexpression of CsBCH2 could cause feedback on other carotenogenic genes and regulate the expression level of these genes to varying degrees, which jointly affects the accumulation of carotenoids in citrus callus. The upregulation of PSY1 led to the increased total carotenoid content in transgenic callus Rm, whereas the total carotenoid content in Rm33 transgenic lines was decreased by 28% compared with that in EV, which might be attributed to the conversion of carotenes into xanthophylls and downstream hormone synthesis. These results were consistent with the previous reports that violaxanthin, rather than zeaxanthin, was accumulated with CrtR-b2 overexpression in tomato [20], and that the contents of β-carotene and total carotenoid were decreased in leaves of AcBCH-overexpressing transgenic lines of kiwifruit [23]. Ultrastructure observations by TEM showed the coexistence of amyloplasts and chromoplasts in CsBCH2-overexpressing transgenic callus with an increase in xanthophylls, supported by the reported findings that xanthophylls could be deposited in amyloplasts, which could convert into chromoplasts in non-photosynthetic tissues [29]. The change in carotenoid composition and content in transgenic callus could affect plastid morphology and the abundance of PGs, indicating that overexpression of CsBCH2 simultaneously influenced carotenoid storage ability at the cellular level. Although the citrus callus validation platform could not reflect the real regulation in natural fruit tissues, it simulates the composition of carotenoids in fruits and provides an abundant β-carotene substrate for β-carotene hydroxylation validation. Our study on the functional exploration of CsBCH2 in vivo and in vitro provided evidence for the pleiotropic effects of CsBCH2 on citrus carotenoid biosynthesis.
Hydroxylation of CsBCH2 expands the synthetic biology toolkit for carotenoid metabolic engineering
The functional complementation assay based on metabolic synthetases from various sources has turned out to be a feasible approach to diversifying metabolic biosynthesis as in nature [35–37]. With E. coli used as a heterologous host, the functional complementation assay of carotenogenic genes can characterize the enzymes encoded by carotenogenic genes and synthesize various carotenoids as enzyme substrates and standards [37]. Compared with that of CsBCH1, hydroxylation of CsBCH2 only converted β-carotene into β-cryptoxanthin, based on which we could construct β-cryptoxanthin-accumulating E. coli by expressing CsBCH2 and analyze the functions of β-cryptoxanthin-related enzymes. Therefore, this study provides the resources for functional complementation assay-based carotenoid metabolism research, especially research on β-cryptoxanthin.
Despite being time-consuming, the callus transgenic system provides an effective platform for research on carotenoid metabolism-related genes in perennial woody trees. Callus-generating carotenoid substrates such as ECMs can be employed for carotenoid studies [29]. The citrus callus used in our study provided a useful in vivo system for carotenoid research, and indicated that the overexpression of CsBCH2 in citrus callus could not only regulate the expression level of other carotenogenic genes to change carotenoid biosynthesis, but also influence the number and size of plastoglobules at the cellular level. Our findings revealed the vital role of CsBCH2 in citrus carotenoid metabolism, which could be utilized to expand the synthetic biology toolbox for carotenoid metabolism.
Roles of two paralogous CsBCH genes in β-carotene hydroxylation and carotenoid metabolism
Paralogous homologs generated from gene duplication events during genome evolution facilitate genetic redundancy and phenotypic robustness [38]. Most duplicated genes are retained during evolution through multiple mechanisms such as neofunctionalization, subfunctionalization, dosage amplification, and backup compensation [39–42]. After the monocot–dicot split, BCH gene duplication events occurred in higher plants, such as the horticultural crops kiwifruit [23], potato [24], tomato [26], and pepper [28], containing two BCH paralogous genes with different tissue-specific expression patterns [5, 43]. The AcBCH genes (AcBCH1 and AcBCH2) exhibit a constitutive expression pattern and are highly expressed in kiwifruit leaves and fruits [23]. BCH1 (also named CHY1) is expressed preferentially in potato leaves, whereas BCH2 (CHY2) is expressed preferentially in potato flowers [24]. In tomato, CrtR-b1 is expressed in leaves and sepals and has low expression in petals, while CrtR-b2 is highly expressed in the petals and anthers of flowers, where large amounts of yellow xanthophylls are accumulated [26]. Our results revealed that the expression pattern of CsBCH1 was consistent with the high xanthophyll accumulation level during the fruit ripening process in citrus. In contrast, CsBCH2 displayed a different expression pattern from CsBCH1 in fruits during the color-change stage, and CsBCH2 was preferentially expressed in flowers (Fig. 2).
Since xanthophylls play crucial roles in photosynthesis and act as precursors of abscisic acid, maintaining xanthophyll synthesis is vital to the plants themselves [6]. When β-CHX (BCH1) was silenced, β,β-xanthophylls (mainly 9-Z-violaxanthin) remained as the primary carotenoid in citrus [25]. Comparison of β-ring hydroxylation between the b1 b2 double mutant and the b1 mutant has demonstrated that BCH2 can largely compensate for the absence of BCH1 activity in mutant Arabidopsis [19]. Besides, a recent study has reported that overexpressing one AcBCH of a paralogous gene pair can negatively regulate the expression of the other AcBCH of this pair in kiwifruit [23]. Our data showed that in carotenoid-deficient callus Rm, the overexpression of CsBCH2 upregulated CsBCH1 to stimulate the increase in carotenoid flux, whereas in transgenic line carotenoid-sufficient callus Rm33 the overexpression of CsBCH2 downregulated CsBCH1 to balance the carotenoid biosynthesis of the α-branch and β-branch (Figs 4C and 5E). Overall, the β-cryptoxanthin synthesis capability of CsBCH1 and CsBCH2 was associated with the considerable accumulation of β-cryptoxanthin in citrus fruits, and the distinct gene expression pattern of CsBCH2 (from CsBCH1) could enable it to compensate for the function of CsBCH1 by providing different enzyme activity. Based on these findings, we speculated that gene compensation might be the evolutionary mechanism for maintaining phenotypic stability when genetic variation or external factors cause the inactivation of CsBCH1. Our speculation was supported by the previous report that most duplicated genes are retained during evolution with functional redundancy [39]. The CRISPR/Cas9 system has been successfully used for gene function research in citrus [44]. Considering this, we suggest that the CRISPR/Cas9 system should be employed for future research on loss-of-function mutants of CsBCH1 or CsBCH2 to reveal the possible retaining mechanism of the two CsBCH genes.
In conclusion, our study indicated that, just like CsBCH1, CsBCH2 could hydroxylate the β-ring of β-carotene, while CsBCH2 exhibited a different and weaker catalytic activity compared with CsBCH1, which could be utilized to provide β-cryptoxanthin substrate in E. coli to expand the synthetic biology toolkit for carotenoid engineering. This study revealed the distinct expression pattern and functional characteristics of two CsBCH paralogous genes, and illustrated the potential compensatory role of CsBCH2 for CsBCH1 in xanthophyll biosynthesis. Our findings provide insights into the mechanism of duplicate β-carotene hydroxylase gene retention during evolution.
Materials and methods
Plant materials
The citrus materials Ponkan mandarin (C. reticulata Blanco), Hamlin sweet orange (C. sinensis Osbeck), and Shatian pummelo (C. maxima (Burm.) Merr.) were collected from trees in the orchard of the National Citrus Breeding Center at Huazhong Agricultural University. Citrus calli were derived from Marsh grapefruit (C. paradisi Macf. Rm) and ECM Rm33. The calli were subcultured at 25°C on solid Murashige and Tucker (MT) medium. All samples were frozen in liquid nitrogen after collection and stored at −80°C for further analysis.
RNA extraction and qRT–PCR analysis
Total RNA extraction was performed as previously described [45]. cDNA synthesis was conducted using the HiScript II RT SuperMix for qPCR (+gDNA wiper, Vazyme). The qRT–PCR primers used in this study are listed in Supplementary Data Table S2. The endogenous reference gene was named CsActin. qRT–PCR was performed on the Roche LightCycler 480 system (Roche, https://www.roche.com). The qRT–PCR procedure and calculation of the relative expression of genes were carried out as previously described [45, 46]. Each experiment was performed independently with three replicates.
Gene cloning and sequence analysis
The reference gene sequences of CsBCH1 (Cs9g19270) and CsBCH2 (Cs5g03200) were obtained from the Citrus Pan-genome to Breeding Database (http://citrus.hzau.edu.cn/). The full-length coding sequence of CsBCH1 and CsBCH2 from the fruits of Ponkan mandarin, Hamlin sweet orange, and Shatian pummelo were amplified using PCR with the primers listed in Supplementary Data Table S1. Multiple sequence alignments were performed using MEGA 6 software, and alignment results were visualized in GeneDoc software.
Heterologous expression and functional characterization of CsBCH1 and CsBCH2
The ORFs of CsBCH1 and CsBCH2 were cloned into vector pHIS8, which was derived from pET28a (+) with modification [47]. To investigate the hydroxylase activity of CsBCH1 and CsBCH2 in vivo, pHIS8-CsBCH1, pHIS8-CsBCH2a, pHIS8-CsBCH2a-s, or pHIS8-CsBCH2b-s was co-transformed into E. coli BL21 (DE3) with pACCAR16∆crtX. The pHIS8 empty vector was co-transformed with pACCAR16∆crtX into E. coli BL21 (DE3) to accumulate β-carotene in host cells as a negative control. Colonies were cultured in Luria–Bertani (LB) medium containing kanamycin (50 mg/ml) and chloramphenicol (25 mg/ml) at 37°C in the dark until the optical density (OD600) reached 0.6. A final concentration of 0.3 mmol/l isopropyl β-d-thiogalactoside (IPTG) was added to induce the expression of CsBCH for 16 hours at 16°C by shaking at 160 rpm. The 50-ml culture was centrifuged at 6000 g for 10 minutes, and the cells were stored at −80°C for further analysis. The experiments were performed with at least three replicates for each sample.
Carotenoid extraction and analysis
Citrus calli were lyophilized and ground into powder for carotenoid analysis using a Labconco FreeZone lyophilizer. The separation and analysis of carotenoid pigments were performed using HPLC, as previously described [48]. Carotenoids were determined by their specific retention time, absorption spectra, and comparison with authentic standards as previously described [49]. Peak areas were recorded for phytoene, phytofluene, and the other carotenoids at 286, 348, and 450 nm, respectively. The content of carotenoids was quantified using calibration curves for the appropriate standards [48]. The experiment was conducted in triplicate.
Callus transformation
The coding sequence of CsBCH2 from the fruits of Ponkan mandarin was cloned into the MT-GFP vector with CaMV35S promoter and enhanced GFP. The stable transformation of citrus callus and growth conditions were as described previously [50]. We performed at least six rounds of callus subculturing on culture media, accompanied by appropriate antibiotics selection. The transgenic calli were collected every 20 days, frozen in liquid nitrogen, and stored at −80°C until further analysis.
Transmission electron microscopy observation
Calli were subcultured on MT medium for 20 days and collected from transgenic lines. Samples were observed on the electron microscopy platform of Huazhong Agricultural University for subsequent analyses, as previously described [29]. The number and area of PGs per plastid were measured using ImageJ software (version 1.53, NIH, USA), based on the images from TEM. Six plastids were selected randomly from each callus transgenic line.
Statistical analysis
All data were expressed as the mean ± standard error of three replicates. The statistical analysis of data was performed using GraphPad Prism v.8. Student’s t-tests were conducted to determine the statistical significance of differences between transgenic lines and EV.
Acknowledgements
The authors are very grateful to Prof. Norihiko Misawa for providing the pACCAR16∆ crtX plasmid and grateful to Prof. Robert M. Larkin and Prof. Pengwei Wang for providing expression vectors. We thank Prof. Ping Liu for language improvement. This research was supported by the National Natural Science Foundation of China (No. 31930095 and 32172527) and the Modern Agro-industry Technology Research System (CARS-26).
Author contributions
X.X.D. supervised the research; Y.Z.Z. and X.X.D. designed the experiments; Y.Z.Z. performed the experiments with contributions from J.J.J.; S.C.Z. and Z.Z.X. provided the plant materials. Y.Z.Z. and X.X.D. wrote the manuscript; Q.S., Y.Z., and J.L.Y. provided critical comments on manuscript editing.
Data availability
All relevant data are included in the paper and its supplementary files.
Conflict of interest
The authors declare no competing interests.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Yingzi Zhang, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
Jiajing Jin, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
Shenchao Zhu, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
Quan Sun, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
Yin Zhang, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
Zongzhou Xie, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
Junli Ye, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
Xiuxin Deng, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, 430070, China.
References
- 1. Moise AR, Al-Babili S, Wurtzel ET. Mechanistic aspects of carotenoid biosynthesis. Chem Rev. 2014;114:164–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fraser PD, Bramley PM. The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res. 2004;43:228–65. [DOI] [PubMed] [Google Scholar]
- 3. Krinsky NI, Johnson EJ. Carotenoid actions and their relation to health and disease. Mol Asp Med. 2005;26:459–516. [DOI] [PubMed] [Google Scholar]
- 4. Kato M. Mechanism of ß-cryptoxanthin accumulation in citrus fruits. Acta Hortic. 2016;1:1–10. [Google Scholar]
- 5. Kim J, Smith JJ, Tian Let al. . The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 2009;50:463–79. [DOI] [PubMed] [Google Scholar]
- 6. Du H, Wang N, Cui Fet al. . Characterization of the β-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 2010;154:1304–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ma G, Zhang L, Yungyuen Wet al. . Expression and functional analysis of citrus carotene hydroxylases: unravelling the xanthophyll biosynthesis in citrus fruits. BMC Plant Biol. 2016;16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma G, Zhang L, Sugiura Met al. . Citrus and health. In: Talon M, Caruso M, Gmitter FG Jr, eds. The Genus Citrus. Elsevier, 2020,495–511. [Google Scholar]
- 9. Zheng X, Zhu K, Sun Qet al. . Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol Plant. 2019;12:1294–307. [DOI] [PubMed] [Google Scholar]
- 10. Ikoma Y, Matsumoto H, Kato M. Diversity in the carotenoid profiles and the expression of genes related to carotenoid accumulation among citrus genotypes. Breed Sci. 2016;66:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alquezar B, Rodrigo M, Zacarías L. Carotenoid biosynthesis and its regulation in citrus fruits. Tree For Sci Biotechnol. 2008;2:23–35. [Google Scholar]
- 12. Kato M, Matsumoto H, Ikoma Yet al. . The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. J Exp Bot. 2006;57:2153–64. [DOI] [PubMed] [Google Scholar]
- 13. Ma G, Zhang L, Matsuta Aet al. . Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of citrus fruit. Plant Physiol. 2013;163:682–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang L, Ma G, Shirai Yet al. . Expression and functional analysis of two lycopene β-cyclases from citrus fruits. Planta. 2012;236:1315–25. [DOI] [PubMed] [Google Scholar]
- 15. Zhu K, Zheng X, Ye Jet al. . Building the synthetic biology toolbox with enzyme variants to expand opportunities for biofortification of provitamin A and other health-promoting carotenoids. J Agric Food Chem. 2020;68:12048–57. [DOI] [PubMed] [Google Scholar]
- 16. Quinlan RF, Shumskaya M, Bradbury LMTet al. . Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol. 2012;160:204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Misawa N, Nakagawa M, Kobayashi Ket al. . Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990;172:6704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun Z, Gantt E, Cunningham FX. Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem. 1996;271:24349–52. [DOI] [PubMed] [Google Scholar]
- 19. Tian L, Magallanes-Lundback M, Musetti Vet al. . Functional analysis of β- and ε-ring carotenoid hydroxylases in Arabidopsis. Plant Cell. 2003;15:1320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D’Ambrosio C, Stigliani AL, Giorio G. Overexpression of CrtR-b2 (carotene beta hydroxylase 2) from S. lycopersicum L. differentially affects xanthophyll synthesis and accumulation in transgenic tomato plants. Transgenic Res. 2011;20:47–60. [DOI] [PubMed] [Google Scholar]
- 21. Kim J-E, Cheng KM, Craft NEet al. . Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a β-carotene ketolase provides insight into in vivo functions. Phytochemistry. 2010;71:168–78. [DOI] [PubMed] [Google Scholar]
- 22. Li T, Deng Y-J, Liu J-Xet al. . DcCCD4 catalyzes the degradation of α-carotene and β-carotene to affect carotenoid accumulation and taproot color in carrot. Plant J. 2021;108:1116–30. [DOI] [PubMed] [Google Scholar]
- 23. Xia H, Zhou Y, Lin Zet al. . Characterization and functional validation of β-carotene hydroxylase AcBCH genes in Actinidia chinensis. Hortic Res. 2022;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diretto G, Welsch R, Tavazza Ret al. . Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 2007;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pons E, Alquézar B, Rodríguez Aet al. . Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol J. 2014;12:17–27. [DOI] [PubMed] [Google Scholar]
- 26. Galpaz N, Ronen G, Khalfa Zet al. . A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell. 2006;18:1947–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li T, Liu J-X, Deng Y-Jet al. . Overexpression of a carrot BCH gene, DcBCH1, improves tolerance to drought in Arabidopsis thaliana. BMC Plant Biol. 2021;21:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bouvier F, Keller Y, d’Harlingue Aet al. . Xanthophyll biosynthesis: molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.). Biochim Biophys Acta. 1998;1391:320–8. [DOI] [PubMed] [Google Scholar]
- 29. Cao H, Zhang J, Xu Jet al. . Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J Exp Bot. 2012;63:4403–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu Q, Huang X, Lv Set al. . Carotenoid profiling of red navel orange ‘Cara Cara’ harvested from five regions in China. Food Chem. 2017;232:788–98. [DOI] [PubMed] [Google Scholar]
- 31. Zhu K, Zheng X, Ye Jet al. . Regulation of carotenoid and chlorophyll pools in hesperidia, anatomically unique fruits found only in citrus. Plant Physiol. 2021;187:829–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li L, Yuan H. Chromoplast biogenesis and carotenoid accumulation. Arch Biochem Biophys. 2013;539:102–9. [DOI] [PubMed] [Google Scholar]
- 33. Wijk KJ, Kessler F. Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu Rev Plant Biol. 2017;68:253–89. [DOI] [PubMed] [Google Scholar]
- 34. Zhu F, Luo T, Liu Cet al. . An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017;216:178–92. [DOI] [PubMed] [Google Scholar]
- 35. Moreno JC, Stange C. Heterologous complementation in bacteria for functional analysis of genes encoding carotenoid biosynthetic enzymes. Methods Enzymol. 2022;671:471–88. [DOI] [PubMed] [Google Scholar]
- 36. Misawa N, Satomi Y, Kondo Ket al. . Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol. 1995;177:6575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cunningham FX, Gantt E. A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth Res. 2007;92:245–59. [DOI] [PubMed] [Google Scholar]
- 38. Huang X, Xiao N, Zou Yet al. . Heterotypic transcriptional condensates formed by prion-like paralogous proteins canalize flowering transition in tomato. Genome Biol. 2022;23:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuzmin E, Taylor JS, Boone C. Retention of duplicated genes in evolution. Trends Genet. 2022;38:59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Force A, Lynch M, Pickett FBet al. . Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nowak MA, Boerlijst MC, Cooke Jet al. . Evolution of genetic redundancy. Nature. 1997;388:167–71. [DOI] [PubMed] [Google Scholar]
- 42. Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–8. [Google Scholar]
- 43. Zhou X, McQuinn R, Fei Zet al. . Regulatory control of high levels of carotenoid accumulation in potato tubers. Plant Cell Environ. 2011;34:1020–30. [DOI] [PubMed] [Google Scholar]
- 44. Zhu C, Zheng X, Huang Yet al. . Genome sequencing and CRISPR/Cas9 gene editing of an early flowering mini-citrus (Fortunella hindsii). Plant Biotechnol J. 2019;17:2199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu S, Zhang Y, Zhu Ket al. . The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018;176:2657–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Ye J, Liu Cet al. . Citrus PH4-Noemi regulatory complex is involved in proanthocyanidin biosynthesis via a positive feedback loop. J Exp Bot. 2020;71:1306–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jez JM, Ferrer J-L, Bowman MEet al. . Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. [DOI] [PubMed] [Google Scholar]
- 48. Zhu K, Chen H, Zhang Yet al. . Carotenoid extraction, detection, and analysis in citrus. Methods Enzymol. 2022;670:179–212. [DOI] [PubMed] [Google Scholar]
- 49. Zheng X, Xie Z, Zhu Ket al. . Isolation and characterization of carotenoid cleavage dioxygenase 4 genes from different citrus species. Mol Gen Genomics. 2015;290:1589–603. [DOI] [PubMed] [Google Scholar]
- 50. Lu S, Zhang Y, Zheng Xet al. . Isolation and functional characterization of a lycopene β-cyclase gene promoter from citrus. Front Plant Sci. 2016;7:1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper and its supplementary files.