Abstract
Sculpins (Family Cottidae) are generally cold-temperate intertidal reef fishes most commonly found in the North Pacific. As part of the California Conservation Genomics Project (CCGP), we sequenced the genome of the Woolly Sculpin, Clinocottus analis, to establish a genomic model for understanding phylogeographic structure of inshore marine taxa along the California coast. These patterns, in turn, should further inform the design of marine protected areas using dispersal models based on genomic data. The small genome of C. analis is typical of marine fishes at less than 1 Gb (genome size = 538 Mb), and our assembly is near-chromosome level (contig N50 = 9.1 Mb, scaffold N50 = 21 Mb, BUSCO completeness = 97.9%). Within the context of the CCGP, the Woolly Sculpin genome will be used as a reference for future whole-genome resequencing projects aimed at enhancing our knowledge of the population structure of the species, and efficacy of marine protected areas across the state.
Keywords: California Conservation Genomics Project, CCGP, marine protected areas
Introduction
Most marine fishes exhibit a bipartite life history, with a pelagic larval stage, which typically lasts from a few days to a few weeks, and a more sedentary adult stage (Leis 1991). Because of the broad dispersal that often results from the larval stage, many marine fishes have, or are presumed to have, relatively modest population structure across coastal seascapes. Variation in dispersal is further influenced by fertilization mode (internal vs. external) and egg type (internal, deposited on the substrate, or pelagic); that latter may be particularly important because fish larvae can swim and position themselves with or against the current, but eggs are transported as passive particles. In California, sculpins (family Cottidae) have internal fertilization and immobile eggs brooded by males in small caves until hatching. Only one other fish family, the monotypic Pantodontidae, is characterized by this unique life history of internal fertilization and male egg guarding. After hatching, larvae remain in the water column for up to 56 d (Davis and Levin 2002). Intertidal sculpins in California are dominated by the genus Clinocottus, which has been shown in multiple molecular studies to be a nonmonophyletic genus comprising 5 species (Ramon and Knope 2008; Knope 2013; Buser and Andrés López 2015). Clinocottus analis, the Wooly Sculpin, is the sister species of the Lavender sculpin (Leiocottus hirundo) and is more closely related to species in the genus Oligocottus than congeners (Fig. 1D).
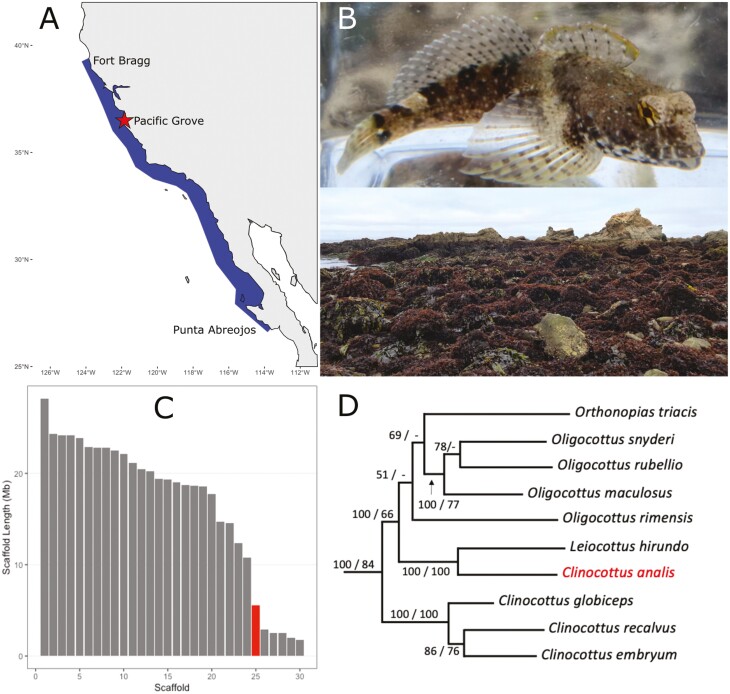
Fig. 1.
A) Distribution of the Woolly Sculpin, Clinocottus analis. Woolly Sculpin are found in the rocky shore intertidal from Fort Bragg, California, USA to Punta Abreojos, Baja California, Mexico. The collection site of the sequenced individual, Pacific Grove, California, is indicated by the red star on the map. B) Top: an image of a Woolly Sculpin (photo by P.R. Blaimont). Bottom: image of Franklin Point, California at low tide, an example of representative intertidal habitat for the Woolly Sculpin. C) Distribution of scaffolds of the genome assembly for the Woolly Sculpin. Only the 30 largest scaffolds are shown in decreasing order of size from left to right. Scaffold size is presented in Mega base pairs (Mb). Scaffold 25, which is discussed in the paper, is highlighted in red. D) Bayesian reconstruction of the nonmonophyletic clade that includes the genus Clinocottus using 2 mitochondrial genes (Cyt b, NADH1) and 1 nuclear gene (S7) redrawn from Ramon and Knope (2008). Bootstrap values are shown for Bayesian inference and maximum likelihood, respectively. Clinocottus analis is highlighted in red.
Woolly Sculpin are small (usually 10 to 12 cm TL) obligatorily intertidal fishes distributed from Fort Bragg, Mendocino County, California, USA to Punta Abreojos, Baja California, Mexico (Fig. 1A; Love 2011). As a result of their unusual life history traits, intertidal species exhibit low levels of dispersal, resulting in a high potential for local adaptation and strong within-species phylogeographic structure (Johnson et al. 2016). Given this, intertidal sculpins can be used as predictors of phylogeographic breaks along the California coast and are important species that help optimize the design and boundaries of marine conservation priorities targeting low-vagility taxa. To further this important goal, which is one of the key marine objectives of the California Conservation Genomics Project (CCGP; Shaffer et al. 2022), we sequenced and assembled a near-chromosome-level reference genome for the Woolly Sculpin, C. analis, within the CCGP framework.
This assembled genome of C. analis will serve as a valuable resource for studying the ecology, life history, adaptation, dispersal capability, and distribution dynamics of this ecologically important species, as well as establish an important model species for the study of evolutionary dynamics in low-vagility taxa across the California Current Large Marine Ecosystem (CCLME).
Methods
Biological materials
One adult Woolly Sculpin, C. analis, was collected at low tide in Pacific Grove California (N 36.6355, W −121.9255) in September 2020 by the senior author under California Department of Fish and Wildlife permit GM-201270003-20134-001 (Fig. 1A). The fish was brought live to the lab, euthanized, and liver, muscle, fin, and gill tissues were immediately harvested and flash frozen in liquid nitrogen. Samples were later transferred to a −80°C freezer until DNA extraction.
High molecular weight genomic DNA isolation
High molecular weight (HMW) genomic DNA (gDNA) was extracted from 33 mg of fin tissue (#09-17) using the Nanobind Tissue Big DNA kit (Pacific Biosciences—PacBio, Menlo Park, California), following the manufacturer’s instructions. We assessed DNA purity using absorbance ratios (260/280 = 1.81 and 260/230 = 2.54) on a NanoDrop ND-1000 spectrophotometer. The DNA yield (158 ng/µL; 18 µg total) was quantified using the Quantus Fluorometer (QuantiFluor ONE dsDNA Dye assay—Promega, Madison, Wisconsin). We estimated the size distribution of the HMW DNA using the Femto Pulse system (Agilent, Santa Clara, California) and found that 56% of the DNA fragments were 50 kb or more.
HiFi library preparation and sequencing
The HiFi SMRTbell library was constructed using the SMRTbell Express Template Prep Kit v2.0 (PacBio, Cat. #100-938-900) according to the manufacturer’s instructions. HMW gDNA was sheared to a target DNA size distribution between 15 and 18 kb. The sheared gDNA was concentrated using 0.45× of AMPure PB beads (PacBio, Cat. #100-265-900) for the removal of single-strand overhangs at 37 °C for 15 min, followed by further enzymatic steps of DNA damage repair at 37 °C for 30 min, end repair and A-tailing at 20 °C for 10 min and 65 °C for 30 min, and ligation of overhang adapter v3 at 20 °C for 60 min. The SMRTbell library was purified and concentrated with 1× Ampure PB beads (PacBio, Cat. #100-265-900) for nuclease treatment at 37 °C for 30 min and size selected using the BluePippin/PippinHT system (Sage Science, Beverly, Massachusetts; Cat #BLF7510/HPE7510) to collect fragments greater than 7 to 9 kb. The 15 to 20 kb average HiFi SMRTbell library was sequenced at University of California Davis DNA Technologies Core (Davis, California) using one 8M SMRT cell, Sequel II sequencing chemistry 2.0, and 30-h movies each on a PacBio Sequel II sequencer.
Omni-C library preparation
The Omni-C library was prepared using the Dovetail Omni-C Kit (Dovetail Genomics, Scotts Valley, California) according to the manufacturer’s protocol with slight modifications. First, specimen tissue was thoroughly ground with a mortar and pestle while cooled with liquid nitrogen. Subsequently, chromatin was fixed in place in the nucleus. The suspended chromatin solution was then passed through 100 and 40 µm cell strainers to remove large debris. Fixed chromatin was digested under various conditions of DNase I until a suitable fragment length distribution of DNA molecules was obtained. Chromatin ends were repaired and ligated to a biotinylated bridge adapter followed by proximity ligation of adapter-containing ends. After proximity ligation, crosslinks were reversed, and the DNA was purified from proteins. Purified DNA was treated to remove biotin that was not internal to ligated fragments, and a NGS library was generated using an NEB Ultra II DNA Library Prep kit (New England Biolabs, Ipswich, Massachusetts) with an Illumina compatible y-adaptor. Biotin-containing fragments were then captured using streptavidin beads. The postcapture product was split into 2 replicates prior to PCR enrichment to preserve library complexity, with each replicate receiving unique dual indices. The library was sequenced at Vincent J. Coates Genomics Sequencing Lab (Berkeley, California) on an Illumina NovaSeq (Illumina, California) to generate approximately 100 million 2 × 150 bp read pairs per GB of genome size.
Nuclear genome assembly
We assembled the Woolly Sculpin genome following the CCGP assembly protocol Version 4.0, which uses PacBio HiFi reads and Omni-C data for the generation of high quality and highly contiguous nuclear genome assemblies (outlined in Table 1). For consistency across assemblies included as part of CCGP, we ran a standardized assembly pipeline while minimizing manual curation. Briefly, we removed remnant adapter sequences from the PacBio HiFi dataset using HiFiAdapterFilt (Sim et al. 2022) and obtained the initial dual or partially phased diploid assembly (http://lh3.github.io/2021/10/10/introducing-dual-assembly) using HiFiasm (Cheng et al. 2021; see Table 1 for relevant software). We tagged output haplotype 1 as the primary assembly, and output haplotype 2 as the alternate assembly, and scaffolded both assemblies using the Omni-C data with SALSA (Ghurye et al. 2017, 2019).
Table 1.
Assembly pipeline and software used.
| Assembly | Software and optionsa | Version |
|---|---|---|
| Filtering PacBio HiFi adapters | HiFiAdapterFilt | Commit 64d1c7b |
| K-mer counting | Meryl (k = 21) | 1 |
| Estimation of genome size and heterozygosity | GenomeScope | 2 |
| De novo assembly (contiging) | HiFiasm (Hi-C Mode, –primary, output p_ctg.hap1, p_ctg.hap2) | 0.16.1-r375 |
| Scaffolding | ||
| Omni-C scaffolding | SALSA (-DNASE, -i 20, -p yes) | 2 |
| Gap closing | YAGCloser (-mins 2 -f 20 -mcc 2 -prt 0.25 -eft 0.2 -pld 0.2) | Commit 0e34c3b |
| Omni-C contact map generation | ||
| Short-read alignment | BWA-MEM (-5SP) | 0.7.17-r1188 |
| SAM/BAM processing | Samtools | 1.11 |
| SAM/BAM filtering | Pairtools | 0.3.0 |
| Pairs indexing | Pairix | 0.3.7 |
| Matrix generation | cCooler | 0.8.10 |
| Matrix balancing | hicExplorer (hicCorrectmatrix correct --filterThreshold -2 4) | 3.6 |
| Contact map visualization | HiGlass | 2.1.11 |
| PretextMap | 0.1.4 | |
| PretextView | 0.1.5 | |
| PretextSnapshot | 0.0.3 | |
| Organelle assembly | ||
| Mitogenome assembly | MitoHiFi (-r, -p 50, -o 1) | 2 Commit c06ed3e |
| Genome quality assessment | ||
| Basic assembly metrics | QUAST (--est-ref-size) | 5.0.2 |
| Assembly completeness | BUSCO (-m geno, -l actinopterygii) | 5.0.0 |
| Merqury | 2020-01-29 | |
| Contamination screening | ||
| Local alignment tool | BLAST+ | 2.1 |
| General contamination screening | BlobToolKit | 2.3.3 |
Software citations are listed in the text.
aOptions detailed for nondefault parameters.
We generated Omni-C contact maps for both assemblies by aligning the Omni-C data against the corresponding assembly with BWA-MEM (Li 2013), identified ligation junctions, and generated Omni-C pairs using pairtools (Goloborodko et al. 2018). We generated a multiresolution Omni-C matrix with cooler (Abdennur and Mirny 2020) and balanced it with hicExplorer [Version 3.6] (Ramírez et al. 2018). We used HiGlass (Kerpedjiev et al. 2018) and the PretextSuite (https://github.com/wtsi-hpag/PretextView; https://github.com/wtsi-hpag/PretextMap; https://github.com/wtsi-hpag/PretextSnapshot) to visualize the contact maps. We checked the contact maps for major misassemblies and cut the assemblies at the gaps where misassemblies were identified. No further joins were made after this step. Using the PacBio HiFi reads and YAGCloser (https://github.com/merlyescalona/yagcloser), we closed some of the remaining gaps generated during scaffolding. We then checked for contamination using the BlobToolKit Framework (Challis et al. 2020).
Mitochondrial genome assembly
We assembled the mitochondrial genome of the Woolly Sculpin from the PacBio HiFi reads using the reference-guided pipeline MitoHiFi (https://github.com/marcelauliano/MitoHiFi; Allio et al. 2020). The mitochondrial sequence of another C. analis (NCBI:FJ848374.1) was used as the starting reference sequence. After completion of the nuclear genome, we searched for matches of the resulting mitochondrial assembly sequence in the nuclear genome assembly using BLAST+ (Camacho et al. 2009) and filtered out contigs and scaffolds from the nuclear genome with a percentage of sequence identity >99% and size smaller than the mitochondrial assembly sequence.
Genome size estimation and quality assessment
We generated k-mer counts (k = 21) from the PacBio HiFi reads using meryl (https://github.com/marbl/meryl). The k-mer database was then used in GenomeScope 2.0 (Ranallo-Benavidez et al. 2020) to estimate genome features including genome size, heterozygosity, and repeat content. To obtain general contiguity metrics, we ran QUAST [Version 5.0.2] (Gurevich et al. 2013). To evaluate genome quality and completeness we used BUSCO (Manni et al. 2021) with the Actinopterygii ortholog database (actinopterygii_odb10) which contains 3,640 genes. Assessment of base level accuracy (QV) and k-mer completeness was performed using the previously generated meryl database and merqury (Rhie et al. 2020). We further estimated genome assembly accuracy via BUSCO gene set frameshift analysis using the pipeline described in Korlach et al. (2017).
Measurements of the size of the phased blocks are based on the size of the contigs generated by HiFiasm on HiC mode. Following data availability and quality metrics established by Rhie et al. (2021), we use the derived genome quality notation x·y·P·Q·C, where x = log10[contig NG50]; y = log10[scaffold NG50]; P = log10 [phased block NG50]; Q = Phred base accuracy QV (quality value); C = % genome represented by the first “n” scaffolds, following a known karyotype of 2n = 48 (Hinegardner and Rosen 1972). Quality metrics for the notation were calculated on the primary assembly.
Results
Mitochondrial assembly
Final mitochondrial genome size was 18,221 bp. The base composition of the final assembly version is A = 30.57%, C = 13.22%, G = 16.88%, T = 39.33%, and consists of 23 unique transfer RNAs and 13 protein-coding genes.
Nuclear assembly
The Omni-C and PacBio HiFi sequencing libraries generated 56.6 million read pairs and 2.3 million reads, respectively. The latter yielded 57.53-fold coverage (N50 read length 12,237 bp; minimum read length 346 bp; mean read length 12,115 bp; maximum read length of 43,596 bp) based on the GenomeScope 2.0 genome size estimation of 484 Mb. Based on PacBio HiFi reads, we estimated 0.164% sequencing error rate and 0.812% nucleotide heterozygosity rate. The k-mer spectrum based on PacBio HiFi reads (Fig. 2A) shows a distribution with 2 peaks at ~27 and 54-fold coverage, where peaks correspond to homozygous and heterozygous states of a diploid species. The distribution presented in this k-mer spectrum supports that of a low heterozygosity profile.
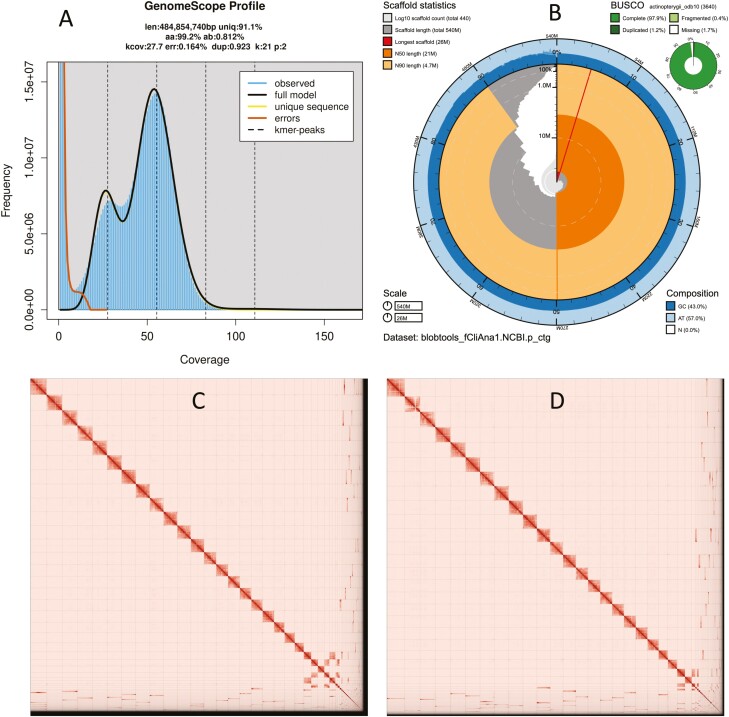
Fig. 2.
Visual overview of genome assembly metrics. A) K-mer spectra output generated from PacBio HiFi data without adapters using GenomeScope 2.0. The bimodal pattern observed corresponds to a diploid genome. K-mers covered at lower coverage and lower frequency correspond to differences between haplotypes, whereas the higher coverage and higher frequency k-mers correspond to the similarities between haplotypes. B) BlobToolKit Snail plot showing a graphical representation of the quality metrics presented in Table 2 for the Clinocottus analis primary assembly. The plot circle represents the full size of the assembly. From the inside-out, the central plot covers length-related metrics. The red line represents the size of the longest scaffold; all other scaffolds are arranged in size-order moving clockwise around the plot and drawn in gray starting from the outside of the central plot. Dark and light orange arcs show the scaffold N50 and scaffold N90 values. The central light gray spiral shows the cumulative scaffold count with a white line at each order of magnitude. White regions in this area reflect the proportion of Ns in the assembly. The dark vs. light blue area around it shows mean, maximum and minimum GC vs. AT content at 0.1% intervals (Challis et al. 2020). Omni-C contact maps for the primary (C) and alternate (D) genome assembly generated with PretextSnapshot. Hi-C contact maps translate proximity of genomic regions in 3D space to contiguous linear organization. Each cell in the contact map corresponds to sequencing data supporting the linkage (or join) between 2 such regions.
The final assembly (fCliAna1) consists of 2 pseudo haplotypes, primary and alternate, both genome sizes in close range to the estimated value from GenomeScope 2.0 (489 Mb, Fig. 2A). The primary assembly consists of 443 scaffolds spanning 538.1 Mb with contig N50 of 9.1 Mb, scaffold N50 of 21 Mb, longest contig of 20 Mb, and largest scaffold of 25 Mb. On the other hand, the alternate assembly consists of 171 scaffolds, spanning 534 Mb with contig N50 of 9.4 Mb, scaffold N50 of 20.4 Mb, largest contig 19 Mb, and largest scaffold of 28 Mb. Assembly statistics are reported in Table 2, and a graphical representation for the primary assembly is presented in Fig. 2B.
Table 2.
Sequencing and assembly statistics, and accession numbers.
| BioProjects and Vouchers | CCGP NCBI BioProject | PRJNA720569 | |||||
| Genera NCBI BioProject | PRJNA765802 | ||||||
| Species NCBI BioProject | PRJNA777156 | ||||||
| NCBI BioSample | SAMN26368113 | ||||||
| Specimen identification | CAN_PGR_092001 | ||||||
| NCBI Genome accessions | Primary | Alternate | |||||
| Assembly accession | JALGQL000000000 | JALGQM000000000 | |||||
| Genome sequences | GCA_023055335.1 | GCA_023055415.1 | |||||
| Genome Sequence | PacBio HiFi reads | Run | 1 PACBIO_SMRT (Sequel II) run: 2.3 M spots, 27.9 G bases, 15 Gb | ||||
| Accession | SRX15223504 | ||||||
| Omni-C Illumina reads | Run | 2 ILLUMINA (Illumina NovaSeq 6000) runs: 56.6 M spots, 17.1 G bases, 5.9 Gb | |||||
| Accession | SRX15223505, SRX15223506 | ||||||
| Genome Assembly Quality Metrics | Assembly identifier (quality codea) | fCliAna1(6.7.P7.Q58.C87) | |||||
| HiFi read coverageb | 57× | ||||||
| Primary | Alternate | ||||||
| Number of contigs | 662 | 393 | |||||
| Contig N50 (bp) | 9,145,431 | 9,433,512 | |||||
| Contig NG50b | 10,429,551 | 9,838,831 | |||||
| Longest contigs | 20,125,327 | 19,750,731 | |||||
| Number of scaffolds | 443 | 171 | |||||
| Scaffold N50 | 21,001,540 | 20,474,872 | |||||
| Scaffold NG50b | 21,652,950 | 21,109,456 | |||||
| Largest scaffold | 25,521,869 | 28,184,607 | |||||
| Size of final assembly | 538,118,947 | 534,899,999 | |||||
| Phased blocks NG50b | 10,429,551 | 9,838,831 | |||||
| Gaps per Gbp (# gaps) | 406 (219) | 415 (222) | |||||
| Indel QV (frameshift) | 47.09853152 | 47.67304317 | |||||
| Base pair QV | 58.6256 | 58.6696 | |||||
| Full assembly = 58.6475 | |||||||
| K-mer completeness | 91.4415 | 91.4643 | |||||
| Full assembly = 99.8107 | |||||||
| BUSCO completeness (actinopterygii) n= 3640 |
C | S | D | F | M | ||
| Pc | 97.90% | 96.70% | 1.20% | 0.40% | 1.70% | ||
| Ac | 98.20% | 97.10% | 1.10% | 0.40% | 1.40% | ||
| Organelles | 1 Partial mitochondrial sequence JALGQL010000443.1 | ||||||
aAssembly quality code x·y·P·Q·C, where x = log10[contig NG50]; y = log10[scaffold NG50]; P = log10 [phased block NG50]; Q = Phred base accuracy QV (quality value); C = % genome represented by the first “n” scaffolds, following a known karyotype of 2n = 48 (Hinegardner and Rosen 1972). BUSCO scores. (C)omplete and (S)ingle; (C)omplete and (D)uplicated; (F)ragmented and (M)issing BUSCO genes. n, number of BUSCO genes in the set/database.
bRead coverage and NGx statistics have been calculated based on the estimated genome size of 484 Mb.
cP(rimary) and (A)lternate assembly values.
We identified a single misassembly on the primary assembly and broke the corresponding join made by SALSA. We were able to close a total of 10 gaps, 3 on the primary and 7 on the alternate assembly. We filtered out a single contig from the primary assembly corresponding to arthropod contaminants. Finally, we filtered out a single contig corresponding to mitochondrial contamination from the alternate assembly. No further contigs were removed.
The primary assembly has a BUSCO completeness score of 97.9% using the Actinopterygii gene set, a per-base quality (QV) of 58.62, a k-mer completeness of 91.44 and a frameshift indel QV of 47.09. The alternate assembly has a BUSCO completeness score of 98.2% using the Actinopterygii gene set, a per-base quality (QV) of 58.66, a k-mer completeness of 91.46 and a frameshift indel QV of 47.67. The Omni-C contact map shows that both assemblies are highly contiguous (Fig. 2C and D). We have deposited scaffolds corresponding to both primary and alternate assemblies on NCBI (see Table 2 and Data availability for details).
Discussion
Relatively little genetic work has been published on C. analis, and most of the genetic work that has been performed used mitochondrial markers and focused broadly on the phylogenetic relationships of sculpin (Kinziger et al. 2005; Ramon and Knope 2008; Knope 2013; Buser and Andrés López 2015). Some research has been completed on the population structure of C. analis across its range, but that early work was limited to a small set of mitochondrial markers and results were confounded by the presence of a mitochondrial genome rearrangement with 2 nearly identical control regions (Ramon 2007). That study assembled a mitochondrial genome with a total length of 18,374 bp, i.e. nearly identical to the 18,221 bp reported here. Early work on the nuclear genome suggested that the genome size was 0.93 pg, based on its estimated c value (where 1 pg to 0.978 Gb) and a karyotype of 2n = 48 (Hinegardner and Rosen 1972).
In this study, we found that the genome size of C. analis is 538 Mb, which is considerably smaller than the 909 Mb c value based estimate of Hinegardner and Rosen (1972). When scaffolds are ordered by length from largest to smallest, the greatest decrease in scaffold size (48.5%) occurs between the 24th and 25th largest scaffolds of our assembly (Fig. 1C). Taken together the largest 24 scaffolds comprise 483 Mb which corresponds to approximately 90% of the full genome. As such, our assembly supports a karyotype of 2n = 48 for C. analis though further work to establish the karyotype for this species is warranted. The mitochondrial genome assembled in this study has a total length of 18,221 bp, slightly shorter than that assembled by Ramon (2007) but longer than typical bony fishes (~16,500).
The high quality of the genome we are presenting here (contig N50 = 9.1 Mb, BUSCO completeness = 97.9%) will allow us to use it as a reference for the medium-coverage whole-genome resequencing project for C. analis that comprises the next phase of the CCGP data collection pipeline (Shaffer et al. 2022). Our long-term goal is to draw a clear picture of the genetic boundaries between adjacent marine regions in California, as well as determine the degree of local adaptation among regions, and to use these data to delineate relevant protected areas that are grounded in strong genetic data. This genome is the first step in an important endeavor that will ultimately result in a sound protection plan for California’s natural marine resources.
Acknowledgments
We would like to thank Marina Ramon for providing expertise in identifying specimens, and time in the field during the sampling effort. PacBio Sequel II library prep and sequencing were carried out at the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. Deep sequencing of Omni-C libraries used the NovaSeq S4 sequencing platforms at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. We thank the staff at the UC Davis DNA Technologies and Expression Analysis Cores and the UC Santa Cruz Paleogenomics Laboratory for their diligence and dedication to generating high-quality sequence data.
Contributor Information
Daniel B Wright, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Merly Escalona, Department of Biomolecular Engineering, University of California Santa Cruz, Santa Cruz, CA, United States.
Mohan P A Marimuthu, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California-Davis, Davis, CA, United States.
Ruta Sahasrabudhe, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California-Davis, Davis, CA, United States.
Oanh Nguyen, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California-Davis, Davis, CA, United States.
Samuel Sacco, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Eric Beraut, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Erin Toffelmier, Department of Ecology & Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States.
Courtney Miller, Department of Ecology & Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States.
H Bradley Shaffer, Department of Ecology & Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States; La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States.
Giacomo Bernardi, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Funding
This work was supported by the California Conservation Genomics Project, with funding provided to the University of California by the State of California, State Budget Act of 2019 [UC Award ID RSI-19-690224].
Data availability
Data generated for this study are available under NCBI BioProject PRJNA777156. Raw sequencing data for sample CAN_PGR_092001 (NCBI BioSample SAMN26368113) are deposited in the NCBI Short Read Archive (SRA) under SRX15223504 for PacBio HiFi sequencing data, and SRX15223505 and SRX15223506 for the Omni-C Illumina sequencing data. GenBank accessions for both primary and alternate assemblies are GCA_023055335.1 and GCA_023055415.1; and for genome sequences JALGQL000000000 and JALGQM000000000. The GenBank organelle genome assembly for the mitochondrial genome is JALGQL010000443.1. Assembly scripts and other data for the analyses presented can be found at the following GitHub repository: www.github.com/ccgproject/ccgp_assembly.
References
- Abdennur N, Mirny LA.. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2020;36(1):311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F.. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 2020;20(4):892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser TJ, Andrés López J.. Molecular phylogenetics of sculpins of the subfamily Oligocottinae (Cottidae). Mol Phylogenet Evol. 2015;86:64–74. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, Cochrane G, Blaxter M.. BlobToolKit—interactive quality assessment of genome assemblies. G3. 2020;10(4):1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Jarvis ED, Fedrigo O, Koepfli K-P, Urban L, Gemmell NJ, Li H.. Robust haplotype-resolved assembly of diploid individuals without parental data. 2021. [accessed 2022 Jul 1]. http://arxiv.org/abs/2109.04785. [DOI] [PMC free article] [PubMed]
- Davis J, Levin L.. Importance of pre-recruitment life-history stages to population dynamics of the woolly sculpin Clinocottus analis. Mar Ecol Prog Ser. 2002;234:229–246. [Google Scholar]
- Ghurye J, Pop M, Koren S, Bickhart D, Chin C-S.. Scaffolding of long read assemblies using long range contact information. BMC Genomics. 2017;18(1):527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, Schmitt A, Selvaraj S, Pop M, Phillippy AM, Koren S.. Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLoS Comput Biol. 2019;15(8):e1007273. doi: 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Abdennur N, Venev S, Brandao HB, Fudenberg G.. mirnylab/pairtools: v0.2.0 (v0.2.0). Zenodo. 2018. doi: 10.5281/zenodo.1490831 [DOI] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinegardner R, Rosen DE.. Cellular DNA content and the evolution of teleostean fishes. Am Nat. 1972;106(951):621–644. [Google Scholar]
- Johnson DW, Freiwald J, Bernardi G.. Genetic diversity affects the strength of population regulation in a marine fish. Ecology. 2016;97(3):627–639. [DOI] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Luber JM, Ouellette SB, Azhir A, Kumar N, et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinziger AP, Wood RM, Neely DA.. Molecular systematics of the genus Cottus (Scorpaeniformes: Cottidae). Copeia. 2005;2005(2):303–311. [Google Scholar]
- Knope ML. Phylogenetics of the marine sculpins (Teleostei: Cottidae) of the North American Pacific Coast. Mol Phylogenet Evol. 2013;66(1):341–349. [DOI] [PubMed] [Google Scholar]
- Korlach J, Gedman G, Kingan SB, Chin CS, Howard JT, Audet JN, Cantin L, Jarvis ED.. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience. 2017;6(10):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J. The pelagic stage of reef fishes. In: Sales P, editor. The ecology of fishes on coral reefs. San Diego (CA): Academic Press Inc.; 1991. p. 182–229. [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, arXiv, arXiv:1303.3997, 2013. doi: 10.48550/arXiv.1303.3997 [DOI] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simao FA, Zdobnov EM.. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes, arXiv, arXiv:2106.11799 [q-bio], 2021. doi: 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T.. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon ML. Molecular ecology and evolution of intertidal sculpin [PhD dissertation]. Santa Cruz, CA:University of California Santa Cruz; 2007. p. 138. [Google Scholar]
- Ramon ML, Knope ML.. Molecular support for marine sculpin (Cottidae; Oligocottinae) diversification during the transition from the subtidal to intertidal habitat in the Northeastern Pacific Ocean. Mol Phylogenet Evol. 2008;46(2):475–483. [DOI] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC.. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11(1):1432. doi: 10.1038/s41467-020-14998-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM.. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21(1):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer HB, Toffelmier E, Corbett-Detig RB, Escalona M, Erickson B, Fiedler P, Gold M, Harrigan RJ, Hodges S, Luckau TK, et al. Landscape genomics to enable conservation actions: the California Conservation Genomics Project. J Hered. 2022;113(6):577–588. doi: 10.1093/jhered/esac020 [DOI] [PubMed] [Google Scholar]
- Sim SB, Corpuz RL, Simmonds TJ, Geib SM. HiFiAdapterFilt, a memory efficient read processing pipeline, prevents occurrence of adapter sequence in PacBio HiFi reads and their negative impacts on genome assembly. BMC Genomics. 2022;23(1):157. doi: 10.1186/s12864-022-08375-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data generated for this study are available under NCBI BioProject PRJNA777156. Raw sequencing data for sample CAN_PGR_092001 (NCBI BioSample SAMN26368113) are deposited in the NCBI Short Read Archive (SRA) under SRX15223504 for PacBio HiFi sequencing data, and SRX15223505 and SRX15223506 for the Omni-C Illumina sequencing data. GenBank accessions for both primary and alternate assemblies are GCA_023055335.1 and GCA_023055415.1; and for genome sequences JALGQL000000000 and JALGQM000000000. The GenBank organelle genome assembly for the mitochondrial genome is JALGQL010000443.1. Assembly scripts and other data for the analyses presented can be found at the following GitHub repository: www.github.com/ccgproject/ccgp_assembly.