Abstract
Rapid, sensitive, and inexpensive point-of-care diagnosis is vital to controlling highly infectious diseases, including COVID-19. Here, we report the design and characterization of a compact fluorimeter called a “Virus Pod” (V-Pod) that enables sensitive self-testing of SARS-CoV-2 viral load in saliva. The rechargeable battery-operated device reads the fluorescence generated by Designer DNA Nanostructures (DDN) when they specifically interact with intact SARS-CoV-2 virions. DDNs are net-shaped self-assembling nucleic acid constructs that provide an array of highly specific aptamer-fluorescent quencher duplexes located at precise positions that match the pattern of spike proteins. The room-temperature assay is performed by mixing the test sample with DNA Net sensor in a conventional PCR tube and placing the tube into the V-Pod. Fluorescent signals are generated when multivalent aptamer-spike binding releases fluorescent quenchers, resulting in rapid (5-min) generation of dose-dependent output. The V-Pod instrument performs laser excitation, fluorescence intensity quantitation, and secure transmission of data to an App via Bluetooth™. We show that the V-Pod and DNA Net assay achieves clinically relevant detection limits of 3.92 × 103 viral-genome-copies/mL for pseudo-typed wild-type SARS-CoV-2 and 1.84 × 104, 9.69 × 104, 6.99 × 104 viral-genome-copies/mL for pathogenic Delta, Omicron, and D614G variants, representing sensitivity similar to laboratory-based PCR. The pocket-sized instrument (∼$294), inexpensive reagent-cost/test ($1.26), single-step, rapid sample-to-answer, and quantitative output represent a capability that is compatible with the needs of frequent self-testing in a consumer-friendly format that can link with medical service systems such as healthcare providers, contact tracing, and infectious disease reporting.
Keywords: Biosensor, Portable fluorometer, Point-of-care test, Designer DNA Nanostructure, Smartphone, COVID-19 testing
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense, single-stranded, enveloped RNA virus (Alexandersen et al., 2020) whose predominant transmission pathway is via inhalation of aerosols and droplets exhaled by infected hosts (Meyerowitz et al., 2021). Like SARS and MERS-CoV (middle east respiratory syndrome coronavirus), SARS-CoV-2 is a member of the coronavirus family with a zoonotic origin that can be transmitted from animal to animal, animal to human, and human to human when directly or indirectly exposed to an etiological agent (Vilcek, 2020). Ever since the initial spillover from an unknown animal reservoir to humans in December 2019, SARS-CoV-2 has spread throughout the world, bringing an unprecedented crisis to society. At the time of this writing, more than 557 million people have been infected, among whom 6.3 million have died (WHO COVID-19 Dashboard, 2022). The global economy is facing the deepest recession since 1945 due to the loss of working hours, disruptions in supply chains, and closures of borders and facilities (Bank, 2021; Mallah et al., 2021). In contrast to past pandemics, the COVID-19 pandemic presents unique clinical and epidemiological characteristics that significantly undermine ongoing global efforts to stop transmission. More than 42% of infected individuals are pre-symptomatic or asymptomatic (Sah et al., 2021). Further, due to accumulating mutations that alter the spike protein, a series of Variants of Concern (VOC) have emerged with increasingly greater infectivity (Thye et al., 2021). Consequently, SARS-CoV-2 has achieved hypermobility despite the interventions of public health authorities, with measures that include masking, social distancing, testing, quarantine, and vaccination.
A key point of failure for COVID-19 containment is the inability of healthcare systems to provide inexpensive, accurate, and real-time detection of an infected person for immediate quarantine and isolation. Identifying and isolating an infected person from new potential hosts is critical to breaking the train of viral transmission. Unfortunately, current detection techniques are largely limited by costly laboratory procedures, high false-negative rates, and lengthy sample-to-result times. The gold standard nucleic acid tests, namely polymerase chain reaction (PCR) and loop-mediated isothermal amplification (LAMP) (Ganguli et al., 2020; Jankelow et al., 2022a), require stringent and technically challenging laboratory protocols, including lysing the viral capsid, RNA extraction, RNA reverse transcription, and enzymatic amplification of specific nucleic acid sequences under precise temperature control (Jankelow et al., 2022b; Afzal, 2020). Even using high throughput automated laboratory-based systems, positive PCR tests do not always represent an active infectious case as non-contagious viral RNA may still reside within samples that have no viable virus present (Makhoul et al., 2022; Byrne et al., 2020; Walsh et al., 2020). Further, 58% of COVID-19 patients have initial false negative RT-PCR results (Pecoraro et al., 2022), which is far less than the minimum recommendation of 80% sensitivity and 97% specificity recommended by the WHO ((WHO) and W.H.O, 2021).
To establish comprehensive disease control, we seek the development of a rapid, accurate, inexpensive, and accessible viral detection method, to enable frequent mass screening and diagnosis at the point-of-care (POC) without the involvement of specialized personnel and laboratory equipment. Based on a recent study conducted by Smith et al., serial screening with testing intervals of at least every 3 days can provide >98% sensitivity for identifying infected individuals when tested with qRT-PCR (Smith et al., 2021). However, achieving population-scale high testing frequency is extremely challenging due to the high cost per test, accessibility to testing/sampling sites, and other infrastructure limitations. Developing countries face further challenges that include a lack of financial resources and underdeveloped systems for healthcare, contact tracing, and reporting to government health authorities. To bridge this pragmatic bottleneck, the hospital-on-chip (HOC) module, which incorporates state-of-the-art technologies in a single frame, has been proposed as a 5th-generation disease control strategy (Chaudhary et al., 2023). By embedding 2D material-based biosensors on a chip with modern technologies, including 5G communication, internet-of-medical-things (IoMT), and artificial intelligence (AI), HOC offers a portable intelligent and multifunctional POC diagnosing platform (Byakodi et al., 2022; Cherusseri et al., 2022). However, its immediate real-life application requires overcoming technological challenges, such as fabrication without corrosive chemicals, scalability, and long-term performance sustainability of the sensor (Chaudhary et al., 2023).
As an alternative to laboratory-based tests, portable POC self-testing devices have become available, including the Abbott BinaxNow COVID-19 antigen self-test kit, the Lucira All-In-One COVID-19 test kit, and the COVID-19 molecular testing kit from Cue Health. Each of these test kits has well-known drawbacks that preclude their use as a primary detection modality. The Abbott BinaxNow antigen self-test provides results in ∼15 min with a nasal swab sample ((FDA), 2022a). However, because the test is a lateral flow immunoassay, the results cannot be accurately quantified while the sensitivity remains inferior to that of the nucleic acid tests. The Lucira All-In-One COVID-19 test kit utilizes lyophilized RT-LAMP reactions for the detection of SARS-CoV-2 RNA in a nasal swab sample with a limit of detection as low as 900 viral genome copies/mL ((FDA), 2022b). However, the cost per test is high (∼$75) and a prescription from a doctor is required for purchasing, which discourages frequent testing. Another isothermal POC nucleic acid test is the Cue COVID-19 Test offered by Cue Health ((FDA), 2022c). Authorized for nonprescription home use, the Cue test kit achieves ∼92% and ∼98% positive and negative percent agreement respectively but requires a specialized instrument along with a single-use cartridge at a price of up to $854 (a reader and 10 tests). The current assay paradigms for SARS-CoV-2 laboratory testing and self-testing do not meet the combined requirements of sensitivity, accuracy, speed, and cost that would reduce the spread of a highly infectious respiratory disease that is largely transmitted by individuals without symptoms.
An alternative paradigm for viral load monitoring is the development of “direct” assays capable of detecting intact infectious virions. By circumventing the need to process a test sample to extract and amplify nucleic acids, direct assays can utilize specific recognition of proteins displayed on the virus outer surface (Yousefi et al., 2021). Most commonly, virus-specific capture molecules, such as antibodies and aptamers, are immobilized upon the surface of a biosensing transducer, upon which accumulated virions are measured after an incubation period with the test sample (Li et al., 2022; Lee et al., 2021). However, direct assays have limitations which include the cost of the testing components (biosensors, antibodies, detection instrument), the shelf life of antibodies, workflow complexity (washing steps), and insufficiently low detection limits (Li et al., 2022; Kevadiya et al., 2021). More specifically, the limit of detection for the direct assay is often greater than indirect assays where more than one labeled secondary antibody is used to bind to the primary antibody to achieve signal amplification (Miftahussurur and Yamaoka, 2016). The specificity of the direct assays is often questioned as the other molecules with similar structures indiscriminately bind to the immobilized capture molecules (Tate and Ward, 2004). Thus, extensive sample purification and preparation to remove interferences from a complex biological sample are sometimes necessary, leading to complex assay procedures and increased costs.
Recently, we described a direct virus detection and quantitation method that utilizes the unique properties of Designer DNA Nanostructures (DDNs). DDNs are comprised of nucleic acid strands designed to self-assemble into precise shapes using Watson-Crick base pairing between joined segments while providing attachment points at specific locations for fluorophores, quenchers, and molecular recognition elements. For example, we designed DDNs that assemble into star-shaped patterns with vertices that precisely match the spatial pattern of outer proteins on the Dengue virus. By integrating Dengue-specific aptamers at the vertices, “DNA stars” make multivalent attachments to their viral targets that provide greater affinity than individual aptamers, which provide excellent selectivity against nontarget viruses (Kwon et al., 2020). We subsequently designed and demonstrated DDNs in a net-shaped configuration (called “DNA Net”) for selective recognition and high-affinity capture of intact SARS-CoV-2 (Chauhan et al., 2022). To provide a means of sensing signal transduction, fluorophore-quencher pairs are integrated with the virus-binding aptamers, which unlock as a consequence of the binding interaction to release quenchers into the sample media, enabling fluorophores to emit. Importantly, DNA Nets provide mechanical flexibility that enables conformation to the curvature of the attached virus, so a single net-virion interaction releases many fluorescence quenchers as a mechanism for signal amplification. Furthermore, the DNA Nets can leverage the unique spatial patterns of spike proteins with quaternary structure, in which each spike presents multiple aptamer binding sites. Through the pattern-matched binding, the binding strength of the aptamer to the spike proteins was demonstrated to increase by 1 × 106 fold when compared to that of the solitary aptamer (Chauhan et al., 2022). While DNA Nets are capable of binding free proteins in the solution, only binding to intact virions unquenches the cascade of fluorophore that generates signals measurable above the background. DDNs have been demonstrated to be non-toxic and biologically stable, while also inexpensive to synthesize at a manufacturing scale, as they are comprised of synthetic nucleic acid strands of pre-designed base sequences that self-assemble by gentle heating and cooling. Using DDNs and commercially available laboratory-based fluorimeters to detect viral targets, the detection of Dengue in human serum and SARS-CoV-2 in saliva with a sensitivity of 1 × 102 p.f.u/mL and 1 × 103 viral genomic copies/mL, respectively, was recently demonstrated (Kwon et al., 2020; Chauhan et al., 2022), achieving detection limits similar to those obtained by laboratory-based qRT-PCR.
In this work, we describe the design and characterization of a compact, inexpensive, pocket-size fluorimeter called a Virus-Pod (V-Pod) to enable DDN-based direct virus detection to be performed in self-testing scenarios. Specifically, the demonstrated system was developed to satisfy the REASSURED (real-time connectivity, ease of specimen collection, affordable, sensitive, specific, user-friendly, rapid and robust, equipment simple, and deliverable to end-user) criteria that describe an ideal test that can be used at POC for detecting infectious diseases. To provide an extremely simple and inexpensive liquid-handling format, we utilize conventional plastic 200 ml PCR tubes pre-filled with 20 μl of DNA Net sensor reagent. A user performs a test by opening the PCR tube, dispensing the test sample into the tube, closing the tube, gently shaking, and then inserting the tube into the form-fitted slot within the V-Pod. The V-Pod is designed to focus laser illumination into the approximate center of the liquid volume at timed intervals while collecting fluorescence emission through an optical filter with a photodiode sensor. Digitized photodiode intensity values are transmitted wirelessly via an encrypted Bluetooth™ connection to a linked smartphone, which runs a custom software application (app). The app enables the smartphone to communicate with a microprocessor circuit board integrated within the V-Pod to send instructions to the optoelectronic components, manage power, and manage the rechargeable battery. From the initiation of a test, results are obtained in 5–10 min, and results are displayed to the user on the smartphone display. The cost per test is ∼$1.26, including the DDN material, buffer, and PCR tube. The plastic body of our V-Pod prototypes was constructed with an inexpensive, 3D-printed material for compact integration of the components (Arduino™ Board, power management circuit, wireless charging module, photodiode, laser with a constant current drive module, and lithium-ion battery). We present a detailed characterization of the V-Pod, including the reproducibility of the illumination source during extended usage of the battery and a direct comparison of the fluorophore detection limit with two commercially available laboratory-based fluorimeters. Furthermore, we demonstrated the detection of pathogenic SARS-CoV-2 variants (including D614G, Delta, and Omicron), along with experimentally obtained corresponding limits of detection. The V-Pod achieved detection limits of 1.84 × 104 viral genome copies/mL for pathogenic D614G, 9.69 × 104 viral genome copies/mL for the Delta variant, and 6.99 × 104 viral genome copies/mL for the Omicron variant, within 5 min of testing in single-pot reactions, representing SARS-CoV-2 viral loads within the clinically relevant range of 104–1010 viral genome copies/mL. The simple testing procedure (single-step, single reagent, room temperature) eliminates the need for complicated training, costly equipment, or trained personnel to enable SARS-CoV-2 self-testing. Furthermore, the low cost/test and familiar form factor of the consumer-friendly instrument facilitate frequent testing. Integration of the detection instrument with the linked mobile device of the test subject enables sharing of test results with cloud-based telehealth providers, contact tracing, and reporting to health authorities. While outside the scope of this report, widespread adoption of simple and frequent testing, combined with anonymous sharing of test results with governmental agencies and streamlined connections to health services, will offer mechanisms for providing effective treatment (such as prescribing antiviral medication), guidance for isolated individuals, and monitoring the pandemic status with longitudinal and geographic precision.
2. Experimental
2.1. Assay workflow for point-of-care diagnosis using V-Pod
Fig. 1 illustrates the assay workflow. First, the Bluetooth™ connection between the smartphone app and V-Pod is initiated by the user. The saliva sample (80 μl) is placed into a PCR tube pre-loaded with the DDN reagent (20 μl). Next, the tube is gently shaken for ∼10 s to allow the DDNs to bind to virion particles. During the binding interactions, the locks (the short oligonucleotides with quencher attached to their ends) are displaced from the structure where the aptamer-reporter duplex (which is fixed at the vertices of the structures with a fluorophore attached to the open end of the nucleotide) becomes unquenched, allowing for fluorescent light generation. A detailed explanation of the design and working principle of the DNA Net sensor for SARS-CoV-2 detection has been fully described in (Chauhan et al., 2022). After mixing, the tube is inserted into its shape-matched slot in the V-Pod, which holds the tube in a precise orientation relative to the laser illumination and photodiode sensor. Measurement is initiated by pressing the “start test” button on the app. While the test is active, the app receives fluorescent intensity measurements in real-time at 1-min intervals and plots the data on a graph that can be viewed by the user. A video showing each step of the workflow is provided in Video S1 within the information supplement. The sample-to-answer time is approximately 6 min; 1 min for collecting and mixing the sample, followed by 5 min for testing. The V-Pod is designed to be rechargeable for repetitive testing as needed.
Fig. 1.
Point-of-care testing workflow of the portable, pocket-sized V-Pod SARS-CoV-2 detection system using the DDN assay. 1. A Bluetooth™ connection is established between the V-Pod and a linked mobile device, followed by saliva sample collection into a PCR tube, pre-loaded with DDN reagents. 2. The PCR tube is gently shaken for 10 s 3. The tube is inserted into the shape-matched slot for fluorescence measurements at 1-min intervals. 4. The app interprets the collected data, giving diagnostic results in 5–10 min.
2.2. V-Pod design, software programming, and fabrication by 3D printing
A schematic drawing of the V-Pod and wireless charging station is shown in Fig. 2 a and b. The design of the V-Pod was inspired by Apple AirPod™ wireless Bluetooth™ earbuds. The performance objectives of the V-Pod are 1. To provide a consistent intensity and steady excitation illumination at a wavelength of 450 nm to the PCR tube throughout the maximum 10-min testing period at a fixed illumination interval; 2. To enable the precise and sensitive collection of the emission intensity (peak wavelength of 517 nm) generated from the DDNs within the PCR tube; and 3. To establish stable and secure wireless communication with the smartphone app for transferring data.
Fig. 2.
Schematic drawing and images of the V-Pod. a) Internal components and housing of the V-Pod unit. The top portion of the fluorimeter is designed to hold the optical components and the PCR tube. Lifting the cover cap exposes the PCR tube insert slot, which firmly holds the tube in a precise position to minimize movement and positional variations between tests. The laser is incident from the side while the photodiode sensor is oriented in an orthogonal direction to collect emission through a long-pass (510 nm) filter. The bottom portion of the fluorimeter incorporates the electrical components, including a power management module, a lithium-ion polymer battery, a microcontroller unit, a wireless charging receiver module, and a push-button module. b) Internal components and housing of the V-Pod charging station. The charging station consists of a wireless charging transmitter circuit and inductor coils to allow recharging of the battery. c) Image of the complete V-Pod instrument. d) Image of the V-Pod with the cover cap open. e) Size comparison image of the V-Pod and an Apple AirPod™ earbud case. f-g) Images of the left and right sides of the fluorimeter.
To achieve the desired optical specifications, the V-Pod includes a power management module (PMM) (PowerBoost 500 Charger, 500 mA+, Adafruit, NY, US) that up-regulates the output voltage of 5 V from a 3.7 V lithium-ion polymer battery (2750, 350 mA, Adafruit, NY, US). The PMM also allows for simultaneous charging of the battery while providing 5 V output. The lithium-ion polymer battery includes protection circuitry to prevent over-charging and discharging of the battery. The PMM provides power to the constant current driver circuit of a laser diode with a peak emission wavelength of 450 nm (Industrial 450 nm 100 mW laser module, AliExpress). The intensity delivered to the test sample is reduced to 10 mW by manual adjustment of the built-in potentiometer during assembly. Laser intensity was verified by using an optical power and energy meter (PM 300E, Thorlabs, NJ, US). To provide laser illumination at fixed time intervals (to prevent photobleaching and excessive heating), the power input of the constant current driver flows through an N-Channel Power MOSFET (IRFZ44n) controlled by a single-board microcontroller unit (Arduino Nano 33 BLE, Arduino LLC, Italy) by pulse width modulation (PWM). The Arduino board not only serves as the means to control the laser illumination duty cycle but also converts the photodiode's analog output signal to digital readings.
A photodiode was selected to meet the second criterion of the V-Pod. The sensitivity of the assay is largely determined by the capability of the light sensor to detect weak fluorescent output from the assay above background light intensity from non-assay sources. For quantitation, it is desirable to differentiate small changes in measured light intensity. The V-Pod utilizes the I2C light sensor module developed by Adafruit (VEML7700 Lux Sensor, 4162, Adafruit, NY, US), which offers a resolution down to 0.0036 lx/ct with a 16-bit dynamic range of up to 120 Klux, and has software-adjustable gain and integration time. Through experimental exploration of detecting 1 μM fluorescent dye, an integration time of 100 ms was selected at a gain of 1x. Increasing the integration time beyond 100 msec did not offer a sensitivity advantage, as the background signal from other sources increased in proportion to the fluorescence signal. To reduce scattered light from the laser from reaching the photodiode, a long pass filter (et510lp, OD 6 at 450 nm, 0 deg AOI, Chroma Technology, VT, US) was placed in front of the sensor. The photodiode is oriented orthogonally to the path of the laser illumination and placed as close as possible to the wall of the inserted PCR tube. In this configuration, the sensing area (0.336 mm2) of the photodiode is located approximately 4.5 mm away from the center of the focal point of the laser. To minimize the cost and size of the instrument, no lens was included to gather and focus fluorescent emission onto the sensor from a larger solid angle. Since the area of the sensing window of the photodiode is 0.336 mm2, we estimate that the solid angle subtended by the photodiode to a point source within the assay volume to be 0.016 Sr, representing a fluorescence photon capture efficiency of approximately 0.1% if we assume that fluorescent emission occurs uniformly in all directions in spherical coordinates. The optical path includes a circular window tunnel (radius of 1.5 mm) between the PCR tube and the photodiode (Fig. S1) that prevents light from reaching the long pass filter at large angles of incidence and prevents extraneous light from internal scattering or leakage through the hinged door from reaching the photodiode.
The third criterion of the V-Pod was addressed by utilizing the Arduino Nano 33 BLE board, which has the Bluetooth™ Low Energy (BLE) chipset incorporated into a single 45 × 18 mm2 circuit board. The BLE technology reduces power consumption while maintaining a similar communication range to that of the conventional Bluetooth™ technology. The Bluetooth™ connection was established by following the programming protocols provided by Arduino. For data security, Curve25519, an elliptic-curve-Diffie-Hellman function suitable for a wide variety of cryptographic applications, is used to drive a shared secret key for advanced encryption standard 256 (AES256). The encryption prohibits unauthorized users from accessing the V-Pod data. The codes for encryption and Bluetooth connection were written with the open-source Arduino IDE (Integrated Development Environment) software and its supporting libraries.
Additional design considerations include the facilitation of repeated and long-term usage of the device, isolation of the PCR tube from ambient light, and battery level indication. Since the V-Pod is a closed system, a means to recharge the enclosed battery must be provided. We utilized a universal Qi wireless charging receiver module (1901, Adafruit, NY, US), connected to the power management module to provide a consistent 5 V at 500 mA when charging. The universal Qi wireless charging transmitter module (2162, Adafruit, NY, US) was selected for the charging station. Additionally, a push-button power switch breakout (1400, Adafruit, NY, US) was used to interrupt the connection between the power management module and the microcontroller unit to conserve energy when not in use. To isolate the PCR tube from ambient light during measurements, the casing was constructed from an opaque black material, and a groove-tongue joint was incorporated into the closure of the hinged door. Lastly, a battery level indicating circuit was connected to the microcontroller unit, enabling the smartphone app to display the battery charge status.
Both the V-Pod body and the wireless charging station were designed using SolidWorks AutoCAD software and fabricated by a Form2 desktop stereolithography 3D printer (Fig. 2c and d). The black resin was printed at a 100-μm resolution. After printing, the parts were immediately washed with isopropyl alcohol to remove excess resin, followed by an isopropyl alcohol bath (10 min), room temperature drying (30 min), and post-curing (1 h, 60 °C, Form Cure, FormLabs, MA, US). Fig. 2e is a photo of the V-Pod next to an Apple AirPod™ case for size comparison, while Fig. 2f and g show the left and right sides of the assembled V-Pod. The optical path of the V-Pod is illustrated in Fig. 3 a, where the laser diode illuminates focused excitation light into the inserted PCR tube, from which the emitted light is collected at an orthogonal angle through a long-pass filter. The overall cost of the device is ∼$294, which includes a long pass filter ($179) and the electronic components ($115). The bill of material for the V-Pod can be found in Table S1. The V-Pod is 6.3 × 2.8 × 4.7 cm3 in size and weighs 75g.
Fig. 3.
Optical pathway of the fluorimeter and schematic overview of the V-Pod system. a) The 450 nm 10 mW laser directs excitation light towards the PCR tube, from which the emission is collected at an orthogonal angle by the photodiode. b) Block diagram of the V-Pod system. The instrument is powered by a lithium-ion polymer battery, which is connected to the power management module to up-convert the 3.7V–5V operating voltage. The push-button module interrupts the 5V power supply line from the power management module to the micro-controller unit to conserve energy when not in use. The microcontroller units supply control signals and log output from the MOSFET switch and the I2C light sensor. The MOSFET switch toggles the laser on and off to provide a zero-power state when the device is not in use. The micro-controller unit incorporates a Bluetooth™ Low Energy (BLE) chipset to establish wireless communication with a linked mobile device. The transmitted data is processed and stored in a cloud-based data service, which can be further utilized for epidemic surveillance, such as contact tracing, and connection to telehealth services.
2.3. Smartphone app design and development
The mobile app was custom-built to operate on Android-based smartphones. In this study, all data were obtained using a Samsung Galaxy Note 11. Upon opening the app for the first time, the user is presented with a general description of the V-Pod, followed by prompts to request permission to use location information and to establish a Bluetooth connection. Once the setup is completed, the testing dashboard appears, where the tests are logged with options to perform additional tests or to add testing results obtained from external facilities. The app is designed to save the test results both locally as well as in cloud-based data services (Firebase, Google, US) with a user-specific unique identifier. The stored data can then be further utilized for epidemic surveillance systems such as contact tracing, infection case identification, and the planning, implanting, and evaluating of public health interventions. A block diagram summarizing the V-Pod hardware and software is shown in Fig. 3b. Several screens from the app interface are shown in Figs. S2–4.
2.4. Device performance evaluation and comparison with commercially available laboratory-based fluorimeters
Initial V-Pod characterization was performed to verify consistent laser illumination intensity and to measure the spectral emission characteristics of DDNs compared to potential sources of background noise. To demonstrate that the laser power provided to the sample remains constant as the battery is depleted, we performed a series of 2 s illuminations (nominally 10 mW) at 1 min intervals until the fully charged battery was completely drained, while measuring the laser output intensity with an optical power meter (PM 300E, Thorlabs, NJ, US) while simultaneously monitoring the battery level with the app. The results are shown in Fig. 4 a. The experiment confirms that the V-Pod is capable of performing 30 consecutive tests on a single battery charge and that the laser power management circuit accurately maintains the desired output intensity even as the battery is depleted. Laser components that passed the above procedure were incorporated into V-Pod assemblies.
Fig. 4.
Characterization of the V-Pod fluorimeter and two commercially available laboratory-based fluorimeters (Qubit Flex and ANDalyze). a) Battery and laser power comparison characterization. b) Spectrum characterization of the optical system of the V-Pod fluorimeter. The excitation wavelength (450 nm) is cut off by the long-pass filter (510 nm). The positive control is tested using a pseudo-typed WT virus at 107 genome copies/mL. The negative control is tested using heat-inactivated pseudo-typed WT virus. c-e) The normalized dose-response curves of the V-Pod, Qubit Flex, and ANDalyze fluorimeters, respectively, using various concentrations of FAM diluted with DI water. The horizontal dotted line represents the limit of detection, while the diagonal dotted line represents the hill slope of the curve. The error bars represent the standard deviation of three independent measurements. f) The combined normalized dose-response curves of the three fluorimeters. The horizontal and vertical dotted lines represent the half-maximal effective concentrations for each dose-response curve. Each measurement is repeated three times. g) Fluid temperature characterization of the three fluorimeters during the 10-min testing period. h) Fluorescence signal percent change characterization of the three fluorimeters. The changes in fluorescent signals of the various FAM concentrations (250 nM, 200 nM, 150 nM, and 125 nM) are measured relative to the baseline concentration of 100 nM FAM.
Next, we studied the spectral characteristics of the light that would be detected at the position of the photodiode by placing the tip of a 400 μm core diameter optical fiber at the sensor position and attaching the distal end of the fiber to a spectrometer (USB2000 Spectrometer, Ocean Optics, FL, US). The long pass filter utilized in the V-Pod was in its position directly in front of the optical fiber tip. The spectra of filtered light from various samples were measured, including the “empty instrument” condition (no PCR tube), PCR tube filled with DI water, positive control (DNA Net sensor with 107 pseudo-typed SARS-CoV-2), and negative control (DNA Net sensor with heat-inactivated pseudo-typed SARS-CoV-2). Results are summarized in Fig. 4b. Without a PCR tube, minimal to no excitation wavelength passed through the long-pass filter and reached the light sensor. However, the insertion of a PCR tube created a wide spectrum of scattered and reflected auto-fluorescence light, which increased the background noise signal. The spectra of the positive and negative controls of the DDN assay are shown in the same figure (Fig. 4b). The positive control included the pseudo-typed wild-type (WT) SARS-CoV-2 virus at a concentration of 107 viral genome copies/mL, while the same concentration of the virus stock, which was heat-inactivated at 75 °C for 30 min, was used as a negative control. Both positive and negative controls generated broad fluorescence spectra with a peak wavelength of 523 nm, which closely corresponds to the peak wavelength of the FAM fluorophore used as the reporter (517 nm). The fluorescence emission observed from the negative control is due to free (not attached to a DDN) unquenched FAM molecules in the solution. However, the positive control showed an approximately 13% increase in light intensity (Fig. S5) with respect to the negative control, due to the interactions between the DNA Net and the intact virions that result in unquenching of the FAM fluorophores on the DNA Net sensors. The background signal resulting from the scattering of the excitation light source and the auto-fluorescence from the PCR tube accounted for approximately 26% of the background signal intensity (Fig. S5).
To characterize the V-Pod sensitivity as a fluorimeter, we prepared calibration standards comprised of FAM fluorophore (the same fluorophore utilized in the DDNs) spiked into a buffer at known concentrations and compared dose-response plots measured with the V-Pod with two commercially available laboratory-based fluorimeters. FAM was serially diluted with deionized (DI) water and 100 μl was transferred into PCR tubes for fluorescent measurement with the V-Pod. For comparison, identical samples were measured using a Qubit Flex (Q33327, Invitrogen, US, $5990) and an ANDalyze fluorimeter (AND1000, ANDalyze, IL, US, $4166). Results are summarized in Fig. 4c–f. Fig. 4c shows the dose-response and a non-linear regression fitting of the data collected using V-Pod, while Fig. 4d and e show that of Qubit Flex and ANDalyze, respectively. The V-Pod was able to detect FAM at concentrations as low as 100 nM, while Qubit Flex and ANDalyze provided empirical detection limits of 50 nM and 250 nM, respectively. The R-square value for both V-Pod and Qubit Flex was 0.99, while ANDalyze had R2 = 0.98, showing an excellent fit to a linear function. The half-maximal effective concentrations (EC50) were 1.528, 0.805, and 0.382 μM for V-Pod, Qubit Flex, and ANDalyze, respectively (Fig. 4f). ANDalyze was dynamic range limited as the fluorescent readings quickly reached a plateau near 5 μM FAM concentration and the hill slope value was 1.58. The V-Pod and Qubit Flex had hill slope values of 1.049 and 0.812, respectively. Qubit Flex exhibited the greatest dynamic range among the devices, while V-Pod showed a similar dynamic range and hill slope as Qubit Flex. Despite being small and inexpensive, the V-Pod showed comparable performance for FAM detection and quantification when compared with commercial fluorometers. A detailed comparison between the instruments is summarized in Table S2.
To monitor for temperature fluctuations in the test sample due to illumination and heating from the instrument itself, a K-type thermocouple probe (TL0260 K-Type Thermocouple, 0.13 mm diameter, Perfect Prime, NY, USA) was inserted and completely submerged in the liquid. The thermocouple was inserted through a hole drilled into a PCR tube cover, which was sealed with adhesive (Super Glue Gel Control, Loctite, Ohio, USA) to prevent evaporation. The submerged thermocouple measured the fluid temperature at 1-s intervals using a thermocouple thermometer (TC0520, Thermocouple Thermometer, Perfect Prime, NY, USA). The temperature characterization results are shown in Fig. 4g. During testing, the V-Pod warmed the fluid to ∼25 °C while the Qubit Flex warmed the solution to ∼30 °C. The heat created from the enclosed electrical components of both the V-Pod and the Qubit Flex contributes to the temperature elevation. However, it was interesting to note that the Qubit Flex always maintained a device temperature of ∼30 °C even in varying ambient temperatures ranging from −4 °C to 25 °C (data not shown). In contrast, the ANDalyze instrument did not show any temperature elevation (fluid temperature remained at a room temperature of 23 °C) being that the tube housing was exposed to the environment, separated from the circuitry.
Lastly, the instruments were characterized to calculate their limits of quantitation. Fig. 4h shows the percentage change in the relative fluorescent signal from various FAM concentrations (250 nM, 200 nM, 150 nM, and 125 nM) collected using each instrument relative to the baseline concentration of 100 nM FAM. As shown in the graph, the V-Pod differentiated the FAM concentration change as low as 50 nM (ANOVA; ****p < 0.0001, **p = 0.0021, ns p = 0.2803), while the Qubit Flex differentiated the FAM concentration of <25 nM (ANOVA; ****p < 0.0001). ANDalyze differentiated the FAM concentration changes of as low as 25 nM but the percentage changes in the signal between 50 nM and 25 nM FAM concentration were not statistically significant (ANOVA; ****p < 0.0001, Δ50nM ***p = 0.0008, Δ25nM **p = 0.002, ns = 0.9961).
2.5. Design, synthesis, and characterization of DNA Net sensors
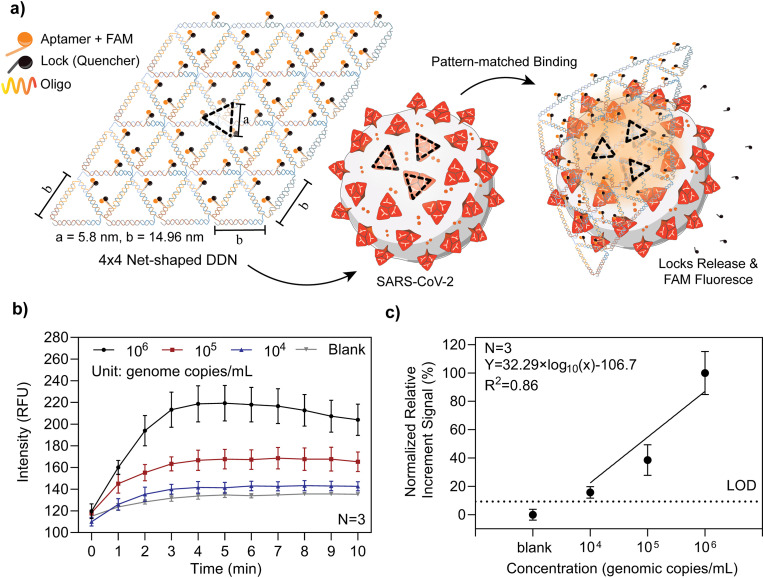
In our previous work, we reported the design, synthesis, and characterization of DDNs that carry multiple trimeric RBD-binding aptamers for high-affinity and selectivity capture of intact SARS-CoV-2 virions (Chauhan et al., 2022) (Fig. 5 a). The spacing between aptamer-RBD binding sites in a trimeric spike cluster is ∼6 nm. The minimal spacing of two adjacent trimeric spike clusters can be ∼15 nm, packaged in the shape of a rhombus for a maximal spike density on the SARS-CoV-2 outer membrane (Chauhan et al., 2022). Based on this structural information, a 4 × 4 net-shaped DDN with repeating units was designed and synthesized to match the spatial pattern and spacing of trimeric spike clusters displayed on the viral surface. Aptamers selected via SELEX targeting of the WT SARS-CoV-2 spike protein's RBD reside on each vertex of the DNA Net to form an array of trimeric clusters for precise matching of the spike protein, resulting in high avidity as proved by SPR binding assays (Chauhan et al., 2022). Each aptamer is FAM-labeled and quenched by a BHQ-1 tagged “lock” DNA that forms a duplex when hybridized with part of the aptamer. When mixed with a sample containing SARS-CoV-2 virions, the DNA Net sensor, with mechanical flexibility, allows it to deform around the convex spherical virus to promote the viral-Net interactions. Binding of aptamers with the spike-RBD will disrupt the aptamer-lock hybridization, resulting in a spontaneous release of the fluorescence quencher, thus switching “on” the FAM reporter to generate a detection signal. We previously demonstrated the limit of detection (LoD) of 103 viral genome copies/mL of WT SARS-CoV-2 in an artificial saliva-containing solution, and a high degree of selectivity against non-target viruses including H1N1, OC43, or Zika using an ANDalyze fluorimeter (Chauhan et al., 2022). Fig. 5a illustrates the binding of the DNA Net sensors to a virion and their subsequent release of the locks for fluorescence emission. By providing a sufficiently high Net sensor concentration in the sample volume, all virions rapidly encounter and bind with DDNs, thus providing viral-load dependent emission intensity within a 5–10 min reaction time. In fact, kinetic monitoring of fluorescence generation shows that the maximal signal is obtained in 5 min. Thus, the detection is performed in a single-step, single reagent, room temperature “mix and read” procedure. Self-assembly of the DDNs was characterized by 1% agarose gel electrophoresis (AGE) (Fig. S7) and further confirmed through atomic force microscopy (AFM) imaging (Chauhan et al., 2022).
Fig. 5.
DDN design and sensing of pseudo-typed SARS-CoV-2 WT virus. a) Schematic of the 4 × 4 DNA Net sensor and illustration of a SARS-CoV-2 virus. The DNA Net with repeating units is designed to match the spatial pattern and spacing of trimeric spike clusters displayed on the SARS-CoV-2 viral surface. Three aptamer-lock pairs are placed at each vertex of the DNA Net sensor. The spike-specific binding aptamer was tagged with a FAM fluorophore, along with a quencher labeled lock DNA that forms a partial duplex with the aptamer. During the virion-Net interactions, the high-affinity binding of the aptamers releases the locks, generating fluorescent signals. b) Raw signal of the 4 × 4 DNA Net sensor with pseudo-typed WT virus generated and collected using V-Pod. c) The normalized detection sensitivity of the 4 × 4 DNA Net sensor with pseudo-typed WT virus. The linear fitting equation is y = 32.29 × log10 (x) - 106.7, where x is the concentration of pseudo-typed SARS-CoV-2 WT in viral genome copies/mL and y is the normalized relative increment signal collected by V-pod. The dashed horizontal line indicates the threshold (LoD = blank signal + 3 standard derivations), Based on the threshold and fitting equation, the limit of detection was determined to be 3.92 × 103 viral genome copies/mL. In b ∼ c), the error bars represent the standard deviation of three independent assays.
It was determined in our previous work that at lower viral loads, the DNA Net sensors demonstrate enhanced performance in the presence of artificial saliva, compared to buffer solutions. This is largely because the SARS-CoV-2 virus is known to be more compatible with saliva medium (Chauhan et al., 2022), thus all the sensing assays were performed in commercially available artificial saliva.
2.6. Detection of pseudo-typed SARS-CoV-2 and pathogenic real SARS-CoV-2 variants using V-Pod
In preparation for assays, 20 μL of 0.1 μM DDN reagent was added to a 200 μL PCR tube and allowed to reach room temperature for at least 10 min. To conduct a test, 80 μL of the viral analyte solution diluted in buffer containing 4.71% vol/vol of artificial saliva was brought to room temperature before being added to the tube. Next, the cap was closed, and the tube was gently shaken for 10 s by hand. The PCR tube was immediately inserted into the V-Pod tube holder, and the hinged door was closed. By pressing the “start test” button on the app, the V-Pod collected data points every minute (2 s “on” time for the laser, for each data point) for 10 min. Photodiode intensity values were uploaded to the cloud-based database for storage and analysis.
The methods for the production and validation of both pseudo-typed SARS-CoV-2 and authentic viruses can be found in the supplementary information within the methods section. Briefly, a multi-plasmid system was used to produce pseudo-typed SARS-CoV-2 in 293T (human kidney) cells, while authentic SARS-CoV-2 was produced by the propagation of authentic viral strains within Calu-3 (human lung) cells. The produced pseudoviral particles display SARS-CoV-2 spike protein arranged on an HIV envelope in a conformation closely mimicking that of authentic SARS-CoV-2. Due to their method of manufacture, these viral particles are replication-incompetent and have a greatly improved lab safety profile as compared to authentic SARS-CoV-2. For each test, a SARS-CoV-2 virus aliquot was thawed on ice. The pseudo-typed SARS-CoV-2 virus dilutions were prepared from their fully concentrated stock solution in 1X TA buffer, pH = 8, while the real pathogenic viruses were diluted in 1X PBS buffer, pH = 7.4. All tests with the pseudo-typed SARS-CoV-2 WT were conducted in a bio-safely level 2 facility at UIUC, and real SARS-CoV-2 Delta, Omicron, and D614G were conducted in a bio-safely level 3 facility at LSU. The V-Pod battery was fully recharged each day prior to performing a set of experiments.
2.7. Data collection and processing
In each test, when the PCR tube is inserted into the instrument, the measurement starts by collecting the initial value at t = 0 followed by reading at 1-min intervals for a duration of 10 min. As the aptamers on the 4 × 4 DNA Net bind to viruses, the release of quenchers results in an increase in the fluorescence signal. To provide a fluorescence intensity baseline and minimize background noise sources such as autofluorescence and non-perfect quenching, a negative control sample (all conditions the same, but no virus present) was measured prior to introducing the test sample. The relative increment signal was obtained by subtracting the t = 0 initial value from the maximum intensity value within the 10-min measurement period. The same procedures are followed to obtain the relative increment signal for both negative and positive control samples, followed by normalizing to the negative control. The resulting graphs are shown in Fig. 5, Fig. 6.
Fig. 6.
DDN sensing of real pathogenic viral particles using V-Pod. a-c) The normalized detection sensitivity of 4 × 4 DNA Net sensor with real pathogenic virus D614G, Delta, and Omicron in PBS buffer. The error bars represent the standard deviation of three independent assays. The linear fitting equation is 16.32 × log10(x) - 45.04 for variant D614G, 18.03 × log10(x) - 82.93 for variant Delta, and 19.31 × log10(x) - 84.27 for Omicron, where x is the concentration of pseudo-typed SARS-CoV-2 WT in viral genome copies/mL and y is the normalized relative increment signal collected by V-Pod. The dashed horizontal line indicates the threshold (LoD = blank signal + 3 standard derivations). Based on the threshold and fitting equation, the limits of detection for D614G, Delta, and Omicron variants were determined to be 1.84 × 104, 9.69 × 104, and 6.99 × 104 viral genome copies/mL. d) The combined normalized dose-response curves of the three real pathogenic viruses. Each measurement is repeated three times.
3. Results and discussion
3.1. Detection of pseudo-typed SARS-CoV-2 WT using V-Pod
We first tested the V-Pod by dose-response characterization of pseudo-typed SARS-CoV-2 WT detection using the DDNs, using spiked-in virus concentrations ranging from 104 to 106 viral genome copies/mL, with results shown in Fig. 5b and c. Kinetic monitoring of the fluorescence during the 10-min measurement time shows that the maximum signal is obtained in 5 min, and additional monitoring does not yield greater fluorescence intensity, as the available virus binding sites become occupied by DDNs. The error bars represent the standard deviation from N = 3 independent tests in separate tubes. Plotting the final fluorescence intensity obtained at t = 10 min demonstrates that the fluorescence intensity scales with concentration from 104 to 106 viral genome copies/mL with an R2 value for linear regression fitting of 0.86. The LoD was calculated by extrapolating the threshold value, which was calculated by summing up the measured negative control signal (where all experimental conditions remained identical, but no viral particles were present) plus three times the standard deviation, resulting in an LoD of 3.92 × 103 viral genome copies/mL.
3.2. Detections of pathogenic real SARS-CoV-2 variants
We next sought to characterize the assay by the detection of recently emerging SARS-CoV-2 variants. Due to the fact that some variants attain increased infectivity through mutations that modify the spike protein (Volz et al., 2021a; Siedner et al., 2022; Boucau et al., 2022), it follows that the described DDNs will be less sensitive to these emerging strains (as these DDNs were designed with WT SARS-CoV-2 in mind). Fig. 6 a–c shows the dose-response characterization and non-linear fitting of the D614G, Delta, and Omicron variants. All variants were diluted to the desired concentration in 1X PBS with 4.7% vol/vol of artificial saliva. Kinetic monitoring of the fluorescence during the 10-min measurement time shows that the maximum signal is obtained at 10 min. Variant D614G and Omicron were measured with a V-Pod containing a 10 mW laser. After completing tests for D614G and Omicron, our initial V-Pod developed a hardware failure, which led us to switch to a backup V-Pod that was fitted with a laser adjusted to 15 mW output power for testing of the Delta variant samples. We observe that the DDN assay can detect all three variants but with a reduced increase in fluorescence intensity compared to the detection of the WT pseudo-typed strain. However, we observe dose-dependent signals for all variants from 105 to 109 viral genome copies/mL. Linear regression fitting of the dose-response curve and determination of LoD by considering a signal “detectable” when its magnitude is greater than 3x the standard deviation of blank controls yields an LoD of 1.84 × 104 viral genome copies/mL for D614G, 9.69 × 104 viral genome copies/mL for Delta, and 6.99 × 104 viral genome copies/mL for Omicron. It should be noted that the presence of residual phenol red from the manufacture of pseudo-typed and authentic SARS-CoV-2 likely increased the background signal for these measurements, though this source of error would not be present in samples of patient saliva.
The results imply that although different SARS-CoV-2 variants may contain multiple mutations that result in subtle changes in the spike protein, the aptamer can still bind with them effectively. Considering that the LoD specified for the FDA-approved RT-PCR test is between 102 and 105 viral genome copies/mL, the sensitivity of our DDN assay measured with the V-Pod meets the requirements for highly sensitive testing. A plot of the normalized dose-response curves of the three variants, shown in Fig. 6d, demonstrates that the V-Pod and DDN assays detect all three variants but with lower sensitivity compared to the Pseudo-typed WT virus, consistent with previous results obtained with a laboratory fluorimeter for detection of pseudo-typed D614G and B.1.1.7 viruses (Chauhan et al., 2022). The D614G variant alters spike trimer hydrogen-bond interactions, reorienting the RBD into an open “up” perfusion conformation (Mansbach et al., 2021) while displaying similar spike protein numbers, spike-to-nucleocapsid ratios, and spike cleavage on individual virion projections when compared to SARS-CoV-2 WT (Hou et al., 2020). These modifications can explain why the detection of D614G loses some sensitivity but still has the best LoD compared to the other two variants. The Delta spike proteins are present predominantly in the open conformation, with higher fusion activity and syncytium formation in contrast to WT (Mlcochova et al., 2021). The Omicron variant contains twice as many mutations (up to a total of 32) compared to the Delta variant (Tian et al., 2022). The spike proteins observed in both the Delta and Omicron variant present with a predominantly “open” conformation (Ye et al., 2022), whereas 60% of the trimers in WT SARS-CoV-2 typically maintain a “closed” perfusion (Ke et al., 2020). Combined, these structural alterations to the SARS-CoV-2 spike protein relative to the WT virus likely lead to the observed reduction in DDN detection sensitivity. We envision the selection of optimized aptamers for variants as they emerge, in which the same DDN net structure can be modified to incorporate new aptamer designs.
3.3. Discussion
Table S3 summarizes Emergency Use Authorized (EUA) COVID-19 diagnostic tests that are currently available for purchase for POC settings. Among 281 molecular SARS-CoV-2 tests and 51 antigen diagnostic tests, only 22 have been authorized for “at-home use” with or without prescriptions. 18 of these tests are based on the lateral flow immunoassay, among which 14 are approved for non-prescription use, one requires a prescription, and the remaining 3 are intended for use by medical professionals or highly trained laboratory personnel. Among the 14 non-prescription use products, only one product incorporates an automatic test result reporting capability, while 13 products recommend voluntary reporting through either an app, QR code, or through a healthcare provider. Besides the lateral flow immunoassay-based tests, three molecular tests, such as RT-LAMP (reverse transcription loop-mediated isothermal amplification), are currently authorized for non-prescription “at-home use”. Among them, two incorporate an automatic reporting system, while one recommends voluntary reporting of the test results through a healthcare provider. The overall average cost per test is $21.64 with a range of $6 to $75. According to a study conducted by Du et al., the target price for SARS-CoV-2 testing should be less than $5 to control rapid viral transmission, assuming a societal willingness to pay (Du et al., 2021). Thus, current SARS-CoV-2 test costs remain too high to achieve effective containment. In this regard, the single-step DDN assay (∼$1.26 reagent cost) measured with a V-Pod could provide a new paradigm for epidemic monitoring and control of a highly infectious respiratory virus. By allowing personalized detection with an inexpensive and portable fluorimeter, frequent testing without the involvement of specialized personnel and laboratory equipment can be achieved with a reduced financial burden in both developing and developed countries. In addition, more accurate and real-time identification of infected individuals is feasible through the implementation of an automatic reporting system and contract tracing in a smartphone app. However, successful pandemic control will require a high degree of civilian compliance, a prevalence of testing devices and assay supply, and strict implementation of epidemiological policies. Moreover, concerns about information security and protection must be addressed. Currently, many efforts are being made to develop strategies for the secure storage of medical data and personal information through the utilization of blockchain technology and artificial intelligence (Ng et al., 2021; Nguyen et al., 2021; Abd-alrazaq et al., 2021). However, prospective solutions remain in the research and development stage, emphasizing the need for timely translation of the technology into clinical and real-life use.
According to a study conducted by Larremore et al., test sensitivity is considered of secondary importance after testing frequency and sample-to-answer time for highly infectious diseases such as COVID-19 (Larremore et al., 2021). However, based on Figs. S6a–b, the average testing time of currently available EUA “at-home” use tests is 17 min, with a range of 10–60 min. It is important to note that the 10-min sample-to-answer time is achieved only with lateral flow assays, which not only fall short in sensitivity but also do not indicate active infectious status. These limitations are overcome by the combination of a DDN assay and the V-Pod instrument. The 5-min sample-to-answer time combined with low testing costs meets the criteria that the modeling studies have identified.
Having the ability to differentiate between different variants of SARS-CoV-2 is considered important in the containment of infectious virus ((CDC) and C.f.D.C.a.P., 2022). This is because different variants of the virus may have different transmission rates, the severity of the disease, or susceptibility to existing treatments and vaccines (Volz et al., 2021b; Lauring and Hodcroft, 2021). However, the current V-Pod system cannot distinguish between the variants due to the overlapping similarities of the spike protein formation of the SARS-CoV-2 variants (Callaway, 2021; Greaney et al., 2021). In this regard, the proposed V-Pod system can further be improved by incorporating aptamers that are highly specific to each variant, which can be generated through introducing the negative selection during the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) process (Kumar Kulabhusan et al., 2020). Moreover, fluorophores with different emission wavelengths can be applied in the assembly of the variant-specific DNA Nets and prepared in a single pot to achieve multiplexing capability, which would also require the addition of multiple photodiodes and optical filters.
4. Conclusion
We have designed, fabricated, and characterized an inexpensive, palm-sized, smartphone-linked fluorimeter (called a “V-Pod”) for the purpose of quantitatively measuring the fluorescent output of a rapid viral load assay for SARS-CoV-2 in saliva. The assay utilizes nucleic acid nanostructures that are precisely engineered to form a net-like structure that places the vertices of the net at the positions of the virus outer spike proteins. Unlike conventional nucleic acid tests or antigen tests, the DDN assay detects intact virions with a high degree of specificity by placing spike protein-recognizing aptamers at the binding locations, where binding interactions between the aptamers and spikes result in the unquenching of many fluorophores for each DDN-virion interaction. Importantly, the DDN assay operates at room temperature, requires only 5 min to generate a signal, and does not require any materials besides the DDN reagent in a conventional PCR tube, to which the test sample is introduced to initiate a test. While previous research demonstrated the effectiveness of the DDN SARS-CoV-2 assay when measured with a laboratory-based fluorimeter, in this report we demonstrate nearly equivalent limits of detection and limits of quantitation using the V-Pod instrument. Furthermore, we demonstrate the detection of three recent SARS-CoV-2 variants. While the sensitivity of the assay for the detection of variants is reduced due to lower affinity between the spike-recognizing aptamer and the mutated spike proteins, the demonstrated limits of detection are still within a clinically relevant range.
Overall, the low costs associated with the items used to conduct the test (DDN reagent and PCR tube) and to measure the test output (V-Pod instrument) are compatible with the goals of frequent SARS-CoV-2 self-testing, while the rapid sample-to-answer time of 5 min from saliva is compatible with the goal of convenience that is necessary for widespread acceptance. Importantly, the instrument links wirelessly and securely to conventional mobile devices to manage the test procedure and to provide a convenient means for sharing data with health authorities, health services, and anonymous contact tracing.
In this work, we demonstrate the feasibility of using a portable and inexpensive fluorimeter for quantitative measurement of fluorescent intensity changes that occur in the DDN assay, where many quenchers are released from associated fluorescent reporters when the nanostructure binds to intact virions in solution. Our results show that a background fluorescent signal is obtained due to incomplete quenching of the DDN reagent, but that the fluorescence increase is dependent upon the dose of the specifically targeted virus. As we consider utilization of the detection capability in real-world point-of-care environments, it is possible to envision the test result to simply provide a binary positive/negative output to the user, in which a predetermined threshold fluorescence increase is utilized as a cut-point that can satisfy a specific sensitivity and selectivity requirement. Should quantitative estimates of viral concentration be desired, it is likely that some form of calibration should be performed either periodically or immediately prior to the detection of test samples with unknown virus concentration. Calibration standards may be most appropriate for the usage of a V-Pod when used in clinical settings, rather than for home-based testing.
We envision the V-Pod instrument as a simple consumer-level device that would ultimately be capable of measuring similar point-of-care diagnostic tests that generate a fluorescent output. For example, DDN-assays may be designed for other highly infectious human viruses transmitted through blood (HIV, HBV) or insect vectors (Dengue, Zika). The DDN detection strategy may also be extended to porcine, poultry, and bovine viruses. Many other diagnostic tests also generate a fluorescent output in liquid media, such as those utilizing molecular beacons, DNAzymes, and others for the detection of analyte classes that include miRNA, drugs, and ions. In future versions of the V-Pod, we envision including the capability to integrate experimental controls for simultaneous measurements of the test sample and a negative control. Additionally, we envision making multiple improvements, including the optimization of optical design, in order to enhance signal-to-noise performance for the detection of weak fluorescence signals. However, an important design criterion for the V-Pod instrument reported here is simplicity, for the sake of ruggedness, miniaturization, and minimization of cost.
CRediT authorship contribution statement
Hankeun Lee: Writing – review & editing, Investigation, Conceptualization, Methodology, Software, Validation, Formal analysis, Visualization. Weijing Wang: Writing – review & editing, Investigation, Methodology, Validation, Formal analysis, Visualization. Neha Chauhan: Resources, Investigation, Methodology, Validation, Formal analysis. Yanyu Xiong: Resources, Investigation, Formal analysis. Nicholas Magazine: Resources, Investigation. Owen Valdescruz: Software. Dong Yeun Kim: Visualization. Tianjie Qiu: Software. Weishan Huang: Resources. Xing Wang: Funding acquisition, Project administration, Supervision, Resources, Writing – review & editing, Conceptualization. Brian T. Cunningham: Funding acquisition, Project administration, Supervision, Resources, Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors are grateful for the support from the NIH Rapid Acceleration of Diagnostics (RADx) program (AA029348), NSF Rapid Response Research (RAPID) program (CBET 20–27778), and Center for pathogen Diagnostics (CPD) DREMES program. H.L. acknowledges the support from the Army Gi Bill. The authors thank Dr. Zhang Tianyi from the Louisiana State University for the valuable help in troubleshooting the pseudo-virus production protocol and producing the initial pseudo-typed WT SARS-CoV-2 stock.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2023.115228.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abd-alrazaq A.A., et al. Blockchain technologies to mitigate COVID-19 challenges: a scoping review. Computer Methods and Programs in Biomedicine Update. 2021;1 doi: 10.1016/j.cmpbup.2020.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersen S., Chamings A., Bhatta T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020;11(1):6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank W. World Bank: © World Bank; Washington, DC: 2021. Global Economic Prospects, January 20221. [Google Scholar]
- Boucau J., et al. medRxiv; 2022. Duration of Viable Virus Shedding in SARS-CoV-2 Omicron Variant Infection. [Google Scholar]
- Byakodi M., et al. Emerging 0D, 1D, 2D, and 3D nanostructures for efficient point-of-care biosensing. Biosens. Bioelectron. X. 2022;12 doi: 10.1016/j.biosx.2022.100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A.W., et al. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open. 2020;10(8):e039856. doi: 10.1136/bmjopen-2020-039856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595(7865):17–18. doi: 10.1038/d41586-021-01696-3. [DOI] [PubMed] [Google Scholar]
- (CDC), C.f.D.C.a.P. 2022. SARS-CoV-2 Variant Classifications and Definitions. [Google Scholar]
- Chaudhary V., et al. Towards hospital-on-chip supported by 2D MXenes-based 5th generation intelligent biosensors. Biosens. Bioelectron. 2023;220 doi: 10.1016/j.bios.2022.114847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N., et al. Net-shaped DNA nanostructures designed for rapid/sensitive detection and potential inhibition of the SARS-CoV-2 virus. J. Am. Chem. Soc. 2022 doi: 10.1021/jacs.2c04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherusseri J., et al. SARS-CoV-2-on-Chip for long COVID management. Biosensors. 2022;12(10):890. doi: 10.3390/bios12100890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., et al. Comparative cost-effectiveness of SARS-CoV-2 testing strategies in the USA: a modelling study. Lancet Public Health. 2021;6(3):e184–e191. doi: 10.1016/S2468-2667(21)00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (FDA) T.F.a.D.A. BINAXNOW COVID-19 AG CARD (PN 195-000) INSTRUCTIONS FOR USE. https://www.fda.gov/media/141570/download [cited 2022 07/17]; Available from:
- (FDA) T.F.a.D.A. LuciraTM COVID-19 all-in-one test kit instructions for use. [cited 2022 07/17] https://www.fda.gov/media/143808/download Available from:
- (FDA) T.F.a.D.A. The CueTM COVID-19 test for home and over the counter (OTC) use instructions for use [cited 2022 07/17] https://www.fda.gov/media/146470/download Available from:
- Ganguli A., et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117(37):22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57. doi: 10.1016/j.chom.2020.11.007. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankelow A.M., et al. Smartphone clip-on instrument and microfluidic processor for rapid sample-to-answer detection of Zika virus in whole blood using spatial RT-LAMP. Analyst. 2022;147(17):3838–3853. doi: 10.1039/d2an00438k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankelow A.M., et al. Smartphone clip-on instrument and microfluidic processor for rapid sample-to-answer detection of Zika virus in whole blood using spatial RT-LAMP. Analyst. 2022 doi: 10.1039/d2an00438k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588(7838):498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B.D., et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021;20(5):593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Kulabhusan P., Hussain B., Yüce M. Current perspectives on aptamers as diagnostic tools and therapeutic agents. Pharmaceutics. 2020;12(7) doi: 10.3390/pharmaceutics12070646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon P.S., et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 2020;12(1):26–35. doi: 10.1038/s41557-019-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore D.B., et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021;7(1) doi: 10.1126/sciadv.abd5393. eabd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325(6):529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., et al. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. Label-free digital detection of intact virions by enhanced scattering microscopy. J. Am. Chem. Soc. 2022;144(4):1498–1502. doi: 10.1021/jacs.1c09579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhoul M., et al. Analyzing inherent biases in SARS-CoV-2 PCR and serological epidemiologic metrics. BMC Infect. Dis. 2022;22(1):458. doi: 10.1186/s12879-022-07425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallah S.I., et al. COVID-19: breaking down a global health crisis. Ann. Clin. Microbiol. Antimicrob. 2021;20(1):35. doi: 10.1186/s12941-021-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansbach R.A., et al. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci. Adv. 2021;7(16) doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz E.A., et al. Transmission of SARS-CoV-2: a review of viral, host, and environmental factors. Ann. Intern. Med. 2021;174(1):69–79. doi: 10.7326/M20-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahussurur M., Yamaoka Y. Diagnostic methods of Helicobacter pylori infection for epidemiological studies: critical importance of indirect test validation. BioMed Res. Int. 2016 doi: 10.1155/2016/4819423. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W.Y., et al. Blockchain applications in health care for COVID-19 and beyond: a systematic review. The Lancet Digital Health. 2021;3(12):e819–e829. doi: 10.1016/S2589-7500(21)00210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.C., et al. Blockchain and AI-based solutions to combat coronavirus (COVID-19)-Like epidemics: a survey. IEEE Access. 2021;9:95730–95753. doi: 10.1109/ACCESS.2021.3093633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization, W.H WHO coronavirus (COVID-19) dashboard. 2022 7/15/22 07/17/22] https://covid19.who.int/ Available from:
- Pecoraro V., et al. Estimate false-negative RT-PCR rates for SARS-CoV-2. A systematic review and meta-analysis. Eur. J. Clin. Invest. 2022;52(2) doi: 10.1111/eci.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P., et al. Asymptomatic SARS-CoV-2 infection: a systematic review and meta-analysis. Proc. Natl. Acad. Sci. USA. 2021;118(34) doi: 10.1073/pnas.2109229118. e2109229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedner M.J., et al. Duration of viral shedding and culture positivity with postvaccination SARS-CoV-2 delta variant infections. JCI Insight. 2022;7(2) doi: 10.1172/jci.insight.155483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.L., et al. medRxiv; 2021. Longitudinal Assessment of Diagnostic Test Performance over the Course of Acute SARS-CoV-2 Infection. 2021.03.19.21253964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J., Ward G. Interferences in immunoassay. Clin. Biochem. Rev. 2004;25(2):105–120. [PMC free article] [PubMed] [Google Scholar]
- Thye A.Y.-K., et al. Emerging SARS-CoV-2 variants of concern (VOCs): an impending global crisis. Biomedicines. 2021;9(10):1303. doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., et al. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022;94(6):2376–2383. doi: 10.1002/jmv.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcek S. SARS-CoV-2: zoonotic origin of pandemic coronavirus. Acta Virol. 2020;64(3):281–287. doi: 10.4149/av_2020_302. [DOI] [PubMed] [Google Scholar]
- Volz E., et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., et al. medRxiv; 2021. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from Linking Epidemiological and Genetic Data. 2020.12.30.20249034. [Google Scholar]
- Walsh K.A., et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020;81(3):357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (WHO), W.H.O Antigen-detection in the diagnosis of SARS-COV-2 infecton. 2021 10/06/2021 [cited 2022 07/17] https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays Available from:
- Ye G., Liu B., Li F. Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat. Commun. 2022;13(1):1214. doi: 10.1038/s41467-022-28882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H., et al. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 2021;143(4):1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.