Abstract
Reverse transcriptases (RTs) are typically assayed using optimized Mg2+ concentrations (~5–10 mM) several-fold higher than physiological cellular free Mg2+ (~0.5 mM). Recent analyses demonstrated that HIV-1, but not Moloney murine leukaemia (MuLV) or avain myeloblastosis (AMV) virus RTs has higher fidelity in low Mg2+. In the current report, lacZα-based α-complementation assays were used to measure the fidelity of several RTs including HIV-1 (subtype B and A/E), several drug-resistant HIV-1 derivatives, HIV-2, and prototype foamy virus (PFV), all which showed higher fidelity using physiological Mg2+, while MuLV and AMV RTs demonstrated equivalent fidelity in low and high Mg2+. In 0.5 mM Mg2+, all RTs demonstrated approximately equal fidelity, except for PFV which showed higher fidelity. A Next Generation Sequencing (NGS) approach that used barcoding to determine mutation profiles was used to examine the types of mutations made by HIV-1 RT (type B) in low (0.5 mM) and high (6 mM) Mg2+ on a lacZα template. Unlike α-complementation assays which are dependent on LacZα activity, the NGS assay scores mutations at all positions and of every type. Consistent with α-complementation assays, a ~four-fold increase in mutations was observed in high Mg2+. These findings help explain why HIV-1 RT displays lower fidelity in vitro (with high Mg2+ concentrations) than other RTs (e.g. MuLV and AMV), yet cellular fidelity for these viruses is comparable. Establishing in vitro conditions that accurately represent RT’s activity in cells is pivotal to determining the contribution of RT and other factors to the mutation profile observed with HIV-1.
Keywords: reverse transcriptase, fidelity, mutation rate, magnesium, retrovirus
Introduction
Retroviral reverse transcriptases (RT) possess RNA- and DNA-dependent DNA polymerase activity and RNase H activity [1]. Magnesium (Mg2+), the most abundant divalent cation in the cell, functions as the physiological co-factor for both activities. Each of the two physically separated active sites is proposed to contain two divalent cation binding sites [2–8].
The enzymatic activities of purified HIV-1 RT have typically been investigated in vitro using ~5–10 mM Mg2+, 25–100 µM dNTPs, and about 80 mM KCl or NaCl. These values approximate the optimal activity levels determined on homopolymeric templates for HIV-1 RT polymerase and RNase H activities [9–12]. However, they are not representative of physiological levels of these components. This is often the case for general enzymatic analysis in vitro which is typically performed under optimized conditions with only required components, in part because it is not possible to completely mimic complex physiological conditions in vitro. Although the use of non-physiological levels of some components may not significancy effect enzyme properties, the concentration of Mg2+ can have profound effects on HIV-1 RT DNA synthesis, drug interaction, and fidelity. While the total Mg2+ concentration in cells is high (typically 10 mM or more [13–15]), most is sequestered by nucleotides and other complex anions, and ‘free’ Mg2+ in lymphocytes and several other cell types has typically been found to be ~0.5 mM [13, 14, 16–21]. RNA-directed ssDNA synthesis reactions performed with HIV-1 RT with low Mg2+ lead to more efficient ssDNA synthesis, despite modestly decreased overall activity [22]. In in vitro reactions, many common nucleoside reverse transcriptase inhibitors (NRTIs) are less effective in lower Mg2+ while non-nucleoside reverse transcriptase inhibitors (NNRTIs) are more effective [23]. Finally, the fidelity of HIV-1 RT is higher, at lower more physiological Mg2+ concentrations [24, 25], while the fidelities of MuLV and AMV RTs are insensitive to Mg2+ [24]. These findings offer a possible explanation for why HIV-1 RT displays lower fidelity in vitro (with high Mg2+ concentrations) than other RTs (e.g. Moloney murine leukaemia virus (MuLV) and avian myeloblastosis virus (AMV)), yet cellular fidelity for these viruses is comparable (for reviews see [26–28]). For example, MuLV RT shows ~five-fold greater fidelity than HIV-1 RT on both RNA and DNA templates across several publications from different groups [26, 28], and even in identical experiments in the same publication [24, 29]. Yet cellular replication data indicates that these viruses have similar mutation rates [27, 28]. Although it has been suggested that these discrepancies may result in part from the exceptionally high mutation frequency of HIV-1 subtype NY5 RT used in some in vitro assay [30], or a presumed high mutation frequency for phage RNA polymerases [31], cellular replication data using the same lacZα template used in many in vitro experiments demonstrates that this issue remains unsolved with the authors noting: “This rate (referring to results in the publication) is about three-fold lower than previously reported for HIV-1 in vivo and is much lower than what has been reported for purified HIV-1 RT in vitro” [32]. The mutation profile observed in cellular assay with HIV-1 is also inconsistent with in vitro results [28, 32, 33]. The lack of correlation with respect to the overall mutation rate and spectrum of mutations observed in vitro vs. cellular replication has made it impossible to determine RT’s contribution to the mutation spectrum observed in cells. This in turn makes it difficult to assess the contributions of mutagenic cellular factors (e.g. APOBEC3G [34–36], UNG [37–40], ADAR [32], and RNA polymerase II [41]) to the observed spectrum. A thorough understanding of the factors that contribute to the genetic diversity of HIV-1 will require a better understanding of HIV-1 RT’s role in the process.
In this report we examined the fidelity of HIV-1 RT (subtype B) wild-type (wt) and several common drug-resistant mutants in high (6 mM) and low (0.5 mM) Mg2+ using a frequently employed lacZα-based α-complementation assay, to see if resistance mutations altered the effect of Mg2+ on RT fidelity. Other RTs including HIV-1 subtype A/E, HIV-2, prototype foamy virus (PFV), MuLV, and AMV were also tested. While confirming previous results with HIV-1, MuLV, and AMV RTs (see above), these experiments demonstrate that MuLV and AMV RTs are outliers as all other tested RTs show greater fidelity in low Mg2+. In addition, a Single Strand Consensus Sequencing (SSCS) Next Generation Sequencing (NGS) analysis was also performed with a lacZα template using wt HIV-1 RT with high and low Mg2+. Unlike α-complementation which can only detect mutations that decrease LacZα activity, the NGS results include all nucleotide sites on the lacZα gene. The NGS analysis produced the most comprehensive mutation profile (~250000 mutations were recovered) to date for a polymerase over the lacZα gene. The results are compared to other RT fidelity assays conducted in vitro and in cells.
Methods
Materials
Deoxyribonucleotide triphosphates (dNTPs) were from Roche. Q5 DNA polymerase, T4 polynucleotide kinase (PNK) calf intestinal phosphatase (CIP), restriction enzymes NlaI, High Fidelity PvuII, EcoRI, and EcoRV, NEBNext Ultra II DNA Library Prep Kit for Illumina (Cat# E7645S), and NEBNext Multiplex Oligos for Illumina (Cat# E7335S) kit were from New England BioLabs. RNase-DNase free was from MilliporeSigma. SYBR Safe DNA gel stain was from Invitrogen. Radiolabeled ATP (γ-32P) was from Perkin-Elmer. G-25 spin columns were from Harvard Apparatus. The RNA purification kit was from GeneJET. All DNA oligonucleotides were from Integrated DNA Technologies (IDT). Wild-type HIV-1 reverse transcriptase (RT) (from HXB2 strain) was prepared as described [42]. Aliquots of HIV-1 RT were stored frozen at −80 °C and fresh aliquots were used for each experiment. Prototype foamy virus reverse transcriptase was kindly provided by Dr Eddy Arnold (Rutgers University). All other enzymes were obtained as clones and kindly provided by Dr Stefan Sarafianos (Emory University), or Dr Stephen Hughes (National Institutes of Health). They were purified and stored using methods similar to wt RT. All other chemicals were from VWR, ThermoFisher Scientific, or MilliporeSigma.
End-labeling of oligonucleotides with T4 PNK
DNA oligonucleotides were 5′ end-labelled in a 50 µl volume containing 50–100 pmol of the oligonucleotide of interest, 1×T4 PNK reaction buffer (provided by manufacturer), 10 U of T4 PNK and 10 µl of (γ-32P) ATP (3000 Ci mmol−1, 10 µCi µl−1). The labelling reaction was done at 37 °C for 30 min according to manufacturer’s protocol. PNK enzyme was heat inactivated by incubating the reaction at 75 °C for 15 min. Excess radiolabeled nucleotides were then removed by centrifugation using a Sephadex G-25 column.
Production of the RNA template for lacZα complementation assay
Plasmid pBS▽EcoRV567 (constructed as described in Supplemental Data) was cleaved with NdeI and T3 RNA polymerase was used to make run-off transcripts ~644 nucleotides in length using the manufacturer’s protocol. RNA was isolated using an RNA purification kit and quantified by UV absorption. The integrity and size of the RNA was confirmed by denaturing gel electrophoresis.
Two round lacZα complementation assay (RNA templated system)
This assay measures two rounds of DNA synthesis by RT. Conditions for extension with RT were: 50 mM Tris-HCl pH 8 (final pH in reactions at 37 °C was ~7.7), 80 mM KCl, 1 mM DTT, 20 µM dNTPs, 0.5 or 6 mM MgCl2 (final free concentration in reactions after correcting for chelation by dNTPs [23]), 37 °C for 45 min. Round one used the RNA template described above to make the first strand of DNA. Four identical reactions were carried out for each condition using 25 nM template and 50 nM primer (round one primer below 5'-32P-labelled at low specific activity). Primer and template were hybridized in reaction buffer without dNTPs and MgCl2 by heating to 65 °C for 5 min then slow cooling to room temperature in a heat block. Reactions were initiated by addition of RT (final concentration 100 nM). After incubations, one microgram of RNase A was then added to each reaction and incubated for 5 min then heated to 65 °C for 2 min. After addition of 2× loading buffer (90% formamide, 10 mM EDTA (pH 8), 0.025% bromophenol blue and xylene cyanol) the samples were run on a 6% denaturing polyacrylamide gel [43]. Fully extended DNA (289 nucleotides) was located with a phosphorimager and excised then isolated by the crush and soak method [43]. The DNA was then used as a template to make a second strand of DNA (252 nucleotides) using the buffer conditions described above and the round two primer shown below. The round two primer was 5ˈ-32P end-labelled at ~ten-fold greater specific activity than the round one primer to make it easier to locate and separate from the 289-nucleotide template. Two pmoles of round two primer was prehybridized (10 µl of 50 mM Tris-HCl pH=8, 80 mM KCl, 1 mM DTT, 80 °C, 3 min, then decrease temperature by 2 °C per minute to 37 °C in a PCR machine) to the recovered DNA from the four round one reactions and round two reactions were carried out as described above. The second round DNA product was separated from the round one template using an 8% denaturing polyacrylamide gel and recovered as described above. The recovered DNA was quantified based on radioactivity and PCR amplified using high fidelity Q5 DNA polymerase. The PCR products were processed by cleavage with EcoRI and EcoRV and purified on native gels. Plasmid pBS▽EcoRV567 was cleaved with the same restriction enzymes and dephosphorylated with PNK. The purified PCR-derived inserts were then ligated into the cleaved plasmid and used to transform competent E. coli capable of α-complementation. Standard blue-white screening was used to score mutations. Details for these steps have been published [24]. The main differences are that plasmid pBS▽EcoRV567 was used here instead of pBS∆PVUII (see Supplemental Data), the insert to vector ratio was 3:1, and restriction enzymes were ‘High Fidelity’ versions of EcoRI and EcoRV while EcoRI and PvuII were used in the previous system. The different restriction enzymes and plasmid sequences required the use of different 5′-32P end-labelled primers to prime the two rounds of RT DNA synthesis: round one: 5′-GATTTAGAGCTTGACGGGGA-3′, producing a full-length extension product of 289 nucleotides; round two: 5′-AGGATCCCCGGGTACCGAGC-3′, producing a full-length extension product of 252 nucleotides. The difference in product lengths was used to separate the products using denaturing gel electrophoresis. These same primers were used to produce the PCR products described above. Bacterial colonies were scored by blue-white screening as previously described [24]. In some cases, the results (Table S1, available in the online version of this article) were from a single experiment while in others they are summed values from more than one independent experiment. Statistical analysis to assess significance was conduct using a Chi-squared tests.
Production of single stranded DNA for single round lacZα complementation assay and single strand consensus sequencing (SSCS) Next Generation Sequencing (NGS)
Single stranded DNA was produced using Q5 DNA polymerase and the manufacture’s recommended protocol. First, a 306 base pair dsDNA product was made by PCR using the primer from round one above and a second primer: 5′- AATTAACCCTCACTAAAGGG. Plasmid pBS▽EcoRV567 (0.1 µg) was used as template. Reactions were in 50 µl with one unit of Q5 DNA polymerase. Cycles were 94 °C, 5 min, then 15 cycles of 94 °C, 55 °C, and 72 °C for 30 s each, followed by 1 cycle of 72 °C for 5 min. Cycling was minimized in order to decrease PCR-derived mutations. Products were run on an 8% non-denaturing polyacrylamide gel [43] using SYBR Safe DNA gel stain which allowed visualization of the products using blue LED light. Excised products were recovered using the crush and soak method [43] and quantified by UV absorbance. One pmole of dsDNA product was used in an asymmetric PCR reaction with 50 pmoles of 5'-32P end-labelled (low specific activity) round one primer above. Conditions were as above but 30 cycles were used. The reactions (typically three reactions were combined) were phenol extracted and precipitated with ethanol using standard procedures. The recovered material was digested with EcoRI (20 unit) for 20 min at 37 °C, then run on a 6 % denaturing polyacrylamide gel along with a size marker to locate the correctly sized DNA (using a phosphorimager) which was excised and recovered as described above. The digestion with EcoRI was used to cleave any double stranded DNA so as not to contaminate the desired 306 nucleotide single stranded product with complementary DNA that could interfere with downstream applications. The ssDNA product was quantified by specific activity.
Single round lacZα-complementation assay (DNA templated system)
The assay was carried out using the round two conditions for the ‘two round lacZα complementation assay’ above. It essentially mimicked the second round of DNA synthesis except that the starting template was the single stranded DNA produced by asymmetric PCR as described above (only a single 25 µl reaction was typically performed for each condition). This DNA was primed with the 5′-32P end-labelled round two primer and the 252 nucleotide fully extended ssDNA product was isolated and processed as described above.
Production of RT DNA products for NGS
Assay conditions for producing the DNA products for NGS were the same as those described above except that the 5′-32P end-labelled primer 5′-AAAAGGTAGTGCTGAATTCGATCACGNNNNNNNNNNNNNNGTGAGTCGTATTACAATTCA-3′ (14 N barcode primer) was used to primer the second synthesis round in the two round assay and the only synthesis round in the single round assay. This primer contains a 14-nucleotide random barcode region that is used for tracking individual NGS products for fidelity analysis (see below and [25]). This corresponds to 414≈2.7×108 different potential barcodes for RT synthesis products. The 3′ terminal nucleotide of the primer is one base upstream of lacZα position ‘1’ in pNLZeoIN-R-E-.LZF/R, allowing a direct comparison of these results with results in cell culture assays using that plasmid [32].
Production of ssDNA control for NGS
The same control was used for the single and two round lacZα assays to set the background mutation rate in the NGS analysis. One pmole of the ssDNA made by asymmetric PCR (see above) was used as a template for a single round of Q5 DNA polymerase extension using 2 pmoles of 14 N NGS index primer from above. One cycle of PCR (95, 50, and 72 °C, for 1 min) with one unit of Q5 DNA polymerase was performed in 50 µl of Q5 supplied buffer and the correct length extension product was isolated from a 6% denaturing polyacrylamide gel as described above. Note that for the two-round assay, this control does not include any mutations generated during the production of the RNA template that was used at the start of the assay (see above). Therefore, background errors from the RNA template are not subtracted away in the assay results.
Production of PCR products and addition of adapters for NGS
For NGS, the final DNA products from the single or two round lacZα complementation assays described under ‘Production of RT DNA products for NGS’ were used to produce PCR products with the following primers: forward, 5′-GACGGGGAAAGCCGATATCTTATTA-3′ and reverse, 5′-32P end-labelled 5′-AAAAGGTAGTGCTGAATTCG-3′. The 3′ nucleotide of the forward primer is one base downstream of nucleotide 162 in pNLZeoIN-R-E-.LZF/R. Therefore, the PCR products produced contain nucleotides 1–162 of the lacZα gene from pNLZeoIN-R-E-.LZF/R and, in our system, these nucleotides were derived from one or two rounds of synthesis with RT. Conditions for PCR reactions with Q5 DNA polymerase were 94 °C, 5 min, then 94 °C, 55 °C, and 72 °C for 30 s each, followed by 1 cycle of 72 °C for 2 min. Reactions were conducted for 20, 22 or 24 cycles with aliquots being removed after the 72 °C for 2 min extension step. Fifty pmoles of each primer were used in a 50 µl final volume and duplicate PCR reactions were performed for each condition in order to get a higher yield of products. For all conditions, 1.5×106 (calculated from specific activity of the radiolabeled primer) molecules of template DNA from RT reactions was used in each 50 µl PCR reaction. PCR products (244 nucleotides) were separated on an 8% nondenaturing polyacrylamide gel. PCR products of the correct size were typically excised from either the 20, 22, or 24 cycle lanes, depending on which yielded the most material. Recovered products were processed using the ‘NEBNext Ultra II DNA Library Prep Kit for Illumina (Cat# E7645S)’ while adapters used were from the “NEBNext Multiplex Oligos for Illumina” (Cat# E7335S) kit. Six different indices were mixed for MiSeq analysis, three each from the DNA templated or RNA templated assays described above. These included controls (described under ‘Production of controls for NGS’, two separate identical controls made at different times were used for DNA and RNA templated assays), and products derived from 0.5 or 6 mM MgCl2 assays from both the DNA and RNA assays.
Analysis of NGS results
The basic approach used for analysis of mutations is described in [25] and is broadly termed Single Strand Consensus Sequencing (SSCS). Briefly, MiSeq analysis produced ~1.6×107 individual reads from the six different conditions (see above) in the analysis. Reads that differed by more than five nucleotides from the predicted length were excluded as the methodologies used to isolate DNA and PCR products (i.e. excision from gels based on size) should have eliminated products that were significantly longer or shorter than ‘full-length’ and these products were therefore assumed to be derived from PCR errors or sequencing errors. Consequently, this approach does not account for long insertion or deletion mutations. However, insertions or deletions of a few nucleotides were scored. After applying various filters to exclude truncated or hypermutated sequences (individual reads with more than ten errors were excluded), recovered reads were grouped based on the 14 random nucleotide barcode sequence (see ‘Production of RT DNA products for NGS’ above). Reads with the same barcode were assumed to be derived from a single RT extension event. Only barcodes recovered five or more times [25] were evaluated and are referred to as a ‘tag family’ in the data. Using three rather than five as the lower limit for recovered sequences with identical tags did not significantly affect the analysis. On average, there were ~230000 tag families recovered for each condition. The sequence region analysed was 155 nucleotides in length (see Fig. 1) which yielded an average number of nucleotides evaluated for a specific condition for the RNA templated system (two rounds of RT synthesis) of ~7×107 (230000×155×2), and ~3.5×107 (230000×155) for the DNA templated system (one round of RT synthesis). In order for a particular tag family to register a mutation, the same error had to occur at the same nucleotide position in all of the recovered reads. As expected, based on the error rate of HIV-1 RT, most evaluated tag families had no registered mutations. The MiSeq analysis was conducted twice using independent experiments. Similar results were obtained in the two separate experiments (see Results). To evaluate mutations in the sequence data, we wrote a versatile programme that allowed analysis of several parameters including potential errors derived from the MiSeq sequencing process and RT-derived error. This programme, along with all the data from these experiments is available from three repositories: (1) output logs and some information regarding the process is at: https://github.com/abelew/error_rate_quantification; (2) software which handles the reading of the raw data and creation of the initial tables of mutants per sample is at: https://github.com/abelew/errrt; and (3) postprocessing software at: https://githubcom/abelew/Rerrrt/.
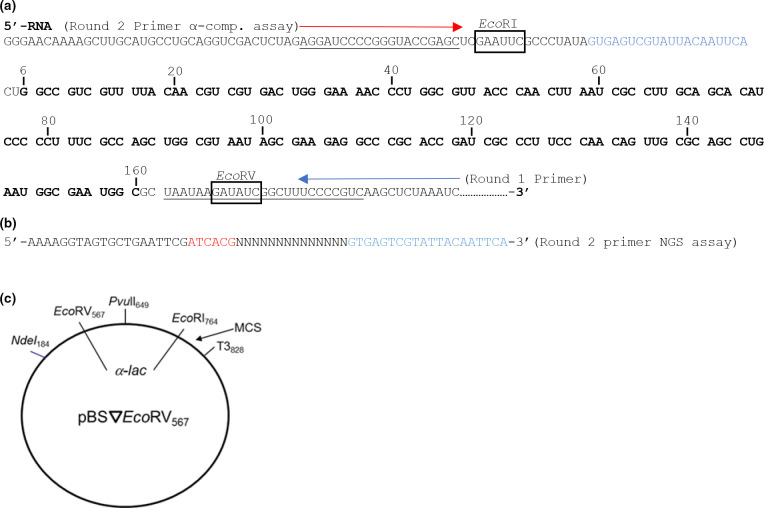
Fig. 1.
Constructs used to examine reverse transcriptase fidelity. (a) The relevant region of the RNA template made from T3 RNA polymerase is shown. A similar region was used for the DNA templated assay (see Methods). Numbering is only shown in the area of the lacZα gene and corresponds to the number used by Abram et al., 2010 [32]. Codons for producing the LacZα peptide are in three base sets. The region of the sequence scored for mutations in the NGS assay are bolded. The α-complementation assay scored the region between the EcoRI and EcoRV sites. Sites of round one and two primers for the two rounds of RT synthesis in the RNA templated assays are depicted by arrows. The DNA template assay, which had only a single round of RT synthesis, used only the round two primer. (b) The sequence of the indexing primer for the NGS assay is shown. This primer was used as the round two primer in the RNA templated NGS assay and the round one primer (only round) in the DNA templated NGS assay. The primer contains a 14-nucleotide random region (N) that was used for indexing (see Methods) and a 6-nucleotide barcode (in red) used to identify the particular condition. The region in blue corresponds to the region shown in the same color in panel a. (c) Plasmid construct used in assays. Plasmid pBS▽EcoRV567 was constructed as described (see Supplementary Data). Numbering is based on the parent plasmid, pBSM13+.
Results
System used to test the fidelity of RTs
The plasmid-based system used for in vitro fidelity analysis was similar to a previous system used in this lab [24]. Construction of the new plasmid (pBS▽EcoRV567) for this system and the sources of mutation in the system are described in the Supplemental Data. The region of the plasmid coding for the lacZα gene is shown in Fig. 1 along with the basic design of the system. This new system evaluates fidelity over a longer region of the lacZα gene that matches up better with others used in the literature [32, 44]. The restriction sites used to insert RT-derived DNA include an EcoRI site in the plasmid’s MCS and an inserted EcoRV site. Unlike the original system, both sites are outside the lacZα gene region, minimizing the potential for white or faint blue colonies resulting from ligation artefacts, although deletion or insertion errors at the EcoRI site could still lead to frameshifts in lacZα. This system also truncates the lacZα gene relative to the previous by adding double stop codons before the EcoRV site. Background error rates for the system were assessed by cleaving pBS▽EcoRV567 with EcoRV and EcoRI, isolating the insert, and reinserting it into plasmids prepared for fidelity analysis, then transforming bacteria in α-complementation assays (see Methods). This approach produced ~one white or faint blue colony per 1000 total colonies (plasmid preparations that produced significantly higher proportions of white and faint blue colonies were discarded). Note that this number did not change significantly compared to inserting DNA produced with Q5 DNA polymerase which was the control for the fidelity assays (Table S1, 16/18371 and 10/11078, for experiments started with RNA and DNA templates, respectively). This is likely due to the very high fidelity of Q5 DNA polymerase [45]. For a detailed explanation of the sources of mutations in the RNA and DNA templated versions of this assay see ‘Sources of mutations in the α-complementation assays’ in the Supplemental Data.
With the exception of MuLV and AMV RTs, all RTs showed significantly higher mutation levels with 6 vs. 0.5 mM Mg2+ in α-complementation assays
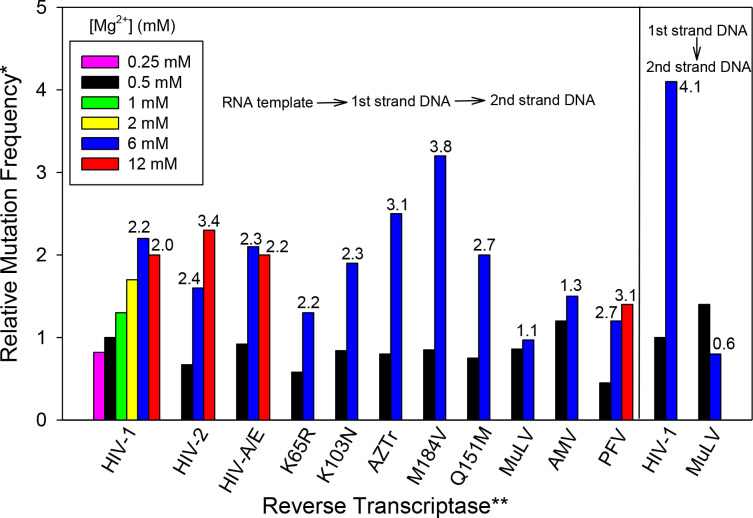
Detailed results for analyses with several RTs are shown in Table S1, and the relative mutation frequencies compared to the mutation frequency for wt HIV-1 RT at 0.5 mM Mg2+ (represented by coloured bars), or to the values for the same RT at 0.5 mM Mg2+ (number above the bars for 6 and 12 mM Mg2+) are shown in Fig. 2. Note that for the RNA templated assay, these values represent the minimum fold differences between the conditions as they do not corrected for mutations present in the starting RNA template (see Supplemental Data: ‘Sources of mutations in the α-complementation assays’). The actual fold differences would be greater as these ‘background’ mutations would have a greater relative effect on mutations frequency values at 0.5 mM Mg2+ since they are considerably smaller than those at 6 mM. For wt HIV-1 RT with the RNA templated system, the analysis was performed at several Mg2+ concentrations, while other RTs were with 0.5 and 6, or 0.5, 6, and 12 mM Mg2+ only (all free Mg2+ concentrations). In the RNA templated system, mutation frequencies were typically 2–4-fold higher with 6 vs. 0.5 mM Mg2+ and increasing to 12 mM did not result in a significant increase above 6 mM except for HIV-2 RT which showed a 2.4-fold higher mutation rate at 6 mM Mg2+ and 3.4-fold at 12 mM. Using P<0.05 as a cutoff, all HIV enzymes, including HIV-2 RT, showed significantly greater mutation frequencies at 6 vs. 0.5 mM Mg2+ (Table S1, second column from right). In general, mutation frequencies of drug resistant mutants did not differ significantly from wt HIV-1 RT under that same Mg2+ condition (Table S1, last column on right). Two notable exceptions were K65R and M184V. At 6 mM Mg2+, the former was significantly lower while the latter was significantly higher than wt. The K65R results are consistent with previous results demonstrating that this mutation results in higher fidelity [46]. Like HIV-1 RT, PFV RT was also less accurate in higher Mg2+, however, this enzyme showed significantly higher fidelity than HIV-1 RT in both 0.5 and 6 mM Mg2+. We note that unlike our observations, studies from others did not find that PFV RT was more accurate than HIV-1 RT [47]. Finally, MuLV and AMV RTs behaved differently than other RTs as neither showed a statistically significant difference in mutation frequencies in 0.5 vs. 6 mM Mg2+, and only MuLV RT at 6 mM Mg2+ was significantly different than HIV-1 RT wt under that condition, demonstrating greater fidelity.
Fig. 2.
Relative fidelity of various reverse transcriptases (RT) in different Mg2+ concentrations. The α-complementation assay (see Methods) was used to estimate the fidelity of the indicated RT with different concentrations of free Mg2+ (as indicated). The value for HIV-1 wild-type (HXB2 clone) at 0.5 mM Mg2+, after background subtraction, was set to “1” (see below) and higher values indicate a higher mutation frequency. Assays conducted with the RNA template, which go through two rounds of RT DNA synthesis, are shown on the left while those using the single round DNA templated assay are on the right. Data used to construct this graph is shown in Table S1. *Relative mutation frequency was based on the colony mutation frequency and was calculated from the proportion of bacterial colonies that were white or faint blue in the α-complementation assay divided by the total number of colonies (i.e.: (white colonies + faint blue colonies) /(white colonies + faint blue colonies + blue colonies)). The background value (see Methods) was subtracted from each condition to get the final value for comparisons. All values represented by colored bars are relative to the value for HIV-1 RT at 0.5 mM Mg2+ which was set to 1. **Numbers above 6 and 12 mM Mg2+ conditions are the fold difference between that value and the value for the same reverse transcriptase at 0.5 mM Mg2+. All fold changes represent the minimum fold change and actual fold changes are likely higher (see Results). ***Reverse transcriptases with amino acid changes are drug-resistant forms of HIV-1 RT; AZTr: D67N/K70R/T215F/K219Q.
HIV-1 and MuLV RT were also evaluated in the DNA templated assay. In this case, the mutation frequency difference was greater for HIV-1 RT than in the RNA templated assay, with 6 mM Mg2+ producing an ~four-fold greater mutation frequency than 0.5 mM, while this difference was about two-fold in the RNA assay. In contrast, and in agreement with the RNA templated system, MuLV RT values were essentially the same at both 0.5 and 6 mM Mg2+, as this enzyme demonstrated significantly higher fidelity than HIV-1 RT only at 6 mM Mg2+. The greater difference for HIV-1 RT at 0.5 vs. 6 mM Mg2+ in the DNA templated assay could result from differences in the fidelity of RT on DNA vs. RNA, however, they could also result from the starting RNA template having a higher mutation frequency than the starting DNA template, which would under-represent the fold differences in the RNA system (see Discussion). Further, the background control better accounts for errors in the DNA vs. RNA templated assay (see above). Therefore, the fold differences in that assay more accurately represent the actual differences.
In general, these results are in agreement with previous results from this lab [24] and others [25] demonstrating that the fidelity of HIV-1 RT is greater in vitro when physiological free Mg2+ (i.e.~0.5 mM, see Introduction) conditions are used. The results also support the notion that HIV-1 RT demonstrates lower fidelity than MuLV and AMV RTs in vitro only when non-physiological high Mg2+ conditions are used [24].
Analysis of mutations made by HIV RT in 0.5 and 6 mM Mg2+ using NGS
A SSCS NGS approach was used to determine the mutation profile of HIV-1 RT wt using 0.5 or 6 mM Mg2+. This approach allowed the analysis of several thousand mutations from material analysed in both the RNA and DNA templated assays described above. While we used an established approach that allows each RT synthesis event to be tracked using a random barcode [25], a new programme to mine the data was generated (see Methods).
Analysis of insertion/deletion (indel) mutations by NGS
The region analysed for mutations in the NGS assay is shown in Fig. 1. Nucleotides 6–160 of the sequence (155 total nucleotides) were evaluated for mutations. Six conditions (BKG control, and 0.5 or 6 mM Mg2+ for both the RNA and DNA templated system (Table 1)) were tested (see Methods for a description of how mutations were scored). This report emphasizes data from one experiment (Exp. 1), while a second set from an independent experiment (Exp. 2) yielded similar results (Table S2). The vast majority of mutations recovered by NGS were substitutions as opposed to indels (Table 1). We should note, however, that our protocol would likely eliminate large insertions and deletions as the material for analysis is excised from gels (see Methods). For this reason, only products that differed from the expected length by five nucleotides or less were evaluated in the NGS analysis, so even modestly large indels would have been excluded. As expected, indels occurred preferentially within runs of the same nucleotide on both the RNA and DNA templated samples and the positions mostly matched hotspots from previous mutational analysis with lacZα in cells [32]. Fig. S1a shows the frequency of all substitution mutations for the RNA templated assay (Fig. S1b shows results for the DNA templated assay) with 0.5 and 6 mM Mg2+ in Exp. 1 (excluding G>T and C>A mutations (see below)). Positions and mutations rates for the strongest (those in which at least one of the two conditions had a mutation frequency ≥1×10−6, these comprised great than 90% of all recovered insertions and about 60% of deletions) indel mutations are also shown (numbers on graphs placed above the specific base position with deletions underlined and insertions not underlined). Indel hotspots were almost exclusively in runs of the nucleotides, as expected. Most recovered insertions with both 0.5 (~91 %) and 6 mM Mg2+ (~79%) occurred at positions 35–38 (insertion of A, 5′-GGGAAAACCC-3′, 35–38 underlined) or 81–83 (insertion of T, 5′-ACCCCCCTTTCGCC-3′). Note that the exact position of the insertions within the run cannot be determined and the numbers in Fig. S1a are placed above the first base of the run. Deletions were more distributed with the two strongest sites being nucleotides 14–17 (deletion of T, 5′-CGTTTTAC-3′) and 75–80 (deletion of C, 5'−5′-ACCCCCCTTTCGCC-3′).
Table 1.
Data from NGS Exp. 1
|
Condition* |
Control DNA |
DNA 0.5 mM Mg2+ |
DNA 6 mM Mg2+ |
Control RNA |
RNA 0.5 mM Mg2+ |
RNA 6 mM Mg2+ |
|---|---|---|---|---|---|---|
|
Total tag families† |
258 515 |
177 278 |
212 279 |
210 220 |
255 341 |
254 006 |
|
Total nucleotides‡ |
4.007×107 |
2.748×107 |
3.290×107 |
6.517×107 |
7.916×107 |
7.874×107 |
|
Substitution Type |
Number of recovered substitutions (substitution frequency per nucleotide x 106)§ |
|||||
|
A>C |
287 (7.163) |
581 (21.14) |
1227 (37.29) |
231 (3.545) |
1216 (15.36) |
2238 (28.42) |
|
A>G |
332 (8.286) |
1500 (54.59) |
8835 (268.5) |
207 (3.176) |
4156 (52.50) |
10 817 (137.4) |
|
A>T |
134 (3.344) |
323 (11.75) |
1037 (31.52) |
41 (0.6291) |
1108 (14.00) |
2049 (26.02) |
|
*C>A |
3459 (86.32) |
3310 (120.5) |
6051 (183.9) |
3597 (55.19) |
7043 (88.97) |
11 804 (149.9) |
|
C>G |
231 (5.765) |
1133 (41.23) |
8441 (256.6) |
226 (3.468) |
2040 (25.77) |
4427 (56.22) |
|
C>T |
749 (18.69) |
1788 (65.07) |
12 460 (378.7) |
540 (8.286) |
4796 (60.59) |
22 619 (287.3) |
|
G>A |
744 (18.57) |
5376 (195.6) |
8126 (247.0) |
371 (5.693) |
4231 (53.45) |
23 202 (294.7) |
|
G>C |
153 (3.818) |
253 (9.207) |
378 (11.49) |
54 (0.8286) |
386 (4.876) |
775 (9.843) |
|
*G>T |
4773 (119.1) |
4212 (153.3) |
8069 (245.3) |
3245 (49.79) |
3274 (41.36) |
4261 (54.11) |
|
T>A |
74 (1.847) |
280 (10.19) |
684 (20.79) |
50 (0.7672) |
1002 (12.66) |
1484 (18.85) |
|
T>C |
321 (8.011) |
478 (17.39) |
1193 (36.26) |
181 (2.777) |
1760 (22.23) |
13 537 (171.9) |
|
T>G |
465 (11.60) |
623 (22.67) |
1564 (47.54) |
417 (6.399) |
1236 (15.61) |
3181 (40.40) |
|
Insertion Type |
Number of recovered insertions (insertion frequency per nucleotide x 106)d |
|||||
|
A |
0 |
16 (0.5822) |
47 (1.429) |
0 |
447 (5.647) |
500 (6.350) |
|
C |
0 |
6 (0.2183) |
6 (0.1824) |
0 |
213 (2.691) |
333 (4.229) |
|
G |
0 |
0 |
10 (0.3040) |
0 |
31 (0.3916) |
23 (0.2921) |
|
T |
0 |
22 (0.8006) |
29 (0.8815) |
0 |
2692 (34.01) |
2605 (33.08) |
|
Deletion Type |
Number of recovered deletions (deletion frequency per nucleotide x 106)§ |
|||||
|
A |
75 (1.872) |
86 (3.130) |
437 (13.28) |
27 (0.4143) |
273 (3.449) |
461 (5.855) |
|
C |
37 (0.9234) |
49 (1.783) |
789 (23.98) |
23 (0.3529) |
745 (9.411) |
1082 (13.74) |
|
G |
28 (0.6988) |
10 (0.3639) |
272 (8.267) |
23 (0.3529) |
201 (2.539) |
400 (5.080) |
|
T |
74 (1.847) |
112 (4.076) |
485 (14.74) |
54 (0.8286) |
540 (6.822) |
1044 (13.26) |
|
Mutation Type |
Percent of various types of mutations for each condition (mutation frequency x 104)|| |
|||||
|
Substitutions¶ |
98.4 (3.618) |
95.6 (12.49) |
79.3 (2.415) |
92.8 (10.35) |
||
|
Insertions |
0.4 (0.0147) |
0.2 (0.0261) |
14 (0.4263) |
3.9 (0.4349) |
||
|
Deletions |
1.2 (0.0440) |
4.2 (0.5485) |
6.7 (0.2040) |
3.2 (0.3568) |
||
|
Overall mutation frequency |
3.677×10−4 |
1.306×10−3 |
3.045×10−4 |
1.115×10−3 |
||
*Refer to Methods for details. DNA assays included one round of RT DNA-template-directed DNA synthesis while RNA assays include an RT RNA-template-directed round and a second RT DNA-template-directed DNA synthesis round.
†Tag families are defined in Methods. To qualify as a ‘tag family’, the same 14-nucleotide barcode had to be recovered in five or more reads in NGS. Specific mutations were recorded when they appeared at a specific position in all the reads that constituted a particular tag family.
‡Total number of RT-directed nucleotides catalysis events evaluated in the assay. For DNA templated assay, this was the number of tag families x 155 nucleotide positions (see Fig. 1). For RNA templated assays, the tag family number was multiplied by 310 because this assay had an additional round of RT-directed synthesis (see Materials and Methods).
§Insertion frequency per nucleotide was calculated by dividing the raw number of mutations of the specified type by the total number of nucleotides that were assessed for that condition. Unlike the raw number of mutations, this value allows a direct relative comparison of the data in the Table with higher numbers representing a greater frequency for that mutation. Note that the DNA and RNA templated assays cannot be directly compare with this number as the templates was synthesized by Q5 DNA polymerase or T3 RNA polymerase in the DNA and RNA assays, respectively. The Q5 synthesis product was used as the Control for both assays. Given the very low mutation rate of Q5, it is likely the RNA assays contain a significant proportion of mutations derived from T3 RNA polymerase that are not accounted for by the Control (see Results for further discussion).
||Values were determined after subtracting background Control values from each mutation type using the frequency per nucleotide data above. These values were then summed for each of the three categories (i.e. substitutions (S), insertions (I), and deletions (D)). The percent values listed were calculated by dividing individual values by the sum of the three categories and multiplying by 100 (e.g. for Substitutions: (S/(S+I+D)) x 100). Mutation frequencies in parentheses were the individual S, I, and D values and the Overall Mutation Frequency was S+I+D for each category.
¶Due to high numbers in the Controls, C>A and G>T substitutions were excluded from data calculations (see Results).
For the RNA templated assay, the mutation frequency for indels did not change much between 0.5 and 6 mM Mg2+ while indels made up a greater percentage of total mutation in low Mg2+ due to the large increase in the substitution mutation rate in high Mg2+ (Table 1). Notable differences in the DNA templated assay were a much lower indel mutation frequency with low Mg2+, and a significant increase in deletions in high Mg2+. In Exp. 2 (Table S2), both insertions and deletions increased significantly at higher Mg2+ with DNA. Overall, in Exp. 1, indels comprised ~21% of all mutations at 0.5 mM Mg2+ in the RNA templated assay, but only 1.6 % in the DNA assay under that condition. A similar trend was observed in Exp. 2 (Table S2). These results are consistent with the T3 RNA polymerase-derived RNA template containing a significantly higher level of indels than the Q5-derived DNA template [31] (see Discussion).
Analysis of substitution mutations by NGS
Unlike indels, the substitution frequency increased ~four-fold in both RNA and DNA templated samples at 6 vs. 0.5 mM Mg2+ (Table 1). Background error frequencies, although relatively low for most mutation types, were much higher for C>A and G>T mutations. High C>A and G>T mutation rates are common in NGS analysis and are exacerbated by heating or other steps that damage or oxidize DNA [48–52]. Duplex-based NGS approaches as opposed to the SSCS approach used here, can more effectively correct for these errors, but these approaches are costly and can dramatically decrease coverage and yields [53, 54]. Because of the high background, G>T and C>A mutations were not included in calculations for mutation rates or proportions of the various mutation types shown in Tables 1 and S2, or Figs 3 and 4, and Figs S1–S3. This is unlikely to significantly affect the mutation rate as G>T and C>A, like most other transversions, are low frequency mutations for RT and DNA polymerases in general.
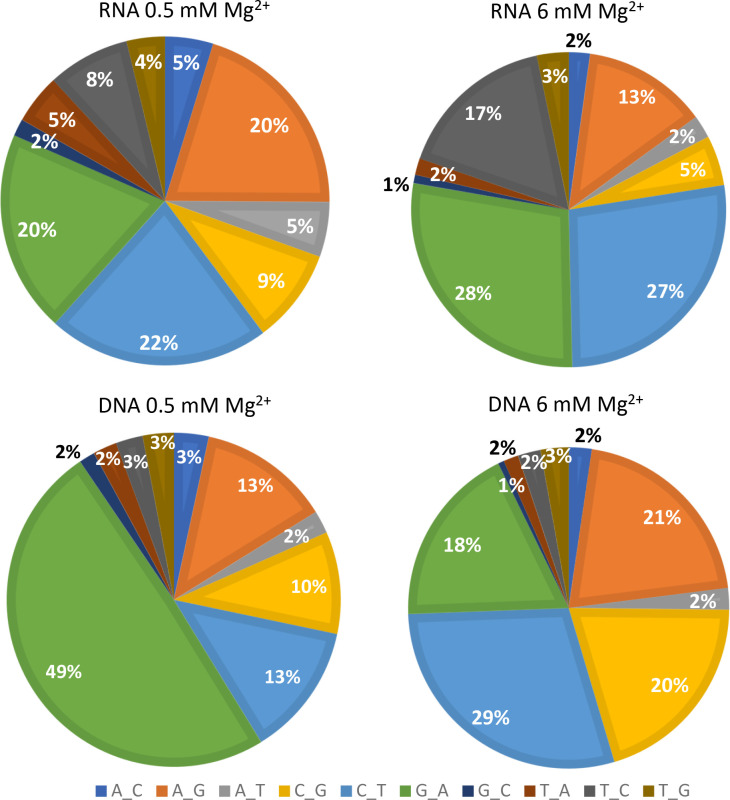
Fig. 3.
Proportion of various substitution mutations in the NGS assay (Exp. 1). The proportions of substitution mutations recovered using the indicated conditions in the NGS assay are shown.G>T and C>A mutations are excluded from the analysis (see Results). See Table 1 for more information on the mutation rates for the various mutations.
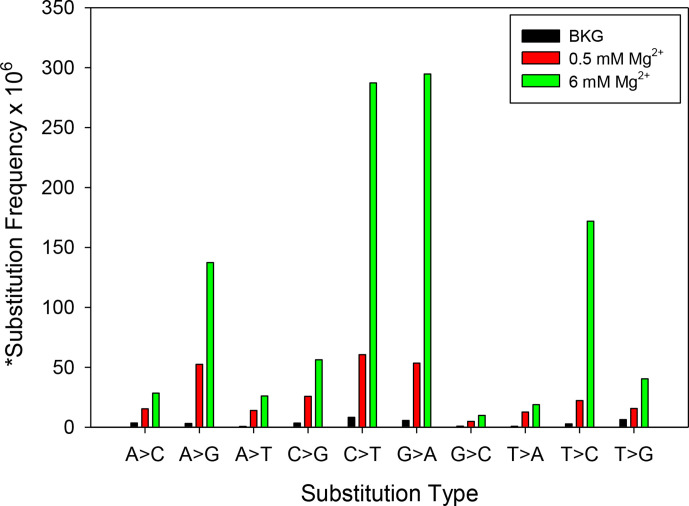
Fig. 4.
Mutation frequencies of different types of substitution mutations in the RNA templated assay at 0.5 or 6 mM Mg2+ using NGS analysis (Exp. 1). A plot of background, 0.5 mM and 6 mM Mg2+ results are shown for the RNA templated NGS assay. The substitution frequency numbers were taken directly from Table 1 and exclude C>A and G>T mutations (see Results). *This parameter is defined in Table 1 and represents the substitution frequency per nucleotide for the particular type of substitution indicated.
As expected, transitions were more common than transversions for all conditions (Tables 1 and S2, Figs 3 and S2 for Exp. 1 and Exp. 2, respectively). It is important to note that for the RNA templated assays, a particular mutation could have arisen during RNA or DNA-directed RT synthesis. For example, a G>A mutation could have resulted from a G coding for a T during RNA-directed DNA synthesis (the T would subsequently code for an A in the DNA-directed second round), or a C coding for an A during DNA-directed DNA synthesis (see Fig. 1). In contrast, RT-derived G>A mutations in the DNA templated assay arise from a G coding for a T during DNA-directed RT synthesis as this is the only RT round in that assay.
For the RNA templated assay, the relative proportion of various mutations was similar between the 0.5 and 6 mM Mg2+ conditions (Figs 3 and S2 for analysis of Exp. 2). There was a modest increase in the relative proportion of G>A, C>T and T>C mutations with 6 mM Mg2+. As these transitions are among the most common mutation types for RT and many polymerases, it is possible that high Mg2+ has a greater effect on mutations that are easier to make.
For the DNA templated assay, the profile of mutation types showed a greater difference at 0.5 vs. 6 mM Mg2+ compared to the RNA templated assays (Fig. 3). The major difference between the results was the higher proportion of G>A mutations with 0.5 compared to 6 mM Mg2+. Also, there was a modest increase in A>G, C>T, and C>G mutations at 6 mM Mg2+, along with the corresponding decrease in the proportion of G>A mutations.
A comparison between mutation proportions at 6 mM Mg2+ between the RNA and DNA templated assay showed that it was similar, with the exception of T>C and C>G mutations (Fig. 3). The latter was relatively high in the DNA templated assays while the former was higher in the RNA templated assays. For the 0.5 mM Mg2+ assays, the most notable difference, once again, was the high proportion of G>A mutations in the DNA vs. RNA templated assay.
Substitution mutation profile for the RNA templated assay at 0.5 and 6 mM Mg2+
An analysis of mutations with the RNA templated assays at individual nucleotides on the template suggested that, for many positions, the main effect of high Mg2+ was to magnify error-prone sites on the template rather than create new ones (see Figs 4 and S1a for Exp. 1, and Fig. S3c for Exp. 2). DNA templated assays showed a similar trend (see Figs S1b and S3a for Exp. 1, and S3b for Exp. 2) although it was more difficult to assess, as the number of recovered mutations was not significantly above background at many sites (background was taken as the frequency of the same mutation type at the same position of the template in the control samples), especially for the 0.5 mM Mg2+ condition. An advantage of the RNA templated assay is that two rounds of RT synthesis are performed (Fig. 1), giving RT two chances to produce a mutation at each nucleotide position. Several of the strongest substitution mutations observed were A>G, G>A, C>T, T>C mutations, both in low and high Mg2+ (Figs 4 and S3a–c). These substitution types are common with HIV-1 RT in vitro [55, 56] and cellular replication assays [32, 44, 57].
Calculation of overall mutation rates in NGS assays for 0.5 and 6 mM Mg2+
The overall mutation frequencies and frequencies for particular types of mutations were calculated as described in the legend to Table 1. Overall mutation rates were modestly higher in the DNA templated assay, a trend that was also observed in Exp. 2 (Table S2). On a per nucleotide basis, in 0.5 mM Mg2+, HIV-1 RT made approximately one mutation for every 2720 (1/(3.677×10−4)) and 3284 (1/(3.045×10−4)) nucleotides in the DNA and RNA templated assay, respectively. This rate rose to about one mutation for every 766 (1/(1.306×10−3)) and 897 (1/(1.115×10−3)) nucleotides in 6 mM Mg2+ for DNA and RNA templated assays, respectively. These rates would exclude G>T and C>A mutations as well as larger insertions/deletions that cannot be tested by this approach (see Methods). In all cases, indel mutation rates were relatively low, although they made up ~21 % of total mutations (after BKG subtraction) in the 0.5 mM RNA templated assay (Table 1). Exp. 2 demonstrated a similar trend although mutation rates in DNA assays were higher than those in RNA assays to a greater extent than in Exp. 1 (Table S2).
Comparison of the NGS results with other results in cellular assays
In general, the mutation rate of HIV-1 is lower when measured during replication of HIV in cells than using HIV-1 RT in vitro. The sensitivity of HIV-1 RT to Mg2+ concentration and the fact that most in vitro assays have been done in high Mg2+ is likely part of the reason for this (see Introduction). In contrast to in vitro assays, most cell culture experiments have identified a relatively small number of mutations, however, more recent SSCS NGS cellular approaches identified larger mutation sets [57], while an extensive analysis of mutation types and frequencies using lacZα complementation in cells was also performed by Abram et al. [32]. Our experiments used a region of lacZα nearly identical to Abram et al. (Fig. 1), allowing a direct comparison of the results. The numbering in Fig. 1 is consistent with numbering in Abram et al. (see Methods). Abram et al. [32] recovered a total of ~600 substitution mutations (including those recovered with lacZα in the forward and reverse orientations) at 71 of the 174 nucleotide lacZα region in their assay (‘single nucleotide substitutions or ‘class 1’ mutations in that report), and indels at a much lower rate (~5% of recovered mutations, ‘class 2’ mutations). Since the assay was based on α-complementation, only nucleotide changes resulting in detectable changes in lacZα activity were recoverable, unless the mutation was coupled with another detectable mutation as part of a lacZα gene with multiple mutations. Although Abram et al. [32] also recovered several mutations as part of multiple nucleotide substitutions in the same lacZα gene (‘class 3’ mutations in that report), we choose to compare our NGS mutations to those recovered as single nucleotide substitutions, or single nucleotide frameshifts (class 1 and 2 mutations, respectively), which constituted the bulk of indel mutations recovered by Abram et al. [32].
There was a strong correlation for signal nucleotide insertion mutations with the two predominant hotspots from Abram et al. [32] constituting a large proportion of the indels recovered with NGS (nts 35–38 and 81–83 discussed above). Unlike Abram et al. [32] where deletions were fewer compared to insertions, our NGS assay recovered a similar level of insertions and deletions (Tables 1 and S2). Perhaps most notable, was the low indel mutation rates in both our assay and the Abrams et al. [32] assay. Only the 0.5 mM RNA templated assays showed indel mutations rates greater than ~8% of total mutations in the NGS assays, comparable to the ~5% for ‘class 2’ mutations in Abrams et al. [32]. Many in vitro assays using lacZα demonstrate higher indel rates [55, 58, 59].
Substitution mutations were more difficult to correlate as our NGS assay does not depend on lacZα activity, essentially detecting all mutations. But since the assay in Abrams et al. [32] was based on α-complementation, only nucleotide changes resulting in detectable changes in LacZα activity were recovered, unless the mutation was coupled with another detectable mutation as part of a lacZα gene with multiple mutations. Unlike substitutions, indels, especially those closer to the start of the lacZα gene, always inactivate the gene leading to a white colony in the α-complementation assay. We took the approach of comparing only the highest frequency mutations from NGS with those from Abram et al. [32]. In the NGS assay for the RNA templated sample with 0.5 mM Mg2+ (this condition was used as it most closely matches cellular replication), a total of 332 different types of mutations were present above background at 155 total positions, with at least one mutation type recovered at each position that was scored. For ‘high-frequency mutations’, an arbitrary mutation frequency cut-off for specific mutation types at a specific nucleotide position of 1.25×10−6 was used, which constituted the 61 highest frequency substitution (about 18% (61/332) of all the types of recovered mutations) in the NGS assay (see Fig. S1a). These were compared to substitution mutations from Abram et al. [32] using the following criteria: only mutations recovered at least one time in the Abram et al. [32] α-complementation assay were classified as ‘detectable’ in that assay. Of the 61 highest frequency mutations in the NGS assay, 39 were not detected in the Abrams et al. [32] assay, and therefore could not be compared between the two assays. Of the remaining twenty-two, seven, six, three, and six, were recovered 1–2, 3–5, 6–11, and 11 or more times, respectively, in the Abram et al. [32] assay. For perspective, there were 33 mutations (of the ~120 different mutation types recovered as class one mutations) in the Abram et al. [32] assay that were recovered six or more times and 16 recovered 11 or more times.
Discussion
This report confirms and expands upon previous results showing that HIV-1 RT, but not MuLV and AMV RTs, demonstrate greater fidelity in Mg2+ concentrations that more closely match the free Mg2+ concentrations in cells. Common drug resistance mutations in HIV-1 RT demonstrated a similar behaviour as did HIV-1 subtype A/E RT, HIV-2 RT and PFV RT, although the latter showed statistically higher fidelity than the other RTs used in this report (Table S1). All HIV-1 drug resistance mutants were less accurate in higher Mg2+ and, consistent with previous results [46], only K65R RT demonstrated a statistically lower mutation rate than wt HIV-1 RT (Table S1). Although some drug resistant mutants (M184V in particular), showed a greater decrease in fidelity than wt in high Mg2+, none of the mutants were dramatically more affected by Mg2+ than wt. We have previously shown that the potency of NRTIs diminished in 0.5 mM Mg2+ compared to 6 mM [23], while the opposite occurs for NNRTIs [22, 23]. This suggests that interactions involving Mg2+ are important for modulating the potency of these drugs, and advocates for the possibility of resistance mutations arising to exploit these properties to further diminish drug potency. Although the experiments here did not directly test this, they do show that the tested mutants retain the pronounced shift to lower fidelity at higher Mg2+ concentration. Therefore, none of the mutants tested here suggest an altered interaction with Mg2+ relative to wt, at least in the context of fidelity.
We previously postulated that the enhanced fidelity of HIV-1 RT at lower Mg2+ could result from the lower velocity of the reaction, which increases the residence time on each template base and therefore affords better discrimination [24]. More recent results indicate that the putative A (catalytic Mg+2) and B (nucleotide bound Mg2+) divalent cation binding sites in the polymerase domain of HIV-1 RT have dramatically different affinities for Mg2+ [60], with sites A and B binding with Kds of 3.7 mM and 29 µM, respectively. The authors propose that “weak binding of the catalytic Mg2+ contributes to fidelity by sampling the correctly aligned substrate without perturbing the equilibrium for nucleotide binding at physiological Mg2+ concentrations”. These findings are consistent with our previous hypothesis. Further, the range of Mg2+ concentrations explored here (from 0.25 to 12 mM) for wt HIV-1 RT (Fig. 2) and in our previous work [24] span the proposed Kd for site A. This mechanism is also consistent with there being little change in fidelity between 6 mM and 12 mM Mg2+ for HIV-1 RT (Fig. 2) as the occupancy of site A would likely change little over this range, assuming the Kd value for this site is close to the predicted value of 3.7 mM. Since MuLV and AMV RTs do not demonstrate large fidelity changes between 0.5 mM and 6 mM Mg2+ (Fig. 2), this may suggest that the binding affinity for Mg2+ at sites A and B differ for these enzymes compared to HIV-1 RT, but this remains to be determined.
The mutation frequencies in the α-complementation assay were similar for most of the enzymes tested. Only PFV RT at 0.5 mM and 6 mM Mg2+, K65R at 6 mM Mg2+, and MuLV RT at 6 mM Mg2+ (in both the RNA and DNA templated assays) showed statistically greater fidelity than HIV-1 RT wt at the same Mg2+ concentration, while M184V RT was statistically less accurate at 6 mM Mg2+ (Table S1). The α-complementation assay examined mutations over a modestly larger sequence range than the NGS assay (Fig. 1). Ignoring this small difference and using the same 155 nucleotide scored range (see Results), the overall mutation frequency for HIV-1 wt on the RNA templated system after background subtraction for 0.5 mM and 6 mM would be approximately 1.2×10−5 and 2.5×10−5 ((colony mutation frequency-BKG)/310 nucleotides (two rounds of RT synthesis over the scored region)), respectively. In the DNA templated assay these values would be 0.64×10−5 and 2.6×10−5 (note there is only a single round of RT synthesis in this assay so the total nucleotides would be 155), respectively. These number are likely to be significantly lower than the real mutation rate as most mutations made by RT are not detected in the α-complementation assay (see Results). The numbers are in the same range as number we previously calculated using a similar assay [24]. Interestingly, these mutations rates are approximately the same as those calculated for HIV-1 infection in cell culture by Abram et al. [32] using an α-complementation assay.
The NGS analysis allowed for a direct comparison of the frequency and types of mutations made in both RNA and DNA templated assays with 0.5 mM or 6 mM Mg2+. Only wt HIV-1 RT was examined with NGS so it is not possible to determine if RT drug resistance mutations affected the types of mutations that were made. An interesting finding was that unlike substitutions which increased about four-fold overall, there was no significant increase in indel mutations between 0.5 mM and 6 mM Mg2+ in the RNA templated assay, and indels made up a lower proportion of the total mutations at 6 mM Mg2+, owing to that condition having more substitutions (Table 1). In contrast, indels increased significantly at the higher Mg2+ concentration in the DNA templated assay. There was more mutation data for the RNA vs. DNA templated assay, probably owing to two vs. one round of RT synthesis in this assay. Also, the data between the two experiments (see Table S2 for second exp.) matched better with the RNA, especially for indel mutations. These discrepancies with the DNA data lend some uncertainty to the results. However, one thing that was especially clear was the much lower indel rate with DNA vs. RNA at 0.5 mM Mg2+ (Tables 1 and S2). This may be due to the different mechanism for indel vs. substitution mutations. The former result mainly from slippage of the template or primer strand while the latter can result from slippage and other mechanism [61, 62]. Indels typically occur by addition of the ‘correct’ nucleotide on a transiently misaligned primer-template, followed by continued elongation prior to realignment. Substitutions can occur by this mechanism when realignment occurs prior to elongation, or by direct incorporation of an incorrect nucleotide. Indels could be more pronounced on RNA, especially if slippage occurs more frequently. An alternative explanation, and one we favour, is that the RNA template in the RNA assays may have more indels (and substitutions) to start, due to the lower fidelity of T3 RNA polymerase used to produce the RNA template vs. Q5 DNA polymerase used to produce the DNA template. The substitution mutation rate of Q5 DNA polymerase has been reported to be ~5×10−7, lower than any other thermostable polymerase [45]. Q5 contributions to errors in the DNA templated assays should be very small as it has ~two orders of magnitude greater fidelity than HIV-1 RT. As noted in the Results, there are many steps in the NGS assay and manipulations such as heating and running material on gels as well as nucleic acid oxidation and PCR cycling could contribute to errors observed in the control assays (see [45] for a detailed analysis of error sources in PCR assays), beyond errors that were produced by Q5 which we expect would be very low. Although the fidelity of T3 and other RNA polymerase has not been directly measured, recent experiments suggest phage RNA polymerases contribute a significant level of mutations to reverse transcriptase fidelity assays that use phage polymerase-derived RNAs as the template [31]. Note that the above discussion would also predict that the mutation rate in the RNA templated assay should be higher than the DNA templated assay since the starting template had more mutations. Despite this, in the NGS experiments, DNA templated assays showed small, but consistently higher mutation rates (Tables 1 and S2). This was not observed in the α-complementation assay, but those results were from a much smaller data set. One possibility is that HIV-1 RT is less accurate when copying DNA than RNA, as has been suggested previously [31, 63]. Since the numbers in our assays were not dramatically different with RNA and DNA, more data and a better understanding of the mutation rate of phage RNA polymerase would be required to address this question.
With respect to the proportion and positions of indel mutations, the NGS results compared reasonably well with results from Abram et al. using the same sequence in HIV-1 cell replication assays [32]. Unlike some previous in vitro results with HIV-1 RT [55, 58, 59], indels made up a relatively low proportion of the observed mutation in our NGS results (Tables 1 and S2), as they did in Abram et al. [32]. Notably, other in vitro NGS results with HIV-1 RT have yielded a low proportion of indels as well [56]. A possible explanation for previous high indel rates in vitro is that most of those assays were performed using α-complementation and indels are easier to detect in those assays (see Results), although this is not consistent with the low indel rates observed by Abram et al. [32]. Perhaps the subjective nature of the α-complementation assay can lead to variation in the results, especially if all the colonies scored as mutations are not sequenced.
Substitution mutations were more difficult to compare between our NGS assay and other cellular assays. Like other NGS assays which use HIV-1 RT in vitro [25, 56], as well as other cellular replication assays [32, 44, 57], our NGS results showed a strong tendency for transitions vs. transversions, and a high proportion of G>A and C>T mutations (Figs 3 and S2). However, comparisons to the Abram et al. [32] results indicated that the majority (39 of 61 (see Results)) of the high frequency mutations (arbitrarily set as those with mutation frequencies≥1.25×10−6) observed in the RNA templated assay with 0.5 mM Mg2+ were not found by Abram et al. using α-complementation and, therefore, could not be compared. Nine of the remaining 22 mutations from that group were found six or more times in the Abram et al. [32] results while the other 13 were recovered less than six times. A complete correlation would not be expected even if our in vitro conditions perfectly mimicked cellular conditions as there are factors in the cell (discussed in Abram et al. [32]) that are likely to modify the mutation spectrum. It would be interesting to use NGS over the same sequence to directly compare both in vitro and cellular results. This would allow a comparison of thousands of mutations and allow a more direct analysis of the contribution of RT vs. cellular factors in the mutation spectrum of HIV-1.
The mutation rate observed with NGS was also somewhat higher than expected based on results with α-complementation. The estimated rate of 1.2×10−5 in the α-complementation in the 0.5 mM Mg2+ RNA templated assay would necessarily underestimate the actual mutation rate as this assay only scores a fraction of the substitution mutations (see Results). However, the rate for the same condition in the NGS assay which detects ‘all’ mutations, was ~20-fold higher (3.045×10−4 and 1.827×10−4, Exp. 1 and 2, respectively (Tables 1 and S2)). Abram et al. [32] recovered about 110 different types of substitution mutation (class one mutations in their data) over the same range we used in our NGS assay. Presumably our assay could have recovered 465 (155×3) different types of substitutions over that range. Using this logic, which does not take into account the mutation frequency at specific nucleotide locations, the NGS assay should detect roughly three times more substitutions. This number is likely to be higher as nearly all detected mutations were in the first two-thirds of the scored region in the Abram et al. [32] assay, suggesting that α-complementation cannot detect many mutations near the lacZα C-terminus. Still, a five-fold increase in NGS would seem more likely than the observed 20-fold. The overall mutation frequency from our results using 0.5 mM Mg2+ compared well to other NGS results with HIV-1 RT [25], but those were conducted with higher Mg2+ concentrations, while NGS results with lower Mg2+ produced a lower mutation rate than we found [25, 56]. An NGS analysis of HIV-1 and HIV-2 replication in cells over a region of the HIV integrase gene has also been conducted using a SSCS approach similar to our NGS approach to correct for errors derived from amplification and sequencing [57]. Only transition mutations were recovered at high enough frequencies above background to analyse. The transition mutation rate for NL4-3 HIV-1 was ~1×10−4, while other HIV-1 subtypes showed lower mutation rates. Since this is the rate for transitions only (which likely constitute a significant portion of the total mutations), the rate for all mutations would be higher. It would be interesting to test the RTs from some of the other subtypes to see if they also demonstrate higher fidelity in vitro compared to NL4-3 RT. Again, an analysis of the same gene region in vitro and in cellular replication using NGS approaches that decrease background mutations like the SSCS used here or duplex sequencing analysis [53, 54] would be very helpful for these comparisons.
In conclusion, these results show that RTs from several retroviruses (HIV-1, HIV-2, and PFV) have higher fidelity in low, more physiological Mg2+ concentrations, although this is not true for all RTs (e.g, MuLV and AMV RTs). Drug resistant HIV-1 RTs also demonstrated this property although there may be quantitative differences between some drug resistant enzymes and wt RT as well as differences in mutation profiles. Addressing this question would likely require investigating these enzymes in a more quantitative assay that can score more mutations like the NGS assay used here for wt HIV-1 RT rather than the α-complementation assay that was used with the drug resistant mutants. Finally, the results demonstrate the utility of the SSCS NGS assay used here or other similar NGS assays that allow thousands of mutations to be evaluated at all positions on the template. These approaches bring a level of quantification and statistical vigour to mutation analysis that has not previously been available and should allow significant advancements in understanding the roles that different mutagenic factors play in generating genetic diversity.
Supplementary Data
Funding information
This work was supported by the National Institute of Allergy and Infectious Disease grant R01AI150480 to JJD.
Acknowledgements
We thank Drs Stefan Sarafianos (Emory University), Eddy Arnold (Rutgers University), and Stephen Hughes (National Institutes of Health) for supplying RTs or RT plasmid clones.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMV, avian myeloblastosis virus; CIP, calf-intestinal phosphatase; HIV, human immunodeficiency virus; MuLV, moloney murine leukemia virus; NGS, next generation sequencing; PNK, T4 polynucleotide kinase; RT, reverse transcriptase; SSCS, single-strand consensus sequencing.
Two supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Telesnitsky A, Goff SP. Reverse Transcriptase. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [PubMed] [Google Scholar]
- 2.Cowan JA, Ohyama T, Howard K, Rausch JW, Cowan SM, et al. Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J Biol Inorg Chem. 2000;5:67–74. doi: 10.1007/s007750050009. [DOI] [PubMed] [Google Scholar]
- 3.Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura H, Katayanagi K, Morikawa K, Ikehara M. Structural models of ribonuclease H domains in reverse transcriptases from retroviruses. Nucleic Acids Res. 1991;19:1817–1823. doi: 10.1093/nar/19.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz SJ, Champoux JJ. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 2008;134:86–103. doi: 10.1016/j.virusres.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, Steitz TA. Recombining the structures of HIV integrase, RuvC and RNase H. Structure. 1995;3:131–134. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- 8.Oda Y, Nakamura H, Kanaya S, Ikehara M. Binding of metal ions to E. coli RNase HI observed by 1H-15N heteronuclear 2D NMR. J Biomol NMR. 1991;1:247–255. doi: 10.1007/BF01875518. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman AD, Banapour B, Levy JA. Characterization of the AIDS-associated retrovirus reverse transcriptase and optimal conditions for its detection in virions. Virology. 1985;147:326–335. doi: 10.1016/0042-6822(85)90135-7. [DOI] [PubMed] [Google Scholar]
- 10.Rey MA, Spire B, Dormont D, Barre-Sinoussi F, Montagnier L, et al. Characterization of the RNA dependent DNA polymerase of a new human T-lymphotropic retrovirus (lymphadenopathy associated virus) Biochem Biophys Res Commun. 1984;121:126–133. doi: 10.1016/0006-291x(84)90696-x. [DOI] [PubMed] [Google Scholar]
- 11.Starnes MC, Cheng YC. Human immunodeficiency virus reverse transcriptase-associated RNase H activity. J Biol Chem. 1989;264:7073–7077. [PubMed] [Google Scholar]
- 12.Wondrak EM, Löwer J, Kurth R. Functional purification and enzymic characterization of the RNA-dependent DNA polymerase of human immunodeficiency virus. J Gen Virol. 1986;67 (Pt 12):2791–2797. doi: 10.1099/0022-1317-67-12-2791. [DOI] [PubMed] [Google Scholar]
- 13.Delva P, Pastori C, Degan M, Montesi G, Lechi A. Intralymphocyte free magnesium and plasma triglycerides. Life Sci. 1998;62:2231–2240. doi: 10.1016/s0024-3205(98)00201-x. [DOI] [PubMed] [Google Scholar]
- 14.Delva P, Pastori C, Degan M, Montesi G, Lechi A. Catecholamine-induced regulation in vitro and ex vivo of intralymphocyte ionized magnesium. J Membrane Biol. 2004;199:163–171. doi: 10.1007/s00232-004-0686-7. [DOI] [PubMed] [Google Scholar]
- 15.Moomaw AS, Maguire ME. The unique nature of mg2+ channels. Physiology (Bethesda) 2008;23:275–285. doi: 10.1152/physiol.00019.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delva P, Pastori C, Degan M, Montesi G, Brazzarola P, et al. Intralymphocyte free magnesium in patients with primary aldosteronism: aldosterone and lymphocyte magnesium homeostasis. Hypertension. 2000;35:113–117. doi: 10.1161/01.hyp.35.1.113. [DOI] [PubMed] [Google Scholar]
- 17.Delva PT, Pastori C, Degan M, Montesi GD, Lechi A. Intralymphocyte free magnesium in a group of subjects with essential hypertension. Hypertension. 1996;28:433–439. doi: 10.1161/01.hyp.28.3.433. [DOI] [PubMed] [Google Scholar]
- 18.Morelle B, Salmon JM, Vigo J, Viallet P. Measurement of intracellular magnesium concentration in 3T3 fibroblasts with the fluorescent indicator Mag-indo-1. Anal Biochem. 1994;218:170–176. doi: 10.1006/abio.1994.1156. [DOI] [PubMed] [Google Scholar]
- 19.Gout E, Rébeillé F, Douce R, Bligny R. Interplay of Mg2+, ADP, and ATP in the cytosol and mitochondria: unravelling the role of Mg2+ in cell respiration. Proc Natl Acad Sci U S A. 2014;111:E4560–7. doi: 10.1073/pnas.1406251111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy E, Freudenrich CC, Levy LA, London RE, Lieberman M. Monitoring cytosolic free magnesium in cultured chicken heart cells by use of the fluorescent indicator Furaptra. Proc Natl Acad Sci U S A. 1989;86:2981–2984. doi: 10.1073/pnas.86.8.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Touyz RM, Yao G. Modulation of vascular smooth muscle cell growth by magnesium-role of mitogen-activated protein kinases. J Cell Physiol. 2003;197:326–335. doi: 10.1002/jcp.10393. [DOI] [PubMed] [Google Scholar]
- 22.Goldschmidt V, Didierjean J, Ehresmann B, Ehresmann C, Isel C, et al. Mg2+ dependency of HIV-1 reverse transcription, inhibition by nucleoside analogues and resistance. Nucleic Acids Res. 2006;34:42–52. doi: 10.1093/nar/gkj411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achuthan V, Singh K, DeStefano JJ. Physiological Mg2+ conditions significantly alter the inhibition of HIV-1 and HIV-2 reverse transcriptases by nucleoside and non-nucleoside inhibitors in vitro . Biochemistry. 2017;56:33–46. doi: 10.1021/acs.biochem.6b00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achuthan V, Keith BJ, Connolly BA, DeStefano JJ. Human immunodeficiency virus reverse transcriptase displays dramatically higher fidelity under physiological magnesium conditions in vitro . J Virol. 2014;88:8514–8527. doi: 10.1128/JVI.00752-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okano H, Baba M, Hidese R, Iida K, Li T, et al. Accurate fidelity analysis of the reverse transcriptase by a modified next-generation sequencing. Enzyme Microb Technol. 2018;115:81–85. doi: 10.1016/j.enzmictec.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Menéndez-Arias L. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses. 2009;1:1137–1165. doi: 10.3390/v1031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svarovskaia ES, Cheslock SR, Zhang W-H, Hu W-S, Pathak VK. Retroviral mutation rates and reverse transcriptase fidelity. Front Biosci. 2003;8:d117–34. doi: 10.2741/957. [DOI] [PubMed] [Google Scholar]
- 29.Ji JP, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro . Biochemistry. 1992;31:954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- 30.Rezende LF, Prasad VR. Nucleoside-analog resistance mutations in HIV-1 reverse transcriptase and their influence on polymerase fidelity and viral mutation rates. Int J Biochem Cell Biol. 2004;36:1716–1734. doi: 10.1016/j.biocel.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 31.Sebastián-Martín A, Barrioluengo V, Menéndez-Arias L. Transcriptional inaccuracy threshold attenuates differences in RNA-dependent DNA synthesis fidelity between retroviral reverse transcriptases. Sci Rep. 2018;8:627. doi: 10.1038/s41598-017-18974-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol. 2010;84:9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/JVI.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 35.Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 37.Guenzel CA, Hérate C, Le Rouzic E, Maidou-Peindara P, Sadler HA, et al. Recruitment of the nuclear form of uracil DNA glycosylase into virus particles participates in the full infectivity of HIV-1. J Virol. 2012;86:2533–2544. doi: 10.1128/JVI.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansky LM. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology. 1996;222:391–400. doi: 10.1006/viro.1996.0436. [DOI] [PubMed] [Google Scholar]
- 39.Mansky LM, Le Rouzic E, Benichou S, Gajary LC. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J Virol. 2003;77:2071–2080. doi: 10.1128/jvi.77.3.2071-2080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 In vivo mutation rate. J Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Neil PK, Sun G, Yu H, Ron Y, Dougherty JP, et al. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J Biol Chem. 2002;277:38053–38061. doi: 10.1074/jbc.M204774200. [DOI] [PubMed] [Google Scholar]
- 42.Hou EW, Prasad R, Beard WA, Wilson SH. High-level expression and purification of untagged and histidine-tagged HIV-1 reverse transcriptase. Protein Expr Purif. 2004;34:75–86. doi: 10.1016/j.pep.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 44.Abram ME, Ferris AL, Das K, Quinoñes O, Shao W, et al. Mutations in HIV-1 reverse transcriptase affect the errors made in a single cycle of viral replication. J Virol. 2014;88:7589–7601. doi: 10.1128/JVI.00302-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potapov V, Ong JL. Correction: Examining Sources of Error in PCR by Single-Molecule Sequencing. PLoS One. 2017;12:e0181128. doi: 10.1371/journal.pone.0181128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah FS, Curr KA, Hamburgh ME, Parniak M, Mitsuya H, et al. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2000;275:27037–27044. doi: 10.1074/jbc.M002881200. [DOI] [PubMed] [Google Scholar]
- 47.Boyer PL, Stenbak CR, Hoberman D, Linial ML, Hughes SH. In vitro fidelity of the prototype primate foamy virus (PFV) RT compared to HIV-1 RT. Virology. 2007;367:253–264. doi: 10.1016/j.virol.2007.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn EH, Lee SH. Detection of Low-Frequency Mutations and Identification of Heat-Induced Artifactual Mutations Using Duplex Sequencing. IJMS. 2019;20:199. doi: 10.3390/ijms20010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arbeithuber B, Makova KD, Tiemann-Boege I. Artifactual mutations resulting from DNA lesions limit detection levels in ultrasensitive sequencing applications. DNA Res. 2016;23:547–559. doi: 10.1093/dnares/dsw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Liu P, Ettwiller LM. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science. 2017;355:752–756. doi: 10.1126/science.aai8690. [DOI] [PubMed] [Google Scholar]
- 52.Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoler N, Arbeithuber B, Povysil G, Heinzl M, Salazar R, et al. Family reunion via error correction: an efficient analysis of duplex sequencing data. BMC Bioinformatics. 2020;21:96. doi: 10.1186/s12859-020-3419-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang TT, Abelson S, Zou J, Li T, Zhao Z, et al. High efficiency error suppression for accurate detection of low-frequency variants. Nucleic Acids Res. 2019;47:e87. doi: 10.1093/nar/gkz474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993;268:10324–10334. doi: 10.1016/S0021-9258(18)82205-0. [DOI] [PubMed] [Google Scholar]
- 56.Yasukawa K, Iida K, Okano H, Hidese R, Baba M, et al. Next-generation sequencing-based analysis of reverse transcriptase fidelity. Biochem Biophys Res Commun. 2017;492:147–153. doi: 10.1016/j.bbrc.2017.07.169. [DOI] [PubMed] [Google Scholar]
- 57.Rawson JMO, Gohl DM, Landman SR, Roth ME, Meissner ME, et al. Single-strand consensus sequencing reveals that HIV type but not subtype significantly impacts viral mutation frequencies and spectra. J Mol Biol. 2017;429:2290–2307. doi: 10.1016/j.jmb.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyer JC, Bebenek K, Kunkel TA. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc Natl Acad Sci U S A. 1992;89:6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bebenek K, Beard WA, Casas-Finet JR, Kim HR, Darden TA, et al. Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J Biol Chem. 1995;270:19516–19523. doi: 10.1074/jbc.270.33.19516. [DOI] [PubMed] [Google Scholar]
- 60.Gong S, Kirmizialtin S, Chang A, Mayfield JE, Zhang YJ, et al. Kinetic and thermodynamic analysis defines roles for two metal ions in DNA polymerase specificity and catalysis. J Biol Chem. 2021;296:100184. doi: 10.1074/jbc.RA120.016489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- 62.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 63.Kerr SG, Anderson KS. RNA dependent DNA replication fidelity of HIV-1 reverse transcriptase: evidence of discrimination between DNA and RNA substrates. Biochemistry. 1997;36:14056–14063. doi: 10.1021/bi971385+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.