Abstract
Botulinum neurotoxins (BoNTs) are among the most lethal toxins known to humans, comprising seven established serotypes termed BoNT/A–G encoded in two types of gene clusters (ha and orfX) in BoNT-producing clostridia. The ha cluster encodes four non-toxic neurotoxin-associated proteins (NAPs) that assemble with BoNTs to protect and enhance their oral toxicity. However, the structure and function of the orfX-type NAPs remain largely unknown. Here, we report the crystal structures for OrfX1, OrfX2, and an OrfX1–OrfX3 complex, which are encoded in the orfX cluster of a BoNT/E1-producing Clostridium botulinum strain associated with human foodborne botulism. These structures lay the foundation for future studies on the potential roles of OrfX proteins in oral intoxication and pathogenesis of BoNTs.
Keywords: botulinum neurotoxin, botulinum, bacterial toxin, progenitor toxin complex, ha gene cluster, orfX gene cluster, OrfX1, OrfX2, OrfX3, P47
Graphical Abstract
We report the crystal structures of OrfX1, OrfX2, and an OrfX1–OrfX3 complex, which are encoded in the orfX gene cluster of a BoNT/E1-producing Clostridium botulinum strain associated with human foodborne botulism. These structures lay the foundation for future studies of the potential roles of OrfX proteins in oral intoxication and pathogenesis of BoNTs and other orfX-related bacterial toxins.
Introduction
Botulinum neurotoxins (BoNTs) that are classified as the Tier 1 select agents by the CDC cause botulism, a severe neurological disease presenting as flaccid paralysis [1-4]. Seven serotypes of BoNT (termed BoNT/A–G) are well established, while new BoNTs (e.g., HA, X) and several BoNT-like proteins in non-clostridial species (e.g., eBoNT/J or BoNT/En, PMP1) have been recently identified [5-11]. Paradoxical to their life-threatening toxicity, BoNT/A- and BoNT/B-based medicines to treat symptoms caused by neuronal over activity and cosmetics are successfully used [12]. While all current pharmaceuticals act long term, the short-acting BoNT/E with quick onset is being tested in clinical trials [13,14].
One of the major forms of botulism, foodborne botulism, can occur as a consequence of ingesting food or drink that were contaminated with C. botulinum spores that subsequently germinate and produce BoNTs in situ, with BoNT/A, B, and E being the most common serotypes [4,15-17]. The 150-kDa BoNT molecules are intrinsically vulnerable to the harsh environment of the gastrointestinal (GI) tract. For example, BoNT/A is readily degraded by digestive proteases and inactivated at pH 3 or less [18]. In fact, all BoNTs are naturally produced by bacteria in the form of progenitor toxin complexes (PTCs). These PTCs are high-molecular-weight multi-protein complexes composed of BoNT and several non-toxic neurotoxin-associated proteins (NAPs) [4,19-24].
The composition of the PTC depends on the neurotoxin gene cluster (NGC). Two major NGCs, the hemagglutinin ha cluster and the orfX cluster, have been recognized among BoNT-producing strains. While both NGC types carry an operon encoding BoNT (bont) and the non-toxic non-hemagglutinin (NTNHA) protein (ntnha) [19,20] (Fig. 1A), the ha cluster contains a residing operon of three HA genes, ha17, ha33, and ha70, and the orfX cluster carries a different set of genes namely orfX1, orfX2, orfX3, and p47 [19,22,25,26].
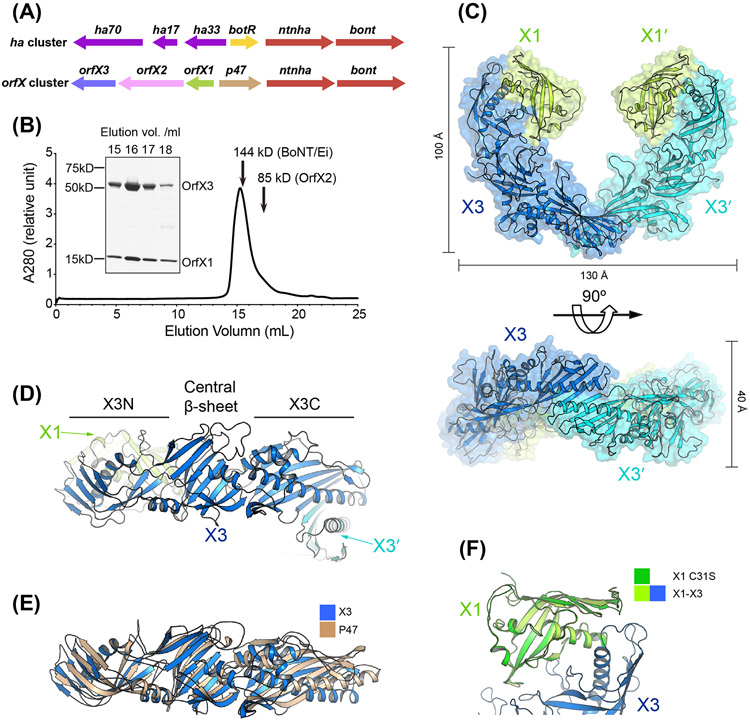
Figure 1. Crystal structure of the tetrameric OrfX1–OrfX3 complex.
(A) Composition of the ha and orfX neurotoxin gene clusters. (B) SEC analysis of the recombinant OrfX1–OrfX3 complex (~142 kDa) (n=3, a representative result is shown). The 144 kDa catalytically inactivated BoNT/Ei and 85 kDa OrfX2 were used as molecular weight references and their peaks were indicated by arrows. The purity of the selected peak fractions was examined by SDS-PAGE. (C) Ribbon and surface representations of the OrfX1–OrfX3 complex in two different views. The two OrfX1 molecules are colored in lime green, while the two OrfX3 are colored in blue and cyan, respectively. (D) Ribbon representation of OrfX3 that has two TULIP-like domains (X3N and X3C) connected by a central β-sheet. (E) Structural superimposition of OrfX3 and P47. (F) Structure superimposition of OrfX1C31S into the WT OrfX1 in the OrfX1–OrfX3 complex.
The ha gene cluster is found in bacterial strains that produce BoNT/ A1, A5, B, C, CD, DC, D, and G along with NTNHA and three HA proteins (HA17, HA33, and HA70). The architecture of the large PTC (L-PTC) of BoNT/A is reminiscent of an Apollo lunar module, whereby the ~300 kDa ovoid-shaped BoNT/A–NTNHA/A complex (the M-PTC) mimics the “ascent stage”, which is docked on top of a ~500 kDa three-arm “descent stage” composed of HA17, HA33, and HA70 [18,21,23,27,28]. Each L-PTC has 14 subunits with BoNT, NTNHA, HA70, HA17, and HA33 in a 1:1:3:3:6 stoichiometric ratio. The overall architecture of the L-PTC is likely conserved across HA-carrying BoNT serotypes [29]. Extensive structural and functional studies of the HA-type PTC represented by BoNT/A demonstrate that the NAPs not only protect the inherently fragile BoNT/A against the hostile environment of the GI tract, but also interact with host receptors to help BoNT/A pass through the intestinal epithelial barrier before it is released into circulation [18,27,28,30-39].
The orfX gene cluster is found in strains producing BoNT/ A1–A4, A6–A8, E, F, X, and HA, eBoNT/J and PMP1 as well as NTNHA and four other proteins (OrfX1, OrfX2, OrfX3, and P47). While the M-PTC of the OrfX-type PTC is believed to be similar to that of the HA-type PTC [24,27,40], there is no sequence or structural similarity between OrfX1–3/P47 and the HA proteins, and the physiological functions of OrfX1–3 and P47 are largely unknown. However, similar locations next to the bont-ntnha operon and similar gene expression patterns between the orfx and ha operons support a hypothesis that OrfX1–3/P47 might also play a role in PTC formation and BoNT pathogenesis [41,42]. Intriguingly, a recent study indicated that OrfX1–3 proteins enhanced the toxicity of the BoNT-like protein PMP1 towards anopheline mosquito larvae [6]. An earlier study also shows that OrfX1–3 and P47 could be detected in association with BoNT immune-extracted from C. botulinum cultures or with some commercially available BoNTs based on mass spectrometry analysis [43]. However, the presence of these proteins varied significantly between samples and no stable OrfX-containing PTCs have been identified.
Another intriguing finding showed that genes putatively encoding OrfX1–3 and P47 are found in genomes of a wide range of non-BoNT-producing bacterial species, such as Alphaproteobacteria, Bacilli, Betaproteobacteria, Cytophagia, and Gammaproteobacteria [44]. These orfX-p47-containing gene clusters show large diversity in their gene arrangement and gene content, and some are neighboring genes encoding oral insecticidal toxins, such as delta-endotoxins (Cry toxin), binary toxins (VIP toxin), or ABC toxins. This finding suggests that homologues of OrfXs and P47 may play a general role in oral toxinogenesis and pathogenesis of some bacteria, as opposed to being specific to BoNTs [44].
In contrast to a wealth of information on the HA proteins and the HA-type PTC, the structure and function of the OrfX-type PTC remain largely uninvestigated. The recent crystal structures of P47 from BoNT/E1 and A2, and OrfX2 from BoNT/A2 reveal that their structures are completely different from the HAs [45,46]. Furthermore, both of P47 and OrfX2 contain a structural fold that has the tubular lipid-binding (TULIP) domain topology, whereas the TULIP-containing proteins have diverse functions that are usually related to lipid binding [47,48].
Here, we report the crystal structures for OrfX1, OrfX2, and an OrfX1–OrfX3 complex from the BoNT/E1-producing C. botulinum strain Beluga. Interestingly, the structural analyses show that all three proteins carry the TULIP domain fold. OrfX2 has two N-terminal TULIP-like domains, which are positioned side-by-side while both interacting with its C-terminal domain. OrfX3 is composed of three parts with a central 6-stranded β-sheet flanked by two TULIP domains at its N- and C- termini. OrfX3 uses its C-terminal TULIP-like domain to form a tail-to-tail homodimer, while its N-terminal TULIP domain interacts with OrfX1 that also adopts a TULIP-like fold. Unexpectedly, we notice that the overall structure of OrfX2 is similar to the OrfX1–OrfX3 complex, with the N-terminal TULIP domain of OrfX2 mimicking OrfX1 and the rest of it mimicking OrfX3. These findings thus lay the structural foundation for future studies of the potential roles of OrfX proteins in oral intoxication and pathogenesis of BoNTs and other orfX-related bacterial toxins.
Materials and Methods
Cloning, protein expression and purification
The genes encoding OrfX1, OrfX2 and OrfX3 were amplified by PCR using the genomic DNA of Clostridium botulinum E1 strain Beluga (Accession code: ACSC01000002) as template. The orfX1, orfX2, and orfX3 genes were cloned into the pQE (Qiagen) expression vector with an N-terminal 6xHis tag fused to a thrombin cleavage site and a C-terminal Strep-tag (H6tOrfX1-ES, H6tOrfX2-ES, H6tOrfX3-ES). The orfX1 gene was also cloned into the pRSFDuet-1 dual expression vector (Novagen) with an N-terminal 6xHis tag and a thrombin cleavage site. This OrfX1 expression construct was further modified by inserting the orfX3 gene into the other expression cassette with no affinity tag for co-expression and purification of these two proteins (H6tOrfX1-E OrfX3-E). We also made another variant of the OrfX1–OrfX3 co-expression plasmid based on this one by inserting a C-terminal Strep-tag to OrfX3 for mutagenesis studies (H6tOrfX1-E OrfX3-ES). The K244A/K245A mutation of OrfX3 was designed by Surface Entropy Reduction prediction server [49]. This OrfX3 mutant in the H6tOrfX1-E OrfX3-E expression plasmid and the C31S mutation of OrfX1 in the H6tOrfX1-ES plasmid were generated by QuikChange site-directed mutagenesis. Site-directed mutagenesis of OrfX3 was conducted employing the H6tOrfX1-E OrfX3-ES as the template DNA by the GeneTailor method (Invitrogen GmbH, Karlsruhe, Germany) to generate V377E/I379E/Y381S (VIY), L390E/V394D/L401E/L405E (LVLL), VIYLVLL, D57A/N60A/R62A (DNR), D152A/R162A (DR), and D57A/N60A/R62A/Q93A/R162A (DNRQR). Nucleotide sequences of all generated plasmids were verified by DNA sequencing.
For structural studies, all proteins were expressed in the BL21 Star (DE3) strain of Escherichia coli (Invitrogen). Bacteria were initially cultured at 37°C in LB (Luria-Bertani) medium. When OD600 reached 0.4–0.6, the temperature was decreased to 18°C and protein expression was induced with 0.5 mM isopropyl-β-D- thiogalactopyranoside (IPTG). The cells were harvested 16 hrs post induction by centrifugation and stored at −20°C until use. The selenomethionine (SeMet) labeled proteins were expressed as previously described [50]. All the 6xHis-tagged proteins were firstly purified using a Ni-NTA (nitrilotriacetic acid, Qiagen) affinity column. Subsequently, the 6xHis-tag was removed through thrombin protease cleavage, followed by ion exchange chromatography using a MonoQ column (GE Healthcare) for the OrfX1–OrfX3 complex and OrfX2, and a MonoS column for OrfX1. The tag-free proteins were further purified through size-exclusion chromatography (SEC) on a Superdex-200 column (GE Healthcare) in a buffer containing 20 mM Tris pH7.5, 150 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). The purities of the proteins were examined by SDS-PAGE gels. The purified proteins were concentrated and stored at −80°C for future use.
For biochemical and mutagenesis studies as well as antigen production for immunization, cultures of E. coli M15 transformed with pQE plasmids or BL21 (DE3) transformed with pRSF plasmids were grown in 600 ml 2YT medium (16 g/l tryptone, 10 g/l yeast extract, 5 g/l NaCl) at 37°C until the OD600 of the culture reached 0.6 to 0.8. After induction with 0.2 mM and 1 mM IPTG (Roth), respectively, cultures were incubated overnight at 22°C until harvest. Cell pellets were resuspended in 23 ml of 0.1 M Tris-HCl, pH 8.0, containing the protease inhibitors pepstatin A (1 mM), benzamidine (5 mM), and phenylmethylsulfonylflouride (PMSF, 0.5 mM) and lysed by ultrasonic treatment for 7 min using 0.5 cycles with 100% amplitude (Hielscher UP200S). The clear lysate was applied to either Co2+-Talon matrix (Takara Bio Europe S.A.S., France) or StrepTactin-Sepharose resin (IBA GmbH, Germany). Matrix was washed twice in resuspension buffer and eluted with the buffer containing 250 mM imidazol (Roth) for Talon matrix or 10 mM desthiobiotin (IBA) for elution of StrepTactin-Sepharose matrix. In case of tandem purification, immobilized metal affinity chromatography (IMAC) was followed by StrepTactin chromatography. N-terminal 6xHis tag was removed by thrombin digestion overnight with 0.02 U/μg 6xHis-tagged protein at room temperature. For further purification and analysis proteins were subjected to gel filtration on Superdex-200 16/60PG column (GE Healthcare, Germany) in 100 mM Tris-HCl, pH 8.0 yielding OrfX2-ES and OrfX1-E/OrfX3-ES. Fractions were analysed via SDS-PAGE and Western blotting. All purified proteins were frozen in liquid nitrogen, and stored at −80°C. Protein concentration was determined by the A280 method using the theoretical absorption coefficients.
Crystallization, data collection, and structure determination
Initial crystallization screens for OrfX1, OrfX2, and the OrfX1–OrfX3 complex were carried out using a Gryphon crystallization robot (Art Robbins Instrument) and commercial high-throughput crystallization screen kits from Hampton Research or Qiagen. After extensive manual optimization, the best crystals of OrfX1C31S were obtained when the protein (~3.5 mg/ml) was mixed in 1:1 (v/v) ratio with a reservoir solution containing 25 % polyethylene glycol (PEG) 3350, 0.2 M ammonium acetate, and 0.1 M Bis-tris pH 5.5. The best crystals of the OrfX1–OrfX3K244A/K245A complex were obtained when the protein (~12 mg/ml) was mixed in 1:1 (v/v) ratio with a reservoir solution containing 13 % PEG3350, 0.3 M ammonium citrate dibasic, and 0.1 M sodium acetate, pH 4.5. The crystals of OrfX2 were obtained when the protein (~4 mg/ml) was mixed in 1:1 (v/v) ratio with a reservoir solution containing 14% PEG3350, 0.2M lithium sulfate, and 0.1M Bis-Tris, pH 6. All crystals were grown by the hanging-drop vapor diffusion method at 18°C. The crystals were cryoprotected in the original mother liquor supplemented with 20 % glycerol for OrfX1C31S, 15% glycerol for the OrfX1–OrfX3K244A/K245A complex, and 20% ethylene glycol for OrfX2, and immediately flash-frozen in liquid nitrogen for data collection.
The X-ray diffraction datasets for SeMet-OrfX1C31S, the SeMet-OrfX1–OrfX3K244A/K245A complex and the native complex, and SeMet-OrfX2 were collected at 100 K at the NE-CAT beamline 24-ID-E, Advanced Photon Source (APS), Argonne National Laboratory using detector ADSC Q315. The data were processed using XDS [51]. All the SeMet derivative crystal structures were solved by single-wavelength anomalous dispersion (SAD) using PHENIX [52]. Structure of the native OrfX1–OrfX3K244A/K245A complex was solved using PHENIX.Phaser-MR [53] with the SeMet-OrfX1–OrfX3K244A/K245A structure serving as a search model. Further refinements of the structures were performed by PHENIX.refine [52] and COOT [54] in an iterative manner. All the refinement progress was monitored with the free R value using a 5% randomly selected test set [55]. The structures were validated by MolProbity [56]. The statistics of data collection and structural refinement for all the structures reported in this manuscript are summarized in Table S1. All structure figures were prepared by PyMOL (http://www.pymol.org). Sequence alignment was made using Clustal Omega [57] and ESPript 3 [58].
Antibody production and purification
Polyclonal rabbit anti-OrfX antibody KOrf154 was purified as IgG from serum of an immunized 1-year-old male New Zealand rabbit (Bundesinstitut für Risikobewertung, Berlin, Germany). Animal experiments were performed in compliance with the German Animal Welfare Act and European legislation for the protection of animals used for scientific purposes (Directive 2010/63/EU), overseen and authorized under license H109/03 by the local state authorities (Landesamt für Soziales und Gesundheit, Berlin, Germany), and supervised by the institutional Animal Welfare Officer. The rabbit had ad libitum access to water and feed and was immunized subcutaneously with a mixture of 20 μg recombinant OrfX2-ES, and 45 μg recombinant OrfX1-E/OrfX3-ES complex, in Freund’s complete adjuvant for the first immunization, and Freund’s incomplete adjuvant for subsequent two booster injections at intervals of four weeks. Serum reactivity was monitored by indirect ELISA using goat anti-rabbit IgG-coupled HRP (Dianova, Hamburg, Germany) and TMB slow substrate (Senova, Weimar, Germany) to confirm serum reactivity against immobilized recombinant OrfX proteins. For the final pAb KOrf154 preparation, IgG were purified from a pool of sera taken 12 days after first and 33 days after second boost by affinity chromatography using protein G matrix (HiTrap® Protein G HP column, Cytiva, Freiburg, Germany) on an HPLC device (ÄKTAexplorer TM 100, Cytiva, Freiburg, Germany). The IgG preparation was dialyzed against PBS at 4°C followed by photometric determination of protein content and analysis in ELISA to confirm specific binding to the targeted recombinant OrfX proteins.
Western blot
Proteins were separated by 12.5% SDS-PAGE and transferred onto a PVDF membrane (Roti®-PVDF, pore size 0.45 μm, ROTH, Karlsruhe, Germany). PVDF membranes were blocked with 5% milk powder in PBST (PBS, 0.05% Tween) for at least 1 h. The antibody KOrf154 was diluted 1:20,000 in 5% milk powder in PBST and incubated overnight at 4°C. The HRP-coupled secondary antibody (goat-anti-rabbit, Rockland, Limerick, PA) was diluted 1:50,000 and incubated at room temperature for 3 h. Bands were visualized employing SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) and the ChemoStar ECL Imager (Intas, Göttingen, Germany).
Results
Structure of the OrfX1–OrfX3 complex and OrfX1 of BoNT/E
During recombinant production and characterization of OrfX1, OrfX2, and OrfX3, we noticed that OrfX1 and OrfX2 could be expressed at high yields in E. coli and purified to high homogeneity, whereas the expression of OrfX3 was very low despite extensive optimization trials. This is not unexpected, especially for recombinant production of some proteins that are naturally part of a multi-protein complex. In such cases, some parts of the target protein that are otherwise protected by its binding partners in the complex are exposed, which may cause problematic protein folding when it is expressed by itself. This was previously observed for HA17 that had to be co-expressed either with its binding partners HA70 or HA33 [28]. As the potential binding partners of OrfX3 were unknown, we tested co-expression of OrfX3 with OrfX1, OrfX2, and P47, separately, and were excited to observe robust expression when OrfX1 and OrfX3 were co-expressed. Furthermore, these two proteins could be co-purified as a stable complex as judged by its SEC profile and co-crystallized (Fig. 1B). However, the crystals of the OrfX1–OrfX3 complex diffracted X-ray weakly with the best crystals exhibiting ~3.5 Å resolution. At that point, we carried out systematic surface entropy reduction (SER) screening by replacing small clusters of two to three flexible surface residues, such as Lys, Glu, and Gln in OrfX3, with Ala in order to minimize the loss of conformational entropy upon crystallization, which has been proven to be an effective method to improve the crystal packing and thus its diffraction capability [49,59]. The best results were obtained when two lysine residues (K244/K245) of OrfX3 were mutated to alanine (termed as OrfX3K244A/K245A) (Fig. S1A).
We first used the selenomethionine (SeMet) derived OrfX1–OrfX3K244A/K245A crystals to obtain phases for structure determination by SAD method. Subsequently, we solved the crystal structure of the OrfX1–OrfX3K244A/K245A complex at 2.05 Å resolution. One heterotetrameric complex composed of two molecules of OrfX1 and two OrfX3 was identified in the asymmetric unit, which has an elongated crescent shape (~130 Å × 100 Å × 40 Å) (Fig. 1C). More specifically, two OrfX3 form a tail-to-tail homodimer in the middle with each of their N-terminal domains bound with an OrfX1. This structure is consistent with the SEC and SDS-PAGE analysis, which also suggests that OrfX1 and OrfX3 predominantly form heterotetramers with a 2:2 stoichiometry in solution (Fig. 1B).
The structure reveals that both OrfX1 and OrfX3 contain a type of incomplete β-barrel fold, which is common to proteins in the TULIP superfamily. The TULIP fold mainly consists of a long α-helix that is wrapped by a twisted β-sheet composed of anti-parallel β-strands. OrfX3 is composed of three parts with a central 6-stranded β-sheet flanked by two TULIP domains located at its N- and C- termini, respectively (Fig. 1D). In the context of the OrfX1–OrfX3 complex, the OrfX3-N domain of each OrfX3 interacts with OrfX1 that also adopts a TULIP fold, while two OrfX3 molecules using their OrfX3-C domains form a homodimer (Fig. 1C-D). Interestingly, the overall structure of OrfX3 is similar to P47 and the superimposition of them yielded a root-mean-square deviation (RMSD) of ~6.058 Å over 262 aligned Cα atom pairs [46] (Fig. 1E).
We also determined the crystal structure of the standalone OrfX1. The wild-type OrfX1 exists as a mixture of monomer and dimer in solution due to a surface exposed residue Cys31, which was mutated to a Ser (C31S) to block Cys-mediated aggregation and thus improve its crystal quality (Fig. S1B). Superimposition of the OrfX3-bound WT OrfX1 and the standalone OrfX1C31S gives a RMSD of ~0.293 Å over 111 aligned Cα atom pairs (Fig. 1F). The negligible difference shows the structural rigidity of OrfX1 regardless of interacting with OrfX3.
Interactions between OrfX1 and OrfX3
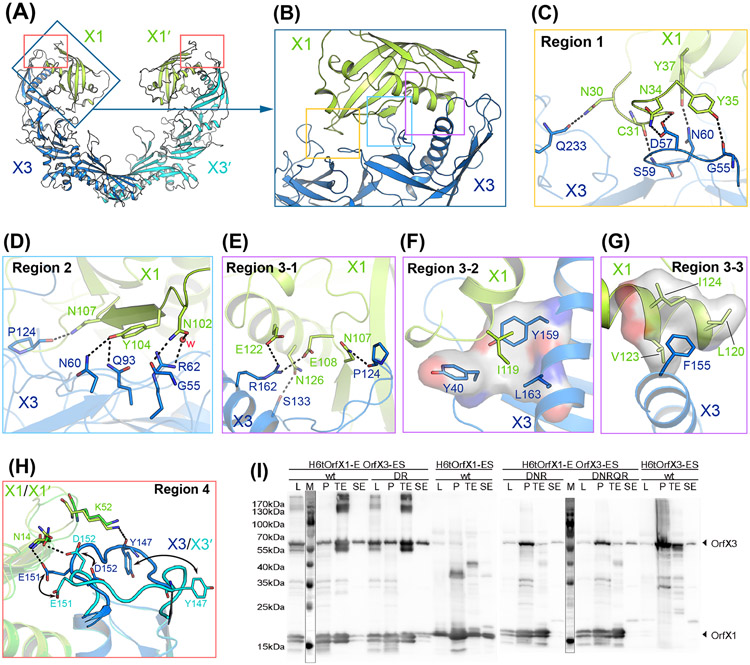
OrfX1 and the OrfX3-N domain, both adopting a TULIP fold, form a face-to-face heterodimer with their long α-helixes facing each other in the middle and the β-sheets on the opposite sides. There is an extensive interacting network at the OrfX1–OrfX3 interface involving both polar and non-polar interactions, which buries a large interface area of ~1,380 Å2 in average (Fig. 2A). The interactions between OrfX1 and OrfX3 can be grouped into four major areas (Fig. 2A-B). In region 1, OrfX1 residues C31, N34, Y35 and Y37 located on a loop and a neighboring β-strand interact with OrfX3 residues S59, D57, G55, and N60, respectively, through hydrogen-bonding interactions. In addition, the side chain of OrfX1 N30 forms a hydrogen bond with Q233 of OrfX3 that is located on a loop in the central β-sheet region (Fig. 2C). In region 2, OrfX1 N102 interacts with both G55 and R62 in OrfX3 via direct and water-mediated hydrogen-bonding interactions, respectively; Y104 of OrfX1 forms hydrogen bond with OrfX3 N60 and Q93; the side chain of OrfX1 N107 interacts with the backbone carbonyl group of OrfX3 P124 through a hydrogen-bonding interaction (Fig. 2D). In region 3, the long amphiphilic α-helixes of OrfX1 and OrfX3-N interact with each other through both polar and hydrophobic interactions. For example, OrfX3 S133 forms a hydrogen bond with OrfX1 N126, while OrfX3 R162 forms salt bridges with OrfX1 E122 and E108 (Fig. 2E). On the hydrophobic side of the helixes, the side chain of OrfX1 I119 inserts into a pocket formed by Y40, Y159 and L163 of OrfX3 (Fig. 2F), while F155 of OrfX3 sticks into a shallow groove formed by L120, V123 and I124 of OrfX1 (Fig. 2G). The large interface between OrfX1 and OrfX3 involving extensive interacting residues indicates a strong binding affinity between them, permitting a successful co-purification of the complex. All residues involved in interactions are summarized in Table S2.
Figure 2. Interactions between OrfX1 and OrfX3.
(A) Overall structure of the OrfX1–OrfX3 complex. (B) Close-up view of the OrfX1–OrfX3 heterodimer interface with the three major interacting areas highlighted in boxes. (C-G) Stick representations of key interacting residues at the OrfX1–OrfX3 interface. Hydrogen bonds are shown as dashed lines and hydrophobic interactions are shown as surface models. (H) The E144–D153 loop in the two OrfX3 molecules in the context of the OrfX1–OrfX3 complex, colored blue and cyan, displays different conformations. (I) Co-expression analysis of the WT H6tOrfX1-ES, WT H6tOrfX3-ES, H6tOrfX1-OrfX3-ES WT complex and the respective OrfX3 mutants D152R/R162A (DR), D57A/N60A/R62A (DNR), and D57A/N60A/R62A/Q93A/R162A (DNRQR) (n=2-4). L, clear lysate; P, cell pellet after lysis; TE, eluate from Co2+-Talon matrix; SE, eluate from StrepTactin matrix. Samples were analyzed by immunoblot using anti-OrfX1-3 specific polyclonal rabbit IgG KOrf154. The corresponding SDS-PAGE and Coomassie staining result is shown in Fig. S2.
It is noteworthy that the two pairs of the OrfX1–OrfX3 complexes in the 2:2 heterotetrameric complex in this crystal form are independent of each other as they are not restricted by crystallographic symmetry. When comparing these two independent pairs of the OrfX1–OrfX3 complexes, we found them largely identical and all interactions in regions 1–3 described above are conserved. However, we noticed that a loop in OrfX3-N (E144–D153) that could adopt two different conformations to interact with OrfX1 (region 4). In one conformation, this loop of OrfX3-N turns towards OrfX1 and establishes two pairs of hydrogen bonds between E151 and Y147 of OrfX3 and N14 and K52 of OrfX1, respectively. In another conformation, this loop of OrfX3-N flips away from OrfX1 and only forms one hydrogen bond between its D152 and N14 of OrfX1 (Fig. 2H). The different conformations of this OrfX3 loop observed here are likely caused by crystal packing, while also indicating its structural flexibility.
To validate our structural observations, we selected some key residues located at the OrfX1–OrfX3 interface for site-directed mutagenesis studies. Since the expression level of the standalone OrfX3 is very low and it can be drastically improved by co-expression of its binding partner OrfX1, we hypothesized that mutations in OrfX3 that disrupt the OrfX1–OrfX3 interaction would lead to a poor expression of the mutated OrfX3 even when OrfX1 is co-expressed, because OrfX1 could no longer help. We then designed three mutants of OrfX3: D57A/N60A/R62A (DNR) mutant with residues in interface regions 1 and 2; D57A/N60A/R62A/Q93A/R162A (DNRQR) mutant that includes the aforementioned 3 residues, Q93A in region 2, and R162A in region 3; D152R/R162A (DR) mutant with residues in region 3 and 4. We co-expressed these three OrfX3 mutants with the wild-type OrfX1, respectively, which were then purified in parallel either with Co2+-Talon affinity purification targeting the His-tagged OrfX1 or StrepTactin resins targeting the Strep-tagged OrfX3. Protein samples were examined by SDS-PAGE Coomassie staining and Western blotting using a polyclonal antibody targeting both OrfX1 and OrfX3 (Fig. S2, S3A-D, and Fig. 2I). For the DNR and DNRQR mutants of OrfX3, the Co2+-Talon-purified samples were further subjected to StrepTactin purification (Fig. S3G). Using this assay, we found that the expression of the soluble folded DNR and DNRQR mutants of OrfX3 decreased significantly in comparison with the wild-type protein, suggesting that the interaction between OrfX1 and OrX3 was abolished by the mutations. Nevertheless, the DR mutant of OrfX3 showed a WT-like expression and SEC profile (Fig. S3A-B and S3E-F), suggesting mutating these two residues were not sufficient to disrupt the interactions.
Interactions between the homodimeric OrfX3
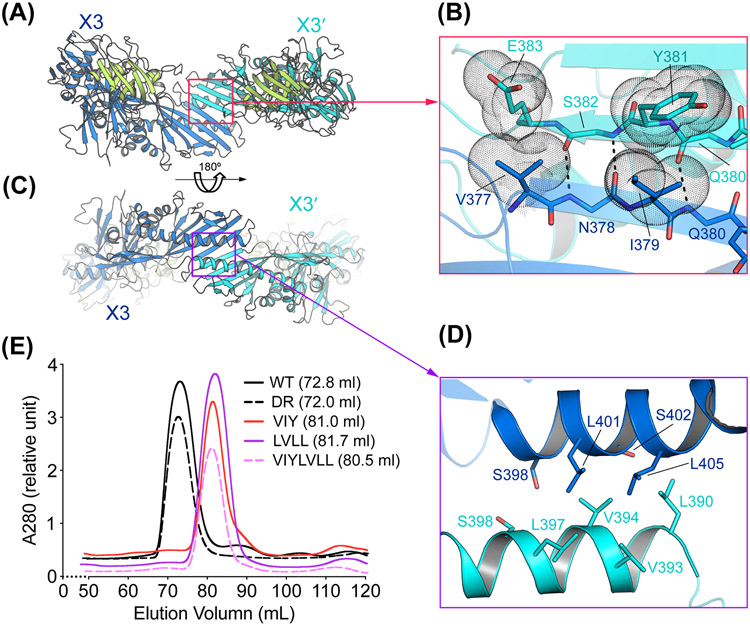
The OrfX3 homodimer interface in the OrfX1–OrfX3 complex is formed between their TULIP-like OrfX3-C domains burying ~666 Å2 interface area. More specifically, two OrfX3 use one open edge of the TUPLIP β-sheet and its central α-helix in OrfX3-C domain to stick to each other, generating an interface with a ~180° rotational symmetry (Fig. 3A, C, and Table S3). The antiparallel edge-to-edge interactions between the two equivalent β-strands, each from one OrfX3, assemble into a continuous 12-stranded β-sheet across two OrfX3-C domains. Interactions in this area are dominated by backbone–backbone hydrogen bonds involving residues N378, Q380 and S382 in each OrfX3 in an antiparallel manner, which are further strengthened by Van der Waals interactions between V377 and I379 in one OrfX3, and E383 and Y381 in another OrfX3, respectively, and vice versa (Fig. 3B). The second OrfX3 homodimeric interface is formed between the long α-helix from each OrfX3-C domain, which is stabilized by mostly hydrophobic interactions involving S398, L401, S402, and L405 from one OrfX3 and L390, V393, V394, L397 and S398 from the other OrfX3 unit (Fig. 3D).
Figure 3. Interactions between the homodimeric OrfX3 in the OrfX1–OrfX3 complex.
(A, C) Two different views of the OrfX1–OrfX3 complex focusing on the OrfX3 dimer interface. (B, D) Close-up views of key interacting residues around the antiparallel edge-to-edge β-strand interface between the two OrfX3-C units (B) and the interface between the long α-helix from each OrfX3-C domain (D). Hydrogen bonds are shown as dashed lines. Residues V377, I379, Y381 that were subjected for mutagenesis study and E383 are show as dotted spheres. Please note that another set of homodimeric OrfX3 interactions related to that shown in panels B and D by a ~180° rotational symmetry are not shown for clear illustration. (E) The WT (n=3) and four mutant OrfX1–OrfX3 complexes (n=1) were analyzed by size exclusion chromatography using a Superdex-200 16/60PG column with peak retention volumes indicated.
We carried out structure-based mutagenesis to validate the homodimerization of OrfX3 in solution. We designed three OrfX3 mutants: V377E/I379E/Y381S (VIY) that carries mutations at the antiparallel β-sheet interface, L390E/V394D/L401E/L405E (LVLL) that carries mutations at the helix–helix interface, and VIY/LVLL that was a combination of the former two. We envisioned that if these mutations in OrfX3 could disrupt its homodimerization, they would lead to an OrfX1–OrfX3 heterodimer (~71 kDa), which could be easily distinguished from the WT OrfX1–OrfX3 heterotetrameric complex (~142 kDa) using size-exclusion chromatography. Again, these three OrfX3 mutants were co-expressed with the wild-type OrfX1, respectively, which were purified employing first IMAC targeting the His-tagged OrfX1, subsequent proteolytic removal of the N-terminal His tag and SEC. Protein samples were examined by SDS-PAGE Coomassie staining (Fig. S4). Using this assay, we found that all three mutants of OrfX3 showed WT-like or even better expression levels, suggesting that these mutations did not disrupt the structural integrity of OrfX3, and might even improve its solubility. The use of a Superdex-200 16/60PG column allowed to analyze for OrfX3 homodimerization, where the WT OrfX1–OrfX3 complex was eluted at ~72.8 ml (Fig. 3E and Fig. S3E). As a negative control, the above described OrfX3-DR mutant that carried mutations outside the OrfX3 dimer interface was also eluted around 72.0 ml in the presence of OrfX1 (Fig. 3E and Fig. S3F). SEC analysis of OrfX3 VIY, LVLL, and VIY/LVLL mutants co-expressed with the WT OrfX1 showed that the OrfX1–OrfX3 complexes carrying these three OrfX3 mutants were eluted around 80.5–81.7 ml, suggesting that these mutations disrupted the assembly of tetrameric complexes (Fig. 3E and Fig. S4D-F). Similar results were observed when using a Superdex-75 16/60PG column, where the WT OrfX1–OrfX3 complex eluted at 50.2 ml and the OrfX1- bound OrfX3-VIY/LVLL mutant at 55.8 ml. These data nicely confirmed the homodimerization of OrfX3 in solution.
Crystal structure of OrfX2 of BoNT/E
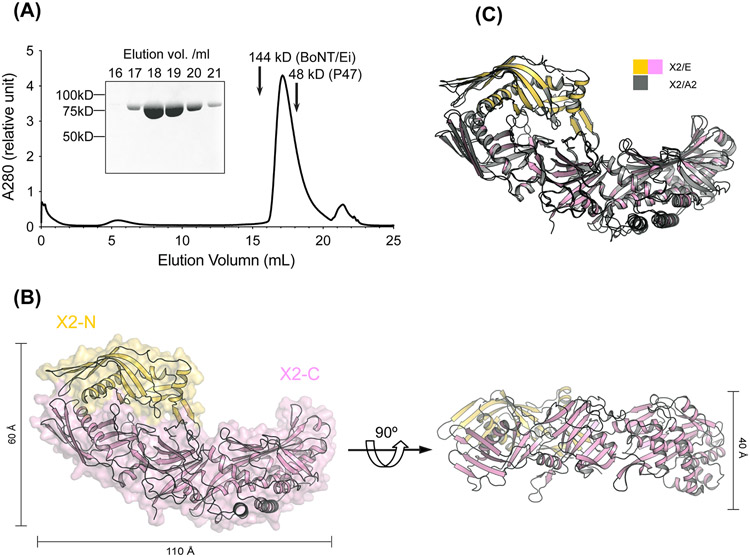
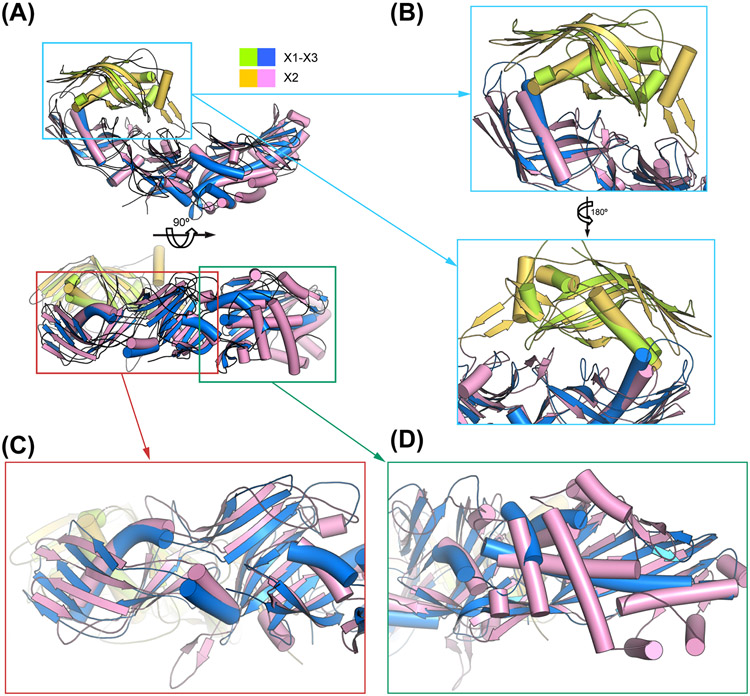
OrfX2 exists predominantly as a homogeneous monomer as examined by SEC and SDS-PAGE analysis (Fig. 4A). Nevertheless, we could observe a small amount of dimeric OrfX2 during protein purification, which could be minimized by including reducing agent in the buffer. We determined the crystal structure of OrfX2 at 1.85 Å resolution using experimental phases derived from the SeMet substituted OrfX2 crystals using the SAD method before the structure of OrfX2 of BoNT/A2 (PDB: 6EKV) was available [45]. The overall structure of OrfX2 reveals an elongated shape with a dimension of ~110 Å × 60 Å × 40 Å, which comprises an N-terminal domain (residues 1-167, OrfX2-N) and a C-terminal domain (residues 168-748, OrfX2-C) (Fig. 4B). Both of these domains consist of TULIP-like folds, where OrfX2-N adopts a single TULIP fold and OrfX2-C contains two TULIP-like domains linked by a central β-sheet that shares a similar topology with P47 and OrfX3. The OrfX2-N domain and the N-terminal TULIP module of OrfX2-C assemble in a face-to-face manner with their long α-helixes facing each other burying a large contacting surface of ~1,440 Å2. The overall structure of OrfX2 of BoNT/E shows a high structural resemblance with the previously reported structure of OrfX2 of BoNT/A2 (PDB: 6EKV) [45]. Superimposition of those two structures yielded ~1.134 Å RMSD over 624 Cα atoms (Fig. 4C), despite they only share ~50.2% sequence identity (Fig. S5). The high structural similarity of OrfX2 produced in the E1 and A2 strains suggests common functional features across different strains.
Figure 4. Crystal structure of BoNT/E OrfX2.
(A) SEC analysis of the recombinant OrfX2 (n=3, a representative result is shown). Purity of the peak fractions is examined by SDS-PAGE. The peak positions of two reference proteins with molecular weights of 144 kDa and 48 kDa are indicated by arrows. (B) Ribbon and surface representations of OrfX2 in two different views. The OrfX2-N domain is colored yellow and the OrfX2-C domain in pink. (C) Structure alignment of OrfX2 of BoNT/E (shown in yellow and pink) and a previously reported OrfX2 of BoNT/A2 (PDB: 6EKV, shown in grey).
Structural similarities between the OrfX1–OrfX3 complex and OrfX2
Unexpectedly, we noticed that the overall structure of OrfX2 is similar to the OrfX1–OrfX3 complex, with OrfX2-N mimicking OrfX1 and OrfX2-C mimicking OrfX3 (Fig. 5A). Both OrfX1 and OrfX2-N contains one TULIP-like domain with the two displaying ~5.58 Å RMSD over 74 Cα pairs, while OrfX3 and OrfX2-C share a similar P47-like fold with ~3.206 Å RMSD over 303 Cα pairs (Fig. 5A). Structural comparison reveals that the way OrfX2-N interacts with the N-terminal TULIP-like domain of OrfX2-C is surprisingly similar to how OrfX1 engages the N-terminal TULIP-like domain of OrfX3 (Fig. 5B). Another intriguing finding is that the TULIP-like OrfX3-N domain highly resembles the N-terminal TULIP-like domain of OrfX2-C with ~1.786 Å RMSD over 130 Cα pairs (Fig. 5C), while the structures of the two C-terminal TULIP-like domains in OrfX3-C and OrfX2-C are more diverse (Fig. 5D). This may explain why OrfX3 could form a homodimer with its C-terminal TULIP-like domain, but OrfX2-C does not.
Figure 5. Structural comparison between the OrfX1–OrfX3 complex and OrfX2.
(A) Superimposition of the OrfX1 (lime green)–OrfX3 (blue) complex and OrfX2 (yellow and pink). (B) Close-up views of the superimposed OrfX1–OrfX3 interface and OrfX2-N–OrfX2-C interface. (C-D) Close-up views of the superimposed OrfX3 and OrfX2-C focusing on the N-terminal TULIP-like domain and the connecting central β-sheet (C) and the C-terminal TULIP-like domain (D).
Discussion
In this work, we report the crystal structures of OrfX1, OrfX2, and the OrfX1–OrfX3 complex, which are encoded in the orfX gene cluster of BoNT/E1 producing C. botulinum strain Beluga. This strain along with other C. botulinum Group II type E strains are associated with foodborne botulism with a particularly high risk linked to consumption of aquatic food in the northern hemisphere. Together with our previously reported structure of P47 (PDB code 5WIX) [60] and the structure of the M-PTC of BoNT/E (PDB code 4ZKT) [24], the structures of all the NAPs encoded in the orfX gene cluster with BoNT/E have been determined. Interestingly, structural analyses show that all three OrfX proteins and P47 contain the TULIP domain topology, which is common to proteins in the TULIP superfamily [47,48]. More specifically, OrfX1 and the OrfX2-N domain consist of a single TULIP module, while P47, OrfX3, and the OrfX2-C domain display a common P47 fold that is composed of two TULIP folds connected by a central β-sheet. Moreover, the P47 fold topology is similar to members in the bactericidal/permeability-increasing (BPI)-like protein family [46,61-63]. BPI-like proteins are known to be able to bind lipids, and prior studies also suggested that OrfX1, OrfX2, and P47 could interact with lipids [45,46]. Although the details of OrfX/P47-lipid interactions remain unknown, these findings suggest possible roles of these OrfX proteins and/or P47 in facilitating interactions of the PTC with host and/or bacterial membrane during oral intoxication.
Our structural findings also raise another intriguing hypothesis that the OrfX proteins and P47 may use the TULIP fold as building blocks to interact with each other, NTNHA, and/or BoNT to form a larger PTC, since the TULIP domain could also mediate protein-protein interactions to form higher-order complexes [64]. In fact, this has been supported by the observations that OrfX1 and OrfX3 form a TULIP-domain-mediated complex, OrfX3 forms a homodimer with its C-terminal TULIP domain, and the two TULIP modules in OrfX2-N and OrfX2-C assemble with each other. It is worth noting that the TULIP-domain-mediated interacting mode between OrfX1 and OrfX3 and that between OrfX2-N and OrfX2-C are unexpectedly similar. However, we have yet to observe interactions among OrfX2, the OrfX1–OrfX3 complex, P47, and the M-PTC in solution. Therefore, the structural basis underlying the assembly of the OrfX type PTC of BoNTs still remains elusive and awaits future investigation.
Note:
After completion of this work and during the final preparation of the manuscript for submission, a paper by Košenina et. al. was published [65]. Our report confirms aspects of their results on the OrfX1–OrfX3 complex from Paraclostridial mosquitocidal protein 1 (PMP1) gene cluster. Our manuscript comprises the structures of OrfX1, OrfX2, and the OrfX1–OrfX3 complex that are encoded in the orfX gene cluster of the BoNT/E1-producing Clostridium botulinum strain Beluga, as well as site-directed mutagenesis studies validating the structural finding regarding the OrfX1–OrfX3 and the homodimeric OrfX3 interfaces.
Supplementary Material
Figure S1. The locations of the K244A/K245A mutations on the OrfX1–OrfX3 complex and the C31S mutation on OrfX1.
Figure S2. Validation of interactions between OrfX1 and OrfX3 in the OrfX1–OrfX3 complex.
Figure S3. Validation of interactions between OrfX1 and OrfX3 in the OrfX1–OrfX3 complex.
Figure S4. Validation of interactions between the homodimeric OrfX3 in the OrfX1–OrfX3 complex.
Figure S5. Sequence alignment between BoNT/E1 OrfX2 and BoNT/A2 OrfX2.
Table S1. Data collection and refinement statistics.
Table S2. Interacting residues at the OrfX1–OrfX3 heterodimer interface.
Table S3. Interacting residues at the OrfX3 homodimer interface in the OrfX1–OrfX3 complex.
Acknowledgements
This work was partly supported by National Institute of Allergy and Infectious Diseases (NIAID) grants R01AI125704 and R01AI091823 to R.J., by the Swiss Federal Office for Civil Protection BABS #353005630 to A.R., by the Deutsche Forschungsgemeinschaft (DFG Exzellenzinitiative GSC 108) to J.L., and by the European Research Council (ERC-CoG 683099) to M.L. This work is based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on 24-ID-E is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Abbreviations
- BoNT
botulinum neurotoxin
- HA
hemagglutinin
- PTC
progenitor toxin complexes
- L-PTC
large PTC
- M-PTC
minimal PTC
- NAPs
neurotoxin-associated proteins
- NGC
neurotoxin gene cluster
- NTNHA
non-toxic non-hemagglutinin
- orf
open reading frame
- PMP1
Paraclostridial mosquitocidal protein 1
- TULIP
tubular lipid-binding
- RMSD
root-mean-square deviation
Footnotes
Conflict of interest
The authors report no conflict of interest
Data accessibility
The coordinates and structure factors for the OrfX1–OrfX3K244A/K245A complex, SeMet-OrfX1C31S, and SeMet-OrfX2 have been deposited in the Protein Data Bank under accession codes 8FBD, 8FBE, and 8FBF, respectively. All other relevant data are within the manuscript and the Supplemental information.
References
- [1].Hatheway CL (1990). Toxigenic clostridia. Clin Microbiol Rev 3, 66–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Montecucco C and Schiavo G (1995). Structure and function of tetanus and botulinum neurotoxins. Q Rev Biophys 28, 423–72. [DOI] [PubMed] [Google Scholar]
- [3].Sugiyama H (1980). Clostridium botulinum neurotoxin. Microbiol Rev 44, 419–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rossetto O, Pirazzini M and Montecucco C (2014). Botulinum neurotoxins: genetic, structural and mechanistic insights. Nat Rev Microbiol 12, 535–49. [DOI] [PubMed] [Google Scholar]
- [5].Brunt J, Carter AT, Stringer SC and Peck MW (2018). Identification of a novel botulinum neurotoxin gene cluster in Enterococcus. FEBS letters 592, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Contreras E et al. (2019). A neurotoxin that specifically targets Anopheles mosquitoes. Nat Commun 10, 2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dover N, Barash JR, Hill KK, Xie G and Arnon SS (2014). Molecular characterization of a novel botulinum neurotoxin type H gene. J Infect Dis 209, 192–202. [DOI] [PubMed] [Google Scholar]
- [8].Lam KH, Sikorra S, Weisemann J, Maatsch H, Perry K, Rummel A, Binz T and Jin R (2018). Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA. Pathogens and Disease 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yao G, Lam KH, Perry K, Weisemann J, Rummel A and Jin R (2017). Crystal Structure of the Receptor-Binding Domain of Botulinum Neurotoxin Type HA, Also Known as Type FA or H. Toxins (Basel) 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang S et al. (2018). Identification of a Botulinum Neurotoxin-like Toxin in a Commensal Strain of Enterococcus faecium. Cell host & microbe 23, 169–176.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang S et al. (2017). Identification and characterization of a novel botulinum neurotoxin. Nat Commun 8, 14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bigalke H (2013). Botulinum toxin: application, safety, and limitations. Curr Top Microbiol Immunol 364, 307–17. [DOI] [PubMed] [Google Scholar]
- [13].Pons L, Vilain C, Volteau M and Picaut P (2019). Safety and pharmacodynamics of a novel recombinant botulinum toxin E (rBoNT-E): Results of a phase 1 study in healthy male subjects compared with abobotulinumtoxinA (Dysport(R)). J Neurol Sci 407, 116516. [DOI] [PubMed] [Google Scholar]
- [14].Yoelin SG, Dhawan SS, Vitarella D, Ahmad W, Hasan F and Abushakra S (2018). Safety and Efficacy of EB-001, a Novel Type E Botulinum Toxin, in Subjects with Glabellar Frown Lines: Results of a Phase 2, Randomized, Placebo-Controlled, Ascending-Dose Study. Plast Reconstr Surg 142, 847e–855e. [DOI] [PubMed] [Google Scholar]
- [15].Lindstrom M and Korkeala H (2006). Laboratory diagnostics of botulism. Clin Microbiol Rev 19, 298–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rasetti-Escargueil C, Lemichez E and Popoff MR (2020). Toxemia in Human Naturally Acquired Botulism. Toxins (Basel) 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rasetti-Escargueil C, Lemichez E and Popoff MR (2020). Human Botulism in France, 1875–2016. Toxins (Basel) 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gu S et al. (2012). Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science 335, 977–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hill KK et al. (2009). Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hill KK and Smith TJ (2013). Genetic Diversity Within Clostridium botulinum Serotypes, Botulinum Neurotoxin Gene Clusters and Toxin Subtypes. Curr Top Microbiol Immunol 364, 1–20. [DOI] [PubMed] [Google Scholar]
- [21].Gu S and Jin R (2013). Assembly and function of the botulinum neurotoxin progenitor complex. Curr Top Microbiol Immunol 364, 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].East AK and Collins MD (1994). Conserved structure of genes encoding components of botulinum neurotoxin complex M and the sequence of the gene coding for the nontoxic component in nonproteolytic Clostridium botulinum type F. Curr Microbiol 29, 69–77. [DOI] [PubMed] [Google Scholar]
- [23].Lam KH and Jin R (2015). Architecture of the botulinum neurotoxin complex: a molecular machine for protection and delivery. Curr Opin Struct Biol 31, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eswaramoorthy S, Sun J, Li H, Singh BR and Swaminathan S (2015). Molecular assembly of clostridium botulinum progenitor M complex of type e. Scientific Reports 5, 17795–17795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].East AK, Bhandari M, Hielm S and Collins MD (1998). Analysis of the botulinum neurotoxin type F gene clusters in proteolytic and nonproteolytic Clostridium botulinum and Clostridium barati. Curr Microbiol 37, 262–8. [DOI] [PubMed] [Google Scholar]
- [26].Fujii N, Kimura K, Yokosawa N, Yashiki T, Tsuzuki K and Oguma K (1993). The complete nucleotide sequence of the gene encoding the nontoxic component of Clostridium botulinum type E progenitor toxin. J Gen Microbiol 139, 79–86. [DOI] [PubMed] [Google Scholar]
- [27].Benefield DA, Dessain SK, Shine N, Ohi MD and Lacy DB (2013). Molecular assembly of botulinum neurotoxin progenitor complexes. Proc Natl Acad Sci U S A 110, 5630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee K et al. (2013). Structure of a bimodular botulinum neurotoxin complex provides insights into its oral toxicity. PLoS Pathog 9, e1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Amatsu S, Sugawara Y, Matsumura T, Kitadokoro K and Fujinaga Y (2013). Crystal structure of Clostridium botulinum whole hemagglutinin reveals a huge triskelion-shaped molecular complex. J Biol Chem 288, 35617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sakaguchi Y et al. (2001). Clostridium botulinum type A haemagglutinin-positive progenitor toxin (HA+-PTX) binds to oligosaccharides containing Galβ1–4GlcNAc through one subcomponent of haemagglutinin (HA1). Microbiology 147, 811–819. [DOI] [PubMed] [Google Scholar]
- [31].Fujinaga Y, Inoue K, Watanabe S, Yokota K, Hirai Y, Nagamachi E and Oguma K (1997). The haemagglutinin of Clostridium botulinum type C progenitor toxin plays an essential role in binding of toxin to the epithelial cells of guinea pig small intestine, leading to the efficient absorption of the toxin. Microbiology 143 ( Pt 12), 3841–7. [DOI] [PubMed] [Google Scholar]
- [32].Fujinaga Y et al. (2004). Molecular characterization of binding subcomponents of Clostridium botulinum type C progenitor toxin for intestinal epithelial cells and erythrocytes. Microbiology 150, 1529–38. [DOI] [PubMed] [Google Scholar]
- [33].Kojima S, Eguchi H, Ookawara T, Fujiwara N, Yasuda J, Nakagawa K, Yamamura T and Suzuki K (2005). Clostridium botulinum type A progenitor toxin binds to Intestine-407 cells via N-acetyllactosamine moiety. Biochem Biophys Res Commun 331, 571–6. [DOI] [PubMed] [Google Scholar]
- [34].Lee K, Lam KH, Kruel AM, Perry K, Rummel A and Jin R (2014). High-resolution crystal structure of HA33 of botulinum neurotoxin type B progenitor toxin complex. Biochem Biophys Res Commun 446, 568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nakamura T, Takada N, Tonozuka T, Sakano Y, Oguma K and Nishikawa A (2007). Binding properties of Clostridium botulinum type C progenitor toxin to mucins. Biochim Biophys Acta 1770, 551–5. [DOI] [PubMed] [Google Scholar]
- [36].Lee K et al. (2014). Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex. Science 344, 1405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Matsumura T, Jin Y, Kabumoto Y, Takegahara Y, Oguma K, Lencer WI and Fujinaga Y (2008). The HA proteins of botulinum toxin disrupt intestinal epithelial intercellular junctions to increase toxin absorption. Cell Microbiol 10, 355–64. [DOI] [PubMed] [Google Scholar]
- [38].Sugawara Y, Yutani M, Amatsu S, Matsumura T and Fujinaga Y (2014). Functional Dissection of the Clostridium botulinum Type B Hemagglutinin Complex: Identification of the Carbohydrate and E-Cadherin Binding Sites. PLoS One 9, e111170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sugawara Y, Matsumura T, Takegahara Y, Jin Y, Tsukasaki Y, Takeichi M and Fujinaga Y (2010). Botulinum hemagglutinin disrupts the intercellular epithelial barrier by directly binding E-cadherin. J Cell Biol 189, 691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sagane Y et al. (2012). Small-angle X-ray scattering reveals structural dynamics of the botulinum neurotoxin associating protein, nontoxic nonhemagglutinin. Biochem Biophys Res Commun 425, 256–60. [DOI] [PubMed] [Google Scholar]
- [41].Chen Y, Korkeala H, Linden J and Lindstrom M (2008). Quantitative real-time reverse transcription-PCR analysis reveals stable and prolonged neurotoxin cluster gene activity in a Clostridium botulinum type E strain at refrigeration temperature. Appl Environ Microbiol 74, 6132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bradshaw M, Dineen SS, Maks ND and Johnson EA (2004). Regulation of neurotoxin complex expression in Clostridium botulinum strains 62A, Hall A-hyper, and NCTC 2916. Anaerobe 10, 321–33. [DOI] [PubMed] [Google Scholar]
- [43].Kalb SR, Baudys J, Smith TJ, Smith LA and Barr JR (2017). Characterization of Hemagglutinin Negative Botulinum Progenitor Toxins. Toxins (Basel) 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nowakowska MB, Douillard FP and Lindstrom M (2019). Looking for the X Factor in Bacterial Pathogenesis: Association of orfX-p47 Gene Clusters with Toxin Genes in Clostridial and Non-Clostridial Bacterial Species. Toxins (Basel) 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gustafsson R, Berntsson RP, Martinez-Carranza M, El Tekle G, Odegrip R, Johnson EA and Stenmark P (2017). Crystal structures of OrfX2 and P47 from a Botulinum neurotoxin OrfX-type gene cluster. FEBS Lett 591, 3781–3792. [DOI] [PubMed] [Google Scholar]
- [46].Lam K. h., Qi R, Liu S, Kroh A, Yao G, Perry K, Rummel A and Jin R (2018). The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein. Toxicon 147, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kopec KO, Alva V and Lupas AN (2011). Bioinformatics of the TULIP domain superfamily. Biochem Soc Trans 39, 1033–8. [DOI] [PubMed] [Google Scholar]
- [48].Alva V and Lupas AN (2016). The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim Biophys Acta 1861, 913–923. [DOI] [PubMed] [Google Scholar]
- [49].Goldschmidt L, Cooper DR, Derewenda ZS and Eisenberg D (2007). Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci 16, 1569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL and Clardy J (1993). Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol 229, 105–24. [DOI] [PubMed] [Google Scholar]
- [51].Kabsch W (2010). XDS. Acta Crystallogr D Biol Crystallogr 66, 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Adams PD et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Emsley P and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32. [DOI] [PubMed] [Google Scholar]
- [55].Brunger AT (1992). Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–5. [DOI] [PubMed] [Google Scholar]
- [56].Chen VB et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Madeira F et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50, W276–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Robert X and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Derewenda ZS (2004). Rational protein crystallization by mutational surface engineering. Structure 12, 529–35. [DOI] [PubMed] [Google Scholar]
- [60].Lam KH, Qi R, Liu S, Kroh A, Yao G, Perry K, Rummel A and Jin R (2018). The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein. Toxicon 147, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Beamer LJ, Carroll SF and Eisenberg D (1999). The three-dimensional structure of human bactericidal/permeability-increasing protein: implications for understanding protein-lipopolysaccharide interactions. Biochem Pharmacol 57, 225–9. [DOI] [PubMed] [Google Scholar]
- [62].Beamer LJ (2003). Structure of human BPI (bactericidal/permeability-increasing protein) and implications for related proteins. Biochem Soc Trans 31, 791–4. [DOI] [PubMed] [Google Scholar]
- [63].Lloyd DB, Bonnette P and Thompson JF (2006). Protein fusions of BPI with CETP retain functions inherent to each. Biochemistry 45, 12954–9. [DOI] [PubMed] [Google Scholar]
- [64].AhYoung AP, Jiang J, Zhang J, Khoi Dang X, Loo JA, Zhou ZH and Egea PF (2015). Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci U S A 112, E3179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kosenina S and Stenmark P (2022). Crystal structure of the OrfX1-OrfX3 complex from the PMP1 neurotoxin gene cluster. FEBS Lett [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The locations of the K244A/K245A mutations on the OrfX1–OrfX3 complex and the C31S mutation on OrfX1.
Figure S2. Validation of interactions between OrfX1 and OrfX3 in the OrfX1–OrfX3 complex.
Figure S3. Validation of interactions between OrfX1 and OrfX3 in the OrfX1–OrfX3 complex.
Figure S4. Validation of interactions between the homodimeric OrfX3 in the OrfX1–OrfX3 complex.
Figure S5. Sequence alignment between BoNT/E1 OrfX2 and BoNT/A2 OrfX2.
Table S1. Data collection and refinement statistics.
Table S2. Interacting residues at the OrfX1–OrfX3 heterodimer interface.
Table S3. Interacting residues at the OrfX3 homodimer interface in the OrfX1–OrfX3 complex.
Data Availability Statement
The coordinates and structure factors for the OrfX1–OrfX3K244A/K245A complex, SeMet-OrfX1C31S, and SeMet-OrfX2 have been deposited in the Protein Data Bank under accession codes 8FBD, 8FBE, and 8FBF, respectively. All other relevant data are within the manuscript and the Supplemental information.