Background:
Low density lipoprotein receptor-related protein 11 (LRP11) was involved in the progression of several tumors. However, its role in cervical cancer still remains uncertain.
Methods:
The original tumor data was downloaded from the Cancer Genome Atlas and genotype-tissue expression databases. The expression of LRP11 in normal tissues, tumor tissues and adjacent tissues were evaluated. In addition, we also explored the genetic alteration, prognostic value, and gene function of LRP11. We deeply assessed the interaction between LRP11 and tumor immunity at the pan-cancer level. Finally, research on the association between LRP11 and the resistance of anti-tumor drugs was carried out.
Results:
LRP11 was highly expressed and played a risk prognostic factor in cervical cancer and a variety of tumors. Enrichment analysis revealed that LRP11 was involved in multiple tumor malignant pathways. Our research also pointed out the unique role between LRP11 and tumor immune microenvironment. The tumor immune microenvironment of patients with high expression of LRP11 are lack of most immune cells, indicating a immune desert tumor microenvironment. The final drug resistant analysis suggested that patients with high expression of LRP11 may be related to the resistance of many anti-tumor drugs.
Conclusion:
LRP11 was a potential oncogene and prognostic marker in cervical cancer and pan-cancer. Patients with high LRP11 expression may have immune desert tumor microenvironment.
Keywords: cervical cancer, immune desert tumor microenvironment, LRP11, TCGA
1. Introduction
Cervical cancer is one of the gynecological tumor with high incidence rate and mortality rates worldwide. The most common pathological type of cervical cancer is cervical squamous cell carcinoma (CESC).[1] When patients with CESC are diagnosed and treated in the early stage of the tumor, the chance of survival can be increased and may push the 5-year survival rate to 90%.[2] However, the early symptoms of cervical cancer are not obvious. Seventy percent of cervical cancer patients are in the middle and late stage of the disease when they are diagnosed with cervical cancer, which will be very unfavorable to the treatment of patients. Currently, the diagnosis of cervical squamous intraepithelial lesions and cervical cancer is based on cervical cytology/human papilloma virus (HPV) testing, colposcopy, and cervical biopsy. Cervical cytology and HPV testing are the basic methods of screening, but HPV testing has low specificity, cytology has low sensitivity, and colposcopy is somewhat subjective, easily missing lesions in the cervical canal and affecting the accuracy of diagnosis. Therefore, highly sensitive and specific tests are needed to improve the efficiency of early diagnosis of cervical cancer. In addition, about 31% of cervical cancer recurred after treatment, and most recurrences occurred within 2 years after the first diagnosis.[3,4] The treatment of cervical cancer is very difficult and the prognosis is very poor. The 1-year survival rate is only 8.12%.[5] The prognosis of cervical cancer is closely related to many factors, including tumor size, lymph node metastasis, depth of interstitial infiltration, lymph-vascular space invasion, medical diseases such as diabetes and obesity, and serum biomarkers. Scientific and accurate assessment of these factors and selection of appropriate treatment options can effectively reduce the mortality rate of CESC. Thus, the search for meaningful therapeutic targets and prognostic markers of cervical cancer has attracted more and more attention.
With the deepening of research, the understanding of tumors is constantly updated. Among them, the study on tumor microenvironment (TME), especially tumor immune microenvironment (TIME) has opened a new door for tumor research particularly.[6] The components of TME involve a variety of cells, most of which are immune cells and stromal cells.[7] Although great progress has been made in the research of TME in recent years, its internal cell interaction still needs to be further studied. At the same time, the study of TIME also gave birth to a new treatment method, namely immune checkpoint inhibitor.[8] However, only a small number of patients are sensitive to immune checkpoint inhibitors. Immune desert microenvironment is a kind of immune microenvironment, which is characterized by the extreme lack of all kinds of immune cells in tumor tissues. Patients with immune desert TIME were resistant to immunotherapy.[9,10] Thus, finding new genes regulating immune microenvironment is of great significance to prompt the efficacy of immunotherapy and improve the prognosis of patients.
Low density lipoprotein receptor-related protein 11 (LRP11) is a member of LRP family, which was involved in the progression of several tumors. For example, LRP11 was reported to activate beta-catenin signaling to evelate the expression of PD-L1 in prostate cancer.[11] In cervical cancer, LRP11 plays important roles in proliferation, migration and invasion of patients with high-grade squamous intraepithelial lesions and cervical cancer.[12] However, the role of LRP11 in CESC and pan-cancer, especially the regulation of immune microenvironment, is still completely unknown.
In this study, we comprehensively analyzed the expression, genetic alteration, prognostic value, and gene function of LRP11 in CESC and pan-cancer. We evaluated the relationship between LRP11 and TIME based on multiple databases, aiming to elaborate on the relationship between LRP11 and tumor immunity. Finally, the relationship between LRP11 and tumor drug sensitivity was also explored.
2. Material and methods
2.1. Data collection
The RNA-seq and clinical data of 33 tumors were downloaded from the Cancer Genome Atlas (TCGA) database and genotype-tissue expression (GTEx) database to assess the expression of LRP11. The genetic alteration, including copy number alteration (CNA) and methylation of LRP11 were downloaded from cBioPortal database (http://www.cbioportal.org/). The HALLMARK pathways used in enrichment analysis were downloaded from MsigDB database (http://www.gsea-msigdb.org/gsea/index.jsp).
2.2. Survival analysis of LRP11
We conducted a univariate regression analysis, including overall survival (OS), disease specific survival (DSS), disease free interval (DFI), and progression free interval (PFI) indicators to evaluate the prognostic value of LRP11. For Kaplan–Meier OS analysis, patients were divided into high and low expression groups according to the best cutoff value of LRP11, calculated by R package “survminer” and “suivival,” in various tumors.
2.3. Enrichment analysis
In order to explore the gene function of LRP11 and its biological effects, gene set variation analysis (GSVA) was performed to calculate the score of HALLMARK pathways using R package “GSVA.” Among all cancers, we paid special attention to cervical cancer and conducted further gene set enrichment analysis (GSEA) of LRP11 in cervical cancer using R package “clusterProfiler.”
2.4. TME analysis
In order to explore the correlation of LRP11 with TME, R package “ESTIMATE” was used to calculate the Stromalcore, ImmuneScore, ESTIMATEScore, and TumorPurity of each tumor tissues from TCGA cohort. The correlation of LRP11 with these scores was further analyzed.
2.5. TIME analysis
The immune cell infiltration information of TCGA pan-cancer were obtained from ImmuCellAI database (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/resource) and TIMER2.0 database (http://timer.cistrome.org/). We calculated the correlation between LRP11 expression and infiltration level of indicated immune cells.
2.6. Drug resistance analysis
The Cancer Drug Sensitivity Multi-omics Database (GDSC, https://www.cancerrxgene.org/) is the largest public resource for tumor cell drug sensitivity and tumor treatment genome data. The GDSC2 data of GDSC database was downloaded, which contains 809 cell lines, 192 anticancer compounds, and 135,242 drug response IC50 (half inhibitory concentration) values. We assessed the correlation of LRP11 with IC50 values of each drug.
2.7. Statistical analysis
Student t test was performed for difference between 2 groups. All analyses were performed using R software (version 4.1.1). The results of P < .05 were considered statistically significant.
3. Results
3.1. The expression of LRP11
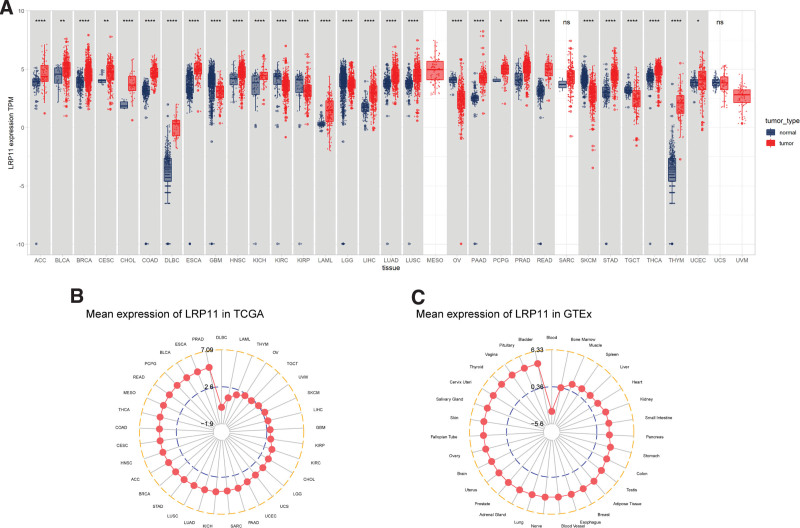
We first evaluated the LRP11 expression in CESC and pan-cancer. Detailed sample sizes and tumor abbreviations were shown in Table S1, Supplemental Digital Content, http://links.lww.com/MD/I625. We found that compared with normal tissues, LRP11 was highly expressed in 22 of 33 tumors, including ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, HNSC, KICH, LAML, LIHC, LUAD, LUSC, PAAD, PCPG, PRAD, READ, STAD, THCA, THYM, and UCEC (Fig. 1A). In tumor tissues from TCGA, LRP11 expression was highest in PRAD and lowest in DLBC (Fig. 1B). In normal tissues from GTEx, LRP11 expression was highest in bladder and lowest in blood (Fig. 1C).
Figure 1.
LRP11 expression. (A) LRP11 expression in pan-cancer. (B) LRP11 expression in TCGA tumor tissues. (C) LRP11 expression in normal tissues of GTEx. GTEx = genotype-tissue expression, LRP11 = low density lipoprotein receptor-related protein 11, TCGA = Cancer Genome Atlas.
3.2. Genetic alteration of LRP11
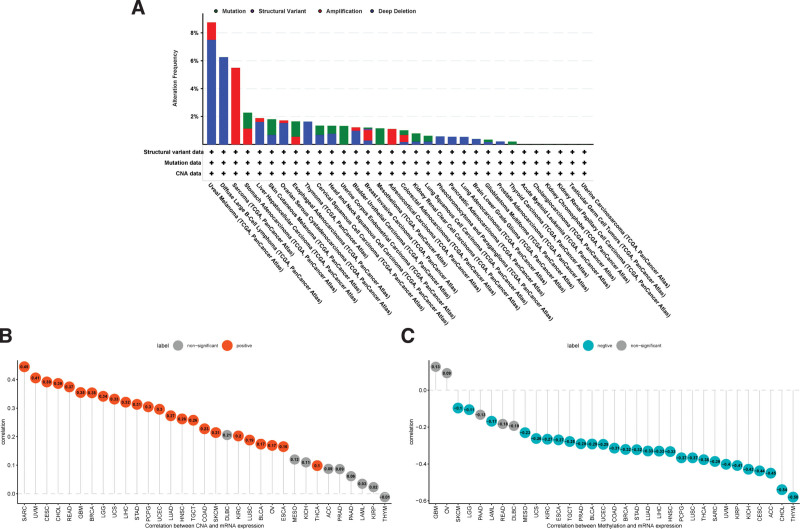
Gene mutation, amplification, methylation, and CNA are often closely related to the mRNA expression. Therefore, we further explored the Genetic alteration of LRP11. The results showed that uveal melanoma have the highest Genetic alteration of LRP11, in which “Deep Deletion” accounts for the largest proportion (Fig. 2A). The frequency of genetic alteration of LRP11 in CESC was less than 2%. In addition, we also found that the CNA was positively correlated with mRNA expression of LRP11 in CESC (r = 0.39, P < .05) and pan-cancer (Fig. 2B). For the methylation analysis, the methylation level was generally negatively associated with mRNA expression of LRP11 in CESC (r = −0.44, P < .05) and pan-cancer (Fig. 2C).
Figure 2.
Genetic alteration of LRP11. (A) Genetic alterations of LRP11 in pan-cancer. (B) Correlation between LRP11 expression and CNA level. (C) Correlation between LRP11 expression and methylation level. CAN = copy number alteration, LRP11 = low density lipoprotein receptor-related protein 11.
3.3. Prognostic value of LRP11
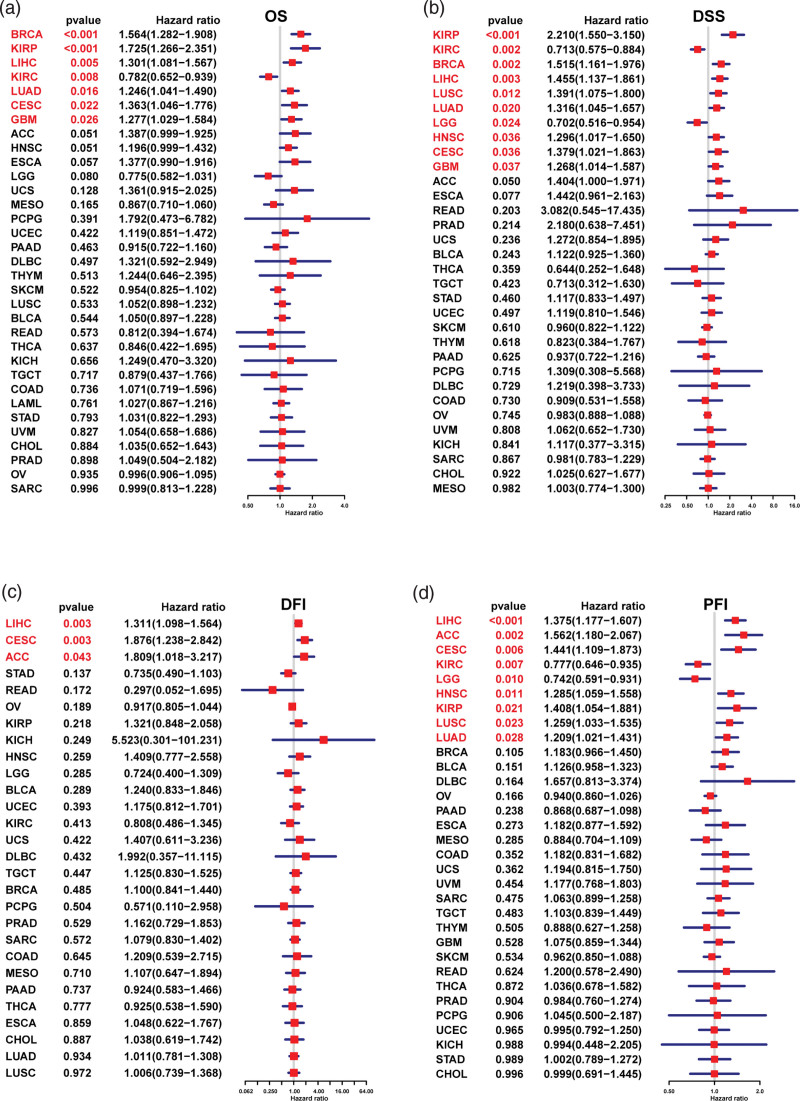
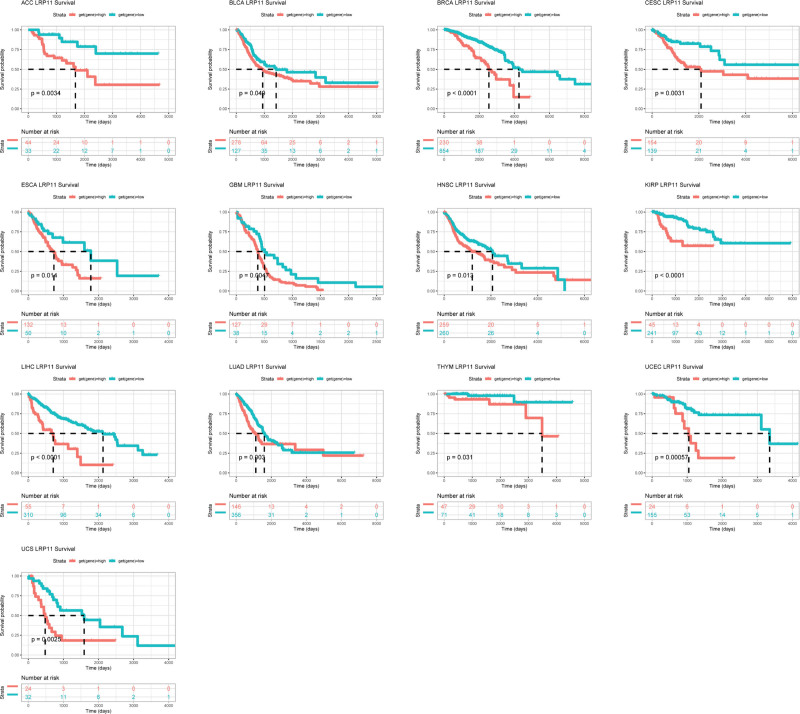
We used forest plots to show the results of univariate regression analysis of LRP11 in pan-cancer. For OS, LRP11 was a risk factor in BRCA, KIRP, LIHC, LUAD, CESC, and GBM, and a protective factor in KIRC (Fig. 3A). For DSS, LRP11 was a risk factor in KIRP, BRCA, LIHC, LUSC, LUAD, HNSC, CESC, and GBM, and a protective factor in KIRC and LGG (Fig. 3B). For DFI, LRP11 was a risk factor in LIHC, CESC, and ACC (Fig. 3C). For PFI, LRP11 was a risk factor in LIHC, ACC, CESC, HNSC, KIRP, LUSC, and LUAD, and a protective factor in KIRC and LGG (Fig. 3D). In short, the above analysis results showed that LRP11 was a risk factor for the OS, DSS, DFI, and PFI of patients with CESC or LIHC. We further conducted Kaplan–Meier OS analysis of LRP11 in pan-cancer. Results indicated that high expression of LRP11 predicted worse survival in ACC, BLCA, BRCA, CESC, ESCA, GBM, HNSC, KIRP, LIHC, LUAD, THYM, UCEC, and UCS (Fig. 4).
Figure 3.
Univariate regression analysis of LRP11. Univariate regression analysis of LRP11 in pan-cancer, the results marked in red were meaningful (P < .05). (A) OS, (B) DSS, (C) DFI, and (D) PFI respectively. DFI = disease free interval, DSS = disease specific survival, LRP11 = low density lipoprotein receptor-related protein 11, OS = overall survival, PFI = progression free interval.
Figure 4.
Kaplan–Meier survival analysis of LRP11. Kaplan–Meier results of LRP11 gene in indicated tumors. The best cutoff value was used to distinguish high and low expression groups. LRP11 = low density lipoprotein receptor-related protein 11.
3.4. Enrichment analysis of LRP11
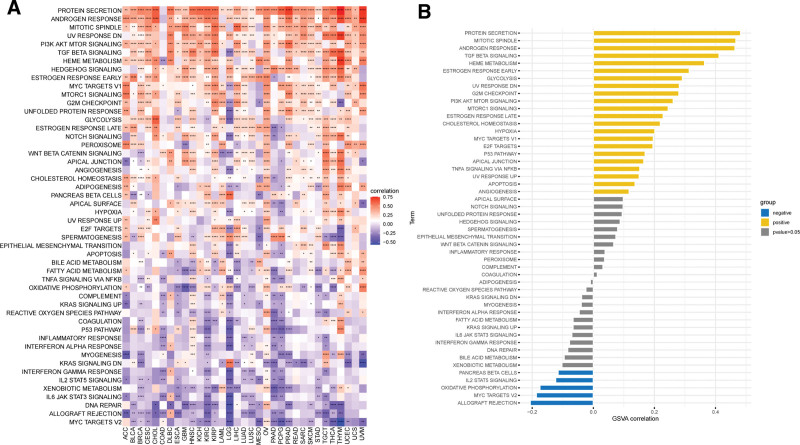
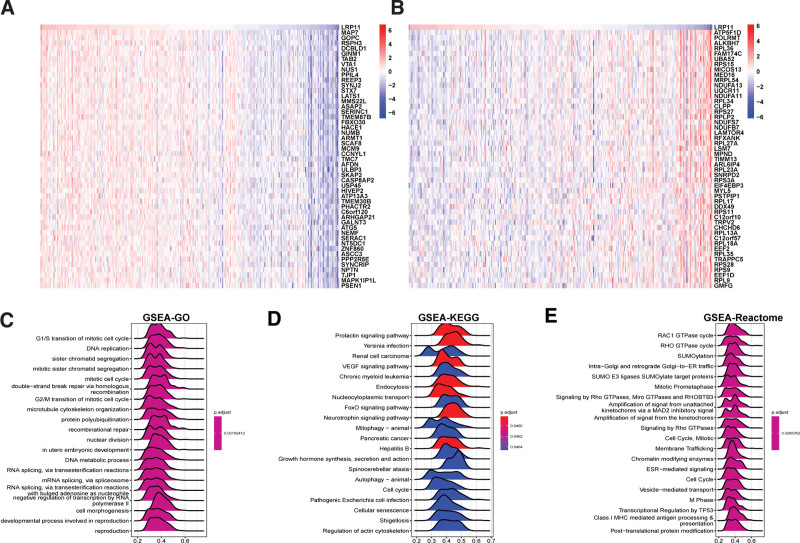
In order to analyze the function of LRP11, we first performed a GSVA in CESC and pan-cancer. Results indicated that LRP11 was positively associated with protein secretion, mitotic spindle, androgen response, transforming growth factor (TGF) beta signaling, hemoglobin metabolism, estrogen response early, and glycolysis pathways in pan-cancer (Fig. 5A) and CESC (Fig. 5B). We also conducted GSEA of LRP11 in CESC. The top 50 genes most positively or negatively correlated with LRP11 expression in CESC were displayed in Figure 6A and B. The GSEA results based on gene ontology revealed that LRP11 was closely associated with DNA replication-related pathways (Fig. 6C). The GSEA results based on Kyoto Encyclopedia of Genes and Genomes revealed that LRP11 was closely associated with Prolactin signaling pathway, vascular endothelial growth factor signaling pathway, Endocytosis, and FoxO signaling pathway (Fig. 6D). The GSEA results based on Reactome revealed that LRP11 was closely associated with RAC1 GTPase cycle, SUMOylation, and Cell Cycle-related pathways (Fig. 6E).
Figure 5.
GSVA of LRP11. Correlation of LRP11 with the score of 50 HALLMARK pathways in pan-cancer (A) and CESC (B). CESC = cervical squamous cell carcinoma, GSVA = gene set variation analysis, LRP11 = low density lipoprotein receptor-related protein 11.
Figure 6.
GSEA of LRP11. (A) Heatmap displayed the expression of top 50 genes with the highest positive correlation with LRP11 in CESC. (B) Heatmap displayed the expression of top 50 genes with the highest negative correlation with LRP11 in CESC. (C–E) Top 20 results of GSEA based on GO (C), KEGG (D), and Reactome (E) in CESC. CESC = cervical squamous cell carcinoma, GO = gene ontology, GSEA = gene set enrichment analysis, KEGG = Kyoto Encyclopedia of Genes and Genomes, LRP11 = low density lipoprotein receptor-related protein 11.
3.5. TME analysis
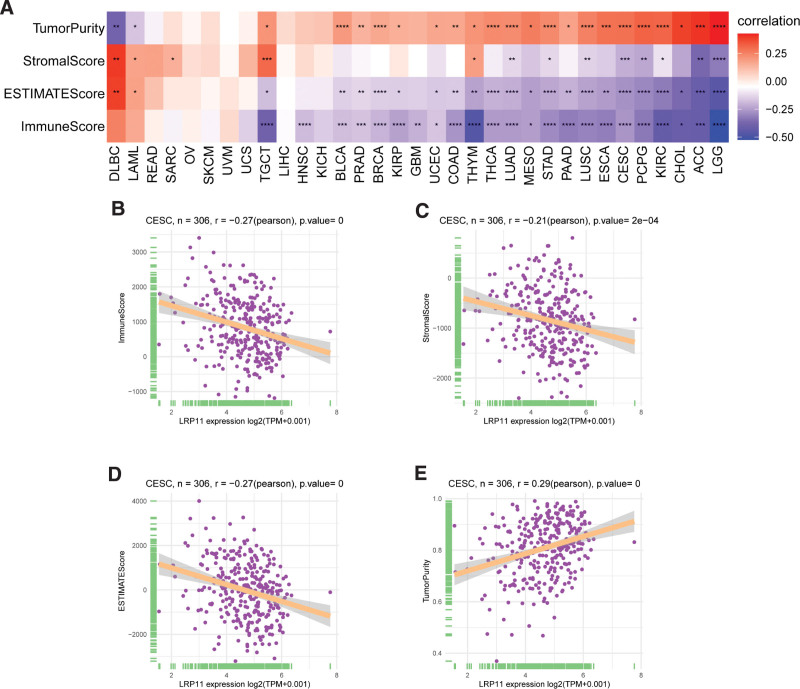
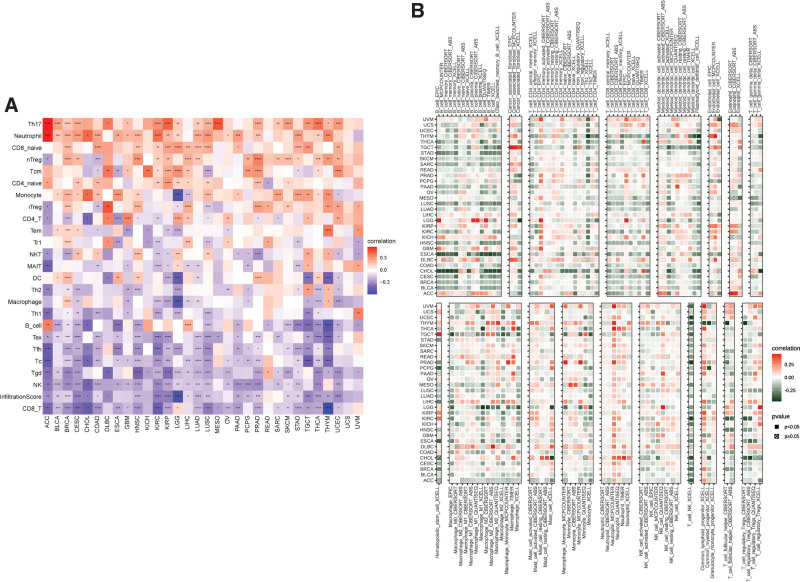
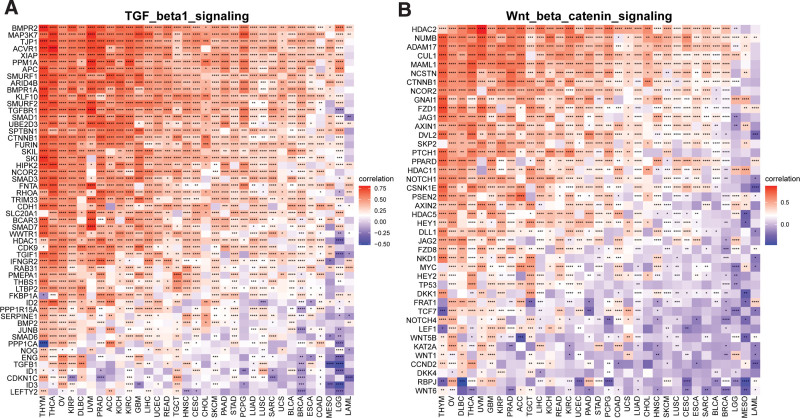
The relationship between LRP11 and ImmuneScore, Stromalcore, ESTIMATEScore and tumor purity in pan-cancer and CESC were explored (Fig. 7A). We found that LRP11 expression was negatively correlated with ImmuneScore, Stromalcore, and ESTIMATEScore, while positively correlated with tumor purity in CESC (Fig. 7B–E). These result indicated that patients with high expression of LRP11 may have immune desert tumor microenvironment. To further prove this result, we downloaded immune cell infiltrate data from ImmuCellAI and TIMER2.0 database and analyzed the relationship of LRP11 with various immune cells. Based on the ImmuCellAI database, we found that at pan-cancer level, the expression of LRP11 is negatively correlated with infiltration level of most immune cells, such as CD8 T cells, NK cells, gamma delta T cells (Fig. 8A). The correlation of immune cell infiltration based on the TIMER2.0 database also illustrated the negative correlation between LRP11 and immune cells in pan-cancer and CESC, including B cells, CD8T cells, and NK cells (Fig. 8B). These results further proved that patients with high LRP11 expression may have immune desert TME. It was reported that the activation of TGF beta1 signaling and Wnt/beta-catenin signaling could induce the immune desert tumor microenvironment.[13,14] We found that LRP11 expression was positively correlated with genes involved in TGF beta1 signaling and Wnt/beta-catenin signaling in pan-cancer and CESC (Fig. 9A and B). The above results all indicated that patients with high expression of LRP11 gene are in a status of immune desert TME, which may be the potential tumor-promoting reason of LRP11.
Figure 7.
ESTIMATE analysis of LRP11. (A) The relationship between LRP11 and Stromalcore, ImmuneScore, ESTIMATEScore and tumor purity in pan-cancer. (B–E) The relationship between LRP11 and Stromalcore, ImmuneScore, ESTIMATEScore and tumor purity in CESC. CESC = cervical squamous cell carcinoma, LRP11 = low density lipoprotein receptor-related protein 11.
Figure 8.
Correlation between LRP11 and immune cell infiltration. (A) Results of immune infiltration analysis based on ImmuCellAI database. (B) Results of immune infiltration analysis based on TIMER2.0 database. LRP11 = low density lipoprotein receptor-related protein 11.
Figure 9.
Correlation between LRP11 and immune-related genes. (A) The correlation of LRP11 with genes involved in TGF beta1 signaling. (B) The correlation of LRP11 with genes involved in Wnt/beta-catenin signaling. LRP11 = low density lipoprotein receptor-related protein 11, TGF = transforming growth factor.
3.6. Drug resistance analysis of LRP11
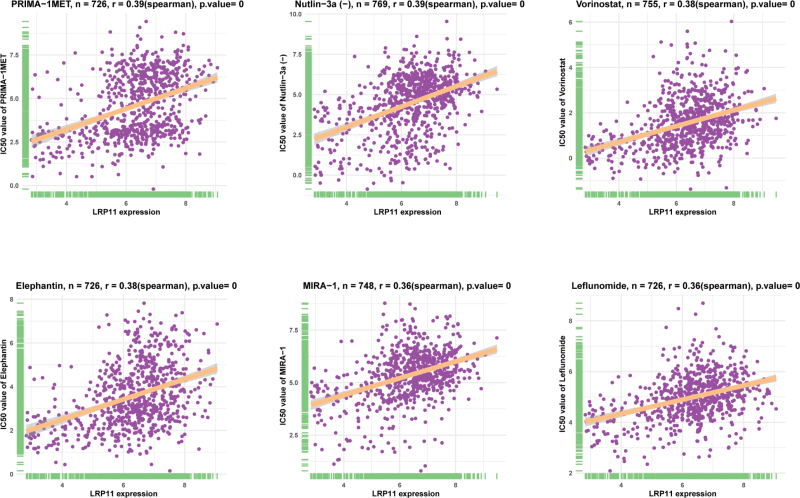
We further explored the effect of LRP11 on sensitivity of anti-tumor drugs. In the tumor drug sensitivity analysis based on the GDSC database, we found that the IC50 values of 159 anti-tumor drugs were positively correlated with LRP11 expression, such as PRIMA-1MET, Nutlin-3a (-), Vorinostat, Elephantin, MIRA-1 and Leflunomide (Fig. 10, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I626). These results suggested that patients with high expression of LRP11 may be resistant to most anti-tumor drugs.
Figure 10.
Correlation of LRP11 with IC50 of indicated anti-tumor drugs. IC50 = half inhibitory concentration, LRP11 = low density lipoprotein receptor-related protein 11.
4. Discussion
The low-density lipoprotein receptor-related protein family is a kind of transmembrane protein, which mainly regulates cholesterol homeostasis through receptor-mediated lipoprotein particle endocytosis.[15] However, more and more experimental evidence shows that members of the gene family have rich functions. Many studies have reported that LRP1 and LRP1B play an important carcinogenic role in COAD, BRCA, THCA, BLCA and KIRC.[16–24] LRP11 is a newly discovered member of LRP family. Fatima’s study found that LRP11 affects lipid metabolism through exosomes, thus promoting the occurrence and development of head and neck cancer.[25] However, the function LRP11 in most tumors is not clear.
In this study, based on the analysis of 33 types of tumor and normal tissues sample data from the TCGA and GTEx databases, we found that LRP11 is highly expressed in 22 of 33 tumors, including ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, HNSC, KICH, LAML, LIHC, LUAD, LUSC, PAAD, PCPG, PRAD, READ, STAD, THCA, THYM, and UCEC, indicating a tumor-promoting role of LRP11. The results of enrichment analysis further illustrated the role of LRP11 in tumorigenesis and development. We found that LRP11 has a significant correlation with a variety of malignant pathways including protein secretion, mitotic spindle, androgen response, TGF beta signaling, hemoglobin metabolism, estrogen response early, and glycolysis pathways in pan-cancer and CESC.
The study on TIME has opened a new door for tumor research particularly.[6] The components of TME involve a variety of cells, most of which are immune cells and stromal cells. Although great progress has been made in the research of TME in recent years, its internal cell interaction still needs to be further studied. At the same time, the study of TIME also gave birth to a new treatment method, namely immune checkpoint inhibitor.[8] However, only a small number of patients are sensitive to immune checkpoint inhibitors. Immune desert microenvironment is a kind of immune microenvironment, which is characterized by the extreme lack of all kinds of immune cells in tumor tissues. Patients with immune desert TIME were resistant to immunotherapy.[9,10] Thus, finding new genes regulating immune microenvironment is of great significance to prompt the efficacy of immunotherapy and improve the prognosis of patients. In this study, we found that LRP11 expression was negatively correlated with ImmuneScore, Stromalcore, and ESTIMATEScore, while positively correlated with tumor purity in CESC. These results indicated that patients with high expression of LRP11 may have immune desert TME. To further prove this, we downloaded immune cell infiltrate data from ImmuCellAI and TIMER2.0 database and analyzed the relationship of LRP11 with various immune cells. We observed that LRP11 is negatively correlated with infiltration level of most immune cells, such as CD8 T cells, NK cells, gamma delta T cells, and B cells. These results proved that patients with high LRP11 expression may have immune desert tumor microenvironment. It was reported that the activation of TGF beta1 signaling and Wnt/beta-catenin signaling could induce the immune desert tumor microenvironment. We found that LRP11 expression was positively correlated with genes involved in TGF beta1 signaling and Wnt/beta-catenin signaling in pan-cancer and CESC. The above results all indicated that patients with high expression of LRP11 gene are in a status of immune desert TME, which may be the potential tumor-promoting mechanism of LRP11.
5. Conclusion
Overall, our research revealed the potential oncogene of LRP11 in CESC and pan-cancer, and deeply analyzed the cross-linking of LRP11 with TIME. Patients with high LRP11 expression have immune desert TME, which may be resistant to immunotherapy. Targeting LRP11 may activate the TME, increase the sensitivity of immunotherapy and improve the prognosis of patients.
Acknowledgments
We thank the Cancer Genome Atlas and Genotype-Tissue Expression for providing the data for this study.
Author contributions
Data curation: Fangyun Gu, Zimeng Pan, Jinglu Yu.
Formal analysis: Fang Xu.
Funding acquisition: Fang Xu, Miao Sun.
Methodology: Fangyun Gu, Fang Xu, Zimeng Pan, Lin Shi, Feifei Song.
Project administration: Feifei Song.
Resources: Fangyun Gu, Fang Xu, Zimeng Pan.
Software: Fangyun Gu, Lin Shi, Jinglu Yu.
Supervision: Lin Shi, ShuFeng Huang, Miao Sun.
Validation: Fangyun Gu, ShuFeng Huang, Miao Sun.
Visualization: Zimeng Pan, Jinglu Yu, Feifei Song, ShuFeng Huang.
Writing – original draft: Fangyun Gu, Fang Xu.
Writing – review & editing: ShuFeng Huang, Miao Sun.
Supplementary Material
Abbreviations:
- CAN
- copy number alteration
- CESC
- cervical squamous cell carcinoma
- DFI
- disease free interval
- DSS
- disease specific survival
- GDSC
- Cancer Drug Sensitivity Multi-omics Database
- GSEA
- gene set enrichment analysis
- GSVA
- gene set variation analysis
- GTEx
- genotype-tissue expression
- HPV
- human papilloma virus
- IC50
- half inhibitory concentration
- LRP11
- low density lipoprotein receptor-related protein 11
- OS
- overall survival
- PFI
- progression free interval
- TCGA
- Cancer Genome Atlas
- TGF
- transforming growth factor
- TIME
- tumor immune microenvironment
- TME
- tumor microenvironment
FG and FX contributed equally to this work.
This study was supported by the National Natural Science Foundation of China (grant numbers 81804138, 82174195, and 82004401).
All the data in this study were retrieved, collated and counted from open databases, and did not involve the ethical approval process.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
Supplemental Digital Content is available for this article.
How to cite this article: Gu F, Xu F, Pan Z, Shi L, Yu J, Song F, Huang S, Sun M. An integrative pan-cancer analysis illustrating the key role of LRP11 in cervical cancer. Medicine 2023;102:11(e33201).
Contributor Information
Fangyun Gu, Email: gufangshi@gmail.com.
Fang Xu, Email: xufang1217507@163.com.
Zimeng Pan, Email: panzimeng0727@sina.com.
Lin Shi, Email: dgushilin@163.com.
Jinglu Yu, Email: 505057595@qq.com.
Feifei Song, Email: 961657897@qq.com.
ShuFeng Huang, Email: hsf0101@126.com.
References
- [1].Zhao Z, Zhou S, Li W, et al. AIB1 predicts tumor response to definitive chemoradiotherapy and prognosis in cervical squamous cell carcinoma. J Cancer. 2019;10:5212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Das M. WHO launches strategy to accelerate elimination of cervical cancer. Lancet Oncol. 2021;22:20–1. [DOI] [PubMed] [Google Scholar]
- [3].Zhao F, Qiao Y. Cervical cancer prevention in China: a key to cancer control. Lancet. 2019;393:969–70. [DOI] [PubMed] [Google Scholar]
- [4].Brisson M, Kim JJ, Canfell K, et al. Impact of HPV vaccination and cervical screening on cervical cancer elimination: a comparative modelling analysis in 78 low-income and lower-middle-income countries. Lancet. 2020;395:575–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cohen PA, Jhingran A, Oaknin A, et al. Cervical cancer. Lancet. 2019;393:169–82. [DOI] [PubMed] [Google Scholar]
- [6].Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–33. [DOI] [PubMed] [Google Scholar]
- [7].Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pitt JM, Marabelle A, Eggermont A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27:1482–92. [DOI] [PubMed] [Google Scholar]
- [9].Pansy K, Uhl B, Krstic J, et al. Immune regulatory processes of the tumor microenvironment under malignant conditions. Int J Mol Sci. 2021;22:13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marzagalli M, Ebelt ND, Manuel ER. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin Cancer Biol. 2019;59:236–50. [DOI] [PubMed] [Google Scholar]
- [11].Gan S, Ye J, Li J, et al. LRP11 activates β-catenin to induce PD-L1 expression in prostate cancer. J Drug Target. 2020;28:508–15. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y, Han S, You X, et al. The role of low density lipoprotein receptor-related protein 11 as a tumor promoter in cervical cancer. Cancer Manag Res. 2019;11:8081–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jati S, Sarraf TR, Naskar D, et al. Wnt signaling: pathogen incursion and immune defense. Front Immunol. 2019;10:2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Perrotti F, Rosa C, Cicalini I, et al. Advances in lipidomics for cancer biomarkers discovery. Int J Mol Sci. 2016;17:E1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dasgupta N, Kumar Thakur B, Chakraborty A, et al. Butyrate-induced in vitro colonocyte differentiation network model identifies ITGB1, SYK, CDKN2A, CHAF1A, and LRP1 as the prognostic markers for colorectal cancer recurrence. Nutr Cancer. 2019;71:257–71. [DOI] [PubMed] [Google Scholar]
- [17].Feng C, Ding G, Ding Q, et al. Overexpression of low density lipoprotein receptor-related protein 1 (LRP1) is associated with worsened prognosis and decreased cancer immunity in clear-cell renal cell carcinoma. Biochem Biophys Res Commun. 2018;503:1537–43. [DOI] [PubMed] [Google Scholar]
- [18].Langbein S, Szakacs O, Wilhelm M, et al. Alteration of the LRP1B gene region is associated with high grade of urothelial cancer. Lab Invest. 2002;82:639–43. [DOI] [PubMed] [Google Scholar]
- [19].Ni S, Hu J, Duan Y, et al. Down expression of LRP1B promotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling in renal cell cancer. Cancer Sci. 2013;104:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Prazeres H, Torres J, Rodrigues F, et al. Chromosomal, epigenetic and microRNA-mediated inactivation of LRP1B, a modulator of the extracellular environment of thyroid cancer cells. Oncogene. 2017;36:146. [DOI] [PubMed] [Google Scholar]
- [21].Wang Z, Sun P, Gao C, et al. Down-regulation of LRP1B in colon cancer promoted the growth and migration of cancer cells. Exp Cell Res. 2017;357:1–8. [DOI] [PubMed] [Google Scholar]
- [22].Benes P, Jurajda M, Zaloudík J, et al. C766T low-density lipoprotein receptor-related protein 1 (LRP1) gene polymorphism and susceptibility to breast cancer. Breast Cancer Res. 2003;5:R77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kang HS, Kim J, Lee HJ, et al. LRP1-dependent pepsin clearance induced by 2’-hydroxycinnamaldehyde attenuates breast cancer cell invasion. Int J Biochem Cell Biol. 2014;53:15–23. [DOI] [PubMed] [Google Scholar]
- [24].Boulagnon-Rombi C, Schneider C, Leandri C, et al. LRP1 expression in colon cancer predicts clinical outcome. Oncotarget. 2018;9:8849–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Qadir F, Aziz MA, Sari CP, et al. Transcriptome reprogramming by cancer exosomes: identification of novel molecular targets in matrix and immune modulation. Mol Cancer. 2018;17:97. [DOI] [PMC free article] [PubMed] [Google Scholar]