ABSTRACT
CRISPR systems are often encoded by many prokaryotes as adaptive defense against mobile genetic elements (MGEs), but several MGEs also recruit CRISPR components to perform additional biological functions. Type IV-A systems are identified in Klebsiella plasmids, yet the distribution, characterization, and role of these plasmids carrying CRISPR systems in the whole Klebsiella genus remain unclear. Here, we performed large-scale comparative analysis of these plasmids using publicly available plasmid genomes. CRISPR-harboring plasmids were mainly distributed in Klebsiella pneumoniae (9.09%), covering 19.23% of sequence types, but sparse in Klebsiella species outside Klebsiella pneumoniae (3.92%). Plasmid genome comparison reiterated that these plasmids often carried the cointegrates of IncFIB and IncHI1B replicons, occasionally linked to other replicons, such as IncFIA, IncFII, IncR, IncQ, and IncU. Comparative genome analysis showed that CRISPR-carrying Klebsiella plasmids shared a conserved pNDM-MAR-like conjugation module as their backbones and served as an important vector for the accretion of antibiotic resistance genes (ARGs) and even virulence genes (VGs). Moreover, compared with CRISPR-negative IncFIB/IncHIB plasmids, CRISPR-positive IncFIB/IncHIB plasmids displayed high divergences in terms of ARGs, VGs, GC content, plasmid length, and backbone structures, suggesting their divergent evolutionary paths. The network analysis revealed that CRISPR-positive plasmids yielded fierce competitions with other plasmid types, especially conjugative plasmids, thereby affecting the dynamics of plasmid transmission. Overall, our study provides valuable insights into the role of CRISPR-positive plasmids in the spread of ARGs and VGs in Klebsiella genus.
KEYWORDS: Klebsiella, plasmid, CRISPR, antibiotic resistance, virulence
INTRODUCTION
Klebsiella spp., a member of the Enterobacteriaceae family, are commonly found in the nose, throat, skin, and intestinal tract of humans and animals (1). The Klebsiella genus is composed of a wide diversity of species, including Klebsiella pneumonia, Klebsiella oxytoca, Klebsiella aerogenes, and other genetically related species (2). The last few years have witnessed the rapid evolution of members of the genus Klebsiella, leading to the emergence of notorious organisms that simultaneously harbor both multidrug resistance and hypervirulence (MDR-hv) phenotypes (3). Such organisms are responsible for a series of hospital- and community-associated infections, ranging from mild pneumoniae to life-threatening diseases, such as pyogenic liver abscesses and septicemia (2). Recent data indicated that MDR-hv Klebsiella spp. have been reported in over 10 countries spanning five continents, which poses serious challenges to public health (1). Horizontal gene transfer (HGT) is deemed as the most important mechanism driving the rapid rise of MDR-hv Klebsiella species (4). Klebsiella spp. can acquire resistance and virulence-related genes via HGT mediated by mobile genetic elements (MGEs) and evolve into MDR-hv strains, enabling themselves to survive in some specific and extreme conditions (e.g., antibiotic pressure) (4). Among these MGEs, conjugative and mobilizable plasmids are the most significant contributors (5).
Clustered regularly interspaced short palindromic repeats (CRISPR) coupled with their associated genes (cas) constitute an adaptive immunity system in prokaryotes, providing protection against plasmids, viruses, and other MGEs (6, 7). Generally, CRISPR systems consists of two main parts: (i) CRISPR array that is characterized by alternating repeat sequences separated by a spacer sequence of regular length; and (ii) cas genes that encode proteins essential for adaptive immunity, including adaptation, expression, and interference (6). These systems have been identified in almost half of bacteria and most archaea since they were first discovered in Escherichia coli (8). Despite the canonical defense role, CRISPR systems exhibit remarkable diversity in CRISPR locus architecture and Cas protein organization (9). Updated classification for CRISPR variants showed that there are two major classes (class 1 and 2), six types (type I, II, III, IV, V, and VI) and over 45 subtypes (9). Previous work has focused primarily on the diversity and evolution of chromosome-derived CRISPR systems, although CRISPR systems are frequently found in different types of MGEs, including virus, plasmids, transposons, and integrative and conjugative elements (ICEs) (10, 11). CRISPR systems encoded by MGEs have been known to be involved in multiple additional biological functions, such as RNA-mediated DNA transposition, the conflicts between or within MGEs, and the escape of host immunity (10).
In Klebsiella spp., two types of CRISPR systems have been identified, including type I (type I-E, I-E*, and I-F) and IV (mainly type IV-A) systems (12). The type I CRISPR system is mainly present in chromosomes, whereas the type IV system is only found in plasmids (13, 14). Because the CRISPR system is resistant to invading MGEs, it is assumed that chromosome-encoded CRISPR systems function in limiting the HGT of antibiotic resistance and virulence mediated by plasmid. Increasing evidence has suggested that there was a negative association between the presence of the chromosome-borne type I system and antibiotic resistance in Klebsiella pneumonia (15). Moreover, it has been experimentally documented that the type I-E system interfered the dissemination of blaKPC-IncF plasmids, which may result in the uneven distribution of antibiotic resistance in different phylogenetic lineages of Klebsiella pneumonia (16). Given the positioning preference of the type IV-A CRISPR system for MGEs, Klebsiella plasmids might repurpose the type IV-A system to benefit their own HGT, thus aiding in the prevalence of MDR-hv Klebsiella. Multiple studies have shown that CRISPR-positive Klebsiella plasmids (here referred to plasmids carrying type IV-A system) contained a series of antibiotic resistance genes (ARGs) that conferred resistance to clinically available antibiotics (13, 17). Besides, the coexistence of ARGs and virulence genes (VGs) has been identified in a CRISPR-positive Klebsiella pneumoniae plasmid isolated from a patient with bacteremia (18). However, our current understanding of the contribution of these CRISPR-positive plasmids to the expansion of MDR-hv Klebsiella strains is still limited due to an insufficient plasmid number. With development of high-throughput DNA sequencing technology, many genome and plasmid sequencing data have been delivered to public databases (19). This provides an opportunity to perform a large-scale plasmid analysis to further clarify the relationship between these plasmids and the propagation of ARGs or VGs.
Here, we performed a comparative genomic analysis of CRISPR-positive plasmids in Klebsiella species, using publicly available genome and plasmid sequences. We elaborated the incidence and characterization of CRISPR-positive plasmids in Klebsiella spp., compared their genetic configurations and the profiles of antibiotic resistance and virulence determinants, and delineated the network of plasmid-plasmid competition based on protospacer-spacer matches.
RESULTS
Global distribution of CRISPR-harboring plasmids in the Klebsiella genus.
A collection of 1,606 Klebsiella strains spanning 12 species were screened for the presence of type IV-A CRISPR systems. Based on search results, 146 type IV-A systems were identified in plasmids from 146 (9.09%, 146/1,606) Klebsiella strains, including 135 complete and 11 degenerated systems (partial or truncated cas gene clusters) (Fig. S1A and Table S3). Specifically, out of 1,300 Klebsiella pneumoniae genomes covering 208 defined sequence types (STs), 134 plasmids in 134 Klebsiella pneumoniae genomes spanning 40 known STs (19.23%, 40/208) carried the type IV-A system (Fig. S1B). The ration of CRISPR-positive plasmids in Klebsiella pneumoniae (10.31%, 134/1,300) was significantly higher than non-Klebsiella pneumoniae species (3.92%, 12/306, P < 0.05). Additionally, among 1,941 completely sequenced Klebsiella plasmids that were not included in NCBI genome database, 57 plasmids harbored type IV-A systems, covering 53 complete and 4 degenerated plasmids. Accordingly, there were a total of 203 type IV-A systems found in this study. Most of type IV-A systems were located next to the umuD gene (Fig. S1A and Table S3). The group II intron reverse transcriptase/maturase gene and insertion sequence (IS) were frequently inserted near type IV-A systems (Table S3).
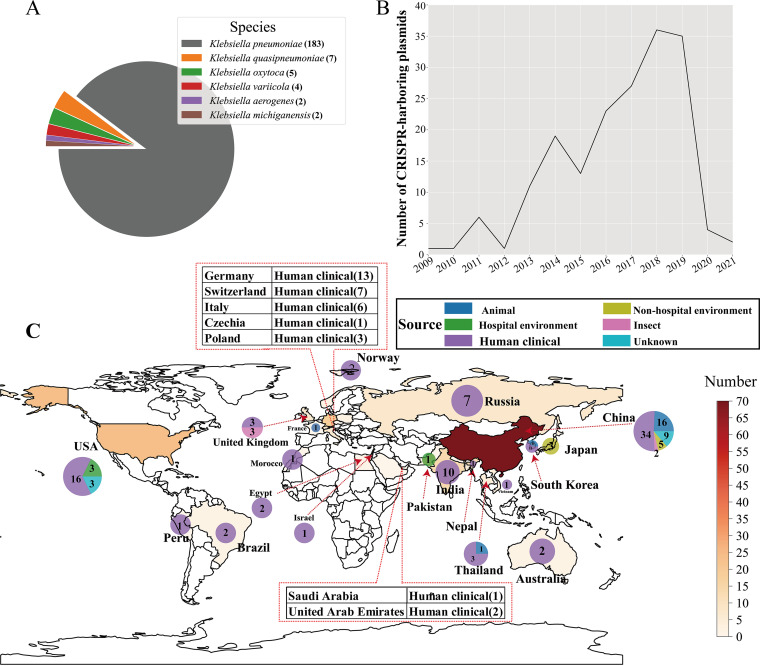
Further analysis showed that the 203 CRISPR-harboring plasmids spanned multiple Klebsiella species, including 183 Klebsiella pneumoniae, 7 Klebsiella quasipneumoniae, 5 Klebsiella oxytoca, 4 Klebsiella variicola, 2 Klebsiella michiganensis, and 2 Klebsiella aerogenes (Fig. 1A). Where metadata were available, geographical analysis showed that these CRISPR-carrying plasmids were identified in 28 countries across six continents (Fig. 1C and Table S3). The top three countries with the highest number of CRISPR-positive plasmids were China (n = 66), USA (n = 22), and Germany (n = 13) (Fig. 1C). Besides, CRISPR-positive plasmid exhibited diverse isolation sources, including human clinical samples (n = 139), animals (n = 25), hospital environments (n = 4), nonhospital environments (n = 12), and insects (n = 2). The distribution, coupled with the wide range of isolation year (Fig. 1B), suggested that CRISPR-positive plasmids might have globally spread for at least 1 decade.
FIG 1.
Global distribution of CRISPR-harboring plasmids in the Klebsiella genus. (A) Species distribution of CRISPR-harboring plasmids in the Klebsiella genus. Each species is represented by a different color. (B) Line chart of isolation years of the strains. The x axis indicates the isolated year, and the y axis represents the number of CRISPR-positive plasmids per year. (C) Global distribution of CRISPR-harboring plasmids in a world map. The red color gradient represents the sample size of CRISPR-positive plasmids in each country. The host information is denoted by different colors (animal: royal blue; hospital environment: green; human clinical: dark orchid; non-hospital environment: yellow; insect: rosy brown; unknown: deep sky blue). The host number of CRISPR-positive plasmids per host in each country is shown.
Most CRISPR-harboring plasmids carried a pNDM-MAR-like conjugation module.
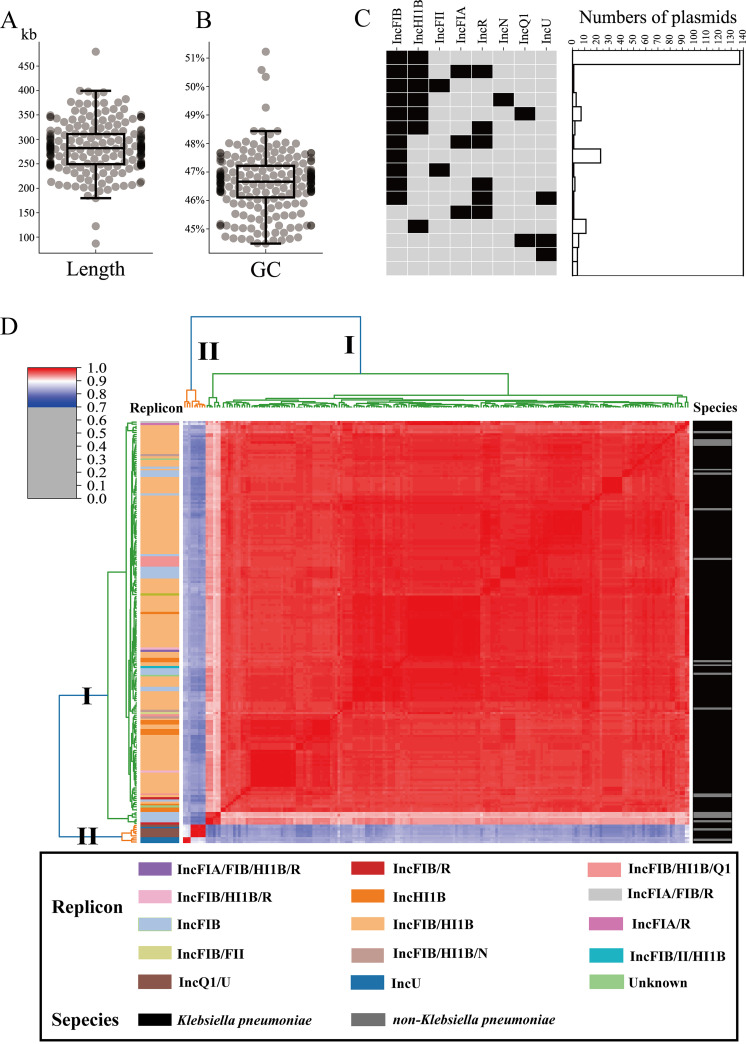
To gain further insights into genetic diversity of CRISPR-harboring plasmids, we analyzed the characterization in detail. These plasmids had various lengths (86 to 479 kb; mean, 284 kb) and GC contents (44.49% to 51.22%; mean, 46.61%) (Fig. 2A and B). Plasmid replicon typing showed 200 out of 203 CRISPR-positive plasmids could be designated a defined incompatibility group, with IncFIB/HI1B (n = 137) being the most prevalent group, followed by IncFIB (n = 23), IncHIB (n = 11), and other replicon types (Fig. 2C). Based on average nucleotide identity (ANI), 203 CRISPR-positive plasmids were grouped into two major clusters, cluster I and II, with each cluster exhibiting high sequence similarities (Fig. 2D and Table S4). As shown in Fig. 2D, cluster I contained 195 plasmids, with ANI ranging from 90.98% to 100%. Most plasmids (96.91%, 188/194) in cluster I harbored IncFIB, IncHI1B replicon, or their combination with other replicons. Nine plasmids with IncQ1 or IncU replicon were assigned to cluster II, with ANI varying from 86.26% to 100%. These findings suggested that the CRISPR-positive plasmids in the Klebsiella genus were almost limited to a highly homologous plasmid group, especially IncFIB/IncHI1B replicon.
FIG 2.
Basic characteristics of CRISPR-positive plasmids in Klebsiella genus. (A) Box plot of the length distribution. (B) Box plot of the GC content distribution. (C) Distribution of different multireplicon plasmids. The matrix on the left indicates different replicon profiles. The histograms on the right represent the numbers of plasmids with the corresponding replicon profiles. (D) Heatmap diagram of paired ANI between the 203 CRISPR-positive plasmids plotted using the seaborn module in python. The blue/red gradient indicates the estimated paired ANI value, and the corresponding legend is shown on the top left. The plasmid replicon types and isolated species are annotated with different colors, and corresponding legends are shown at the bottom.
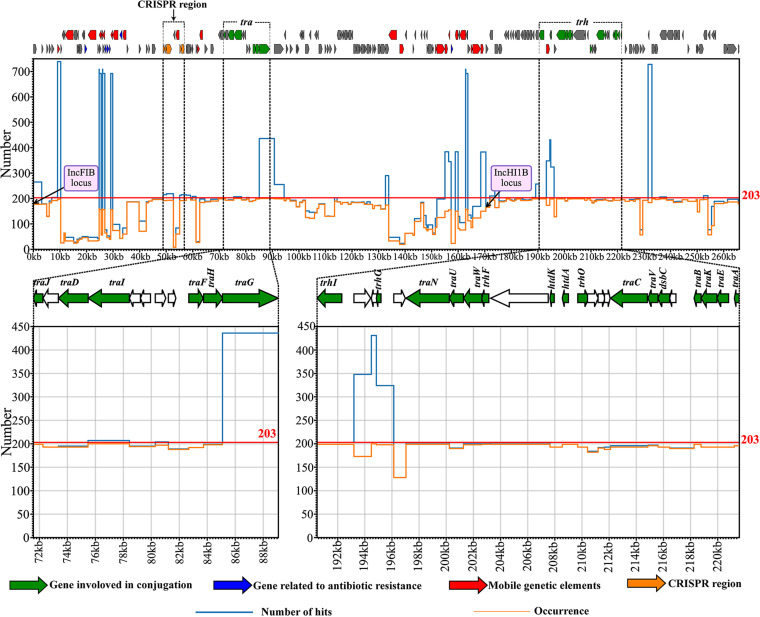
Plasmid pNDM-MAR (GenBank accession no. JN420336.1) was a well-characterized CRISPR-harboring IncFIB/HI1B plasmid, which was conjugative and harbored a series of antibiotic resistance genes (20). To explore the genetic configuration of CRISPR-positive plasmids in this study, we aligned the reference plasmid pNDM-MAR against each of the CRISPR-harboring plasmids. As shown in Fig. 3, more than 123 kb of the coding sequence (CDS) region (46.07%, 123/267) on plasmid pNDM-MAR (length: 267 kb) were shared by 84.72% (172/203) of CRISPR-harboring plasmids, thereby supporting that CRISPR-carrying plasmids harbored a backbone structure similar to pNDM-MAR. Additionally, the conjugation transfer module (tra and trh locus) on the pNDM-MAR backbone were shared by at least 97.54% (198/203) of CRISPR-carrying plasmids, regardless of plasmids belonging to cluster I and II (Fig. S2 and Table S5). Moreover, most proteins involved in conjugation exhibited >90% amino acid identities to their counterparts on pNDM-MAR, except for trhG in trh locus (Fig. S2 and Table S5). Gene truncation or inactivation due to point mutation, insertion sequence, and disruption were found. Some single proteins were occasionally absent, such as TraJ, TraH, and TraG in the tra locus, TraU, TrhF, TraB, and TraK in the trh locus. The entire tra and trh loci were missing in 3 and 2 plasmids, respectively. Apart from 3 plasmids lacking the tra locus, the other 200 plasmids were predicted to be conjugative by mob_typer software. The 200 conjugative plasmids carried genes encoding relaxases of MOBH family (Table S3).
FIG 3.
Alignment plot of CRISPR-positive plasmids against plasmid pNDM-MAR. pNDM-MAR (GenBank accession no. JN420336.1) is used as reference for alignment. The conjugation module (transfer region), ARGs, MGEs, and CRISPR region are denoted by green, blue, red, and orange, respectively. The orange line in the chart represents the corresponding region of pNDM-MAR that occurred and in how many plasmids (occurrence), while the blue line represents how many hits of the corresponding region of pNDM-MAR are identified among CRISPR-harboring plasmids (hits). The horizontal axis represents the coordinates of pNDM-MAR opened in the repA gene of the IncFIB replicon. A zoom-in view of the conjugation module is shown at the bottom.
CRISPR-positive plasmids encode both antibiotic resistance and virulence genes.
Plasmids were known as the important vectors of antibiotic resistance genes (ARGs). We observed that 73.40% (149/203) of the CRISPR-positive plasmids contained at least one ARG or remnant (Fig. S3). Moreover, almost all the CRISPR-positive plasmids carrying ARGs (95.30%, 142/149) were deemed as putative multidrug-resistant (MDR) plasmids, as they encoded resistance to at least three different antibiotic classes. Besides, over three-fifths of CRISPR-positive plasmids (67.49%, 137/203) harbored at least one β-lactam resistance gene or remnant. The carriage of carbapenem resistance genes was particularly concerning. The blaNDM-1 genes were identified in 21.18% (43/203) of CRISPR-positive plasmids. Notably, two plasmids carried mcr-1, mcr-2 or mcr-3 genes that conferred resistance to the last-resort antibiotic colistin.
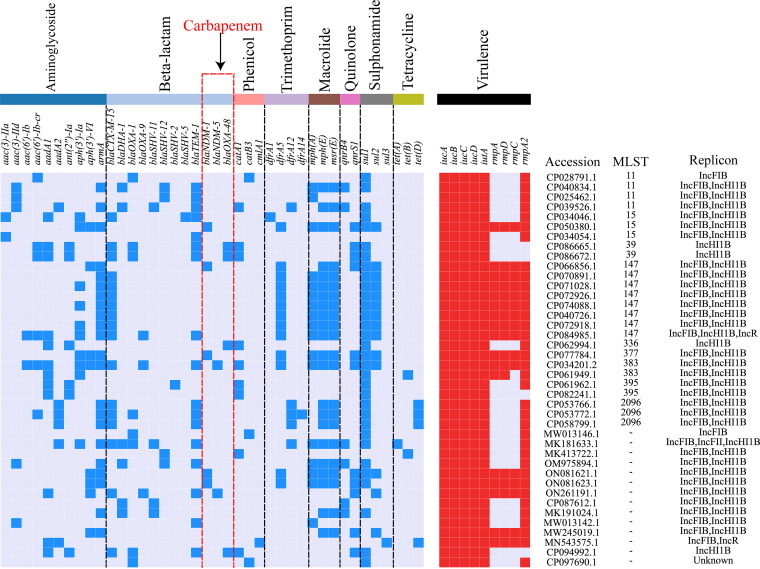
Interestingly, we found that 40 CRISPR-positive plasmids (19.70%, 40/203) not only carried ARGs, but also harbored a series of VGs (Fig. 4). Among these plasmids encoding ARGs and VGs, nearly one-third (30%, 12/40) harbored carbapenem resistance genes, including blaNDM-1, blaNDM-5, and blaOXA-48 (Fig. 4). Moreover, 36 plasmids carried rmpA/rmpA2, iucABCD, and iutA, which were deemed as potential virulence genes associated with hypervirulence phenotypes (21). A total of 10 plasmids carried both the carbapenem resistance genes and hypervirulence-related VGs.
FIG 4.
ARG and VG profiles of 25 CRISPR-harboring plasmids. The antibiotic classes of ARGs are denoted by different colors. GenBank accession, MLST, and replicon types for each plasmid are shown on the right. The blue and red blocks indicate the presence of ARGs and VGs, respectively, whereas the white blocks represent the absence of ARGs and VGs.
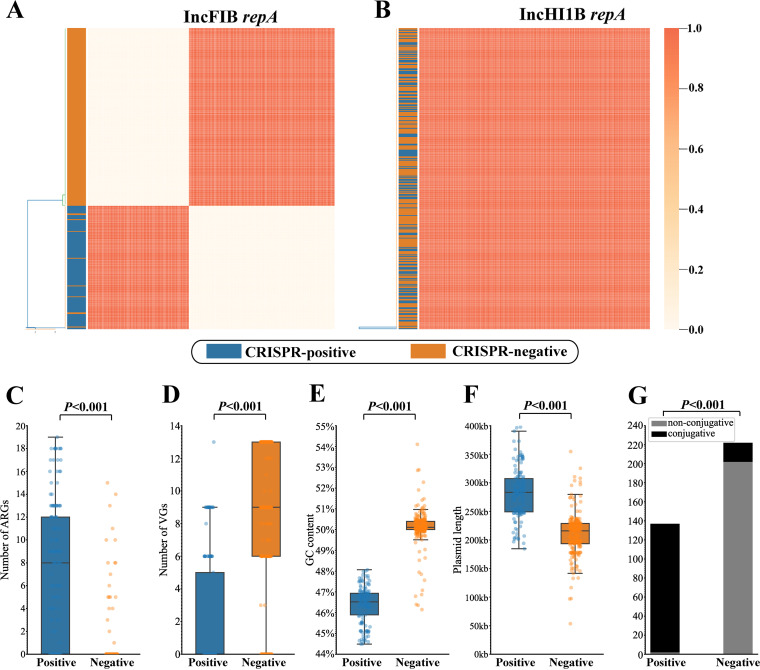
Comparative analysis of IncFIB/HIB plasmids with and without CRISPR.
According to the above findings, most CRISPR-positive plasmids carried IncFIB/HI1B replicons. To determine whether there was a genetic relationship between plasmids with and without CRISPR, we collected 359 IncFIB/HI1B plasmids (covering 137 CRISPR-positive and 222 CRISPR-negative) to further compare their gene compositions. As shown in Fig. 5A and B, the repA marker gene of IncFIB exhibited high differences between CRISPR-positive and -negative plasmids, with most nucleotide identities being 0% (Table S6), whereas the repA gene of IncHI1B was highly similar, with nucleotide identities ranging from 99.83% to 100% (Table S7). Moreover, CRISPR-positive plasmids carried more ARGs than CRISPR-negative, but the distribution of VGs showed a completely opposite trend (Fig. 5C and D) (P < 0.001). There were also significant differences in GC content and plasmid length among them (Fig. 5E and F) (P < 0.001). Conjugative transfer ability prediction showed CRISPR-negative plasmids were rarely conjugative (Fig. 5G).
FIG 5.
Comparative analysis of CRISPR-positive with CRISPR-negative IncFIB/HI1B plasmids. (A) Nucleotide similarity matrix of the repA gene in the IncFIB replicon. CRISPR-positive and -negative plasmids are marked in blue and orange, respectively. (B) Nucleotide similarity matrix of the repA gene in the IncHI1B replicon. (C) Comparison of number of ARGs. (D) Comparison of number of VGs. (E) Comparison of GC content. (F) Comparison of plasmid length. (G) Comparison of conjugative transfer ability.
To further clarify the evolutionary differences between plasmids with and without CRISPR, the genetic configuration of all CRISPR-negative IncFIB/HI1B plasmids were also compared with pNDM-MAR. As shown in Fig. S4, there were only a few genes shared between CRISPR-positive and -negative plasmids.
The type IV-A CRISPR system reflects the plasmid-plasmid competition.
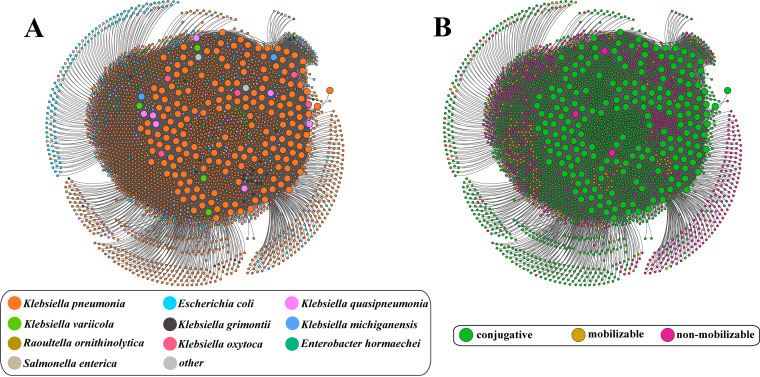
A total of 2,859 spacers were identified in 203 CRISPR-positive plasmids, which was composed of 179 nonredundant and distinct spacers (Table S9). The homology search revealed that over one-third of the spacers (35.75%, 64/179) were significantly homologous to plasmids or phages (Table S10). The spacer sequences exhibited strong targeting preference for plasmid rather than phage (32.40%, 58/179 versus 3.35, 6/179). The network analysis of plasmid-plasmid interaction found that there was an intense competition among plasmids in Klebsiella pneumoniae (Fig. 6A). Also, we observed that the spacers from Klebsiella plasmids matched the plasmids from other non-Klebsiella species, such as Escherichia coli, Raoultella ornithinolytica, Enterobacter hormaechei, and so on. Surprisingly, most plasmids targeted by spacers (67.99%, 1,804/2,653) were conjugative. A total of 8 spacers were found to be directly homologous to genes involved in conjugation transfer (Table S4). These spacers were distributed in 181 CRISPR-harboring plasmids, accounting for 89.16% (181/203) of CRISPR-carrying plasmids.
FIG 6.
Network of plasmid-plasmid competition based on protospacer-spacer matches. (A) Network of plasmid-plasmid competition colored at the host species level. (B) Network of plasmid-plasmid competition colored according to conjugation transmissibility. Nodes indicate individual plasmids and edges represent CRISPR spacer targeting based on spacer-protospacer matches. The presence and absence of type IV-A CRISPR systems are represented by large and small nodes, respectively.
DISCUSSION
We have performed a large-scale analysis of CRISPR-positive plasmids to gain further insights into their distribution and characterization. CRISPR-positive plasmids displayed a biased species distribution. They were mainly distributed in Klebsiella pneumoniae but sparse in other Klebsiella species. Other investigation also reported the rarity of CRISPR-positive plasmids in Enterobacteriacea outside Klebsiella species (17, 22, 23). The above-mentioned findings highlight that these plasmids are strictly narrow-host range. Previous investigation has suggested a strong association between IncFIB/IncHI1B cointegrates and CRISPR-positive plasmids (23). The multireplicon status contributes to broadening the host range of plasmids by merging broad-host-range replicons (24). Unfortunately, the incompatible groups IncF and IncH both have a narrow host range (24), which further explains why CRISPR-carrying plasmids are largely restricted to the Klebsiella genus. Based on ANI values, these plasmids were further divided into two subclusters. Cluster I was mainly composed of plasmids with IncFIB/IncHI1B, whereas cluster II harbored IncQ1/IncU and IncU plasmids. In general, the IncQ and IncU plasmids are characterized by their relatively small size and broad host range (25, 26). However, in the current study, all CRISPR-positive IncQ1 and IncU plasmids were relatively large (29 kb to 37 kb), which were mosaic plasmids that shared similar conjugation transfer region with pNDM-MAR. The presence of broad-host-range replicons in CRISPR-positive plasmids suggest their potential ability to transfer into other species. Furthermore, we observed that CRISPR-harboring plasmids were widespread in different Klebsiella pneumoniae STs, although there was an inevitable sample bias by prevalent clinical clones and clones from outbreak in the current database (ST11, ST258, and ST147 were overrepresented). The random distribution of CRISPR-positive plasmids in Klebsiella pneumoniae further underscored their HGT within this species.
The misuse and abuse of carbapenems have resulted in the selection, evolution and spread of MDR Klebsiella strains that harbor carbapenemase-encoding plasmids (27). The first identified plasmid-borne carbapenemase in Klebsiella strains was blaIMP-1, which was isolated as early as 1991 in Japan (28). Subsequently, other plasmid-mediated carbapenem genes were continuously reported in the Klebsiella genus, such as blaKPC-1 (29), blaVIM-1 (30), blaOXA-48 (31), and blaNDM-1 (32). In the current study, almost a quarter of CRISPR-positive plasmids (24.63%, 50/203) carried at least one carbapenem resistance gene, including blaNDM-1, blaNDM-5, blaNDM-7, and blaOXA-48. The enrichment of carbapenem resistance genes in CRISPR-positive plasmids reaffirmed the prominent carrier role of these plasmids in the prevalence of MDR Klebsiella strains. The phenotypes of MDR and hypervirulence in Klebsiella strains had been nonoverlapping for a long time as MDR genes are often carried by classical but not hypervirulent Klebsiella strains (33). However, the convergence of carbapenem and hypervirulence genes was identified in 10 CRISPR-positive plasmids, suggesting that these plasmids can confer Klebsiella strains both MDR and hypervirulence phenotypes at one step. The evolution process of hybrid plasmids carrying both ARGs and VGs follows two distinct paths. The first is the acquisition of ARGs by a virulent plasmid, and the second is the insertion of VGs into a resistant plasmid. Obviously, the second mechanism is more convincing for the evolution from CRISPR-positive plasmids to hybrid plasmids, because these plasmids were more open to ARGs than VGs (Fig. 5C and D). Moreover, these hybrid plasmids exhibited similar backbone structure to antibiotic-resistant plasmid pNDM-MAR, which further corroborated the above hypothesis. Multiple investigations reported that the hybrid plasmids coding for resistance and virulence were typically cointegrates with two plasmid backbones, which creates a scenario where ARGs and VGs were located on a single plasmid (34, 35). Similarly, we observed that 30 hybrid plasmids were cointegrate plasmids carrying IncFIB and IncHI1B replicons. Thus, it is plausible that the segments of virulence plasmids are integrated into these CRISPR-positive plasmids for persistence under strong stress.
Comparative genome analysis showed that CRISPR-positive plasmids were highly variable but commonly share a pNDM-MAR-like conjugation module as their backbones. The pNDM-MAR-like conjugation module consisted of a tra gene cluster and a trh gene cluster. Plasmid pNDM-MAR and other plasmids bearing this transfer region have been reported to be conjugative (17, 20). Combined with prediction results by mob_typer software, most CRISPR-positive plasmids were capable of conjugation transfer. This further supports the view that conjugative plasmids facilitate HGT of type IV-A CRISPR systems in Klebsiella species. Nevertheless, the conserved pNDM-MAR-like conjugation module was not common in CRISPR-negative plasmids with IncFIB/IncHI1B replicons. The result hints that CRISPR-positive and -negative plasmids may have gone through different evolution trajectories or recombination events, which is also evidenced by the difference of ARGs, VGs, and the IncFIB repA gene. In addition to ARGs and VGs, the gene rearrangement of diverse MGEs (e.g., ISs, integrons, and transposons) constitutes highly variable regions in plasmids (36). It has been well known that MGEs usually have a relatively low GC content (37). We observed that CRISPR-positive plasmids exhibited lower GC content and higher plasmid length than CRISPR-negative plasmids, thereby suggesting the higher genome plasticity of CRISPR-carrying plasmids. Accordingly, it can be deduced that the type IV-A CRISPR system, pNDM-MAR-like conjugation module, and replication initiation proteins related to pNDM-MAR together formed a unique backbone structure, which served as the important platform for MGEs accretion, especially ARGs and VGs. Considering the global dissemination of CRISPR-positive plasmids and their frequent occurrence in clinical environments (Fig. 1C), tracking this plasmid lineage will be very crucial for surveillance of MDR-hv Klebsiella.
Spacers are the product of invading genetic elements, which reflects the exposure of the host to invading genetic elements. Exploring the origin of the spacers in the type IV-A system will provide insights into the interaction of CRISPR-carrying plasmids with other mobile genetic elements. Our analysis showed that only a small fraction of spacers displayed significant matches to protospacer sequences, consistent to previous investigations (13). This relatively low match is attributed to multiple reasons, including the paucity of MGE sequences in current public databases and the frequent escape mutation of MGE protospacers (38, 39). The recruitment of type IV-A CRISPR systems by plasmids has been reported to be involved in plasmid-plasmid warfare dynamics (22). We found that type IV-A CRISPR in Klebsiella species tend to carry a larger fraction of spacers that targeted other plasmids. The plasmid-plasmid competitions from closely related species were more frequent than that from distantly related species. This is consistent to the community ecology view that similar entities inhabiting in overlapping niches will compete more strongly for overlapping cellular resources (40, 41). Another interesting finding was that CRISPR-positive plasmids biasedly targeted conjugative plasmids. There were two possible underlying mechanisms: (i) the self-transmissible properties of conjugative plasmids bring conceivably higher rates of encounters with CRISPR-positive plasmids in cells; and (ii) conjugative plasmids pose more threats to plasmid-host balance already established in a single cell than other plasmid types, such as more metabolic burden. Expectedly, genes involved in conjugation transfer were frequently targeted, which provided direct evidence for limiting the HGT of conjugative plasmids. Conjugation by plasmids is a common mechanism of HGT in bacteria that is instrumental in the spread of antibiotic resistance (42). The direct targeting to conjugation transfer proteins implies a very important role of CRISPR-positive plasmids in shaping the ARGs profiles of Klebsiella strains.
Conclusion.
Our study demonstrates that plasmids that carry the type IV-A CRISPR system in the Klebsiella genus harbor a pNDM-MAR-like backbone structure, which plays an important role in the spread of ARGs and VGs. Further surveillance of this plasmid lineage is very necessary to prevent and control the prevalence of MDR-hv Klebsiella strains.
MATERIALS AND METHODS
Data collection.
All the Klebsiella genomes that are annotated as “chromosome” or “complete” at assembly level were retrieved from National Center for Biotechnology Information (NCBI) genome database (https://ftp.ncbi.nih.gov/genomes/) as of 31 December 2021. These genome sequences were downloaded after reconfirming the species by kleborate v2.1.0 (43). For genomes of repeatedly recorded strains, the one with a higher sequencing quality was taken as applicable or otherwise taken randomly. A total of 1,606 unique strains spanning 12 Klebsiella species from the NCBI genome database were included in this study. Among these completely sequenced genomes, 1,294 contained 4,760 plasmid sequences, while others contained none. Detailed information for 1,606 K. pneumoniae genomes is shown in Table S1. Besides, 1,941 fully sequenced Klebsiella plasmids that were not present in the NCBI genome database were collected from the plasmid database of the NCBI RefSeq database (https://ftp.ncbi.nih.gov/refseq/release/plasmid/) (Table S2).
CRISPR/Cas system identification.
The identification of CRISPR arrays was performed by CRISPRCasFinder v4.2.20 using default parameters (44). The high confidence arrays predicted by CRISPRCasFinder (evidence level 4) were automatically kept. Subsequently, the low confidence arrays predicted by CRISPRCasFinder (evidence level <4) were deemed as putative arrays. These putative arrays were reused for subsequent analysis if they were located within 1 kb to a predicted cas gene or matched with any repeat sequence (95% coverage and 95% identity) from already defined high-confidence CRISPR arrays. The classification and subtyping of CRISPR/Cas systems were implemented by CRISPRCasType v1.6.0 using default parameters (45).
MLST typing and phylogenetic analysis.
In silico MLST typing was performed with mlst v2.1 (identity = 100% and coverage = 100%) using the seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) as queries (https://github.com/tseemann/mlst). Subsequently, the sequences of seven housekeeping genes were concatenated to create a maximum likelihood (ML) tree to estimate the phylogenetic relationship of all Klebsiella pneumoniae strains using iqtree v2.1.4 with 1,000 bootstraps (model GTR+F+R3) (46).
Comparative genomics of CRISPR-positive and -negative plasmids.
Paired average nucleotide identity (ANI) between CRISPR-positive plasmids was calculated by Python script pyani (https://github.com/widdowquinn/pyani). All CRISPR-positive or -negative plasmids were aligned against pNDM-MAR by using Mega BLAST (E value ≤ 0.0001) (47). The number of hits of different regions of pNDM-MAR were counted to identify conserved regions. The conjugation module of each CRISPR-positive plasmid was compared with that of pNDM-MAR, gene by gene.
Plasmid conjugative transfer function and incompatibility group prediction.
The conjugative transfer function and relaxase type for each plasmid were predicated by MOB-suite v3.0.3 using the mob_typer function and default parameters (48). The incompatibility group for each plasmid was determined by PlasmidFinder v2.0.1 using default parameters (49).
Identification of ARGs and VGs.
The identification of ARGs was performed using ResFinder software by default parameters (coverage ≥ 60%, identity ≥ 90%) (50). A series of virulence genes involved in yersiniabactin, aerobactin, and other siderophore production were confirmed by kleborate v2.1.0 (43).
Spacer-protospacer match analysis.
The putative origin of CRISPR spacers was analyzed by the CRISPRTarget web tool (51). A strong protospacer was considered when two compared sequences showed ≥85% identity. The matches to CRISPR sequence were ruled out for subsequent analysis. The network of plasmid-plasmid competitions was visualized in Gephi with the layout generated by a combination of Fruchterman Reingold and Noverlap algorithms (https://github.com/gephi/gephi). Each pair of plasmids was connected by at least one spacer-protospacer match.
Statistical analysis.
Statistical analysis was performed with SPSS 21.0. Differences of the numbers of ARGs, VGs, plasmid length, and GC content between CRISPR-positive and -negative plasmids were assessed using unpaired Student's t test (normal distribution) or Mann-Whitney test (nonnormal distribution). Chi square was used for the comparison of conjugation transmissibility between CRISPR-positive and -negative plasmids. In all cases, a P value lower than 0.05 was deemed as be statistically significant.
ACKNOWLEDGMENTS
The work was supported by the China Postdoctoral Science Foundation (2022M712859) and National Science and Technology Specific Projects (2018ZX10301407). We thank National Supercomputing Center in Zhengzhou for its technical supports.
J.L. and G.D. designed the study. J.L., Y.X., and Jiangfeng Zhang analyzed data and wrote the paper. H.Y. and Jiaxue Zhao helped collect and analyze some data. All authors read and approved the final manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dong N, Yang X, Chan EW, Zhang R, Chen S. 2022. Klebsiella species: taxonomy, hypervirulence and multidrug resistance. EBioMedicine 79:103998. 10.1016/j.ebiom.2022.103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 3.Zhu W, Liu Y, Chen F, Chen S, Zhu Y, Li H, Wang J, Liu J, Li Y, Yu J, Guan H, Yu J, Shen L. 2022. Cooccurrence of antibiotic resistance and hypervirulence in high-risk carbapenem-resistant K14.K64 and Wzi209 Klebsiella pneumoniae strains driven by plasmids and their derivatives. Microbiol Spectr 10:e0254121. 10.1128/spectrum.02541-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Dong N, Chan EW, Zhang R, Chen S. 2021. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol 29:65–83. 10.1016/j.tim.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Prensky H, Gomez-Simmonds A, Uhlemann AC, Lopatkin AJ. 2021. Conjugation dynamics depend on both the plasmid acquisition cost and the fitness cost. Mol Syst Biol 17:e9913. 10.15252/msb.20209913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 7.Garneau JE, Dupuis M-È, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 8.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faure G, Shmakov SA, Yan WX, Cheng DR, Scott DA, Peters JE, Makarova KS, Koonin EV. 2019. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol 17:513–525. 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Vliet AHM, Charity OJ, Reuter M. 2021. A Campylobacter integrative and conjugative element with a CRISPR-Cas9 system targeting competing plasmids: a history of plasmid warfare? Microb Genom 7:000729. 10.1099/mgen.0.000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Lv L, Wang X, Xiu Z, Chen G. 2017. Comparative analysis of CRISPR-Cas systems in Klebsiella genomes. J Basic Microbiol 57:325–336. 10.1002/jobm.201600589. [DOI] [PubMed] [Google Scholar]
- 13.Kamruzzaman M, Iredell JR. 2019. CRISPR-Cas system in antibiotic resistance plasmids in Klebsiella pneumoniae. Front Microbiol 10:2934. 10.3389/fmicb.2019.02934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H-Y, Kao C-Y, Lin W-H, Zheng P-X, Yan J-J, Wang M-C, Teng C-H, Tseng C-C, Wu J-J. 2018. Characterization of CRISPR-Cas systems in clinical Klebsiella pneumoniae isolates uncovers its potential association with antibiotic susceptibility. Front Microbiol 9:1595. 10.3389/fmicb.2018.01595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Y, Fu P, Zhou Y, Xie Y, Jin J, Wang B, Yu L, Huang Y, Li G, Li M, Liang W, Ou H-Y, Jiang X. 2020. Absence of the type I-E CRISPR-Cas system in Klebsiella pneumoniae clonal complex 258 is associated with dissemination of IncF epidemic resistance plasmids in this clonal complex. J Antimicrob Chemother 75:890–895. 10.1093/jac/dkz538. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Tang Y, Fu P, Tian D, Yu L, Huang Y, Li G, Li M, Wang Y, Yang Z, Xu X, Yin Z, Zhou D, Poirel L, Jiang X. 2020. The type I-E CRISPR-Cas system influences the acquisition of bla(KPC)-IncF plasmid in Klebsiella pneumonia. Emerg Microbes Infect 9:1011–1022. 10.1080/22221751.2020.1763209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng YH, Chou SH, Huang PH, Yang TC, Juan YF, Kreiswirth BN, et al. 2021. Characterization of a mcr-1 and CRISPR-Cas system co-harboring plasmid in a carbapenemase-producing high-risk ST11 Klebsiella pneumoniae strain. Front Microbiol 12:762947. 10.3389/fmicb.2021.762947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shankar C, Vasudevan K, Jacob JJ, Baker S, Isaac BJ, Neeravi AR, Sethuvel DPM, George B, Veeraraghavan B. 2022. Hybrid plasmids encoding antimicrobial resistance and virulence traits among hypervirulent Klebsiella pneumoniae ST2096 in India. Front Cell Infect Microbiol 12:875116. 10.3389/fcimb.2022.875116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–45. 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother 67:1645–1650. 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 21.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR, Backer M, Bajwa R, Catanzaro AT, Crook D, de Almeda K, Fierer J, Greenberg DE, Klevay M, Patel P, Ratner A, Wang J-T, Zola J. 2018. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 56. 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinilla-Redondo R, Mayo-Muñoz D, Russel J, Garrett RA, Randau L, Sørensen SJ, Shah SA. 2020. Type IV CRISPR-Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res 48:2000–2012. 10.1093/nar/gkz1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newire E, Aydin A, Juma S, Enne VI, Roberts AP. 2020. Identification of a Type IV-A CRISPR-Cas System Located Exclusively on IncHI1B/IncFIB Plasmids in Enterobacteriaceae. Front Microbiol 11:1937. 10.3389/fmicb.2020.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H, Yano H, Brown CJ, Top EM. 2010. Predicting plasmid promiscuity based on genomic signature. J Bacteriol 192:6045–6055. 10.1128/JB.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlings DE, Tietze E. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev 65:481–496, table of contents. table of contents. 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tartari DC, Zamparette CP, Martini G, Christakis S, Costa LH, Silveira ACdO, Sincero TCM. 2021. Genomic analysis of an extensively drug-resistant Pseudomonas aeruginosa ST312 harbouring IncU plasmid-mediated bla(KPC-2) isolated from ascitic fluid. J Glob Antimicrob Resist 25:151–153. 10.1016/j.jgar.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haruta S, Yamaguchi H, Yamamoto ET, Eriguchi Y, Nukaga M, O'Hara K, Sawai T. 2000. Functional analysis of the active site of a metallo-beta-lactamase proliferating in Japan. Antimicrob Agents Chemother 44:2304–2309. 10.1128/AAC.44.9.2304-2309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giakkoupi P, Xanthaki A, Kanelopoulou M, Vlahaki A, Miriagou V, Kontou S, Papafraggas E, Malamou-Lada H, Tzouvelekis LS, Legakis NJ, Vatopoulos AC. 2003. VIM-1 Metallo-beta-lactamase-producing Klebsiella pneumoniae strains in Greek hospitals. J Clin Microbiol 41:3893–3896. 10.1128/JCM.41.8.3893-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C-R, Lee JH, Park KS, Jeon JH, Kim YB, Cha C-J, Jeong BC, Lee SH. 2017. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol 7:483. 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turton J, Davies F, Turton J, Perry C, Payne Z, Pike R. 2019. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying bla(NDM-5). Microorganisms 7:326. 10.3390/microorganisms7090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie M, Dong N, Chen K, Yang X, Ye L, Chan EW-C, Zhang R, Chen S. 2020. A hybrid plasmid formed by recombination of a virulence plasmid and a resistance plasmid in Klebsiella pneumoniae. J Glob Antimicrob Resist 23:466–470. 10.1016/j.jgar.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 37.Nishida H. 2012. Comparative analyses of base compositions, DNA sizes, and dinucleotide frequency profiles in archaeal and bacterial chromosomes and plasmids. Int J Evol Biol 2012:342482. 10.1155/2012/342482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. 2017. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio 8. 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050. 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 40.Burns JH, Strauss SY. 2011. More closely related species are more ecologically similar in an experimental test. Proc Natl Acad Sci USA 108:5302–5307. 10.1073/pnas.1013003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russel J, Roder HL, Madsen JS, Burmolle M, Sorensen SJ. 2017. Antagonism correlates with metabolic similarity in diverse bacteria. Proc Natl Acad Sci USA 114:10684–10688. 10.1073/pnas.1706016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graf FE, Palm M, Warringer J, Farewell A. 2019. Inhibiting conjugation as a tool in the fight against antibiotic resistance. Drug Dev Res 80:19–23. 10.1002/ddr.21457. [DOI] [PubMed] [Google Scholar]
- 43.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russel J, Pinilla-Redondo R, Mayo-Munoz D, Shah SA, Sorensen SJ. 2020. CRISPRCasTyper: automated identification, annotation, and classification of CRISPR-Cas loci. Crispr J 3:462–469. 10.1089/crispr.2020.0059. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson J, Nash JHE. 2018. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom 4:e000206. 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biswas A, Gagnon JN, Brouns SJ, Fineran PC, Brown CM. 2013. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol 10:817–827. 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.01189-22-s0001.xlsx, XLSX file, 1.8 MB (1.8MB, xlsx)
Supplemental material. Download aac.01189-22-s0002.pdf, PDF file, 2.1 MB (2.1MB, pdf)