Abstract

While oral desensitization is capable of alleviating peanut allergen anaphylaxis, long-term immune tolerance is the sought-after goal. We developed a liver-targeting lipid nanoparticle (LNP) platform to deliver mRNA-encoded peanut allergen epitopes to liver sinusoidal endothelial cells (LSECs), which function as robust tolerogenic antigen-presenting cells that induce FoxP3+ regulatory T-cells (Tregs). The mRNA strand was constructed by including nucleotide sequences encoding for nonallergenic MHC-II binding T-cell epitopes, identified in the dominant peanut allergen, Ara h2. These epitopes were inserted in the mRNA strand downstream of an MHC-II targeting sequence, further endowed in vitro with 5′ and 3′ capping sequences, a PolyA tail, and uridine substitution. Codon-optimized mRNA was used for microfluidics synthesis of LNPs with an ionizable cationic lipid, also decorated with a lipid-anchored mannose ligand for LSEC targeting. Biodistribution to the liver was confirmed by in vivo imaging, while ELISpot assays demonstrated an increase in IL-10-producing Tregs in the spleen. Prophylactic administration of tandem-repeat or a combination of encapsulated Ara h2 epitopes induced robust tolerogenic effects in C3H/HeJ mice, sensitized to and subsequently challenged with crude peanut allergen extract. In addition to alleviating physical manifestations of anaphylaxis, there was suppression of Th2-mediated cytokine production, IgE synthesis, and mast cell release, accompanied by increased IL-10 and TGF-β production in the peritoneum. Similar efficacy was demonstrated during LNP administration postsensitization. While nondecorated particles had lesser but significant effects, PolyA/LNP-Man lacked protective effects. These results demonstrate an exciting application of mRNA/LNP for treatment of food allergen anaphylaxis, with the promise to be widely applicable to the allergy field.
Keywords: peanut allergy, lipid nanoparticles, mRNA delivery, liver-targeting, anaphylaxis
Food allergy affects 15 million people in the United States, including children and adults.1,2 Of particular concern is the steady rise in the prevalence of peanut allergy in children, with impact on lifestyle and potentially life-threatening allergic reactions. We asked whether a mRNA approach could be used to achieve immune tolerance for the prevention and treatment of peanut anaphylaxis, a highly prevalent food allergy that impacts one in 50 children and one in 200 adults.1,2 As a front-runner of this study, we have previously shown that the delivery of an egg-white protein, ovalbumin (OVA), or a dominant peanut allergen, Arachis hypogaea protein 2 (Ara h2), can alleviate anaphylaxis if these proteins are encapsulated in poly(lactic-co-glycolic acid) (PLGA) nanoparticles, followed by targeted delivery to the stabilin scavenger receptor expressed on liver sinusoidal endothelial cells (LSECs).3−5 The basis of this tolerogenic effect is the ability of LSECs to act as specialized antigen-presenting cells (APCs), which generate regulatory T-cells (Tregs), capable of suppressing T-helper 2 (Th2) immunity, IgE production, mast cell release, and anaphylaxis resulting from allergen sensitization (Figure 1).6,7 Noteworthy, it has also been shown that short nonallergenic peptide sequences, present in these food allergens, can be selected for class II major histocompatibility (MHC-II) binding and presentation to naïve CD4+ T-cell precursors of Tregs.5 This is in keeping with the discovery of so-called immune-regulatory T-cell epitopes capable of suppressing allergic responses to cat, house dust mite, and grass pollen in preclinical and human studies.8 In fact, the most robust T-cell epitope identified in our NIAID Immune Epitope Database (IEDB) search for MHC-II binding Ara h2 peptides was more effective for anaphylaxis prevention than the intact, purified Ara h2 protein.5 The same applies to an MHC-II-presented OVA peptide compared to the whole egg-white protein.3−5
Figure 1.
Schematic to explain liver targeting by a tolerogenic mRNA/LNP platform for treatment of peanut anaphylaxis. (A) Liver microanatomy to delineate the localization of liver sinusoidal endothelial cells (LSECs) that are targeted by ionizable cationic LNPs to deliver mRNA. (B) Hypothesis for considering the use of a LNP carrier to target tolerogenic LSEC premised on previous discovery that the delivery of ovalbumin and peanut allergen (Ara h2) epitopes by a PLGA nanocarrier is capable of Treg generation, with the capability of suppressing allergic inflammation and anaphylaxis.5 We propose that the same outcome is accomplishable with mRNA constructs that express single-tandem or a combination of peanut allergen epitopes routed to the MHC-II compartment for CD4+ T-cell activation. These are also known as T-cell epitopes.8 We list the advantages and challenges of mRNA delivery, with a proposal to test the outcome in animal models of oral peanut sensitization and anaphylaxis induction.
The advantages of developing a tolerogenic mRNA platform instead of using a peptide-delivery platform include facile carrier design, reducing the cost of peptide synthesis, and ease of obtaining nucleic acid incorporation into the nanocarrier, in contradistinction to the challenge of loading peptides with heterogeneous charge and solubility (Figure 1). Moreover, mRNA presents a versatile platform for coexpression and loading of multiple antigens or epitopes, in addition to adapting the nucleotide backbone for tuning the immune response in a desired way.9 Thus, while unmodified mRNA is capable of inducing type I interferon production through the activation of endosomal toll-like receptors (e.g., TLR-7) or inflammasomes (e.g., MDA5, RIG-I, NOD2, and PKR), it is advantageous to substitute uridine with methyl-pseudouridine to prevent excessive innate immune activation.9 Not only is this beneficial for tolerance induction but also the uridine modification enhances protein expression as well as cost-efficient mRNA production, using in vitro cell-free transcription to prepare the expression strand. Moreover, the mRNA construct can be endowed with an endoplasmic reticulum (ER) membrane targeting sequence to ensure epitope routing to the MHC-II compartment, essential for T-cell epitope presentation to CD4+ T-cells.10,11 It has also been established that peptide expression, reliant on small amounts of mRNA, is very effective for the high level of antigen expression by APC, without the danger of nuclear entry or genome integration, resulting from the use of DNA constructs.12,13
In spite of these listed advantages, it is important to consider that most mRNA immune-modulatory approaches currently are aimed at generating antibody or cytotoxic T-cell responses for the treatment of infectious diseases (e.g., COVID-19) or cancer immunotherapy.14,15 In addition, most of these applications are designed for intact proteins or protein subunits, rather than considering the adaptability and diversity of epitopes for tolerogenic immunotherapy (Figure 1B). A further advantage is that the lipid nanoparticles (LNPs) used for mRNA encapsulation will distribute to the liver after intravenous injection, where a large LSEC capture bed can participate in cellular uptake, further enhanced by particle surface decoration with a lipid-anchored mannose ligand targeting the mannose receptor in these APCs (Figure 1A).16 We will refer to these particles as mRNA/LNP.
Against this background, we investigated the possibility of targeted delivery of epitope-delivering mRNA/LNP to LSECs, capable of suppressing anaphylaxis to a crude peanut protein extract. To accomplish this, we developed codon-optimized mRNA constructs for the expression of immune dominant Ara h2 epitopes, routed to the MHC-II compartment for purposes of Treg generation. We demonstrate successful encapsulation of in vitro-synthesized mRNA constructs in mRNA/LNP, designed for liver biodistribution and LSEC uptake. Administration of these tolerogenic NPs was successful in preventing anaphylaxis in a murine peanut anaphylaxis model. Alleviation of the anaphylaxis response was accompanied by evidence of suppressing Th2 immunity, allergen sensitization, IgE production, and mast cell release. These features also render the platform usable for additional allergic disorders. This demonstrates another use application for mRNA delivering nanoparticles.
Results
Synthesis and Characterization of LNP to Deliver mRNA Constructs
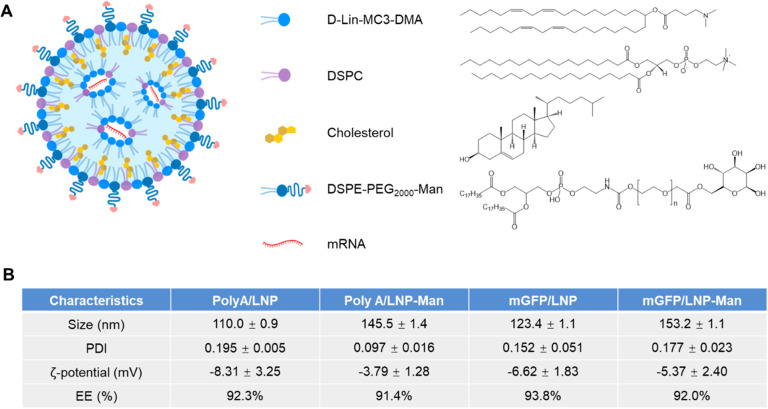
Our primary study goal was to develop an epitope-expressing mRNA carrier for peanut allergen delivery and presentation by tolerogenic LSECs. The selection of mRNA instead of protein or peptide sequences is the ease of producing nucleic acid constructs to conduct immunotherapy, as demonstrated for COVID vaccination as well as cancer immunotherapy.17 However, in order to accomplish antigen expression by LSECs, it is important to use LNP compositions that favor liver biodistribution. This was accomplished by including an ionizable cationic lipid, 4-(dimethylamino)butanoic acid (DLin-MC3-DMA, aka MC3) and helper lipids [distearoylphosphatidylcholine (DSPC) and cholesterol] as well as a surface-stabilizing lipid component, dimyristoyl glycerol polyethylene glycol 2000 (DMG-PEG2000) into our particle design (Figure 2A).18 To demonstrate that the capability of such a particle design to deliver a green fluorescence protein (mGFP) reporter-encoding mRNA construct to the liver, a microfluidic synthesis process was used (Figure 2A).19 This involved the use of a NanoAssemblr Benchtop for blending and mixing the aqueous mGFP phase (prepared in RNase-free sodium acetate buffer) with an ethanol-dissolved lipid phase that included DLin-MC3-DMA, DSPC, cholesterol and DMG-PEG2000 at molar ratios of 50, 10, 38.5, 1.5 (Figures 2A and Supporting Information Figure S1).20 PolyA was used as a nonfluorescent nucleic acid strand, serving as control, while also providing an inexpensive source to fine-tune optimum volume and flow rate settings for the NanoAssemblr (Figure S1). Following the synthesis of mGFP/LNP and PolyA/LNP particle batches, the materials were dialyzed against PBS (pH 7.4) and fully characterized.21 This included the performance of dynamic light scattering (DLS) (Figure 2B) and cryo-EM (Figure 2C). The cryo-EM images confirmed the formation of multilamellar, electron-dense particle cores obtained through the nanoprecipitation of the mRNA by MC3. DLS demonstrated the synthesis PolyA and mGFP particles of sizes 110 and 127 nm and corresponding ζ potentials of −11.4 and −5.5 mV, respectively.
Figure 2.
Preparation and characterization of unmodified LNP. (A) Schematic to explain microfluidics mixing for construction of LNPs, which are composed of an ionizable cationic lipid (DLin-MC3-DMA), helper lipids (cholesterol and DSPE), and DSPE-PEG2000. (B) Physicochemical characterization of LNP size, polydispersity index (PDI), and ζ-potential. mRNA encapsulation efficiency (EE) was determined by the RiboGreen assay. (C) Cryo-EM images of PolyA/LNP and mGFP/LNP. Scale bars are 100 nm. (D) Confocal images to show GFP expression, following LSEC incubation with 0.5 μg/mL PolyA/LNP or mGFP/LNP for 24 h. The images were obtained by staining nuclei and the surface membrane with Hoechst33342 and WGA594, respectively. Scale bar is 40 μm. (E) In vivo and ex vivo IVIS images following IV injection of 1 mg/kg DiR/mGFP/LNP. The ex vivo images of the explanted organs were used to show that most of particles are localized in the liver and spleen. The lower panel shows a digital fluorescence scanning view of a liver slice to show the sinusoidal distribution of GFP expression in relation to the more scattered distribution of Kupffer cells. GFP staining was performed with anti-GFP antibody (AF488), and Kupffer cells were detected using an anti-F4/80 antibody (AF647). Left scale bar, 500 μm; right scale bar, 200 μm, c.v., central vein (n = 3).
To determine if mGFP delivery can achieve GFP expression in vitro, confocal imaging was performed in a liver sinusoidal endothelial cell line incubated with mGFP/LNP for 24 h (Figure 2D). This confirmed GFP expression, compared to the inability of PolyA/LNP to generate a fluorescent product. To determine whether GFP expression can be obtained in vivo, an mGFP/LNP was synthesized in which 0.3% of the cholesterol content was replaced by the carbocyanine iodide dye, DiOC18(7) (DiR). Physicochemical characterization of the DiR/mGFP/LNP is shown in Figure S1. These particles were intravenously (IV) injected in C3H/HeJ mice at 1 mg/kg (n = 3), followed by IVIS image collection 6 h later to determine in vivo biodistribution (Figure 2E). Image analysis demonstrated a strong DiR signal in an upper abdominal location, which represents the liver. This was confirmed by subsequent animal sacrifice and organ explantation to obtain ex vivo images. These data show most LNPs distributed to the liver, with some particles also trafficking to the spleen (Figure 2E and Figure S2). Following liver tissue sectioning, digital fluorescence microscopy was performed after staining with AlexaFluor488-conjugated anti-GFP as well as anti-F4/80 antibodies (the latter to identify Kupffer cells). Visualization under an Aperio AT Turbo scanner demonstrated widespread GFP expression in a sinusoidal distribution fashion around the central vein, compared to the more disperse punctate distribution of Kupffer cells through the liver lobules (Figure 2E). This agrees with the previous demonstration that LNPs that contain ionizable cationic DLin-MC3-DMA and DMG-PEG2000 are capable of distributing to all major liver cell types that exhibit tolerogenic properties, including Kupffer cells, LSECs, and hepatocytes.22 Digital fluorescence microscopy was also used for the spleen, where it could be shown that GFP expression is mostly localized to the white pulp, where lymphoid elements such as lymphocytes surround splenic blood vessels (Figure S2).
Enhancement of LSEC Targeting Efficiency by LNP Surface Decoration with a Mannose Ligand
The widespread sinusoidal distribution of mRNA-expressing LNPs in the liver has been ascribed to the formation of an apolipoprotein E (ApoE) protein corona in the circulation, which is assisted by the dissociation of the 14-carbon lipid tail in DMG-PEG2000 from the particle surface.23 ApoE targets the low-density lipoprotein receptor (LDLR).24 Although Kupffer cells and hepatocytes contribute to the tolerogenic capabilities of the liver, the more robust Treg-inducing effects of LSECs are preferred for use in our application.3−5 Thus, to improve LSEC uptake of mRNA/LNP, a commercially available lipid DSPE-PEG2000-mannose was used for particle surface decoration, with a view to enhance LSEC uptake through binding to the CD206 mannose receptor (Figure 3A).25 Not only does this preserve the one-step synthesis approach for LNP production but also introduces an 18-carbon DSPE lipid tail, which is more stably associated with the LNP surface, compared to the DMG-PEG2000 lipid tail. Thus, the LNP composition shown in Figure S3A was used for successful synthesis of a series of mannose-decorated (mGFP/LNP-Man, PolyA/LNP-Man), as well as nondecorated (mGFP/LNP, PolyA/LNP) LNPs. Particle characterization is shown in Figures 3B and S3B,C. While the spherical particle morphology and slight negative ζ potential are maintained, the addition of the mannose ligand leads to a 25–35 nm size increase, which could also be advantageous to promote LSEC uptake by slowing particle egress through the endothelial fenestrae into the space of Disse. Aliquots of the PolyA/LNP or PolyA/LNP-Man was also used to assess particle stability in storage at 4 °C over 15 days. This demonstrated slight increase in the particle size, while maintaining PDI and the percent encapsulation efficacy (EE) of nucleic acid loading (Figure S3D).
Figure 3.
Preparation and characterization of mannose-modified LNP (LNP-Man) for enhancing LSEC uptake. (A) Composition of LNP-Man, including mannose contained lipid DSPE-PEG2000-Man. (B) Particle physicochemical characterization, including size, polydispersity (PDI), ζ potential, and encapsulation efficiency (EE) (n = 3).
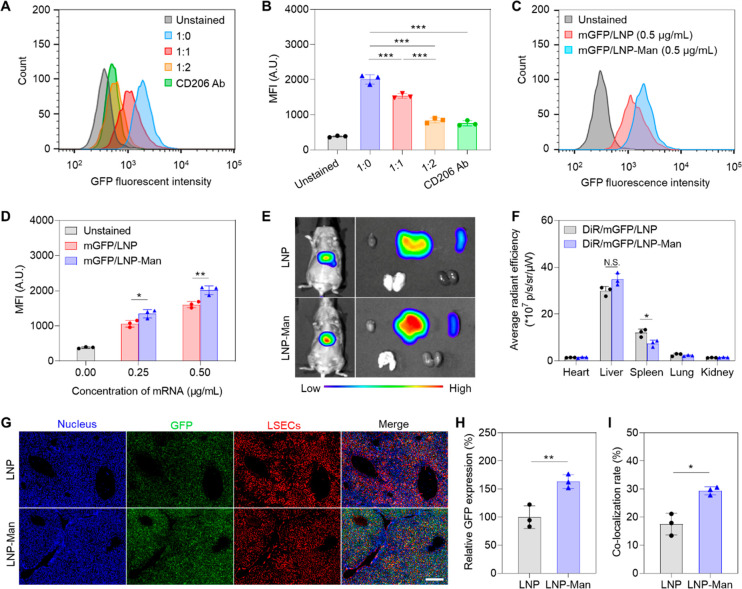
In order to assess the specificity of the ligand/receptor interaction and abundance of GFP expression, a series of flow cytometry experiments were performed in a LSEC line (Figure 4). The first experiment looked at the effect of cellular incubation with a mixture of mGFP/LNP-Man in the presence of incremental amounts of PolyA/LNP-Man (1:0, 1:1, and 1:2) for 24 h. This allowed assessment of nonfluorescent particles interfering in the binding of GFP-expressing particles, when the former is present in molar axis. The binding competition using this approach takes into consideration the appropriate stereospecific and density of the ligand display on the particle surface, as compared to free ligand. The competition of bound mannose was also compared to the impact of a blocking anti-CD206 antibody (Figure 4A,B). While mannose surface decoration increased GFP expression in the absence of binding competition, incremental amounts of nonfluorescent LNPs or anti-CD206 could be seen to decrease GFP expression (Figure 4A). Additional display of the data as a mean fluorescence intensity (MFI) was used for statistical comparison (Figure 4B). A second experiment compared the level of GFP expression of the nondecorated mGFP/LNP with decorated mGFP/LNP-Man, using 24 h particle exposure. Nontreated cells served as control (Figures 4C,D). This demonstrated increased GFP expression of decorated versus nondecorated LNPs (Figure 4C). Moreover, MFI calculations showed that in spite of increased GFP expression by both delivery vehicles, these expression levels were significantly increased for mannose-decorated particles (Figure 4D).
Figure 4.
Assessment of mannose-decorated LNP that deliver reporter-encoding GFP mRNA. The LNPs described in Figure 3 were used to perform these studies: (A, B) Flow cytometry to assess GFP expression in LSECs, exposed to 0.5 μg/mL mGFP/LNP-Man, with or without co-incubation with competitive binding PolyA/LNP-Man at ratios of 1:0, 1:1, and 1:2 for 24 h (n = 3). CD206 binding specificity was determined by including a blocking antibody. The statistically significant decline in GFP fluorescence as a result of binding competition was confirmed by assessing mean fluorescence intensity (MFI). (C, D) Flow cytometry to compare GFP fluorescence intensity in LSEC, following uptake of 0.25 and 0.5 μg/mL mGFP/LNP or mGFP/LNP-Man (n = 3) over 24 h. (E) In vivo and ex vivo IVIS imaging following IV injection of 1 mg/kg DiR-labeled nondecorated or mannose-decorated LNPs for 6 h (n = 3). Particles were synthesized as in Figure 3, with composition shown in Figure S5. (F) Average DiR radiant efficiency analysis of the main organs (n = 3). (G) Representative digital fluorescence scanning images to compare the intrahepatic distribution of DiR/mGFP/LNP and DiR/mGFP/LNP-Man. Nuclei were stained with DAPI, while GFP and LSEC were detected by anti-GFP (AF488) and anti-LYVE1 antibody (AF594), respectively. Scale bar is 200 μm. (H) Relative GFP fluorescence intensity compared using ImageJ analysis. (I) Fluorescence overlap to determine colocalization of GFP with LSEC, using ImageJ analysis (n = 3); N.S., no significant difference; * p < 0.05; ** p < 0.01; and *** p < 0.001.
Successful therapeutic cargo delivery and mRNA expression in LSECs depend on endosomal escape of the LNP, where lipid composition and the presence of the ionizable lipid play important roles in destabilizing the endosomal lipid bilayer (Figure S4A).26 This escape is dependent on formation of a particle hexagonal (HII) lipid phase, a property associated with ionizable cationic lipids, not observed in other lipid nanocarriers, such as liposomes. Thus, while cylindrical cone lipids (e.g., DSPC) favor the formation of lipid bilayers, protonated ionizable lipids (e.g., DLin-MC3-DMA) form inverted cone-shaped lipid structures (hexagonal phase), which have a destabilizing effect on the endosomal membrane, leading to mRNA release. To assess whether the surface decoration of a mannose ligand interfere with the membrane destabilizing effects of the hexagonal phase, the surrogate lipid bilayer of red blood cells (RBC) was used in an assay to assess the impact of the LNP lipid phase.27 This was accomplished by suspending RBC in pH 7.4 or pH 5.5 buffers in tissue culture plates, followed by the addition of different concentrations of PolyA/LNP or PolyA/LNP-Man. PBS served as a blank, while 0.1% Triton X-100 provided 100% hemolysis. The analysis demonstrated that while there was no hemolysis during particle addition at pH 7.4, both LNPs induced roughly equivalent membrane damage at pH 5.5 (Figure S4B,C). These results confirm that mannose decoration does not impact the hexagonal lipid phase activity.
We also assessed the impact of mannose surface ligation on particle distribution and GFP expression in vivo (Figure 4). This was accomplished by synthesizing DiR-labeled mGFP/LNP and mGFP/LNP-Man. Particle physicochemical characterization is shown in Figure S5. These particles were IV injected into C3H/HeJ mice, followed by IVIS imaging 6 h later (Figure 4E). The appearance of decorated and nondecorated particles in an abdominal location was confirmed to be directed to the liver and spleen after organ explantation and ex vivo imaging (Figure 4E). Assessment of DiR radiation efficiency confirmed 71.6% distribution to the liver with 17.2% and 4.7% of particles appearing in the spleen and lung, respectively (Figure 4F). No significant differences were noted for DiR/mGFP/LNP compared to DiR/mGFP/LNP-Man. To assess intrahepatic distribution, liver sections were stained with DAPI (nuclei), as well as antibodies recognizing GFP or the lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1), expressed on LSEC surfaces (Figure 4G). Fluorescence microscopy demonstrated that while both carrier types induced widespread GFP expression in the liver, the relative fluorescence intensity of mGFP/LNP-Man was 1.7-fold higher than that for mGFP/LNP (Figure 4H). Moreover, when comparing fluorescence overlap between GFP and LYVE1, the colocalization index in animals treated with DiR/mGFP/LNP was 17%, compared to 28% for DiR/mGFP/LNP-Man (Figure 4I). These data confirmed that mannose decoration was accompanied by increased GFP expression in LSECs.
Constructing mRNA Nanoparticles to Deliver Dominant Peanut Allergen Epitopes, With Generation of Tregs
We have previously described use of the NIAID Immune Epitope Database (IEDB) and Analysis Resource for identifying MHC-II binding epitopes in the dominant peanut allergen, Ara h2.5 This peanut allergen is responsible for IgE generation in >80% of peanut-allergic human subjects (Figure 5A).28 Four top-ranking 15-mer T-cell epitopes with affinity binding to murine MHC-II alleles (H2-IAb, H2-IEd, and H2- IAd) were identified (Figure S6A).5 Among them, epitope 4 was the most robust inducer of CD25+FoxP3+ T-cells in the spleen of mice receiving IV injection with a LSEC-targeting PLGA nanoparticle (Figure S6B).5 To design an equivalent mRNA construct, epitope 4 was endowed with natural flanking sequences, followed by the reverse translation (Figure 5B). Tandem-repeat epitope sequences were inserted in the mRNA construct downstream of an invariant Ii chain sequence (aa 1–80), specifically designed to allow the hybrid peptide to be inserted into the endoplasmic reticulum (ER) membrane of the MHC-II compartment (Figure 5C). Figure S7 explains the role of the Ii chain, including insertion into MHC-II vesicles, where proteolytic processing of hybrid peptides allows insertion into the peptide groove and trafficking to the cell surface.29 The design of the mRNA strand was completed by the addition of start (ATG) and stop codons (TAA), followed by codon optimization (Figure 5C). This sequence was sent to TriLink for in vitro cell-free transcription during which the mRNA was further endowed with 5′ and 3′ UTR sequences and a PolyA tail, as described in Figures 5D and S6C.30,31 All uridine nucleotides in the strand were substituted with N1-methyl-pseudouridine before RNA purification and returning a 624-nucleotide strand (Figure S6C). This mRNA construct was used to synthesize mAra h2#4/LNP and mAra h2#4/LNP-Man carriers, capable of epitope 4 delivery. Particle construction proceeded as described in Figure 3, yielding spherical LNP of sizes 95.3 and 148.7 nm, respectively (Figure 5E). The corresponding cryo-EM images are shown in Figure S6D.
Figure 5.
Design and encapsulation of a codon-optimized mRNA strand for expression of peanut allergen epitopes. (A) Epitope selection in the NIAID Immune Epitope Database during a search for non-IgE binding Ara h2 T-cell epitopes that can be presented by the MHC-II complex. Our previous studies employing these peptide epitopes demonstrated the best tolerogenic effect with epitope 4.5 (B) For proof-of-principle testing, epitope 4 was incorporated as a tandem-repeat nucleotide sequence, following the addition of flanking amino acids. (C) mRNA expression construct designed by including two repeats of epitope 4 downstream of an Ii invariant chain sequence that inserts into the ER membrane, allowing epitope incorporation into the MHC-II complex, as explained in Figure S7. Start and stop codons were added. (D) In vitro synthesis, being contracted with TriLink, of a codon-optimized expression strand, also endowed with 5′ and 3′ UTR sequences as well as substituting all uridine bases with N1-methyl-pseudouridine (Figure S6). (E) Physicochemical characterizations of mAra h2#4/LNP and mAra h2#4/LNP-Man, including size, polydispersity (PDI), ζ potential, and encapsulation efficiency (EE).
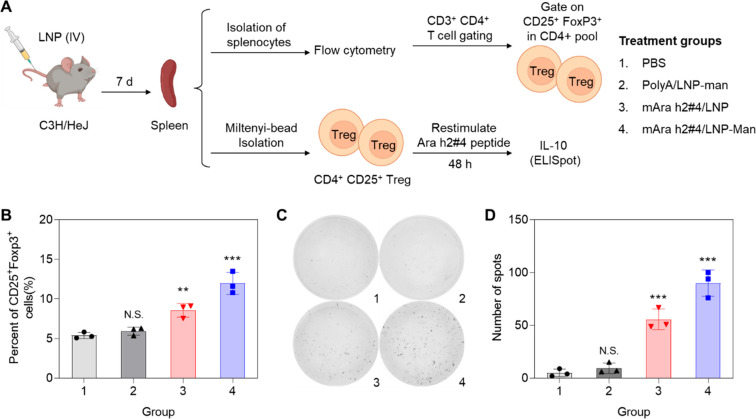
To determine whether epitope 4 delivery is capable of generating CD25+FoxP3+ T-cells in C3H/HeJ mice, animals (n = 3) were IV injected with 1.25 mg/kg LNP before spleen harvesting on day 7, preparation of splenocyte suspensions and staining with antibodies recognizing CD3, CD4, CD25, and FoxP3 (Figure 6A).5,32 Following gating on CD3+CD4+ cells, the percentages of CD25+FoxP3+ cells in each study group were recorded (Figure S8). This demonstrated significant (p < 0.01; p < 0.001) increases above the 5.4% CD25+FoxP3+ T-cells of untreated animals, reaching 8.6% and 11.1% after treatment with mAra h2#4/LNP and mAra h2#4/LNP-Man, respectively (Figure 6B). This includes a significant (p < 0.05) increase as a result of mannose decoration. PolyA/LNP-Man injection had no impact. We also conducted ELISpot assays to quantify the number of IL-10-producing cell clusters. Splenocyte suspensions were used to collect CD4+CD25+ cells by magnetic beads before the addition of the epitope 4 peptide for 48 h, followed by assessment of the number of IL-10-producing colonies (Figure 6C).33 This demonstrated a significant increase in the number of IL-10 spots in mAra h2#4/LNP and mAra h2#4/LNP-Man treated animals compared to control (Figure 6D). Meanwhile, the increase was significantly higher for mannose-decorated particles.
Figure 6.
mAra h2#4/LNP-Man generating IL-10-producing CD25+FoxP3+ T-cells in the spleen. (A) Experimental design to assess the impact of PolyA/LNP-Man, mAra h2#4/LNP, and mAra h2#4/LNP-Man on splenic generation of T-cells with phenotypic Treg characteristics and the ability to produce IL-10 (n = 3). (B) Spleens harvested 7 days after IV injection of 1.25 mg/kg LNPs and used to obtain splenocyte populations for staining with an antibodies combination recognizing CD3, CD4, CD25, and FoxP3 (n = 3). Flow cytometry gating, as explained in Figure S8A, was used to quantify the percent of CD25+FoxP3+ T-cells. (C, D) ELISpot assays performed in separate experiment to show the generation of IL-10-producing CD4+CD25+ T-cells prepared by magnetic bead separation of the splenocyte population. Cells were stimulated with the Ara h2#4 peptide before counting the number of IL-10 cellular spots. N.S., no significant difference; * p < 0.05; ** p < 0.01; and *** p < 0.001.
Prophylactic mAra h2#4/LNP Administration for Anaphylaxis Prevention in a Peanut Sensitization Model
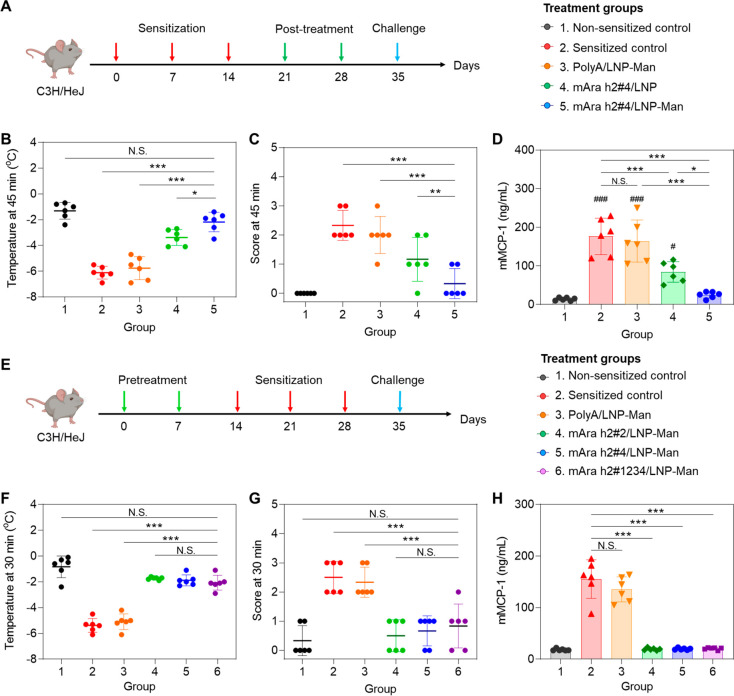
6–8 weeks old C3H/HeJ mice were orally sensitized with crude peanut extract (CPE), administered three times before peritoneal challenge to induce anaphylaxis (Figure 7A).5,34 Anaphylaxis was monitored by recording rectal temperatures, as well as scoring physical manifestations of anaphylaxis every 15 min, using the following criteria: 0 = no symptoms; 1 = ear and mouth scratching; 2 = puffiness around the eyes and mouth, pilar erection, labored breathing; 3 = prolonged motionlessness; 4 = severely reduced motility, tremors, severe respiratory distress; 5 = death.35 To assess the impact of prophylactic LNP administration, animals (n = 6) received two IV injections of 1.25 mg/kg mAra h2#4/LNP or mAra h2#4/LNP-Man 7 days apart, before the performance of 3 rounds of oral CPE sensitization, and finally CPE challenge (Figure 7A). While CPE challenge in nonsensitized animals did not induce hypothermia, noticeable temperature drops in untreated sensitized mice were observed, with a maximal and statistically significant decline at 45 min, before gradual recovery (by 120 min) (Figure 7B). Essentially similar results were obtained in animals pretreated with PolyA/LNP-Man, with no evidence of protection. However, prophylactic administration of mAra h2#4/LNP or mAra h2#4/LNP-Man prevented hypothermia onset, with the impact of mannose-decorated particles highly significant (p < 0.001) compared with nondecorated LNP (p < 0.01) (Figure 7C). The hypothermic response also agrees with the anaphylaxis scores, showing significant declines for mAra h2#4 pretreatment, with the response to mAra h2#4/LNP-Man being more significant (p < 0.05) than that for nondecorated LNP (Figure 7D,E). All considered, these data confirmed the significant tolerogenic effect of the Ara h2 epitope 4 TriLink nucleotide version.
Figure 7.
Prophylactic administration of decorated and nondecorated LNP delivering mAra h2#4, preventing peanut anaphylaxis. (A) Schematic to explain the experimental outline, making use of IV injection of 1.25 mg/kg PolyA/LNP-Man, mAra h2#4/LNP, and mAra h2#4/LNP-Man on two occasions in female C3H/HeJ mice (n = 6). These animals were sensitized by oral administration of 100 mg/kg CPE plus 0.5 mg/kg cholera toxin on three occasions before peritoneal challenge with CPE on day 35. Rectal temperatures and anaphylaxis scores were obtained every 15 min for 2 h. Blood, peritoneal lavage fluid, and spleen tissue were collected for further analysis. Nonsensitized mice were used as a negative control, while sensitized animals receiving CPE challenge without any co-treatment served as positive control. (B, C) Time-dependent rectal temperature scores, with statistical comparisons at the time of maximum hypothermia (45 min). (D, E). Time-dependent anaphylaxis scoring and statistical comparisons at the point of maximum impact (45 min). The scoring criteria are provided in Materials and Methods. (F) Total serum IgE assessed by ELISA. (G) ELISA measurement of mouse mast cell protease 1 (mMCP-1) in the serum. (H) Peritoneal fluid IL-4 concentration determined by ELISA. (I) Percent of CD25+FoxP3+ T-cells in the spleen. (J, K) Peritoneal fluid IL-10 and TGF-β concentrations determined by ELISA Additional data for quantification of antigen-specific IgG1 and IgE, IL-5, and IL-13 appear in Supporting Information Figure S9. N.S., no significant difference; * p < 0.05; ** p < 0.01; and *** p < 0.001.
Apart from the impact on physical disease manifestations, preadministration of mAra h2#4/LNP-Man, and to a lesser (yet statistically significant) extent for mAra h2#4/LNP, suppressed the rise in total serum IgE, antigen-specific IgE, and murine mast cell protease-1 (mMCP-1) release (Figures 7F, S9, and 7G, respectively).36 The same impact was also seen for peanut-specific IgG1, which, as for IgE, identifies an IL-4 mediated immunoglobulin class switching event (Figure S9D,E). This was also confirmed by showing mAra h2#4/LNP-Man suppression of IL-4 release in the peritoneal fluid, in addition to impacting IL-5 and IL-13 (Figures 7H and S9F,G). Noteworthy, PolyA/LNP-Man had no impact.
In order to assess the tolerogenic effects of mAra h2#4 on the percent of CD25+FoxP3+ T-cells in the spleen, flow cytometry was used as described in Figure S9H, demonstrating a highly significant increase of the Treg-associated phenotype during treatment with both particles, including a higher level of increase for mannose-decorated LNP (Figure 7I). The same was true for the quantification of IL-10 (Figure 7J) and TGF-β (Figure 7K) in the peritoneal lavage fluid. This is compatible with Treg action in the peritoneal cavity.
Postsensitization mRNA Epitope Delivery to Suppress Peanut-Induced Anaphylaxis
While prophylactic interference in allergic sensitization is accomplishable (Figure 7), a key question is whether Treg generation by mAra h2#4/LNP can prevent anaphylaxis once allergic sensitization is in progress. This was accomplished by adopting the protocol in Figure 8A, allowing 2 doses of PolyA/LNP-Man, mAra h2#4/LNP, and mAra h2#4/LNP-Man to be administered IV in C3H/HeJ mice (n = 6) after 3 rounds of oral sensitization (Figure 8A). Peritoneal CPE challenge triggered hypothermia and onset of anaphylaxis in nontreated animals or animals injected with PolyA/LNP-Man (Figures 8B,C). Noteworthy, the administration of both particle types to deliver epitope 4 interfered in the generation of anaphylaxis, with the impact of mAra h2#4/LNP-Man significantly better (p < 0.001) than mAra h2#4/LNP (p < 0.01). The time-dependent response profiles are shown in Figure S10B,C. Suppression of the physical manifestations of anaphylaxis was accompanied by a reduction in mast cell protease release (Figure 8D), serum IgE elevation (Figure S10D), serum IgG1 elevation (Figure S10E,F) and IL-4 increase in peritoneal lavage fluid (Figure S10G). Moreover, the tolerogenic effect of mAra h2 delivery postsensitization was accompanied by an increase in the percent of CD25+FoxP3+ T-cells in the spleen (Figure S10H) as well as by elevation of IL-10 (Figure S10I) and TGF-β (Figure S10J) levels in peritoneal fluid. Mannose-decorated LNP was more effective than nondecorated particles in all response outcomes.
Figure 8.
Therapeutic effects of mAra h2#4/LNP postsensitization as well as comparison of additional mRNA epitopes in a prophylaxis model. (A) Experimental timeline to assess the therapeutic effect of different nanoparticles (PolyA/LNP-Man, mAra h2#4/LNP, mAra h2#4/LNP-Man), administered postsensitization in the same CPE-induced anaphylaxis model described in Figure 7 (n = 6). (B). Comparison of body temperatures at the time of maximum hypothermia (45 min) postchallenge. (C) Comparison of the anaphylaxis scores at 45 min. (D) mMCP-1 release to the serum in the same experiment, with additional biological response parameters shown in Figure S10. (E) Experimental timeline for comparing the prophylactic effects of decorated LNP delivering mRNA constructs encoding for Ara h2 epitope 2 and epitope 4 as well as an epitope combination (1, 2, 3, and 4) (n = 6). The designs of the different mRNA-encoding strands are shown in Figure S11A–C, with the physiochemical characterization of the corresponding LNP-Man in Figure S11D. (F) Comparison of the anaphylactic response at the point of maximal hypothermia (30 min). (G) Comparison of the anaphylaxis scores at 30 min. (H) mMCP-1 serum concentration. The time-dependent hypothermia and anaphylaxis scores are shown in Figure S12. N.S., no significant difference; * p < 0.05; ** p < 0.01; and *** p < 0.001.
Single- versus Multiple-Epitope Comparison in the Prophylactic Model
Previous analysis of Ara h2 peptide delivery by PLGA nanoparticles demonstrated that epitope 4 exerted the most robust tolerogenic effect, in addition to exhibiting the most favorable solubility features that allows the best loading capacity.5 We were interested, therefore, in whether the delivery of mRNA constructs encoding epitope 2 or a combination of epitopes 1, 2, 3, and 4 can exert comparable effects to epitope 4. Utilizing the synthesis and design features outlined in Figure 5, two additional codon-optimized mRNA constructs were ordered from TriLink. These constructs included a dual repeat of Ara h2 epitope 2 as well as the combination of epitopes 1, 2, 3, and 4, as outlined in Figure S11A–C. Both constructs were designed to be expressed downstream of the invariant Ii chain with the required linkers, start and stop codon, and design modifications as shown in Figure 5. These mRNA constructs were used to synthesize two additional mannose-decorated LNPs (mAra h2#2/LNP-Man and mAra h2#1234/LNP-Man), which were characterized in comparison to mAra h2#4/LNP-Man. Figure S11D demonstrates the particle physicochemical characteristics, showing particle sizes ∼ 150 nm, PDI < 0.2, slight-negative surface charge, and EE > 90%.
These particles were used to assess the effect of prophylactic administration on hypothermia and anaphylactic scores, using the same approach as described in Figure 7A. Noteworthy, the results demonstrate that the prophylactic efficacy of mAra h2#2/LNP-Man and mAra h2#1234/LNP-Man was comparable to mAra h2#4/LNP-Man, with no significant differences in alleviating anaphylaxis (Figure 8E–G and Figure S12). In contrast, PolyA/LNP-Man had no protective effect. All of the Ara h2 epitopes prevented the release of mMCP-1 into the serum (Figure 8H).
LNP Safety Analysis in Nonsensitized Animals
The structural headgroup of cationic lipids have the potential to generate cytotoxicity by engaging pro-apoptotic cascades.37 LSECs were used to conduct a cytotoxicity study in which incremental doses of PolyA/LNP and PolyA/LNP-Man particles were used to perform a MTS assay, following 24 h of exposure (Figure S13A). The results showed that while both PolyA/LNP and PolyA/LNP-Man exert little cytotoxicity at doses < 1 μg/mL, a decrease in cell viability was observed at a concentration of 2.5 μg/mL (Figure S13A). Utilizing the same particles for IV injection into 6-week-old mice (n = 3) at 2 mg/kg, studies were undertaken to look for toxicity in the liver and other organs (Figure S13B). The concentrations of ALT, AST, BUN, and creatinine were measured in the serum, 24 and 48 h after injection (Figure S13C). There were no significant changes in liver or kidney biochemical parameters. Animals were subsequently sacrificed and H&E staining was performed on tissue sections from the heart, liver, spleen, lung, and kidney (Figure S13D). No evidence was obtained of any histological abnormalities in these organs.
Discussion
We demonstrate the development of a liver-targeting cationic LNP platform to deliver mRNA-encoded peanut allergen epitopes to LSECs, capable of acting as tolerogenic APC for interference in anaphylaxis, including through the generation of regulatory T-cells. The encoding mRNA strand was constructed by including nucleotide sequences that express T-cell epitopes, identified in an IEDB search of the dominant peanut allergen protein, Ara h2. Single-tandem- or combined-epitope sequences were inserted downstream of an MHC-II routing sequence. Following codon optimization, in vitro strand synthesis proceeded by the addition of 5′ and 3′ UTR sequences as well as uridine replacement by N1-methyl-pseudouridine. The mRNA was used for the preparation of LNPs, including a variation in which the particle surface is decorated with a lipid-anchored mannose ligand. IV injection of the mAra h2#4/LNP-Man increased the generation of IL-10-producing CD25+FoxP3+ T-cells in the spleen. Prophylactic administration of these particles also induced robust tolerogenic effects in an oral sensitization anaphylaxis model to CPE proteins. In addition to alleviating the physical manifestations of anaphylaxis, the tolerogenic effects were accompanied by Treg generation, IL-10 and TGF-β release as well as suppression of Th2-mediated immunity, allergen-specific IgE synthesis, and mast cell release. Similar efficacy was demonstrated using mAra h2#4/LNP-Man delivery after sensitization. Nondecorated LNP exerted lesser but still significant effects, while PolyA/LNP-Man lacked tolerogenic effects. While it has previously been demonstrated that oral delivery of chitosan-DNA nanoparticles can generate immune protection in a murine model of peanut allergy, we could not find any other literature evidence of mRNA epitope delivery to treat peanut allergy or other forms of food anaphylaxis.38 All considered, our results demonstrate the feasibility of using a tolerogenic mRNA/LNP to treat anaphylaxis, introducing a technology platform that can be applied for the treatment of multiple allergic disorders as well as autoimmune diseases.
The most significant finding of this study is the demonstration that mRNA delivery of dominant peanut allergen epitopes can induce a tolerogenic response, capable of interfering in the generation of anaphylaxis by CPE. Noteworthy, this outcome is accomplishable by single-tandem as well as a combination of mAra h2 epitopes, in keeping with our previous demonstration that the encapsulated delivery of MHC-II-binding Ara h2 or ovalbumin peptides to LSECs can suppress allergic inflammation and anaphylaxis by initiating a tolerogenic pathway that involves Treg generation in the liver.4,5 Moreover, the tolerogenic effect could be sustained for at least 60 days, demonstrated by experimentally increasing the duration of time between prophylactic particle administration up to the day of challenge.5 Our finding with the use of the peanut allergen epitopes is also in keeping with a growing interest and utility of short MHC-II binding T-cell epitopes for immunotherapy of cat, house dust mite, grass pollen, and food allergies, as demonstrated in proof-of-concept studies in animal models as well as the emergence of randomized, double-blind, placebo-controlled clinical trials involving short synthetic immune-regulatory T-cell epitopes.8,39,40
Confirmation of the efficacy of mRNA-based T-cell epitope delivery to the liver introduces a number of advantages over a more expensive tolerogenic therapy that makes use of peptides, in addition to offering a versatile combinatorial platform for coexpression of multiple epitopes and immunomodulatory molecules. Nonetheless, the use of mRNA constructs for epitope delivery by LNPs requires careful consideration of design features. A core requirement for mRNA introduction into the endosomal compartment in LSECs is the cytosolic release that is reliant on the hexagonal (HII) lipid phase of the LNP (Figure S4A), as well as subsequent routing of the epitopes to the MHC-II compartment by the Ii invariant chain, which is able to insert into the ER membrane (Figure S6).41 The replacement of uridine with N1-methyl-pseudouridine in the nucleotide backbone constitutes another important design feature to avoid skewing of the immune response, which could negatively impact the tolerogenic effects of LSECs.6 An additional design feature of the LNP was the lipid selections to favor biodistribution to the liver as well as the addition of a mannose ligand, which facilitates endocytic uptake after docking onto CD206 scavenger receptor on LSECs.16 This was accomplished by making use of the 18-carbon lipid tail of DSPE-PEG2000-mannose, which provides stable attachment to the particle surface.23 This design modification improved LSEC uptake in the liver compared to DMG-PEG2000 (Figure 4H) and was also associated with a significant increase in the tolerogenic effect of decorated LNP. Collectively, these LNP design features are of particular significance for mRNA expression in the liver in contrast to other lipid-based immunomodulatory nanocarriers that can be used for cancer immunotherapy, for example, successful implementation of mannose-modified liposomes to co-deliver papillomavirus peptide plus a CpG oligodeoxynucleotide adjuvant for enhancing antitumor immunity.42
Importantly, although we ascribe the tolerogenic effects of mRNA epitope delivery to Treg generation, we do not rule out a contribution by other tolerogenic cell types, including anergic T-cells, type I regulatory T-cells (Tr1), regulatory B-cells, etc. What is clear from our data is that the tolerogenic effects of LNP include the ability to interfere with Th2 immunity, as evidenced by the decrease in total and antigen-specific IgE production, mast cell protease release, and limiting IL-4, IL-5, and IL-13 production. The clampdown on Th2 immunity could involve indirect (e.g., TGF-β, IL-10, and IL-35) or direct immune suppressive pathways (e.g., cell–cell contact). This is in keeping with the major immune suppressive mechanisms used by Tregs, including (i) Treg cytolysis by granzyme A, granzyme B, and perforin; (ii) IL-2 depletion and apoptosis through Treg expression of CD25+; (iii) checkpoint receptor expression (e.g., CTLA-4, LAG-3, and PD-L1), interfering in CD80/CD86 co-stimulation, dendritic cell maturation, or indoleamine 2,3-dioxygenase (IDO) production; (iv) ectonucleotidase expression (e.g., CD39 and CD73), leading to ATP conversion to adenosine; (v) inhibition of cell migration and tissue inflammation.43
An intriguing question is how the use of a single Ara h2 epitope exerts tolerogenic control over a potentially large number of participating IgE-binding epitopes, displayed among 17 peanut allergens in the extract. One possibility is that Ara h2 T-cell epitopes make a dominant contribution to the alleviation of allergic inflammation induced by the accompanying IgE-binding epitopes in the same allergen.8,39,40 Another possibility is the occurrence of infectious tolerance, denoting tolerance transfer from one cell population to another.44,45 This could include a role for FoxP3+ Tregs, capable of converting conventional T-cells into regulatory T-cells, either indirectly (e.g., TGF-β, IL-10, or IL-35) or through direct interference (e.g., checkpoint receptor expression).44 Growing evidence for the participation of infectious tolerance in the suppression of allergic disease has been provided previously.8 For instance, a dominant T-cell epitope in the major house dust mite allergen, Der p 1, is capable of inducing tolerance to whole Der p 1 4A, or a crude house dust mite extract.46,47 Also, administration of selected Fel d 1 peptides has been shown to alter T-cell responses to cat allergen extract in humans, including non-Fel d 1 related peptides.48 Similar findings of linked suppression with alteration of cytokine profiles have also been demonstrated by using dominant T-cell epitopes in murine allergy models for tree pollen (e.g., birch, cedar, and olive), bee and hornet venom, egg-white protein, and Timothy grass pollen.8,39,40 Based on these findings, we propose that tolerogenic mRNA lipid nanoparticles could also be useful for treating a wide range of allergies, including epitope combinations derived from more than one allergen to treat mixed food allergies, e.g., peanut plus egg-white proteins. An important requirement for the clinical translation of mRNA epitope delivery for each disease application necessitates studies that address response duration, frequency of administration, and the required nucleic acid dose to assess efficacy outcome.
mRNA epitope therapy could also be applicable to the treatment of autoimmune diseases. For example, it has been demonstrated that a mRNA-based construct expressing a myelin oligodendrocyte glycoprotein peptide, MOG35-55, is capable of inducing Tregs that prevent the generation of experimental autoimmune encephalomyelitis (EAE) in response to the same peptide in a mouse model resembling multiple sclerosis.49 These results also agree with the previous demonstration that the delivery of a MOG35-55 nucleic acid construct to the liver by an adenovirus-associated vector can prevent EAE or reverse neurological damage in animals challenged with the immune epitope.50 Based on the finding that multiple antigens and epitopes play a role in autoimmune disease processes such as type I diabetes, the availability of multiple-epitope mRNA vaccines could also make inroads into the treatment of these disease processes.
Conclusions
In summary, we developed a mannose-modified LNP that encapsulates mRNA designed for expressing peanut epitopes for biodistribution to the liver, LSEC uptake and ability to induce a tolerogenic effect, capable of suppressing an anaphylactic response to a crude peanut allergen extract. The tolerogenic effect was accompanied by the suppression of Th2 immunity, IgE production, and mast cell release. Since this platform is highly adaptable for targeted delivery of mRNA epitopes, it provides a promising approach for therapeutic intervention in the range of allergic disorders as well as possibly also autoimmune disease.
Materials and Methods
Materials
DLin-MC3-DMA was purchased from MedKoo Bioscience, Inc. DSPC, cholesterol, and DMG-PEG2000 were obtained from Avanti Polar Lipids, Inc. DSPE-PEG2000-mannose was purchased from Biopharma PEG Scientific Inc., and PolyA was acquired from Sigma-Aldrich, Inc. GFP mRNA (mGFP) and the Ara h2 epitope-expressing mRNA constructs were ordered from TriLink Biotechnologies. DiR and RiboGreen RNA Assay Kit, mouse mMCP-1 uncoated ELISA Kit, and mouse IgE ELISA kit were purchased from Thermo Fisher Scientific. Mouse IL-10 ELISpot kit (HRP) was purchased from Mabtech Inc. CD4+CD25+ Regulatory T-cell isolation kit was purchased from Miltenyi Biotec. Mouse IL-4, IL-5, IL-10, IL-13, and TGF-β ELISA kits were purchased from DuoSet. Goat antimouse IgG1 secondary antibody (HRP) was purchased from Invitrogen.
mRNA Design for Expressing a Single or Combination of Ara h2 Epitopes
Epitope selection was performed by an Immune Epitope Database (IEDB) database search of Ara h2, a dominant peanut IgE-generating allergen in humans (Figure S7). The selection criteria are discussed in the legend of Figure S7. The initial mRNA construct was designed for expression of epitope 4, the best-performing tolerogenic peptide included in our PLGA nanocarrier.5 Subsequently, another mRNA construct was designed to encode epitope 2 as well as an epitope combination (i.e., epitopes 1–4). All epitopes were endowed with linker sequences, allowing proteolytic processing and MHC-II association. Epitope routing to the MHC-II compartment was accomplished by upstream insertion of a nucleotide sequence for expressing amino acids 1–80 of the invariant (Ii) chain, which targets the ER membrane (Figure S6). Start and (ATG) and stop (TAA) codons were added before codon optimization, using the GenSmartTM Codon Optimization tool (https://www.genscript.com/gensmart-free-gene-codon-optimization.html). This sequence was sent to TriLink for in vitro synthesis of a mRNA construct with full substitution of uridine by N1-methyl-pseudouridine, in addition to adding 5′ and 3′ UTRs and PolyA (120) tails, as described in Figure S7C.
Synthesis and Characterization of LNPs with and without Mannose Surface Decoration
LNPs were synthesized by a microfluidics approach, using a NanoAssemblr Benchtop (Precision NanoSystems Inc.) to blend and mix the lipid-containing organic phase with an aqueous phase containing the mRNA strand. The organic phase was prepared by mixing the ionizable cationic lipid, DLin-MC3-DMA, with DSPC, cholesterol, and DMG-PEG2000 or DSPE-PEG2000-mannose in ethanol at molar ratios of 50:10:38.5:1.5. The aqueous phase was prepared by diluting the mRNA constructs (PolyA, mGFP, or mAra h2) in RNase-free sodium acetate buffer (pH 4.0). Particles in the effluent phase were dialyzed against PBS (pH 7.4) to remove ethanol, before filtering (0.2 μm) to remove large aggregates. The LNP size, polydispersity index (PDI), and ζ potential were determined by dynamic light scattering (DLS) measurement, using a Zetasizer Nano ZS (Malvern Instruments Ltd.). mRNA encapsulation efficiency (EE) was determined by a RiboGreen (Thermo Scientific) assay to assess nucleic acid content. Cryogenic electron microscopy (Cryo-EM) images were obtained in a TF20 FEI Tecnai-G2 instrument to assess particle morphology. For the performance of in vivo imaging, we also synthesized DiOC18(7) (DiR)-labeled LNPs (DiR/mGFP/LNP and DiR/mGFP/LNP-Man), in which 0.3% of the cholesterol content was substituted to yield a lipid composition of DLin-MC3-DMA:DSPC:cholesterol:DMG-PEG2000 or DSPE-PEG2000-Man:DiR in the molar ratio 50:10:38.2:1.5:0.3. Particle stability was assessed by storing the PolyA/LNP and PolyA/LNP-Man at 4 °C, with the regular removal of aliquots to assess particle size, polydispersity, and RiboGreen content over 14 days.
GFP Expression and LSEC Targeting
LSEC were exposed to mGFP/LNP (0.5 μg/mL mGFP) at 37 °C for 24 h, followed by fixing in 4% paraformaldehyde and incubation with Hoechst33342 and WGA594 dyes at room temperature. GFP expression was observed by confocal microscopy (Leica SP8 STED). In order to compare GFP expression of nondecorated with mannose-decorated LNP, cells were incubated with mGFP/LNP or mGFP/LNP-Man at mGFP concentrations of 0.25 or 0.5 μg/mL for 24 h at 37 °C. Cells were treated with 0.25% trypsin and washed three times before assessing GFP mean fluorescence intensity (MFI) in a LSR Fortessa X-20 SORP flow cytometer. To assess the impact of mannose ligation on cellular uptake and transfection efficiency, a competitive inhibition experiment was carried out by mixing mGFP/LNP-Man with PolyA/LNP-Man at mGFP/PolyA ratios of 1:0, 1:1, and 1:2. These mixtures were incubated with LSEC for 24 h at 37 °C, keeping the mGFP concentration at 0.5 μg/mL. Flow cytometry was conducted to assess the MFI of GFP expression in LSECs, using the same instrument. Nontreated LSECs served as a blank, while anti-CD206 antibody was used as the positive control.
Hemolysis Assay
mRNA release in the cytosol is dependent on the LNP’s hexagonal lipid phase, which allows destabilization of the endosomal lipid membrane, as depicted in Figure S4A. Because it is difficult to assess the impact of mannose decoration on the function of the hexagonal lipid phase inside cells, a surrogate mouse red blood cell (RBC) hemolysis assay was used to assess lipid bilayer destabilization under acidic endosomal conditions. Purified murine RBC, suspended in PBS at a pH of 5.5 or 7.4, was mixed with different quantities (0.5, 1.0, and 2.0 μg/mL) of PolyA/LNP or PolyA/LNP-Man for 1 h at 37 °C. Plates were centrifuged at 3000 rpm to collect supernatants for assessing hemoglobin release in a microplate reader at 575 nm. RBC lysis with 0.1% Triton X-100 was used as the positive control (100% hemolysis).
LNP In Vivo Biodistribution
Animal experimentation was carried out in 6-week-old female C3H/HeJ mice, using 3 animals per group (n = 3), receiving tail vein injection with DiR/mGFP/LNP or DiR/mGFP/LNP-Man at a mGFP dose of 1 mg/kg. IVIS imaging (PerkinElmer Lumina II) was performed 6 h later to assess in vivo DiR distribution. Animals were sacrificed for ex vivo IVIS imaging of the explanted organs, as shown in Figures 2 and 4. We also used the liver tissue for paraformaldehyde fixation and sectioning. The mounted slides were stained with DAPI, as well as antibodies recognizing GFP and the LSEC surface receptor, LYVE1. Digital fluorescence microscopy was performed in a Leica SP8-STED/FLIM/FCS in the Electron Microscopy Services Center at UCLA.
CD25+FoxP3+ T-Cell Analysis in the Spleen
6-weeks-old female C3H/HeJ mice were randomly assigned into 4 groups, each including 3 animals. These animals were IV injected once with PBS, PolyA/LNP-Man, mAra h2#4/LNP, and mAra h2#4/LNP-Man, to deliver 1.25 mg/kg PolyA or mAra h2#4. Spleens were harvested 7 days later and used to prepare splenocyte populations by tissue mincing and straining. RBCs were removed by a lysis buffer. The splenocyte populations were stained with BV421-conjugated anti-CD3 (2 μg/mL), Alexa 488-conjugated anti-CD4 (5 μg/mL), PE-conjugated anti-CD25 (5 μg/mL), and APC-conjugated anti-FoxP3 (5 μg/mL) antibodies, according to the manufacturer’s instructions. Flow cytometry analysis was carried out in an Attune NxT, used for sequential gating of single cells, CD3+CD4+ and CD25+FoxP3+ populations to quantify the percent of CD25+FoxP3+ T-cells in the spleen (Figure S8) and statistical comparison between the groups.
IL-10 ELISpot Assay
6-weeks-old female C3H/HeJ mice were randomly assigned among 4 groups, each including 3 animals. These animals were IV injected with PBS, PolyA/LNP-Man, mAra h2#4/LNP, and mAra h2#4/LNP-Man at the dose of 1.25 mg/kg. The spleens were harvested 7 days after injection and used to prepare splenocyte populations in which non-CD4+ cells were magnetically labeled with a cocktail of biotin-conjugated antibodies against CD8, CD11b, CD45R, CD49b, Ter-119, and antibiotin microbeads, before flow-through in a MACS Column to collect CD4+ cells. These cells were treated with anti-CD25 PE and labeled with anti-PE microbeads for obtaining magnetic separation of CD4+CD25+ cells, considered phenotypically similar to Tregs. One million CD4+CD25+ cells were added to each well of an ELISpot plate, precoated with IL-10 capture antibody and blocked with BSA. Cells were stimulated with 1 μg/mL Ara h2 peptide (epitope 4) for 48 h. The supernatants were discarded, and HRP-conjugated antimouse IL-10 antibodies were added to each well for 2 h before removal of the supernatant and adding the TMB substrate for 30 min. After addition of a stop solution, plates were washed, dried, and imaged in an ELISpot reader (CTL Immunospot), with software for calculating the number of spots.
Preparation of a Crude Peanut Allergen Extract
The crude peanut extract (CPE) was prepared from peanut powder, obtained from Anthony’s Goods. Briefly, the powder was dissolved in 9% sodium chlorate and stirred at room temperature for 4 h. The suspension was processed through 0.45 and 0.22 μm filters, before dialysis against ddH2O and freeze-drying. The freeze-dried material was dissolved in PBS for further use.
Prophylactic LNP Administration to Prevent Anaphylaxis to a CPE
To study the prophylactic effect of different LNPs, 6-weeks-old female C3H/HeJ mice were randomly assigned to 5 groups (n = 6): nonsensitized control, sensitized control without pretreatment, PolyA/LNP-Man pretreatment, mAra h2/LNP pretreatment, and mAra h2/LNP-Man pretreatment. mAra h2 refers to the use of single-tandem mRNA epitopes (i.e., epitopes 2 and epitopes 4) or a combination of epitopes 1–4. Animals received IV injection of 1.25 mg/kg of each LNP on days 0 and 7 before oral gavage on 3 occasions, using a CPE extract (100 mg/kg), supplemented with cholera toxin (0.5 mg/kg) (Figure 7A and Figure 8E). Animals were challenged by peritoneal administration with 100 mg/kg CPE extract on day 35. Rectal temperatures and anaphylaxis scores were recorded every 15 min for 2 h. Anaphylaxis scoring was done using the following criteria: 0 = no symptoms; 1 = scratching of ear and mouth; 2 = puffiness around eyes and mouth, pilar erection, labored breathing; 3 = prolonged period of motionlessness; 4 = severely reduced motility, tremors, severe respiratory distress; and 5 = death. Blood and peritoneal fluid were collected 3 h after challenge and used for quantification of mouse mast cell protease 1 (mMCP-1), total serum IgE, peanut-specific IgE and IgG1, peritoneal lavage fluid Th2 cytokine levels (IL-4, IL-5. and IL-13), and peritoneal IL-10 and TGF-β levels. Spleens were harvested to assess the percent of CD25+FoxP3+ T-cells in the CD3+CD4+-gated population, as described above. All animal experiments were performed with permission from the UCLA Animal Research Committee, using animal welfare guidelines established by the USDA, AVMA, and the U.S. government.
Assessment of mAra h2#4/LNP-Man Tolerogenic Effects in a Postsensitization Mouse Model
Therapeutic intervention in already-sensitized animals (n = 6) followed the same outline as for the prophylactic administration experiments, except that administration of epitope 4 mRNA, incorporated into mannose-decorated or nondecorated LNP as well as PolyA/LNP-Man, occurred after oral sensitizations (days 0, 7, and 14). This was done by IV injection of the same particle dose on days 21 and 28 (Figure 8A). Anaphylaxis was induced by peritoneal challenge on day 35. Assessment of anaphylaxis manifestations and quantification of immunological end points proceeded as described for the prophylactic model.
Use of ELISA to Assess Antibody Levels, mMCP-1, Cytokines, and TGF-β
Antipeanut IgG1 and IgE antibody titers were determined by ELISA. 96 well plates were used for coating with CPE (20 μg/mL) for 16 h before incubation with serum dilutions (20–12500-fold) for 2 h. Bound IgG1 or IgE were detected by HRP-conjugated anti-IgG1 or anti-IgE, followed by washing, addition of TMB and OD assessment in a microplate reader at 450 nm. The concentration of serum mMCP-1, serum IgE, peritoneal fluid IL-4, IL-5, IL-10, IL-13, and TGF-β levels were determined by ELISA in 96 well plates, as previously described by us.4,5
Nanoparticle Safety Assessment
In vitro cytotoxicity was studied by using LSECs to conduct a MTS assay. Briefly, the cells were incubated with a range of PolyA/LNP or PolyA/LNP-Man concentrations, amounting to 0, 0.1, 0.25, 0.5, 1.0, and 2.5 μg/mL PolyA at 37 °C. After 24 h, the MTS was added and the absorbance was recorded in the microplate reader, using a 490 nm wavelength. Because their cytotoxic events at high concentrations, the LNPs also underwent in vivo safety assessment in nonsensitized C3H/HeJ mice. The experiment included 3 animal groups, each including 3 mice. These mice received IV administration of PBS or 2 mg/kg each of PolyA/LNP or PolyA/LNP-Man. Blood was collected after 24 and 48 h to assess the impact on liver enzymes (ALT, AST) and kidney function (BUN, creatinine). Animals were sacrificed after 48 h for organ harvesting and histological analysis.
Statistics
Comparative analysis of differences between groups was performed using 1-way ANOVA followed by Tukey’s multiple comparisons. Values were expressed as mean ± SD of multiple determinations. For all statistical analyses, a p-value <0.05 was considered statistically significant
Acknowledgments
Initial discovery of liver-based APC responses to nanoparticles was obtained with support from the National Institute of Environmental Health Sciences of the National Institutes of Health (U01 ES027237). Subsequent studies looking at the therapeutic implications of allergen-encapsulating particles were supported by the Marlin Miller, Jr. Family Foundation and the Noble Family Innovation award (made by the CNSI). We acknowledge the Advanced Light Microscopy/Spectroscopy (ALMS) Shared Facility at UCLA and the Electron Imaging Center for Nanomachines (EICN) for assisting LNP characterization. We also acknowledge the infrastructure in the Preclinical Imaging Technology Center, the Translational Pathology Core Laboratory (TPCL), and the Molecular Instrumentation Center for technical assistance for biological experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.2c12420.
Design parameters for LNP-Man synthesis; LNP-Man characterization; GFP expression demonstration; hemolysis assay to demonstrate LNP-Man hexagon lipid phase; design schematic illustration for in vitro mRNA expression strand; mRNA design and characterization; flow cytometry analysis; additional results showing the prophylactic and therapeutic effects; biosafety of LNP-Man (PDF)
Author Contributions
X.X. and X.W. contributed equally to this work. The original liver-targeting tolerogenic platform was conceptualized by A.E.N., with additional conceptual advances for using mRNA therapy for epitope delivery by A.E.N., T.X., X.W., and X.X. Drafting of the manuscript was introduced by A.E.N. with input from X.X., X.W., and T.X. Preparation and characterization of the nondecorated LNP and LNP-Man were performed by X.W. and X.X., respectively. The design of the mRNA strand and microfluidics incorporation into LNP involved T.X., X.W., and A.E.N. Animal experiments were performed X.X., X.W., Y.-P.L., and L.L. Data analysis and discussion were carried out by X.X., X.W., T.X., and A.E.N.
The authors declare the following competing financial interest(s): A.E.N. is co-founder and equity holder in Westwood Biosciences Inc. and Nammi Therapeutics. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Warren C.; Lei D.; Sicherer S.; Schleimer R.; Gupta R. Prevalence and Characteristics of Peanut Allergy in US Adults. J. Allergy Clin. Immunol. 2021, 147, 2263–2270. 10.1016/j.jaci.2020.11.046. [DOI] [PubMed] [Google Scholar]
- Gupta R. S.; Warren C. M.; Smith B. M.; Blumenstock J. A.; Jiang J.; Davis M. M.; Nadeau K. C. The Public Health Impact of Parent-Reported Childhood Food Allergies in the United States. Pediatrics 2018, 142, e20181235. 10.1542/peds.2018-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Wang X.; Liu X.; Kumar S.; Gochman G.; Ji Y.; Liao Y. P.; Chang C. H.; Situ W.; Lu J.; Jiang J.; Mei K. C.; Meng H.; Xia T.; Nel A. E. Use of Polymeric Nanoparticle Platform Targeting the Liver To Induce Treg-Mediated Antigen-Specific Immune Tolerance in a Pulmonary Allergen Sensitization Model. ACS Nano 2019, 13, 4778–4794. 10.1021/acsnano.9b01444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Wang X.; Liu X.; Liao Y. P.; Chang C. H.; Mei K. C.; Jiang J.; Tseng S.; Gochman G.; Huang M.; Thatcher Z.; Li J.; Allen S. D.; Lucido L.; Xia T.; Nel A. E. Antigen- and Epitope-Delivering Nanoparticles Targeting Liver Induce Comparable Immunotolerance in Allergic Airway Disease and Anaphylaxis as Nanoparticle-Delivering Pharmaceuticals. ACS Nano 2021, 15, 1608–1626. 10.1021/acsnano.0c09206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Wang X.; Liao Y. P.; Chang C. H.; Li J. L.; Xia T.; Nel A. E. Use of a Liver-Targeting Nanoparticle Platform to Intervene in Peanut-Induced Anaphylaxis Through Delivery of an Ara h2 T-cell Epitope. Nano Today 2022, 42, 101370. 10.1016/j.nantod.2021.101370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. L.; Tian Z. G. The Liver Works As a School to Educate Regulatory Immune Cells. Cell. Mol. Immunol. 2013, 10, 292–302. 10.1038/cmi.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst A. K.; Neumann K.; Diehl L.; Tiegs G. Modulation of Liver Tolerance by Conventional and Nonconventional Antigen-Presenting Cells and Regulatory Immune Cells. Cell. Mol. Immunol. 2016, 13, 277–292. 10.1038/cmi.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett S. R.; Rolland J. M.; O’Hehir R. E. Immunoregulatory T Cell Epitope Peptides: the New Frontier in Allergy Therapy. Clin. Exp. Allergy 2015, 45, 1015–1026. 10.1111/cea.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K.; Muramatsu H.; Welsh F. A.; Ludwig J.; Kato H.; Akira S.; Weissman D. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.; Vera M.; Gandin V.; Singer R. H.; Tutucci E. Intracellular mRNA Transport and Localized Translation. Nat. Rev. Mol. Cell Biol. 2021, 22, 483–504. 10.1038/s41580-021-00356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J. S. Roles of Endoplasmic Reticulum Stress in Immune Responses. Mol. Cells 2018, 41, 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zhang Z. Q.; Luo J. W.; Han X. J.; Wei Y. Q.; Wei X. W. mRNA Vaccine: a Potential Therapeutic Strategy. Mol. Cancer 2021, 20, 33. 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake T.; Thess A.; Thran M.; Jordan I. mRNA as Novel Technology for Passive Immunotherapy. Cell. Mol. Life Sci. 2019, 76, 301–328. 10.1007/s00018-018-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P. S.; Scott M. K. D.; Hagan T.; Li C. F.; Feng Y. P.; Wimmers F.; Grigoryan L.; Trisal M.; Edara V. V.; Lai L. L.; Chang S. E.; Feng A. L.; Dhingra S.; Shah M. H.; Lee A. S.; Chinthrajah S.; Sindher S. B.; Mallajosyula V.; Gao F.; Sigal N.; Kowli S.; Gupta S.; Pellegrini K.; Tharp G.; Maysel-Auslender S.; Hamilton S.; Aoued H.; Hrusovsky K.; Roskey M.; Bosinger S. E.; Maecker H. T.; Boyd S. D.; Davis M. M.; Utz P. J.; Suthar M. S.; Khatri P.; Nadeau K. C.; Pulendran B. Systems Vaccinology of the BNT162b2 mRNA Vaccine in Humans. Nature 2021, 596, 410–416. 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N.; Hogan M. J.; Weissman D. Recent Advances in mRNA Vaccine Technology. Curr. Opin. Immunol. 2020, 65, 14–20. 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Kim M.; Jeong M.; Hur S.; Cho Y.; Park J.; Jung H.; Seo Y.; Woo H. A.; Nam K. T.; Lee K.; Lee H. Engineered Ionizable Lipid Nanoparticles for Targeted Delivery of RNA Therapeutics into Different Types of Cells in the Liver. Sci. Adv. 2021, 7, eabf4398. 10.1126/sciadv.abf4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M. J.; Pardi N. mRNA Vaccines in the COVID-19 Pandemic and Beyond. Annu. Rev. Med. 2022, 73, 17–39. 10.1146/annurev-med-042420-112725. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Maier M. A.; Manoharan M.; Fitzgerald K.; Jayaraman M.; Barros S.; Ansell S.; Du X. Y.; Hope M. J.; Madden T. D.; Mui B. L.; Semple S. C.; Tam Y. K.; Ciufolini M.; Witzigmann D.; Kulkarni J. A.; van der Meel R.; Cullis P. R. The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- Leung A. K. K.; Tam Y. Y. C.; Chen S.; Hafez I. M.; Cullis P. R. Microfluidic Mixing: A General Method for Encapsulating Macromolecules in Lipid Nanoparticle Systems. J. Phys. Chem. B 2015, 119, 8698–8706. 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- Maeki M.; Uno S.; Niwa A.; Okada Y.; Tokeshi M. Microfluidic Technologies and Devices for Lipid Nanoparticle-Based RNA Delivery. J. Controlled Release 2022, 344, 80–96. 10.1016/j.jconrel.2022.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q.; Wei T.; Farbiak L.; Johnson L. T.; Dilliard S. A.; Siegwart D. J. Selective Organ Targeting (SORT) Nanoparticles for Tissue-Specific mRNA Delivery and CRISPR-Cas Gene Editing. Nat. Nanotechnol. 2020, 15, 313–320. 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzigmann D.; Kulkarni J. A.; Leung J.; Chen S.; Cullis P. R.; van der Meel R. Lipid Nanoparticle Technology for Therapeutic Gene Regulation in the Liver. Adv. Drug Delivery Rev. 2020, 159, 344–363. 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani F.; Yanez Arteta M.; Lerche M.; Porcar L.; Lang C.; Bragg R. A.; Elmore C. S.; Krishnamurthy V. R.; Russell R. A.; Darwish T.; Pichler H.; Waldie S.; Moulin M.; Haertlein M.; Forsyth V. T.; Lindfors L.; Cardenas M. Apolipoprotein E Binding Drives Structural and Compositional Rearrangement of mRNA-Containing Lipid Nanoparticles. ACS Nano 2021, 15, 6709–6722. 10.1021/acsnano.0c10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva Sanchez A. J.; Dobrowolski C.; Cristian A.; Echeverri E. S.; Zhao K.; Hatit M. Z. C.; Loughrey D.; Paunovska K.; Dahlman J. E. Universal Barcoding Predicts In Vivo ApoE-Independent Lipid Nanoparticle Delivery. Nano Lett. 2022, 22, 4822–4830. 10.1021/acs.nanolett.2c01133. [DOI] [PubMed] [Google Scholar]
- Gracia-Sancho J.; Caparros E.; Fernandez-Iglesias A.; Frances R. Role of Liver Sinusoidal Endothelial Cells in Liver Diseases. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 411–431. 10.1038/s41575-020-00411-3. [DOI] [PubMed] [Google Scholar]
- Schlich M.; Palomba R.; Costabile G.; Mizrahy S.; Pannuzzo M.; Peer D.; Decuzzi P. Cytosolic Delivery of Nucleic Acids: The Case of Ionizable Lipid Nanoparticles. Bioeng. Transl. Med. 2021, 6, e10213. 10.1002/btm2.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L.; Lin J. Q.; Huang Y. X.; Li L. X.; Delcassian D.; Ge Y. F.; Shi Y. H.; Anderson D. G. Synergistic Lipid Compositions for Albumin Receptor Mediated Delivery of mRNA to the Liver. Nat. Commun. 2020, 11, 2424. 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings O.; Du Toit G.; Radulovic S.; Lack G.; Santos A. F. Ara h 2 is the Dominant Peanut Allergen Despite Similarities with Ara h 6. J. Allergy Clin. Immunol. 2020, 146, 621–630.e5. 10.1016/j.jaci.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.; Anna F.; Authie P.; Pawlik A.; Ku M. W.; Blanc C.; Souque P.; Moncoq F.; Noirat A.; Hardy D.; Sougakoff W.; Brosch R.; Guinet F.; Charneau P.; Majlessi L. A Lentiviral Vector Encoding Fusion of Light Invariant Chain and Mycobacterial Antigens Induces Protective CD4(+) T cell Immunity. Cell Rep. 2022, 40, 111142. 10.1016/j.celrep.2022.111142. [DOI] [PubMed] [Google Scholar]
- Jia L. F.; Qian S. B. Therapeutic mRNA Engineering from Head to Tail. Acc. Chem. Res. 2021, 54, 4272–4282. 10.1021/acs.accounts.1c00541. [DOI] [PubMed] [Google Scholar]
- Kim S. C.; Sekhon S. S.; Shin W. R.; Ahn G.; Cho B. K.; Ahn J. Y.; Kim Y. H. Modifications of mRNA Vaccine Structural Elements for Improving mRNA Stability and Translation Efficiency. Mol. Cell. Toxicol. 2022, 18, 1–8. 10.1007/s13273-021-00171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Chen C. L.; Yu D.; Liu Z. Roles of Follicular Helper and Regulatory T Cells in Allergic Diseases and Allergen Immunotherapy. Allergy 2021, 76, 456–470. 10.1111/all.14639. [DOI] [PubMed] [Google Scholar]
- Boonpiyathad T.; Sozener Z. C.; Akdis M.; Akdis C. A. The Role of Treg Cell Subsets in Allergic Disease. Asian Pac. J. Allergy Immunol. 2020, 38, 139–149. 10.12932/AP-030220-0754. [DOI] [PubMed] [Google Scholar]
- Orgel K.; Smeekens J. M.; Ye P.; Fotsch L.; Guo R. S.; Miller D. R.; Pardo-Manuel de Villena F.; Burks A. W.; Ferris M. T.; Kulis M. D. Genetic Diversity Between Mouse Strains Allows Identification of the CC027/GeniUnc Strain as an Orally Reactive Model of Peanut Allergy. J. Allergy Clin. Immunol. 2019, 143, 1027–1037.e7. 10.1016/j.jaci.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempski J. W.; Lama J. K.; Iijima K.; Kobayashi T.; Matsunaga M.; Kita H. A Mouse Model of the LEAP Study Reveals a Role for CTLA-4 in Preventing Peanut Allergy Induced by Environmental Peanut Exposure. J. Allergy Clin. Immunol. 2022, 150, 425–439. 10.1016/j.jaci.2022.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anvari S.; Miller J.; Yeh C. Y.; Davis C. M. IgE-Mediated Food Allergy. Clin. Rev. Allergy Immunol. 2019, 57, 244–260. 10.1007/s12016-018-8710-3. [DOI] [PubMed] [Google Scholar]
- Cui S.; Wang Y.; Gong Y.; Lin X.; Zhao Y.; Zhi D.; Zhou Q.; Zhang S. Correlation of the Cytotoxic Effects of Cationic Lipids with Their Headgroups. Toxicol Res. (Camb) 2018, 7, 473–479. 10.1039/C8TX00005K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K.; Mao H. Q.; Huang S. K.; Leong K. W. Oral Gene Delivery with Chitosan--DNA Nanoparticles Generates Immunologic Protection in a Murine Model of Peanut Allergy. Nat. Med. 1999, 5, 387–91. 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- O’Hehir R. E.; Prickett S. R.; Rolland J. M. T Cell Epitope Peptide Therapy for Allergic Diseases. Curr. Allergy Asthma Rep. 2016, 16, 14. 10.1007/s11882-015-0587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani R.; Hossenbaccus L.; Ellis A. K. Immunoregulatory T Cell Epitope Peptides for the Treatment of Allergic Disease. Immunotherapy 2021, 13, 1283–1291. 10.2217/imt-2021-0133. [DOI] [PubMed] [Google Scholar]
- Vyas J. M.; Van der Veen A. G.; Ploegh H. L. The Known Unknowns of Antigen Processing and Presentation. Nat. Rev. Immunol. 2008, 8, 607–618. 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Wang H.; Yang Y.; Jia W.; Su T.; Che Y.; Feng Y.; Yuan X.; Wang X. Mannose-Modified Liposome Co-Delivery of Human Papillomavirus Type 16 E7 Peptide and CpG Oligodeoxynucleotide Adjuvant Enhances Antitumor Activity Against Established Large TC-1 Grafted Tumors in Mice. Int. J. Nanomedicine 2020, 15, 9571–9586. 10.2147/IJN.S275670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares O.; Yaman G.; Azkur A. K.; Akkoc T.; Akdis M.; Akdis C. A. Role of Treg in Immune Regulation of Allergic Diseases. Eur. J. Immunol. 2010, 40, 1232–40. 10.1002/eji.200940045. [DOI] [PubMed] [Google Scholar]
- Gravano D. M.; Vignali D. A. A. The Battle Against Immunopathology: Infectious Tolerance Mediated by Regulatory T Cells. Cell. Mol. Life Sci. 2012, 69, 1997–2008. 10.1007/s00018-011-0907-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdis M.; Blaser K.; Akdis C. A. T Regulatory Cells in Allergy: Novel Concepts in the Pathogenesis, Prevention, and Treatment of Allergic Diseases. J. Allergy Clin. Immunol. 2005, 116, 961–968. 10.1016/j.jaci.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Higgins J. A.; Lamb J. R.; Marsh S. G. E.; Tonks S.; Hayball J. D.; Rosenbronson S.; Bodmer J. G.; Ohehir R. E. Peptide-Induced Nonresponsiveness of Hla-Dp Restricted Human T-Cells Reactive with Dermatophagoides Spp. J. Allergy Clin. Immunol. 1992, 90, 749–756. 10.1016/0091-6749(92)90098-M. [DOI] [PubMed] [Google Scholar]
- Hoyne G. F.; O'Hehir R. E.; Wraith D. C.; Thomas W. R.; Lamb J. R. Inhibition of T-Cell and Antibody-Responses to House-Dust Mite Allergen by Inhalation of the Dominant T-Cell Epitope in Naive and Sensitized Mice. J. Exp. Med. 1993, 178, 1783–1788. 10.1084/jem.178.5.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. D.; Buckland K. F.; McMillan S. J.; Kearley J.; Oldfield W. L. G.; Stern L. J.; Gronlund H.; van Hage M.; Reynolds C. J.; Boyton R. J.; Cobbold S. P.; Kay A. B.; Altmann D. M.; Lloyd C. M.; Larche M. Peptide Immunotherapy in Allergic Asthma Generates IL-10-Dependent Immunological Tolerance Associated with Linked Epitope Suppression. J. Exp. Med. 2009, 206, 1535–1547. 10.1084/jem.20082901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienke C.; Kolb L.; Diken E.; Streuber M.; Kirchhoff S.; Bukur T.; Akilli-Ozturk O.; Kranz L. M.; Berger H.; Petschenka J.; Diken M.; Kreiter S.; Yogev N.; Waisman A.; Kariko K.; Tureci O.; Sahin U. A Noninflammatory mRNA Vaccine for Treatment of Experimental Autoimmune Encephalomyelitis. Science 2021, 371, 145–153. 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- Keeler G. D.; Kumar S.; Palaschak B.; Silverberg E. L.; Markusic D. M.; Jones N. T.; Hoffman B. E. Gene Therapy-Induced Antigen-Specific Tregs Inhibit Neuro-inflammation and Reverse Disease in a Mouse Model of Multiple Sclerosis. Mol. Ther. 2018, 26, 173–183. 10.1016/j.ymthe.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.