Abstract
With over 15 FDA approved drugs on the market and numerous ongoing clinical trials, RNA therapeutics, such as small interfering RNAs (siRNAs) and antisense oligonucleotides (ASOs), have shown great potential to treat human disease. Their mechanism of action is based entirely on the sequence of validated disease-causing genes without the prerequisite knowledge of protein structure, activity or cellular location. In contrast to small molecule therapeutics that passively diffuse across the cell membrane's lipid bilayer, RNA therapeutics are too large, too charged, and/or too hydrophilic to passively diffuse across the cellular membrane and instead are taken up into cells by endocytosis. However, endosomes are also composed of a lipid bilayer barrier that results in endosomal capture and retention of 99% of RNA therapeutics with 1% or less entering the cytoplasm. Although this very low level of endosomal escape has proven sufficient for liver and some CNS disorders, it is insufficient for the vast majority of extra-hepatic diseases. Unfortunately, there are currently no acceptable solutions to the endosomal escape problem. Consequently, before RNA therapeutics can be used to treat widespread human disease, the rate-limiting delivery problem of endosomal escape must be solved in a nontoxic manner.
Keywords: ASOs, RNA therapeutics, delivery, endosomal escape, siRNAs
INTRODUCTION
During the first year of our laboratory some 29 years ago, we indirectly stumbled into the endosomal escape problem (Ezhevsky et al. 1997), and we have been working directly on the problem for the last 15-plus years (Wadia et al. 2004; Lönn et al. 2016). Given the magnitude of impact that solving the endosomal escape problem would have on the entire RNA therapeutics field (and perhaps others), if it was easy, it would have already been solved. Unfortunately, endosomal escape has remained a highly recalcitrant problem (Dowdy et al. 2022). In fact, the more we work on it, the greater the appreciation I have for how difficult it will be to successfully overcome while maintaining a low level of cytotoxicity. My guess is that it will take years of significant effort from multiple groups before we devise a clinically acceptable approach to endosomal escape.
Built on ∼50 years of oligonucleotide chemistry that has resulted in an increased on-target activity and metabolic stability, while decreasing off-target activity and immunogenicity (Dowdy 2017; Khvorova and Watts 2017; Crooke et al. 2021), there are currently more than 15 FDA approved combined siRNAs, phosphorothioate backbone ASOs and neutral phosphorodiamidate morpholino oligomer (PMO) RNA therapeutics targeting disease-causing genes in the liver, muscle and CNS (Hammond et al. 2021; Corey et al. 2022). As one example, inclisiran, a GalNAc–siRNA conjugate targeting the PSCK9 gene in liver to treat hypercholesterolemia, has a single-dose 6 mo duration of response (Fitzgerald et al. 2017). Not surprisingly, due to these clinical successes, there has been significant interest and investment in RNA therapeutics by biotechs and large pharmaceutical companies resulting in numerous ongoing early- and late-stage clinical trials.
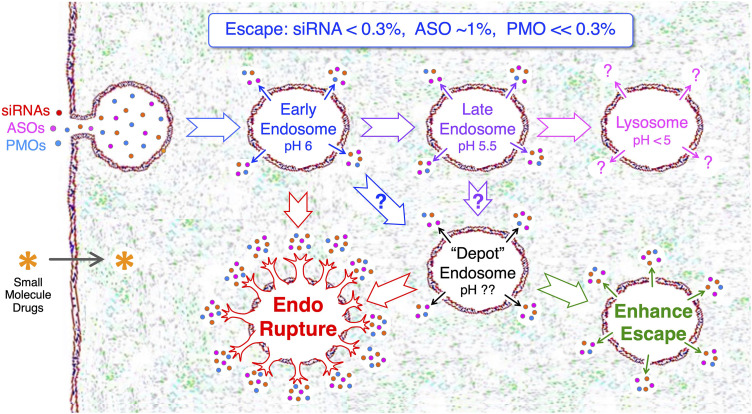
The mechanism of action of oligonucleotide RNA therapeutics is based entirely on targeting the mRNA sequence of validated disease-causing genes and does not a priori require the structure of the protein product, knowing the protein activity and/or its cellular location (Dowdy 2017). In comparison to small molecule drugs that, based on an extracellular concentration gradient, are capable of passively diffusing across the cell membrane (Lipinski 2004), RNA therapeutics are too charged, too large and/or too hydrophilic to diffuse across the cell membrane and instead are taken up by endocytosis into cells (Fig. 1). However, endosomes contain a lipid bilayer that entrap and retain ∼99% of RNA therapeutics (Juliano 2021; Dowdy et al. 2022). Indeed, using quantitative NanoSIMS microscopy, Haibo Jiang's laboratory at the University of Hong Kong determined that only 1%–2% of GalNAc-ASO conjugates escape from endosomes in hepatocytes in vivo (He et al. 2021). Due to siRNA degradation both in the cytoplasm and inside the endosome, Vasant Jadhav's group from Alnylam Pharmaceuticals experimentally found that only 0.3% of endocytosed GalNAc–siRNA conjugate was present in the cytoplasm in vivo at any given time (Brown et al. 2020). Although endosomally trapped RNA therapeutics serve as a depot, enabling long single-dose duration of responses, this also results in ∼99% of endocytosed RNA therapeutics failing to enter the cytoplasm. Consequently, whereas escape from endosomes remains the rate-limiting delivery problem to solve before RNA therapeutics can be widely applied to treat human disease, this will need to be balanced to maintain a partial depot effect for long duration of responses.
FIGURE 1.
Where do RNA therapeutics escape from endosomes? Unlike small molecule drugs, RNA therapeutics (siRNAs, ASOs, and PMOs) are too large, too charged and/or too hydrophilic to passively diffuse across the cell membrane lipid bilayer, but instead are taken up into cells by various forms of endocytosis. Endosomes are also composed of a lipid bilayer barrier that prevents the vast majority (∼99%) of RNA therapeutics from entering the cytoplasm. Following endocytosis, early endosomes mature into late endosomes and then fuse with lysosomes. Although it remains unclear where RNA therapeutics escape from endosomes, they likely escape from multiple types of endosomes. Depot endosomes occur in vivo, but their origins and composition are uncharacterized. RNA therapeutics escaping from depot endosomes results in long duration of responses (3 to 6 mo or more). Increasing the efficiency of endosomal escape can be thought of as two generalized approaches: (i) endosomal rupture; or (ii) enhanced endosomal escape.
Unlike small molecule therapeutics that require 100,000 to 500,000 intracellular molecules to achieve therapeutic activity, siRNAs appear to only require ∼2000 cytoplasmic molecules and ASOs require ∼50,000 for maximal activity (Wittrup et al. 2015; Buntz et al. 2019). Surprisingly, single-dose GalNAc–siRNA conjugates require 2–3 wk before achieving a maximal RNAi response in human clinical trials (Fitzgerald et al. 2017), suggesting a very slow rate of spontaneous endosomal escape. Whereas insects have functional RNA transporters, mammals do not. In fact, it remains entirely unknown mechanistically how endosomally trapped FDA approved RNA therapeutics escape into the cytoplasm (Fig. 1). We hypothesize that there is a spontaneous, short lived, small breach (<10 nm) of the endosomal lipid bilayer that repeatedly occurs over time and that if an RNA therapeutic were proximal to the breach, it would be drawn into the cytoplasm. Alternatively, fusion events between endosomes, multivesicular bodies and lysosomes potentially generate a temporary breach of the lipid bilayer for RNA therapeutics to leak into the cytoplasm (Dowdy et al. 2022). Lastly, there is incomplete, but suggestive data, that RNA therapeutics may also escape via retro-transport from the Golgi (Juliano 2021). Due to the ∼1% of RNA therapeutics that productively escapes from endosomes, definitively determining if any, or all, of these mechanisms are involved in endosomal escape has been technically challenging. However, understanding the spontaneous mechanism of endosomal escape would offer the potential to selectively enhance it.
CURRENT STRATEGIES FOR ENHANCING ENDOSOMAL ESCAPE OF RNA THERAPEUTICS
Although there has been a litany of approaches put forward to address the endosomal escape problem, unfortunately, none of these has yet sufficiently succeeded in disengaging enhanced endosomal escape from toxicity, especially in vivo. There are two generalized mechanisms: (i) endosomal rupture; and (ii) enhancing endosomal escape (Fig. 1). Unfortunately, to date, the majority of effort has (likely) resulted in endosomal rupture with its inherent concomitant cytotoxicity due to release of a wide array of endosomally compartmentalized proteins and molecules into the cytoplasm, resulting in activation of the innate immune system and other toxic pathways (Dowdy et al. 2022). Rudy Juliano from the University of North Carolina has recently put forth a series of in-depth reviews describing these approaches and detailing the endocytotic routes of RNA therapeutics (Juliano et al. 2018; Juliano 2018, 2021).
Endolytic small molecule agents, typified by chloroquine, an antimalarial agent, embody some of the earliest direct attempts to enhance endosomal escape of RNA therapeutics (Dowdy 2017; Juliano et al. 2018). Chloroquine and related compounds are membrane permeable and freely diffuse into cells; however, once they diffuse into the low pH environment of endosomes they become protonated (positively charged) and trapped inside. Due to this sink effect, the endosomal concentration increases logarithmically resulting in endosomal rupture due to insertion of chloroquine's bicyclic aromatic rings into the endosomal lipid bilayer (Dowdy et al. 2022). Unfortunately, other endolytic agents show similar patterns of enhanced endosomal escape, but also at cytotoxic concentrations, which is a class pharmacological feature. Moreover, endolytic agents not only rupture endosomes containing RNA therapeutic cargo, but also rupture many other endosomes in target and nontarget cells. Consequently, although chloroquine treatment results in enhanced endosomal escape, it concomitantly results in significant and unacceptably high cytotoxicity at the effective concentrations.
Cationic peptides and related synthetic peptidomimetics have been used to enhance endosomal escape of RNA therapeutics. Due to the anionic phosphodiester and phosphorothioate backbones, cationic peptides and derivatives form ionic aggregates with siRNAs and ASOs and are therefore restricted to delivery of neutral backbone PMOs and peptide nucleic acid (PNAs). Impressively, the team of Mike Gait (University of Cambridge) and Matthew Wood (University of Oxford) as well as Sarepta Therapeutics have shown significant enhanced exon skipping activity in mouse models by conjugating neutral PMOs to cationic peptide domains (Hammond et al. 2016; Gan et al. 2022). Indeed, Sarepta currently has a cationic peptide PMO (PPMO) in clinical trials that at monthly dosing shows 18-fold more activity than weekly dosing of the FDA approved noncationic eteplirsen PMO for the treatment of Duchenne muscular dystrophy (DMD) (Sarepta 2022). We have previously shown that cationic peptides strongly bind to the anionic surface of cells, including all blood cells (Ho et al. 2001), stimulate endocytosis, specifically macropinocytosis, and facilitate endosomal escape (Wadia et al. 2004). The difficult aspect that both groups have mastered is in balancing just the right amount and type of cationic charge to allow cell adhesion, but not too tightly. Based on more than 25 years of work on the cationic TAT peptide and determining its endosomal escape rate of ∼1% (Lönn et al. 2016), my guess is that these cationic peptide PMO conjugates also have a similar rate of endosomal escape. Although the exact molecular mechanism that cationic peptides use to escape the endosomes is unknown, we speculate that they carpet the endosomal membrane by binding to anionic phospholipid head groups and thereby disorganize the lipid bilayer architecture leading to endosomal membrane disruption and leakage into the cytoplasm. Although still early in clinical trials, the cationic peptide delivery approach looks to be a game changer for neutral PMOs (and potentially for neutral PNAs), but not for anionic siRNA and ASOs.
Endolytic peptides and derivatives from insect toxins and enveloped viruses have been utilized to enhance endosomal escape of RNA therapeutics with mixed results (Dowdy 2017; Hammond et al. 2021). A derivative of influenza's hemagglutinin 2 (HA2) fusogenic domain, called INF7 (Mastrobattista et al. 2002), when conjugated to GalNAc and codelivered in vivo with GalNAc–siRNA conjugates resulted in a 20-fold increase in activity in liver hepatocytes, from enhanced endosomal escape (Brown et al. 2020). However, this required a 30-fold molar excess of GalNAc–INF7 peptide to GalNAc–siRNA and was cytotoxic, likely due to endosomal rupture. Bee venom melittin peptide, which contains a hydrophobic amino terminus and cationic carboxyl terminus, forms pores through membranes that siRNAs can pass through (Houa et al. 2015). Not surprisingly, melittin is exceedingly cytotoxic, difficult to control and ultimately too immunogenic to use for the clinical delivery of RNA therapeutics. In an attempt to tame the cytotoxicity of toxin peptides, we synthesized short hydrophobic endosomal escape domain (EED) peptides containing two or three aromatic Trp or Phe residues and found that we could enhance endosomal escape eightfold (Lönn et al. 2016). Although hydrophobic EED peptides pointed us in the right direction, due to the exposed aromatic hydrophobicity, they inserted into the cell membrane, causing cytotoxicity. Consequently, at the current level of development, it is difficult to envision any of these viral or insect toxin peptide approaches moving forward to clinical trials.
Tamping down the extent of hydrophobicity to reduce cytotoxicity, while still maintaining enhanced endosomal escape, has shown significantly more success than toxin peptides. Front and center, the addition of the clinically validated phosphorothioate (PS) backbone to ASOs, where one of the nonbridging oxygens is substituted with a biochemically more “hydrophobic” sulfur atom, confers an increased metabolic stability and protein binding that results in increased pharmacokinetics, delivery and enhanced endosomal escape (Crooke et al. 2021). The extent of PS conferred hydrophobicity is on the far weak side of the spectrum (vs. the extreme hydrophobicity of toxin peptides), resulting in a clinically tolerable cytotoxicity profile and 1%–2% endosomal escape of PS ASOs (He et al. 2021). Likewise, to increase metabolic stability from exo-RNases, the current most advanced siRNA chemistry utilizes one or two PS bonds on the ends of each strand (Khvorova and Watts 2017). This also likely has the added benefit of enhancing endosomal escape due to the more hydrophobic PS biophysical properties versus highly anionic phosphodiester bonds. The conjugation of more hydrophobicity to both ASOs and siRNAs, such as cholesterol or lipids, results in both semiselective delivery to tissues, especially the liver and neurons of the CNS likely due to insertion into LDL particles, and an enhanced endosomal escape (Soutschek et al. 2004; Biscans et al. 2019; Wang et al. 2019; Brown et al. 2022). However, this level of increased hydrophobicity can also dominate the RNA therapeutic's overall biophysical properties and thereby limit or nullify incorporation of selective targeting domains.
HOW DO WE SOLVE THE RATE-LIMITING ENDOSOMAL ESCAPE PROBLEM?
Although, we currently have more than 15 FDA approved RNA therapeutics, surprisingly, we have not yet ascertained the mechanism through which they escape from endosomes. RNA therapeutics are too large, too charged and/or too hydrophilic to passively diffuse across lipid bilayer membranes. There are also no known mammalian RNA therapeutic transporters and RNA therapeutics do not contain pore forming abilities, yet they clearly escape across the endosomal membrane and enter the cytoplasm of cells. To this end, there has been significant effort to address this mechanism for ASOs from Stan Crooke's group at Ionis Pharmaceuticals (Crooke et al. 2021). In contrast, there has been a dearth of understanding for escape of siRNA therapeutics, though it is likely a similar mechanism as for ASOs involving the limited number of PS bonds on the ends of each siRNA strand. A detailed comprehensive understanding of the molecular mechanism could open up development of approaches capable of further enhancing it in a highly selective and nontoxic manner. However, this will require significantly more effort than is currently being put forth.
We speculate that the hydrophobic backbone PS, and cholesterol and lipid conjugates aid with enhancing endosomal escape by concentrating RNA therapeutics to the luminal surface of the endosomal membrane via binding to membrane proteins and various degrees of interaction with the hydrophobic lipid bilayer. Likewise, cationic peptides concentrate PMOs onto the endosomal surface via ionic interactions with anionic phospholipid head groups. This is followed by a rare, though consistent, spontaneous, yet exceedingly temporary, breach of the endosomal lipid bilayer through which proximal RNA therapeutics are drawn into the cytoplasm, followed by rapid repair of the lipid bilayer breach.
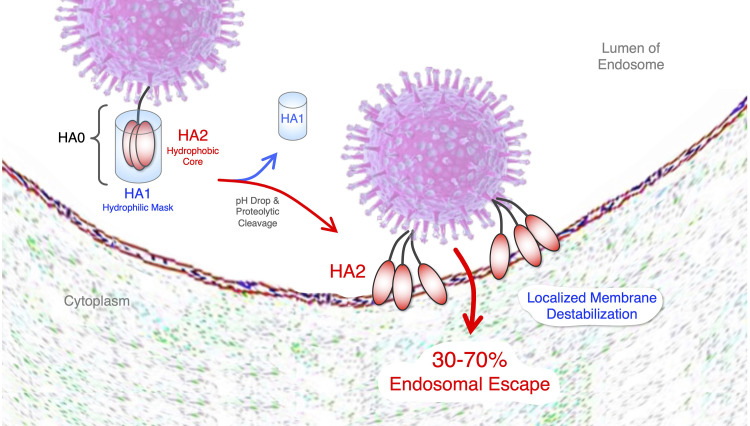
In looking for clues from Mother Nature on how to dramatically increase RNA therapeutic endosomal escape, viruses stand out for already having efficiently solved this problem (Cohen 2016). Indeed, endocytosed enveloped viruses demonstrate a profoundly efficient ability to escape endosomes with a calculated estimate of 30% to 70% (Lagache et al. 2012). Of the enveloped viruses, influenza virus is the most well studied. Influenza's surface hemagglutinin (HA) homotrimer protein is composed of two subdomains: HA1 and HA2. HA1 is the receptor binding domain (RBD) that both stimulates endocytosis by binding cell surface sialic acid and it also masks HA2's hydrophobic endosomal escape domain (Fig. 2). Mechanistically, upon entrance into the low pH of endosomes, HA1 is shed off of HA2 allowing it to undergo a gymnastic conformational change resulting in the insertion of HA2's fusion peptide into the endosomal lipid bilayer membrane, driving capsid entrance into the cytoplasm in a nontoxic manner (Staring et al. 2018). Thus, in our view, HA is the bar that all endosomal escape approaches should be held up to.
FIGURE 2.
Enveloped virus mechanism of endosomal escape. Viruses also have an endosomal escape problem. Enveloped viruses, including influenza, have evolved a similar approach to escape endosomes that involves selectively converting a parental trimeric protein, hemagglutinin (HA), into two domains inside of endosomes. In plasma, the outer shell HA1 domain contains the receptor binding domain (RBD) and serves to mask the hydrophobic HA2 endosomal escape fusion domain. After stimulating endocytosis, due to low pH and endosomal proteolytic activity, HA1 is shed to expose HA2 that buries its hydrophobic fusion peptide into the hydrophobic interior of the endosomal lipid bilayer membrane, resulting in a highly efficient insertion of the viral capsid into the cytoplasm.
Not surprisingly, as one of nature's most evolutionarily efficient mechanisms of endosomal escape, HA causes a localized endosomal membrane disruption, not endosomal rupture. However, attempting to directly pirate this approach to deliver RNA therapeutics is decidedly not straightforward. HA and related proteins from other enveloped viruses, such as Env from HIV, GP from ebola and spike from SARS-CoV-2 (Cohen 2016), serve as vaccine immunogens and will likely stimulate an adaptive immune response after repeated dosing. Therefore, they cannot merely be produced in vitro and conjugated to RNA therapeutics. Moreover, HA is composed of a complex protein architecture that has been recalcitrant to reduction into short functionalized peptide motifs, which, even if possible, could also ultimately become immunogenic after repeated dosing. However, the overall mechanism and design of HA with an external hydrophilic domain masking an interior hydrophobic endosomal escape domain is a highly appealing strategy. The creative trick will be to devise a synthetic biomimetic (not composed of peptides) that is inert in plasma, but selectively activated inside of endosomes and where the byproducts are not inherently toxic.
For more than 40 years, escape from endosomes has remained the most significant problem for delivery of RNA therapeutics that is preventing us from treating cancer, pandemic viruses, and many other systemic indications (Dowdy et al. 2022). In our way of thinking of potential solutions, a successful endosomal escape domain (EED) should incorporate the following parameters:
10-fold or greater enhanced endosomal escape in the absence of cytotoxicity;
Covalently linked to RNA therapeutic, hydrophilic and inert in plasma, but selectively activated inside of endosomes to expose hydrophobicity and/or cationic charge;
Causes a localized endosomal lipid bilayer disruption, not endosomal rupture;
Biophysical properties do not interfere with targeting domains.
Whereas this sounds straightforward and achievable, to date, developing EEDs capable of fulfilling all four parameters has remained elusive. Indeed, many, if not most, of the approaches that dramatically enhance endosomal escape appear to result in endosomal rupture, which even if engineered to be highly controllable, will likely result in clinically unacceptable toxicity. Consequently, significantly more resources, effort and especially creativity focused on enhancing endosomal escape are going to be required before we ultimately achieve widespread use of RNA therapeutics to treat human disease. In my opinion, we are currently only at the beginning of the beginning for effectively enhancing endosomal escape.
ACKNOWLEDGMENTS
I am grateful to Curt Bradshaw, Chris Brown, Satish Jadhav, Art Levin, Mano Manoharan, Laura Sepp-Lorenzino, Punit Seth, and Matt Stanton for numerous discussions on delivery and endosomal escape of RNA therapeutics. Work on this topic in the author's laboratory was supported by ASAP (Michael J. Fox Foundation), National Institutes of Health (NCI and NINDS), and an Epstein Family Alzheimer’ Research Collaboration Award.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079507.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Biscans A, Coles A, Haraszti R, Echeverria D, Hassler M, Osborn M, Khvorova A. 2019. Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res 47: 1082–1096. 10.1093/nar/gky1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Gupta S, Qin J, Racie T, He G, Lentini S, Malone R, Yu M, Matsuda S, Shulga-Morskaya S, et al. 2020. Investigating the pharmacodynamic durability of GalNAc-siRNA conjugates. Nucleic Acids Res 48: 11827–11844. 10.1093/nar/gkaa670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Nair JK, Janas MM, Anglero-Rodriguez YI, Dang LTH, Peng H, Theile CS, Castellanos-Rizaldos E, Brown C, Foster D, et al. 2022. Expanding RNAi therapeutics to extrahepatic tissues with lipophilic conjugates. Nat Biotechnol 40: 1500–1508. 10.1038/s41587-022-01334-x [DOI] [PubMed] [Google Scholar]
- Buntz A, Killian T, Schmid D, Seul H, Brinkmann U, Ravn J, Lindholm M, Knoetgen H, Haucke V, Mundigl O. 2019. Quantitative fluorescence imaging determines the absolute number of locked nucleic acid oligonucleotides needed for suppression of target gene expression. Nucleic Acids Res 47: 953–969. 10.1093/nar/gky1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen FS. 2016. How viruses invade cells. Biophys J 110: 1028–1032. 10.1016/j.bpj.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DR, Damha MJ, Manoharan M. 2022. Challenges and opportunities for nucleic acid therapeutics. Nucleic Acid Ther 32: 8–13. 10.1089/nat.2021.0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST, Baker BF, Crooke RM, Liang XH. 2021. Antisense technology: an overview and prospectus. Nat Rev Drug Discov 20: 427–453. 10.1038/s41573-021-00162-z [DOI] [PubMed] [Google Scholar]
- Dowdy SF. 2017. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol 35: 222–229. 10.1038/nbt.3802 [DOI] [PubMed] [Google Scholar]
- Dowdy SF, Setten RL, Cui XS, Jadhav SG. 2022. Delivery of RNA therapeutics: the great endosomal escape!. Nucleic Acid Ther 32: 361–368. 10.1089/nat.2022.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhevsky SA, Nagahara H, Vocero-Akbani AM, Gius DR, Wei MC, Dowdy SF. 1997. Hypo-phosphorylation of the retinoblastoma protein (pRb) by cyclin D:Cdk4/6 complexes results in active pRb. Proc Natl Acad Sci 94: 10699–10704. 10.1073/pnas.94.20.10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K, White S, Borodovsky A, Bittencourt BR, Strahs A, Clausen V, Wijngaard P, Horton JD, Taubel J, Brooks A, et al. 2017. A highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med 376: 41–51. 10.1056/NEJMc1703361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Wu LCL, Wood JA, Yao M, Treleaven CM, Estrella NL, Wentworth BM, Hanson GJ, Passini MA. 2022. A cell-penetrating peptide enhances delivery and efficacy of phosphorodiamidate morpholino oligomers in mdx mice. Mol Ther Nuc Acids 30: 17–27. 10.1016/j.omtn.2022.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Hazell G, Shabanpoor F, Saleh AF, Bowerman M, Sleigh JN, Meijboom KE, Zhou H, Muntoni F, Talbot K, et al. 2016. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc Natl Acad Sci 113: 10962–10967. 10.1073/pnas.1605731113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Aartsma-Rus A, Alves S, Borgos SE, Buijsen RAM, Collin RWJ, Covello G, Denti MA, Desviat LR, Echevarría L, et al. 2021. Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol Med 13: e13243. 10.15252/emmm.202013243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Migawa MT, Chen K, Weston TA, Tanowitz M, Song W, Guagliardo P, Iyer KS, Bennett CF, Fong LG, et al. 2021. High-resolution visualization and quantification of nucleic acid-based therapeutics in cells and tissues using nanoscale secondary ion mass spectrometry (NanoSIMS). Nucleic Acid Res 49: 1–14. 10.1093/nar/gkaa1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A, Schwarze SR, Mermelstein SJ, Waksman G, Dowdy SF. 2001. Synthetic protein transduction domains: enhanced transduction potential in vitro and in vivo. Cancer Res 61: 474–477. [PubMed] [Google Scholar]
- Houa KK, Panb H, Schlesingerc PH, Wickline SA. 2015. A role for peptides in overcoming endosomal entrapment in siRNA delivery: a focus on melittin. Biotechnol Adv 33: 931–940. 10.1016/j.biotechadv.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano RL. 2018. Intracellular trafficking and endosomal release of oligonucleotides: what we know and what we don't. Nucleic Acids Ther 28: 166–177. 10.1089/nat.2018.0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano RL. 2021. Chemical manipulation of the endosome trafficking machinery: implications for oligonucleotide delivery. Biomed 9: 512. 10.3390/biomedicines9050512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano RL, Wang L, Tavares F, Brown EG, James L, Ariyarathna Y, Ming X, Mao C, Suto M. 2018. Structure-activity relationships and cellular mechanism of action of small molecules that enhance the delivery of oligonucleotides. Nucleic Acids Res 46: 1601–1613. 10.1093/nar/gkx1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Watts JK. 2017. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35: 238–248. 10.1038/nbt.3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagache T, Danos O, Holcman D. 2012. Modeling the step of endosomal escape during cell infection by a nonenveloped virus. Biophys J 102: 980–989. 10.1016/j.bpj.2011.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA. 2004. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1: 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Lönn P, Kacsinta AD, Cui XS, Hamil AS, Kaulich M, Gogoi K, Dowdy SF. 2016. Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci Rep 6: 32301. 10.1038/srep32301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrobattista E, Koning GA, van Bloois L, Filipe AC, Jiskoot W, Storm G. 2002. Functional characterization of an endosome-disruptive peptide and its application in cytosolic delivery of immunoliposome-entrapped proteins. J Biol Chem 277: 27135–27143. 10.1074/jbc.M200429200 [DOI] [PubMed] [Google Scholar]
- Sarepta Therapeutics. https://investorrelations.sarepta.com/news-releases/news-release-details/sarepta-therapeutics-announces-fda-has-lifted-its-clinical-hold (accessed Sept. 26, 2022).
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432: 173–178. 10.1038/nature03121 [DOI] [PubMed] [Google Scholar]
- Staring J, Raaben M, Brummelkamp TR. 2018. Viral escape from endosomes and host detection at a glance. J Cell Sci 131: jcs216259. 10.1242/jcs.216259 [DOI] [PubMed] [Google Scholar]
- Wadia JS, Stan RV, Dowdy SF. 2004. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med 10: 310–315. 10.1038/nm996 [DOI] [PubMed] [Google Scholar]
- Wang S, Allen N, Prakash TP, Liang XH, Crooke ST. 2019. Lipid conjugates enhance endosomal release of antisense oligonucleotides into cells. Nucleic Acid Ther 29: 245–255. 10.1089/nat.2019.0794 [DOI] [PubMed] [Google Scholar]
- Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K, Manoharan M, Kirchhausen T, Lieberman J. 2015. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol 33: 870–876. 10.1038/nbt.3298 [DOI] [PMC free article] [PubMed] [Google Scholar]