Abstract
RNA interference is almost always associated with post-transcriptional silencing in the cytoplasm. MicroRNAs (miRNAs) and critical RNAi protein factors like argonaute (AGO) and trinucleotide repeat binding containing 6 protein (TNRC6), however, are also found in cell nuclei, suggesting that nuclear miRNAs may be targets for gene regulation. Designed small duplex RNAs (dsRNAs) can modulate nuclear processes such as transcription and splicing, suggesting that they can also provide leads for therapeutic discovery. The goal of this Perspective is to provide the background on nuclear RNAi necessary to guide discussions on whether nuclear RNAi can play a role in therapeutic development programs.
Keywords: RNA interference, nuclei, TNRC6, GW182, alternative splicing, transcription, Argonaute
INTRODUCTION
The ability of double-stranded RNAs (dsRNAs) and antisense oligonucleotides (ASOs) to control gene expression and contribute to new therapies is now well established (Shen and Corey 2018). Chemical modifications to stabilize nucleic acid drugs, optimize their activity, and minimize toxicity have been well characterized (Khvorova and Watts 2017). The pharmacological properties of oligonucleotides are understood. Delivery of ASOs and dsRNAs to the liver is established, but progress has also been made with delivery to the central nervous system (Bennett 2019; Bennett et al. 2019), the eye (Garanto 2022), lungs (Shin et al. 2022), muscle (Desjardins et al. 2022), and other organs. Applications range from N = 1 therapies for ultra-rare diseases (Kim et al. 2019; Dhuri et al. 2020) to the potential for broad use of dsRNAs to be used to reduce high cholesterol levels (Migliorati et al. 2022).
Although this progress has established oligonucleotides as a growing class of successful drugs, important questions remain. How successful will oligonucleotides become? Will they remain a relatively niche drug category providing important benefits to a relative handful of patients, or will they expand to a major category of therapeutics that are commonly used in the clinic? Exploiting the full potential of oligonucleotides will require clever identification of disease targets, which, in turn, will require a strong fundamental understanding of what cellular RNAs might be targets.
For dsRNAs, targets are typically thought to be messenger RNAs (mRNAs) that reside in the cytoplasm. The assumption is that fully complementary dsRNAs, in complex with argonaute 2 (AGO2) protein, will bind mRNA and induce the cleavage of the target RNA, leading to lower levels of protein. This mechanism is well understood and is the basis of all six currently approved dsRNA drugs.
What if dsRNA action was not limited to cleavage of mRNA in the cytoplasm? What if dsRNAs could also function in cell nuclei? What if they could affect transcription, splicing, or other biological processes? What if they could bind noncoding RNAs that are known to be the root causes of several inherited diseases? (Pandolfo 2009; Wheeler et al. 2009; DeJesus-Hernandez et al. 2011; Renton et al. 2011; Gattey et al. 2014; Mootha et al. 2017) What if miRNAs had nuclear roles and these roles could also be modulated? It is possible that new targets for drug discovery could be identified that go beyond the targets possible using dsRNAs that act in the cytoplasm. This Perspective examines the basic science of nuclear RNAi and how that understanding can be applied to therapeutic discovery.
Discovery of nuclear RNAi
Morris et al. (2004) reported that duplex RNAs that target gene promoters could modulate gene expression by inducing DNA methylation and blocking transcription. Although the control of transcription by small RNAs was well known in plants (Wassenegger et al. 1994; Castel and Martienssen 2013; Holoch and Moazed 2015), these data suggesting mammalian nuclear RNAi ran counter to established dogma that mammalian RNAi was restricted to inhibition of translation in the cytoplasm.
Our laboratory had been studying the inhibition of endogenous gene transcription by single-stranded peptide nucleic acids (PNAs) (Janowski et al. 2005a) and it was a simple matter to adapt our assays to use promoter-targeted dsRNAs. To our surprise (because first experiments almost never succeed), we immediately observed potent and reproducible inhibition of gene expression by duplex RNAs that target sequences immediately adjacent to the most upstream experimentally determined start site (Janowski et al. 2005b). Subsequent experiments showed that inhibition occurred at the level of transcription and that one strand of the inhibitory dsRNA was binding to an antisense transcript (not the mRNA nor chromosomal DNA) (Schwartz et al. 2008). Inhibition required expression of argonaute (AGO) protein (Chu et al. 2010).
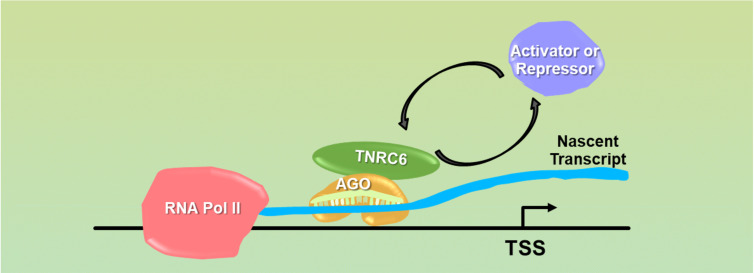
The inhibition of transcription by promoter-targeted RNA was reminiscent of the mechanism of action of protein transcription factors (Fig. 1). Since protein transcription factors are known to inhibit expression in some contexts while activating expression in others, we examined whether gene activation was also possible. We had achieved transcriptional inhibition by dsRNA in a cell line, T47D, where expression of our progesterone receptor target gene was high (Janowski et al. 2005b). We reasoned that we might be able to see activation in a cell line, MCF7, where progesterone receptor expression was low. As with inhibition of transcription, activation of transcription was easily achieved, efficient, and reproducible (Janowski et al. 2007). The sequences of the activating or inhibitory strands were similar in that they targeted the same region of the promoter antisense transcript, but we observed that shifting the sequence just one or two bases could affect whether a strand was capable of potent gene activation or inhibition. Once again, the molecular target was an antisense promoter transcript and expression of AGO2 protein was necessary (Schwartz et al. 2008).
FIGURE 1.
Model showing a potential mechanism for transcriptional activation or repression. The RNA:AGO:TNRC6 complex binds at a promoter to a nascent transcript (not directly to chromosomal DNA) and alters the mix of proteins necessary for gene activation or repression.
Cyclooxygenase-2 (COX-2), a model for activating gene transcription
Just as proteins have the potential to control transcription through many different mechanisms, it is possible that dsRNAs may also act through many different mechanisms. We examined the mechanism at one gene locus in detail—the control of cyclooxygenase-2 (COX-2) expression by dsRNAs (Fig. 2A; Matsui et al. 2013). COX-2 was chosen because it had low- and high-expression states that were responsive to a variety of environmental inputs.
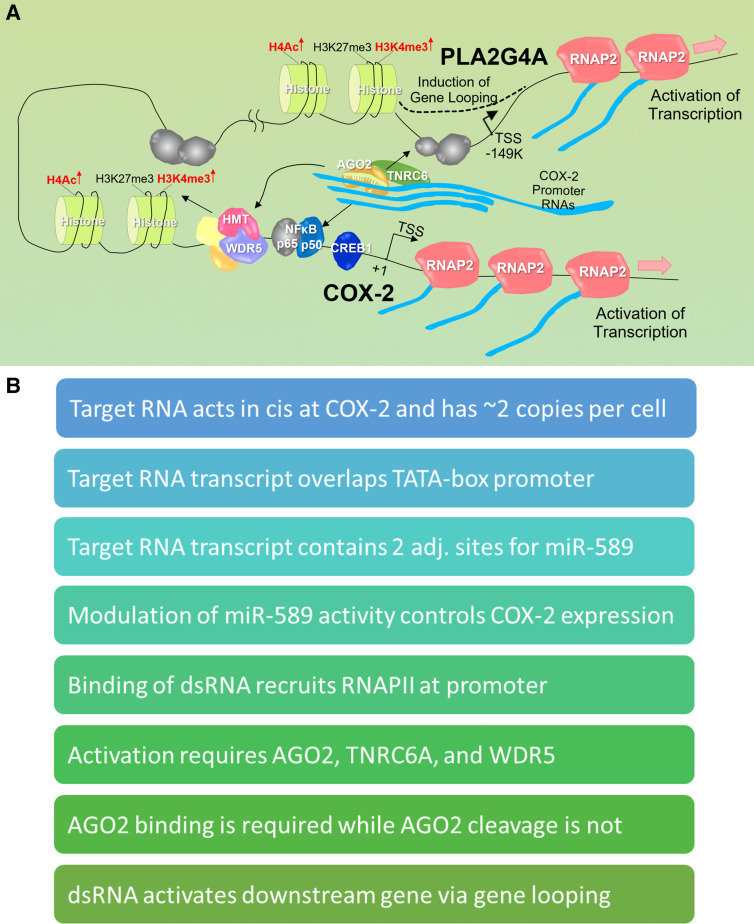
FIGURE 2.
Model of promoter-mediated activation of transcription of COX-2. (A) In the basal state, promoter RNAs are expressed at the COX-2 promoter in the sense direction. COX-2 and PLA2G4A are expressed at low levels. The sense promoter RNA is a platform for recognition by a small complementary RNA (endogenous or synthetic) and recruitment of AGO2 and TNRC6. For gene activation, binding of a small RNA in complex with AGO2 and TNRC6 leads to association of transcription factors and WDR5 and an increase in activating histone marks. Gene looping that juxtaposes the COX-2 and PLA2G4A promoters enables activation of both genes. (B) Summary of some of the key mechanistic findings from RNA-mediated COX-2 activation.
COX-2 proved to be a rich source of insights into mechanism (Fig. 2B; Matsui et al. 2013). The molecular target of the duplex RNA was a rare, approximately one to two copies per cell, sense transcript that overlapped the highly defined TATA-box promoter of COX-2. There were two binding sites for miR-589 within that transcript immediately upstream of the promoter's transcription start site and we found that miR-589 bound that site and activated transcription. Designed synthetic RNAs complementary to the transcription start site also activated transcription and were sensitive to mismatches within the sequence. AGO2-mediated cleavage of the target transcript was not necessary—introduction of a mismatch at position 9, a mismatch that inactivates cleavage, does not reduce gene activation. Activation required RNAi factors AGO2 and TNRC6. Levels of activation were substantial—20- to 30-fold with just the dsRNA and 90- to 100-fold when combined with known pro-inflammatory activators, IL1β or TNFα.
Phospholipase A2 (PLA2G4A) is the enzyme that makes arachidonic acid, the substrate for COX-2. The gene encoding PLA2G4A is the nearest gene to COX-2 (149 kB distant), which led us to test whether RNA-mediated control of COX-2 might influence expression of PLA2G4A. We found that both miR-589 and synthetic designed RNAs complementary to the COX-2 promoter also controlled PLA2G4A transcription. As with regulation of COX-2, RNA-mediated regulation of PLA2G4A expression required expression of AGO2 and TNRC6. How could RNAs complementary to the COX-2 promoter also control expression of a gene separated by 149,000 bases? Chromosome conformation capture (3C) analysis showed that the COX-2 and PLA2G4A transcription start regions contact one another, suggesting gene looping and the possibility of common mechanisms for gene regulation. These data suggested that RNA is instrumental to the regulation of a mammalian gene operon consisting of COX-2 and PLA2G4A.
Taken together, the data on transcriptional regulation from our studies focusing on the control of PR and COX2 can best be explained by the hypothesis that the small RNA/AGO2 complex acts like a transcription factor. The complex binds a nascent noncoding transcript that overlaps the gene promoter while the transcript remains tethered to the chromosome. Given the correct context of basal gene expression (low basal expression makes activation more achievable, high basal expression is better suited for gene repression), RNA-mediated modulation of gene expression can be achieved.
Controlling gene splicing with duplex RNAs
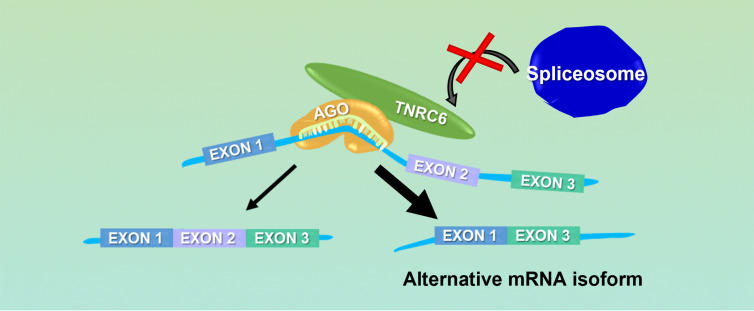
ASOs that are designed to bind and disrupt the recognition of RNA sequences by the protein splicing machinery are robust agents for controlling splicing (Dominski and Kole 1993; Havens and Hastings 2016). We reasoned that duplex RNAs that target similar sites might also be able to block critical regulatory elements and affect splicing (Fig. 3). To test this hypothesis, we targeted duplex RNAs to known sequences within a well-characterized luciferase-based splicing model (Kang et al. 1998) and intronic sequences within established therapeutic targets, survival motor neuron 2 (SMN2, spinal muscular atrophy) (Lim and Hertel 2001; Miyajima et al. 2002; Cartegni and Krainer 2003; Skordis et al. 2003; Hua et al. 2008), and dystrophin (Duchenne muscular dystrophy) (Aartsma-Rus et al. 2003; Arechavala-Gomeza et al. 2007; van Deutekom et al. 2007; Lu et al. 2011; Voit et al. 2014).
FIGURE 3.
Model showing a potential mechanism for RNAi-mediated control of alternative splicing. The RISC complex binds within intronic regions near a splice site and blocks association of the spliceosome and alters exon inclusion.
In all three cases, the dsRNAs modulated splicing (Liu et al. 2012). The Kornblihtt Laboratory has observed regulation of splicing by a different mechanism that involves modulation of histone modification (Allo et al. 2009; Naftelberg et al. 2015). These data from different splice-modulation strategies suggest that dsRNAs may offer an alternative to the use of ASOs for modulating the splicing of genes for therapeutic discovery and development. It is important to note that sequences for active double-stranded RNAs cannot be directly inferred from the sequences of active ASOs and will need to be determined empirically.
RNAi factors are in mammalian cell nuclei
Despite the observation of robust control of transcription and splicing in multiple experimental systems by our laboratory and others (Kawasaki and Taira 2004; Morris et al. 2004; Ting et al. 2005; Janowski et al. 2005b, 2006, 2007; Kim et al. 2008; Schwartz et al. 2008; Allo et al. 2009; Chu et al. 2010; Matsui et al. 2010; Younger and Corey 2011; Ameyar-Zazoua et al. 2012; Liu et al. 2012; Matsui et al. 2013; Allo et al. 2014; Salmanidis et al. 2014; Naftelberg et al. 2015; Kalantari et al. 2016a), many researchers continued to assume that critical RNAi factors like AGO were restricted to cell cytoplasm. These assumptions slowed dissemination of research results and progress investigating the biochemical and mechanistic basis of nuclear RNAi. After realizing that disagreement over the existence of nuclear RNAi factors was an obstacle, we reexamined experimental data and approaches related to cellular distribution of AGO and other RNAi factors (Gagnon et al. 2014a,b).
We first developed a nuclear purification protocol that removes the endoplasmic reticulum (ER), eliminating the possibility that RNAi factors in the ER might be contaminating nuclear preparations. We later learned that this protocol may need to be revised depending on the cell line or tissue being used in a study (Fig. 4). Fractionation of nuclei followed by western analysis showed that RNAi factors including AGO, Dicer, TRBP, and TNRC6A can be detected in cell nuclei. In parallel experiments, AGO2 and TNRC6A can be detected in nuclei by microscopy and appear to overlap. Size exclusion chromatography of nuclear extract reveals the formation of high molecular weight complexes that include AGO2, TNRC6A, and other RNAi factors.
FIGURE 4.
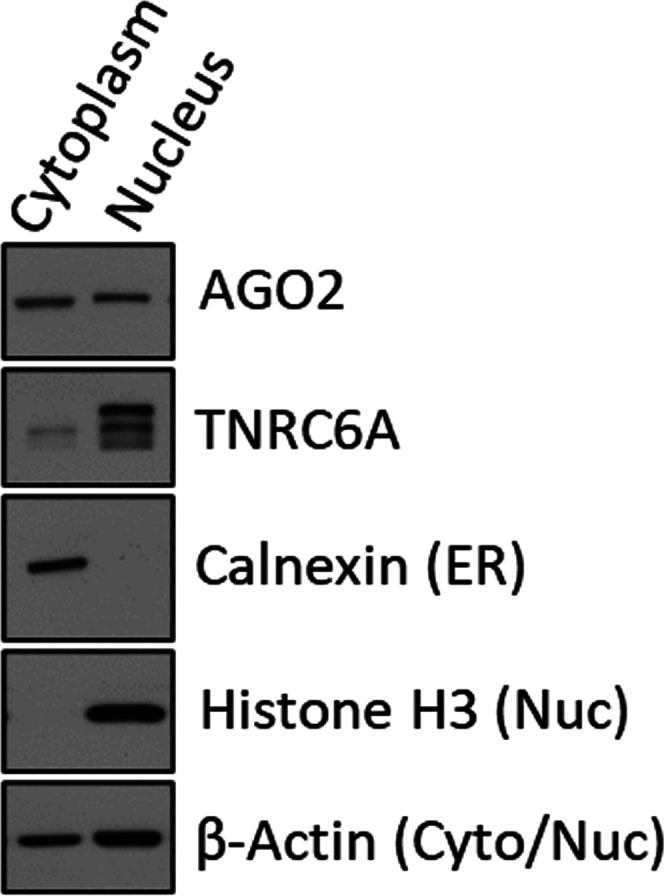
Western blot of cytoplasm and nuclear fractionation of HCT116 cells. Cellular localization of AGO2, TNRC6A, and fraction purity markers shown. Calnexin is a marker for endoplasmic reticulum (ER) (Gagnon et al. 2014a,b).
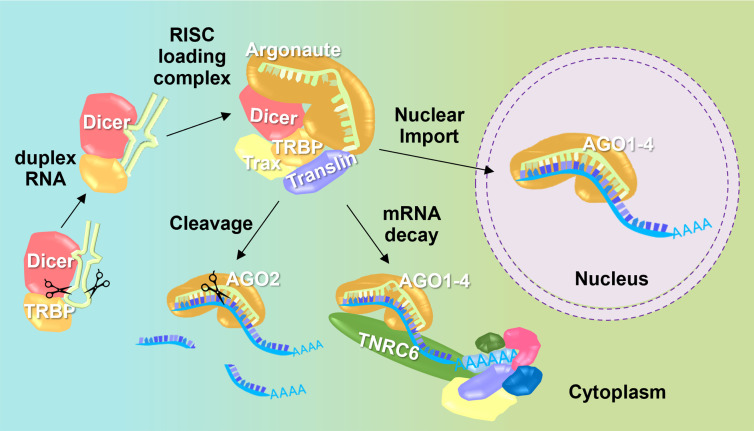
Loading of miRNAs into AGO2 occurs in the cytoplasm, but recognition and cleavage of target RNAs can occur in the nucleus as well as in the cytoplasm (Fig. 5). Loading in the cytoplasm is consistent with the observation that the loading factors Trax and Translin are restricted to the cytoplasm and are not observed in cell nuclei. miRNAs can also be found in cell nuclei, and their distribution is like the distribution observed in cell cytoplasm. These multiple, independent lines of evidence build a strong case that strand loading occurs in the cytoplasm, but that miRNAs and protein RNAi factors can enter cell nuclei and promote recognition of RNA.
FIGURE 5.
Model showing small RNA loading into RNA-induced silencing complex (RISC) in the cytoplasm prior to cleavage, mRNA decay, or import into the nucleus (Gagnon et al. 2014a,b).
Nuclear RNAi, same mechanism as cytoplasmic RNAi, or different?
Applying nuclear RNAi for therapy requires an understanding of how complexes of RNA and protein RNAi factors act in concert to modulate gene expression. We have a relatively mature understanding of how fully complementary designed RNAs recognize mRNA in cell cytoplasm. The proliferation of dsRNA drugs is evidence of the value of these insights into mechanism.
Are the rules governing nuclear RNAi and cytoplasmic RNAi the same? Can an understanding of cytoplasmic RNAi be directly applied to nuclear RNAi? It is obvious that cytoplasmic RNAi offers lessons for RNAi. Specifically, the complex of AGO:RNA has the potential to efficiently recognize complementary sequences within cellular RNA. The basic fact that AGO:RNA drives recognition is the starting point for any study. However, there is more to the story.
The pools of RNA in the cytoplasm and the nucleus are not the same. Cytoplasmic RNA is mostly mature mRNA. Nuclear RNAi offers the potential to recognize intronic sequences, RNAs that are retained in the nucleus, or are associated with chromosomal DNA. So, at the basic level of potential RNA targets, the two processes, nuclear and cytoplasmic, differ.
The molecular mechanisms of RNA recognition may also differ. Most uses of dsRNAs as gene silencing tools or therapeutics rely on dsRNAs promoting cleavage of fully complementary RNA targets in the cytoplasm. This routine cleavage is assumed in most gene silencing experiments commonly used in laboratories.
Do fully complementary duplex RNAs also cause cleavage of RNA targets in cell nuclei? Lennox and Behlke examined this question using panels of dsRNAs and ASOs targeting RNAs known to have different cellular localizations (Lennox and Behlke 2016). They concluded that the answer was “maybe.” They found that ASOs were more reliable and potent knockdown tools in cell nuclei, whereas dsRNAs were more reliable in cell cytoplasm. The greater potency of ASOs in cell nuclei is consistent with the integral role played by RNase H in the mechanism of gapmer-mediated cleavage of target transcripts.
In our experience with RNAs complementary to gene promoters, fully complementary dsRNAs that were designed to target promoter transcripts did not induce cleavage of the promoter transcripts. Consistent with the results of Behlke and Lennox, gapmer ASOs targeting the promoter transcripts cause transcript cleavage. dsRNAs complementary to intron/exon junctions that modulated splicing did not cause detectable cleavage of the target RNA (Liu et al. 2012)
However, again consistent with data from Behlke and Lennox, we observed cleavage of the well-expressed nuclear RNA MALAT-1 (Gagnon et al. 2014a). 5′-RACE even detected cleavage products on isolated chromatin. At this stage, we have too little mechanistic data to rationalize why the efficient cleavage in the cytoplasm is much less predictable in the nucleus. Robust cleavage of nuclear targets by dsRNAs cannot be assumed. It must be demonstrated experimentally.
Differences in mechanism of action may arise from assembly of distinct protein complexes. Critical RNAi factors, AGO and TNRC6, interact with different protein partners in cytoplasm and nuclei. Using immunoprecipitation coupled with mass spectrometry in purified cytoplasm and nuclear lysates, we have examined the shell of interacting proteins associated with AGO2 and TNRC6A. Proteins necessary for loading small RNAs into AGO protein interact with AGO2 proteins in the cytoplasm but are not enriched in association with nuclear AGO2 (Kalantari et al. 2016b). Most of AGO2's interacting proteins in the cytoplasm and nucleus are core components of the RNAi machinery such as the TNRC6 proteins.
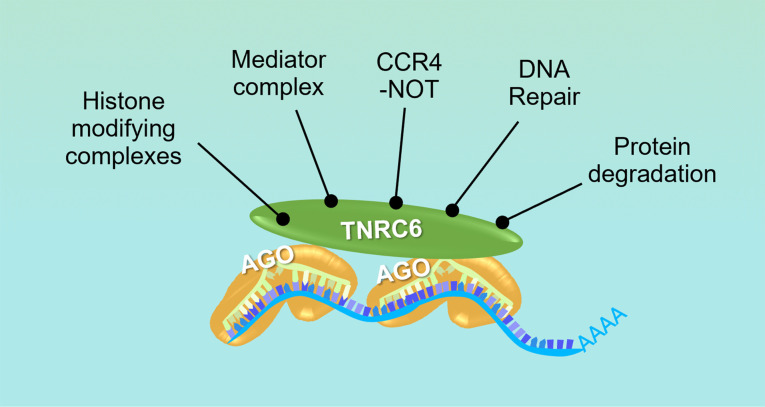
In stark contrast to this narrow group of AGO2-mediated interactions, TNRC6A functions as a scaffolding protein and bridges interactions with many more proteins. TNRC6A interacts with proteins involved in protein degradation, RNAi, the CCR4-NOT complex, the mediator complex, histone modifying complexes, and DNA repair (Fig. 6; Hicks et al. 2017). These protein interaction profiles reinforce the primary role of AGO proteins in recognition of RNA targets regardless of cellular compartment and expand the functional repertoire of TNRC6 proteins in scaffolding and recruitment of noncanonical, nuclear-specific effector protein complexes.
FIGURE 6.
Model showing assembly of different protein complexes in the nucleus based on recruitment of cofactors by RNAi scaffolding protein, TNRC6. AGO loaded with small RNA guide facilitates recognition of RNA targets, and TNRC6 bridges diverse protein interactions (Hicks et al. 2017).
Nuclear regulation by endogenous microRNAs
Although nuclear regulation of splicing by synthetic RNAs can be robust, the endogenous role of microRNAs in the nucleus remains unclear. Using enhanced crosslinking immunoprecipitation sequencing (eCLIP-seq), we mapped AGO2 protein binding sites in isolated cytoplasm and nuclei. Comparing the AGO2–eCLIP sequencing data from the cytoplasm (Chu et al. 2020) and the nucleus (Chu et al. 2021) revealed that the most abundant miRNAs were distributed similarly between both cellular compartments under normal growth conditions. In agreement with the canonical microRNA mechanism, most AGO2 binding sites within RNA in the cytoplasm were mapped to 3′UTR regions (Chu et al. 2020). The majority of AGO2 binding sites in the nucleus, however, were found within intronic sequences, with almost 10-fold more AGO2 occupancy within intronic RNA than within the 3′UTR (Chu et al. 2021).
We examined the hypothesis that AGO2 binding with miRNAs along intronic regions may contribute to alternative splicing. We analyzed splicing changes from RNA-sequencing in WT HCT116 and AGO knockout cells along genes with AGO2–eCLIP-sequencing binding sites, yet even the deletion of three AGO paralogs in the triple knockout cell line, AGO1/2/3−/−, revealed less than fifty alternative splicing events associated with AGO2 binding sites within introns (Chu et al. 2021). Under normal growth conditions, AGO:miRNA complexes can target intronic regions and affect a small number of validated endogenous alternative splicing events. This number may increase under different cellular contexts in physiology and disease and examining cells under conditions that are more extreme than normal cell culture will be an important priority for future research.
Practical considerations
We note that rigorous experimental tests for nuclear RNAi are not always simple. A nuclear RNA may be primarily in the nucleus, but “nuclear” RNAs like MALAT-1 can also be detected in the cytoplasm. It is possible to imagine a scenario in which the minor portion of “nuclear” RNA is in the cytoplasm, is cleaved there, and the fragments recycled into the nucleus. To ensure that cleavage can take place in the nucleus, experiments can be performed in nuclear extract (Gagnon et al. 2014b).
Obtaining nuclear extracts that rigorously exclude almost all cytoplasmic contamination is also not always straightforward. We find that a protocol which leads to pure nuclei for one cell line cannot be relied on to produce pure nuclei for other cell lines or tissues. Detergent, physical extraction method, or other steps may need to be optimized. All purifications need to be verified by western analysis of appropriate compartment-specific control proteins.
The endogenous distribution of RNAi machinery varies widely depending on cellular and environmental context. The choice of cell line, growth condition, and disease state can have a profound impact on cellular localization of miRNAs, their protein partners, and the availability of RNA targets.
Finally, we note that miRNAs are likely to control gene expression through complex mechanisms. Even for cytoplasmic RNAi, which has been studied for two decades, the understanding of miRNA action and gene regulation is incompletely understood (Kilikevicius et al. 2022). Studies of nuclear RNAi mechanism, recognition, and regulation should be well controlled and transparent. Whereas this Perspective describes examples of RNA-mediated control of transcription and splicing, it is necessary to consider whether RNAi factors may also play a role in the modulation of other nuclear processes that involve RNA.
The clinic is the ultimate test for all hypotheses related to nucleic acid therapeutics. Currently, MiNA Therapeutics is testing a duplex RNA, MTL-CEBPA, in clinical trials to treat hepatocellular carcinoma (Hashimoto et al. 2021). The compound is designed to activate CCAAT/enhancer binding protein alpha (C/EMP-α) and enhance the efficacy of cancer therapies for solid tumors. MTL-CEBPA is currently being tested in Phase 1 and Phase 2 trials in combination with Atezolizumab/Bevacizumab and Sorafenib, respectively.
Conclusions
dsRNAs are proving to be a successful strategy for drug development. Optimized chemistries, efficient large-scale synthesis protocols, and new conjugation strategies for delivery to a broader range of tissues will likely combine to increase the positive impact of dsRNAs for patients. Soon, the limitation will be the identification of disease targets where dsRNAs have a competitive advantage relative to gene therapy, small molecule drugs, or antibodies. The nucleus offers many targets that do not exist in cell cytoplasm and the broad potential for nuclear regulation is clear. The challenge for applying this gene control mechanism to therapy is to learn more about the basic science of nuclear RNAi and apply those insights to identify and regulate therapeutic target genes.
COMPETING INTEREST STATEMENT
D.R.C. has licensed technology related to gene activation to MiNA Therapeutics.
ACKNOWLEDGMENTS
D.R.C. was supported by the National Institutes of Health (NIH; GM118103) and the Robert Welch Foundation (I-1244). D.R.C. holds the Rusty Kelley Professorship in Medical Science. K.C.J. is supported by an NIH predoctoral fellowship (1F31GM137591). Author contributions: K.C.J. and D.R.C. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079500.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, van Ommen GJ, van Deutekom JC. 2003. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet 12: 907–914. 10.1093/hmg/ddg100 [DOI] [PubMed] [Google Scholar]
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724. 10.1038/nsmb.1620 [DOI] [PubMed] [Google Scholar]
- Allo M, Agirre E, Bessonov S, Bertucci P, Gomez Acuna L, Buggiano V, Bellora N, Singh B, Petrillo E, Blaustein M, et al. 2014. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc Natl Acad Sci 111: 15622–15629. 10.1073/pnas.1416858111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. 2012. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004. 10.1038/nsmb.2373 [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Graham IR, Popplewell LJ, Adams AM, Aartsma-Rus A, Kinali M, Morgan JE, van Deutekom JC, Wilton SD, Dickson G, et al. 2007. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum Gene Ther 18: 798–810. 10.1089/hum.2006.061 [DOI] [PubMed] [Google Scholar]
- Bennett CF. 2019. Therapeutic antisense oligonucleotides are coming of age. Annu Rev Med 70: 307–321. 10.1146/annurev-med-041217-010829 [DOI] [PubMed] [Google Scholar]
- Bennett CF, Krainer AR, Cleveland DW. 2019. Antisense oligonucleotide therapies for neurodegenerative diseases. Annu Rev Neurosci 42: 385–406. 10.1146/annurev-neuro-070918-050501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. 2003. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol 10: 120–125. 10.1038/nsb887 [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112. 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. 2010. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res 38: 7736–7748. 10.1093/nar/gkq648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Kilikevicius A, Liu J, Johnson KC, Yokota S, Corey DR. 2020. Argonaute binding within 3'-untranslated regions poorly predicts gene repression. Nucleic Acids Res 48: 7439–7453. 10.1093/nar/gkaa478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Yokota S, Liu J, Johnson K, Kilikevicius A, Corey D. 2021. Argonaute binding within human nuclear RNA and its impact on alternative splicing. RNA 27: 991–1003. 10.1261/rna.078707.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Yao M, Hall J, O'Donnell E, Venkatesan R, Spring S, Wen A, Hsia N, Shen P, Russo R, et al. 2022. Enhanced exon skipping and prolonged dystrophin restoration achieved by TfR1-targeted delivery of antisense oligonucleotide using FORCE conjugation in mdx mice. Nucleic Acids Res 50: 11401–11414. 10.1093/nar/gkac641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhuri K, Bechtold C, Quijano E, Pham H, Gupta A, Vikram A, Bahal R. 2020. Antisense oligonucleotides: an emerging area in drug discovery and development. J Clin Med 9: 2004. 10.3390/jcm9062004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Kole R. 1993. Restoration of splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci 90: 8673–8677. 10.1073/pnas.90.18.8673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. 2014a. RNAi factors are present and active in human cell nuclei. Cell Rep 6: 211–221. 10.1016/j.celrep.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Janowski BA, Corey DR. 2014b. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc 9: 2045–2060. 10.1038/nprot.2014.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garanto A. 2022. Delivery of antisense oligonucleotides to the mouse retina. Methods Mol Biol 2434: 321–332. 10.1007/978-1-0716-2010-6_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattey D, Zhu AY, Stagner A, Terry MA, Jun AS. 2014. Fuchs endothelial corneal dystrophy in patients with myotonic dystrophy: a case series. Cornea 33: 96–98. 10.1097/ICO.0000000000000018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A, Sarker D, Reebye V, Jarvis S, Sodergren MH, Kossenkov A, Sanseviero E, Raulf N, Vasara J, Andrikakou P, et al. 2021. Upregulation of C/EBP inhibits suppressive activity of myeloid cells and potentiates antitumor response in mice and patients with cancer. Clin Cancer Res 27: 5961–5978. 10.1158/1078-0432.CCR-21-0986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens MA, Hastings ML. 2016. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res 44: 6549–6563. 10.1093/nar/gkw533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JA, Li L, Matsui M, Chu Y, Volkov O, Johnson KC, Corey DR. 2017. Human GW182 paralogs are the central organizers for RNA-mediated control of transcription. Cell Rep 20: 1543–1552. 10.1016/j.celrep.2017.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D. 2015. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16: 71–84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848. 10.1016/j.ajhg.2008.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. 2005a. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol 1: 216–222. 10.1038/nchembio725 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR. 2005b. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat Chem Biol 1: 210–215. 10.1038/nchembio724 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. 2006. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol 13: 787–792. 10.1038/nsmb1140 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. 2007. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol 3: 166–173. 10.1038/nchembio860 [DOI] [PubMed] [Google Scholar]
- Kalantari R, Chiang CM, Corey DR. 2016a. Regulation of mammalian transcription and splicing by nuclear RNAi. Nucleic Acids Res 44: 524–537. 10.1093/nar/gkv1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari R, Hicks JA, Li L, Gagnon KT, Sridhara V, Lemoff A, Mirzaei H, Corey DR. 2016b. Stable association of RNAi machinery is conserved between the cytoplasm and nucleus of human cells. RNA 22: 1085–1098. 10.1261/rna.056499.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Cho MJ, Kole R. 1998. Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry 37: 6235–6239. 10.1021/bi980300h [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431: 211–217. 10.1038/nature02889 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Watts JK. 2017. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35: 238–248. 10.1038/nbt.3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilikevicius A, Meister G, Corey DR. 2022. Reexamining assumptions about miRNA-guided gene silencing. Nucleic Acids Res 50: 617–634. 10.1093/nar/gkab1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O Jr, Rossi JJ. 2008. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci 105: 16230–16235. 10.1073/pnas.0808830105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, Larson A, Pendergast MK, Goldkind SF, Lee EA, Kuniholm A, et al. 2019. Patient-customized oligonucleotide therapy for a rare genetic disease. N Engl J Med 381: 1644–1652. 10.1056/NEJMoa1813279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA. 2016. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 44: 863–877. 10.1093/nar/gkv1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SR, Hertel KJ. 2001. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J Biol Chem 276: 45476–45483. 10.1074/jbc.M107632200 [DOI] [PubMed] [Google Scholar]
- Liu J, Hu J, Corey DR. 2012. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic Acids Res 40: 1240–1250. 10.1093/nar/gkr780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Yokota T, Takeda S, Garcia L, Muntoni F, Partridge T. 2011. The status of exon skipping as a therapeutic approach to Duchenne muscular dystrophy. Mol Ther 19: 9–15. 10.1038/mt.2010.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. 2010. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem Biol 17: 1344–1355. 10.1016/j.chembiol.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. 2013. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res 41: 10086–10109. 10.1093/nar/gkt777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorati JM, Jin J, Zhong XB. 2022. siRNA drug Leqvio (inclisiran) to lower cholesterol. Trends Pharmacol Sci 43: 455–456. 10.1016/j.tips.2022.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima H, Miyaso H, Okumura M, Kurisu J, Imaizumi K. 2002. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J Biol Chem 277: 23271–23277. 10.1074/jbc.M200851200 [DOI] [PubMed] [Google Scholar]
- Mootha VV, Hansen B, Rong Z, Mammen PP, Zhou Z, Xing C, Gong X. 2017. Fuchs’ endothelial corneal dystrophy and RNA foci in patients with myotonic dystrophy. Invest Ophthalmol Vis Sci 58: 4579–4585. 10.1167/iovs.17-22350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292. 10.1126/science.1101372 [DOI] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G, Kornblihtt AR. 2015. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem 84: 165–198. 10.1146/annurev-biochem-060614-034242 [DOI] [PubMed] [Google Scholar]
- Pandolfo M. 2009. Friedreich ataxia: the clinical picture. J Neurol 256: 3–8. 10.1007/s00415-009-1002-3 [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268. 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmanidis M, Pillman K, Goodall G, Bracken C. 2014. Direct transcriptional regulation by nuclear microRNAs. Int J Biochem Cell Biol 54: 304–311. 10.1016/j.biocel.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. 2008. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol 15: 842–848. 10.1038/nsmb.1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Corey DR. 2018. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res 46: 1584–1600. 10.1093/nar/gkx1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Chan IL, Cao Y, Gruntman AM, Lee J, Sousa J, Rodriguez TC, Echeverria D, Devi G, Debacker AJ, et al. 2022. Intratracheally administered LNA gapmer antisense oligonucleotides induce robust gene silencing in mouse lung fibroblasts. Nucleic Acids Res 50: 8418–8430. 10.1093/nar/gkac630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. 2003. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci 100: 4114–4119. 10.1073/pnas.0633863100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, Schuebel KE, Herman JG, Baylin SB. 2005. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet 37: 906–910. 10.1038/ng1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, et al. 2007. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med 357: 2677–2686. 10.1056/NEJMoa073108 [DOI] [PubMed] [Google Scholar]
- Voit T, Topaloglu H, Straub V, Muntoni F, Deconinck N, Campion G, De Kimpe SJ, Eagle M, Guglieri M, Hood S, et al. 2014. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol 13: 987–996. 10.1016/S1474-4422(14)70195-4 [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576. 10.1016/0092-8674(94)90119-8 [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. 2009. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 325: 336–339. 10.1126/science.1173110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger ST, Corey DR. 2011. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res 39: 5682–5691. 10.1093/nar/gkr155 [DOI] [PMC free article] [PubMed] [Google Scholar]