Abstract
Glycol nucleic acid (GNA) is an acyclic nucleic acid analog connected via phosphodiester bonds. Crystal structures of RNA–GNA chimeric duplexes indicated that nucleotides of the right-handed (S)-GNA were better accommodated in the right-handed RNA duplex than were the left-handed (R)-isomers. GNA nucleotides adopt a rotated nucleobase orientation within all duplex contexts, pairing with complementary RNA in a reverse Watson–Crick mode, which explains the inabilities of GNA C and G to form strong base pairs with complementary nucleotides. Transposition of the hydrogen bond donor and acceptor pairs using novel (S)-GNA isocytidine and isoguanosine nucleotides resulted in stable base-pairing with the complementary G and C ribonucleotides, respectively. GNA nucleotide or dinucleotide incorporation into an oligonucleotide increased resistance against 3′-exonuclease-mediated degradation. Consistent with the structural observations, small interfering RNAs (siRNAs) modified with (S)-GNA had greater in vitro potencies than identical sequences containing (R)-GNA. (S)-GNA is well tolerated in the seed regions of antisense and sense strands of a GalNAc-conjugated siRNA in vitro. The siRNAs containing a GNA base pair in the seed region had in vivo potency when subcutaneously injected into mice. Importantly, seed pairing destabilization resulting from a single GNA nucleotide at position 7 of the antisense strand mitigated RNAi-mediated off-target effects in a rodent model. Two GNA-modified siRNAs have shown an improved safety profile in humans compared with their non-GNA-modified counterparts, and several additional siRNAs containing the GNA modification are currently in clinical development.
Keywords: glycol nucleic acid, GNA, oligonucleotide therapeutics, RNA therapeutics, RNAi therapeutics, siRNAs
INTRODUCTION
RNA interference (RNAi) is a natural mechanism of post-transcriptional gene silencing that was discovered in C. elegans in 1998 (Fire et al. 1998). The first RNAi-based human therapeutic, patisiran (ONPATTRO), received approval by the U.S. Food and Drug Administration (FDA) in 2018 (Akinc et al. 2019). Patisiran is indicated for the treatment of polyneuropathy of hereditary transthyretin-mediated amyloidosis and is a small interfering RNA (siRNA) composed of 21 nt antisense and sense strands. This siRNA mediates targeting of the RNAi machinery to the TTR mRNA, which encodes transthyretin. Both strands have 2′-deoxy-TT overhangs at their 3′ ends and 2′-O-methyl (2′-O-Me) ribonucleotides at selected positions to improve metabolic stability and reduce stimulation of an innate immune response. The siRNA drug is administered intravenously and encapsulated in a multicomponent lipid nanoparticle (LNP) that mediates delivery to the liver (Soutschek et al. 2004; Zimmermann et al. 2006; Akinc et al. 2019).
In the past four years, four additional siRNA therapeutics have received FDA approval: givosiran (GIVLAARI) for treatment of acute hepatic porphyria, lumasiran (OXLUMO) for treatment of primary hyperoxaluria type 1, inclisiran (LEQVIO), an adjunct therapy to lower LDL-C in heterozygous familial hypercholesterolemia or clinical atherosclerotic cardiovascular disease, and vutrisiran (AMVUTTRA) for treatment of the polyneuropathy of ATTR amyloidosis (Egli and Manoharan 2023). The 3′-termini of the sense strands of these siRNAs are conjugated to a triantennary N-acetylgalactosamine (GalNAc), a ligand that specifically binds to the asialoglycoprotein receptor on the surface of liver hepatocytes, resulting in efficient internalization, delivery, and eventual endosomal release of the siRNA into the cytosol (Nair et al. 2014, 2017; Matsuda et al. 2015; Rajeev et al. 2015; Foster et al. 2018; Janas et al. 2018; Willoughby et al. 2018). Targeted delivery, such as that mediated by GalNAc, holds enormous promise for the future of oligonucleotide therapeutics and is expected to overcome limitations with regards to extrahepatic delivery and the LNP platform (Seth et al. 2019; Roberts et al. 2020; Hammond et al. 2021).
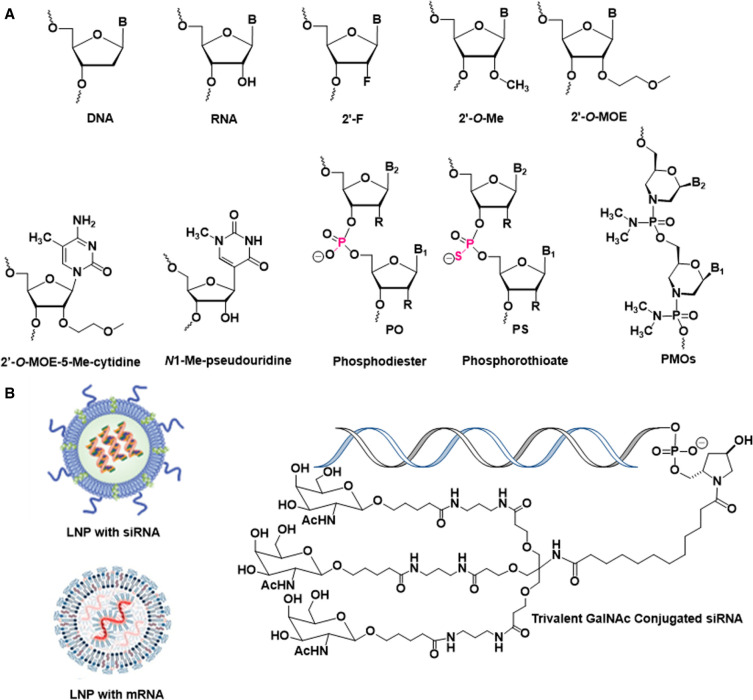
All 18 nucleic acid therapeutics that have reached the market to date, including the five siRNAs described above, four antisense oligonucleotides, five splice site switching oligonucleotides, an anti-VEGF aptamer, an HBV vaccine adjuvant, and the two COVID-19 mRNA vaccines have chemical modifications (Deleavey and Damha 2012; Agrawal and Gait 2019; Egli and Manoharan 2023). The chemical modifications utilized in these therapeutics are the first-generation 2′-deoxy-2′-fluororibonucleotides (2′-F) and 2′-OMe sugar modifications and the phosphorothioate (PS) backbone linkage and the second-generation 2′-O-(2-methoxyethyl) (2′-O-MOE) and 5-methyl-C-2′-O-MOE sugar modification and the phosphorodiamidate morpholino (PMO) backbone modification. Both COVID-19 mRNA vaccines contain N-methyl-pseudouridine in place of U to mitigate immune stimulation and 2′-OMe in their 5′ cap structures (Fig. 1).
FIGURE 1.
(A) Structures of building blocks used in nucleic acid therapeutics. (B) Schematics of lipid nanoparticle (LNP) formulations and GalNAc conjugate.
The four more recently approved siRNAs contain no native ribonucleotides, instead they are fully modified using 2′-F- and 2′-OMe nucleotides. In addition, at the ends of the strands (except for the 3′-end of the sense strand, which carries the GalNAc ligand), the terminal and penultimate bridging phosphate groups are replaced by PS moieties, which provide protection from exonuclease. A recent investigation demonstrated that stereopure PS-modification at terminal phosphates of the antisense strand improves pharmacology in vivo compared to diastereoisomeric mixtures of PS linkages (Jahns et al. 2022). For each siRNA, the placement of 2′-F and 2′-OMe ribonucleotides requires optimization, and the two analogs are carefully balanced for favorable metabolic stability and protein binding. For example, a 2′-OMe modification at the second position of the siRNA antisense strand hampers loading of the antisense strand into RNA-induced silencing complex (RISC). The reason for this limitation is that the methyl group results in a steric conflict with an α-helical curl of the Ago2 MID domain (Egli and Manoharan 2019). Conversely, the 2′-F modification is well tolerated at this site. Similarly, in the seed region of the antisense strand (positions 2–8), the 2′-O-MOE modification results in clashes with side chains of the PIWI endoribonuclease domain of Ago2, but the smaller 2′-F and 2′-OMe chemistries are both tolerated in this region (Manoharan et al. 2011; Egli and Manoharan 2019). Unlike the uniform modifications used in splice site switching oligonucleotides, nusinersen (SPINRAZA), which is uniformly modified with 2′-O-MOE and PS, and eteplirsen (EXONDYS 51) and golodirsen (VYONDYS 53), which are PMOs, siRNA strand design requires a regiospecific modification strategy (Egli and Manoharan 2019). Regiospecific modification is also important for some antisense oligonucleotides, for example in the gapmer design of oligonucleotides such as mipomersen sodium (KYNAMRO) and inotersen (TEGSEDI), which involves 2′-O-MOE flanking a central PS DNA region.
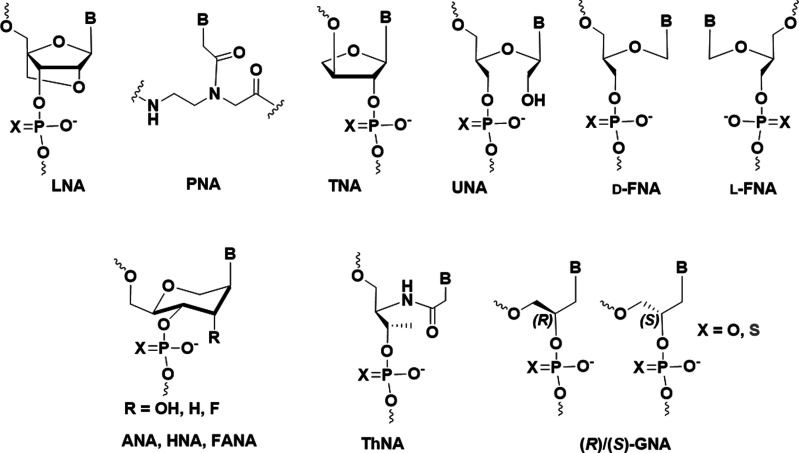
siRNAs with 2′-F, 2′-OMe, and PS modifications in combination with GalNAc conjugation have proven successful in the clinic against a range of diseases; however, the complex interactions between siRNA strands and Ago2, the key player in the RNAi pathway, demand that we keep an open mind regarding alternative modification strategies. In addition to familiar ribose modification sites, such as O2′, C4′, O4′, and C5′, the search for novel modifications that will result in siRNAs with better pharmacodynamic, pharmacokinetic, and metabolic properties as well as low toxicity and limited immunogenic and off-target profiles have been expanded to chemistries that abandon the standard DNA, RNA, or bridged or locked nucleic acid (LNA) sugar-phosphate framework (Fig. 2). These so-called xeno nucleic acids (XNAs) feature alternative backbone linkages and/or nucleobases (Egli and Herdewijn 2012; Anosova et al. 2016). Chaput and Herdewijn proposed a definition of XNA and describe what sets XNAs apart from unnatural nucleic acids or chemically modified DNA and RNA (Chaput and Herdewijn 2019). For example, LNA should not be classified as XNA because it only contains a modified ribose moiety. However, peptide nucleic acid (PNA), which has a neutral backbone that lacks both the sugar and phosphate linkages, is an XNA (Fig. 2). Other XNAs include L-α-threofuranosyl nucleic acid (TNA, which has a tetrose sugar), glycol nucleic acid (GNA, which is based on a glycol monomer), unlocked nucleic acid (UNA, which lacks the ribose C2′-C3′ bond), flexible nucleic acid (FNA, which lacks C2′ and O2′), altritol nucleic acid (ANA, which has a D-altritol sugar), hexitol nucleic acid (HNA, which has a 1′,5′-anhydrohexitol sugar), and threoninol nucleic acid (ThNA), combine alternative sugars or acyclic moieties with phosphodiester linkages and in most cases cross-pair with DNA and RNA (Fig. 2; Anosova et al. 2016).
FIGURE 2.
Structures of XNAs with potential for use in nucleic acid–based therapeutics.
FLEXIBLE NUCLEIC ACIDS
The RNA backbone consists of β-d-ribofuranose units that are linked by phosphodiester moieties. The cyclic sugar is constrained to adopt one of two conformations, C3′-endo or C2′-endo. The former is more common and uniformly adopted in A-form RNA duplexes (Egli 2022). Removal of C2′ and O2′ yields an acyclic or flexible nucleic acid (FNA), an XNA, without the constraints of the cyclic sugar (Fig. 2). The synthesis of this “glycerol” nucleoside analog was first reported by Schneider and Benner (1990). FNA contains a single chiral center (C4′) and is an isostere (i.e., a molecule with similar shape and electronic properties) of RNA in that it retains six bonds per nucleotide unit in its backbone. However, incorporation of a single FNA T into a DNA oligonucleotide caused a loss of more than 10°C in the thermal stability of the duplex. At first sight this seemed a disappointing result, and it was tempting to conclude that the greater than expected entropic loss upon formation of a duplex with a single FNA precluded a role in prebiotic chemistry and perhaps also in therapeutic applications of nucleic acids. Conformational preorganization is important for pairing stability; a longer strand composed entirely of FNA appeared unlikely to form a stable duplex opposite DNA or RNA in aqueous solution.
Not surprisingly, FNA stability and structure are somewhat insensitive to chirality as demonstrated by analyses of D- (=S) vs. L- (=R) FNA (Fig. 2). FNA has significantly increased resistance to degradation by snake venom phosphodiesterase compared to DNA: A 12-mer of FNA has a half-life of 110 h compared to 3 min for dT12. Moreover, an FNA oligomer serves as a template for E. coli DNA polymerase I Klenow fragment (Merle et al. 1995). However, the stability of duplexes between an FNA A 12-mer and the complementary DNA strand, dT12, is low (Tm < 24°C). Despite the inability of FNA to form stable duplexes with DNA, FNA triphosphates are substrates for DNA polymerases such as Therminator, Vent Exo−, and Deep Vent Exo− (Heuberger and Switzer 2008; Zhang et al. 2010; for review, see D'Alonzo et al. 2011). Both (S)- and (R)-FNA triphosphates are accepted with a slight advantage for the “unnatural” (R)-isomer. In addition, nonenzymatic templated synthesis of DNA on repeating units of FNA that lack regular stereochemical configuration was also demonstrated (Chaput and Switzer 2000).
UNLOCKED NUCLEIC ACID
UNA is an alternative acyclic nucleic acid that lacks the C2′–C3′ ribose bond (Fig. 2; Nielsen et al. 1995). Oligonucleotides modified with UNA at their 3′ end are significantly more stable against 3′-exonuclease degradation compared to DNA. UNA residues placed in a central position render an RNA duplex less stable by 4–6 kcal/mol at 37°C. When placed at the termini, they destabilize the duplex by 0.5–1.5 kcal/mol. The observed effects are additive and affect stacking unfavorably (Pasternak and Wengel 2010). Chimeric UNA and DNA and chimeric UNA and 2′-fluoroarabino nucleic acid (FANA) strands elicit cleavage of complementary RNA by RNase H (Mangos et al. 2002). UNA monomers in siRNAs reduce off-target effects (Bramsen et al. 2010). Of note is a particularly favorable influence on the reduction of the off-target effect when a UNA residue is placed at position 7 of the antisense strand, a position within the seed region (positions 2–8) while the potency remained high. siRNAs with UNA at position 1, 2, or 3 of the antisense strand are not phosphorylated by the mammalian Clp1 kinase; a 5′ phosphate is necessary for the interaction of the antisense strand with Ago2 (Kenski et al. 2010). When 5′-phosphorylated antisense strands containing UNA were synthesized, RNAi activity in vitro and in vivo was shown to be comparable to the parent siRNA when UNA was located at position 1 but not at positions 2 or 3 (Pasternak and Wengel 2011). Placement of UNA at the 5′ terminus of the sense strand is expected to preclude its incorporation into RISC, therefore eliminating this type of off-target effect.
ORIGIN, DESIGN, AND SYNTHESIS OF CHIRAL GLYCOL NUCLEIC ACID
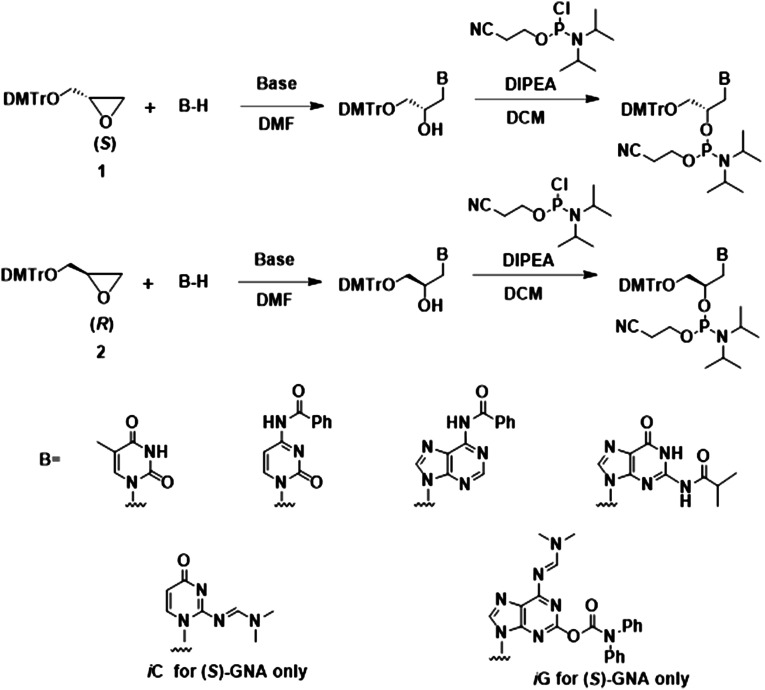
GNA is an XNA with a 3′–2′ linked glycol-phosphate backbone and is the simplest phosphodiester-based oligomer building block (Fig. 2). Compared to DNA and RNA, the GNA backbone is shortened by one atom and hence one bond (Fig. 3A). Racemic GNA nucleosides were first synthesized by the groups of Ueda (Ueda et al. 1971) and Imoto (Seita et al. 1972); enantiomerically pure compounds were synthesized by the Holý group (Holý and Ivanova 1974). The first GNA phosphoramidites and GNA-containing oligonucleotides were synthesized by Cook et al. (1995, 1999) and the Wengel group (Nielsen et al. 1995). Meggers and colleagues subsequently published improved and simplified methods for the synthesis of GNA phosphoramidites and GNA oligonucleotides (Zhang et al. 2006; Schlegel and Meggers 2009). To further reduce the total number of synthetic steps for GNA phosphoramidites and allow for a more facile synthesis on a kilogram scale, our group at Alnylam developed a procedure that allows for the ring opening of enantiopure DMT-glycidol using protected purine nucleobases (Scheme 1; Schlegel et al. 2017, 2021). Whereas previous reports mention failure in the direct alkylation of protected purines we found that the reaction in the presence of N,N-diisopropylethylamine results in the desired products while preserving the exocylic protecting groups. The alkylation of purines in contrast to alkylation using unprotected purines in the presence of NaH (Zhang et al. 2006; Schlegel and Meggers 2009), also resulted in the formation of a significant amount of the N7 regioisomer, which required separation from the N9 product via silica gel chromatography. This method was applied to the pyrimidine nucleobases resulting in yields that were similar to those in previous reports utilizing NaH for deprotonation of the nucleobase. Subsequently, the iso-C and iso-G (S)-nucleoside regioisomers were also synthesized in a similar manner. Phosphoramidites of both the (S)- and (R)-isomers of GNA were synthesized from these nucleosides using previously reported procedures (Zhang et al. 2006; Schlegel and Meggers 2009; Schlegel et al. 2017, 2021, 2022). A general overview of the current methods for the synthesis of various GNA building blocks is summarized in Scheme 1.
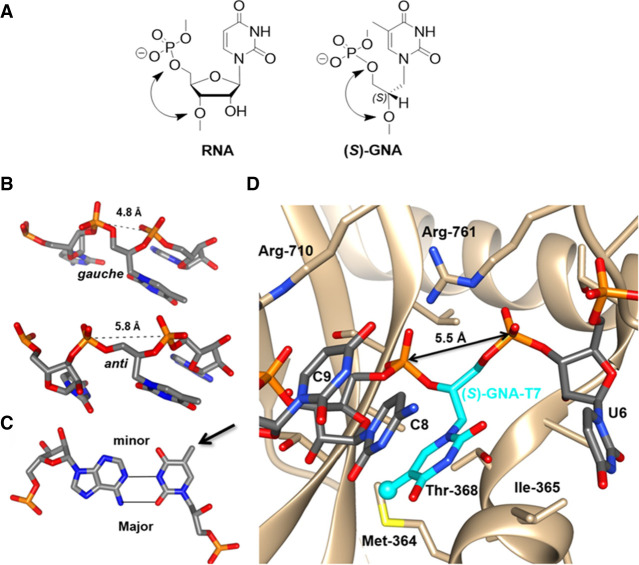
FIGURE 3.
Structure, conformations, base-pairing, and Ago2 binding of (S)-GNA. (A) RNA-U and (S)-GNA-T nucleotides. (B) GNA nucleotides adopt both gauche and anticonformations when incorporated into RNA. (C) Example of an (S)-GNA-T:RNA-A base pair showing a rotated nucleobase conformation for the GNA nucleotide (arrow). (D) An (S)-GNA-T residue can seamlessly and with optimal geometry replace an RNA nucleotide at position 7 of the antisense strand RNA bound to human Ago2. The RNA strand assumes a kink at that site that is associated with Ile-365 of Ago2 and results in unstacking of bases of nucleotides 6 and 7.
SCHEME 1.
General procedures for the synthesis of (S)-GNA (top) and (R)-GNA (middle) phosphoramidites starting from enantiopure glycidol. Various bases and base analogs (B) can be used to functionalize GNA (bottom). Detailed protocols for syntheses have been reported (Zhang et al. 2005, 2006; Schlegel and Meggers 2009; Schlegel et al. 2017, 2021).
GNA PAIRING STABILITY AND SPECIFICITY
(S)-GNA 18-mer oligomers consisting entirely of A or entirely of T form stable antiparallel duplexes with a Tm of 63°C (Zhang et al. 2005). A stable duplex did not form in the parallel orientation. A single T-T mismatch within the GNA duplex lowered the Tm to 55°C, and a duplex with an A-A and a T-T mismatch had a Tm of 44°C. The thermal stability of fully complementary GNA 18-mers exceeds stabilities of the corresponding DNA (Tm = 40.5°C) and RNA (Tm = 42.5°C) duplexes by 22.5°C and 20.5°C, respectively! These results stand in stark contrast to the earlier experiences with duplexes carrying a single flexible nucleoside monomer (Schneider and Benner 1990) or oligomers of 1′,2′-seco-DNA, an analog that lacks the sugar C1′–C2′ bond (Peng and Roth 2004). Seco-DNA oligonucleotides with sequence (A)10 and (T)10 failed to pair, and the analog does not cross-pair with DNA. Four-helix junctions composed of either all (S)-GNA or all (R)-GNA 18-mers were also significantly more stable than the corresponding DNA four-helix junction. The Tm of the latter was 37°C compared to Tms > 70°C for the GNA junctions (Zhang et al. 2008).
The circular dichroism CD spectrum of an (S)-GNA duplex has a strong negative Cotton effect, which is a characteristic change in optical rotatory dispersion and/or circular dichroism for an optically active compound inside its absorption region, at 275 nm; the opposite effect is seen with (R)-GNA. Similarly, the CD spectra of four-helix junctions derived from (S)-GNA and (R)-GNA oligomers exhibit mirror image symmetry: The (S)-GNA junction shows a strong negative peak at 280 nm and the (R)-GNA junction shows a strong positive peak at 280 nm (Zhang et al. 2008). (R)-GNA and (S)-GNA oligomers do not pair and neither cross-pairs with DNA. However, (S)-GNA cross-pairs with RNA in an antiparallel manner, and the resulting hybrid duplex (an 18-mer with an RNA polyU strand) has a Tm of about 35°C. Thus, it is lower than those of the corresponding RNA:RNA (Tm = 42.5°C) and (S)-GNA:(S)-GNA duplexes (Tm of 44°C). In their initial report, Meggers and coworkers concluded that GNA forms highly stable duplexes of antiparallel strand orientation that follow the Watson–Crick base-pairing rules (Zhang et al. 2005). However, GNA—the simplest phosphodiester-based pairing system—had a surprise in store. (R)-GNA does not form a hybrid duplex with RNA. The initial finding that only (S)-GNA pairs with RNA implied that this isomer would be potentially of interest as an siRNA modification.
ENZYMATIC SYNTHESIS OF DNA ON GNA TEMPLATES AND GNA ON DNA TEMPLATES
GNA consists of a 3-carbon unit that is fused with a nucleobase and could have been a simple carrier of genetic information prior to the proposed RNA World. The 3-carbon unit could be contributed by glycerol, glyceraldehyde, or glycidol. The latter is used to synthesize GNA monomers (Zhang et al. 2005). From the prebiotic perspective, it is interesting that glycidol can be produced from glycerol carbonate in a zeolite-catalyzed reaction (Yoo and Mouloungui 2001). Further, both GNA self-pairing and cross-pairing with RNA are stereo-specific, which is of interest in the context of the rise of homochirality in the origin of life (Tsai et al. 2007).
The A-family DNA polymerase from Bacillus stearothermophilus is the most proficient of those polymerases tested in carrying out DNA synthesis on a GNA template (Tsai et al. 2007). Remarkably, template-directed synthesis by the B. stearothermophilus DNA polymerase proceeds despite the absence of formation of a stable duplex between GNA and DNA. The synthesis of GNA on DNA templates appears to be more challenging for existing polymerases than DNA synthesis on a GNA template. Therminator DNA polymerase carries out primer extension by incorporating two GNA residues; however, extension with additional GNA monomers was much less efficient (Chen et al. 2009). Based on steady-state kinetic experiments, Therminator-catalyzed GNA synthesis was stifled both by low catalytic rates and weak substrate binding. Attempts to synthesize GNA on GNA templates were unsuccessful even when the primer and primer-binding region were composed of DNA. In addition to the higher flexibility of the GNA backbone relative to DNA and RNA, the conformation of the GNA strand is significantly different from those of the natural counterparts, and this likely limits recognition by DNA polymerases.
CRYSTAL STRUCTURES OF GNA DUPLEXES
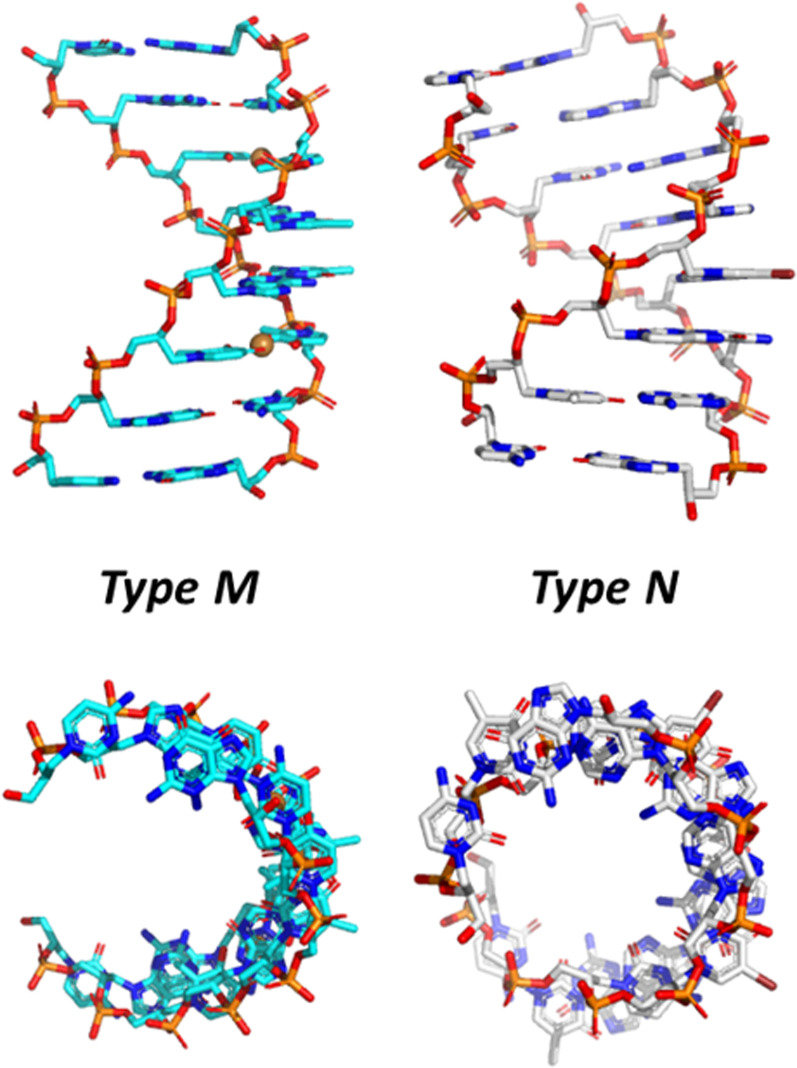
Crystal structures of three different GNA duplexes have been determined at resolutions between 0.97 and 1.83 Å (Fig. 4). The initial 1.3-Å structure was of the (S)-GNA octamer 3′-g(CGHATHCG)-2′ with two Cu(II)-mediated hydroxypyridone (H) nucleobase pairs (PDB ID 2JJA; Schlegel et al. 2008). Hallmarks of this so-called M-type helix are 16 residues per helical turn, a 3.8-Å rise, and 60-Å helical pitch, a twist of 24°, and an average phosphate-phosphate distance of 5.4 Å. The structure of the (S)-GNA hexamer GNA 3′-g(GBrCGCGC)-2′ was determined at 0.97-Å resolution (PDB ID 2WNA; Schlegel et al. 2010), and the (S)-GNA octamer 3′-g(CTCBrUAGAG)-2′ structure was solved at 1.83-Å resolution (PDB ID 2XC6; Johnson et al. 2011). The two latter structures are of the so-called N-type with an A-form DNA/RNA-like backbone curvature and the following geometric hallmarks: 10 residues per turn, a 2.6-Å rise, a 26-Å helical pitch, a twist of 36°, and an average phosphate-phosphate distance of 5.4 Å (Fig. 3). Individual nucleosides in the backbones of these duplexes show variations in the torsion angle around the C3′–C2′ bond that either falls into the negative synclinal (sc−) or the antiperiplanar (ap) range. The torsion angle around the C2′–C1′ bond is limited to the ap range.
FIGURE 4.
Crystal structures of (S)-GNA octamer duplexes, PDB ID 2JJA (left) and PDB ID 2XC6 (right), viewed across the two grooves (top) and along the helical axis (bottom).
(S)-GNA forms a right-handed duplex with a pronounced x-displacement of base pairs and a strong negative backbone-base inclination angle (Pallan et al. 2007; Egli and Pallan 2010). The latter is a fundamental property of RNA strands in canonical A-form duplexes. The (R)-GNA duplex has a left-handed helical twist and a positive backbone-base inclination angle. These observations help explain how (S)-GNA and RNA can pair, but RNA and (R)-GNA cannot. However, there are two findings that the structural data cannot explain. First, in the CD spectra, the right-handed (S)-GNA duplex displays a strong negative peak at ∼280 nm; in the same wavelength range, right-handed DNA and RNA duplexes show a positive signal. Second, GNA sequences that contain G or C nucleosides do not cross-pair with RNA (Meggers and Zhang 2010).
CRYSTAL STRUCTURES OF RNA DUPLEXES WITH SINGLE GNA RESIDUES PER STRAND
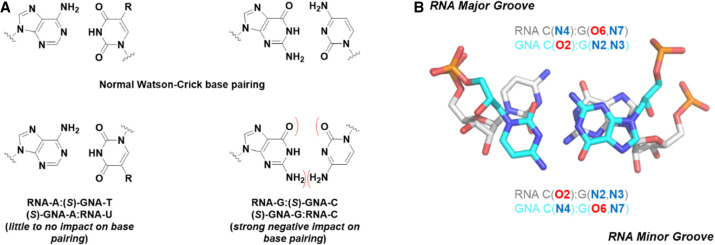
To analyze the conformation of GNA in an RNA duplex environment and to better understand (S)-GNA:RNA pairing and the lack thereof for (R)-GNA and RNA, we determined four crystal structures of RNA duplexes with a single GNA residue per strand. These crystal structures are of 5′-BrCGAA[(S)-GNA T]UCG-3′ (solved to 1.08 Å, PDB ID 5V2H), 5′-CGCGAAU[(S)-GNA T]AGCG-3′ (solved to 1.2 Å, PDB ID 5V1L), 5′-CGCGAABrU[(R)-GNA T]AGCG-3′ (solved to 1.18 Å, PDB ID 5V1K) (Schlegel et al. 2017), and 5′-CGCG[(S)-GNA A]ABrUUAGCG-3′ (solved to 1.78 Å, PDB ID 7LO9) (Schlegel et al. 2021). These structures demonstrate the considerable local conformational flexibility of GNA residues inside RNA strands (Fig. 3B). In the crystal structure of the 12-mer with a single (S)-GNA T, the trimer containing the modification is observed in six different conformations with variations in the puckers of ribonucleotides, backbone torsion angles of the GNA residue, and the distance between the 3′- and 2′-phosphates of the GNA (4.78 to 5.83 Å range; Egli and Manoharan 2019). Remarkably, in all these structures, GNA residues assume a conformation with a flipped nucleobase relative to the standard base orientation of ribonucleotides (Fig. 3C). Thus, the 5-methyl group of the (S)-GNA T points into the minor groove; the N6 amino group of the (S)-GNA A in the duplex containing this residue also points into the minor groove (Fig. 5). It was surprising that this flipped orientation is adopted by both (S)-GNA and (R)-GNA residues! This peculiar behavior of GNA does not affect base-pairing between A and T. Thus, both (S)-GNA T and (R)-GNA T pair with A by forming two hydrogen bonds, except that it is O2 of the GNA T that is positioned opposite N6 of A rather than O4 of T (or U) in a pair formed by DNA (or RNA) (Fig. 5). The (R)-GNA T residue inside the RNA duplex disrupts stacking and base-pairing. In one-half of the duplex formed by the modified, self-complementary dodecamer, the U adjacent to (R)-GNA T is inserted into the major groove and establishes a base triple with the A:(R)-GNA T pair, orphaning the A immediately opposite. The presence of a left-handed (R)-GNA residue inside the right-handed RNA duplex introduces a kink in the backbone (Schlegel et al. 2017; Liczner et al. 2021).
FIGURE 5.
Various base-pairing modes between GNA and RNA contrasted with standard Watson–Crick base pairs.
The flipped nucleobase orientation of GNA inside RNA explains why G:C pairs are not tolerated in RNA:(S)-GNA duplexes. Unlike A and T that can establish two hydrogen bonds in both the Watson–Crick and reverse Watson–Crick pairing modes, reverse Watson–Crick G:C pairs have only two hydrogen bonds, and their formation requires a sheared orientation of the two bases (Fig. 5). These changes result in inferior stability due to loss of stacking and hydrogen bonding, thus precluding G:C pairing between GNA and RNA. Of note, (S)-GNA:RNA hybrids, even those that contain only A:T or A:U pairs, are of lower thermal stability than GNA:GNA and RNA:RNA duplexes (Zhang et al. 2005). However, it is possible to overcome this limitation by introducing isonucleotides such as iso-C and iso-G (Fig. 5). Indeed, pairing between GNA and RNA that involves G and C can be rescued by the introduction of either (S)-iso-GNA C opposite G or (S)-iso-GNA G opposite C (Schlegel et al. 2021).
Probably the most stunning insight from these structures that revealed a flipped base orientation of GNA bases inside an otherwise RNA duplex is that this behavior is not an exception but rather the rule. Thus, the backbone curvature of (S)-GNA duplexes closely resembles that of A-form DNA and RNA duplexes. However, base pairs in the GNA duplex are inverted relative to DNA and RNA; H-bonding groups such as N6 of A, N4 of C, O6 of G, and O4 of T map to the “convex surface” of the GNA duplex, which corresponds to the shallow minor groove of an RNA duplex. Conversely, C2 of A, O2 of C and T, and N2 of G are located inside the deep “minor groove” of the duplex, which corresponds to the deep major groove of an RNA duplex (Schlegel et al. 2021). This inverted orientation of base pairs inside the GNA duplex had been overlooked for 15 years! These structures suggested that a simple approach to overcome this pairing limitation is through the utilization of iso base pairs of GNA G and GNA C (Fig. 5; Scheme 1), which resulted in an improved thermal stability when paired with complementary RNA of 2.5°C and 4.6°C, respectively. The enhanced pairing resulted in an siRNA structure more stabilized against nuclease degradation (Schlegel et al. 2021).
Rather than the simple Watson–Crick pairing system that it is often referred to, GNA behaves in a unique way by combining properties of right-handed A-form DNA and RNA duplexes (backbone geometry) and the left-handed Z-DNA duplex (base-pair orientation). Incidentally, this also helps rationalize the surprising negative Cotton effect at 280 nm seen in CD spectra of right-handed (S)-GNA duplexes (Zhang et al. 2005) that is reminiscent of Z-DNA. Nevertheless, our attempts to crystallize Z-DNA duplexes of sequence 5′-d(CGCGCG)-3′ with a single left-handed (R)-gC residue per strand have not resulted in viable crystals.
IMPLICATIONS OF SHORT INTERNUCLEOTIDE LINKAGES: GNA AND TNA
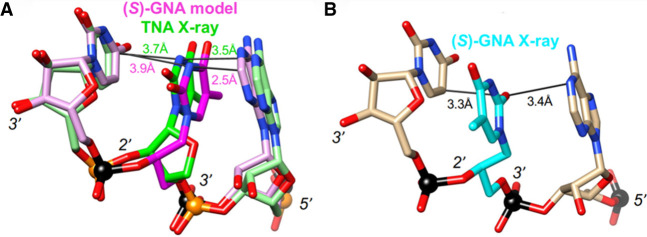
An additional puzzling finding concerns the lack of pairing between GNA and TNA (Yang et al. 2007), which both feature a five-atom repeat unit that is one atom (or bond) shorter relative to DNA and RNA (Fig. 2). The threose sugars uniformly adopt a C4′-exo pucker in TNA:TNA duplexes (Ebert et al. 2008) and inside A-form (Pallan et al. 2003) and B-form DNA duplexes (Wilds et al. 2002) as well as in a TNA strand paired opposite DNA (Chim et al. 2017). However, unlike GNA, TNA does not exhibit the flipped nucleobase orientation, and its right-handed backbone curvature, nucleobase orientation, and backbone-base inclination angle are similar to those of RNA. Because of these features, TNA pairs stably with itself, and it cross-pairs with both DNA and RNA (Schöning et al. 2000) but not with GNA. In fact, if one models a nonrotated nucleobase orientation of GNA by changing the torsion angles around the C2′–C1′ and C1′–N1 bonds of the GNA residue to flip the thymine base to resemble that of TNA without generating any eclipsed bonds, the distance between bases is <2.5 Å on average (Fig. 6). This suggests that in the standard orientation, flexing of the GNA backbone cannot push apart the two bases to the optimal stacking distance of ∼3.4 Å, thus resulting in the preferred flipped base orientation.
FIGURE 6.
(A) Superimposed (S)-GNA T and TNA T inside RNA, demonstrating that GNA with the thymine base flipped (i.e., matching the TNA base orientation) results in a clash with the adjacent base (suboptimal stacking). (B) Crystal structure of RNA with an incorporated (S)-GNA T residue that displays the rotated orientation of the thymine base.
IMPACT OF GNA MODIFICATION ON siRNA POTENCY
The likelihood that GNA would improve RNAi-mediated gene silencing was grounded in several observations regarding the favorable performance of chemically modified siRNAs containing acyclic, thermally destabilizing nucleotides. Modifications can enhance the chemical and metabolic stability of siRNA and increase down-regulation via diverse mechanisms (Kenski et al. 2010; Laursen et al. 2010; Kamiya et al. 2014; Alagia et al. 2015). Modulating the thermal stability of the siRNA duplex via site-specific modification affects the discrimination between the strands (i.e., antisense vs. sense) when the siRNA duplex is loaded into Ago2. The enzyme favors loading the 5′ end of the strand from the end of the duplex that exhibits lower thermal stability, and modification can enhance strand bias (Khvorova et al. 2003; Sano et al. 2008; Bramsen et al. 2009; Addepalli et al. 2010). Another potentially beneficial consequence of introducing a thermally destabilizing modification into an siRNA duplex is that this can facilitate strand separation necessary for an active RISC (Meister et al. 2004).
Incorporation of (S)-GNA or (R)-GNA residues into an RNA duplex strongly lowers the thermal stability with the effect more pronounced for the (R)-isomer (a reduction of between 7.6°C and 18.2°C) and is strongest in the case of incorporation of a single GNA G:C pair [reductions of 20.1°C for (S)-GNA and 27.6°C for (R)-GNA] (Schlegel et al. 2017). A single (S)- or (R)-GNA residue at the 3′ end of a strand also protects against degradation from the 3′-end by snake venom phosphodiesterase to a greater extent than a PS-linkage in place of the last phosphate. When placed at the penultimate position at the 3′ end, GNA does not shield against degradation, but combining the GNA and PS modifications resembles the effect seen when placing two (S)- or (R)-GNA residues at the 3′ end. Optimal protection is achieved by placing two (S)-GNA residues combined with a PS linkage at the 3′ terminus: Relative to the half-life of the oligonucleotide with two Ts at the 3′ end (<1 h), the half-life of the (S)-GNA- and PS-protected oligonucleotide was 27.5 h (Schlegel et al. 2017).
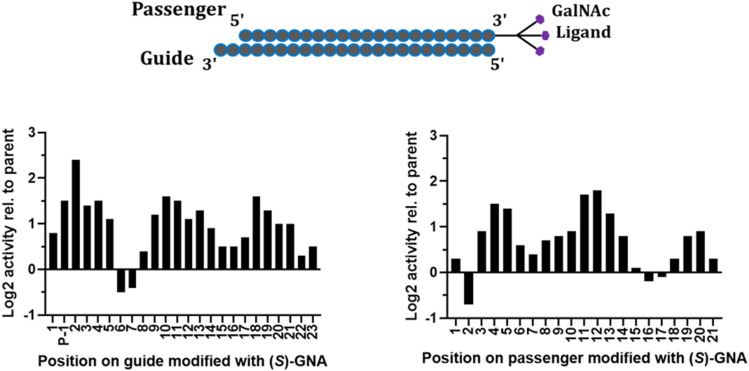
To investigate the positional impact of GNA incorporation on in vitro silencing activity, siRNAs with single (S)-GNA residues along the antisense and sense strands of an siRNA targeting mouse Ttr were tested in a cell-based assay (Fig. 7). Compared to the parent siRNA, GNA was not well tolerated in the antisense strand at positions 1, 2, or 4. The siRNA with a GNA at position 3 of the antisense strand had similar activity to the parent siRNA. GNA at positions 6 and 7 of the antisense strand improved activity relative to the parent (Schlegel et al. 2017). The in vitro potency of the siRNA with an (S)-GNA base pair at position 7 of the antisense strand was approximately threefold better than that of the parent. Importantly, the activity was maintained in mice following subcutaneous injection of this siRNA conjugated to GalNAc (Schlegel et al. 2017).
FIGURE 7.
Position-dependent effect of (S)-GNA on in vitro potency of siRNA-mediated silencing of the Ttr gene in an in vitro assay; antisense (guide) strand on left and sense (passenger) strand on right.
The increased potency of siRNA with an (S)-GNA residue incorporated at position 7 of the antisense strand is particularly noteworthy. Inspection of crystal structures of Ago2 bound either to a microRNA (miR-20a) single strand (PDB ID 4F3T; Elkayam et al. 2012) or a seed duplex (PDB ID 4W5T; Schirle et al. 2014) revealed that the miRNA and the antisense strand of the siRNA have kinks between positions 6 and 7; this kink is particularly pronounced in the miRNA complex (Fig. 8). Bending or kinking of the nucleic acid sugar-phosphate backbone results in compressed intra-strand phosphate-phosphate distances. In the Ago2 complexes the phosphate-phosphate distance at the site of the kink is only 5.5 Å (Schlegel et al. 2017). This distance is considerably shorter than the distance between adjacent phosphate groups in an A-form duplex, but the same as the distance between the 3′- and 2′-phosphate groups in GNA (Fig. 3B). We speculate that the enhanced potency of siRNAs with (S)-GNA at position 7 of the antisense strand is due to the modification facilitating the kink that is imposed by Ago2 binding (Fig. 3D; Schlegel et al. 2022). Favorable effects by GNA modification on RNAi-induced silencing may therefore be the result of two synergistic effects: thermal rebalancing and conformational preorganization.
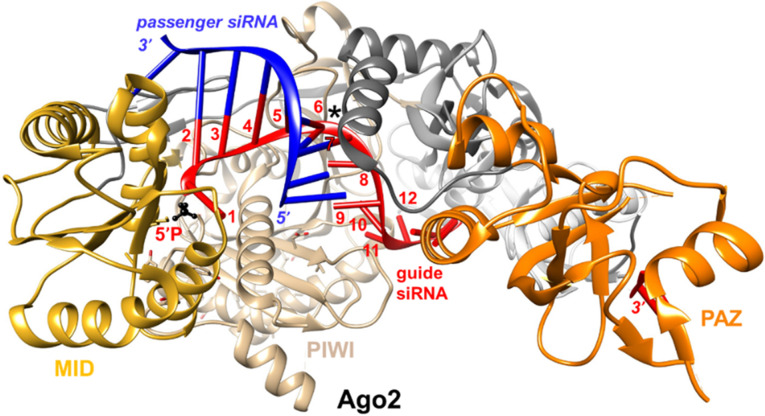
FIGURE 8.
View of the crystal structure of Ago2 bound to an RNA duplex (PDB ID 4W5T) kinked between positions 6 and 7 of the antisense (guide) strand (asterisk). Ago2 domains are highlighted and labeled, and the siRNA (antisense and sense) strands are colored in red and blue, respectively. Antisense strand residues 1 to 12 are numbered, and the 5′-phosphate group is highlighted in black and ball-and-stick mode.
GNA MODIFICATION OF siRNAS CAN MITIGATE RNAi OFF-TARGET EFFECTS
Preclinical mechanistic studies have suggested that liver hepatotoxicity of GalNAc-siRNA conjugates is largely driven by seed-mediated off-target effects (Janas et al. 2018). We were motivated by the aforementioned data and other previous reports to assess the potential of GNA to mitigate seed-mediated repression of off-target gene transcripts, a common limitation in the development of some siRNAs (Janas et al. 2018, and references cited). In such scenarios, XNA modification of siRNAs can provide a straightforward seed-pairing destabilization approach to limit the ability of antisense strand-loaded Ago2 to engage through seed-only binding to off-target gene transcripts. Unlike similar approaches using, for example, UNA to mitigate off-target binding, which has only been demonstrated in vitro (Bramsen et al 2010), GNA has been shown to improve the off-target profile both in vitro and in vivo. A single GNA modification in the seed region results in a six- to eightfold improved therapeutic index in rats (Schlegel et al. 2022). Here again, the unique structure of GNA incorporated at position 7 may disfavor seed-only base-pairing of the Ago2-antisense strand with off-target mRNAs but allow on-target activity through full-length pairing of the antisense strand and the on-target mRNA. This approach and class of siRNAs, featuring high metabolic stability, long duration, and high specificity, was termed ESC+ (Enhanced Stability Chemistry Plus).
CLINICAL STATUS OF GNA-MODIFIED (ESC+) RNAi THERAPEUTICS
Clinical experience with GalNAc-siRNAs to date has shown that these potential therapeutics elicit a robust and dose-responsive silencing of the intended target in the liver and are generally well-tolerated. Two siRNAs, ALN-AAT (targeting Alpha-1 antitrypsin gene) and ALN-HBV (targeting Hepatitus B virus), had dose-dependent, transient, and asymptomatic liver enzyme elevations in a subset of patients in the clinic. These transient elevations in liver enzymes appeared to be the result of a sequence-specific effect since other development candidates with a similar chemistry and design had not demonstrated such liver enzyme elevations (Schlegel et al. 2022). A similar observation was made in preclinical rodent studies where hepatotoxicity was deemed the result of sequence and not chemistry. In addition, the timing of liver function test (LFT) increases in the clinic coincided with the point at which maximal knockdown and RISC loading occurred, further suggestive of a seed-mediated, off-target effect. Presenting a unique opportunity to evaluate the ability of GNA substitution to impact efficacy and safety, we redesigned ALN-AAT and ALN-HBV to have a single (S)-GNA in the seed region, providing novel candidates ALN-AAT02 and ALN-HBV02, respectively. These two candidates showed an improved preclinical safety profile and were subsequently evaluated in a phase 1 study in healthy volunteers. Neither siRNA, in contrast to the respective parent siRNAs of the same sequence, caused liver enzyme elevations at the highest dose tested (6 or 10 mg/kg), providing positive human proof that incorporation of a single GNA improves safety of siRNA and representing a rare example of clinical studies with nucleic acid–based therapeutics incorporating an XNA modification and designated as the ESC+ approach (Schlegel et al. 2022). Several other ESC+ candidate siRNAs that contain GNA residues are currently in clinical development, with the most advanced candidates currently in phase II trials.
CONCLUSIONS
The unique properties of (S)-GNA have been exploited to improve the clinical profile of siRNA. GNA is a thermally destabilizing modification due to its glycol-phosphate backbone, which is shorter than that of DNA and RNA. GNA is well tolerated at most positions of the antisense and sense strands when tested individually. Most importantly, when placed at position 7 of the antisense strand, (S)-GNA efficiently mitigates off-target effects. At this position, (S)-GNA is hypothesized to have two synergistic effects that benefit off-target mitigation in the context of the seed complex with RISC Ago2: seed region destabilization and a preorganized kinked conformation (Schlegel et al. 2017, 2021, 2022). Modeling studies support this hypothesis, but a detailed picture of Ago2–GNA interactions is missing. Goals of our laboratories are to solve crystal and solution structures of Ago2 in complex with GNA-modified duplexes and antisense strands. The synthesis of (S)-GNA, which requires no protection and deprotection of the free hydroxyl group, is simpler than syntheses of UNA and 2′–5′ linked RNA, making it attractive for use in clinical candidates. In fact, (S)-GNA was the first XNA to be tested clinically in a therapeutic that acts through an RNAi mechanism. Future studies will focus on an assessment of the effect of placing multiple GNAs in the same strand of siRNAs and an evaluation of pharmacology and metabolic stability.
ACKNOWLEDGMENTS
We want to thank Dhrubajyoti Datta for help with figures and manuscript preparation, and Cindy Courtney and Tim Long for review and comments. We are grateful to all our colleagues at Alnylam who contributed to this exciting journey on GNA modified siRNAs whose names have been mentioned in the cited publications, in particular Alex Ke'lin and Shigeo Matsuda for GNA monomer synthesis and Maja Janas for GNA mediated off-target mitigation studies.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079526.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Addepalli H, Meena, Peng CG, Wang G, Fan Y, Charisse K, Jayaprakash KN, Rajeev KG, Pandey RK, Lavine G, et al. 2010. Modulation of thermal stability can enhance the potency of siRNA. Nucleic Acids Res 38: 7320–7331. 10.1093/nar/gkq568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Gait MJ. 2019. Advances in nucleic acid therapeutics. Royal Society of Chemistry, Cambridge, UK. [Google Scholar]
- Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, Ansell S, Du X, Hope MJ, Madden TD, et al. 2019. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol 14: 1084–1087. 10.1038/s41565-019-0591-y [DOI] [PubMed] [Google Scholar]
- Alagia A, Terrazas M, Eritja R. 2015. Modulation of the RNA interference activity using central mismatched siRNAs and acyclic threoninol nucleic acids (aTNA) units. Molecules 20: 7602–7619. 10.3390/molecules20057602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anosova I, Kowal EA, Dunn MR, Chaput JC, Van Horn WD, Egli M. 2016. The structural diversity of artificial genetic polymers. Nucleic Acids Res 44: 1007–1021. 10.1093/nar/gkv1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Højland T, Abramov M, Van Aerschot A, et al. 2009. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res 37: 2867–2881. 10.1093/nar/gkp106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjær N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, et al. 2010. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res 38: 5761–5773. 10.1093/nar/gkq341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput JC, Herdewijn P. 2019. What is XNA? Angew Chem Int Ed Engl 58: 1570–11572. 10.1002/anie.201905999 [DOI] [PubMed] [Google Scholar]
- Chaput JC, Switzer C. 2000. Nonenzymatic oligomerization on templates containing phosphodiester-linked acyclic glycerol nucleic acid analogues. J Mol Evol 51: 464–470. 10.1007/s002390010109 [DOI] [PubMed] [Google Scholar]
- Chen JJ, Tsai C-H, Cai X, Horhota AT, McLaughlin LW, Szostak JW. 2009. Enzymatic primer-extension with glycerol-nucleoside triphosphates on DNA templates. PLoS ONE 4: e4949. 10.1371/journal.pone.0004949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim N, Shi C, Sau SP, Nikoomanzar A, Chaput JC. 2017. Structural basis for TNA synthesis by an engineered TNA polymerase. Nat Commun 8: 1810. 10.1038/s41467-017-02014-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PD, Acevedo OL, Davis PW, Ecker DJ, Hebert N. 1995. Synthesis of acyclic oligonucleotides as antiviral and antiinflammatory agents and inhibitors of phospholipase A2. WO patent 9518820. [Google Scholar]
- Cook PD, Acevedo OL, Davis PD, Ecker DJ, Hebert N. 1999. Phosphate linked oligomers. U.S. patent no. 5,886,177. [Google Scholar]
- D'Alonzo D, Guaragna A, Palumbo G. 2011. Exploring the role of chirality in nucleic acid recognition. Chem Biodiv 8: 373–413. 10.1002/cbdv.201000303 [DOI] [PubMed] [Google Scholar]
- Deleavey GF, Damha MJ. 2012. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol 19: 937–954. 10.1016/j.chembiol.2012.07.011 [DOI] [PubMed] [Google Scholar]
- Ebert M-O, Mang C, Krishnamurthy R, Eschenmoser A, Jaun B. 2008. The structure of a TNA−TNA complex in solution: NMR study of the octamer duplex derived from α-(L)-threofuranosyl-(3'-2')-CGAATTCG. J Am Chem Soc 130: 15105–15115. 10.1021/ja8041959 [DOI] [PubMed] [Google Scholar]
- Egli M. 2022. DNA and RNA structure. In Nucleic acids in chemistry and biology, 4th ed. (ed. Blackburn GM, et al.), pp. 20–95. Royal Society of Chemistry, Cambridge, UK. [Google Scholar]
- Egli M, Herdewijn P. 2012. Chemistry and biology of artificial nucleic acids. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- Egli M, Manoharan M. 2019. Re-engineering RNA molecules into therapeutic agents. Acc Chem Res 52: 1036–1047. 10.1021/acs.accounts.8b00650 [DOI] [PubMed] [Google Scholar]
- Egli M, Manoharan M. 2023. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res 51 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M, Pallan PS. 2010. The many twists and turns of DNA: template, telomere, tool and target. Curr Opin Struct Biol 20: 262–275. 10.1016/j.sbi.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn C-D, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. 2012. The structure of human Argonaute-2 in complex with miR-20a. Cell 150: 100–110. 10.1016/j.cell.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Foster DJ, Brown CR, Shaikh S, Trapp C, Schlegel MK, Qian K, Sehgal A, Rajeev KG, Jadhav V, Manoharan M, et al. 2018. Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol Ther 26: 708–717. 10.1016/j.ymthe.2017.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Aartsma-Rus A, Alves S, Borgos SE, Buijsen RAM, Collin RWJ, Covello G, Denti MA, Desviat LR, Echevarría L, et al. 2021. Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol Med 13: e13243. 10.15252/emmm.202013243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuberger BD, Switzer C. 2008. A pre-RNA candidate revisited: both enantiomers of flexible nucleoside triphosphates are DNA polymerase substrates. J Am Chem Soc 130: 412–413. 10.1021/ja0770680 [DOI] [PubMed] [Google Scholar]
- Holý A, Ivanova GS. 1974. Aliphatic analogs of nucleotides. Synthesis and affinity towards nucleases. Nucleic Acids Res 1: 19–34. 10.1093/nar/1.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns H, Taneja N, Willoughby JLS, Akabane-Nakata M, Brown CR, Nguyen T, Bisbe A, Matsuda S, Hettinger M, Manoharan RM, et al. 2022. Chirality matters: stereo-defined phosphorothioate linkages at the termini of small interfering RNAs improve pharmacology in vivo. Nucleic Acids Res 50: 1221–1240. 10.1093/nar/gkab544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas MM, Schlegel MK, Harbison CE, Yilmaz VO, Jiang Y, Parmar R, Zlatev I, Castoreno A, Xu H, Shulga-Morskaya S, et al. 2018. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat Commun 9: 723. 10.1038/s41467-018-02989-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AT, Schlegel MK, Meggers E, Essen LO, Wiest O. 2011. On the structure and dynamics of duplex GNA. J Org Chem 76: 7964–7974. 10.1021/jo201469b [DOI] [PubMed] [Google Scholar]
- Kamiya Y, Takai J, Ito H, Murayama K, Kashida H, Asanuma H. 2014. Enhancement of stability and activity of siRNA by terminal substitution with serinol nucleic acid (SNA). ChemBioChem 5: 2549–2555. 10.1002/cbic.201402369 [DOI] [PubMed] [Google Scholar]
- Kenski DM, Cooper AJ, Li JJ, Willingham AT, Haringsma HJ, Young TA, Kuklin NA, Jones JJ, Cancilla MT, McMasters DR, et al. 2010. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5'-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res 38: 660–671. 10.1093/nar/gkp913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216. 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- Laursen MB, Pakula MM, Gao S, Fluiter K, Mook OR, Baas F, Langklaer N, Wengel SL, Wengel J, Kjems J, et al. 2010. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol BioSyst 6: 862–870. 10.1039/b918869j [DOI] [PubMed] [Google Scholar]
- Liczner C, Duke K, Juneau G, Egli M, Wilds CJ. 2021. Beyond ribose and phosphate: selected nucleic acid modifications for structure-function investigations and therapeutic applications. Beilstein J Org Chem 17: 908–931. 10.3762/bjoc.17.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangos MM, Min K-L, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. 2002. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2′F-ANA constructs containing acyclic nucleotide inserts. J Am Chem Soc 25: 654–661. 10.1021/ja025557o [DOI] [PubMed] [Google Scholar]
- Manoharan M, Akinc A, Pandey RK, Qin J, Hadwiger P, John M, Mills K, Charisse K, Maier MA, Nechev L, et al. 2011. Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew Chem Int Ed Engl 50: 2284–2288. 10.1002/anie.201006519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Keiser K, Nair JK, Charisse K, Manoharan RM, Kretschmer P, Peng CG, Kel'in AV, Kandasamy P, Willoughby JLS, et al. 2015. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem Biol 10: 1181–1187. 10.1021/cb501028c [DOI] [PubMed] [Google Scholar]
- Meggers E, Zhang L. 2010. Synthesis and properties of the simplified nucleic acid glycol nucleic acid. Acc Chem Res 43: 1092–1102. 10.1021/ar900292q [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197. 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Merle Y, Bonneil E, Merle L, Sági J, Szemzö A. 1995. Acyclic oligonucleotide analogues. Int J Biol Macromol 17: 239–245. 10.1016/0141-8130(95)98150-W [DOI] [PubMed] [Google Scholar]
- Nair JK, Willoughby JLS, Chan A, Charisse K, Alam MR, Wang Q, Hoekstra M, Kandasamy P, Kel'in AV, Milstein S, et al. 2014. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136: 16958–16961. 10.1021/ja505986a [DOI] [PubMed] [Google Scholar]
- Nair JK, Attarwala H, Sehgal A, Wang Q, Aluri K, Zhang X, Gao M, Liu J, Indrakanti R, Schofield S, et al. 2017. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res 45: 10969–10977. 10.1093/nar/gkx818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P, Dreiøe LH, Wengel J. 1995. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg Med Chem 3: 19–28. 10.1016/0968-0896(94)00143-Q [DOI] [PubMed] [Google Scholar]
- Pallan PS, Wilds CJ, Wawrzak Z, Krishnamurthy R, Eschenmoser A, Egli M. 2003. Why does TNA pair more strongly with RNA than with DNA?: an answer from X-ray analysis. Angew Chem Int Ed Engl 115: 5893–5895. 10.1002/anie.200352553 [DOI] [PubMed] [Google Scholar]
- Pallan PS, Lubini P, Bolli M, Egli M. 2007. Backbone-base inclination as a fundamental determinant of nucleic acid self- and cross-pairing. Nucleic Acids Res 35: 6611–6624. 10.1093/nar/gkm612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A, Wengel J. 2010. Thermodynamics of RNA duplexes modified with unlocked nucleic acid nucleotides. Nucleic Acids Res 38: 6697–6706. 10.1093/nar/gkq561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A, Wengel J. 2011. Unlocked nucleic acid—an RNA modification with broad potential. Org Biomol Chem 9: 3591–3597. 10.1039/c0ob01085e [DOI] [PubMed] [Google Scholar]
- Peng L, Roth H-J. 2004. Synthesis and properties of 2′-deoxy-1′,2′-seco-D-ribosyl (5′→3′) oligonucleotides (=1′,2′-seco-DNA) containing adenine and thymine. Helv Chim Acta 80: 1494–1512. 10.1002/hlca.19970800513 [DOI] [Google Scholar]
- Rajeev KG, Nair JK, Jayaraman M, Charisse K, Taneja N, O'Shea J, Willoughby JLS, Yucius K, Nguyen T, Shulga-Morskaya S, et al. 2015. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem 16: 903–908. 10.1002/cbic.201500023 [DOI] [PubMed] [Google Scholar]
- Roberts TC, Langer R, Wood MJA. 2020. Advances in oligonucleotide drug delivery. Nat Rev Drug. Disc 19: 673–694. 10.1038/s41573-020-0075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M, Sierant M, Miyagishi M, Nakanishi M, Takagi Y, Sutou S. 2008. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res 36: 5812–5821. 10.1093/nar/gkn584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ. 2014. Gene regulation. Structural basis for microRNA targeting. Science 346: 608–613. 10.1126/science.1258040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel MK, Meggers E. 2009. Improved phosphoramidite building blocks for the synthesis of the simplified nucleic acid GNA. J Org Chem 74: 4615–4618. 10.1021/jo900365a [DOI] [PubMed] [Google Scholar]
- Schlegel MK, Essen LO, Meggers E. 2008. Duplex structure of a minimal nucleic acid. J Am Chem Soc 130: 8158–8159. 10.1021/ja802788g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel MK, Essen L-O, Meggers E. 2010. Atomic resolution duplex structure of the simplified nucleic acid GNA. Chem Commun (Camb) 46: 1094–1096. 10.1039/B916851F [DOI] [PubMed] [Google Scholar]
- Schlegel MK, Foster DJ, Kel'in AV, Zlatev I, Bisbe A, Jayaraman M, Lackey JG, Rajeev KG, Charisse K, Harp J, et al. 2017. Chirality dependent potency enhancement and structural impact of glycol nucleic acid modification on siRNA. J Am Chem Soc 139: 8537–8546. 10.1021/jacs.7b02694 [DOI] [PubMed] [Google Scholar]
- Schlegel MK, Matsuda S, Brown CR, Harp JM, Barry JD, Berman D, Castoreno A, Schofield S, Szeto J, Manoharan M, et al. 2021. Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs. Nucleic Acids Res 49: 10851–10867. 10.1093/nar/gkab916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel MK, Janas MM, Jiang Y, Barry JD, Davis W, Agarwal S, Berman D, Brown CR, Castoreno A, LeBlanc S, et al. 2022. From bench to bedside: improving the clinical safety of GalNAc-siRNA conjugates using seed-pairing destabilization. Nucleic Acids Res 50: 6656–6670. 10.1093/nar/gkac539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider KC, Benner SA. 1990. Oligonucleotides containing flexible nucleoside analogs. J Am Chem Soc 112: 453–455. 10.1021/ja00157a073 [DOI] [Google Scholar]
- Schöning KU, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A. 2000. Chemical etiology of nucleic acid structure: the α-threofuranosyl-(3'→2') oligonucleotide system. Science 290: 1347–1351. 10.1126/science.290.5495.1347 [DOI] [PubMed] [Google Scholar]
- Seita T, Yamauchi K, Kinoshita M, Imoto M. 1972. Condensation polymerization of nucleotide analog. Makromol Chem 154: 255–261. 10.1002/macp.1972.021540123 [DOI] [Google Scholar]
- Seth PP, Tanowitz M, Bennett CF. 2019. Selective tissue targeting of synthetic nucleic acid drugs. J Clin Invest 129: 915–925. 10.1172/JCI125228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432: 173–178. 10.1038/nature03121 [DOI] [PubMed] [Google Scholar]
- Tsai C-H, Chen J, Szostak JW. 2007. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc Natl Acad Sci 104: 14598–14603. 10.1073/pnas.0704211104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda N, Kawabata T, Takemoto K. 1971. Synthesis of N-(2,3-dihydroxypropyl) derivatives of nucleic bases. J Heterocycl Chem 8: 827–829. 10.1002/jhet.5570080527 [DOI] [Google Scholar]
- Wilds CJ, Wawrzak Z, Krishnamurthy R, Eschenmoser A, Egli M. 2002. Crystal structure of a B-form DNA duplex containing (L)-α-threofuranosyl (3'→2') nucleosides: a four-carbon sugar is easily accommodated into the backbone of DNA. J Am Chem Soc 124: 13716–13721. 10.1021/ja0207807 [DOI] [PubMed] [Google Scholar]
- Willoughby JLS, Chan A, Sehgal A, Butler JS, Nair JK, Racie T, Shulga-Morskaya S, Nguyen T, Qian K, Yucius K, et al. 2018. Evaluation of GalNAc-siRNA conjugate activity in pre-clinical animal models with reduced asialoglycoprotein receptor expression. Mol Ther 26: 105–114. 10.1016/j.ymthe.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YW, Zhang S, McCullum EO, Chaput JC. 2007. Experimental evidence that GNA and TNA were not sequential polymers in the prebiotic evolution of RNA. J Mol Evol 65: 289–295. 10.1007/s00239-007-9017-9 [DOI] [PubMed] [Google Scholar]
- Yoo JW, Mouloungui Z. 2001. Zeolites and mesoporous materials at the dawn of the 21st century. In Proceedings of the 13th International Zeolite Conference, Montpellier, France, p. 238. (ed. Galarneau A, et al.). Abstract # 23-P-32. Elsevier, Amsterdam. eBook ISBN: 9780080543918 [Google Scholar]
- Zhang L, Peritz A, Meggers E. 2005. A simple glycol nucleic acid. J Am Chem Soc 127: 4174–4175. 10.1021/ja042564z [DOI] [PubMed] [Google Scholar]
- Zhang L, Peritz AE, Carroll PJ, Meggers E. 2006. Synthesis of glycol nucleic acids. Synthesis (Mass) 4: 645–653. [Google Scholar]
- Zhang RS, McCullum EO, Chaput JC. 2008. Synthesis of two mirror image 4-helix junctions derived from glycerol nucleic acid. J Am Chem Soc 130: 5846–5847. 10.1021/ja800079j [DOI] [PubMed] [Google Scholar]
- Zhang S, Switzer C, Chaput JC. 2010. The resurgence of acyclic nucleic acids. Chem Biodiv 7: 245–258. 10.1002/cbdv.200900281 [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. 2006. RNAi-mediated gene silencing in non-human primates. Nature 441: 111–114. 10.1038/nature04688 [DOI] [PubMed] [Google Scholar]