Abstract
Adenosine deamination by the ADAR family of enzymes is a natural process that edits genetic information as it passes through messenger RNA. Adenosine is converted to inosine in mRNAs, and this base is interpreted as guanosine during translation. Realizing the potential of this activity for therapeutics, a number of researchers have developed systems that redirect ADAR activity to new targets, ones that are not normally edited. These site-directed RNA editing (SDRE) systems can be broadly classified into two categories: ones that deliver an antisense RNA oligonucleotide to bind opposite a target adenosine, creating an editable structure that endogenously expressed ADARs recognize, and ones that tether the catalytic domain of recombinant ADAR to an antisense RNA oligonucleotide that serves as a targeting mechanism, much like with CRISPR-Cas or RNAi. To date, SDRE has been used mostly to try and correct genetic mutations. Here we argue that these applications are not ideal SDRE, mostly because RNA edits are transient and genetic mutations are not. Instead, we suggest that SDRE could be used to tune cell physiology to achieve temporary outcomes that are therapeutically advantageous, particularly in the nervous system. These include manipulating excitability in nociceptive neural circuits, abolishing specific phosphorylation events to reduce protein aggregation related to neurodegeneration or reduce the glial scarring that inhibits nerve regeneration, or enhancing G protein-coupled receptor signaling to increase nerve proliferation for the treatment of sensory disorders like blindness and deafness.
Keywords: ADARs, RNA editing, RNA therapies, site-directed RNA editing, adenosine deamination
RNA EDITING BY ADENOSINE DEAMINATION
Enzymatic systems that can manipulate genetic information have been the foundation of some of the most powerful tools for biological research and, more recently, modern medicine. RNA interference and CRISPR-Cas genome editing are prime examples used to control genetic information in RNA and DNA, respectively. RNA editing through adenosine deamination is another enzymatic system that has recently begun to attract attention related to its potential as a therapeutic tool (Montiel-Gonzalez et al. 2018; Merkle and Stafforst 2021). This process is catalyzed by ADARs (adenosine deaminase that acts on RNA), a family of enzymes that is expressed in all multicellular metazoans. ADARs possess two stereotypical domains: a variable number of double-stranded RNA binding domains (dsRBDs) at their amino terminus, followed by a carboxy-terminal deaminase domain (DD). The dsRBDs bind to higher-order structures in RNAs, positioning the DD next to a target adenosine (A); the DD then catalyzes the hydrolytic deamination of the A to inosine (I), a biological mimic of guanosine (G). A → I conversions influence base-pairing, and when they occur at nonsynonymous positions in mRNAs, they recode codons. Mammalian genomes encode two catalytically active ADARs, ADAR1 and ADAR2. There are two forms of ADAR1, the constitutively expressed ADAR1 p110 and the interferon inducible p150. Although ADAR1 is thought to be involved in regulating innate immunity and combatting transposable genetic elements (Mannion et al. 2014; Liddicoat et al. 2015), ADAR2 is the main codon recoder (Tan et al. 2017; Chalk et al. 2019). By converting an A → I, ADARs can recode 28 codons, changing over half of the possible amino acids, and all stop codons, to a different amino acid. Thus, if this activity could be controlled, it would prove useful for research and therapeutic applications.
SITE-DIRECTED RNA EDITING SYSTEMS
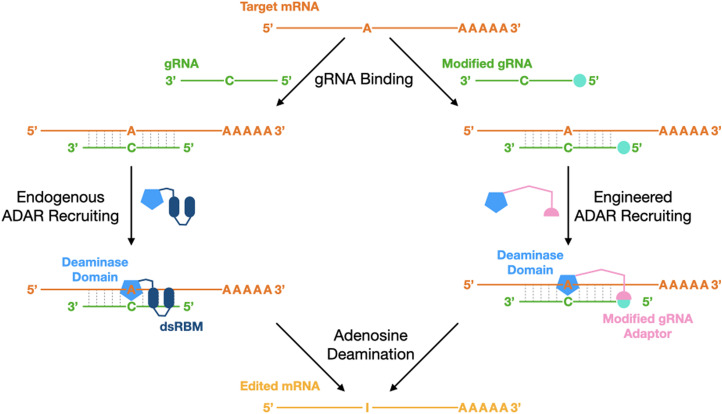
Soon after the first ADARs were discovered, Woolf et al. demonstrated the ability to redirect ADAR to a new target—in this case an A within a premature termination codon in a dystrophin mRNA (Woolf et al. 1995). Editing recoded this position to a tryptophan, allowing read-through, both in cell extracts and Xenopus embryos. Although this was the first demonstration of the potential for therapeutic RNA editing, the concept laid dormant for over 17 years until the Stafforst and Rosenthal groups revisited the idea (Stafforst and Schneider 2012; Montiel-Gonzalez et al. 2013). By this point, the crystal structure of ADAR's DD had been solved and this, along with accompanying data, demonstrated that this domain on its own is catalytically active (Macbeth et al. 2005). Accordingly, the new approaches sought to couple the DD to an antisense RNA oligonucleotide (ASO) that would serve the dual purpose of replacing the dsRBDs as a targeting mechanism and, once bound to their target sequence, creating the dsRNA structure required by ADARs to edit (Fig. 1). Different approaches were used to join the oligonucleotide to the DD. The Stafforst group used a SNAP-tag, which creates a covalent linkage between an engineered O-6-methylguanine-DNA methyltransferase fused to DD and an O6-benzylguanine on the 5′ end of the oligonucleotide. The Rosenthal group used the λN peptide-BoxB hairpin interaction (Keppler et al. 2002; Lazinski et al. 1989), which creates a noncovalent interaction between the λN peptide fused to DD and a BoxB hairpin fused to the oligonucleotide. Subsequently, other linkages have been used, such as the MS2 system (Azad et al. 2017; Montiel-Gonzalez et al. 2018). A common problem with delivering the naked DD with a gRNA is that it tends to create many off-target edits, not only within the targeted message but also in unrelated messages across the transcriptome (Vallecillo-Viejo et al. 2018; Buchumenski et al. 2021). Approaches such as localizing the DD to the nucleus, or splitting it into two units that only form a functional deaminase enzyme when they come together, have significantly reduced unwanted edits (Vallecillo-Viejo et al. 2018; Katrekar et al. 2022a). These engineered systems have been used to good effect in vivo. The Mandel lab has shown that a genetic mutation in the MECP2 protein that underlies Rett syndrome can be corrected in mice, with symptoms of the disorder alleviated, using the λN peptide-BoxB system (Sinnamon et al. 2017, 2020, 2022). In these experiments, both the DD and the gRNA were delivered by adeno associated virus (AAV) to the central nervous system, either through direct injection into the hippocampus or a systemic injection of the AAV PHP.B variant that crosses the blood-brain barrier. One particularly encouraging aspect of these studies is the fact that editing persisted for a month after the administration of the virus and this alleviated disease symptoms and, in some cases, extended the life of the treated animals. Editing at later times after virus injection was not examined.
FIGURE 1.
Strategies for SDRE. There are two basic strategies for directing adenosine deamination to a specific adenosine within a mRNA, and both require gRNAs. The left side of the figure depicts the recruitment of endogenously expressed ADAR enzymes, and the right side represents the delivery of an engineered deaminase domain from ADAR. For the second approach, the gRNA contains an element at its 5′ end (cyan ball) that specifically binds to a protein tag appended to the deaminase domain (pink line and half circle), and together they form the linkage between the gRNA and the deaminase domain.
Despite the promise of engineered SDRE systems (i.e., those that utilize the DD and a gRNA), far more attention has been paid to developing systems designed to recruit endogenous ADARs to new targets (Fig. 1). These systems are attractive because of their simplicity: only an antisense gRNA needs to be delivered. When bound to its target message, the gRNA creates an editable dsRNA structure surrounding a selected adenosine. The key challenge confronting these systems is designing an effective gRNA. Naked RNA degrades rapidly in a cellular environment and perfectly duplexed RNA structures are poor substrates for ADARs, which prefer some mismatches and bulges. The challenge is that there are no known rules for positioning the mismatched areas. The first studies on redirecting ADARs to new sites were performed by the Stafforst group, using gRNAs which partially mimicked natural ADAR structures (Schneider et al. 2014; Wettengel et al. 2017). They also demonstrated the effectiveness of chemically modifying oligonucleotides to protect them from degradation inside the cell (Vogel et al. 2014). gRNA design has varied tremendously between studies and no consensus has emerged. Some have used gRNAs which produce long, near perfect duplexes (Qu et al. 2019). Others have used gRNAs that form large clusters of base-paired regions (Reautschnig et al. 2022a), and others have used circular gRNAs (Katrekar et al. 2022b; Yi et al. 2022). A promising recent approach has focused on chemical modifications. By making chimeric backbones using stereopure phosphorothioate and nitrogen-containing linkages, in vivo recruitment of ADAR was greatly enhanced (Monian et al. 2022). Thus, currently there are no universal rules for gRNA design. In addition, some As are much more difficult to edit than others, and this probably relates to the identity of the residues that surround the A, higher-order structures within a message, and/or competing RNA binding proteins that might limit access of ADAR or the gRNA, or the ADAR expression profile within a targeted cell. In addition, it is difficult to limit editing to the target A when neighboring As are present.
Given the challenges related to the recruitment of endogenous ADARs, it is reasonable to conclude that in some cases the use of engineered SDRE might be more desirable. For example, some As are poorly edited by endogenous ADAR enzymes (Eggington et al. 2011; Matthews et al. 2016). It is well known that a 5′ G inhibits editing. In addition, codons like lysine and asparagine have As at the first two positions and often it is desirable to only edit one of them to produce the desired codon. In these cases, using the endogenous recruitment strategy, one can only manipulate the gRNA to induce more precise and efficient editing. In addition, the targeted cells might not express ADAR at a sufficient level or the diversion of ADAR may reduce its activity at its natural substrates. With the engineered ADAR strategies, one can manipulate the DD of ADAR itself to boost context specific editing (e.g., with the E488Q mutation of ADAR2) (Pokharel and Beal 2006; Montiel-Gonźalez et al. 2016), and the necessary enzymatic activity is delivered exogenously. Another important consideration is the desired duration of the editing activity. In most cases, an antisense oligonucleotide delivered systemically will be cleared from the body fairly rapidly, and thus the duration of the effect would depend on the turnover of the edited RNAs and the proteins that they encode—which could be brief. A notable exception is within the central nervous system, where oligonucleotides can persist far longer (Bennett et al. 2019b). Genetically encoded SDRE systems have the potential for far longer editing durations because target mRNAs will continue to be edited so long as the enzyme and/or gRNA continue to be expressed, an outcome that is desirable when targeting genetic disorders which require continual correction. For example, editing by the λN peptide-BoxB system was shown to persist for over a month when delivered by AAV to mice (Sinnamon et al. 2020, 2022). A significant drawback to the engineered systems, however, is their propensity to generate off-target edits, most likely due to the delivery of additional catalytic activity uncoupled to ADAR's natural targeting architecture (Vallecillo-Viejo et al. 2018; Buchumenski et al. 2021). A good compromise for precise, longer duration editing may be to deliver gRNAs for endogenous recruitment via AAV. SDRE has some clear advantages over conventional gene therapy using AAV. First, the ∼4.5 KB packaging limit of AAV excludes many genes. Second, gene delivery risks both over- and underexpression, while SDRE works within the endogenous expression level of a transcript.
FUTURE DIRECTIONS FOR SDRE: DESIGN CHALLENGES
There are a number of immediate obstacles that must be overcome for SDRE applications. First, with the engineered systems, off-target editing needs to be reduced. This can presumably be accomplished by generating less active forms of the DD and then selecting specific gRNAs that create structures that are selected for these engineered forms. These specific structures would naturally occur infrequently (or never) across the transcriptome. This idea was recently utilized by the Beal group. Position E488 of ADAR forms the tip of the base-flipping loop and occupies the space vacated by the target A while it is undergoing catalysis. When this position was mutated to a tyrosine, a relatively bulky residue, editing activity was severely reduced unless the gRNA position opposite the target A was made abasic (Monteleone et al. 2019). This approach significantly reduced off-target edits. Another important consideration for the development of this system is the fact that the consequences of off-target edits in RNA are not well understood. Due to their transience, and the fact that they would not transform cells, one would guess that they are less consequential than those in DNA. On the other hand, they might cause elevated levels of protein misfolding, causing a metabolic drag on the cell. In addition, specific mRNA off-targets might prove toxic, depending on the protein that is encoded. Thus, the tolerable limits of off-target RNA edits need further examination. Another consideration is the fact that a mutant codon cannot always be edited to encode the wild-type amino acid. Premature termination codons are good examples, which can only be edited to tryptophan, even though the wild-type codon may have been something else. In these cases, the effect of the new amino acid on protein function must be determined empirically.
A significant issue facing the therapeutic use of SDRE relates to the size of the gRNAs that have been developed. For both the engineered ADAR and endogenous ADAR recruitment strategies, the effective gRNAs that have been produced thus far are large, some reaching lengths of hundreds of nucleotides. In contrast, all therapeutic ASOs currently approved by the FDA are small, ranging in size from 18–30 nt. Thus, it is likely that the size of gRNAs for SDRE therapeutics will need to be significantly reduced in order to be tolerated and for ease of manufacturing. This will undoubtedly pose a major challenge, as most natural structures edited by ADARs are large (Higuchi et al. 1993; Reenan 2005) and some of the approaches to making gRNAs, such as the addition of ADAR recruiting domains, require large sequences. However, it is not unreasonable to think that small gRNAs, when carefully selected, may direct efficient editing. Some of the smallest naturally occurring structures and their derivatives, which are on par with the dsRNA structures generated from 20–30 nt gRNAs, drive editing at some of the highest catalytic rates (Eifler et al. 2013; Wang et al. 2018). Thus, small structures are possible but rare, and selection assays may be required to identify them. As more small gRNAs are identified, perhaps general rules regarding their design may become apparent.
FUTURE DIRECTIONS FOR SDRE: NEW CLASSES OF TARGETS
Virtually all of the targets for therapeutic SDRE applications have been genetic mutations that underlie human disease (Montiel-Gonzalez et al. 2013; Wettengel et al. 2017; Qu et al. 2019; Reautschnig et al. 2022b; Sinnamon et al. 2022). This is understandable. When a G is mutated to an A, a directed edit can unequivocally restore function to the encoded protein. However, from a theoretical perspective, genetic mutations, which are permanent, are not ideal targets for SDRE because RNA is transient; any RNA-level intervention must be repeatedly administered. In addition, when delivered systemically, oligonucleotides are rapidly cleared from most of the body (Shadid et al. 2021). The potential targets for SDRE, however, extend far beyond genetic mutations. The only theoretical limitations relate to the functional changes that can be generated by changing an A to an I. We believe that one of the next frontiers for SDRE will be its use to transiently regulate cell physiology. Using this approach, the duration of effects can be far longer than with conventional therapeutics (e.g., small molecules). For these applications, the consequences of edits can be more nuanced than the simple correction of a genetic “mistake” and will require partnerships between experts in SDRE and specific physiological systems. Below, we provide some examples of biological processes that might be exploited.
Voltage- and ligand-dependent ion channels are the molecular machines that produce electrical excitability. They control essential neural and muscular functions such as respiration, locomotion, cognition, along with numerous other functions. Mutations in ion channels underlie countless pathologies: Examples in Na+ channels alone lead to various paralyses, epilepsies, migraines and abnormal sensitivities to pain (Mantegazza et al. 2021). The essential function of an ion channel is to open and close over carefully prescribed timescales—a process known as gating—to permit select ions to pass through their pore. Voltage-dependent channels, like those that pass Na+, K+, or Ca2+, are gated by transient changes in transmembrane potential, and ligand-gated channels, like ionotropic glutamate or GABA (gamma-aminobutyric acid) receptors, are gated by chemical signals. There are multiple isoforms for most channels and the precise voltage range, or neurotransmitter concentration, that gates a specific subtype is a critical feature that defines its identity. For example, to create an action potential, the voltage sensitivities of Na+ and K+ channels must be perfectly balanced. In fact, mutations that shift the voltage dependence of Na+ channels by just a few millivolts have been shown to underlie various pain disorders, such as hypersensitivity to temperature (Dib-Hajj et al. 2005; Rush et al. 2006; Samuels et al. 2008; Faber et al. 2012; Bennett et al. 2019a). The structure and function of voltage-dependent ion channels have been intensively studied for decades, leading to an in-depth understanding of how they operate. By consequence, there is a rich literature on how individual codon changes affect function. Naturally, many of these changes can be introduced by A → I RNA editing. Thus, SDRE carries tremendous potential for manipulating neural circuits for therapeutically advantageous outcomes. Channel voltage sensitivities or ligand affinities are natural targets for tuning excitability. NaV1.7, encoded by SCN9A, is a good example. This channel is highly expressed in nociceptive neurons and mutations within it are tightly associated with pain disorders (Cummins et al. 2004; Bennett et al. 2019a; Hameed 2019). As a validated pharmacological target for pain, much effort has been applied to identifying selective, small molecule blockers for this channel (Kushnarev et al. 2020). As an alternative, SDRE could have significant advantages, because the gating machinery could be recoded to produce shifts in voltage dependence or gating kinetics and these would translate into relatively long duration effects coupled with more precise, nonsystemic delivery.
The treatment of various neurodegenerative disorders, such as Alzheimer's disease (AD), might be approached via the selective regulation of phosphorylation by SDRE. In general, codons encoding phosphorylated residues, or the lysine frequently found in phosphorylation motifs, can be recoded by adenosine deamination. Among other phenotypes, AD is characterized by the formation of neurofibrillary tangles of Tau protein in cholinergic neurons (Long and Holtzman 2019). Hyperphosphorylation of Tau is thought to drive its aggregation, causing microtubule instability which leads to transport defects and general neuronal dysfunction (Salcedo-Tello et al. 2011; Sayas and Ávila 2021). Tau phosphorylation is driven by the β isoform of the glycogen synthase kinase-3 (GSK3), a ubiquitous serine/threonine kinase. Many studies have linked the activity of GSK3-β with the progression of AD. For example, in AD mouse models, inhibition of GSK3-β reduces neuritic plaque formation and Tau phosphorylation. Furthermore, the activity of GSK3-β itself is up-regulated by the phosphorylation of tyrosine at codon 216, and in the frontal cortex of AD patients the phosphorylation of this residue is elevated (Salcedo-Tello et al. 2011; Sayas and Ávila 2021). Tyrosine 21 can be converted to a cysteine (Y216C) through RNA editing. The delivery of SDRE reagents to the mouse brain can lead to durable editing (Sinnamon et al. 2020, 2022) and thus could prove useful in reducing Tau tangles and the progression of AD.
There are innumerable other examples where the selective manipulation of phosphorylation sites could prove beneficial for alleviating medical conditions, many of which are in the nervous system. A good example relates to the process of reinnervation by axonal regrowth following traumatic injuries. Reactive gliosis is a process where astrocytes proliferate, become hypertrophic and up-regulate the expression of intermediate filaments like glial fibrillary acidic protein and vimentin (Hol and Pekny 2015; Pekny and Pekna 2016); it is stimulated by traumatic nerve injury in both the peripheral and central nervous systems and reactive gliosis promotes the formation of a glial scar which helps to stabilize the lesion site and prevent further tissue damage (Diaz Quiroz and Echeverri 2013; Pekny and Pekna 2016). A negative consequence of the scar is that it prevents axonal regrowth and is thought to be one of the main barriers to axonal regeneration. Furthermore, reactive gliosis in response to injury in the peripheral nervous system contributes to the development of chronic pain (Dominguez et al. 2008; Tsuda et al. 2011). Following nerve injury, the signal transducer and activators of transcription 3 (STAT3) pathway is up-regulated in astrocytes and its activity has been associated with reactive gliosis. Similar to GSK3-β, the phosphorylation of Y705 in STAT3 activates its signaling. This tyrosine can be converted to cysteine by SDRE, thus reducing STAT3 signaling. There are good reasons to believe that this could reduce reactive gliosis as it was attenuated in mice lacking this phosphorylation site (Herrmann et al. 2008), and chemical inhibition of STAT3 in wt mice reduced reactive gliosis and hypersensitivity to innocuous stimuli after peripheral nerve injury (Dominguez et al. 2008; Tsuda et al. 2011). Thus, whereas reactive gliosis is a necessary component of healing, its attenuation could provide a more favorable environment for nerve regeneration.
Outside of the removal of phosphorylation sites, the manipulation of G protein-coupled receptor (GPCR) signaling is another attractive target for SDRE. They are an enormous class of receptors, regulating exceptionally diverse cellular functions. Over a third of all FDA approved drugs target GPCRs, most in the form of small molecule agonists or antagonists (Hauser et al. 2018). SDRE could be used to manipulate the intrinsic architecture of GPCRs in order to modulate agonist or G protein affinity. A good example is with the tropomyosin receptor kinase B (TrkB), which can be activated by brain-derived neurotrophic factor (BDNF) to stimulate nerve cell proliferation. In fact, the administration of BDNF has shown some promise in promoting nerve cell regrowth to treat deafness and blindness (Shibata et al. 2010; Khalin et al. 2015); however, a major challenge has been the targeted and durable delivery of BDNF. An alternative would be to manipulate the receptor itself. When searching for point mutations that result in receptor activation and cell proliferation, those that are associated with cancer are a good place to start. The neurotrophic receptor tyrosine kinase (ntrk) genes, which encode the Trk receptors, are oncogenes. A mutation in the ntrk2 gene that encodes TrkB-R458G renders the receptor constitutively active via noncanonical mechanisms that may involve altering interactions with lipids in the bilayer (Joshi et al. 2020). This mutation has been associated with hematologic cancers. It is reasonable to postulate that the introduction of this mutation within mRNA might stimulate a controlled proliferation and not lead to cellular transformation resulting in neoplasms. In theory, R458G could be introduced by SDRE. When compared to BDNF administration, this route would have the benefit of a longer lasting effect because receptor stimulation would persist as long as the edited mRNA and the receptors that they encode last.
In the preceding paragraphs we have provided some examples of how SDRE may be extended into new areas, beyond the correction of genetic mutations. These cases and many others illustrate the essential difference between targeting RNA versus DNA. For the life of a cell, changes in DNA are permanent, and for long-lived cells like neurons, this often equates to the life of an organism. Genetic mutations would preferably be corrected in DNA, but this is not always possible due to the current limitations of DNA editing systems (e.g., low efficiency in nondividing cells like neurons and relatively dangerous off-target edits). As with all RNA-based therapeutics, SDRE is best matched with the transient nature of RNA expression. Although the changes that it directs will turn over, they may last substantially longer than conventional drugs. Furthermore, some of the required changes are also transient (e.g., stimulating initial cell proliferation or resolving reactive gliosis). SDRE is in its infancy, without any ongoing clinical trials, to our knowledge. There are significant challenges to overcome, including optimizing gRNA design in terms of sequence and chemistry, improving delivery to specific tissues and cell types within those tissues, and limiting off-target edits for the engineered SDRE systems. However, the upsurge of basic research and biotech startups invested in advancing these technologies (e.g., ADARx, Shape Therapeutics, Korro Bio, Wave Therapeutics, ProQR, Beam Therapeutics, and Vico Therapeutics) draws attention to their promise. Close collaborations between the developers of SDRE systems and experts in disorders of cell physiology should lead to novel uses.
COMPETING INTEREST STATEMENT
J.J.C.R. is a founder scientific advisor and shareholder of Korro Bio and receives remuneration from them.
ACKNOWLEDGMENTS
Work on this topic in the authors’ laboratory was supported by NIH/NINDS/HEAL 1 U19 NS 126038, NIH/NINDS 1R21NS125495, and a sponsored research agreement by Korro Bio (Cambridge, Massachusetts).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079518.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Azad MTA, Bhakta S, Tsukahara T. 2017. Site-directed RNA editing by adenosine deaminase acting on RNA for correction of the genetic code in gene therapy. Gene Ther 24: 779–786. 10.1038/gt.2017.90 [DOI] [PubMed] [Google Scholar]
- Bennett DL, Clark XAJ, Huang J, Waxman SG, Dib-Hajj SD. 2019a. The role of voltage-gated sodium channels in pain signaling. Physiol Rev 99: 1079–1151. 10.1152/physrev.00052.2017 [DOI] [PubMed] [Google Scholar]
- Bennett CF, Krainer AR, Cleveland DW. 2019b. Antisense oligonucleotide therapies for neurodegenerative diseases. Annu Rev Neurosci 42: 385. 10.1146/annurev-neuro-070918-050501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchumenski I, Roth SH, Kopel E, Katsman E, Feiglin A, Levanon EY, Eisenberg E. 2021. Global quantification exposes abundant low-level off-target activity by base editors. Genome Res 31: 2354–2361. 10.1101/gr.275770.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalk AM, Taylor S, Heraud-Farlow JE, Walkley CR. 2019. The majority of A-to-I RNA editing is not required for mammalian homeostasis. Genome Biol 20: 268. 10.1186/s13059-019-1873-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins TR, Dib-Hajj SD, Waxman SG. 2004. Electrophysiological properties of mutant Nav1.7 sodium channels in a painful inherited neuropathy. J Neurosci 24: 8232–8236. 10.1523/JNEUROSCI.2695-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Quiroz JF, Echeverri K. 2013. Spinal cord regeneration: Where fish, frogs and salamanders lead the way, can we follow? Biochem J 451: 353–364. 10.1042/BJ20121807 [DOI] [PubMed] [Google Scholar]
- Dib-Hajj SD, Rush AM, Cummins TR, Hisama FM, Novella S, Tyrrell L, Marshall L, Waxman SG. 2005. Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain 128: 1847–1854. 10.1093/brain/awh514 [DOI] [PubMed] [Google Scholar]
- Dominguez E, Rivat C, Pommier B, Mauborgne A, Pohl M. 2008. JAK/STAT3 pathway is activated in spinal cord microglia after peripheral nerve injury and contributes to neuropathic pain development in rat. J Neurochem 107: 50–60. 10.1111/j.1471-4159.2008.05566.x [DOI] [PubMed] [Google Scholar]
- Eggington JM, Greene T, Bass BL. 2011. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun 2: 319. 10.1038/ncomms1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler T, Pokharel S, Beal PA. 2013. RNA-seq analysis identifies a novel set of editing substrates for human ADAR2 present in Saccharomyces cerevisiae. Biochemistry 52: 7857–7869. 10.1021/bi4006539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber CG, Hoeijmakers JG, Ahn HS, Cheng X, Han C, Choi JS, Estacion M, Lauria G, Vanhoutte EK, Gerrits MM, et al. 2012. Gain of function NaV1.7 mutations in idiopathic small fiber neuropathy. Ann Neurol 71: 26–39. 10.1002/ana.22485 [DOI] [PubMed] [Google Scholar]
- Hameed S. 2019. Nav1.7 and Nav1.8: role in the pathophysiology of pain. Mol Pain 15: 1744806919858801. 10.1177/1744806919858801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Chavali S, Masuho I, Jahn LJ, Martemyanov KA, Gloriam DE, Babu MM. 2018. Pharmacogenomics of GPCR drug targets. Cell 172: 41–54.e19. 10.1016/j.cell.2017.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JE, Imura T, Song B, Qi J, Ao Y, Nguyen TK, Korsak RA, Takeda K, Akira S, Sofroniew M V. 2008. STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J Neurosci 28: 7231–7243. 10.1523/JNEUROSCI.1709-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. 1993. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75: 1361–1370. 10.1016/0092-8674(93)90622-W [DOI] [PubMed] [Google Scholar]
- Hol EM, Pekny M. 2015. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr Opin Cell Biol 32: 121–130. 10.1016/j.ceb.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Joshi SK, Qian K, Bisson WH, Watanabe-Smith K, Huang A, Bottomly D, Traer E, Tyner JW, Mcweeney SK, Davare MA, et al. 2020. Discovery and characterization of targetable NTRK point mutations in hematologic neoplasms. Blood 135: 2159–2170. 10.1182/blood.2019003691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrekar D, Xiang Y, Palmer N, Saha A, Meluzzi D, Mali P. 2022a. Comprehensive interrogation of the ADAR2 deaminase domain for engineering enhanced RNA editing activity and specificity. Elife 11: e75555. 10.7554/eLife.75555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrekar D, Yen J, Xiang Y, Saha A, Meluzzi D, Savva Y, Mali P. 2022b. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat Biotechnol 40: 938–945. 10.1038/s41587-021-01171-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. 2002. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 21: 86–89. 10.1038/nbt765 [DOI] [PubMed] [Google Scholar]
- Khalin I, Alyautdin R, Kocherga G, Bakar MA. 2015. Targeted delivery of brain-derived neurotrophic factor for the treatment of blindness and deafness. Int J Nanomedicine 10: 3245–3267. 10.2147/IJN.S77480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnarev M, Pirvulescu IP, Candido KD, Knezevic NN. 2020. Neuropathic pain: preclinical and early clinical progress with voltage-gated sodium channel blockers. Expert Opin Investig Drugs 29: 259–271. 10.1080/13543784.2020.1728254 [DOI] [PubMed] [Google Scholar]
- Lazinski D, Grzadzielska E, Das A. 1989. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 59: 207–218. 10.1016/0092-8674(89)90882-9 [DOI] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349: 1115–1120. 10.1126/science.aac7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JM, Holtzman DM. 2019. Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179: 312–339. 10.1016/j.cell.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, VanDemark AF, Lingam AT, Hill CP, Bass BL. 2005. Structural biology: inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539. 10.1126/science.1113150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9: 1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantegazza M, Cestèle S, Catterall WA. 2021. Sodium channelopathies of skeletal muscle and brain. Physiol Rev 101: 1633–1689. 10.1152/physrev.00025.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews MM, Thomas JM, Zheng Y, Tran K, Phelps KJ, Scott AI, Havel J, Fisher AJ, Beal PA. 2016. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat Struct Mol Biol 23: 426–433. 10.1038/nsmb.3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle T, Stafforst T. 2021. New frontiers for site-directed RNA editing: harnessing endogenous ADARs. Methods Mol Biol 2181: 331–349. 10.1007/978-1-0716-0787-9_19 [DOI] [PubMed] [Google Scholar]
- Monian P, Shivalila C, Lu G, Shimizu M, Boulay D, Bussow K, Byrne M, Bezigian A, Chatterjee A, Chew D, et al. 2022. Endogenous ADAR-mediated RNA editing in non-human primates using stereopure chemically modified oligonucleotides. Nat Biotechnol 40: 1093–1102. 10.1038/s41587-022-01225-1 [DOI] [PubMed] [Google Scholar]
- Monteleone LR, Matthews MM, Palumbo CM, Thomas JM, Zheng Y, Chiang Y, Fisher AJ, Beal PA. 2019. A bump-hole approach for directed RNA editing. Cell Chem Biol 26: 269–277.e5. 10.1016/j.chembiol.2018.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJC. 2013. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci 110: 18285–18290. 10.1073/pnas.1306243110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel-Gonźalez MF, Vallecillo-Viejo IC, Rosenthal JJC. 2016. An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Res 44: e157. 10.1093/nar/gkw738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel-Gonzalez MF, Diaz Quiroz JF, Rosenthal JJCC. 2018. Current strategies for site-directed RNA editing using ADARs. Methods 156: 16–24. 10.1016/j.ymeth.2018.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Pekna M. 2016. Reactive gliosis in the pathogenesis of CNS diseases. Biochim Biophys Acta 1862: 483–491. 10.1016/j.bbadis.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Pokharel S, Beal PA. 2006. High-throughput screening for functional adenosine to inosine RNA editing systems. ACS Chem Biol 1: 761–765. 10.1021/cb6003838 [DOI] [PubMed] [Google Scholar]
- Qu L, Yi Z, Zhu S, Wang C, Cao Z, Zhou Z, Yuan P, Yu Y, Tian F, Liu Z, et al. 2019. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat Biotechnol 37: 1059–1069. 10.1038/s41587-019-0178-z [DOI] [PubMed] [Google Scholar]
- Reautschnig P, Wahn N, Wettengel J, Schulz AE, Latifi N, Vogel P, Kang TW, Pfeiffer LS, Zarges C, Naumann U, et al. 2022a. CLUSTER guide RNAs enable precise and efficient RNA editing with endogenous ADAR enzymes in vivo. Nat Biotechnol 40: 759–768. 10.1038/s41587-021-01105-0 [DOI] [PubMed] [Google Scholar]
- Reautschnig P, Wahn N, Wettengel J, Schulz AE, Latifi N, Vogel P, Kang TW, Pfeiffer LS, Zarges C, Naumann U, et al. 2022b. CLUSTER guide RNAs enable precise and efficient RNA editing with endogenous ADAR enzymes in vivo. Nat Biotechnol 40: 759–768. 10.1038/s41587-021-01105-0 [DOI] [PubMed] [Google Scholar]
- Reenan RA. 2005. Molecular determinants and guided evolution of species-specific RNA editing. Nature 434: 409–413. 10.1038/nature03364 [DOI] [PubMed] [Google Scholar]
- Rush AM, Dib-Hajj SD, Liu S, Cummins TR, Black JA, Waxman SG. 2006. A single sodium channel mutation produces hyper- or hypoexcitability in different types of neurons. Proc Natl Acad Sci 103: 8245–8250. 10.1073/pnas.0602813103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo-Tello P, Ortiz-Matamoros A, Arias C. 2011. GSK3 function in the brain during development, neuronal plasticity, and neurodegeneration. Int J Alzheimers Dis 2011: 189728. 10.4061/2011/189728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, te Morsche RHM, Lynch ME, Drenth JPH. 2008. Compound heterozygosity in sodium channel Nav1.7 in a family with hereditary erythermalgia. Mol Pain 4: 21. 10.1186/1744-8069-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayas CL, Ávila J. 2021. GSK-3 and tau: a key duet in Alzheimer's disease. Cells 10: 721. 10.3390/cells10040721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MF, Wettengel J, Hoffmann PC, Stafforst T. 2014. Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Res 42: e87. 10.1093/nar/gku272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadid M, Badawi M, Abulrob A. 2021. Antisense oligonucleotides: absorption, distribution, metabolism, and excretion. Expert Opin Drug Metab Toxicol 17: 1281–1292. 10.1080/17425255.2021.1992382 [DOI] [PubMed] [Google Scholar]
- Shibata SB, Cortez SR, Beyer LA, Wiler JA, Di Polo A, Pfingst BE, Raphael Y. 2010. Transgenic BDNF induces nerve fiber regrowth into the auditory epithelium in deaf cochleae. Exp Neurol 223: 464. 10.1016/j.expneurol.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Kim SY, Corson GM, Song Z, Nakai H, Adelman JP, Mandel G. 2017. Site-directed RNA repair of endogenous Mecp2 RNA in neurons. Proc Natl Acad Sci 114: E9395–E9402. 10.1073/pnas.1715320114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Kim SY, Fisk JR, Song Z, Nakai H, Jeng S, McWeeney SK, Mandel G. 2020. In vivo repair of a protein underlying a neurological disorder by programmable RNA editing. Cell Rep 32: 107878. 10.1016/j.celrep.2020.107878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Jacobson ME, Yung JF, Fisk JR, Jeng S, McWeeney SK, Chan CN, Yee SP, Mandel G, Parmelee LK. 2022. Targeted RNA editing in brainstem alleviates respiratory dysfunction in a mouse model of Rett syndrome. Proc Natl Acad Sci 119: e2206053119. 10.1073/pnas.2206053119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafforst T, Schneider MF. 2012. An RNA-deaminase conjugate selectively repairs point mutations. Angew Chem Int Ed Engl 51: 11166–11169. 10.1002/anie.201206489 [DOI] [PubMed] [Google Scholar]
- Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, et al. 2017. Dynamic landscape and regulation of RNA editing in mammals. Nature 550: 249–254. 10.1038/nature24041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Kohro Y, Yano T, Tsujikawa T, Kitano J, Tozaki-Saitoh H, Koyanagi S, Ohdo S, Ji RR, Salter MW, et al. 2011. JAK-STAT3 pathway regulates spinal astrocyte proliferation and neuropathic pain maintenance in rats. Brain 134: 1127–1139. 10.1093/brain/awr025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallecillo-Viejo ICIC, Liscovitch-Brauer N, Montiel-Gonzalez MFMF, Eisenberg E, Rosenthal JJCJJC. 2018. Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol 15: 104–114. 10.1080/15476286.2017.1387711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P, Schneider MF, Wettengel J, Stafforst T. 2014. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew Chem Int Ed Engl 53: 6267–6271. 10.1002/anie.201402634 [DOI] [PubMed] [Google Scholar]
- Wang Y, Park SH, Beal PA. 2018. Selective recognition of RNA substrates by ADAR deaminase domains. Biochemistry 57: 1640–1651. 10.1021/acs.biochem.7b01100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettengel J, Reautschnig P, Geisler S, Kahle PJ, Stafforst T. 2017. Harnessing human ADAR2 for RNA repair: recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res 45: 2797–2808. 10.1093/nar/gkw911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf TM, Chase JM, Stinchcomb DT. 1995. Toward the therapeutic editing of mutated RNA sequences. Proc Natl Acad Sci 92: 8298–8302. 10.1073/pnas.92.18.8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z, Qu L, Tang H, Liu Z, Liu Y, Tian F, Wang C, Zhang X, Feng Z, Yu Y, et al. 2022. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat Biotechnol 40: 946–955. 10.1038/s41587-021-01180-3 [DOI] [PubMed] [Google Scholar]