Abstract
Conserved oligomeric Golgi (COG) complex orchestrates intra-Golgi retrograde trafficking and glycosylation of macromolecules, but the detailed mechanism of COG action is unknown. Previous studies employed prolonged protein knockout and knockdown approaches which may potentially generate off-target and indirect mutant phenotypes. To achieve a fast depletion of COG subunits in human cells, the auxin-inducible degradation system was employed. This method of protein regulation allows a very fast and efficient depletion of COG subunits, which provides the ability to accumulate COG complex dependent (CCD) vesicles and investigate initial cellular defects associated with the acute depletion of COG complex subunits. This protocol is applicable to other vesicle tethering complexes and can be utilized to investigate anterograde and retrograde intracellular membrane trafficking pathways.
Keywords: COG complex, Membrane trafficking, Auxin, Golgi, Glycosylation, Vesicle tethering
1. Introduction
The Golgi apparatus (GA) is the central hub of the secretory and endocytic pathways. It is composed of flattened membranes organized in a cis-medial-trans fashion [1-5]. Cargo proteins and lipid passing the GA are undergoing post-translational modifications such as N- and O-linked glycosylation and then sorted and trafficked to specialized compartments within the cell or secreted. Recycling of Golgi enzymes and cargo receptors within the Golgi is essential for proper modification and sorting of those proteins [6-9]. The conserved oligomeric Golgi (COG) complex is the key component of the GA trafficking machinery that orchestrates trafficking events and tethers cargo containing transport vesicles [5, 10-12]. COG is an evolutionally conserved Golgi-associated tethering complex that consists of eight subunits named COG1 through COG8, which can be divided into two structurally and functionally distinct subcomplexes, lobe A (COG1–4) and lobe B (COG5–8) [2, 13-16]. Previous studies revealed important interactions of COG with several different families of trafficking regulatory proteins (SNAREs, SNARE-interacting proteins, Rab GTPases, coiled-coil tethers, and COPI subunits) [5, 16-20]. COG is required for the proper recycling of Golgi localized glycosylation enzymes to maintain the proper glycosylation of secretory proteins; mutations in COG subunits result in a class of human rare and incurable diseases known as congenital disorders of glycosylation (CDG) type II [5, 10, 21, 22]. The cellular phenotypes and clinical presentations of COG-CDGs are heterogeneous which makes it difficult to compare contributions of different subunits with the overall function in human cells. It is of paramount importance to know the molecular mechanism of COG function to facilitate the molecular study of the COG-CDGII as well.
Loss-of-function (LOF) analyses are an important tool to analyze the function of the protein. LOF methods can reduce gene function by targeting the DNA, RNA, or protein. The protein function can be elucidated by analyzing the phenotypes that are caused by those disturbances [23]. The common problem with targeting RNA or DNA is incomplete or chronic protein depletion [24]. The immediate phenotypes could be obscured with gene knockouts and knockdowns as the cells still contain the protein that needs to be degraded and/or deleted [25]. To study the function of COG, we initially applied siRNA mediated knockdown (KD) approach and also created a complete set of HEK293T cell lines lacking individual COG subunits (COG knockout, KO) to characterize them and study the phenotypes [12, 26]. The characterization of COG KO cell lines revealed defects in Golgi morphology, retrograde trafficking, sorting, abnormal secretion, and accumulation of enlarged endolysosomal structures (EELSs) [9, 16, 20]. One of the major limitations of those approaches is chronic depletion of COG which provides enough time to produce secondary defects to cover the primary phenotypes resulting from the acute COG depletion. Moreover, long-time depletion of critical functions provides cells enough time to adapt and skew the phenotypes by activating compensatory pathways [27]. Another challenge with knockout and knockdown is unpredicted consequences that confound interpretation of results such as off-target effects [28]. Therefore, it is very important to look at the effect of acute depletion of COG to elucidate the primary phenotypes by avoiding long-term secondary/indirect defects.
The advancement in chemical genetics has initiated the possibility of regulating classically nondruggable targets by tagging proteins of interest with a degron, where proteins can be stabilized or targeted for degradation through the introduction of small molecules [29-31]. Among those techniques, the auxin-inducible degron (AID) technology has been recently utilized to help acute and rapid depletion of the targeted protein to study its primary function [27, 32, 33].
In this chapter, we have described how to create the hTERT RPE1 COG4-mAID-mCherry cell line to perform fast auxin-inducible rapid knockdown of COG4 (Fig. 1). First, an auxin receptor F-box protein (Transport Inhibitor Responce1, OsTIR1) is exogenously expressed to form a chimeric E3 ubiquitin ligase complex (SCFTIR1) in hTERT RPE1 COG4KO cell line (Fig. 1a). Second, the resulting cell line is transduced with the lentivirus expressing COG4 tagged with mAID and fluorescent marker under the control of the endogenous COG4 promoter for a stable expression of COG4-mAID-mCherry (Fig. 1b). Third, the addition of indole-3-acetic acid (IAA) induces proteasomal degradation of COG4-mAID-mCherry via the SCFTIR1-dependent pathway (Fig. 1c). It is important to test the functionality of expressed hybrid protein. COG4 depletion results in mislocalization and degradation of multiple Golgi proteins [9, 22]. Therefore, to confirm the functionality of COG4-mAID-mCherry, resulting cells should be tested for their ability to stabilize COG-sensitive proteins. Here, we used B4GalT1 as an example of COG-sensitive protein whose stability is restored in hTERT RPE1 COG4 KO cells expressing COG4-mAID-mCherry (Fig. 3a). After selecting the rescued clone, the testing for inducible depletion of COG4-mAID-mCherry is performed using Western blot (WB) (Fig. 2a, b) and immunofluorescence (IF) analysis (Fig. 2c). Similar WB and IF analyses are applied to determine the effect of acute COG4 depletion on the total level and localization of B4GalT1 (Fig. 3a, c). The chapter provides a description of all the materials and procedures involved in the manipulation of the COG4KO cell line expressing OsTIR1 and COG4-mAID-mCherry.
Fig. 1.
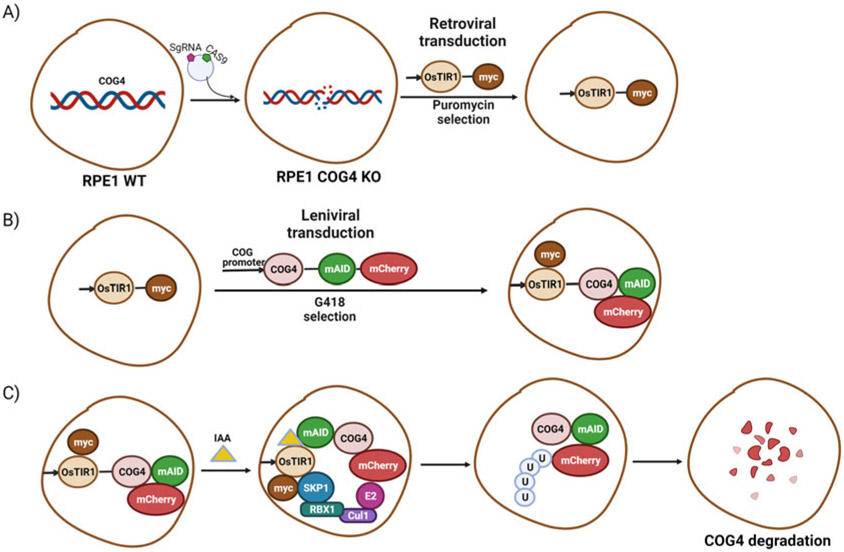
Generation of hTERT RPE1 COG4 KO cell line expressing OsTIR1-9myc and COG4-mAID-mCherry: Cartoon image showing (A) The development of hTERT RPE1 cell line depleted for COG4 subunit by CRISPR-Cas9 approach and retroviral transduction resulting COG4 KO cells to express OsTIR1-9myc. (B) Creation of COG4-mAID stable cell line by rescuing the COG4 KO/OsTIR1-9myc cells by the lentiviral transduction to express COG4-mAID-mCherry under the control of COG4 promoter. (C) The experimental approach for COG4-mAID-mCherry rapid degradation by IAA (auxin) treatment
Fig. 3.
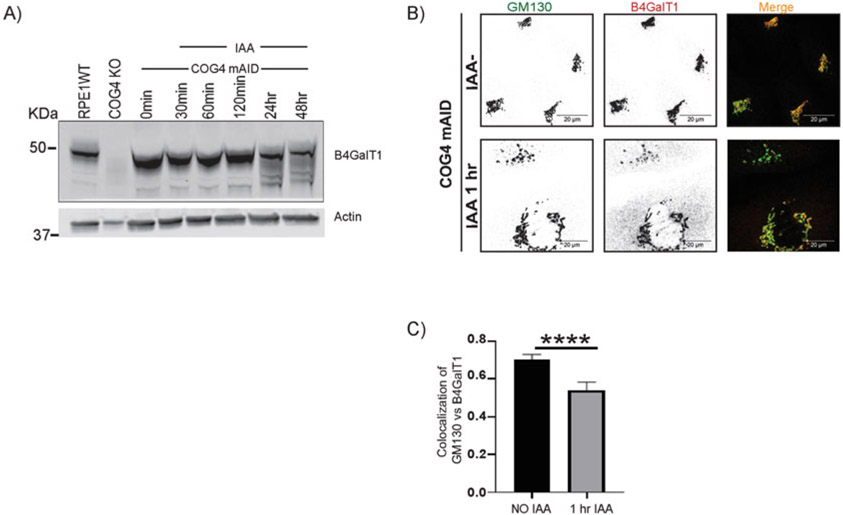
Acute depletion of COG4-mAID-mCherry displaces the B4GalT1 enzyme from the Golgi into CCD vesicles. (A) The hTERT RPE1 COG4 KO/OsTir1-9myc/hCOG4-mAid-mCherry cells were treated with 0.5 mM IAA for different times as indicated. Cells were collected and lysed, and the expression of B4GalT1 and actin was analyzed by WB. No depletion of B4GalT1 was observed within 2 h of treatment, but the B4GalT1 band was more diffuse at 24 h IAA treatment, indicating protein degradation. (B) IF analysis of untreated and IAA treated cells. The COG4-mAID clone was treated 0.5 mM IAA for 1 h. The cells were fixed and stained for GM130 (green) and B4GalT1 (as red). Note that the acute depletion of COG4-mAID-mCherry displaced the enzyme from the Golgi into vesicle-like structures. Scale bars, 20 μm. (C) Quantification of colocalization between GM130 and B4GalT1 from n = 2 independent experiment with >30 cells analyzed. ****, P < 0.0001, significant. Error bar represents mean ± SD. The green and red channels are presented in inverted black and white mode, whereas the merged view is shown in RGB mode. Scale bars, 20 μm
Fig. 2.
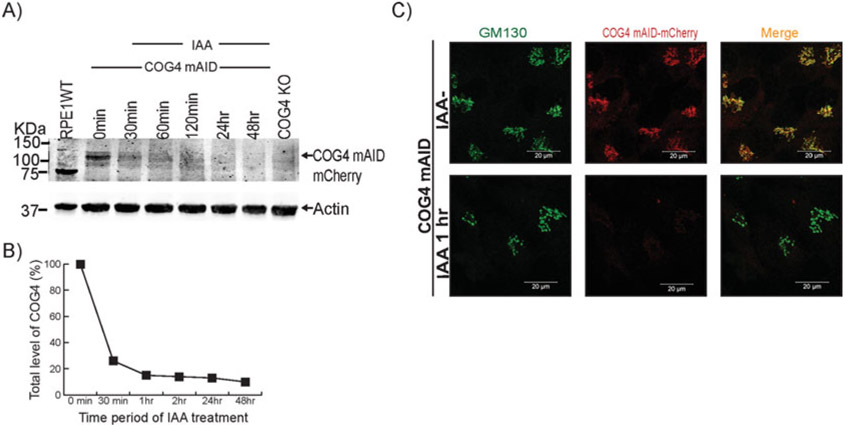
Auxin (IAA) treatment induces rapid depletion of hCOG4-mAID-mCherry. (A) The hTERT RPE1 COG4 KO/OsTir1-9myc/hCOG4-mAid-mCherry cells were treated with 0.5 mM IAA for different times as indicated. Cells were collected and lysed, and the expression of wild-type and hybrid COG4 protein was analyzed by WB using an anti-COG4 antibody. (B) Quantification of the COG4 protein level at different time points after IAA treatment. Three independent experiments have been performed. (C) IF analysis of COG4-mAID-mCherry depletion after IAA treatment. The COG4 mAID clone was treated with IAA for 1 h. Untreated cells were used as a control. The cells were fixed and stained for GM130 (green). Scale bars, 20 μm
2. Materials
2.1. Cell Culture
hTERT RPE1 cells (retinal pigment epithelial) (ATCC CRL-4000).
hTERT RPE1 COG4 KO cells [22].
HEK293FT cells (Thermo Fisher Scientific R70007).
DMEM/F12: Dulbecco’s Modified Eagle’s Medium (DMEM) containing nutrient mixture F-12 (Corning 10–092-CV).
Serum-reduced Opti-MEM.
Complete medium: DMEM/F12 with 10% fetal bovine serum.
Trypsin, 0.25%.
GlutaMAX 100×.
Chloroquine 25 mM in water.
Poly-L-lysine 0.1 mg/mL in water.
Dulbecco’s phosphate-buffered saline (DPBS).
Tissue culture-treated dishes: (a) 35 mm and 100 mm tissue culture dish, (b) 6-well, 24-well, 12-well, and 96-well sterile tissue culture plate.
1.5 mL microcentrifuge tube
12 mm round glass coverslips (#1.5, 0.17 mm thickness)
Beckman Coulter Vi-Cell XR cell counter.
Pipette sets (P10, P20, P200, P1000) with pipette tips.
Disposable sterile 10 mL syringes.
Centrifuge with the rotor for 15 mL conical tubes.
Tissue culture CO2 incubator.
Inverted microscope with 5× objective.
2.2. Expression Plasmid
pBabe OsTIR1–9myc (PuroR) (Addgene #47328) [34].
pMK292 mAID-mCherry2-NeoR (Addgene #72830) [35].
pUMVC (Addgene #8449) (packaging plasmid for producing MNuLV retroviral particles) [36].
pMD2.G (VSV-G envelope expressing plasmid) (a gift from Didier Trono (Addgene # 12259; http://n2t.net/addgene:12259; RRID: Addgene_12259)).
pRSV-Rev (Addgene #12253) [37].
pMDLg/pRRE (Addgene #12251) [37].
COG4-2xGFP in pEntral A[22].
pLenti COG4 Neo DEST plasmid with COG4 promoter [22].
2.3. DNA Preparation and Recombination
NEB 10-beta competent E. coli.
SOC (super optimal broth) media.
LB (Luria–Bertrani) broth.
LB agar plates with ampicillin (100 μg/mL).
LB agar plates with kanamycin (30 μg/mL).
Cell spreaders.
Disposable sterile plastic loops.
Kanamycin stock solution (1000×): 150 mg kanamycin sulfate in 5 mL water.
Ampicillin stock solution (1000×): 1 g ampicillin in 5 mL of water.
Buffer 1 (10 mM Bis-Tris-Propane-HCl, 10 mM MgCl2, 1 mM DTT, pH 7) (NEB).
Kpnl (20 units/μL) (NEB).
Xhol (20 units/μL) (NEB).
T4 DNAligase (NEB).
10x ligase buffer (NEB)
QIAprep Spin Miniprep DNA extraction kit (QIAGEN).
Gateway LRClonase II Enzyme Mix (Thermo Fisher).
Zymoclean gel DNA recovery kit for gel purification.
TAE running buffer: 40 mM Tris, 20 mM acetate, 1 mM EDTA, pH 8.6.
1% agarose gel: 1.5 g of agarose in 150 mL of TAE buffer (40 mM Tris, 20 mM acetate, 1 mM EDTA, pH 8.6) containing 0.1 μg/mL ethidium bromide.
Gel-loading dye loading buffer: 60 mM Tris-HCl, pH 8.6, 100 mM NaCl, 2 mM ß-mercaptoethanol.
1 kB DNA ladder
Horizontal gel electrophoresis system.
NanoDropTM Lite Spectrophotometer.
Table-top minicentrifuge.
Chemidoc Imaging System.
UV lamp.
Water bath.
Incubating orbital shaker.
2.4. Polymerase Chain Reaction (PCR)
- Primers used to amplify pMK292 mAID-mCherry2-NeoR:
- Forward primer: AATTGGTACCGGATCCGGTGCAGGCGCCAAG.
- Reverse primer: GCGCCTCGAGTTACTTGTACAGCTCGTCGTCCAT.
DMSO.
Nuclease-free sterile water.
dNTPs.
10× PrimeSTAR HS DNA polymerase reaction buffer (Takara Bio).
PrimeSTAR HS DNA polymerase (Takara Bio).
0.2 mL PCR tubes.
QIAquick PCR purification kit (QIAGEN)
0.2 mL PCR tubes.
2.5. Transfection and Transduction Reagents
Lipofectamine 3000 transfection reagent (Thermo Fisher).
- Transfection medium: – Serum-reduced Opti-MEM.
- 25 μM chloroquine and
- GlutaMAX (100×).
Puromycin stock solution: sterile-filtered 0.1% puromycin dihydrochloride in water.
G418 (Geneticin) stock solution: sterile-filtered 5% G418 sulfate in water.
10 mL sterile syringes
0.45 μm polypropylene (PES) syringe filter.
Polybrene transfection reagent (optional).
0.22 μm PES filter
Sodium butyrate: 50× stock solution. 50 mM sodium butyrate in PBS.
2.6. Validation (Lysate Preparation, Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis, and Western Blot Components)
SDS: 2% sodium dodecyl sulfate in water.
Heat blocks set to 70 °C.
1.5 mL microcentrifuge tubes.
Sonicator homogenizer.
SDS-PAGE gel (see Note 1).
Vertical mini-gel electrophoresis unit.
SDS-PAGE running buffer: 25 mM Tris, 0.192 M glycine, 0.1% SDS in water.
0.2 μm Whatman Protran nitrocellulose blotting membranes.
Blotting or filter paper.
Pierce G2 Fast Blotter.
1-Step Transfer Buffer (Thermo Scientific).
WB blocking buffer: 5% non-fat dry milk in PBS (see Note 2).
Antibodies: mentioned (see Subheading 2.8).
2× Laemmli sample buffer: 4% SDS, 5% β-mercaptoethanol, 20% glycerol, 0.004% bromophenol blue, 0.125 M Tris- HCl, pH 6.8.
Ponceau S stain: 0.1% Ponceau S in 5% acetic acid.
PBST: 0.05% Tween 20 in PBS.
Membrane incubation box.
Plastic forceps.
2.7. Validation (Immunofluorescence Experiment Reagents and Confocal Microscopy)
Fixative: 4% paraformaldehyde (PFA) (freshly made from 16% stock solution in PBS0.1% Triton X-100 in PBS.
50 mM ammonium chloride in PBS
IF blocking buffer: 1% bovine serum albumin (BSA) and 0.1% saponin in PBS.
IF antibody buffer: 1% cold fish gelatin, 0.1% saponin in PBS.
DAPI: 1 mg/mL in water.
Mounting reagent: Prolong® Gold Antifade reagent (Life Technologies).
Glass slides: frosted microscope slides (precleaned).
Parafilm.
Metal fine point high precision forceps.
Kimwipe.
Clear nail polish.
Fluorescent microscope (see Note 3).
2.8. Antibodies
Primary
Rabbit polyclonal anti-hCOG4 (C-terminal) (Sigma SAB4200469) (1:1000 for WB).
Mouse monoclonal anti-beta-Actin (Sigma, A5541) (1: 2000 for WB).
Rabbit polyclonal anti-GM130 (CalBiochem, CB1008) (1: 2000 for IF).
Mouse monoclonal anti-myc-tag (Cell Signaling 9B11) (1: 1000 for WB and 1:2000 for IF).
Goat polyclonal anti-B4GalT1 (R & D Systems AF-3609) (1: 500 both for WB and IF).
Secondary
Donkey anti-rabbit Alexa Fluor 647 (Jackson Immuno Research/705605-152) (1:4000 for WB and 1:500 for IF).
Donkey anti-mouse Alexa Fluor 647 (Jackson Immuno Research/705605-151) (1:4000 for WB and 1:500 for IF).
Donkey anti-rabbit Alexa Fluor 488 (Jackson Immuno Research/705605-151) (1:4000 for WB and 1:500 for IF).
Goat anti-mouse IRDye 800 (LiCOR/5-32210) (1:2000 for WB).
Donkey anti-goat Alexa Fluor 647 (Jackson Immuno Research/705605-147) (1:4000 for WB and 1:500 for IF).
2.9. Materials for Degradation System
3. Methods
Three consecutive modifications are required to generate cells in which COG4 protein level is regulated by the AID system: (1) KO of wild-type COG4, (2) stable expression of the OsTIR1 F-box protein, and (3) stable wild-type level expression of the target protein with AID tag. The creation of the COG4 KO cells using CRISPR/Cas9 approach was described in detail previously
[12, 22]. An outline of the procedure used to generate hTERT RPE1 COG4 KO cells stably expressing OsTir1-9myc/ COG4-mAID-mCherry is shown in Fig. 1.
3.1. Generation of COG4 KO Cell Line Expressing OsTIR1
OsTIR1 recognizes a degron sequence (the AID tag) in the presence of phytohormone auxin for degradation of target protein via the ubiquitin–proteasome pathway [38]. Therefore, the expression of OsTIR1 is an essential step (see Note 5).
3.1.1. Plasmid Transformation and Preparation
Add 1 μL of pBabe OsTIR1-9myc (PuroR) plasmid to the tube containing competent E.coli cells. Incubate for 30 min on ice (see Note 6).
Place tube with cells in a 42 °C water bath for 40 s.
Put tube back on ice for 1 min for recovery.
Add 250 μL SOC media to cells.
Incubate in 37 °C water bath for 1 h.
Spin down for 1 min (5000 × g).
Remove 200 μL and resuspend pellet in remaining liquid.
Use cell spreader to spread 100 μL of cells on a LB agar plate with ampicillin.
Place plate in incubator at 37 °C overnight.
Next day, pick up a single colony with sterile plastic loop and place it in 5 mL of LB broth (add ampicillin to concentration).
Incubate the bacteria in a shaker set at 200 rpm and 37 °C overnight.
Sediment cells using table-top centrifuge and isolate plasmid DNA using the QIAprep Spin Miniprep Kit and following the manufacturer’s instructions.
Measure the DNA concentration using NanoDropTM Lite Spectrophotometer (see Note 7).
Load the 0.5 μg of plasmid DNA along with 1kB DNA ladder on 1% agarose gel and run-in horizontal gel electrophoresis unit containing TAE buffer.
Check the DNA band in Chemidoc imaging system (see Note 8).
3.1.2. Production of OsTIR1-9myc Retrovirus
Seed 2.5 × 106 HEK293FT cells in 10-cm dish with 10 mL of complete medium (see Note 9).
Incubate cells at 37 °C and 5% CO2 overnight.
Next day, change the media to 10 mL serum-reduced Opti-MEM with 25 μM chloroquine and 100 μL GlutaMAX.
-
Prepare the transfection cocktail following the lipofectamin-3000 protocol.
First cocktail- 750 μL of Opti-MEM
- 30 μL of p3000
- 5.2 μg of pUMVC
- 2.6 μg of pMD2.G
- 7.2 μg pBabe OsTIR1-9myc
Second cocktail- 750 μL of Opti-MEM
- 45 μL of lipofectamin-3000
Mix the two cocktails gently and incubate 20 min at room temperature.
Add mixture dropwise to existing media on HEK293FT cells in 10-cm dish. Incubate cells at 37 °C and 5% CO2.
Next day, replace cell media with fresh Opti-MEM with 1xGlutaMAX.
Incubate cells at 37 °C and 5% CO2 for 48 h.
Collect virus-containing media into 15 mL conical tubes and remove remaining cell debris by centrifugation (1000 × g, 5 min).
Transfer the supernatant into 10 mL syringe and filter through a 0.45 μm filter (fixed to syringe) to remove cells (see Note 10).
Save the viral supernatant as 1 mL aliquots in −80 °C (see Note 11).
3.1.3. Retroviral Transduction of OsTIR1-9myc into RPE1 COG4 KO Cells
Plate 6 × 105 hTERT RPE1 COG4 KO cells in two wells of a 6-well plate in complete medium. One of these wells will serve as a control for antibiotic selection.
Incubate cells at 37 °C and 5% CO2 overnight.
The next day, remove media and add back 1.5 mL of complete media. Add 500 μL of viral supernatant to one of the wells and 500 μL of complete media to another well. Place back in the incubator and allow 2 days for expression of OsTIR1-9myc (see Notes 12, 13, 14, and 15).
3.1.4. Selection and Isolation of RPE1 COG4 KO Single Clones Expressing OsTIR1
After 2 days of transduction add puromycin (10 μg/mL final concentration, selection dose) to transduced and control cells and maintain selection for 48 h until all the control cells have died (see Note 16).
After 48 h, replace the media with complete media containing 5 μg/mL of puromycin (maintenance dose). Incubate cells at 37 °C and 5% CO2 for 48 h or longer if required.
After puromycin selection, isolate the single-cell clones by serial dilution.
Trypsinize cells for 3 min and resuspend in complete medium with puromycin.
Count the cells to prepare 10,000 cells/1 mL suspension, dilute it in complete medium with puromycin first 1:100 and then 1:10 to final concentration 10 cells/mL (see Note 17).
Add 100 μL of the cell suspension to each well of the 96-well plate and place in incubator.
Next day, use an inverted microscope with 5× objective to identify wells with single colonies (see Note 18).
After 1 week of growth add 100 μL of fresh complete medium with puromycin.
After 2 weeks in the incubator collect cell colonies by trypsin digestion and expand each colony into 12-well plate (see Notes 19 and 20).
After 2 days, expand cells to 6-well plate and to 12-well plate with 12 mm round glass coverslips for WB and IF analysis, respectively (see Note 21).
3.1.5. Screening for OsTIR1-9myc Expressing Clones by WB Analysis
After the clonal expansion of transduced hTERT-RPE1 COG4 KO clones, those should be screened to identify the clone with OsTIR1–9myc expression. The procedure is described below.
Identify the fast-growing monoclonal cell lines from the 96-well plate and expand them into 12-well plate using complete media containing 5 μg/mL of puromycin. Allow cells to grow to confluence (see Note 22).
Trypsinize cells, resuspend in 2 mL of complete media. Leave 5 mL in the original well and transfer 1 mL of cells on the well in 6-well plate and another 0.5 mL on the well in 12-well plate with 12 mm round glass coverslip (see Subheading 3.1.6). Allow the cells grow to reach 90–100% confluence.
Transfer SDS solution to the 1.5 mL tube and heat to 70 ° C using heat block for 5 min.
When the cells become confluent in 6-well plate, wash them with PBS for 3 times and collect cells in 0.1 mL of 2% hot SDS. Transfer cell lysates to 1.5 mL microcentrifuge tube.
Heat the sample at 70 °C for 10 min (see Note 23).
Keep the sample tube in ice. Sonicate sample with a micro-tip probe at 24 KHz for two times 5 s each (see Note 24).
Add 0.1 mL of 2× SDS sample buffer; mix and heat the sample at 70 °C for another 3 min.
Load 15 μL lysates on an SDS-PAGE gel, and run at 180 V until the dye front reaches the bottom of the gel.
Soak 2 thick sheets of filter paper and the nitrocellulose membrane in 1-Step Transfer Buffer.
Remove the gel from the chamber and pry open glass. Place gel in double-distilled water.
Assemble the transfer stack on the anode side of the Pierce G2 Fast Blotter cassette. Gently remove air bubbles using a roller on every layer of the transfer sandwich: filter paper, membrane, gel, filter paper.
Place cathode portion on top and press to secure the cassette. Place cassette into Fast Blotter machine.
Select Mixed Range MW protocol and press start to run the transfer.
Stain the membrane with Ponceau S stain for 5 min to ensure the transfer went well and to assess overall protein levels in each lysate.
Rinse the membrane in PBS (3 times for 4 min each) at room temperature while rocking on a table rocker.
Block the membrane in WB blocking buffer (see Note 25) for 20 min while rocking at room temperature and incubate with primary antibody against myc-tag diluted 1:2000 in WB blocking buffer for 1 h at RT or overnight at 4 °C with gentle rocking.
Wash the membrane four times with PBST (5 min for each wash) and incubate it with secondary Goat anti-Mouse IRDye 800 antibody diluted 1: 2000 in WB blocking buffer for 1 h.
Wash the membrane three times with PBST and once with PBS (5 min for each wash).
Image the blot using the Odyssey imaging system. Process the image using the LI-COR Image Studio software.
3.1.6. Screening for Single Clones That Uniformly Express OsTIR1 by IF Analysis
Perform IF analysis and confocal microscopy for screening myc-positive clones selected in Subheading 3.1.5 for the uniform expression of OsTIR1-myc. The procedure is given below (see Note 26).
Grow cells on 12 mm round coverslips to 80–90% confluence in a tissue culture dish.
Take coverslips out of the dish with forceps and place them on parafilm in a drop of PBS.
Wash gently with PBS at room temperature (2 times).
Fix cells with fixative for 15 min (see Note 27).
Permeabilize with 0.1% Triton X-100 in PBS for 1 min.
Incubate the coverslip with 50 mM NH4Cl in PBS for 5 min.
Rinse in PBS 2 times.
Incubate in IF blocking buffer 2 times 10 min each.
Incubate with primary anti-myc-tag antibody diluted 1:2000 in IF antibody buffer for 45 min.
Wash with PBS four times, 2 min each.
Incubate with secondary donkey anti-rabbit Alexa Fluor 647 antibody diluted 1:500 in IF antibody buffer for 30 min.
Wash with PBS four times 2 min each.
Incubate with DAPI diluted 1:2000 in PBS for 1 min.
Rinse in PBS four times.
Using forceps, pick up the coverslip and dunk it 10 times with PBS in a beaker (see Note 28).
Dunk coverslips 10 times with double-distilled water in a beaker.
Dry off excess liquid with a kimwipe by gently touching the edge of the coverslip to it.
Mount coverslips in mounting media (put a small ~10 μL drop of Prolong Gold media on slide and mount coverslip with cells upside-down).
Cure overnight in the dark.
Next day, seal the edges with clear nail polish (see Note 29).
Image the cell to observe myc-signal using LSM880 Zeiss Laser inverted microscope (see Note 30).
Select colonies uniformly expressing OsTIRI-myc, expand, and freeze them.
3.2. Generation of COG4 KO-OsTIR1 Expressing AID-Tagged COG4 Stable Cell Line
The target protein should be tagged with AID protein to promote the degradation of AID-tagged proteins through the binding of OsTIR1 (see Notes 31 and 32).
3.2.1. PCR of the mAID-mCherry
- Place tubes in a thermocycler and use the following program
- 95 °C, 1.00 min
- 95 °C, 10 s
- 60 °C, 10 s
- 72 °C, 40 s
- Repeat ii-iv, 35× (see Note 35)
- 72 °C, 2.00 min
- 4 °C, ∞
Check the PCR yield and expected size using 1% agarose gel. Load 8 μL (6 μL of each reaction product plus 2 μL of gel-loading dye) on the agarose gel. Load 1 kB DNA ladder on a separate lane. Electrophorese the samples for 30 min and visualize the PCR products on a UV transilluminator and document the results.
Perform purification of mAID-mCherry PCR product using QIAquick PCR purification kit (QIAGEN): Follow standard protocol for the kit.
Measure DNA concentration using nanodrop spectrophotometer (see Note 36).
3.2.2. Cloning COG4-mAID-mCherry in pEntra1A
Previously, we have created COG4-2xGFP in pEntra1A (for further details check [22]) To create the COG4-mAID-mCherry in pEntra1A, perform subcloning of a PCR product (mAID-mCherry) into COG4-2xGFP in pEntra1A.
- Digestion of COG4-2xGFP in pEntra1A and PCR product (pMK292 mAID-mCherry2-NeoR DNA)
- Prepare restriction digestion of each DNA with KpnI and XhoI
- Nuclease-free water (up to 20 μL)
- 10× buffer 2 μL
- DNA 1 μg
- KpnI 1 μL (20 unit/μL)
- XhoI 1 μL (20 unit/μL)
- Incubate the reaction mix at 37 °C overnight.
- Next day, add 4 μL loading dye to 20 μL of each digested reaction and load on 1% agarose gel.
- Electrophorese the samples for 30 min.
- Visualize DNA using a hand-held UV lamp and cut digested PCR product (959 bp) and plasmid (4670 bp) using sterile surgical blade.
- Gel purification of DNA fragments
- Transfer agarose pieces to 1.5 mL Eppendorf tubes and purify DNA using the Zymoclean gel DNA recovery kit and following the manufacturer’s instructions. Elute DNA from the column in 16 μL of nuclease-free water.
- Analyze 3 μL of gel-purified products by agarose gel electrophoresis to test the yield and the size of purified DNA fragments.
- DNA ligation. Ligate the vector COG4-pEntra1A and insert mAID-mCherry minimum at 1:5 molar ratio (see Note 37). Use two ligation mixes for each reaction. In one reaction, omit the mAID-mCherry DNA; this reaction serves as a control. Combine the following in a 0.2 mL tube.
- H2O up to total of 20 μL
- Ligase buffer 2 μL (10× buffer) (see Note 38)
- KpnI/XhoI digested COG4-pEntra1A
- KpnI/XhoI digested mAID-mCherry or water
- T4 DNA Ligase 1 μL
- Incubate reactions at room temperature for 2 h, or at 16 °C overnight.
- Use 2 μL of ligation reactions for bacterial transformation (Discussed above in Subheading 3.1.1).
- Next day, pick up six single colonies with sterile plastic loop (see Note 39) and place them in 5 mL of LB broth. Add kanamycin to the LB broth (add 5 μL of stock solution to 5 mL LB to final concentration 30 μg/mL).
- Incubate the bacteria in a shaker set at 200 rpm and 37 ° C overnight.
- Extract plasmid DNA using the QIAprep Spin Miniprep Kit and following the manufacturer’s instructions.
3.2.3. Creation of Expression Clone COG4-mAID-mCherry in pLenti
Recombine the COG4-mAID-mCherry in pEntra1A with pLenti COG4 promoter Neo DESTconstruct [22] using Gateway LR Clonase II Enzyme Mix according to the manufacturer’s instructions.
Perform the bacterial transformation into Stbl3 competent cells and DNA preparation as discussed in Subheading 3.1.1 but using LB with ampicillin (add 5 μL of stock solution to 5 mL LB to final concentration 100 μg/mL).
Isolate plasmid DNA with QIAprep Spin Miniprep DNA extraction Kit.
Measure DNA concentration using nanodrop spectrophotometer.
Validate by transfecting the cells following WB analysis (see Note 40).
3.2.4. Production of COG4-mAID-mCherry Lentivirus
Seed 1.0 × 106 HEK293FT cells in 10-cm dish with 10 mL of complete media. Grow cells for 2 days to become 90% confluent.
In the day of transfection, change the media with 10 mL serum-reduced Opti-MEM with 25 μM chloroquine and 100 μL GlutaMAX.
-
Prepare the transfection cocktail following the lipofectamin-3000 protocol.
First cocktail- 750 μL of Opti-MEM
- 30 μL of p3000
- 3.75 μg of pRSV-Rev 3.75 μg of pMDLg/pRRE
- 3.75 μg of pMD2.G
- 3.75 μg of COG4-mAID-mCherry pLenti.
Second cocktail- 750 μL of Opti-MEM
- 30 μL of lipofectamin-3000
Mix the two cocktails gently and incubate 20 min at room temperature.
Add the transfection mix dropwise to cells.
After 5 h add 1:10 dilutions of 50 mM of Na-butyrate.
Next day, replace remaining media with fresh media containing Opti-MEM and 1 x GlutaMAX.
Incubate for 48 h in a CO2 incubator at 37 °C.
Collect virus-containing media by pipette into the syringe and filter through a 0.45 μm PES filter (attached with the syringe) to remove cells.
Save the viral supernatant as 1 mL aliquots and store in −80 °C.
3.2.5. Lentiviral Transduction of COG4-mAID-mCherry into RPE1 COG4 KO TIR1-9myc Cells
Plate 6 × 105 hTERT RPE1 COG4 KO OsTir1-9myc cells in two wells of a 6-well plate in complete medium. One of these wells will serve as a control for antibiotic selection.
Incubate cells at 37 °C and 5% CO2 overnight.
The next day, remove media and add back 1.5 mL of complete media and add 500 μL of viral supernatant to one of the wells. Add 500 μL of complete media to another well. Place back in the incubator and allow 2 days for expression of COG4-mAID-mCherry.
3.2.6. Selection and Isolation of RPE1 COG4 KO Tir1-9myc Single Clone Expressing COG4-mAID-mCherry
The strategy is similar as discussed in Subheading 3.1.4. Here, antibiotic G418 is used for selection.
After two days of transduction, add G418 (600 μg/mL final concentration, selection dose) to transduced and control cells and maintain selection for 48 h until all the control cells have died (see Note 41).
Replace the media with complete media containing 200 μg/mL of G418 (maintenance dose). Incubate cells at 37 °C and 5% CO2 for 48 h or longer if required.
- After G418 selection, isolate the single-cell clones by serial dilution.
- Trypsinize cells for 3 min and resuspend in complete medium with G418.
- Count the cells to prepare 10,000 cells/mL suspension, and dilute it in complete medium with G418 first 1:100 and then 1:10 to final concentration 10 cells/mL.
- Add 100 μL of the cell suspension to each well of the 96-well plate and place in incubator.
- Next day use an inverted microscope with 5× objective to identify wells with single colonies.
- After one week, add 100 mL of fresh complete medium with G418.
- After another week, collect cells by trypsin digestion and expand each colony into 12-wells plate.
After 2 days, expand each colony to 6-well plates and to 12-well plate with 12 mm round glass coverslips for WB and IF analysis, respectively.
3.2.7. Screening for COG4-mAID-mCherry Expressing Clones by WB Analysis
Similar procedure as described in Subheading 3.1.5. Use primary rabbit anti-hCOG4 and secondary donkey anti-rabbit Alexa Fluor 647 to select clones expressing COG4-mAID-mCherry. Use wild-type hTERT RPE1 cells as a control. Clones showing wild-type expression of COG4-mAID-mCherry are chosen for IF analysis.
3.2.8. Screening for COG4-mAID-mCherry Expressing Single Clones by IF Analysis
Grow cells on 12 mm round coverslips to 80–90% confluence in a tissue culture dish.
Take coverslips out of the dish with forceps and place them on parafilm in a drop of PBS.
Wash gently with PBS at room temperature (2 times).
Fix cells with fixative for 15 min.
Permeabilize with 0.1% Triton X-100 in PBS for 1 min.
Incubate the coverslip with 50 mM NH4Cl in PBS for 5 min.
Rinse in PBS 2 times.
Incubate in IF blocking buffer 2 times 10 min each.
Incubate with primary rabbit GM130 antibody diluted 1:2000 in IF antibody buffer for 45 min.
Wash with PBS four times 2 min each.
Incubate with secondary antibody donkey anti-rabbit Alexa Fluor 647 diluted 1:500 in IF antibody buffer for 30 min.
Wash with PBS four times 2 min each.
Incubate with DAPI diluted 1:2000 in PBS for 1 min.
Rinse in PBS four times.
Using forceps, pick up the coverslip and dunk it 10 times with PBS in a beaker.
Dunk coverslips 10 times with water in a beaker.
Dry off excess liquid with a kimwipe by gently touching the edge of the coverslip to it.
Mount coverslips in mounting media (put small drop of Prolong Gold media on slide and mount coverslip with cells upside-down).
Cure overnight in dark.
Next day seal the edges with clear nail polish.
Image the cell to observe mCherry and GM130 signals using a 63 x oil 1.4 numerical aperture (NA) objective of LSM880 Zeiss Laser inverted microscope with Airyscan. Colocalization of mCherry signal with GM130 will confirm correct expression of COG4 (see Note 42).
Select colonies uniformly expressing COG4-mAID-mCherry, and expand and freeze them.
3.2.9. Screening for the Clones Rescuing the COG4 KO Phenotypes
To select the desired clone, perform WB and IF analysis using the antibody to B4GalT1 or other COG-sensitive proteins (see Note 43). Use the COG4 KO clone as a negative control and hTERT RPE1 cells as a positive control. In the case of the COG4 KO cell line, the WB should show the depletion of the B4GalT1. This phenotype should be rescued by COG4KO TIRI-9myc expressing COG4-mAID-mCherry (Fig. 3a).
3.3. Testing Inducible Depletion of COG4-mAID Protein by Auxin (IAA)
Auxin-inducible depletion of COG4-mAID-mCherry is assayed by WB and IF assays. Recommended procedures are described below.
3.3.1. WB Assay
Plate 6 × 105 COG4-mAID (hTERT RPE1 COG4 KO expressing TIR1–9myc and COG4-mAID-mCherry) cells into each well of 6-well plate.
Next day, prepare an auxin (IAA) from the fresh frozen aliquot and perform time course treatment with 500 μM IAA for 30 min, 1 h, 2 h, 24 h, and 48 h in 37 °C cell culture incubator. Leave one well without auxin as the untreated control.
At the end of the time point treatment, remove the media containing auxin, wash cells with 1× PBS, and add 100 μL of hot 2% SDS to each well to harvest lysates for WB analysis.
Heat samples at 70 °C for 10 min. Sonicate (as discussed in Subheading 3.1.5) and add 0.1 mL of 2× Laemmli sample buffer.
Perform SDS-PAGE and WB as discussed in Subheading 3.1.5. Use the primary antibody against COG4 diluted 1:1000 in WB antibody dilution buffer and quantify the signal across different time points. Reduction in the intensity of COG4-mAID-mCherry band indicates degradation of COG4 hybrid protein (Fig. 2a, b). Incubate the blot with a primary antibody against the trans-Golgi enzyme B4GalT1 diluted 1:500 in WB blocking buffer. This will reveal the effect of COG4 depletion on the total level of B4GalT1 protein (Fig. 3a, b) (see Notes 44 and 45).
As a loading control, incubate the blot with a primary antibody against actin diluted at 1:2000 in antibody buffer for 1 h. After the incubation with the primary antibodies, incubate the blots with corresponding secondary antibodies (described in the Subheading 2.8).
3.3.2. IF Assay
Grow validated COG4-mAID clone on 12 mm round coverslips to 80–90% confluence in a 6-well plate.
Next day, prepare an auxin (IAA) in fresh complete media and perform time point treatment: Add in 500 μM IAA to treat cells for a total of 30 min, 1 h, and 2 h in 37 °C cell culture incubator. Use one coverslip without adding auxin as the untreated control.
At the end of the time course treatment, rinse cells with PBS, and fix in Fixative for 20 min at room temperature.
Proceed with standard immunostaining procedures as discussed in Subheading 3.1.6. Check the mCherry signal to confirm the auxin-mediated COG4-mAID-mCherry depletion (Fig. 2c). Co-stain cells on coverslips for COG-insensitive Golgi marker GM130, and COG-sensitive B4GalT1 and quantify their colocalization across conditions to check the change in colocalization of those markers upon auxin-mediated rapid COG4 depletion (antibodies are mentioned in the Subheading 2.8) (Fig. 3c) (see Notes 44 and 45).
4. Notes
We used pre-casted 4–15% SDS-PAGE mini-gels (Bio-Rad) or 8–16% SDS-PAGE mini-gels (GenScript) in our experiments, but any 10% linear or gradient homemade or precast gel can be used.
Blocking buffer from LiCor or 5% BSA in PBST can be used at this stage to improve the detection.
COG4-mAID-mCherry fluorescent signal is weak; therefore, for these studies LSM880 Zeiss Laser inverted microscope with Airyscan was used, but any high-end fluorescent microscope with 63× or 100× high NA oil objective is suitable.
Filter–sterilize the solution and freeze aliquots in opaque tubes and store at −20 °C. Keep protected from light.
To achieve the stable expression of OsTIR1-9myc, transduce RPE1 COG4 KO cells with retroviral OsTIR-9myc particle followed by puromycin selection and isolation of individual cellular clones. Then, validate several clones by WB and select a fast-growing and high-expressing OsTIR1 clone.
Retroviral and lentiviral vectors may undergo recombination after transformation. Stbl3 competent cells are recommended for amplification of pBabe and pLenti plasmids.
As DNA adsorbs at 260 nm, any compatible spectrophotometer can be used to measure DNA concentration.
Any system to handle DNA gels can be used. The majority of plasmid DNA should run on DNA gel as a supercoiled DNA.
To improve the efficiency of the attachment of HEK293FT cells to plastic dishes, add 0.01% poly-L-lysine (0.1 mg/mL in water) to the dish and coat it for 2 h. Then, wash with PBS and seed the cells.
Virus can be collected after 48, 72, or 96 h post-transfection.
The virus-contaminated waste must be carefully collected in 100% bleach and sealed before discarding.
The exact amount of viral supernatant used for cell transduction should be tested experimentally to ensure that the final titer of the active virus will be less than 1 MOI. As an example, the ELISA technique could be applied to measure the concentration of the virus [39]. More than one transduction may be performed to determine optimal transduction efficiency.
Polybrene (8 μg/mL final concentration) could be used to increase the transduction efficiency. The concentration of poly-brene can be increased or decreased according to the cell type.
Check the cell confluence the next day. It should be near 90–100%.
The duration of transduction can range from 4 to 48 h. If the target cells cannot tolerate the virus, shorter incubation times should be used.
hTERT RPE1 cells are partially resistant to puromycin, and therefore, the exact puromycin concentration should be selected based on cell sensitivity.
The serial cell dilution can be done in 10 mL of media in each step. As we need to plate 100 μL of media (containing single cell) in each well of 96-well plates, the 10 mL media volume is convenient to use.
hTERT RPE1 cells are highly movable, so it is important to identify single colonies 1–2 days after serial dilution procedure.
Cells could be contaminated during serial dilution and cell sorting procedure. For this reason, we recommend sorting cells into wells containing media with 1% antibiotic/antimycotics.
Cells generally grow on the edge of wells; for this reason, it is difficult to see colonies before 10 days of plating.
Keep cells on the original 12-well plate for maintenance.
Due to the random insertion of pLenti DNA into chromosomes of hTERT RPE1 cells, some of clones may show growth defects. Discard these clones.
Cell lysates for WB analysis could be quite viscous due to chromosomal DNA. To prevent this, use a higher volume of 2% SDS, sonicate the lysate briefly, or boil the lysate for an additional minute or two.
Place the sonicator probe at the frequency of 24 KHz. Then, gently move the tube under the tip of the sonicator probe for 5 s. Check the viscosity of the sample with tips. Sonicate for another 5 s if the sample remains viscous.
5% BSA in PBS or commercial membrane blocking buffers from LiCor or Bio-Rad can be used at this stage to improve sensitivity and decrease the non-specific background.
The procedure is performed at room temperature.
Prepare the fixative (4% PFA) immediately before the experiment by diluting 16% PFA in PBS.
Remember the side of the coverslip with growing cells.
Let nail polish dry before imaging. This step is necessary to prevent coverslips from sliding.
Depending on the microscope system laser intensity and expo-sure time should be adjusted.
We describe procedures for making DNA construct for the expression of COG4-mAID-mCherry and for producing hTERT-COG4 KO cells lines co-expressing OsTIR1–9myc and COG4-mAID-mCherry.
It is important to ensure that tagging of COG complex subunit does not impair its localization, stability, and the function. This should be tested by WB, IF, and functional analysis as described in [22].
During the preparation of PCR mixture, be sure to mix well between each step.
Other proofreading polymerase can be used in this PCR reaction.
Expanding cycling to 35 cycles could give a higher PCR product yield.
Any UV spectrophotometer can be used.
Usually, 3:1 insert to vector molar ratio is sufficient, but the amount of insert and vector can be optimized to improve ligation efficiency in situations where the 3:1 ratio is not working or when doing more complicated cloning.
The ligase buffer contains ATP, which can be degraded by freeze/thaw cycles. Prepare single-use 10 μL aliquots and store them at −20 °C.
Sterile wooden toothpicks or sterile metal loop can be used instead of disposable plastic loops.
To check the resulting construct, transfect HEK293T cells with the recombined pLenti COG4-mAID-mCherry DNA using lipofectamine 3000 transfection protocol (following manufacturer instruction). Then, perform WB analysis using antibody against hCOG4 to check the transient expression of COG4-mAID-mCherry protein.
If the control cells are not dying with 600 μg/mL of G418, the concentration of antibiotic can be increased up to 800 μg/mL in both wells. The exact concentration and time period of antibiotic treatment should be selected based on the specific cell line sensitivity.
Imaging of the cell has been controlled using ZEN software. Colocalization of COG4-mAID-mCherry with GM130 has been analyzed using Pearson’s correlation coefficient [40].
We have reported previously that the stability of Golgi enzymes (B4GalT1, MGAT1) and glycosylation of the Golgi glycoprotein TMEM165 and lysosomal glycoprotein Lamp2 are altered in RPE1 and HEK293T COG4 KO cells [16, 22]. For this setup, COG4-mAID-mCherry-expressing clones which can rescue the COG4 KO cellular phenotypes completely should be selected. In the case of the COG4 KO cell line, the WB should show an increase in the electrophoretic mobility of Lamp2 and TMEM165 and depletion of the Golgi enzymes. These phenotypes should be rescued by COG4KO TIR1–9myc expressing COG4-mAID-mCherry.
Continuous high expression of OsTir1 may cause significant degradation of COG4 hybrid protein even without IAA addition. This is known as “basal degradation” which may create altered auxin-inducible depletion in a context- and target-specific manner [41]. To resolve this problem, an OsTIR1 inhibitor auxinole could be used [41]. Auxinole should be prepared in media (200 μM final concentration) to treat cells overnight before the auxin treatment.
To assure uniform depletion of COG4-mAID-mCherry protein in cell population, it is important to maintain cells in a complete media supplemented with puromycin (2 μg/mL) and G418 (200 μg/mL).
Acknowledgements
This work was supported by the National Institute of Health grant R01GM083144.
References
- 1.Jackson CL (2009) Mechanisms of transport through the Golgi complex. J Cell Sci 122: 443–452. 10.1242/jcs.032581 [DOI] [PubMed] [Google Scholar]
- 2.Willett R, Ungar D, Lupashin V (2013) The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem Cell Biol 140:271–283. 10.1007/s00418-013-1117-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang S, Wang Y (2017) Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Res 6: 2050. 10.12688/f1000research.11900.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makhoul C, Gosavi P, Gleeson PA (2019) Golgi dynamics: the morphology of the mammalian Golgi apparatus in health and disease. Front Cell Dev Biol 7. 10.3389/fcell.2019.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Souza Z, Taher FS, Lupashin VV (1864) Golgi inCOGnito: from vesicle tethering to human disease. Biochim Biophys Acta Gen Subj 2020:129694. 10.1016/j.bbagen.2020.129694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116: 153–166. 10.1016/s0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- 7.Mc D, Pa G (2007) New insights into membrane trafficking and protein sorting. Int Rev Cytol 261:47–116. 10.1016/s0074-7696(07)61002-x [DOI] [PubMed] [Google Scholar]
- 8.Pokrovskaya I, Willett R, Smith R et al. (2011) COG complex specifically regulates the maintenance of Golgi glycosylation machinery. Glycobiology 21:1554–1569. 10.1093/glycob/cwr028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn JB, D’Souza Z, Lupashin VV (2019) Maintaining order: COG complex controls Golgi trafficking, processing, and sorting. FEBS Lett 593:2466–2487. 10.1002/1873-3468.13570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ungar D, Oka T, Krieger M, Hughson FM (2006) Retrograde transport on the COG railway. Trends Cell Biol 16:113–120. 10.1016/j.tcb.2005.12.004 [DOI] [PubMed] [Google Scholar]
- 11.Bröcker C, Engelbrecht-Vandré S, Ungermann C (2010) Multisubunit tethering complexes and their role in membrane fusion. Curr Biol 20:R943–R952. 10.1016/j.cub.2010.09.015 [DOI] [PubMed] [Google Scholar]
- 12.Blackburn JB, Lupashin VV (2016) Creating knockouts of conserved oligomeric Golgi complex subunits using CRISPR-mediated gene editing paired with a selection strategy based on glycosylation defects associated with impaired COG complex function. Methods Mol Biol 1496:145–161. 10.1007/978-1-4939-6463-5_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whyte JRC, Munro S (2001) The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell 1: 527–537. 10.1016/S1534-5807(01)00063-6 [DOI] [PubMed] [Google Scholar]
- 14.Ungar D, Oka T, Brittle EE et al. (2002) Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol 157:405–415. 10.1083/jcb.200202016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotso P, Koryakina Y, Pavliv O et al. (2005) Cog1p plays a central role in the organization of the yeast conserved oligomeric golgi complex*. J Biol Chem 280:27613–27623. 10.1074/jbc.M504597200 [DOI] [PubMed] [Google Scholar]
- 16.Bailey Blackburn J, Pokrovskaya I, Fisher P et al. (2016) COG complex complexities: detailed characterization of a complete set of HEK293T cells lacking individual COG subunits. Front Cell Dev Biol 4:23. 10.3389/fcell.2016.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suvorova ES, Duden R, Lupashin VV (2002) The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol 157:631–643. 10.1083/jcb.200111081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shestakova A, Zolov S, Lupashin V (2006) COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic 7:191–204. 10.1111/j.1600-0854.2005.00376.x [DOI] [PubMed] [Google Scholar]
- 19.Laufman O, Hong W, Lev S (2013) The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic SNARE complexes assembly. J Cell Sci 126(6):1506–1516. 10.1242/jcs.122101 [DOI] [PubMed] [Google Scholar]
- 20.D’Souza Z, Blackburn JB, Kudlyk T et al. (2019) Defects in COG-mediated Golgi trafficking alter endo-lysosomal system in human cells. Front Cell Dev Biol 7:118. 10.3389/fcell.2019.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ondruskova N, Cechova A, Hansikova H et al. (1865) Congenital disorders of glycosylation: still “hot” in 2020. Biochim Biophys Acta Gen Subj 2021:129751. 10.1016/j.bbagen.2020.129751 [DOI] [PubMed] [Google Scholar]
- 22.Sumya FT, Pokrovskaya ID, Lupashin V (2021) Development and initial characterization of cellular models for COG complex-related CDG-II diseases. Front Genet 12: 733048. 10.3389/fgene.2021.733048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Housden BE, Muhar M, Gemberling M et al. (2017) Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet 18:24–40. 10.1038/nrg.2016.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Divekar NS, Horton HE, Wignall SM (2021) Methods for rapid protein depletion in C. elegans using auxin-inducible degradation. Curr Protoc 1:e16. 10.1002/cpz1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MS, Sahlender DA, Foster SD (2010) Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell 18:324–331. 10.1016/j.devcel.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zolov SN, Lupashin VV (2005) Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol 168:747–759. 10.1083/jcb.200412003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Seemann J (2021) Rapid degradation of GRASP55 and GRASP65 reveals their immediate impact on the Golgi structure. J Cell Biol 220:e202007052. 10.1083/jcb.202007052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camlin NJ, Evans JP (2019) Auxin-inducible protein degradation as a novel approach for protein depletion and reverse genetic discoveries in mammalian oocytes†. Biol Reprod 101:704–718. 10.1093/biolre/ioz113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banaszynski LA, Chen L, Maynard-Smith LA et al. (2006) A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126: 995–1004. 10.1016/j.cell.2006.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwamoto M, Björklund T, Lundberg C et al. (2010) A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol 17:981–988. 10.1016/j.chembiol.2010.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambrus BG, Moyer TC, Holland AJ (2018) Chapter 5 – Applying the auxin-inducible degradation system for rapid protein depletion in mammalian cells. In: Maiato H, Schuh M (eds) Methods in cell biology. Academic Press, pp 107–135 [DOI] [PubMed] [Google Scholar]
- 32.Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9:302–308. 10.1016/j.tplants.2004.04.003 [DOI] [PubMed] [Google Scholar]
- 33.Nishimura K, Fukagawa T, Takisawa H et al. (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- 34.Holland AJ, Fachinetti D, Han JS, Cleveland DW (2012) Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci USA 109:E3350–E3357. 10.1073/pnas.1216880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT (2016) Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep 15:210–218. 10.1016/jxelrep.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 36.Stewart SA, Dykxhoorn DM, Palliser D et al. (2003) Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9: 493–501. 10.1261/rna.2192803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dull T, Zufferey R, Kelly M et al. (1998) A third-generation lentivirus vector with a conditional packaging system. J Virol 72:8463–8471. 10.1128/JVI.72.11.8463-8471.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakano R, Ihara N, Morikawa S et al. (2019) Auxin-mediated rapid degradation of target proteins in hippocampal neurons. Neuroreport 30:908–913. 10.1097/WNR.0000000000001299 [DOI] [PubMed] [Google Scholar]
- 39.Wright PF, Nilsson E, Van Rooij EM et al. (1993) Standardisation and validation of enzyme-linked immunosorbent assay techniques for the detection of antibody in infectious disease diagnosis. Rev Sci Tech 12:435–450. 10.20506/rst.12.2.691 [DOI] [PubMed] [Google Scholar]
- 40.Sedgwick P (2012) Pearson’s correlation coefficient. BMJ 345:e4483–e4483. 10.1136/bmj.e4483 [DOI] [Google Scholar]
- 41.Yesbolatova A, Natsume T, Hayashi K-I, Kanemaki MT (2019) Generation of conditional auxin-inducible degron (AID) cells and tight control of degron-fused proteins using the degradation inhibitor auxinole. Methods 164–165:73–80. 10.1016/j.ymeth.2019.04.010 [DOI] [PubMed] [Google Scholar]