Abstract
Literary and archaeological sources have preserved a rich history of southern Europe and West Asia since the Bronze Age that can be complemented by genetics. Mycenaean period elites in Greece did not differ from the general population, and included both people with some steppe ancestry, and others, like the Griffin Warrior, without it. Similarly, people in the central area of the Urartian Kingdom around Lake Van lacked the steppe ancestry characteristic of the kingdom’s northern provinces. Anatolia exhibited extraordinary continuity down to the Roman and Byzantine periods, with its people serving as the demographic core of much of the Roman Empire, including the city of Rome itself. During medieval times, migrations associated with Slavic and Turkic speakers profoundly impacted the region.
One-Sentence Summary:
Polities of the ancient Mediterranean world preserved contrasts of ancestry since the Bronze Age but were linked by migration.
The works of ancient writers provide powerful information about the ancient world, recording information on different groups, their political organization, customs, relations, and military conflicts. The manuscript tradition has been augmented by the archaeological record which also included the discovery of diverse texts of past Mediterranean and West Asian civilizations. Here, we leverage the power of ancient DNA to provide a third source of information about the people inhabiting the states and empires of the past. Many of these aspects have been recorded, or hinted at, in ancient texts composed close to the time of the events they describe. However, no text is fully objective, and is inevitably shaped by authors’ biases and world views. Ancient DNA provides independent evidence, with its own strengths and weaknesses, and cannot paint a picture of the past on its own. Nonetheless, it complements the ancient texts and evidence from archaeology. By using genetic data we can hope to obtain a more nuanced impression of past processes, especially regarding movements of people and biological phenotypes, than would be possible without such data.
This study is a part of a comprehensive archaeogenetic study of the genetic history of the populations of the Southern Arc spanning a trio of papers. For a description of the full dataset and analysis framework and characterization of the population history of the Chalcolithic and Bronze Age periods see (1). For analysis of the population history of the Neolithic, see 2). The present study focuses on peoples for which there is also information from written texts, and a main theme is to test the extent to which textual insights are supported or not by the genetic data, and furthermore to explore what complementary information genetics can provide. When we reference ancient literature, we use standard abbreviations for locating passages in online repositories of texts such as the Perseus Digital Library(3). Our study begins at the end of the Bronze Age, and traces the region’s history via the 1st millennium BCE, through the Roman Empire and to the present, a timespan of more than three thousand years.
The Bronze Age Aegean World
Previous work has demonstrated that the Bronze Age civilizations of Greece of the periods labeled Minoan (on the island of Crete, spanning the entirety of the Bronze Age ~3500-1100BCE) and Mycenaean (on the Greek mainland and its islands, spanning the latter part of the Bronze Age since the last phase of the Middle Helladic period to the end of the Late Helladic period ~1750-1050BCE) (4) were inhabited by genetically similar populations that traced most of their ancestry to the Neolithic inhabitants of the region and who, in turn, were related to the farmers of Anatolia (4–7). We refer to people associated with these archaeological contexts as Minoan and Mycenaean, recognizing that the people themselves would almost certainly not have considered themselves as belonging to two groups defined according to this framework defined by modern archaeology, and that there was in fact extensive genetic variation in ancestry among people associated with such cultures, as we prove here for the first time. Both Mycenaeans and Minoans possessed extra “eastern” Caucasus-related ancestry compared to the Neolithic inhabitants of Greece, but differed from each other in that the Mycenaeans taken as a group had some steppe ancestry that Minoans lacked (4). Here we extend the geographical sampling to multiple new sites, complementing published Mycenaean data from the Peloponnese and Salamis and Minoan data from Lasithi and Moni Odigitria(4). From Crete, we report a Middle Minoan individual from Zakros. From mainland contexts we report the first Mycenaean data from central Greece—that is, the previously unsampled region north of the Isthmus of Corinth—including Attica, Kastrouli near Delphi in Phokis, and Lokris in Phthiotis. South of the Isthmus in the Peloponnese, we report data from many individuals from the “Palace of Nestor” at Pylos and its environs, including the elite “Griffin Warrior”, a young (30-35 years old) male buried in a large stone-built tomb with hundreds of precious artifacts, many of them made in Minoan Crete (8).
To contextualize the transformations in the Bronze Age Aegean, it is critical to characterize the pre-Bronze Age genetic landscape (Fig. 1). We begin with the Neolithic inhabitants (4, 6, 7), estimating proportions of ancestry using a 5-source model that we developed for Southern Arc Holocene populations (1) which includes as proxies for the sources Caucasus hunter-gatherers (9), Eastern European hunter-gatherers (5, 10), Levantine Pre-Pottery Neolithic (11), Balkan hunter-gatherers from the Iron Gates in Serbia (7), and Northwestern Anatolian Neolithic from Barcın (5). We infer that not only Neolithic Greeks from the Peloponnese (7), but also from Northern Greece (6) possessed ~8-10% Caucasus hunter-gatherer-related ancestry (Fig. 1C). Such ancestry was inferred for Southeastern Europe Neolithic populations in general in contrast to central-western Europe (1) and provides proof of multiple streams of migration from different Anatolian Neolithic populations into Europe.
Fig. 1: Genetic heterogeneity in the Aegean.
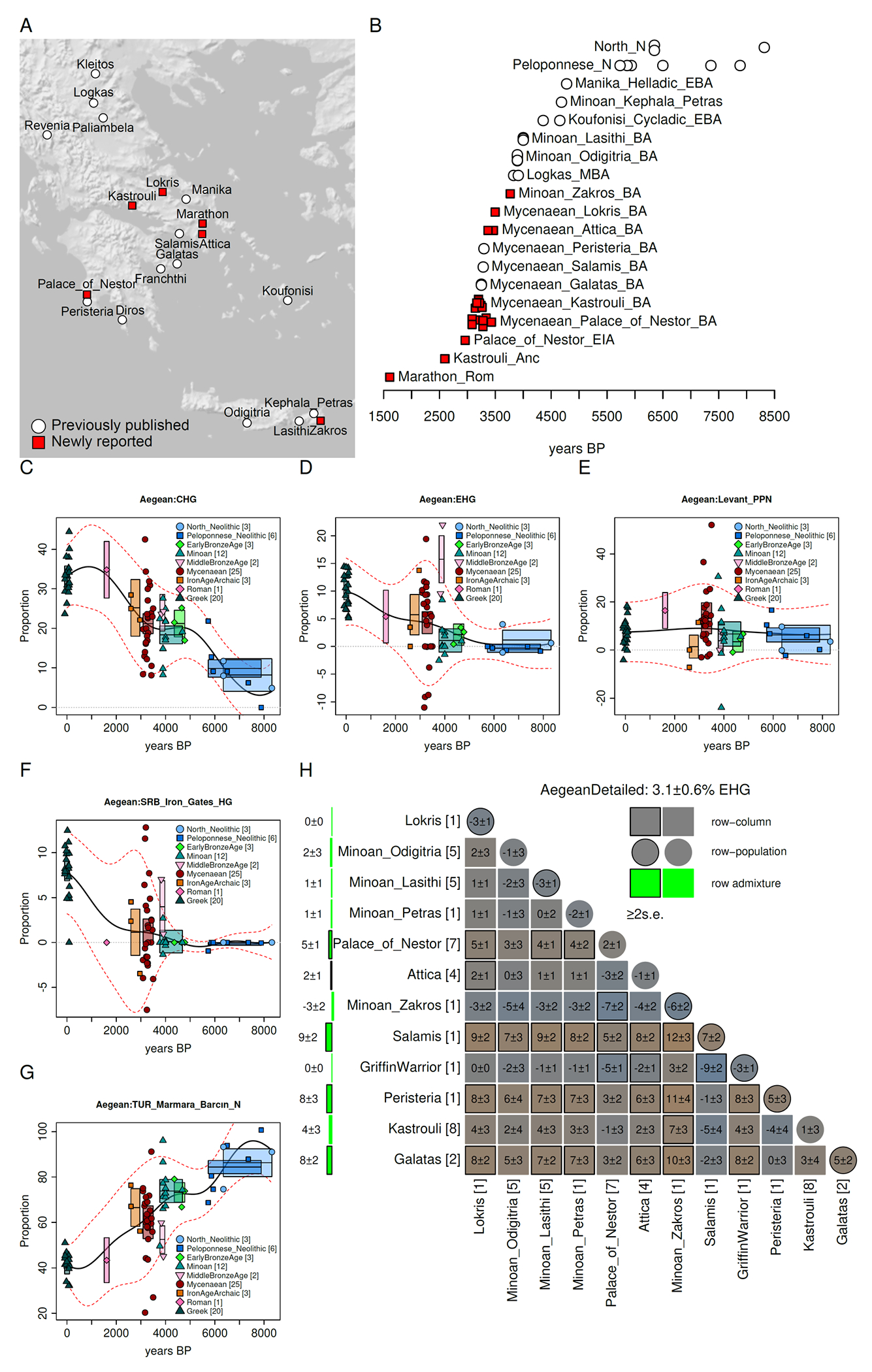
(A) A map of Aegean sites. (B) Timeline of Aegean individuals, with vertical jitter added to distinguish contemporaneous individuals. Ancestry changes of five components (C-G) show an increase of Caucasus hunter-gatherer (CHG) and Eastern European hunter-gatherer (EHG) ancestry over time and a dilution of Anatolian Neolithic ancestry. During the Minoan and Mycenaean periods of the Bronze Age (H) Eastern European hunter-gatherer ancestry was variable, absent in Minoan individuals of Crete and present in most, but not all Mycenaean individuals of the mainland.
Both Caucasus- and Eastern European hunter-gatherer-related ancestry increased in the Bronze Age in the Aegean just as the Anatolian-related ancestry decreased (Fig. 1), with Mycenaean Greeks having 21.2±1.3% for Caucasus hunter-gatherer ancestry and 4.3±1.0% Eastern European hunter-gatherer . Given the evenly balanced proportions of these components in the Yamnaya and the “high steppe” cluster from the Balkans(1), it can be assumed that the Eastern European hunter-gatherer component in the Aegean was not introduced there on its own, but was accompanied by an approximately matching amount of Caucasus hunter-gatherer ancestry, thus leaving a remainder of ~21.2-4.3=16.9% Caucasus hunter-gatherer. This allows us to infer that steppe migrants admixed with a population whose composition must have included or ~18.5% Caucasus hunter-gatherer-related ancestry. Remarkably, the estimated proportion of Caucasus hunter-gatherer ancestry in Minoans is a virtually identical 18.3±1.2%. Thus, our analyses resolve the question of the origins of the Late Bronze Age population by strongly supporting one of two previously proposed hypotheses (4): that Mycenaeans were the outcome of admixture of Yamnaya-like steppe migrants with a Minoan-like substratum, rather than the hitherto plausible alternative scenario of an Anatolian Neolithic-like substratum admixing with an Armenian-like population from the east. This alternative scenario is further contradicted by the fact that pre-Mycenaean period individuals belonging to the Early Bronze Age from the islands of the Cyclades and Euboea in southern Greece ~2,500BCE (12) had 21.2±1.7% Caucasus hunter-gatherer-related ancestry(12), consistent with our inferred proportion and providing direct evidence for the predicted Minoan-like substratum (4).
The fact that Mycenaeans can be modelled as a mixture in an ~1:10 ratio of a Yamnaya-like steppe-derived population and a Minoan/Early Bronze Age-like Aegean population suggests that any contribution of geographically intermediate populations (between the steppe and the Aegean) to the formation of Mycenaeans was minor. This conclusion is further supported by: (i) the lower (~5%) Caucasus hunter-gatherer ancestry in the Neolithic of the Balkans compared to the ~20% inferred for the Aegean substratum(1), (ii) the near absence of Balkan hunter-gatherer (Fig. S1) ancestry in the Aegean in contrast to other Southeastern European populations (~10%) (1), and (iii) the presence of Yamnaya-like individuals with minimal local ancestry, immediately to the north of the Aegean, in Albania and Bulgaria during the Early Bronze Age (1). Whatever the genetic makeup of people mediating the spread of steppe ancestry into the ancestors of Mycenaeans, the genetic impact of steppe on Aegean populations was quantitatively minor. We estimate the Yamnaya-related steppe ancestry proportion in Mycenaeans to be ~1/3 of the level in the Balkans to the north, ~1/2 of that in Armenia in the east, and ~1/5-1/8 of populations of central-northern Europe associated with the Bell Beaker and Corded Ware cultures (1).
Eastern European hunter-gatherer ancestry as a marker for Yamnaya steppe pastoralist ancestry is absent in a newly reported Middle Minoan period individual from Zakros in the eastern edge of Crete, generally similar to those previously published (13), but with significant Levantine ancestry (30.5±9.1%) which is consistent with her either being a migrant to the island from the east or part of a structured Cretan population whose past ethnic diversity was noted as early as the Odyssey of Homer (Hom. Od. 19.172-177).
We show, for the first time, that Eastern European hunter-gatherer ancestry was also absent in some Mycenaean individuals, suggesting that while the contrast between the mainland and Crete was significant (Fig. S1), the penetration of Eastern European hunter-gatherer ancestry did not reach the totality of the mainland population during the Late Bronze Age and was even significantly variable within Mycenaean sites. The Griffin Warrior (14), the earliest individual (~1450 BCE) from the Palace of Nestor in Pylos, is genetically right in the middle of the general population of the Aegean, and was thus plausibly of local Aegean origin. He had no detectable Eastern European hunter-gatherer ancestry (compared to the average of 4.8±1.1% for the rest of the Mycenaean-era individuals sampled at the Palace; Fig. 1H). This finding could be consistent with a Cretan origin of this individual or his ancestors; alternatively, he could be drawn from a mainland population that had not experienced Eastern European hunter-gatherer admixture, as could two later individuals from Pylos, one buried near the Palace in a chamber tomb, and another in a cist grave. Variation in Eastern European hunter-gatherer ancestry is observed at short geographical distance scales and within the same time periods, as we observe that 4 individuals (~1450BCE) of the sample of individuals from Attica buried at Kolikrepi-Spata had only 2±1% Eastern European hunter-gatherer ancestry that was significantly less (by more than 2 standard errors) than those of the neighboring island of Salamis and all sampling locations in the Peloponnese. This suggests that the classical Athenian claim (e.g., Plat. Menex. 237b) of having received fewer migrants than other Greek poleis in the remote past may have had an element of truth, although larger sample sizes will be necessary to establish such geographic patterns definitively.
Northern migrants made an impact throughout mainland Greece even if it was a modest one, which is also evidenced on the male line as shown, for example, by a Y-chromosome match of the rare R-PF7562 haplogroup between a pair of patrilineal relatives from the Palace of Nestor which links Late Bronze Age Mycenaean Greece with an Early Bronze Age individual of the North Caucasus at Lysogorskyja that is genetically similar to Yamnaya Y chromosomes(15). This patrilineal connection to the Yamnaya should not be interpreted as a general association of steppe ancestry with elite burial status, as the common people, making up most of our Mycenaean-era individuals, also had steppe ancestry, while some members of the elite (such as the Griffin Warrior) did not have significant evidence of it. A parallel example of an elite individual with less steppe ancestry than others from the same cultural context during a period of steppe ancestry spread is given by the “Amesbury Archer,” the most well-furnished grave in the Stonehenge mortuary landscape of Great Britain(16); these two examples highlight the pitfalls of conflating genetic ancestry with narratives of social dominance. Whatever the social role of early steppe migrants into the Aegean, they did not establish a system that precluded admixture with locals or prevented them from rising to positions of power. This inclusiveness may explain the substantial dilution of steppe ancestry in the Aegean as migrants and locals blended to form the ancestors of the Mycenaean-era population, and may shed light on the genesis of the Greek language, linked, on one hand with the rest of Indo-European via steppe ancestry(1) and with the people of the Aegean that preceded the Proto-Greek speakers on the other(17).
One of the two patrilineal relatives at Pylos (I13518) was almost certainly the offspring of first cousins; we document such close-kin unions not only in elite Mycenaean society but also in different localities of the Bronze Age Southern Arc (Fig. S2) (18), including an individual from Bezdanjača in Croatia (I18717) who was likely the offspring of an uncle/niece pairing. This documents for the first time the later persistence of the practice of close-kin matings that had started with the Neolithic (19, 20), although whether this is due to the burials we analyzed being a biased subset of a population, or reflects society-wide cultural preferences, cannot be resolved with our available sample. Did descriptions of such unions in classical mythological accounts of the “Heroic Age” reflect practices that persisted to the authors’ own time? Ancient DNA studies from more locations would allow these patterns of mating preferences inferred from a handful of sites to be characterized at higher resolution.
The era of Greek colonization
We report a preliminary look at demographic patterns associated with the Greek colonial period (8th-6th c. BCE) by identifying individuals from both the Southern Arc and outside of it that were genetically similar to Bronze Age individuals of the Mycenaean period (Supplementary Text S1; Fig. S3) (18). This identifies an Archaic period individual from Kastrouli in Phokis in Delphi on the Greek mainland, and individuals at Empúries in northeastern Spain who are genetically very similar to Mycenaean era individuals from the Greek mainland(21). Empúries was an outpost colonized by Phocaeans from western Anatolia, who were themselves said to be colonists from Phokis (Paus. 7.3.10). Thus, we capture the end points of a long chain of transmission, with little admixture, across the Mediterranean. Could the ancestry of the Empúries individuals be traced back to the beginning of this chain or was it drawn from another, genetically similar source? While we do not yet have rich sampling of the peoples of the Greek colonial world, systematic sampling of diverse Greek colonies spread over the Mediterranean and Black Sea coasts would make it possible to test systematically for evidence of specific metropolis-colony connections and document the extent to which migration, admixture with local populations, and genetic heterogeneity played a role in Greek colonization.
Ancestry typical of the Mycenaean period spread also to the eastern Mediterranean as in the case of an individual from Ashkelon associated with a Philistine archaeological context (22). We also show the similarity of some individuals from inland Thrace (at Kapitan Andreevo) with the Mycenaean genetic profile, suggesting that Mycenaeans were genetically similar to some Thracians from the east Balkans, outside the sphere of the Late Bronze Age Aegean. This provides a cautionary tale highlighting the dangers of conflating genetic and cultural similarity.
The coastal regions of Anatolia formed another area of Greek settlement and much of the Anatolian peninsula was incorporated into the Hellenistic kingdoms established by the successors of Alexander the Great, providing opportunity for population transfer from Southeastern Europe to Anatolia. Yet, we do not find Mycenaean-like individuals either at 1st millennium BCE Greek colony sites such as Halicarnassus (modern Bodrum) or Amisos (modern Samsun) in the Aegean and Black Sea regions respectively. This pattern is qualitatively different from that at Empúries in Iberia and is consistent with the account of Herodotus that early Greek colonists of Anatolia married indigenous Carian women of Anatolia when they first settled there (Hdt. 1.146). It is also reminiscent of the marriages of Alexander himself and his companions with local women of the conquered Persian Empire (Arr. An. 7.4.4ff). Clearly, Greeks segregated themselves socially and reproductively from non-Greeks in some parts of the Greek world and not in others; an important topic for future research is to identify the factors that correlated with Greeks mixing with peoples from local communities.
The Urartian Kingdom and its neighbors in Iran and Mesopotamia
We have already seen how the Aegean was an area of limited Eastern European hunter-gatherer penetrance that nonetheless differentiated it from neighboring Anatolia where Eastern European hunter-gatherer ancestry was negligible (1). An even more striking case is that of the Iron Age Kingdom of Urartu situated in the mountainous and geographically fragmented regions of eastern Turkey and Armenia where the linguistic landscape must have been complex in the Bronze and Iron Ages. The people at the center of this kingdom in the Lake Van region of Turkey (Çavuştepe) and its northern extension in Armenia, were strongly connected by material culture, and were buried only ~200km apart, yet formed distinct genetic clusters with little overlap during the kingdom’s early (9th-8th c. BCE) period (Fig. 2). The Van cluster is in continuity with the pre-Urartian population (~1300BCE) at neighboring Muradiye also in the Van region, and is characterized by more Levantine ancestry and the absence of steppe ancestry. It contrasts with the cluster of Urartian period individuals from Armenia which have less Levantine and some steppe ancestry like the pre-Urartian individuals of the Early Iron Age (1). Our genetic results help explain the formation of linguistic relationships in the region. Population continuity of the Lake Van core population with greater “Levantine” ancestry may well correspond to the Hurro-Urartian language family (23) that linked the non-Indo-European Urartian language of the kingdom with the earlier Bronze Age Hurrian language whose more southern distribution encompassed parts of Syria and North Mesopotamia. Into the periphery of this Hurro-Urartian linguistic sphere came a steppe-admixed population from the north, whose presence marks the southern edge of steppe expansion we discussed above and whose proximity to the Urartian speakers would provide a mechanism for the incorporation of Urartian words into the Armenian lexicon.
Fig. 2: The Kingdom of Urartu and its neighbors.
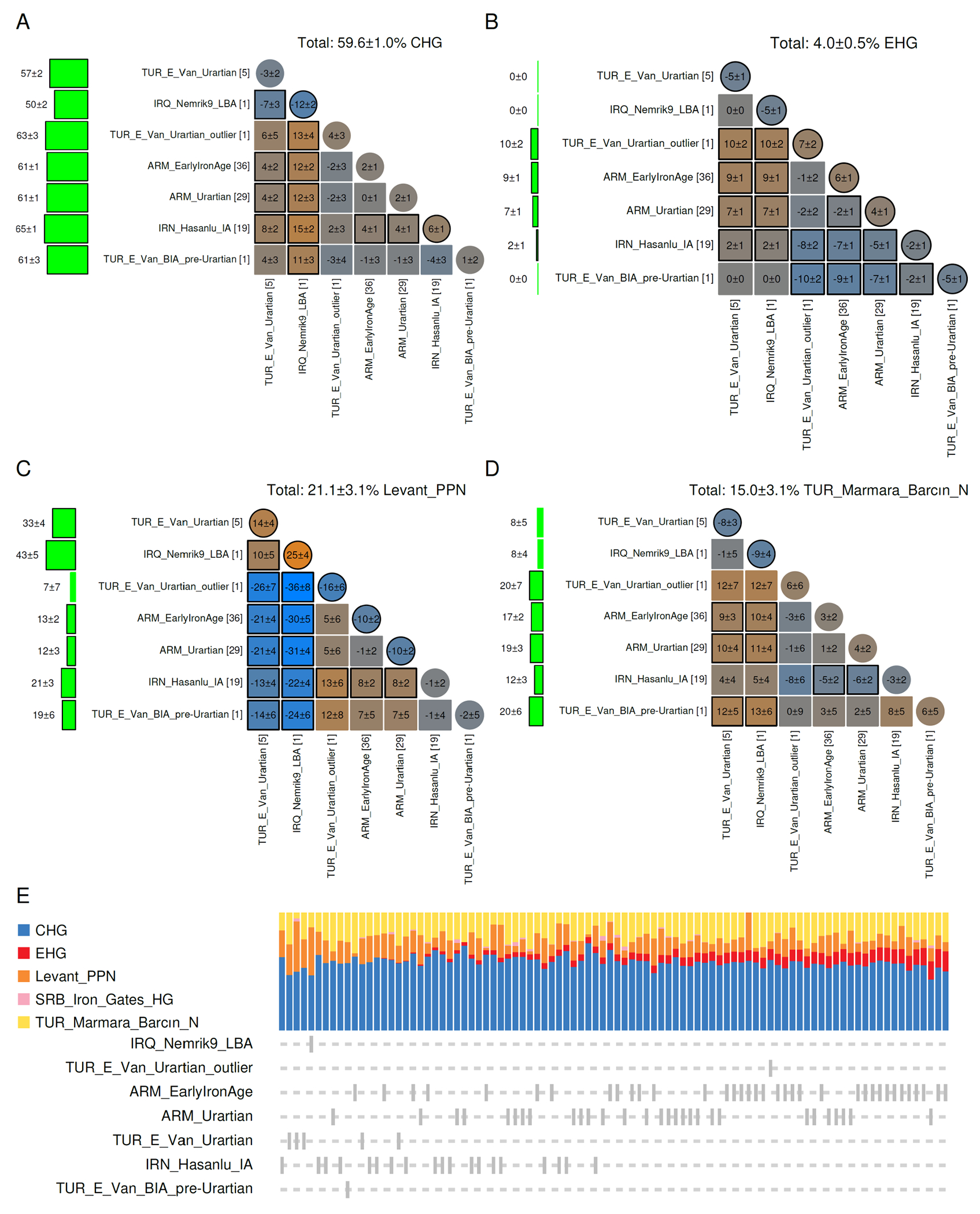
Panels (A-D) show comparisons of ancestry in four ancestral components (SRB_Iron_Gates_HG, the 5th component of the model of (1) is negligible). This analysis shows a stark contrast between Armenia and the other populations in terms of Eastern European hunter-gatherer ancestry (B), and between Van and Assyrian Mesopotamia in terms of Levantine ancestry (C). When unlabeled individuals are ordered in increasing Eastern European hunter-gatherer ancestry, Assyrian Mesopotamia, Van lack this ancestry (except an outlier individual from Van), while individuals from Armenia mostly possess it, and those from Hasanlu have a limited range from zero Eastern European hunter-gatherer ancestry to a maximum level that is less than that seen in Armenia.
When we compare (Fig. 2E) the Urartian individuals with their neighbors at Iron Age Hasanlu in NW Iran (~1000BCE), we observe that the Hasanlu population possessed some of Eastern European hunter-gatherer ancestry, but to a lesser degree than their contemporaries in Armenia. The population was also linked to Armenia by the presence of the same R-M12149 Y-chromosomes (within haplogroup R1b), linking it to the Yamnaya population of the Bronze Age steppe(1). Which language was spoken here is not clear, but the population shows no connection with the high-Eastern European hunter-gatherer, R-Z93 (within haplogroup R1a) haplogroup-bearing groups from Central and South Asia belonging to steppe populations ancestral to Indo-Aryan speakers (24) the closest linguistic relatives of Iranian speakers (25). Present-day Iranians do possess R-Z93 Y-chromosomes (26), or the more general upstream R1a-M17 ones (observed in every one of 19 regional or linguistic subset populations from Iran (27), as do present-day Indians (28), who have <1% of R1b Y-chromosomes). Thus, it appears that R1a-haplogroup Y-chromosomes represent a common link between ancient and modern Indo-Iranians while R1b-haplogroup ones (to which many of the Hasanlu males belonged) do not. The absence of any R1a examples among 16 males at Hasanlu who are, instead, patrilineally related to individuals from Armenia suggests that a non-Indo-Iranian (either related to Armenian or belonging to the non-Indo-European local population) language may have been spoken there, and that Iranian languages may have been introduced to the Iranian plateau from Central Asia only in the 1st millennium. Finally, a single individual from the Late Bronze Age of Assyrian North Mesopotamia (~1250BCE) resembles the Urartian Van individuals in lacking Eastern European hunter-gatherer ancestry, had the highest amount of Levantine autosomal ancestry (42.8±5.3%), and possessed a J-P58 derived Y-chromosome with strong Levantine geographical associations (1) and may have plausibly been a speaker of a Semitic language such as those that have been spoken and recorded in the region for most of its history. Archaeology has furnished a wealth of information about the political geography of the ancient Near East, and future genetic studies will elucidate changes of population through either voluntary migration or policy that occurred between the states and empires that arose in this region.
The Anatolian origins of the population of the Roman-Byzantine Empire
A study of a time transect of the city of Rome in Central Italy (29) identified an ancestry shift towards the Near East during the Imperial period (27 BCE to 300 CE), but was unable to localize the origin of the migrants driving this phenomenon. We sought to identify the geographic sources of these Imperial era Romans by co-analyzing the data from Italy with the new sampling from the Southern Arc of Southeastern Europe and West Asia. Surprisingly, the ancestry of people who lived around Rome in the Imperial period was almost identical to that of Roman/Byzantine individuals from Anatolia in both their mean (Fig. 3A) and pattern of variation (Fig. 3B), while Italians prior to the Imperial period had a very different distribution (29, 30). We clustered diverse Roman/Byzantine/Medieval individuals and immediate predecessors without any knowledge of their population labels and found that the Italian and Anatolian individuals clustered together with those of pre-Roman Anatolia, while pre-Imperial people around the city of Rome were systematically different (Fig. 3C). This suggests that the Roman Empire in both its shorter-lived western part and the longer-lasting eastern centered on Anatolia had a diverse but similar population plausibly drawn to a substantial extent from Anatolian pre-Imperial sources. In an irony of history, though the Roman Republic prevailed in its existential military struggle against the Anatolians rallied by Mithridates VI of Pontus during the 1st c. BCE, the final incorporation of Anatolia into the Roman Empire and the increased connectivity that ensued may have set the stage for the very same Anatolians to become the demographic engine of Imperial Rome itself. This recreated, in historical time, the mythical journey of Aeneas and his Trojan exiles from Anatolia to the shores of Italy.
Fig. 3: The Roman Empire East and West.
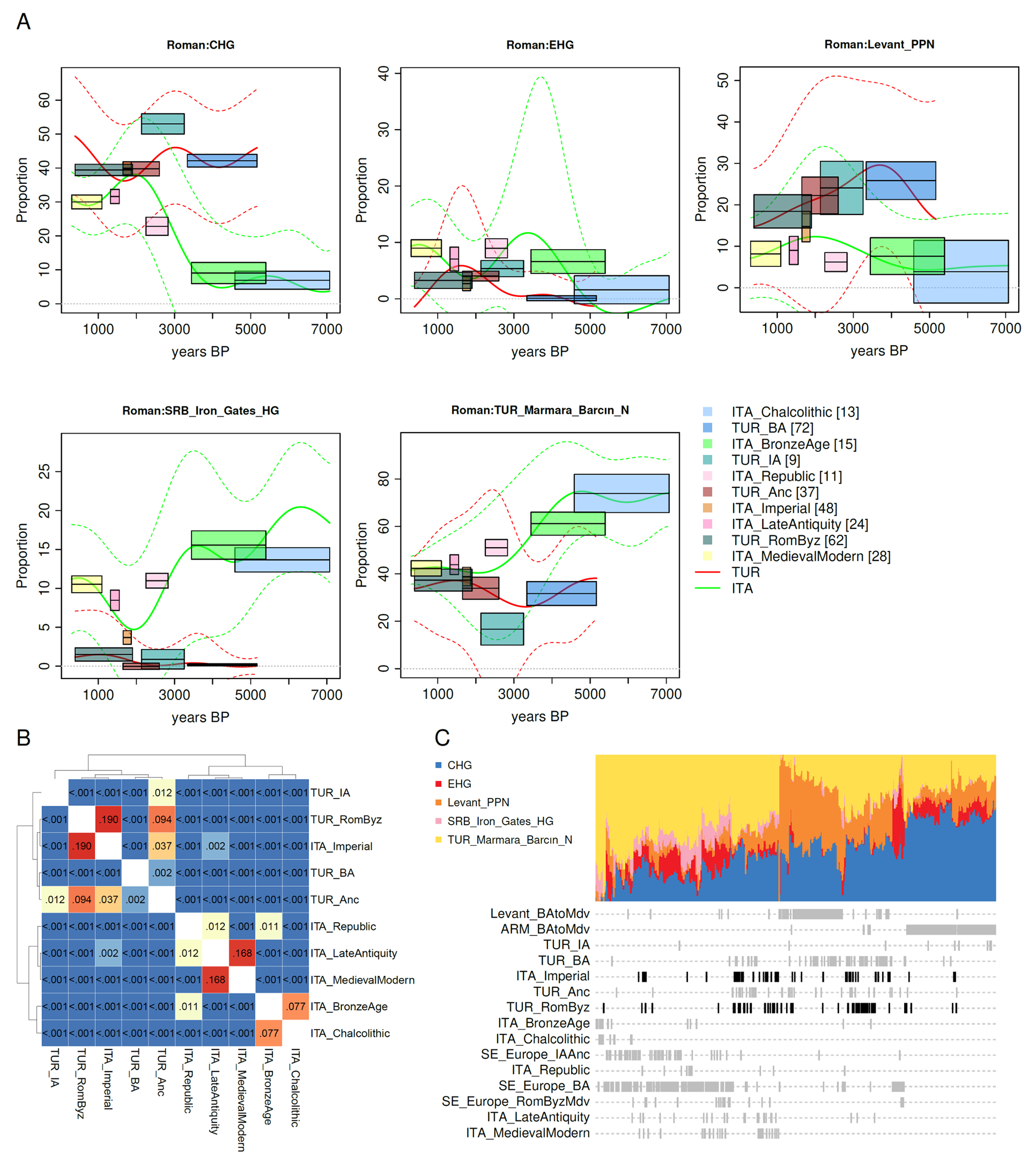
(A) The Imperial period Romans from the vicinity of the city of Rome in Central Italy resembled Roman/Byzantine Anatolians in their average admixture proportions (95% confidence interval (C.I.) of ±1.96 standard errors shown as boxes and a heteroskedastic Gaussian process is fitted to unlabeled Italian and Anatolian individuals; dashed lines indicate 5% and 95% quantiles). (B) P-values of the Baringhaus-Franz multivariate two-sample test(47) for pairs of populations indicate that Imperial Romans can be drawn from the same distribution as Roman/Byzantine ones (p=0.19), but are significantly different (p≤2.16e-03) from all other periods of Italy. (C) Hierarchical clustering of raw ancestry estimates of diverse individuals shows overlapping distributions of Imperial Roman and Anatolian Roman/Byzantine individuals (black) without knowledge of their ancestry labels and differentiated from the distributions of SE Europe, Armenia, and the Levant.
The Southern Arc was also a recipient of many immigrants from outside the region in the historical period, such as two individuals sampled in Samsun in the Black Sea region from the Roman era in the 2nd-3rd centuries CE (18). These individuals have both Eastern European hunter-gatherer and some East Eurasian ancestry that contrasts them with the local population of the Black Sea region that had been stable since the Chalcolithic (31), across the Early Bronze Age transition at Amasya, and down to the time of the Kingdom of Pontus (1st c. BCE). Broad genetic stability in Anatolia during the Roman/Byzantine period did not mean isolation, as outliers of likely Levantine, northern European/Germanic, and Iberian origin are detected in the Marmara region (in the Basilica of Nicaea/present-day Iznik and the Virgin Mary Monastery at Zeytinliada, Erdek) close to the Imperial capital of Constantinople (present-day Istanbul) which may have attracted a more diverse set of foreigners. Other outliers are found at the periphery of the Southern Arc, in Moldova and Romania, in the Iron Age and long after the early steppe migrants previously discussed. These are distinctive because of the East Eurasian admixture of Central Asian Scythian individuals (32–34).
Medieval migrations into Anatolia and the Balkans
East Eurasian ancestry also helps identify an intriguing set of outliers at Çapalıbağ in the Aegean coast of Turkey dating from the 14th-17th centuries (Fig. 4) (18). These have ~18% such ancestry unlike Byzantine-era individuals from Turkey (Fig. 4B), suggesting a Central Asian influence. An admixture date estimate of 12.2±1.4 generations prior to their time using Roman/Byzantine and Central Asian sources (Fig. 4C) suggests that the admixture occurred in the period surrounding the 11th century arrival and expansion of Seljuq Turks to Anatolia. Present-day Turkish individuals have an admixture date estimate of 30.6±1.9 generations (Fig. 4D), and thus from the same early centuries of the 1000s CE which coincided with the transfer of control of Anatolia from the Romans to the Seljuqs and eventually Ottomans. The genetic contribution of Central Asian Turkic speakers to present-day people can be provisionally estimated by comparison of Central Asian ancestry in present-day Turkish people (~9%) and sampled ancient Central Asians (range of ~41-100%) to be between and or ~9-22%. People from Turkey were sampled from eight localities (n=58) (35), representing broadly the present-day population. The genetic data thus point to Turkish people carrying the legacy of both ancient people who lived in Anatolia for thousands of years covered by our study and people coming from Central Asia bearing Turkic languages.
Fig. 4: Central Asian Turkic admixture in Anatolia.
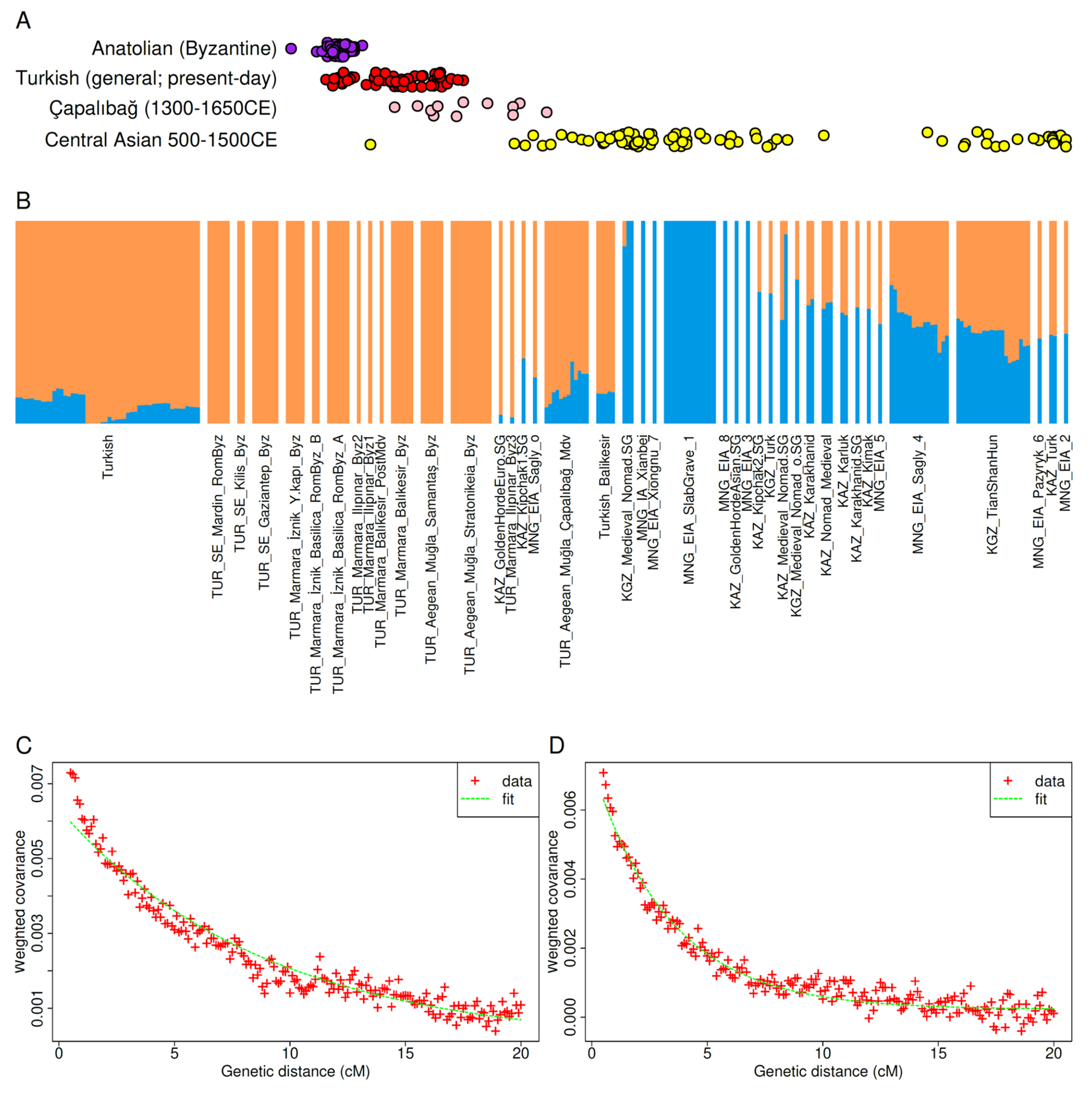
(A) Individuals from Çapalıbağ (1300-1650CE) and present-day Turkish individuals are intermediate between Byzantine Anatolia and 500-1500CE Central Asians along a global principal components analysis distinguishing West from East Eurasians (left-to-right on the horizontal dimension; noise added on the vertical dimension to distinguish points). (B) 2-way unsupervised ADMIXTURE analysis of “eastern” ancestry: Byzantine: (0%), present-day Turkish (9%), Çapalıbağ (18%), Central Asian individuals differ between 100% (in Mongolia) to 43% (some ancient populations of Kazakhstan and Kyrgyzstan). (C) Individuals from Çapalıbağ in Turkey admixed 12.2±1.4 generations (342±39 years) prior to their time using Byzantine Anatolians and Central Asians (from 500-1500CE) as sources. (D) Present-day Turkish people genotyped on the Human Origins array (35) admixed 30.6±1.9 generations ago (857±53 years) using the same sources as in (D).
The medieval period was marked by Slavic migrations into the Balkans on the basis of the genetic analysis of present-day populations (36, 37) and recorded in historical sources such as those of Procopius (38) in the 6th c. BCE when Slavic groups came into contact with the Roman Empire(39). The South Slavs of today in the Balkans are one of the major groups of Slavic speakers and the question of which migrations played a role in their origin is of interest for understanding how this group of languages little-attested until medieval times came to be so widespread across the greater part of Eastern Europe. We highlight Roman, Byzantine, and Medieval individuals from Albania, Bulgaria, Croatia, Greece, North Macedonia, and Serbia, which we studied in conjunction with those that preceded them in the Balkans and with published data from present-day people genotyped on the Human Origins array (35, 40) (Fig. 5). The reduction of Anatolian Neolithic ancestry was a long-term process in Southeastern Europe (1), which allows us to differentiate present-day populations from those preceding the Slavic migrations. When we order individuals along this component of ancestry (Fig. 5), we observe that present-day Slavs outside the Balkans have least, while pre-Slavic inhabitants from the Balkans have the most of this type of ancestry, with present-day people from Southeastern Europe being intermediate between the two extremes. Three individuals from Bulgaria (Samovodene), North Macedonia (Bitola), and an outlier individual from Trogir in Croatia (700-1100CE) have the lowest levels of this ancestry. The majority of individuals from Trogir (a port city of the Adriatic in Croatia that was founded by Ancient Greek colonists and was part of the Byzantine Empire) overlapped with present-day people ~700-900CE, as did 12th c. CE individuals from Veliko Tarnovo and Ryahovets in Bulgaria and a mid-4th c. CE Roman era individual from Marathon in Greece which, however, lacked the Balkan hunter-gatherer ancestry found consistently in the present-day population (Fig. 1). Finally, three medieval individuals from Albania (500-1100CE) and a Late Antique (~500CE) individual from Boyanovo in Bulgaria preceding the Slavic migrations overlapped with the more ancient population, having high levels of Anatolian Neolithic ancestry. Among present-day speakers, Greeks and Albanians have more Anatolian Neolithic ancestry than their South Slavic neighbors. Slavic migrations have some echoes, ~3,000 years later, to the spread of the descendants of Yamnaya steppe pastoralists into Southeastern Europe (1, 7). Although both events were transformative, any analogy should not be pushed too far. The medieval movements were carried out by large organized communities engaging with complex states such as the Avar Khaganate and Byzantine Empire, and no comparable polities existed in Yamnaya times. Collectively, our data suggest that while Balkan groups experienced a shift of ancestry in the medieval period, the fusion of locals and migrants was variable with individuals of diverse ancestry being present in medieval times and persisting up to the present.
Fig. 5: Byzantine and Medieval Southeastern Europe.
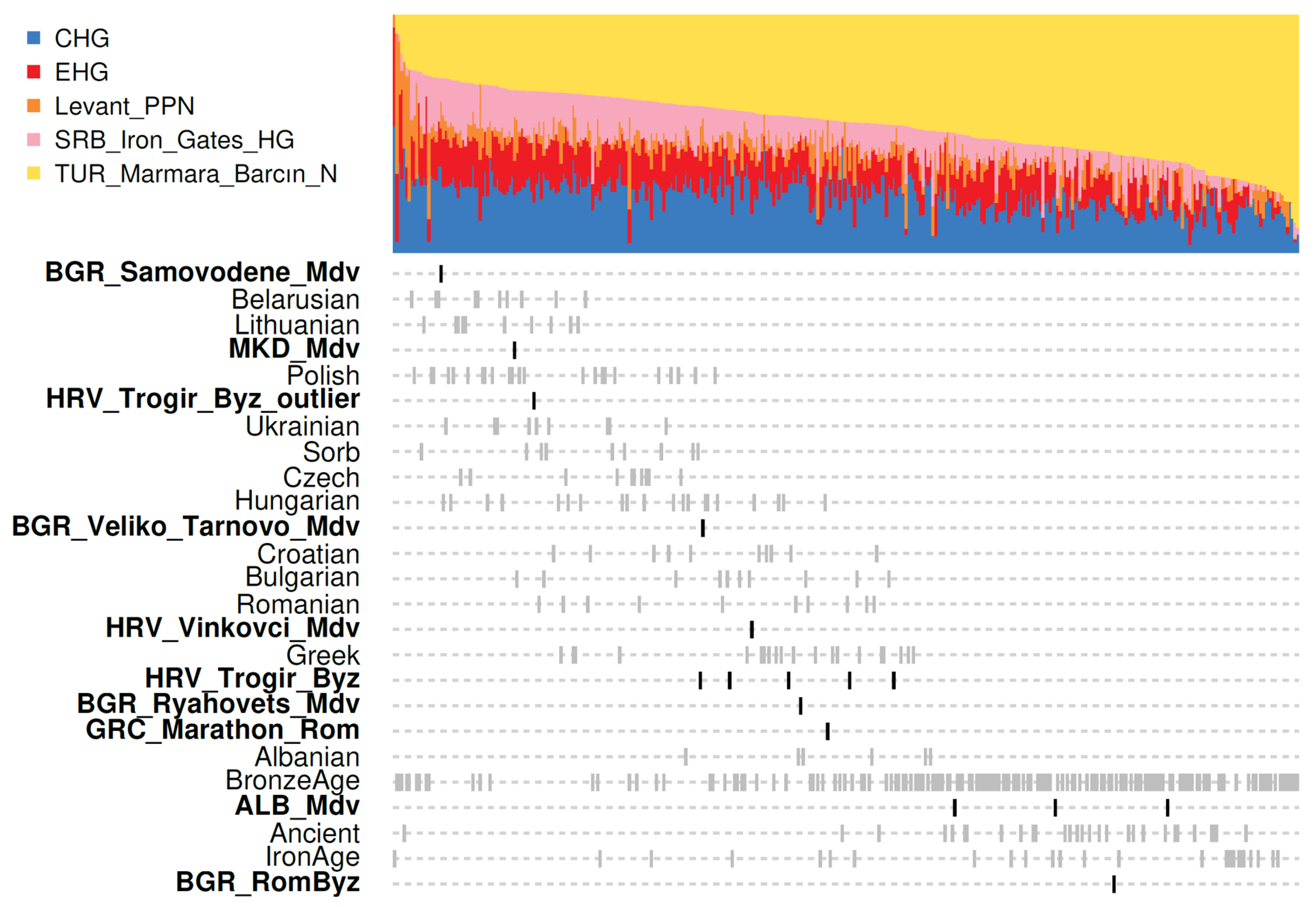
We sort admixture proportions of Anatolian Neolithic ancestry to investigate the dilution of this ancestry in present-day populations from Southeastern Europe. Roman/Medieval/Byzantine-era individuals are indicated in bold. During the Bronze Age the range of this ancestry was immense as observed in (1), but present-day people from the Balkans have less of this ancestry than was the case from the Bronze Age through the Iron Age and down to the classical antiquity (Ancient). Medieval/Byzantine people from the Balkans were diverse, with some (right) continuing the ancient pattern of high Anatolian Neolithic ancestry, several (middle) overlapping with the range of present-day people, and some (left) having as little such ancestry as present-day Balto-Slavic people from Eastern Europe.
Phenotypes of the Southern Arc in their West Eurasian context
Our survey of populations of the Southern Arc focuses on ancestry, but it also illuminates other aspects of biology. Superficial phenotypes such as pigmentation were remarked upon by ancient writers. We carried out a survey of predicted pigmentation and other phenotypes of West Eurasian populations across time (Supplementary Text S3; Fig. 6) (18) to discover the extent to which ancient authors’ perceptions (based on direct observation or through reports of faraway peoples) might correspond to the genetic inference of their appearance (41). We find that the modal phenotype of eye, skin, and hair pigmentation in ancient West Eurasians was brown-eyed, of intermediate complexion, and brown hair—even among Yamnaya steppe pastoralists—contradicting stereotypical characterizations of Steppe peoples as being blue-eyed, pale-skinned, and light-haired (42, 43). Note that when we use categorizations—such as “intermediate”—of the continuous skin tone phenotype, we use the scheme adopted by HIrisPlex-S (41); in that scheme “intermediate” skin tones are commonly found in present-day Mediterranean populations and “pale” ones in present-day Northern European ones. A general depigmentation trend can be seen across time (Fig. 6) with a reduction of black hair and darker skin tones accompanying the increase of brown hair and intermediate skin tones. However, inhabitants of the Southern Arc had significantly darker pigmentation on average than those of the north (defined as Europe outside the Southern Arc and the Eurasian steppe) over all periods (Fig. 6), providing support for the identification by ancient writers of light pigmentation phenotypes as being more common in some groups of the north such as Celts and Scythians. Another contrast made by ancient writers was with people of Africa, such as Egyptians and Ethiopians, who were said to be of darker pigmentation (e.g., Hdt. 2.104); a comparison of people of the Southern Arc with their southern neighbors will become possible when genomic data from people living south of the Mediterranean become available. When examining composite pigmentation phenotypes (Fig. 6D), we observe that while average pigmentation did indeed differentiate between populations of the Southern Arc and the north, light phenotypes were found in both areas at similar early dates, growing in parallel in the more recent millennia of history. Light pigmentation in West Eurasia was the result of selection across time which continued into the Historical period(44, 45), and not the survival of supposed ancient Indo-Europeans phenotypes as some 19th /20th century writers supposed (42, 43) or the product of the direct influence of climate that some Greco-Roman writers hypothesized in order to explain patterns they observed during their own time (18). The malleability of human phenotypes across time, and the presence of diverse ones—whether dark, light, or interemediate—across space undermine prejudiced views of history that overemphasize superficial traits at the expense of the more meaningful aspects of human culture and biology.
Fig. 6: Pigmentation in West Eurasia.
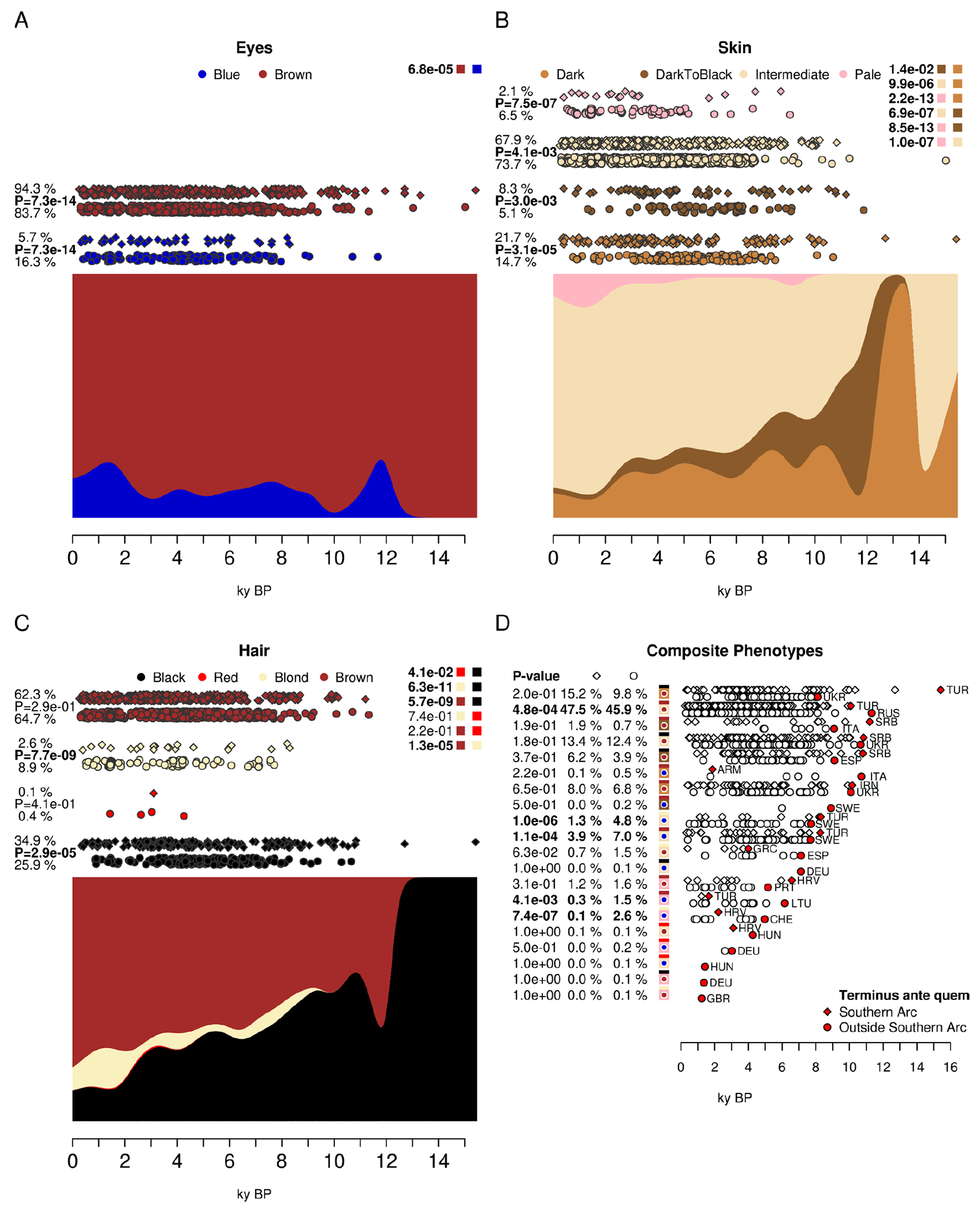
We show the temporal distribution of genetically predicted Eye (A), Skin (B), and Hair (C) color in West Eurasians of the last 16,000 years; each point represents an individual, with the top row for each subphenotype corresponding to Southern Arc and the bottom row corresponding to northern, central and western Europeans and people of the Eurasian steppe. Panel (D) shows composite phenotypes of all three aspects of pigmentation using the same color scheme as A-C and denoted as eye color (circle), hair color (top), and skin color (bottom) in the composite phenotype symbols. The modal phenotype of West Eurasians had brown eyes, intermediate skin pigmentation, and brown hair, with the highest prevalence (Fisher’s exact test) of low pigmentation outside the Southern Arc (in the rest of Europe and the Eurasian steppe).
This study illustrates the potential of archaeogenetic study of people of the civilizations of the ancient world in conjunction with archaeological and textual evidence. Ancient writings are replete with the descriptions of little-known groups, such as the numerous tribes encountered by Xenophon the Athenian at the end of the 5th c. BCE and recorded in his Anabasis, as he and his fellow mercenaries escaped from Mesopotamia northward to the Black Sea. To what extent did these and other named entities of antiquity correspond to ancestral groups that may one day be placed on the genetic landscape of the ancient world? Ancient DNA is bringing some of the stories of these forgotten peoples back to life and paying homage to their legacies.
Supplementary Material
Ethics Statement and Acknowledgments:
This study was carried following the principles for ethical DNA research on human remains laid out in (46). We are grateful to the authorities and sample stewards including museums, museum curators, and archaeologists, for providing written permission to sample each human remain. We acknowledge the ancient individuals whose genetic data we analyzed and whose permission we could not directly ask. We aimed to write a manuscript that was respectful of the ancient individuals, treating samples from them as derived from real people whose memories must be respected. We sought to reflect the perspectives of people from the diverse geographic regions and cultural contexts from which the sampled individuals came by having each sample be represented by at least one co-author who was a sample steward and was part of a network engaged with local communities. We thank J. Bennett, V. Narasimhan, H. Ringbauer; J. Sedig, A. Shaus, L. Vokotopoulos, M. Wiener, and several anonymous reviewers for critical comments.
Funding:
The newly reported dataset is described in detail in an accompanying manuscript where we also acknowledge the funders who supported dataset generation (1). Analysis of data was supported by the National Institutes of Health (NIGMS GM100233), the John Templeton Foundation (grant 61220), by a private gift from Jean-Francois Clin, by the Allen Discovery Center program, a Paul G. Allen Frontiers Group advised program of the Paul G. Allen Family Foundation and the Howard Hughes Medical Institute (DR).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and Materials availability:
Genotype data for individuals included in this study can be obtained from the Harvard Dataverse repository through the following link (doi to be added upon publication). BAM files of aligned reads can be obtained from the European Nucleotide Archive (Accession number PRJEB54831).
References and Notes
- 1.Lazaridis I, Alpaslan-Roodenberg S et al. , The genetic history of the Southern Arc: a bridge between West Asia and Europe (in submission), (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazaridis I, Alpaslan-Roodenberg S et al. , Ancient DNA from Mesopotamia suggests distinct Pre-Pottery and Pottery Neolithic migrations into Anatolia. (in submission), (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perseus Digital Library. Ed. Gregory R. Crane Tufts University. http://www.perseus.tufts.edu (accessed 2021-2022). [Google Scholar]
- 4.Lazaridis I. et al. , Genetic origins of the Minoans and Mycenaeans. Nature 548, 214–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathieson I. et al. , Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofmanová Z. et al. , Early farmers from across Europe directly descended from Neolithic Aegeans. Proceedings of the National Academy of Sciences 113, 6886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathieson I. et al. , The genomic history of southeastern Europe. Nature 555, 197–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis JL, Stocker SR, The Lord of the Gold Rings: The Griffin Warrior of Pylos. Hesperia: The Journal of the American School of Classical Studies at Athens 85, 627–655 (2016). [Google Scholar]
- 9.Jones ER et al. , Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nature Communications 6, 8912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haak W et al. , Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazaridis I et al. , Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemente F. et al. , The genomic history of the Aegean palatial civilizations. Cell, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamov D, Gurianov VM, Karzhavin S, Tagankin V, Urasin V, Defining a New Rate Constant for Y-Chromosome SNPs based on Full Sequencing Data. Russian Journal of Genetic Genealogy 7, 1920–2997 (2015). [Google Scholar]
- 14.Jack LD, Sharon RS, The Lord of the Gold Rings: The Griffin Warrior of Pylos. Hesperia: The Journal of the American School of Classical Studies at Athens 85, 627–655 (2016). [Google Scholar]
- 15.Wang C-C et al. , Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nature Communications 10, 590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson N. et al. , Large-scale migration into Britain during the Middle to Late Bronze Age. Nature 601, 588–594 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakellariou MB, Les proto-grecs. (Ekdotik?? Athenon, Athens, 1980). [Google Scholar]
- 18. Detailed information is provided in the supplementary materials. [Google Scholar]
- 19.Yaka R. et al. , Variable kinship patterns in Neolithic Anatolia revealed by ancient genomes. Current Biology, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ringbauer H, Novembre J, Steinrücken M, Parental relatedness through time revealed by runs of homozygosity in ancient DNA. Nature Communications 12, 5425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olalde I. et al. , The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldman M. et al. , Ancient DNA sheds light on the genetic origins of early Iron Age Philistines. Science Advances 5, eaax0061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diakonoff J, Starostin S. A. e., Hurro-Urartian as an Eastern Caucasian Language. Muenchener Studien zur sprachwissenschaft, (1986). [Google Scholar]
- 24.Vagheesh M. Narasimhan et al. The formation of human populations in South and Central Asia. Science 365, eaat7487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kassian AS et al. , Rapid radiation of the inner Indo-European languages: an advanced approach to Indo-European lexicostatistics. Linguistics 59, 949–979 (2021). [Google Scholar]
- 26.Underhill PA et al. , The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. European Journal of Human Genetics 23, 124–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grugni V. et al. , Ancient Migratory Events in the Middle East: New Clues from the Y-Chromosome Variation of Modern Iranians. PLOS ONE 7, e41252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta S. et al. , Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 78, 202–221 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonio ML et al. , Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science 366, 708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saupe T. et al. , Ancient genomes reveal structural shifts after the arrival of Steppe-related ancestry in the Italian Peninsula. Curr Biol 31, 2576–2591.e2512 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Skourtanioti E. et al. , Genomic History of Neolithic to Bronze Age Anatolia, Northern Levant, and Southern Caucasus. Cell 181, 1158–1175.e1128 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Unterländer M. et al. , Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nature Communications 8, 14615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damgaard P. d. B. et al. , 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Krzewińska M. et al. , Ancient genomes suggest the eastern Pontic-Caspian steppe as the source of western Iron Age nomads. Science Advances 4, eaat4457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazaridis I. et al. , Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralph P, Coop G, The Geography of Recent Genetic Ancestry across Europe. PLOS Biology 11, e1001555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busby George B. J. et al. , The Role of Recent Admixture in Forming the Contemporary West Eurasian Genomic Landscape. Current Biology 25, 2518–2526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.James E, Europe’s Barbarians, AD 200-600. (Pearson Longman, 2009). [Google Scholar]
- 39.Olalde I. et al. , Cosmopolitanism at the Roman Danubian Frontier, Slavic Migrations, and the Genomic Formation of Modern Balkan Peoples. bioRxiv, 2021.2008.2030.458211 (2021). [Google Scholar]
- 40.Patterson N. et al. , Ancient Admixture in Human History. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaitanya L. et al. , The HIrisPlex-S system for eye, hair and skin colour prediction from DNA: Introduction and forensic developmental validation. Forensic Science International: Genetics 35, 123–135 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Poliakov L, Le mythe aryen. essai sur les sources du racisme et du nationalisme (Calmann-Levy, Paris, 1971). [Google Scholar]
- 43.Day JV, Indo-European Origins: The Anthropological Evidence. (Institute for the Study of Man, 2001). [Google Scholar]
- 44.Ju D, Mathieson I, The evolution of skin pigmentation-associated variation in West Eurasia. Proceedings of the National Academy of Sciences 118, e2009227118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilde S. et al. , Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proceedings of the National Academy of Sciences 111, 4832–4837 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alpaslan-Roodenberg S. et al. , Ethics of DNA research on human remains: five globally applicable guidelines. Nature 599, 41–46 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baringhaus L, Franz C, On a new multivariate two-sample test. Journal of Multivariate Analysis 88, 190–206 (2004). [Google Scholar]
- 48.Fu Q et al. , The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van de Loosdrecht M. et al. , Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Lazaridis I. et al. , Paleolithic DNA from the Caucasus reveals core of West Eurasian ancestry. bioRxiv, 423079 (2018). [Google Scholar]
- 51.de Barros Damgaard P et al. , The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 360, eaar7711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narasimhan VM et al. , The formation of human populations in South and Central Asia. Science 365, eaat7487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moorjani P. et al. , A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proceedings of the National Academy of Sciences 113, 5652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amorim CEG et al. , Understanding 6th-century barbarian social organization and migration through paleogenomics. Nature Communications 9, 3547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marcus JH et al. , Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia. Nature Communications 11, 939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panzanelli R, Schmidt ED, Lapatin KDS, Museum JPG, The color of life : polychromy in sculpture from antiquity to the present. (J. Paul Getty Museum : The Getty Research Institute, Los Angeles, 2008). [Google Scholar]
- 57.Mallory JP, Mair VH, Thames Hudson, The Tarim mummies : ancient China and the mystery of the earliest peoples from the West. (Thames & Hudson, New York, 2008). [Google Scholar]
- 58.de Gobineau A, Essai sur l’Inégalité des Races humaines. (F. Didot, 1855). [Google Scholar]
- 59.de Lapouge GV, L’Aryen; son rôle social. (A. Fontemoing, 1899). [Google Scholar]
- 60.Grant M, The passing of the great race or, The racial basis of European history. (1916). [Google Scholar]
- 61.Chamberlain HS, Die Grundlagen des neunzehnten Jahrhunderts. (Bruckmann, München, 1898). [Google Scholar]
- 62.Spiro JP, Defending the master race : conservation, eugenics, and the legacy of Madison Grant. (2009). [Google Scholar]
- 63.Stoddard L, The rising tide of color against white world-supremacy. (Charles Scribner’s Sons, New York, 1920). [Google Scholar]
- 64.Günther HFK, Wheeler GCWC, The racial elements of European history. (E.P. Dutton and Co., New York, 1927). [Google Scholar]
- 65.Günther HFK, Rassenkunde des deutschen Volkes. (1937). [Google Scholar]
- 66.Chapoutot J, Nybakken RR, Greeks, Romans, Germans : how the Nazis usurped Europe’s classical past. (2017). [Google Scholar]
- 67.Peterson R, The Greek Face. Journal of Indo-European Studies 2, 385–406 (1974). [Google Scholar]
- 68.Day JV, Indo-European origins : the anthropological evidence. (Institute for the Study of Man, Washington, D.C., 2001). [Google Scholar]
- 69.Skaarup BO, Physical anthropology and the Aryan question: A historical overview and a few words of warning. in LANGUAGE AND PREHISTORYOF THE INDO-EUROPEAN PEOPLES: A cross-disciplinary perspective, 173–186 (2017). [Google Scholar]
- 70.Walsh S. et al. , Global skin colour prediction from DNA. Hum Genet 136, 847–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh S et al. , Developmental validation of the HIrisPlex system: DNA-based eye and hair colour prediction for forensic and anthropological usage. Forensic Sci Int Genet 9, 150–161 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Harney É et al. , Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nature Communications 9, 3336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allentoft ME et al. , Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Järve M et al. , Shifts in the Genetic Landscape of the Western Eurasian Steppe Associated with the Beginning and End of the Scythian Dominance. Current Biology 29, 2430–2441.e2410 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Schroeder H et al. , Unraveling ancestry, kinship, and violence in a Late Neolithic mass grave. Proceedings of the National Academy of Sciences 116, 10705–10710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mittnik A. et al. , The genetic prehistory of the Baltic Sea region. Nature Communications 9, 442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frost P, European hair and eye color: A case of frequency-dependent sexual selection? Evolution and Human Behavior 27, 85–103 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype data for individuals included in this study can be obtained from the Harvard Dataverse repository through the following link (doi to be added upon publication). BAM files of aligned reads can be obtained from the European Nucleotide Archive (Accession number PRJEB54831).


