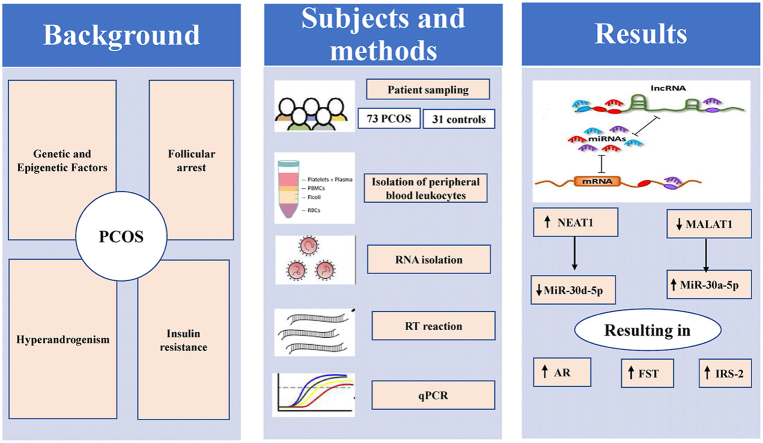
Abstract
Accumulating evidence has shown an abnormal expression of several non-coding RNAs in ovarian tissues which might be closely linked with the pathogenesis of PCOS. The aim of this study was to identify competing endogenous (ce) RNA network: long non-coding RNA (lncRNA), microRNA (miRNA) and their target genes: androgen receptor (AR), follistatin (FST) and insulin receptor substrate-2 (IRS-2), which are relevant to PCOS, to underline the molecular pathogenesis of PCOS and assist in early diagnosis and treatment. Bioinformatic analysis was performed to retrieve a ceRNA network: [lncRNA (NEAT1 and MALAT1) - miRNA (miR-30a-5p and miR-30d-5p) - mRNA (AR, FST and IRS-2)] linked to PCOS. Expression of the selected RNAs was examined by qPCR in peripheral blood leukocytes obtained from 73 PCOS patients (41 obese and 32 non-obese) and 31 healthy controls. PCOS patients showed significantly higher expression levels of NEAT1, miR-30a-5p, AR, FST and IRS-2, with significantly lower expression levels of MALAT1 and miR-30d-5p relative to controls especially in obese versus non-obese patients. Receiver operating characteristic (ROC) curve analysis indicated that most of the selected RNAs could serve as potential early diagnostic markers for PCOS with the highest efficiency obtained upon combining NEAT1 and miR-30d-5p or MALAT1 and miR-30a-5p with either of PCOS target genes. Moreover, all addressed RNAs had been proved as potential predictors of PCOS. The obtained data of ceRNA network raised the possibility that NEAT1 overexpression may increase the expression levels of AR, FST and IRS-2 by sponging miR-30d-5p, while low expression of MALAT1 may allow higher expression of the above genes via increasing miR-30a-5p, suggesting their involvement in PCOS pathogenesis and promising role for future diagnosis and targeted therapy.
Keywords: Polycystic ovary syndrome, NEAT1, MALAT1, Androgen receptor, Follistatin and insulin receptor substrate-2
Graphical abstract
Highlights
-
•
High NEAT1 may increase AR, FST and IRS-2 expression by sponging miR-30d-5p.
-
•
Low MALAT1 may increase expression of the above genes via increasing miR-30a-5p.
-
•
The addressed RNAs were proved as potential PCOS-diagnostic markers and predictors.
1. Introduction
Polycystic ovary syndrome (PCOS) represents one of the most common heterogenous reproductive and metabolic disorders affecting about 5–10% of women during their reproductive age and 75% of the anovulatory infertility worldwide [1,2]. The major clinical features of PCOS include: hyperandrogenism, irregular menstruation, chronic anovulation, polycystic ovarian morphology, obesity, follicular arrest as well as insulin resistance with hyperinsulinemia [3]. PCOS patients have higher risk for afflicting diabetes, dyslipidemia, cardiovascular diseases, infertility and other medical problems.
Among the difficulties for timely treatment and management of PCOS are the complexity of the disease resulting from its multiple underlying mechanisms and lack of sensitive and specific biomarkers for early detection, prognosis and potential therapy. Hence, enhanced understanding of PCOS pathogenesis would aid in the early detection and optimal disease management. In PCOS, large number of biochemical and molecular pathways, causing follicular arrest and ovulatory dysfunction were previously linked to hyperandrogenism, hyperinsulinemia and to an imbalance between ovarian activin and follistatin (FST) [4].
Hyperandrogenism has been arised from high level of testosterone and/or increased androgen receptor (AR) expression and activity [5]. Excessive androgen production is mainly linked to hyperinsulinemia and elevated insulin like growth factor-1 (IGF-1) level which may account for irregularity in pulsatile secretion of hypothalamic gonadotropin releasing hormone (GnRH) with subsequent hypersecretion of luteinizing hormone (LH), leading to ovulatory dysfunction and polycystic ovarian morphology [3,6]. In spite of an increased number of follicles in PCOS compared to normal ovaries, their growth is prematurely arrested, resulting in failure of ovulation [7]. In this respect, Suganthi et al. [8] have demonstrated an imbalance between ovarian activin and FST in their action, where FST can bind to activin suppressing its role in stimulating the secretion of follicle stimulating hormone (FSH) resulting in ovarian follicular arrest.
Moreover, Insulin and IGF-1 initiate their signal transduction through phosphorylation of tyrosine residues on insulin receptor substrate (IRS)-1 and IRS-2 [9]. IRS-1 appears to mediate mainly the metabolic pathways in peripheral tissues such as: skeletal muscle and adipose tissue, whereas IRS-2 has a potential role in ovarian mitogenic and steroidogenic responses [10]. In PCOS, there is a selective impairment in metabolic response, along with enhanced ovarian mitogenic and steroidogenic responses [9]. Thus, it is reasonable to anticipate that the alteration in IRS-2 expression may account for impaired downstream signaling pathways for insulin action and ovarian function in PCOS. Nevertheless, molecular mechanisms responsible for PCOS pathogenesis still deserve further investigation.
Remarkably, genetic and epigenetic variations may contribute considerably to most of the alterations in PCOS [3]. Non-coding RNAs, a class of RNA transcripts without protein-coding capacity have emerged as regulatory molecules in wide variety of biological processes. Lately, they are increasingly appreciated for their involvement in the regulation of PCOS-related molecular processes [11]. A lot of evidence has indicated that multiple microRNAs (miRNAs) (miR-122, miR-140, miR-193 b, miR-320 and miR-592) were abnormally expressed in follicular fluid (F·F.) and serum of PCOS patients suggesting their participation in the regulatory pathways of follicular development, androgen synthesis and insulin sensitivity [12]. Importantly, miR-30 family was identified as an epigenetic regulator of AR, FST and IRS-2 expression in diseases other than PCOS [[13], [14], [15], [16]]. Among the miR-30 family, miR-30a-5p expression has been reported to be increased in clinical [17] and experimental PCOS [18]. To date, reports demonstrating the expression of miR-30d-5p in PCOS patients are still lacking.
On the other side, the function of long non-coding RNAs (lncRNAs) in PCOS pathogenesis is just beginning to be understood. Recently, several lncRNAs have been detected in PCOS and in various biological and pathological processes of granulosa cells (GCs), F·F, peripheral blood and ovaries suggesting their involvement in follicular development [11]. Interestingly, miR-30 family has been demonstrated to form complementary base pairing with these lncRNAs: nuclear enriched abundant transcript 1 (NEAT1) and metastasis associated lung adenocarcinoma transcript 1 (MALAT1) [[19], [20], [21]]. NEAT1 and MALAT1 have been detected in early ovarian follicles, where they can function as miRNA sponges or competing endogenous (ce) RNAs, hence minimizing the availability of miRNAs for their target mRNAs [[22], [23], [24], [25]]. Accordingly, this evidence suggests that NEAT1 and MALAT1 may be involved in PCOS development, possibly via cooperative interaction with a set of miRNAs to modulate PCOS target genes.
Therefore, the aim of this study was directed to develop a network-based system of three types of RNAs: lncRNA (NEAT1 and MALAT1), miRNA (miR-30a-5p and miR-30d-5p), mRNA (AR, FST and IRS-2) and to explore the interactive connections among the three types of RNAs in regulation of PCOS pathogenesis, hoping that such work may aid in early detection and optimal disease management.
2. Subjects and methods
2.1. Studied population
This study included 104 women (age range: 20–35 years) recruited from Kasr Al-Ainy hospital (Department of Obstetrics and Gynecology), Cairo, Egypt between July 2021 to February 2022. Of these 104 women, 73 were PCOS patients and 31 were age-matched healthy women as control group. All women provided informed consent before sample collection and use for research purposes under the protocol approved by the ethical committee of Faculty of Pharmacy, Cairo University (BC 2531) and in accordance with the ethical standard laid down in the Helsinki declaration (2008).
PCOS patients were diagnosed in accordance with the 2003 Rotterdam criteria, which requires the presence of two or more of the following features: clinical or biochemical evidence of hyperandrogenism, oligoanovulation (oligomenorrhoea or amenorrhea) and polycystic ovaries upon ultrasonography (at least 12 follicles measuring 2–9 mm or an ovarian volume >10 cm3). On the other side, controls had no evidence of clinical or biochemical hyperandrogenism and no history of menstrual cycle irregularities or other endocrine disorders related to PCOS.
None of the participants had taken any hormonal drugs, such as oral contraceptives, antiandrogens or insulin sensitizers for at least 3 months before the study. The PCOS patients were divided into women with normal weight (body mass index (BMI) < 25 kg/m2) and obese women (BMI ≥30 kg/m2). Participants were excluded from the study if they had Cushing's syndrome, congenital adrenal hyperplasia, thyroid dysfunction and androgen secreting tumors. Physical examinations were carried out on all participants and data were collected including: age, weight, height and BMI. All patients also received transvaginal ultrasonography to identify the presence of polycystic ovarian morphology.
2.2. Blood and follicular fluid sampling
5 ml intravenous blood samples were collected from all participants after an overnight 12-h fasting at an early follicular phase to minimize hormonal fluctuations from third to fifth days of a spontaneous menstrual cycle or progesterone-induced withdrawal bleed between 8:00 a.m. and 10:00 a.m. These blood samples were divided into 2 portions, one portion was used to obtain serum and stored at −80 °C until further analysis and the other was collected in ethylene diamine tetraacetic acid (EDTA) coated tubes and used for isolation of peripheral blood leukocytes. In addition, F.F. was obtained at the time of oocyte retrieval during in vitro fertilization (IVF) from 10 PCOS patients and 5 healthy women as controls who visited the IVF unit because of male factors related infertility in order to confirm the reliability of peripheral blood leukocytes results.
2.3. Biochemical analysis
Serum was used to determine the following parameters: LH, FSH, estradiol (E2), total testosterone (TT), fasting insulin and glucose, total cholesterol, triglycerides (TGs), high-density lipoprotein (HDL)-cholesterol and low-density lipoprotein (LDL)-cholesterol. Serum concentrations of LH, FSH, E2, TT and fasting insulin were determined using Enzyme linked immunosorbent assay (ELISA) kits (Biovision, USA for LH and FSH, Cat. No, K7426-100 and Cat. No, K7425-100, respectively, Origene, USA for E2, Cat. No, EA100858, Elabscience, USA for TT, Cat. No, E-EL-0155 and Raybiotech, USA for fasting insulin, Cat. No, ELH-Insulin-1) by Tecan, A-5082 ELISA reader at wavelength 450 nm for all of them according to the manufacturer's instructions. Their concentrations were presented as mIU/ml for LH and FSH, pg/ml for E2, ng/ml for TT and μIU/ml for fasting insulin. Serum fasting glucose concentration was measured colorimetrically using glucose oxidase method (Spectrum, Germany, Cat. No, 250 004) by Jenway 6051 at wavelength 520 nm presented as mg/dl. The lipid profile was measured using an automated colorimetric method by Beckman Coulter AU480 chemistry analyzer presented as mg/dl.
2.4. Isolation of peripheral blood leukocytes
For each subject, 2 ml of blood were diluted with an equal volume of phosphate-buffered saline (PBS), and then overlaid onto a Ficoll reagent (Histopaque, Sigma Aldrich, UK, Cat. No, 10 771) in 1:1 ratio. Each sample was then centrifuged at 800×g for 30 min at 4 °C using Hermle Z 323 k cooling centrifuge. After centrifugation, the middle layer containing the mononuclear cells was obtained and washed twice with PBS to remove Ficoll reagent and plasma. Then these samples were stored at −80 °C until further analysis.
2.5. Bioinformatics analysis and rational of lncRNAs, miRNAs and mRNAs selection
Initially, the Gene Atlas [26] was used to identify the relevant protein-coding genes involved in the pathogenesis of PCOS. Second, miR-30a-5p and miR-30d-5p were selected because they have been identified as epigenetic regulators of the AR, FST and IRS-2 genes (Diana tools [27] and TargetScan [28]). Using the Diana tools database [29], the lncRNAs NEAT1 and MALAT1 were selected as ce for miR-30a-5p and miR-30d-5p. Moreover, the association between the lncRNAs and disease was investigated using lncRNA disease database [30].
2.6. RNA isolation and reverse transcription
Total RNA was extracted from isolated peripheral blood leukocytes and follicular fluid samples using Direct-zol RNA Miniprep extraction kit (Zymo research, USA, Cat. No, R2050-1-50) in accordance with the manufacturer's instructions. The concentration and purity of the resultant RNA was then determined using Implen nanophotometer N50 UV–Vis spectrophotometer at 260/280 nm, so that a ratio of more than 1.8 was considered indicative of an acceptable yield and purity of RNA. Next, NEAT1, MALAT1, AR, FST and IRS-2 were reverse-transcribed using Omiscript RT kit (Qiagen, Germany, Cat. No, 205 111) using the following cycle: 60 min at 37 °C. While, miR-30a-5p and miR-30d-5p were reverse-transcribed using miRCURY LNA miRNA PCR starter kit (Qiagen, Germany, Cat. No, 339 320) using the following cycle: 60 min at 42 °C for reverse transcription, 5 min at 95 °C to inactivate the reaction. Reverse transcription reactions were carried out by Step One Real-time PCR system (Applied Biosystems, USA).
2.7. Quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction (qPCR) was carried to measure the expression levels of the following: NEAT1, MALAT1, AR, FST and IRS-2 using QuantiNova SYBR Green PCR kit (Qiagen, Germany, Cat. No, 208 052) in accordance with the manufacturer's instructions with specific primers (eurofins, Germany). The primers' sequences utilized for PCR reactions of NEAT1, MALAT1, AR, FST and IRS-2 are illustrated in Table 1. PCR reactions were carried out by Step One Real-time PCR system with the following cycling conditions: 2 min at 95 °C for PCR initial heat activation step, 5 s at 95 °C for denaturation and PCR amplification for 10 s at 60 °C (40 cycles).
Table 1.
Primers' sequences used in qPCR reactions of the selected lncRNAs and target mRNAs.
| RNA | Primer sequence |
|---|---|
| β-actin | Forward 5′-CCTGTACGGTCCACTGCTTA-3′ |
| Reverse 5′-TGGACTTGCATCCAGGTTCA-3′ | |
| NEAT1 | Forward 5′-TTACCAGCTTCCTCCTGGTG-3′ |
| Reverse 5′-TCTGCTGCGTATGCAAGTCT-3′ | |
| MALAT1 | Forward 5′-ATGCGAGTTGTTCTCCGTCT-3′ |
| Reverse 5′-TATCTGCGGTTTCCTCAAGC-3′ | |
| AR | Forward 5′-CCTGGCTTCCGCAACTTACAC-3′ |
| Reverse 5′-GGACTTGTGCATGCGGTACTCA-3′ | |
| FST | Forward 5′-CTCTGCCAGTTCATGGAGGA-3′ |
| Reverse 5′-TCCTTGCTCAGTTCGGTCTT-3′ | |
| IRS-2 | Forward5′-CAAAAGCCATCTCGGTGTAGT-3′ |
| Reverse 5′-GCTCTCCGACTACATGAACCTC-3′ |
NEAT1: nuclear enriched abundant transcript 1, MALAT1: metastasis associated lung adenocarcinoma transcript 1, AR: androgen receptor, FST: follistatin and IRS-2: insulin receptor substrate-2.
In addition, miR-30a-5p and miR-30d-5p expression levels were measured using miRCURY LNA miRNA PCR starter kit in which ready-made primers were provided in accordance with the manufacturer's instructions. PCR reactions were carried out by Step One Real-time PCR system using the following cycling conditions: 2 min at 95 °C for PCR initial heat activation step, 10 s at 95 °C for denaturation and PCR amplification for 60 s at 56 °C (40 cycles). Melting curve analysis was used to determine the specificity of the reactions. Relative expression levels of the studied RNAs in each sample were then determined by the 2−ΔΔCT method using β-Actin as a house-keeping reference gene for NEAT1, MALAT1, AR, FST and IRS-2, whereas miR-103a-3p was used as a reference gene for miR-30a-5p and miR-30d-5p.
2.8. Statistical analysis
Shapiro-Wilk test was used to check whether the data obey normal distribution. Normally distributed data were compared using either unpaired student's t-test for 2 groups or one way analysis of variance (ANOVA) followed by post hoc Tukey's test for 3 groups. Data which did not follow normal distribution were compared using either Mann-Whitney U test for 2 groups or Kruskal-Wallis followed by post hoc Dunn's test for 3 groups. Spearman's rank correlation test was used to search for linear relationships between the studied RNAs expression and clinical as well as biochemical characteristics. The diagnostic value of RNAs levels in peripheral blood leukocytes was evaluated using a receiver operating characteristic (ROC) curve, which analyzed PCOS patients' data as truly positive and healthy women data as truly negative. The sensitivity and specificity of detection were evaluated by the area under the ROC curve (AUC). The positivity rates for each of the studied RNA were then determined by Chi-square (χ2) test. Univariate analysis with logistic regression was used to evaluate the predictive power of the studied RNAs for PCOS. P < 0.05 was considered statistically significant. All statistical calculations were done using computer program IBM SPSS (Statistical Package for the Social Science; IBM Corp, Armonk, NY, USA) version 26 and Microsoft Windows and GraphPad prism 6.
3. Results
3.1. Anthropometric, biochemical and hormonal characteristics of the study population
No statistically significant differences were observed in age, height, FSH and E2 between controls and PCOS patients as shown in Table 2. In comparison to controls, PCOS patients showed significantly higher weight, BMI, LH, LH/FSH, TT, fasting insulin and glucose,HOMA-IR, total cholesterol, TGs and LDL, with significantly lower HDL. Importantly, most of these changes were mainly contributed by obese PCOS.
Table 2.
Anthropometric, biochemical and hormonal data of the studied participants.
| Parameter | Controls (n = 31) | PCOS (n = 73) |
||
|---|---|---|---|---|
| All | Non-obese PCOS (n = 32) | Obese PCOS (n = 41) | ||
| Age (years) | 28.87 ± 0.77 | 27.9 ± 0.54 | 26.88 ± 0.73 | 28.71 ± 0.76 |
| Weight (kg) | 64.77 ± 2.19 | 78.05 ± 2.007*** | 61.22 ± 0.99 | 91.2 ± 1.56 b |
| Height (m) | 1.61 ± 0.007 | 1.62 ± 0.005 | 1.62 ± 0.008 | 1.63 ± 0.006 |
| BMI (kg/m2) | 24.81 ± 0.72 | 29.44 ± 0.71*** | 23.24 ± 0.22 | 34.27 ± 0.54 b |
| LH (mIU/ml) | 5.76 ± 0.25 | 11.69 ± 0.36*** | 11.55 ± 0.42 | 11.8 ± 0.55 |
| FSH (mIU/ml) | 5.87 ± 0.25 | 5.28 ± 0.16 | 5.47 ± 0.202 | 5.14 ± 0.25 |
| LH/FSH | 0.98 ± 0.01 | 2.22 ± 0.02*** | 2.11 ± 0.02 | 2.31 ± 0.04b |
| E2 (pg/ml) | 46.46 ± 2.36 | 48.79 ± 1.47 | 49.96 ± 2.41 | 47.88 ± 1.83 |
| TT (ng/ml) | 0.303 ± 0.02 | 0.71 ± 0.04*** | 0.61 ± 0.05 | 0.84 ± 0.05 a |
| Fasting insulin (μIU/ml) | 8.203 ± 0.109 | 12.07 ± 0.509*** | 9.33 ± 0.29 | 14.2 ± 0.72 b |
| Fasting glucose (mg/dl) | 81.19 ± 1.56 | 109 ± 4.54*** | 84.73 ± 2.707 | 127.9 ± 6.44 b |
| HOMA-IR | 1.65 ± 0.05 | 3.65 ± 0.34*** | 2.01 ± 0.12 | 4.94 ± 0.51b |
| Total cholesterol (mg/dl) | 154.1 ± 2.73 | 182.7 ± 3.38*** | 160 ± 3.78 | 200.4 ± 31.89 b |
| TGs (mg/dl) | 94.45 ± 2.48 | 136.6 ± 4.16*** | 110.3 ± 4.88 | 157.2 ± 4.14 b |
| HDL (mg/dl) | 50.84 ± 1.209 | 46.04 ± 0.97** | 50.13 ± 1.303 | 42.85 ± 1.2 b |
| LDL (mg/dl) | 84.32 ± 2.26 | 112.9 ± 2.86*** | 90.91 ± 3.289 | 130.1 ± 1.72 b |
Quantitative data are represented as mean ± SE, BMI: body mass index, LH: luteinizing hormone, FSH: follicle stimulating hormone, E2: estradiol, TT: total testosterone, HOMA-IR: homeostasis model assessment for insulin resistance, TGs: triglycerides, HDL: high-density lipoprotein and LDL: low-density lipoprotein. Significantly different at **P < 0.01 and ***P < 0.001 versus controls and significantly different at aP < 0.01 and bP < 0.001 versus non-obese PCOS.
3.2. Relative expression levels of the studied RNAs in peripheral blood leukocytes and F·F. Samples
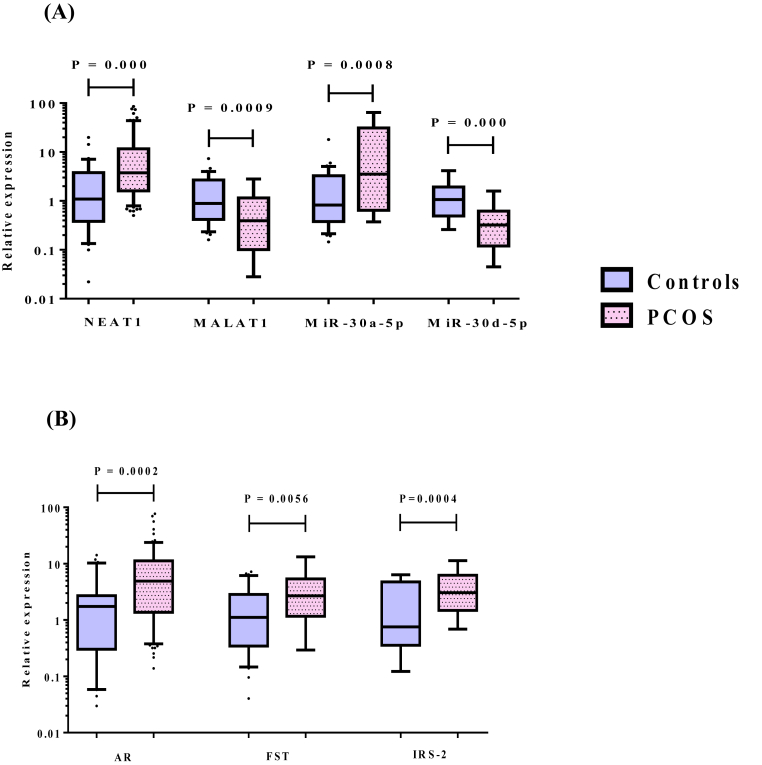
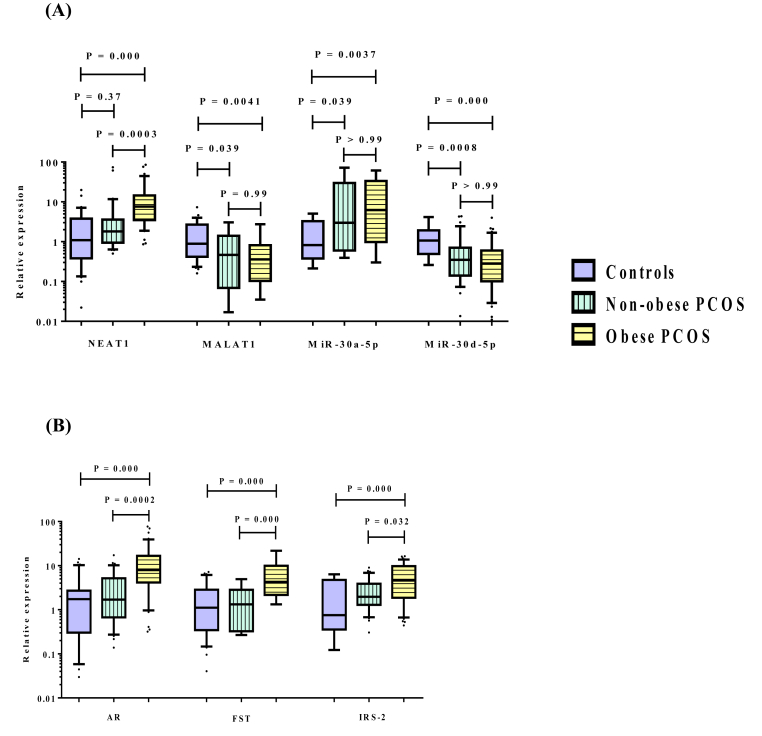
Compared with the healthy controls, PCOS patients demonstrated significantly higher expression levels of lncRNA NEAT1, miR-30a-5p as well as the mRNAs of AR, FST and IRS-2, with lower expression levels of lncRNA MALAT1 and miR-30d-5p in peripheral blood leukocytes as depicted in Fig. 1. Similarly, the expression levels of the above selected RNAs in F.F. samples of PCOS patients revealed remarkable comparable changes to those observed in peripheral blood leukocytes when compared to controls (Supplementary Table 1). Notably, the significant elevations in expression levels of NEAT1, AR, FST and IRS-2 were chiefly observed in obese PCOS group compared to either non-obese or control group. However, the significant higher expression levels of miR-30a-5p, with the lower expression levels of MALAT1 and miR-30d-5p were found in both obese and non-obese patients compared to controls, without any significant differences detected between obese and non-obese patients as demonstrated in Fig. 2.
Fig. 1.
Relative expression levels of (A) lncRNAs (NEAT1, MALAT1) and miRNAs (miR-30a-5p, miR-30d-5p), (B) target mRNAs (AR, FST and IRS-2) in peripheral blood leukocytes obtained from controls (n = 31) and PCOS patients (n = 73). Horizontal lines inside the box plots represent the median; the boxes mark the interval between the 25th and 75th percentiles. The whiskers denote the intervals between the 10th and 90th percentiles. Filled circles indicate data points outside the 10th and 90th percentiles. NEAT1: nuclear enriched abundant transcript 1, MALAT1: metastasis associated lung adenocarcinoma transcript 1, AR: androgen receptor, FST: follistatin and IRS-2: insulin receptor substrate-2. Significant P-values are indicated on graph at p < 0.05.
Fig. 2.
Relative expression levels of (A) lncRNAs (NEAT1, MALAT1) and miRNAs (miR-30a-5p, miR-30d-5p), (B) target mRNAs (AR, FST and IRS-2) in peripheral blood leukocytes obtained from controls (n = 31), non-obese (n = 32) and obese (n = 41) PCOS patients. Horizontal lines inside the box plots represent the median; the boxes mark the interval between the 25th and 75th percentiles. The whiskers denote the intervals between the 10th and 90th percentiles. Filled circles indicate data points outside the 10th and 90th percentiles. NEAT1: nuclear enriched abundant transcript 1, MALAT1: metastasis associated lung adenocarcinoma transcript 1, AR: androgen receptor, FST: follistatin and IRS-2: insulin receptor substrate-2. Significant P-values are indicated on graph at p < 0.05.
3.3. The association of NEAT1, MALAT1, AR, FST and IRS-2 expression levels with clinical and biochemical characteristics in PCOS patients
As shown in Table 3, Spearman's rank correlation analysis was used to explore the potential relationship between the relative expression levels of NEAT1, MALAT1, AR, FST and IRS-2 and clinical as well as biochemical characteristics. The correlation analysis showed that the expression level of NEAT1 was positively correlated with BMI, TT, total cholesterol, TGs and LDL. However, no statistically significant differences were observed between the relative expression level of MALAT1 and any of these characteristics. Furthermore, AR and FST were positively correlated with BMI, TT, fasting insulin and glucose, HOMA-IR, total cholesterol, TGs and LDL; however, they were negatively correlated with HDL. Finally, IRS-2 was positively correlated only with BMI, TT, total cholesterol and TGs.
Table 3.
Spearman's rank correlation analysis of NEAT1, MALAT1, AR, FST and IRS-2 with clinical and biochemical characteristics in PCOS patients.
| Parameter | NEAT1 |
MALAT1 |
AR |
FST |
IRS-2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | P-value | r | P-value | r | P-value | r | P-value | r | P-value | |
| BMI | 0.47 | 0.000 | −0.029 | 0.809 | 0.51 | 0.000 | 0.45 | 0.000 | 0.32 | 0.005 |
| TT | 0.308 | 0.008 | 0.082 | 0.49 | 0.27 | 0.018 | 0.403 | 0.0004 | 0.28 | 0.014 |
| Fasting insulin | 0.19 | 0.108 | −0.105 | 0.37 | 0.302 | 0.009 | 0.33 | 0.004 | 0.18 | 0.12 |
| Fasting glucose | 0.17 | 0.14 | −0.108 | 0.36 | 0.29 | 0.012 | 0.29 | 0.011 | 0.18 | 0.12 |
| HOMA-IR | 0.16 | 0.15 | −0.109 | 0.36 | 0.28 | 0.013 | 0.304 | 0.009 | 0.16 | 0.16 |
| Total cholesterol | 0.48 | 0.000 | −0.05 | 0.65 | 0.37 | 0.001 | 0.41 | 0.0003 | 0.32 | 0.005 |
| TGs | 0.28 | 0.014 | 0.02 | 0.85 | 0.38 | 0.0007 | 0.43 | 0.0001 | 0.26 | 0.023 |
| HDL | −0.12 | 0.309 | −0.12 | 0.27 | −0.301 | 0.009 | −0.31 | 0.007 | −0.1 | 0.39 |
| LDL | 0.48 | 0.000 | −0.07 | 0.53 | 0.35 | 0.002 | 0.41 | 0.0003 | 0.22 | 0.061 |
r: correlation coefficient, NEAT1: nuclear enriched abundant transcript 1, MALAT1: metastasis associated lung adenocarcinoma transcript 1, AR: androgen receptor, FST: follistatin, IRS-2: insulin receptor substrate-2, BMI: body mass index, TT: total testosterone, HOMA-IR: homeostasis model assessment for insulin resistance, TGs: triglycerides, HDL: high-density lipoprotein and LDL: low-density lipoprotein. Significant P-values are indicated in the table.
3.4. Diagnostic values and positivity rates of the relative expression levels of the studied RNAs for PCOS
The potential utility of the selected RNAs as diagnostic biomarkers of PCOS in peripheral blood leukocytes was assessed by performing ROC analysis. As shown in Table 4, the diagnostic power of MALAT1, miR-30a-5p, AR, FST and IRS-2 relative expression levels was comparable to each other with AUC of 0.7, 0.71, 0.73, 0.67 and 0.72, respectively, whereas, the diagnostic power of NEAT1 and miR-30d-5p was more promising with AUC of 0.76 and 0.79, respectively. Interestingly, the highest potential efficiency was obtained upon combination of NEAT1 and miR-30d-5p or MALAT1 and miR-30a-5p with either of the selected PCOS target genes, where AUC values for all NEAT1 combinations increased to 0.8, while those of MALAT1 combinations increased to 0.86, 0.82 and 0.8, respectively. The combination of MALAT1, miR-30a-5p and AR was particularly the most efficient for better diagnosis.
Table 4.
Receiver operating characteristic analysis of the selected RNAs in the studied participants.
| RNA | AUC | P-value | Cut off value | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| NEAT1 | 0.76 | 0.000 | 1.701 | 75.3 | 67.7 | 84.6 | 53.8 |
| MALAT1 | 0.7 | 0.001 | 0.14 | 37 | 100 | 100 | 40.3 |
| MiR-30a-5p | 0.71 | 0.001 | 6.11 | 43.8 | 96.8 | 97 | 42.3 |
| MiR-30d-5p | 0.79 | 0.000 | 0.41 | 65.8 | 87.1 | 92.3 | 51.9 |
| AR | 0.73 | 0.0003 | 3.59 | 61.6 | 83.9 | 90 | 48.1 |
| FST | 0.67 | 0.006 | 3.309 | 45.2 | 83.9 | 86.8 | 39.4 |
|
IRS-2 |
0.72 |
0.0004 |
0.79 |
89 |
51.6 |
81.3 |
66.7 |
| Combination of NEAT1, miR-30d-5p and AR | 0.8 | 0.000 | – | 69.9 | 80.6 | 89.5 | 53.2 |
| Combination of NEAT1, miR-30d-5p and FST | 0.8 | 0.000 | – | 89 | 64.5 | 85.5 | 71.4 |
|
Combination of NEAT1, miR-30d-5p and IRS-2 |
0.8 |
0.000 |
– |
60.3 |
90.3 |
93.6 |
49.1 |
| Combination of MALAT1, miR-30a-5p and AR | 0.86 | 0.000 | – | 79.5 | 83.9 | 92.1 | 63.4 |
| Combination of MALAT1, miR-30a-5p and FST | 0.82 | 0.000 | – | 64.4 | 93.5 | 95.9 | 52.7 |
| Combination of MALAT1, miR-30a-5p and IRS-2 | 0.8 | 0.000 | – | 47.9 | 100 | 100 | 44.9 |
AUC: area under the curve, PPV: positive predictive value, NPV: negative predictive value, NEAT1: nuclear enriched abundant transcript 1, MALAT1: metastasis associated lung adenocarcinoma transcript 1, AR: androgen receptor, FST: follistatin and IRS-2: insulin receptor substrate-2. Significant P-values are indicated in the table.
The positivity rates for each of the selected RNAs were determined using the calculated cut-off values for the detection of PCOS as shown in Table 5. IRS-2 and NEAT1 had the highest positivity rates in PCOS patients reaching 89% and 75.3%, followed by miR-30d-5p, AR, FST, miR-30a-5p and MALAT1 with positivity rates of, 65.8%, 61.6%, 45.2%, 43.8% and 37%, respectively.
Table 5.
Positivity rates of the studied RNAs across the investigated groups.
| RNA | Controls | PCOS | χ2 | P-value |
|---|---|---|---|---|
| NEAT1 | ||||
| No. Of +ve cases (≥ 1.701) | 10 (32.3%) | 55 (75.3%) | 17.23 | 0.000 |
| MALAT1 | ||||
| No. Of +ve cases (≤ 0.14) | 0 (0%) | 27 (37%) | 15.48 | 0.000 |
| MiR-30a-5p | ||||
| No. Of +ve cases (≥ 6.11) | 1 (3.2%) | 32 (43.8%) | 16.56 | 0.000 |
| MiR-30d-5p | ||||
| No. Of +ve cases (≤ 0.41) | 4 (12.9%) | 48 (65.8%) | 24.31 | 0.000 |
| AR | ||||
| No. Of +ve cases (≥ 3.59) | 5 (16.1%) | 45 (61.6%) | 18.05 | 0.000 |
| FST | ||||
| No. Of +ve cases (≥ 3.309) | 5 (16.1%) | 33 (45.2%) | 7.93 | 0.005 |
| IRS-2 | ||||
| No. Of +ve cases (≥ 0.79) | 15 (48.4%) | 65 (89%) | 20.25 | 0.000 |
χ2: chi-square, NEAT1: nuclear enriched abundant transcript 1, MALAT1: metastasis associated lung adenocarcinoma transcript 1, AR: androgen receptor, FST: follistatin and IRS-2: insulin receptor substrate-2. Significant differences were detected between the investigated groups using chi-square (χ2) test. Significant P-values are indicated in the table.
3.5. Univariate regression analysis of the predictive power of the studied RNAs for PCOS
Univariate logistic regression analysis was conducted to evaluate the predictive values of the selected RNAs for PCOS. Data revealed that NEAT1, MALAT1, miR-30a-5p, miR-30d-5p, AR, FST and IRS-2 were verified as significant predictors of PCOS with odds ratios: 1.14, 0.66, 1.11, 0.53, 1.17, 1.23 and 1.28 as shown in Table 6.
Table 6.
Univariate regression analysis of the predictive power of the studied RNAs for PCOS.
| RNA |
Coefficient (β) |
SE |
P-value |
OR |
95% CI |
|---|---|---|---|---|---|
| Univariate analysis | |||||
| NEAT1 | 0.13 | 0.06 | 0.033 | 1.14 | 1.01–1.29 |
| MALAT1 | −0.406 | 0.16 | 0.013 | 0.66 | 0.48–0.91 |
| MiR-30a-5p | 0.11 | 0.05 | 0.028 | 1.11 | 1.01–1.23 |
| MiR-30d-5p | −0.63 | 0.21 | 0.003 | 0.53 | 0.35–0.801 |
| AR | 0.16 | 0.06 | 0.009 | 1.17 | 1.04–1.32 |
| FST | 0.207 | 0.09 | 0.022 | 1.23 | 1.03–1.46 |
| IRS-2 | 0.24 | 0.09 | 0.007 | 1.28 | 1.07–1.53 |
NEAT1: nuclear enriched abundant transcript 1, MALAT1: metastasis associated lung adenocarcinoma transcript 1, AR: androgen receptor, FST: follistatin and IRS-2: insulin receptor substrate-2. Coefficient (β), SE (standard error), P-values, OR (odds ratio) and 95% CI (confidence intervals) were estimated by using binary logistic regression model. Significant P-values are indicated in the table.
4. Discussion
PCOS is a complex endocrine disorder in which etiology remains mostly unclear. Over the past decade, genetic abnormalities have been suspected. Thus, attention has been directed to genetic and epigenetic variations which might have a potential regulatory role in such disorder [3]. Previous studies performed whole genome transcriptomic analysis of PCOS patients and healthy controls and identified many lncRNAs, miRNAs and mRNAs to be differentially expressed in ovarian GCs and plasma of PCOS patients [31,32]. Thus, in an attempt to give new insights into the pathogenesis of PCOS, we have combined a bioinformatic-based selection procedure with clinical validation to identify ceRNA network. The proposed ceRNA network represented a combination of lncRNA, miRNA and mRNA of target genes relevant to PCOS. Studying the interactions between these non-coding RNAs and target genes might explore and uncover mystery in PCOS pathogenesis and help in early diagnosis and treatment.
In the current study, peripheral blood leukocytes seemed to be well-suitable for the analysis of RNA expression. Because PCOS is a well-recognized systemic endocrinopathy of the whole body, therefore the use of peripheral blood leukocytes may indicate the overall state of the whole body at the time of sample collection [33,34]. Importantly, peripheral blood could be used to detect and monitor the disease much earlier, even before symptoms appear. Accordingly, the differential expression levels of selected RNAs: NEAT1, MALAT1, miR-30a-5p, miR-30d-5p and their target genes; AR, FST and IRS-2 were estimated in peripheral blood leukocytes obtained from PCOS patients as well as in F.F. obtained from some participants in order to confirm the reliability of peripheral blood leukocytes results.
In the current study, PCOS patients demonstrated significantly higher expression levels of NEAT1, miR-30a-5p, AR, FST and IRS-2, with significantly lower expression levels of MALAT1 and miR-30d-5p in both peripheral blood leukocytes and F.F. samples. Notably, the upregulation of NEAT1 and the three target genes was mostly observed in obese, not in non-obese patients. However, the differential expression levels of other RNAs were demonstrated in both obese and non-obese patients, without any significant difference in between.
Consistent with our study, previous studies have reported upregulation of NEAT1 in serum and GCs of PCOS patients and ovarian tissues of PCOS patients and experimental model [11,24,25]. On the other side, downregulation of MALAT1 observed in our patients was in line with the finding of Zhang et al. [35] and Li et al. [36] in GCs of PCOS patients as well as with Chen et al. [23] in ovarian tissues of rat model. In these studies, the roles of NEAT1 and MALAT1 in driving the development and progression of PCOS were thought to be mediated through the regulation of expression of related miRNAs and genes. One of the well-characterized regulatory mechanisms of lncRNAs is their action as ceRNAs to sponge complementary miRNAs. In support to the roles of lncRNA-mediated ceRNA, recently, Zhou et al. [21] demonstrated the augmenting role of NEAT1 in H2O2-induced human vascular smooth muscle cell Injury by sponging miR-30d-5p. As well, NEAT1 was competing with miR-30d-5p in diabetic nephropathy [37]. Additionally, MALAT1 was acting as a sponge for miR-30a-5p in hepatocellular carcinoma [19], breast cancer [20] and systemic sclerosis [38]. Such interactions between (NEAT1, MALAT1) and (miR-30a-5p, miR-30d-5p) stimulate our interest to elucidate their regulatory role in development of PCOS.
MiRNAs have been emerged as crucial players in development and progression of PCOS. As predicted by the bioinformatic analysis, miR-30 family was identified as epigenetic regulator of AR, FST and IRS-2 expression in diseases other than PCOS such as breast [16], prostate [15] and colon [14] cancers. Bioinformatic analysis also indicated a putative binding site for miR-30a and d in the 3′-untranslated region of AR mRNA, confirming that AR gene is a direct target of miR-30a and d. In the current study, our patients demonstrated higher expression of miR-30a-5p, with lower expression of miR-30d-5p. Consistent with our work, miR-30a-5p had been overexpressed in F.F. of PCOS women [17] as well as in rat ovaries displaying PCOS with insulin resistance [18]. On the other side, no study has previously demonstrated the relative expression of miR-30d-5p in PCOS patients although it has been implicated in multiple human cancers including ovarian, cervical and breast cancers, in which it had been downregulated [39].
Hyperandrogenism, insulin resistance and follicular arrest are not only the main characteristic features of PCOS, but also are important pathogenic factors for PCOS. Emerging evidence indicated that AR signaling plays key roles in PCOS pathogenesis. Increased AR expression was in line with previous studies showing upregulation of its expression in endometrium and ovary of PCOS women [[40], [41], [42]]. Excessive androgen production is mainly linked to hyperinsulinemia and insulin resistance observed in our patients particularly obese and documented in other studies [43]. In PCOS, Wu et al. [9], reported a selective impairment in insulin metabolic responses together with enhanced ovarian mitogenic and steroidogenic responses. Under such conditions, the authors highlighted a shift of the follicular insulin signaling from IRS-1 to IRS-2, suggesting an association between such changes and ovarian abnormality in PCOS. In line with our findings, He et al. [10] demonstrated a significant upregulation of IRS-2 at both gene and protein level in GCs of PCOS patients.
The last gene addressed is FST, a member of the TGF-β superfamily, expressed in various tissues, including ovarian GCs and acts as an important regulator of follicular development due to its ability to bind and neutralize activins. Supporting our findings, overexpression of FST gene in transgenic mice resulted in infertility and arrest of ovarian folliculogenesis at an early stage, similar to that in PCOS [44]. As well, Raeisi et al. [45] documented a significant increase in circulating FST level in PCOS patients.
In the current study, overexpression of AR, FST and IRS-2 can be explained in the light of action ceRNAs which represent a novel mechanism of gene regulation that mediates aberrant expression of miRNAs and mRNAs. Herein, upregulation of NEAT1 in PCOS patients indirectly promotes the expression of AR, FST and IRS-2 by sponging miR-30d-5p. As miR-30d-5p was found to be a negative regulator of AR expression in prostate cancer [15]. Whereas, downregulation of MALAT1 may contribute to overexpression of AR, FST and IRS-2 through releasing more miR-30a-5p, since AR and IRS-2 were identified as a target for miR-30a in breast [16] and colon [14] cancers. These findings partially may unveil the roles of NEAT1 and MALAT1 in development of PCOS pathogenesis. However, further studies using high-throughput gene-silencing system are needed for the functional assessment and identification of the interactive connection among the three RNAs types within the postulated ceRNA network.
In the current investigation, the expression level of NEAT1 was positively correlated with BMI, TT, total cholesterol, TGs and LDL. Such a correlation may emphasize the preceding work of Gernapudi et al. [46], in which NEAT1 had been noticed to promote adipogenesis in adipocyte-derived stem cells. Remarkably, Wu et al. [25] observed that downregulation of NEAT1 mitigated the metabolic disorders and alleviated the ovarian pathological changes in PCOS mice. In addition, AR and FST were positively correlated with BMI, TT, fasting insulin and glucose, HOMA-IR, total cholesterol, TGs and LDL; however, they were negatively correlated with HDL. Likewise, Diamanti-Kandarakis et al. [47] demonstrated that excessive androgen in PCOS can adversely affect insulin sensitivity, promoting visceral adiposity and insulin resistance. On the other side, the positive correlation of IRS-2 with BMI, TT and total cholesterol may account for the preferential role of IRS-2 in mediating ovarian steroidogenesis.
Data of ROC curve analysis and positivity rates of the selected RNAs posited that all of them may serve as potential diagnostic markers of PCOS with the highest efficiency obtained upon combination of NEAT1 and miR-30d-5p or MALAT1 and miR-30a-5p with either of PCOS target genes. On conducting univariate logistic regression analysis, all RNAs were significant, thus these findings may provide additional competency for their adequate usage as robust biomarkers for PCOS prediction.
The main study limitations are the small sample size and ethnic limitation, since the study was conducted at a single center in Egypt, which might limit the application of the findings to the wider population. Therefore, further large multi-centered studies are recommended to obtain more data with comparable outcomes.
In conclusion, the most prominent innovation of our study is to introduce a multi-level gene regulatory network formed by interactions among lncRNAs, miRNAs and mRNAs associated with PCOS pathogenesis. The obtained data of ceRNA network raised the possibility that NEAT1 overexpression may increase the expression levels of AR, FST and IRS-2 by sponging miR-30d-5p, while low expression of MALAT1 may allow higher expression of the above genes via increasing miR-30a-5p, suggesting their involvement in PCOS development and progression and a promising role for future diagnosis and targeted therapy. However, detailed functional analysis of these ceRNA networks in PCOS requires further study in the future.
Ethics approval and consent to participate
All women provided informed consent before sample collection and use for research purposes under the protocol approved by the ethical committee of Faculty of Pharmacy, Cairo University (BC 2531).
Consent for publication
Not applicable
Availability of data and materials
The data and materials of this study are available from the corresponding authors upon request
Funding
This work was financially supported by Faculty of Pharmacy, Cairo University.
Authors' contributions
Asmaa A. ElMonier collected the patients' samples and data, performed the practical work, bioinformatic and statistical analyses, interpreted the patients' data and prepared the manuscript; Sally A. Fahim helped in the practical work, bioinformatic and statistical analyses; Dina Sabry aided in the practical work; Khaled A. Elsetohy was responsible for diagnosis of patients and supervised the collection of patients' samples; Noha A. El-Boghdady and Amira A. Shaheen contributed to the study design and data interpretation, reviewed and edited the manuscript. All authors read and approved the final manuscript.
CRediT authorship contribution statement
Asmaa A. ElMonier: Methodology, Software, Formal analysis, Investigation, Writing – original draft, Visualization. Noha A. El-Boghdady: Conceptualization, Validation, Data curation, Writing – review & editing, Visualization, Supervision. Sally A. Fahim: Methodology, Software, Formal analysis, Investigation. Dina Sabry: Methodology, Investigation. Khaled A. Elsetohy: Resources. Amira A. Shaheen: Conceptualization, Validation, Data curation, Writing – review & editing, Visualization, Supervision, All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgments
We are grateful to the staff of Kasr Al-Ainy hospital (Department of Obstetrics and Gynecology), Cairo, Egypt for their assistance in sample and data collection. This paper content is published as a part of phD degree thesis of Mrs/Asmaa A. ElMonier.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2023.02.008.
Contributor Information
Asmaa A. ElMonier, Email: asmaa.ahmed@pharma.cu.edu.eg.
Noha A. El-Boghdady, Email: noha.elboghdady@pharma.cu.edu.eg.
Sally A. Fahim, Email: sally.atef@ngu.edu.eg.
Dina Sabry, Email: dinasabry@kasralainy.edu.eg.
Khaled A. Elsetohy, Email: kelsetohy@kasralainy.edu.eg.
Amira A. Shaheen, Email: amira.shaheen@pharma.cu.edu.eg.
List of abbreviations
- ANOVA
Analysis of variance
- AR
Androgen receptor
- AUC
Area under the curve
- BMI
Body mass index
- ce
Competing endogenous
- 95% CI
95% confidence interval
- E2
Estradiol
- EDTA
Ethylene diamine tetraacetic acid
- ELISA
Enzyme linked immunosorbent assay
- F·F.
Follicular fluid
- FSH
Follicle stimulating hormone
- FST
Follistatin
- GCs
Granulosa cells
- GnRH
Gonadotropin releasing hormone
- HDL:
High-density lipoprotein
- IGF-1
Insulin like growth factor-1
- IRS
Insulin receptor substrate
- IVF
In vitro fertilization
- LDL:
Low-density lipoprotein
- LH
Luteinizing hormone
- LncRNA
Long non-coding RNA
- MALAT1
Metastasis associated lung adenocarcinoma transcript 1
- MiRNA
MicroRNA
- mRNA
Messenger RNA
- NEAT1
Nuclear enriched abundant transcript 1
- NPV
Negative predictive value
- OR
Odds ratio
- PBS
Phosphate buffer saline;
- PCOS
Polycystic ovary syndrome
- PPV
Positive predictive value
- qPCR
Quantitative real-time polymerase chain reaction
- r
Correlation coefficient
- ROC:
Receiver operating characteristic
- SE
Standard error
- SPSS
Statistical Package for the Social Science
- TGs
Triglycerides
- TT
Total testosterone
- χ2
Chi-square
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Legro R.S., Arslanian S.A., Ehrmann D.A., Hoeger K.M., Murad M.H., Pasquali R., Welt C.M. Diagnosis and treatment of polycystic ovary syndrome: an endocrine society clinical practice guideline. J. Clin. Endocrinol. Metab. 2013;98(12):4565–4592. doi: 10.1210/jc.2013-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saeed N.A.A.A.H., Hamzah I.H., Al-Gharrawi S.A.R. Polycystic ovary syndrome dependency on mtDNA mutation; copy Number and its association with insulin resistance. BMC Res. Notes. 2019;12(1):455. doi: 10.1186/s13104-019-4453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azziz R., Carmina E., Chen Z., Dunaif A., Laven J.S., Legro R.S., Lizneva D., Natterson-Horowtiz B., Teede H.J., Yildiz B.O. Polycystic ovary syndrome. Nat. Rev. Dis. Prim. 2016;2 doi: 10.1038/nrdp.2016.57. [DOI] [PubMed] [Google Scholar]
- 4.Puttabyatappa M., Padmanabhan V. Ovarian and extra-ovarian mediators in the development of polycystic ovary syndrome. J. Mol. Endocrinol. 2018;61(4):R161–R184. doi: 10.1530/JME-18-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Leo V., Musacchio M.C., Cappelli V., Massaro M.G., Morgante G., Petraglia F. Genetic, hormonal and metabolic aspects of PCOS: an update. Reprod. Biol. Endocrinol. 2016;14(1):38. doi: 10.1186/s12958-016-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly C.J., Stenton S.R., Lashen H. Insulin-like growth factor binding protein-1 in PCOS: a systematic review and meta-analysis. Hum. Reprod. Update. 2011;17(1):4–16. doi: 10.1093/humupd/dmq027. [DOI] [PubMed] [Google Scholar]
- 7.Welt C.K., Taylor A.E., Fox J., Messerlian G.M., Adams J.M., Schneyer A.L. Follicular arrest in polycystic ovary syndrome is associated with deficient inhibin A and B biosynthesis. J. Clin. Endocrinol. Metab. 2005;90(10):5582–5587. doi: 10.1210/jc.2005-0695. [DOI] [PubMed] [Google Scholar]
- 8.Suganthi R., Manonayaki S., Fathima Benazir J.A. Follistatin concentrations in women from Kerala with polycystic ovary syndrome. Int. J. Reprod. Biomed. 2010;8(4):131–134. http://ijrm.ssu.ac.ir/article-1-185-en.html [Google Scholar]
- 9.Wu X., Sallinen K., Anttila L., Makinen M., Luo C., Pollanen P., Erkkola R. Expression of insulin-receptor substrate-1 and -2 in ovaries from women with insulin resistance and from controls. Fertil. Steril. 2000;74(3):564–572. doi: 10.1016/s0015-0282(00)00688-9. 0015-0282/00/$20.00 PII S0015-0282(00)00688-9. [DOI] [PubMed] [Google Scholar]
- 10.He T., Liu Y., Zhao S., Liu H., Wang Z., Shi Y. Comprehensive assessment the expression of core elements related to IGFIR/PI3K pathway in granulosa cells of women with polycystic ovary syndrome. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019;233:134–140. doi: 10.1016/j.ejogrb.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Tu M., Wu Y., Mu L., Zhang D. Long non-coding RNAs: novel players in the pathogenesis of polycystic ovary syndrome. Ann. Transl. Med. 2021;9(2):173. doi: 10.21037/atm-20-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B., Xu P., Wang J., Zhang C. The role of miRNA in polycystic ovary syndrome (PCOS) Gene. 2019:706. doi: 10.1016/j.gene.2019.04.082. [DOI] [PubMed] [Google Scholar]
- 13.Lie S., Morrison J.L., Williams-Wyss O., Suter C.M., Humphreys D.T., Ozanne S.E., Zhang S., MacLaughlin S.M., Kleemann D.O., Walker S.K., Roberts C.T., McMillen I.C. Impact of periconceptional and preimplantation undernutrition on factors regulating myogenesis and protein synthesis in muscle of singleton and twin fetal sheep. Phys. Rep. 2015;3(8) doi: 10.14814/phy2.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q., Tang Q., Qin D., Yu L., Huang R., Lv G., Zou Z., Jiang X.C., Zou C., Liu W., Luo J., Zhao Z., Muhammad S., Wang G., Chen Y.G., Wang X. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol. Cell Biol. 2015;35(6):988–1000. doi: 10.1128/MCB.01242-14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Kumar B., Khaleghzadegan S., Mears B., Hatano K., Kudrolli T.A., Chowdhury W.H., Yeater D.B., Ewing C.M., Luo J., Isaacs W.B., Marchionni L., Lupold S.E. Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget. 2016;7(45):72593–72607. doi: 10.18632/oncotarget.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyu S., Liu H., Liu X., Liu S., Wang Y., Yu Q., Niu Y. Interrelation of androgen receptor and miR-30a and miR-30a function in ER(-), PR(-), AR(+) MDA-MB-453 breast cancer cells. Oncol. Lett. 2017;14(4):4930–4936. doi: 10.3892/ol.2017.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scalici E., Traver S., Mullet T., Molinari N., Ferrières A., Brunet C., Belloc S., Hamamah S. Circulating microRNAs in follicular fluid, powerful tools to explore in vitro fertilization process. Sci. Rep. 2016;6 doi: 10.1038/srep24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C., Yu C., Lin Z., Pan H., Li K., Ma H. MiRNAs expression profiling of rat ovaries displaying PCOS with insulin resistance. Arch. Gynecol. Obstet. 2020;302(5):1205–1213. doi: 10.1007/s00404-020-05730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Y., Tong S., Cui R., Fan J., Liu C., Lin Y., Tang J., Xie H., Lin P., Zheng T., Yu X. Long non-coding MALAT1 functions as a competing endogenous RNA to regulate vimentin expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell. Physiol. Biochem. 2018;50(1):108–120. doi: 10.1159/000493962. [DOI] [PubMed] [Google Scholar]
- 20.Ramzy A., Youness R.A., Manie T., Sebak A., Mansour S. MALAT-1/miR-30a-5p competing endogenous (ceRNA) network releases the brakes of immune surveillance in breast cancer through its quadruple targets: PD-L1, MIF, IL-10 and TNF-α. Ann. Oncol. 2021;32:S1357–S1358. doi: 10.1016/j.annonc.2021.08.2036. [DOI] [Google Scholar]
- 21.Zhou F., Zheng Z., Zha Z., Xiong T., Pan Y. Nuclear paraspeckle assembly transcript 1 enhances hydrogen peroxide-induced human vascular smooth muscle cell injury by regulating miR-30d-5p/a disintegrin and metalloprotease 10. Circ. J. 2022;86(6):1007–1018. doi: 10.1253/circj.CJ-21-0042. [DOI] [PubMed] [Google Scholar]
- 22.Ernst E.H., Nielsen J., Ipsen M.B., Villesen P., Lykke-Hartmann K. Transcriptome analysis of long non-coding RNAs and genes encoding paraspeckle proteins during human ovarian follicle development. Front. Cell Dev. Biol. 2018;6(78):78. doi: 10.3389/fcell.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Chen Y., Cui X., He Q., Li H. Down-regulation of MALAT1 aggravates polycystic ovary syndrome by regulating miR-302d-3p-mediated leukemia inhibitory factor activity. Life Sci. 2021;277 doi: 10.1016/j.lfs.2021.119076. [DOI] [PubMed] [Google Scholar]
- 24.Zhen J., Li J., Li X., Wang X., Xiao Y., Sun Z., Yu Q. Downregulating lncRNA NEAT1 induces proliferation and represses apoptosis of ovarian granulosa cells in polycystic ovary syndrome via microRNA-381/IGF1 axis. J. Biomed. Sci. 2021;28(1):53. doi: 10.1186/s12929-021-00749-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Wu L., Tu Z., Bao Y., Zhai Q., Jin L. Long noncoding RNA NEAT1 decreases polycystic ovary syndrome progression via the modulation of the microRNA-324-3p and BRD3 axis. Cell Biol. Int. 2022;46(12):2075–2084. doi: 10.1002/cbin.11893. [DOI] [PubMed] [Google Scholar]
- 26.The Gene Atlas. http://genatlas.medecine.univ-paris5.fr/. Accessed 15 July 2019.
- 27.Diana tools. http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex. Accessed 15 July 2019.
- 28.TargetScan. https://targetscan.org/vert 71/. Accessed 15 July 2019.
- 29.The Diana tools database. https://diana.e-ce.uth.gr/lncbasev3/interactions. Accessed 15 July 2019.
- 30.LncRNA disease database. http://www.rnanut.net/lncrnadisease/index.php/home. Accessed 15 July 2019.
- 31.Romero-Ruiz A., Pineda B., Ovelleiro D., Perdices-Lopez C., Torres E., Vazquez M.J., Guler I., Jiménez Á., Pineda R., Persano M., Romero-Baldonado C. Molecular diagnosis of polycystic ovary syndrome in obese and non-obese women by targeted plasma miRNA profiling. Eur. J. Endocrinol. 2021;185(5):637–652. doi: 10.1530/EJE-21-0552. [DOI] [PubMed] [Google Scholar]
- 32.Deng Y., Li H., Song Y., Cen J., Zhang Y., Sui Y., Cui D., Li T.C., Xu Y., Wang C.C., Chung P.W.J., Tang T. Whole genome transcriptomic analysis of ovary granulosa cells revealed an anti-apoptosis regulatory gene DLGAP5 in polycystic ovary syndrome. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.781149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z., Hao C., Huang X., Zhang N., Bao H., Qu Q. Peripheral blood leukocyte expression level of lncRNA steroid receptor RNA activator (SRA) and its association with polycystic ovary syndrome: a case control study. J. Gynaecol. Endocrinol. 2015;31(5):363–368. doi: 10.3109/09513590.2014.999763. [DOI] [PubMed] [Google Scholar]
- 34.Sun X., Liu Y., Gao X., Du M., Gao M., Zhong X., Wei X. Analysis of lncRNA-mRNA co-expression profiles in patients with polycystic ovary syndrome: a pilot study. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.669819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D., Tang H.Y., Tan L., Zhao D.M. MALAT1 is involved in the pathophysiological process of PCOS by modulating TGFβ signaling in granulosa cells. Mol. Cell. Endocrinol. 2020;499 doi: 10.1016/j.mce.2019.110589. [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Xiang Y., Song Y., Zhang D., Tan L. MALAT1 downregulation is associated with polycystic ovary syndrome via binding with MDM2 and repressing P53 degradation. Mol. Cell. Endocrinol. 2022;543 doi: 10.1016/j.mce.2021.111528. [DOI] [PubMed] [Google Scholar]
- 37.Chen L., Wu B., Wang S., Xiong Y., Zhou B., Cheng X., Zhou T., Luo R., Lam T.-W., Yan B., Chen J. Identification of cooperative gene regulation among transcription factors, lncRNAs, and microRNAs in diabetic nephropathy progression. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dichev V., Mehterov N., Kazakova M., Karalilova R., Batalov A., Sarafian V. The lncRNAs/miR-30e/CHI3L1 axis is dysregulated in systemic sclerosis. Biomedicines. 2022;10(2) doi: 10.3390/biomedicines10020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Q., Yuan X., Zheng L., Xue M. MiR-30d-5p: a non-Coding RNA with potential diagnostic, prognostic and therapeutic applications. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.829435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apparao K.B.C., Lovely L.P., Gui Y., Lininger R.A., Lessey B.A. Elevated endometrial androgen receptor expression in women with polycystic ovarian syndrome. Biol. Reprod. 2002;66:297–304. doi: 10.1095/biolreprod66.2.297. [DOI] [PubMed] [Google Scholar]
- 41.Yongli C., Yongyu S., Hongyu Q. Study of androgen and androgen receptor in relation to insulin resistance in polycystic ovary syndrome. J. Huazhong Univ. Sci. Technol. - Med. Sci. 2003;23(1):52–54. doi: 10.1007/BF02829462. [DOI] [Google Scholar]
- 42.Younas K., Quintela M., Thomas S., Garcia-Parra J., Blake L., Whiteland H., Bunkheila A., Francis L.W., Margarit L., Gonzalez D., Conlan R.S. Delayed endometrial decidualisation in polycystic ovary syndrome; the role of AR-MAGEA11. J. Mol. Med. 2019;97(9):1315–1327. doi: 10.1007/s00109-019-01809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bulsara J., Patel P., Soni A., Acharya S. A review: brief insight into polycystic ovarian syndrome. Endocrine and Metabolic Science. 2021;3 doi: 10.1016/j.endmts.2021.100085. [DOI] [Google Scholar]
- 44.Chen M., Chen H.F., Chen S.U., Ho H.N., Yang Y.S., Yang W.S. The relationship between follistatin and chronic low-grade inflammation in women with polycystic ovary syndrome. Fertil. Steril. 2009;92(6):2041–2044. doi: 10.1016/j.fertnstert.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Raeisi T., Rezaie H., Darand M., Taheri A., Garousi N., Razi B., Roever L., Mohseni R., Hussien Mohammed S., Alizadeh S. Circulating resistin and follistatin levels in obese and non-obese women with polycystic ovary syndrome: a systematic review and meta-analysis. PLoS One. 2021;16(3) doi: 10.1371/journal.pone.0246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gernapudi R., Wolfson B., Zhang Y., Yao Y., Yang P., Asahara H., Zhou Q. MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol. Cell Biol. 2016;36(1):30–38. doi: 10.1128/MCB.00702-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diamanti-Kandarakis E., Papalou O., Kandaraki E.A. The role of androgen excess on insulin sensitivity in women. Front. Horm. Res. 2019;53:50–64. doi: 10.1159/000494902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials of this study are available from the corresponding authors upon request