Abstract
Background
Anal squamous cell carcinoma (SCCA) is an uncommon malignancy with a rising incidence that has a high cure rate in its early stages. There is an unmet need for a reliable method to monitor response to treatment and assist in surveillance. Circulating tumor DNA (ctDNA) testing has shown great promise in other solid tumors for monitoring disease progression and detecting relapse in real time. This study aimed to determine the feasibility and use of personalized and tumor-informed ctDNA testing in SCCA.
Patients and Methods
We analyzed real-world data from 251 patients (817 plasma samples) with stages I-IV SCCA, collected between 11/5/19 and 5/31/22. The tumor genomic landscape and feasibility of ctDNA testing was examined for all patients. The prognostic value of longitudinal ctDNA testing was assessed in patients with clinical follow-up (N = 37).
Results
Whole-exome sequencing analysis revealed PIK3CA as the most commonly mutated gene, and no associations between mutations and stage. Anytime ctDNA positivity and higher ctDNA levels (MTM/mL) were associated with metastatic disease (P = .004). For 37 patients with clinical follow-up, median follow-up time was 21.0 months (range: 4.1-67.3) post-diagnosis. For patients with stages I-III disease, anytime ctDNA-positivity after definitive treatment was associated with reduced DFS (HR: 28.0; P = .005).
Conclusions
Our study demonstrates the feasibility of personalized and tumor-informed ctDNA testing as an adjunctive tool in patients with SCCA as well as potential use for detection of molecular/minuteimal residual disease, and relapse during surveillance. Prospective studies are needed to better evaluate the use of ctDNA testing in this indication.
Keywords: circulating tumor DNA, anal squamous cell carcinoma, prognostic biomarker, minimal/molecular residual disease
Circulating tumor DNA (ctDNA) testing has shown great promise in solid tumors for monitoring disease progression and detecting relapse in real time. This article reports on the feasibility and use of personalized and tumor-informed ctDNA testing specific to anal squamous cell carcinoma. Exploratory analysis from whole exome sequencing (WES; N=251) is also reported.
Implications for Practice.
This study demonstrated the feasibility and prognostic value of ctDNA testing in SCCA. Incorporation of ctDNA testing into clinical practice can allow providers to identify relapse occurring on the molecular level, which often predates radiological findings. Importantly ctDNA-informed relapse can create opportunities for timely therapeutic intervention. Similar to the utility of tumor-informed ctDNA testing in colorectal cancer in patient prognostication, the study provides the benchmark for trials incorporating escalation and de-escalation of therapy for patients with anal cancer. As IO treatments in early-stage disease are being studied, these trials would help establish ctDNA as a predictive marker in this setting.
Introduction
Anal cancer is a rare malignancy, accounting for approximately 4% of all malignancies of the lower GI tract.1 However, in the past few decades, incidence has been increasing, with an estimate of 9,090 cases diagnosed in the US in 2021.2 Of all cases of anal cancer, ~90% are squamous cell carcinoma (SCCA).2 For patients with locoregional disease, the standard of care is local chemoradiotherapy (CRT), which is usually curative in intent.3,4 Nevertheless, survival outcomes vary widely, with 5-year survival rates of 82% for localized disease, 66% for regional disease and only 34% for metastatic disease.2 For patients with locoregional recurrence, salvage abdominoperineal resection (APR) is recommended, which can achieve disease control in up to 77% of patients.5 Those with stage IV disease rely on platinum-based chemotherapy (carboplatin with paclitaxel; 5-fluorouracil (5-FU), leucovorin and oxaliplatin (FOLFOX); Docetaxel Cisplatin, and 5-FU (DCF); and 5-FU and cisplatin (FOLFCIS)).4 Recently, immunotherapy has been integrated into the standard of care for patients with metastatic disease in the chemotherapy-refractory setting.4 Anti-PD-1 monotherapy has been reported to be effective for the treatment of metastatic disease,6 and as a second-line regimen for patients with chemotherapy-refractory disease.7
Surveillance after definitive treatment is a key challenge for both patients and providers. NCCN guidelines recommend patients undergo extensive surveillance after cessation of definitive treatment to monitor disease recurrence. Approximately 8-12 weeks after chemoradiation is complete, patients are recommended to have digital rectal exams and lymph node exams every 3-6 months for 5 years, and anoscopy every 6-12 months for 3 years. For stages II-IV disease, at least annual CT scan of the chest, abdomen, and pelvis and/or annual MRI of the abdomen and pelvis for 3 years is also recommended. However, imaging results can be difficult to interpret, as anal squamous cell carcinoma can have slow tumor regression after chemoradiation. Furthermore, imaging can miss small foci of residual tumor. While PET/CT can be helpful in some situations, significant false positive results have been reported, up to 40% in some series.3 Increased frequency of imaging can allow for early identification of recurrence, and consequently earlier therapeutic intervention, they are not supported by current guidelines.4 Cost and coverage by insurance of imaging are also of concern for these patients.
To address these unmet clinical needs, biomarkers that can accurately measure response to therapy and identify recurrence ahead of imaging would be useful. Unlike other solid tumors (eg, colorectal, pancreatic, ovarian), where glycoprotein-based markers can be used to monitor disease status (eg, CEA, CA-125, CA-19-9), there are no such established markers for anal cancer.8 Recent data have validated the use of a personalized and tumor-informed circulating tumor DNA (ctDNA)-based approach for patients with colorectal cancer (CRC) and other solid tumors.9-14 Here, we examined the feasibility and utility of personalized and tumor-informed ctDNA testing in a real-world setting.
Materials and Methods
Study Population
We analyzed real-world data in patients with stages I-IV SCCA who were monitored by ctDNA testing. Commercially submitted blood samples collected between November 5, 2019 and May 31, 2022 were evaluated for the presence and levels of ctDNA. Eligible patients were 18 years or older, had successful personalized tumor-informed primer design, and had clinical evidence of SCCA on imaging. Blood samples were collected at the discretion of the treating physician, and ctDNA dynamics were evaluated longitudinally. A total of 251 patients and 817 plasma samples were included in this study. Of the 251 patients, complete clinical information was available for 37 patients. Tumor tissue was collected at resection or at initial diagnosis via biopsy. Patients received treatment and follow-up in compliance with standard clinical practice and at the discretion of the treating physician. For the majority of patients, clinical staging was used in place of pathological staging, as most patients did not undergo surgery in order to be evaluated pathologically. Disease response was evaluated based on RECIST criteria and treating clinician’s evaluation. Informed consent was obtained from patients as part of ordering the assay. This study was approved by the corresponding Ethical and Independent Review Services (protocol # 20-049-ALL) and was conducted in accordance with the Declaration of Helsinki.
Personalized and Tumor-Informed ctDNA Assay
A personalized and tumor-informed ctDNA assay (Signatera bespoke mPCR NGS assay) was used for the detection and quantification of ctDNA, as previously described.10 Briefly, for each patient, tumor tissue and a matched normal whole blood sample were sequenced by whole-exome sequencing (WES). Sequencing results allowed the identification and selection of up to 16 clonal, somatic, single-nucleotide variants (SNVs) present in the tumor but not the matched normal sample. The selected SNVs were used in the design of a multiplex PCR NGS-based ctDNA assay. The designed personalized ctDNA assays were then used to detect ctDNA in patient plasma samples. Plasma samples testing positive for 2 or more SNVs were defined as ctDNA-positive. ctDNA was quantified in mean tumor molecules per milliliter of plasma (MTM/mL). Results were provided in real time to treating clinicians.
Genomic Analysis
To identify genomic profiles for all patients with SCCA (N = 251), we analyzed WES data obtained from our commercial database. Our exploratory genomic analysis examined SNVs and short insertions and deletions. Only non-synonymous mutations were included in the analysis. The genomic assessment was solely conducted as an exploratory analysis and is not routinely offered as a component of clinical Signatera testing.
Statistical Analysis
The primary objective was to measure disease-free survival (DFS), from the time of diagnosis to the first radiologic evidence of disease progression. An exploratory analysis was performed wherein DFS was defined by the first radiologic evidence of disease progression or molecular recurrence by ctDNA (Supplementary Fig. S1). A fisher’s exact test was used to compare categorical variables. The Kaplan-Meier method was used to estimate the survival distribution. Differences between the groups were tested by the log-rank test. All P values were based on 2-sided testing, and differences were considered significant at P ≤ .05.
Results
Patient Demographics
In this study, plasma samples (n = 817) were collected from 251 patients with SCCA (180 females, 71 males; median age 63.5 years; range: 27.9-89.4) at various time points. Of patients with plasma samples collected, 37 had complete clinical information available and had a median follow-up of 21.0 months (range: 4.1-67.3), post-diagnosis. Of the 37 patients analyzed, 3 (8.1%) patients had stage I disease, 10 (27.0%) had stage II disease, 17 (46.0%) had stage III disease, and 7 (18.9%) had stage IV disease. A total of 21 patients had information on HPV status reported, of which 20 were HPV positive (95.2%). A total of 31 patients had information on HIV status available, where 12 (38.7%) were HIV positive. In terms of types of treatments used, including both upfront and sequential therapy, 35 patients (94.6%) received chemoradiotherapy (CRT), 6 (16.2%) received immunotherapy, 2 (5.4%) received radiotherapy, 4 (10.8%) received chemotherapy, 2 (5.4%) received chemoimmunotherapy. A total of 2 patients (5.4%) underwent surgery. The median tumor mutation burden (TMB) was 3.81 and only 1 patient (2.7%) had an MSI-high tumor. Complete demographic information for both the original cohort (N = 251) and a subcohort with clinical follow-up (N = 37) is summarized in Table 1.
Table 1.
Cohort demographics.
| Category | Annotated cohort, N = 37 | Full cohort, N = 251 | ||
|---|---|---|---|---|
| n | % | n | % | |
| Gender | ||||
| Female | 19 | 51.4% | 180 | 71.7% |
| Male | 18 | 48.6% | 71 | 28.3% |
| Overall clinical stage | ||||
| I | 3 | 8.1% | 30 | 12.0% |
| II | 10 | 27.0% | 68 | 27.1% |
| III | 17 | 46.0% | 96 | 38.2% |
| IV | 7 | 18.9% | 49 | 19.5% |
| Unknown | 0 | 0% | 8 | 3.2% |
| Anytime ctDNA-positivity | ||||
| I | 2 | 67.0% | 12 | 41.4% |
| II | 4 | 36.0% | 28 | 41.8% |
| III | 6 | 35.0% | 35 | 38.0% |
| IV | 5 | 72.0% | 34 | 70.8% |
| Treatment received | ||||
| Surgery | 2 | 5.4% | N/A | N/A |
| Chemoradiotherapy | 35 | 94.6% | N/A | N/A |
| Radiotherapy | 2 | 5.4% | N/A | N/A |
| Chemotherapy | 4 | 10.8% | N/A | N/A |
| Immunotherapy | 6 | 16.2% | N/A | N/A |
| Chemoimmunotherapy | 2 | 5.4% | N/A | N/A |
| HIV status | ||||
| Negative | 19 | 51.4% | N/A | N/A |
| Positive | 12 | 32.4% | N/A | N/A |
| Unknown | 6 | 16.2% | N/A | N/A |
| HPV status | ||||
| Negative | 1 | 2.7% | N/A | N/A |
| Positive | 20 | 54.1% | N/A | N/A |
| Unknown | 16 | 43.2% | N/A | N/A |
| MSI status | ||||
| MS-Stable | 36 | 97.3% | 250 | 99.6% |
| MSI-High | 1 | 2.7% | 1 | 0.4% |
| Category | Median (range) | Median (range) | ||
|---|---|---|---|---|
| Age, years | 64.42 (32.6-77.2) | 63.5 (27.9-89.4) | ||
| Number of ctDNA timepoints | 6.0 (1-27) | 2 (1-27) | ||
| Follow-up time, months | 21.0 (4.1-67.3) | N/A | ||
| Tumor mutational burden, muts/Mb | 3.81 (0.46-21.88) | 4.27 (0.17-85.63) | ||
Abbreviations: N/A, not available; ctDNA, circulating tumor DNA; MSI, microsatellite instability; HPV, human papillomavirus; HIV, human immunodeficiency virus.
Genomic Alterations Most Frequently Observed in SCCA Patients
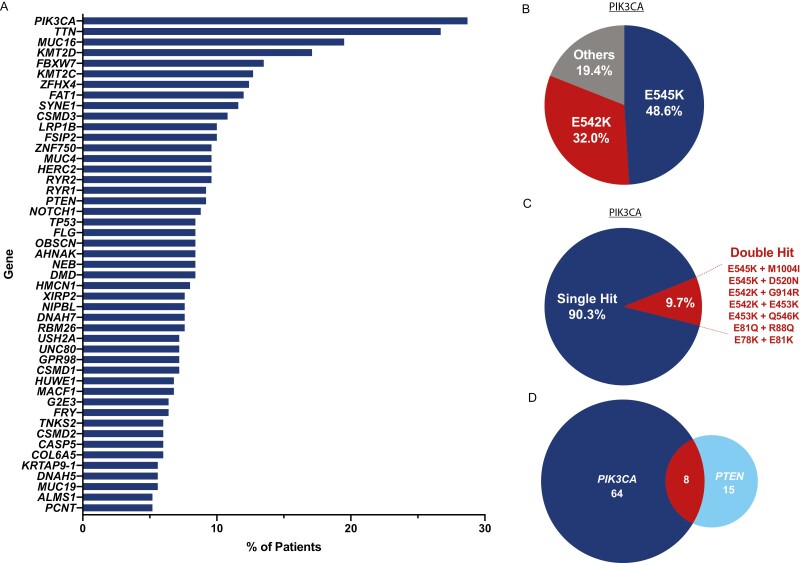
We performed an exploratory analysis on the WES data available from our commercial database to identify genomic profiles and characteristics for all patients (N = 251). WES results revealed that PIK3CA (28.7%), TTN (26.7%), MUC16 (19.5%), KMT2D (17.1%), FBXW7 (13.5%), and KMT2C (12.7%) as the most commonly mutated genes observed, when considering non-synonymous variants. Mutations in PTEN, NOTCH1, and TP53 were also observed in 9.2%, 8.8%, and 8.4% of patients, respectively. No trends were observed between tumor stage and frequency of any genetic mutation. Of the patients PIK3CA mutations, the most common variants observed were activating E545K (48.6%) and E542K (31%) mutations (Fig. 1B). Of note, 9.7% of patients with PIK3CA-mutated tumors had double mutations (Fig. 1C). Mutations in PIK3CA and PTEN tended to occur in patients in a mutually exclusive fashion (Fig. 1D), as expected.
Figure 1.
Comprehensive Genomic Profiling of 251 SCCA Patients. (A) Most frequently altered genes (genes mutated in ≥5% of patients) observed in our cohort (N = 251). (B) Frequency of observed PIK3CA variants. Four patients in the E545K/E542K categories possessed an additional PIK3CA mutation (C). The other category represents patients lacking E545K or E542K mutations. (C) Frequency of single hit versus double hit PIK3CA mutations observed in our cohort. (D) Number of patients with alterations in PIK3CA alone, PTEN alone, or both genes together. Alterations in the 2 genes tended to occur in a mutually exclusive fashion.
Correlation of ctDNA Status and Levels With Disease Stage
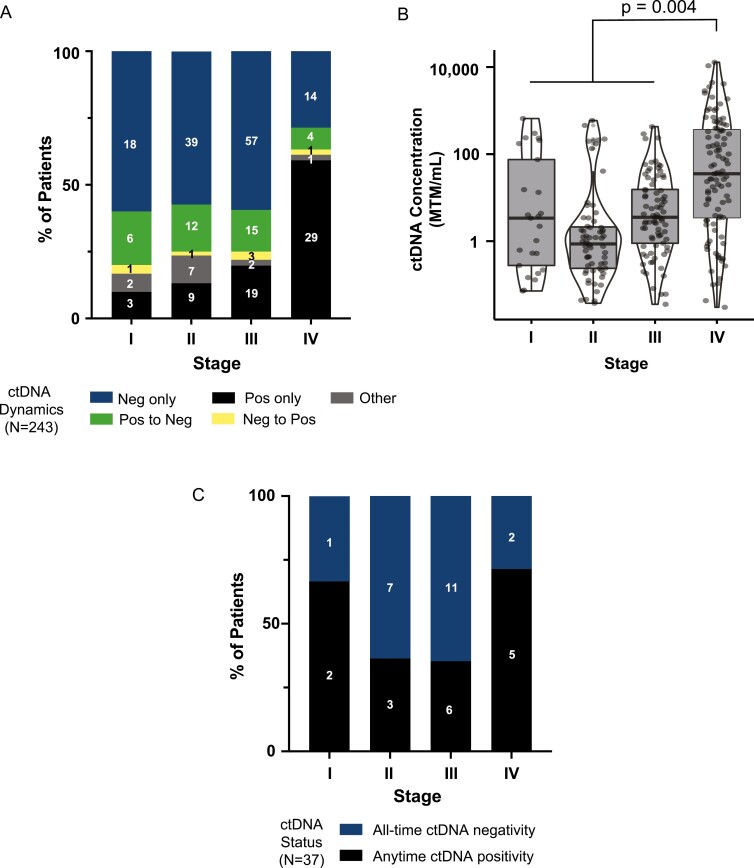
For the 251 patients with successful ctDNA assay design, we sought to examine the correlation between disease stage and ctDNA status and levels across all time points. Eight patients did not have a reported clinical stage. Of the remaining 243 patients, we found that those with metastatic disease were more likely to test ctDNA positive (71.4% vs 40.6%, P = .0002), and less likely to clear ctDNA than those with localized disease (11.4% vs 42.3%, P = .003) (Fig. 2A). In addition, patients with metastatic disease had significantly higher levels of ctDNA than those with localized disease (Fig. 2B; Student’s t test, P = .004). We did not observe any significant differences in ctDNA levels between patients with stage I, II, or III disease (Fig. 2B).
Figure 2.
Feasibility of ctDNA testing in patients with SCCA. (A,B) ctDNA dynamics and levels by tumor stage in 243 patients. Stage IV patients had significantly higher median levels of ctDNA as compared to stages I-III patients (P = .004). (C) ctDNA dynamics by tumor stage in a subcohort of patients (N = 37) who had complete clinical follow-up.
Association of ctDNA Detection With Patient Outcomes
We next examined the association of ctDNA status post-definitive therapy (CRT/immunotherapy/surgery) with radiological relapse. For this analysis, only stages I-III patients (N = 30) who underwent treatment with curative intent, with ctDNA results and confirmatory testing (either anoscopy with biopsy or radiological imaging) were included. Among the patients with stages I-III disease, we further excluded 3 patients: patient 6, who was treated on molecular recurrence; patient 21, who had clinical recurrence prior to the start of ctDNA testing; and patient 30, who had no ctDNA result post-definitive therapy.
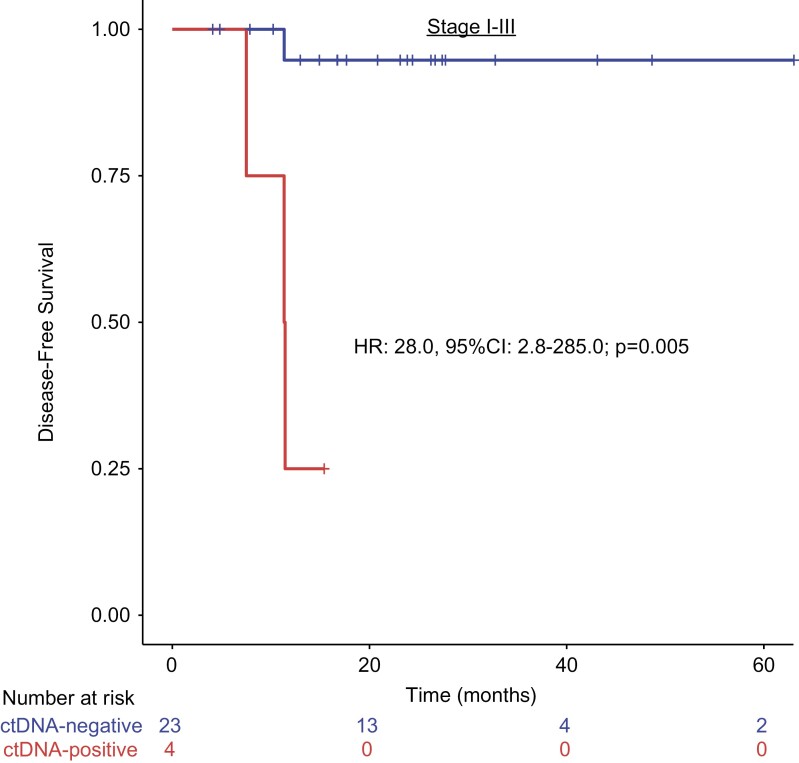
Of the 27 patients analyzed, 23 were ctDNA-negative post-definitive therapy, one of whom (patient 26) was observed to have stable disease on imaging and showed confirmed residual disease by biopsy (Negative Predictive Value of 95.7%; 22/23). Of the 4 ctDNA-positive patients, 3 (patients 11, 23, and 24) experienced disease progression on imaging. The remaining patient (patient 27) had radiological findings suggesting partial responses to therapy. We next examined the associations between ctDNA results and survival outcomes. Stratifying stages I-III patients by anytime ctDNA-positivity or negativity, we found that anytime ctDNA positivity post-definitive therapy was associated with a significantly shorter DFS (median DFS for anytime ctDNA-positive patients 11.4 months, not reached for all time ctDNA-negative patients; HR = 28.0, 95% CI 2.8-285.0; P = .005) (Fig. 3). In addition, we performed an exploratory analysis where DFS was defined as the first radiologic evidence of disease progression or molecular recurrence by ctDNA. Once again, ctDNA positivity was significantly associated with shorter DFS (HR = 109.6, P < .0001) (Supplementary Fig. S1).
Figure 3.
Anytime ctDNA-positivity after completion of definitive therapy is associated with poor disease-free survival. Stages I-III patients were stratified by anytime ctDNA-positivity versus negativity, post-definitive therapy. P-value was calculated by Cox-regression.
Potential Use of Personalized and Tumor-Informed ctDNA Testing in Clinical Practice
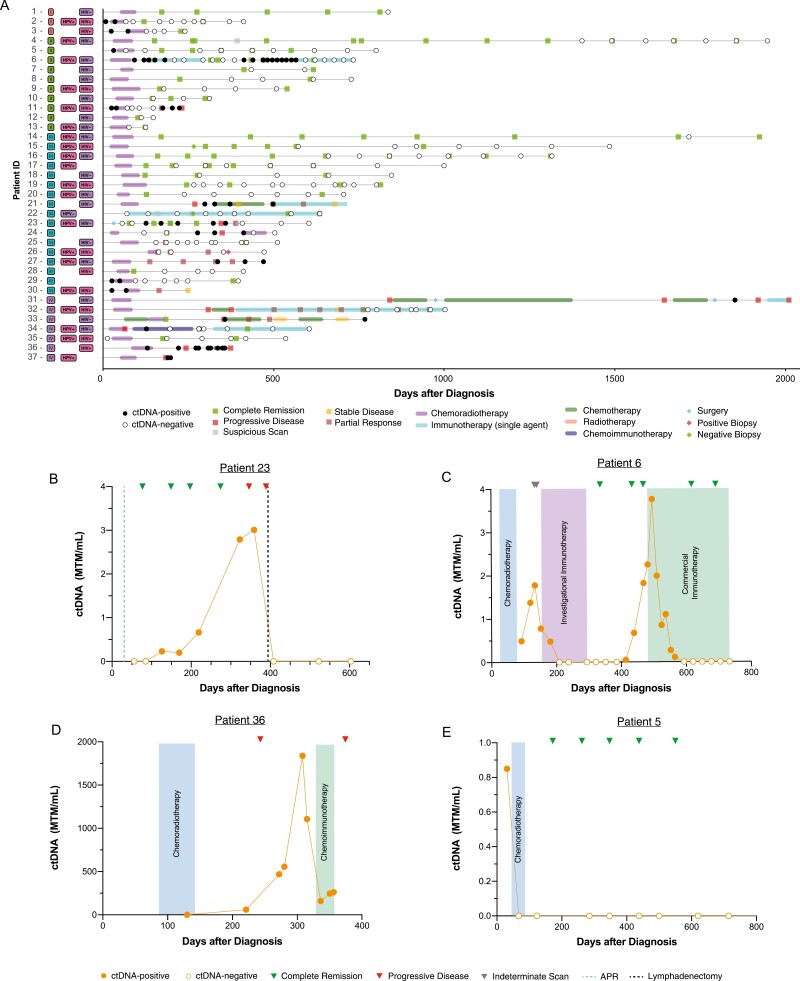
The complete clinical course for the 37 patients with clinical follow-up is summarized in Fig. 4A. Of the 37 patients with clinical follow-up, 91.9% (34/37) had serial ctDNA results available (defined as ≥2 samples taken at different timepoints). In this group, ctDNA positivity was detected in 43.2% (16/37) of patients. Of the ctDNA-positive patients, 2 had stage I, 3 had stage II, 6 had stage III, and 5 had stage IV disease (Fig. 2C). All but 2 patients were treated with definitive chemoradiotherapy (CRT). Patient 22 had a rare MSI-high phenotype, was treated with single-agent immunotherapy (IO). Patient 23 had a history of prostate cancer treated with radiation and was not eligible for additional radiotherapy. A complete breakdown of types of therapy administered among the 37 patients with clinical follow up is detailed in Table 1.
Figure 4.
Case examples: ctDNA Testing provides rationale for initiation of adjuvant therapy. (A) Overview plot of cohort with clinical follow-up (N = 37). (B-D) Case examples of SCCA patients who underwent longitudinal ctDNA testing, wherein ctDNA results were used to inform clinical decision making.
In this study, we identified several case examples where the physician altered the treatment regimen and management, in the context of ctDNA testing (Fig. 4B-E). One patient (#23) underwent abdominal perineal resection (APR). Post-operatively, the patient initially tested ctDNA-negative, but was later found to be ctDNA positive. Based on these results, the provider elected to increase the cadence of imaging. Although the patient initially had no evidence of disease, they were later found to have inguinal lymph node metastasis. Importantly, ctDNA positivity occurred approximately 200 days prior to radiological recurrence. A lymphadenectomy was performed and the patient subsequently cleared ctDNA, providing evidence of complete remission (Fig. 4B). Notably, no additional systemic therapy was given after lymphadenectomy. Another patient (#6) was treated on the basis of molecular recurrence, as evidenced by increasing ctDNA levels, in the absence of radiological recurrence (Fig. 4C). A third patient (#36) tested positive for ctDNA while on CRT, as expected. Subsequent testing after completion of CRT revealed increasing ctDNA levels, along with evidence of liver metastasis on imaging, which prompted the decision to start the patient on chemoimmunotherapy. Although ctDNA levels initially declined, they later increased, and the patient was confirmed to have progressive disease on imaging (Fig. 4D). A final patient of note (#5) who cleared ctDNA on definitive chemoradiotherapy was later confirmed to be disease free by serial imaging (Fig. 4E). Together, these case examples demonstrate the value of longitudinal ctDNA testing for treatment response monitoring, as well as early detection of relapse.
Discussion
Our study is the first real-world experience to demonstrate the feasibility of personalized and tumor-informed ctDNA testing in SCCA and its use for the detection of residual disease, and relapse during surveillance. To date, studies exploring the use of ctDNA in SCCA have been limited. Previous studies examining the performance of ctDNA in SCCA have used ddPCR, to detect and quantify tumor-derived HPV DNA (HPV ctDNA),15 wherein, the presence of residual HPV ctDNA after definitive CRT was associated with significantly shorter DFS (P < .0001).16 Another ancillary study similarly found the presence of residual HPV ctDNA was associated with shorter progression-free survival than patients who tested ctDNA negative (HR 5.5; P < .001).17 Although HPV ctDNA testing is not routinely used in clinical practice at present, these studies further support the potential of ctDNA as a prognostic biomarker in SCCA. Personalized and tumor-informed ctDNA testing may be more broadly applicable to this indication, as it would enable monitoring of patients who are HPV negative.
Our study demonstrates the prognostic value of ctDNA testing, which can detect recurrence early and provide rationale for early therapeutic intervention ahead of imaging. ctDNA positivity at any time after definitive therapy was predictive of DFS in stages I-III patients (HR: 28.0; P = .005). In addition, in patient 23 (Fig. 4B), ctDNA was first detected approximately 200 days prior to positive imaging, confirming metastasis to the inguinal lymph nodes. This information could provide a window of opportunity for more intensive surveillance and early intervention. In patient 6 (Fig. 4C) who became ctDNA-positive after a period of negative testing, the provider chose to treat the patient on the basis of “molecular recurrence”, in the absence of disease progression on imaging. This resulted in a precipitous drop and eventual clearance of ctDNA. Although treating patients solely based on molecular recurrence is not currently recommended by NCCN guidelines, we anticipate an increase in ctDNA-guided treatment decision-making in the clinical setting, given its high predictive and prognostic value across cancers.
Our WES analysis provides insight into the mutational landscape of SCCA. Comprehensive genomic profiling of SCCA using a targeted gene panel has been previously described in stages II-IV patients.18 In our cohort of stages I-IV patients, no significant association between any mutation and SCCA stage was observed for any of the genes, consistent with the previous report.18 Similarly, PIK3CA was the most frequently mutated gene observed in our cohort, with the activating mutations E545K and E542K comprising approximately 80% of all variants detected (Fig. 1A and B). These mutations have been shown to be targetable in various solid tumors by selective PI3Kɑ inhibitors such as alpelisib and taselisib.19-21 Patients harboring double-hit mutations in PIK3CA were also fairly prevalent (~10%) in our cohort (Fig. 1C), which has been previously identified in several cancers, including breast cancer and CRC.21,22 It is speculated that tumors with double-hit mutations in cis may have enhanced sensitivity to PI3K inhibitors22 although we could not confirm these double mutations occurred on the same allele. In addition, approximately 35% of patients possessed alterations in either the oncogene PIK3CA or in the tumor suppressor PTEN, highlighting the potential significance of targeting the PI3K signaling pathway in SCCA. As expected, these mutations tended to be mutually exclusive in patients, with co-occurrence found in only 3% of the genomics cohort (Fig. 1D). This finding is in agreement with results from a prior landscape study of PI3K pathway alterations across multiple solid tumors.23 In addition, FBXW7 inactivating mutations, found in 13.5% of patients in this cohort, have also been linked to increased levels of mTOR and modest benefit from mTOR inhibitors in solid tumors [24, 25]. This finding further confirms the prominent role of PI3K/AKT/mTOR pathway dysregulation in SCCA, and would be of value for future studies and clinical trials. Expected high frequency of mutations in TTN and MUC16, 2 of the largest genes in the human genome, have been associated with higher tumor mutational burden (TMB) and favorable prognosis and may be a predictive biomarker of IO response [26, 27]. Overall, we observed significant genomic heterogeneity in our cohort, further highlighting the potential use of personalized and tumor-informed approach to ctDNA testing, designed to detect clonal variants. Panel-based ctDNA assays designed to detect a handful of variants may fail to detect ctDNA in a large proportion of SCCA patients.
This real-world study also demonstrates the potential use of longitudinal ctDNA testing to monitor tumor response to immunotherapy. In this cohort, 6 patients received single-agent IO and 2 were treated with chemoimmunotherapy combinations, of whom only 2 had progressed on their treatments. Of note, 2 patients with stage IV disease (32 and 34) who had progressed on prior lines of therapy achieved durable ctDNA clearance on single-agent and combination IO and remained in complete remission as of last follow-up. Although studies have demonstrated that some SCCA patients respond well to IO, it is important to note that the majority of patients do not benefit from this form of treatment. A phase II single-arm study of 37 SCCA patients treated with nivolumab found that only 24% of patients responded to nivolumab.6 Another study of 75 metastatic SCCA patients (part of the KEYNOTE-158 phase II study) administered pembrolizumab monotherapy and identified a response in ~15% of patients.7
Our study is limited by being a real-world study, examining a heterogenous group of patients who received ctDNA testing at non-standardized time points, during and after their treatment with majority of patients receiving ctDNA testing after CRT. Although we did not have imaging results for the majority of patients, trends in anytime positivity were similar in the larger and smaller cohort (Fig. 2, Table 1), suggesting the cohort of 37 with clinical follow-up is representative of the larger cohort. Future prospective studies will be needed to validate the relevance of our findings. Due to the rarity of SCCA, at present few clinical studies have been designed to better optimize patient treatment regimens. However, currently a number of ongoing trials are being designed to test the efficacy of novel treatments, including lower doses of CRT (NCT04166318-DECREASE study), IO regimens (NCT02314169, NCT02919969, NCT04444921, NCT03233711), and IO with chemotherapy (NCT03519295-SCARCE study).28
In conclusion, our findings from a real-world cohort of SCCA demonstrated the feasibility of a tumor-informed ctDNA assay in this population. Incorporation of ctDNA testing into the design of clinical studies and/or in clinical practice can help identify patients that would most benefit from more intensive treatment/surveillance, or reduced treatment/surveillance in the case of ctDNA negativity. In addition, stratification of patients by ctDNA status after definitive treatment can help determine those at higher risk of relapse, while potentially decreasing invasive surveillance methods for the lower-risk population. Similar to the utility of tumor-informed ctDNA testing in colorectal cancer in patient prognostication, our study provides the benchmark for trials incorporating ctDNA into physician decision making regarding escalation and de-escalation of therapy for patients with anal cancer. Apart from being prognostic, ctDNA has shown to be predictive of response to IO in patients with advanced stage disease.29 As IO treatments are being studied in early-stage disease as well, these trials would help establish ctDNA as a prognostic as well as a potential predictive marker in this setting.
Supplementary Material
Contributor Information
Georges Azzi, HolyCross Medical Group, Ft. Lauderdale, FL, USA.
Mehrad Tavallai, Natera, Inc., Austin, TX, USA.
Vasily N Aushev, Natera, Inc., Austin, TX, USA.
Allyson Koyen Malashevich, Natera, Inc., Austin, TX, USA.
Gregory P Botta, UC San Diego Health, San Diego, CA, USA.
Mohamedtaki A Tejani, AdventHealth Cancer Institute, Orlando, FL, USA.
Diana Hanna, USC Norris Comprehensive Cancer Center, Los Angeles, CA, USA.
Shifra Krinshpun, Natera, Inc., Austin, TX, USA.
Meenakshi Malhotra, Natera, Inc., Austin, TX, USA.
Adham Jurdi, Natera, Inc., Austin, TX, USA.
Alexey Aleshin, Natera, Inc., Austin, TX, USA.
Pashtoon M Kasi, Weill Cornell Medicine, New York, NY, USA.
Funding
None declared.
Ethics Statement
Informed consent was obtained as part of the ordering assay. This study was approved by the corresponding Ethical and Independent Review Services (protocol# 20-049-ALL) and was conducted in accordance with the Declaration of Helsinki.
Conflict of Interest
Georges Azzi declared ownership interests (stocks) in Halozyme Therapeutics, Illumina, Guardant Health, TG Therapeutics, AptoseBiosciences, CytomxTherapeutics, Zymeworks, Global Blood, Invitae, SangamoTherapeutics, Cellectis, Cohbar, and Fibrogen; Advisory Board relationships with Pfizer and Astellas/Seattle Genetics; and Speaker’s Bureau with Natera, Inc. and Guardant Health. Mehrad Tavallai reported employment and shareholder with Natera. Allyson Koyen Malashevich reported employment and shareholder with Natera. Vasily N. Aushev, Shifra Krinshpun, Meenakshi Malhotra, Adham Jurdi, and Alexey Aleshin are full-time employees of Natera, Inc. with stock options. Gregory P. Botta declared advisory committee and speaking/teaching relationships with Natera, Inc. and consultancy for CEND Therapeutics. Meenakshi Malhotra declared employment and ownership interests with Natera Inc. Adham Jurdi declared employment with Natura. Alexey Aleshin declared employment and ownership interests with Natera Inc. and consultancy/advisory relationships with Notable Labs and Mission Bio. Pashtoon M. Kasi declared consultancy/advisory board relationships with AstraZeneca/Daiiche Sankyo (AZ), Axiom, Bayer, Boston Health Care, Daiiche Sankyo, Delcath, Eli Lilly, Eisai, Exact Sciences, Foundation Medicine, Illumina, Incyte, Ipsen (to institution), IPBA, Merck/MSD Oncology, Natera, QED Therapeutics, SAGA Diagnostics, Servier, Seagen—Seattle Genetics, Tempus, Taiho Oncology (self and to institution). Scientific Advisory Board and Shares: Elicio Therapeutics. Research/Trial Support (to institution), Advanced Accelerator Applications, Amgen, AstraZeneca, BTG/Boston Scientific, Merck and RenovoRx, Seagen, Tersera. The other authors indicated no financial relationships.
Author Contributions
Conception: G.A., M.T., S.K., A.J., A.A., G.P.B., M.A.T., D.H., P.M.K. Provision of study materials or patients: G.A., G.P.B., M.A.T., D.H., A.J., P.M.K. Collection and assembly of data: G.A., M.T., V.N.A. Data analysis and interpretation: G.A., M.T., V.N.A., A.K.M., A.J., M.M., S.K., P.M.K. Manuscript writing: G.A., M.T., V.N.A., A.K.M., M.M., P.M.K. Manuscript editing/approval: All authors.
Data Availability
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.
References
- 1. Pessia B, Romano L, Giuliani A, et al. Squamous cell anal cancer: management and therapeutic options. Ann Med Surg (Lond). 2020;55(7):36-46. 10.1016/j.amsu.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Cancer Society. Anal Cancer. Available at https://www.cancer.org/cancer/anal-cancer.html. Accessed August 4, 2022.
- 3. Eng C, Messick C, Glynne-Jones R.. The management and prevention of anal squamous cell carcinoma. Am Soc Clin Oncol Educ Book. 2019;39(1):216-225. [DOI] [PubMed] [Google Scholar]
- 4. NCCN clinical practice guidelines in oncology: anal carcinoma version 1.2022. National Comprehensive Cancer Center Network, Available at NCCN.org. [DOI] [PubMed]
- 5. Morton M, Melnitchouk N, Bleday R.. Squamous cell carcinoma of the anal canal. Curr Probl Cancer. 2018;42(5):486-492. 10.1016/j.currproblcancer.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 6. Morris VK, Salem ME, Nimeiri H, et al. Nivolumab for previously treated unresectable metastatic anal cancer (nci9673): A multicentre, single-arm, phase 2 study. Lancet Oncol. 2017;18(4):446-453. 10.1016/S1470-2045(17)30104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marabelle A, Cassier PA, Fakih M, et al. Pembrolizumab for advanced anal squamous cell carcinoma (ascc): results from the multicohort, phase ii keynote-158 study. J Clin Oncol. 2020;38(4):1-1. 10.1200/jco.2020.38.4_suppl.1.31682550 [DOI] [Google Scholar]
- 8. Hester R, Advani S, Rashid A, et al. Cea as a blood-based biomarker in anal cancer. Oncotarget. 2021;12(11):1037-1045. 10.18632/oncotarget.27959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henriksen TV, Tarazona N, Frydendahl A, et al. Circulating tumor DNA in stage iii colorectal cancer, beyond minimal residual disease detection, toward assessment of adjuvant therapy efficacy and clinical behavior of recurrences. Clin Cancer Res. 2022;28(3):507-517. 10.1158/1078-0432.CCR-21-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages i to iii colorectal cancer. JAMA Oncol.2019;5(8):1124-1131. 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Powles T, Assaf ZJ, Davarpanah N, et al. Ctdna guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595(7867):432-437. 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
- 12. Magbanua MJM, Swigart LB, Wu HT, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol. 2021;32(2):229-239. 10.1016/j.annonc.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kotaka M, Shirasu H, Watanabe J, et al. Association of circulating tumor DNA dynamics with clinical outcomes in the adjuvant setting for patients with colorectal cancer from an observational galaxy study in circulate-japan. J Clin Oncol. 2022;40(4):9-9. 10.1200/jco.2022.40.4_suppl.009. [DOI] [Google Scholar]
- 14. Oki E. Dynamics of circulating tumor DNA after resection of colorectal cancer: Galaxy study in circulate-japan. Oral Presentation presented at Society of Surgical Oncology, 2022.
- 15. Morris VK. Circulating tumor DNA in advanced anal cancer: A blood biomarker goes viral. Clin Cancer Res. 2019;25(7):2030-2032. 10.1158/1078-0432.ccr-18-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cabel L, Jeannot E, Bieche I, et al. Prognostic impact of residual hpv ctdna detection after chemoradiotherapy for anal squamous cell carcinoma. Clin Cancer Res. 2018;24(22):5767-5771. 10.1158/1078-0432.ccr-18-0922. [DOI] [PubMed] [Google Scholar]
- 17. Bernard-Tessier A, Jeannot E, Guenat D, et al. Clinical validity of hpv circulating tumor DNA in advanced anal carcinoma: An ancillary study to the epitopes-hpv02 trial. Clin Cancer Res. 2019;25(7):2109-2115. 10.1158/1078-0432.ccr-18-2984. [DOI] [PubMed] [Google Scholar]
- 18. Chung JH, Sanford E, Johnson A, et al. Comprehensive genomic profiling of anal squamous cell carcinoma reveals distinct genomically defined classes. Ann Oncol. 2016;27(7):1336-1341. 10.1093/annonc/mdw152. [DOI] [PubMed] [Google Scholar]
- 19. André F, Ciruelos E, Rubovszky G, et al. Alpelisib for pik3ca-mutated, hormone receptor–positive advanced breast cancer. N Engl J Med. 2019;380(20):1929-1940. 10.1056/nejmoa1813904. [DOI] [PubMed] [Google Scholar]
- 20. Juric D, Rodon J, Tabernero J, et al. Phosphatidylinositol 3-kinase α–selective inhibition with alpelisib (byl719) in pik3ca-altered solid tumors: Results from the first-in-human study. J Clin Oncol. 2018;36(13):1291-1299. 10.1200/jco.2017.72.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vasan N, Razavi P, Johnson JL, et al. Double pik3ca mutations in cis increase oncogenicity and sensitivity to pi3kα inhibitors. Science. 2019;366(6466):714-723. 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cecchini M, Sokol E, Vasan N, et al. Molecular characteristics of advanced colorectal cancer and multi-hit pik3ca mutations. J Clin Oncol. 2022;40(16):3535-3535. 10.1200/jco.2022.40.16_suppl.3535. [DOI] [Google Scholar]
- 23. Millis SZ, Ikeda S, Reddy S, Gatalica Z, Kurzrock R.. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol. 2016;2(12):1565-1573. 10.1001/jamaoncol.2016.0891. [DOI] [PubMed] [Google Scholar]
- 24. Mao J-H, Kim I-J, Wu D, et al. Fbxw7 targets mtor for degradation and cooperates with pten in tumor suppression. Science. 2008;321(5895):1499-1502. 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jardim DL, Wheler JJ, Hess K, et al. Fbxw7 mutations in patients with advanced cancers: Clinical and molecular characteristics and outcomes with mtor inhibitors. PLoS One. 2014;9(2):e89388. 10.1371/journal.pone.0089388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jia Q, Wang J, He N, et al. Titin mutation associated with responsiveness to checkpoint blockades in solid tumors. JCI Insight. 2019;4(10):e127901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li X, Pasche B, Zhang W, Chen K.. Association of muc16 mutation with tumor mutation load and outcomes in patients with gastric cancer. JAMA Onco. 2018;4(12):1691-1698. 10.1001/jamaoncol.2018.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamatham S. Metastatic anal cancer. Springer Publishing Company, 2019. [Google Scholar]
- 29. Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nature Cancer. 2020;1(9):873-881. 10.1038/s43018-020-0096-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.