Abstract
Chromodomain helicase DNA-binding protein 5 (Chd5) is an ATP-dependent chromatin remodeler that promotes neuronal differentiation. However, the mechanism behind the action of Chd5 during neurogenesis is not clearly understood. Here we use transcriptional profiling of cells obtained from Chd5 deficient mice at early and late stages of neuronal differentiation to show that Chd5 regulates neurogenesis by directing stepwise transcriptional changes. During early stages of neurogenesis, Chd5 promotes expression of the proneural transcription factor Six3 to repress Wnt5a, a non-canonical Wnt ligand essential for the maturation of neurons. This previously unappreciated ability of Chd5 to transcriptionally repress neuronal maturation factors is critical for both lineage specification and maturation. Thus, Chd5 facilitates early transcriptional changes in neural stem cells, thereby initiating transcriptional programs essential for neuronal fate specification.
Keywords: neural stem cells, chromatin, transcription, differentiation, maturation
Graphical Abstract
Chromatin remodeler Chd5 turns on proneural transcription factor Six3 during early neurogenesis to keep neuron maturation factor Wnt5a in a repressed state at this time point, so that cells can appropriately upregulate its expression when its function is required at a later neurogenesis stage. However, in the absence of Chd5, Six3 is downregulated; consequently, Wnt5a is prematurely upregulated at early neurogenesis, abrogating neuron differentiation and maturation programs later on.
Significance Statement.
Chromatin regulators orchestrate gradual epigenetic and transcriptional changes as stem cells differentiate into various lineages. Here we show that the chromatin remodeler Chd5 regulates expression of the proneural transcription factor Six3, preventing premature differentiation of neural stem cells and providing proper transcriptional maturation at late differentiation stages. This work highlights the importance of transcriptional regulation of stem cells starting from early differentiation stages and provides insight into molecular and cellular processes impacted in diseases caused by deregulated CHD5, as overrepresented in patients with cancer and neurodevelopmental disorders (NDDs).
Introduction
In the developing brain, neural stem cells (NSCs) first proliferate to self-renew and increase the stem cell pool, which later transition to produce neuronal and glial cells of the central nervous system (CNS).[1,2] During this intricate process of neurogenesis, NSCs go through a succession of intermediate and lineage-specific cell states to generate differentiated cell types. To form the robust size and complex cell composition characteristic of the mammalian brain, NSC proliferation and the respective switch to differentiation need to be tightly regulated. These processes are guided by cell intrinsic as well as extrinsic factors; for instance, expression of proneural transcription factors Mash1 and Neurogenin2, as well as morphogens Wnt3a and Wnt7a are sufficient to induce neuronal differentiation.[3–5] In addition, such intrinsic and extrinsic factors can also regulate each other’s expression and/or function. For example, during embryonic neurogenesis, the transcription factor Pax6 represses the expression of Sonic Hedgehog (Shh) in the developing epithalamus to ensure proper formation of the habenula.[6] Furthermore, extracellular signals can also be converted into secondary messengers that ultimately consolidate functional alterations in transcriptional output, such as the Wnt/β-catenin pathway regulating the expression of Neurogenin1/2.[4] In addition, misregulation of Wnt ligands in the progenitors can have profound impacts on neurogenesis, as inhibition of Wnt5a in pluripotent stem cells is essential to generate cerebral organoids.[7] Hence, to fully understand brain organogenesis, the dynamics of cell intrinsic and extrinsic factors regulating differentiation and maturation of neurons and glia need to be elucidated.
When stem cells undergo differentiation, various chromatin modulators such as ATP-dependent chromatin remodelers, histone-modifying enzymes, and lineage-specific transcription factors work together to fine-tune chromatin organization and gene regulation, to progressively achieve the transcriptional state required for differentiated cell types.[8] Early changes during the differentiation process enable the cells to follow a coherent trajectory toward lineage specification; hence, defects in early transcriptional programs can abrogate the process of fate specification and maturation later on. For example, the nucleosome remodeling and deacetylation (NuRD) complex repress a number of proteins in embryonic stem cells (ESCs) to maintain transcriptional homeostasis; however, in the absence of NuRD components Mta1/2/3 or Mbd3, ESCs have excessive transcriptional noise, causing them to deviate from following differentiation cues that dictate cell fate specification.[9–11] Hence, appropriate regulation of chromatin dynamics starting from early differentiation time points is necessary for the generation of fully functional mature cell types. Although the roles of transcription factors in development have been well studied over the past few decades, knowledge about ATP-dependent chromatin remodelers is still very limited.
Chd proteins are ATP-dependent chromatin remodelers that form a nine-member Chd family of proteins. All of them are expressed in neural tissues to regulate various biological processes, including NSC proliferation, differentiation, and maturation of neurons and/or glia.[12] Furthermore, mutations in 8 of 9 CHD proteins have been correlated to 1 or more neurodevelopmental disorders (NDDs).[13] CHD5 is a tumor suppressor that is frequently inactivated in brain malignancies,[14] and mutations in CHD5 have been reported in patients with NDDs, as patients with heterozygous mutations in CHD5 have language deficits, intellectual disability, epilepsy, and motor delay.[15] Hence, CHD5 is suspected to play a role in regulating proliferation and differentiation of cells during brain development. In mice, depletion of Chd5 using shRNAs during embryonic neurogenesis impedes neuronal differentiation and migration; [16,17] however, the mechanism of action of Chd5 during neurogenesis remains unclear. We have previously shown that Chd5 deficiency in NSCs causes increased proliferation and a premature surge of protein synthesis. Furthermore, at 3 h post-differentiation (hpd)—the time point at which enhanced translation is seen in wild-type (WT) cells—Chd5−/− cells have a reciprocal trend with a precipitous drop in nascent peptide synthesis. In addition, fewer neurons are generated by Chd5−/− NSCs.[18]
Chd5−/− cells have defects starting at early neuronal differentiation stages; we therefore sought to investigate transcriptional changes caused by Chd5 deficiency during neuronal differentiation. In this study, we report that loss of Chd5 in NSCs causes premature transcriptional activation of neuronal differentiation programs, despite proneural transcription factors such as Six3 being downregulated. Six3 maintains the progenitor state of NSCs by repressing Wnt5a, a non-canonical Wnt ligand that plays an important role in the formation and/or maintenance of axons and dendrites during neuronal maturation.[19–22] Due to downregulation of Six3 in Chd5−/− cells, Wnt5a is prematurely upregulated early during differentiation. Consequently, Chd5−/− NSCs are unable to follow an appropriate neuronal differentiation trajectory to activate maturation programs later on, including upregulation of Wnt5a.
Materials and Methods
Neural Stem Cell Culture and Differentiation
NSCs were prepared from tissue collected from the periventricular region of the WT and Chd5−/− mouse brain at P1 using established procedures.[23,24] For neuronal differentiation, cells were grown as monolayers and maintained in media without growth factors. Chd5 (NCBI transcript ID: NM_001081376.1) and Six3 (NCBI transcript ID: NM_011381.4) were cloned into PCW (Addgene: 50661) and PHAGE (Addgene: 106281) vectors respectively, and expressed in NSCs using lentiviral transduction method. 12.5 ng/μL of doxycycline (Sigma) was used to induce the expression of Chd5. All animal handling procedures and experimental protocols followed the guidelines of Cold Spring Harbor Institutional Animal Care and Use Committee.
Gene Expression Analysis
Total RNA was extracted from the cells with Trizol (Thermo Fisher) using the manufacturer’s protocol. Superscript III Reverse Transcriptase (Thermo Fisher) was used to convert mRNA into cDNA for qPCR analysis. qPCR was done using the SYBRGreen master mix in the Quantstudio 6 Flex machine (Thermo Fisher) for 40 cycles. For RNA-seq, libraries were prepared using TruSeq Stranded mRNA Sample Preparation Kit from Illumina. Single-end sequencing (75bp) was performed using NextSeq 500 in high-output mode (See Supplementary Methods section for more detail).
Molecular Analysis
Protein level expression was assessed by Western blot as previously described.[25]
BrdU Cell Proliferation Assay
NSCs cultured as monolayers were treated with 10 μM BrdU and incubated for 24 h. BrdU cell proliferation assay kit (Cell Signaling Technology) was used to quantify proliferation.
Immunofluorescence Analysis
Immunofluorescence of adherent cells was done exactly as described.[18] Images were obtained using confocal microscopes (Zeiss LSM710 and Zeiss LSM780). Map2-positive and Gfap-positive cells were quantified using the Cell Counter Plugin of Fiji software.
CUT&RUN
CUT&RUN was performed with anti-FLAG M2 antibody (Sigma; 1:200 dilution) using WT cells transduced with PHAGE-Six3 at 3 hpd as previously described.[26]
Statistical Analysis
All plots show mean with SD. Statistical analyses were performed using ANOVA or Kruskal Wallis followed by a two-tailed Student’s t-test for comparing more than 2 groups. Significance level was set at P < .05.
Results
Chd5 Deficiency Leads to Premature Differentiation of Neural Stem Cells
Chd5 is an ATP-dependent chromatin remodeler that is expressed robustly in neurons (Fig. 1A; Supplementary Fig. S1A), suggesting it may have a cell-type-specific function during normal development. We have previously shown that loss of Chd5 in NSCs causes defects in the dynamics of chromatin compaction and nascent peptide synthesis starting at early differentiation time points (between 0 and 3 hpd), leading to untimely upregulation of the proneural transcription factor Mash1 and generation of fewer neurons.[18] To further investigate the role of Chd5 in the process of neuronal differentiation and to delineate the effects of loss of Chd5 during neurogenesis, we cultured primary NSCs from WT and Chd5−/− mice and differentiated them into neuronal lineages. We collected these cells at 3 hpd (early-stage differentiation) and 48 hpd (late-stage differentiation) and assessed global transcriptional alterations caused by Chd5 deficiency during the neuronal differentiation process (Fig. 1B). Both WT and Chd5−/− NSCs expressed stem cell markers Pax6 and Nestin (Fig. 1C). However, we observed enhanced expression of Nestin, a protein that is predominantly expressed in activated NSCs in Chd5−/− samples, suggesting premature activation of Chd5−/− NSCs (Supplementary Fig. S1B). In the absence of growth factors, ~90% of WT and Chd5−/− cells entered G1/G0 phase by 24 h (Supplementary Fig. S1C). They differentiated into the neuronal lineage and expressed neurogenic transcription factor Neurod1 at 3 hpd (Fig. 1C), and at 48 hpd, 49.9% of WT and 39.1% of Chd5−/− cells expressed differentiated neuron-specific microtubule associated protein Map2 (Fig. 1C; Supplementary Fig. S1D). Furthermore, the expression of Neurod1 at 3 hpd and Map2 at 48 hpd were lower in Chd5−/− samples (Supplementary Fig. S1E–S1F), suggesting defects in neuronal differentiation caused by loss of Chd5.
Figure 1.
Chd5 deficiency causes transcriptional defects during neuronal differentiation, leading to premature differentiation at an early time point and abrogated maturation at a later time point. (A) Normalized expression levels of Chd5 in single cells collected from adult mouse brains. (Data adapted from Allen Brain Map portal—Mouse Whole Cortex and Hippocampus SMART-seq (2019) with 10×-SMART-seq taxonomy (2020) as reference profiles[40]). (B) Schematic diagram of the experimental design for RNA-seq experiment. Samples for RNA-seq were collected at 3 and 48 hpd. (C) Representative immunofluorescent images of WT and Chd5−/− NSCs at 0, 3, and 48 hpd, analyzed for Nestin and Pax6, Neurod1, and Map2, respectively. Scale bar = 100 μm. (D) Heatmap showing scaled expression values of differentially expressed genes in WT and Chd5−/− samples at 3 hpd with their corresponding GO terms (n = 3). Differentially expressed genes were called with FDR < 0.05 and fold change of (±) 1.5. See also Supplementary Fig S2B. (E) Heatmap showing scaled expression values of differentially expressed genes in WT and Chd5−/− samples at 48 hpd with their corresponding GO terms (n = 3). Differentially expressed genes were called with FDR < 0.05 and fold change of (±) 1.5. See also Supplementary Fig S3B.
RNA-seq analyses revealed that at 3 hpd, 752 genes were upregulated, whereas 883 genes were downregulated in Chd5−/− cells (Supplementary Fig. S2A), suggesting Chd5 was not exclusively an activator or repressor. Using gene ontology (GO) analysis of the upregulated genes, we found that factors required for NSC proliferation, neuron differentiation, and synapse organization, including genes encoding Prox1, En2, Eomes, and Nlgn1, were upregulated in Chd5−/− samples (Fig. 1D; Supplementary Fig. S2B), in line with the increased proliferation and premature differentiation phenotype we previously reported.[18] On the other hand, regulators of apoptosis and cell migration were downregulated in these samples (Fig. 1D; Supplementary Fig. S2B), consistent with defects in migration seen after shRNA-mediated knockdown of Chd5 in vivo [17]. Furthermore, gene set enrichment analysis (GSEA) using gene sets containing factors required for cells undergoing neuronal differentiation and calcium signaling pathway showed positive enrichment in Chd5−/− samples at 3 hpd (S2C-D). These observations indicate that Chd5 deficiency causes premature transcriptional upregulation of neurogenic programs during early differentiation.
Chd5 Deficiency Abrogates Proper Transcription of Neuronal Maturation Factors During Neurogenesis
At 48 hpd, a time point at which the NSCs had differentiated, 440 genes were upregulated and 409 genes were downregulated in Chd5−/− samples, highlighting Chd5 as both an activator and a repressor in differentiated cells (Supplementary Fig. S3A). GO analysis revealed that genes required for other lineages such as male genitalia and skeletal system development, as well as morphogens required for embryonic development were upregulated in Chd5−/− cells, but genes encoding factors required for nervous system development and ion transport, such as Map1b, Hes6, Nalcn, and Panx1, were downregulated (Fig. 1E; Supplementary Fig. S3B). Moreover, GSEA enrichment analysis showed a negative enrichment for genes required for neuronal maturation and calcium signaling pathway (Supplementary Fig. S3C–S3D). These results suggest that Chd5−/− neurons have defects in initiating transcriptional programs required for maturation at 48 hpd. The transcriptional analyses at early- and late-differentiation time points indicate that Chd5 deficiency causes premature differentiation of NSCs and abrogation of maturation programs in neurons.
Neurogenic Chromatin Modulators are Misregulated in Chd5 Deficient Cells
To explore downstream pathways responsible for the premature differentiation phenotype of Chd5−/− cells, we examined the expression of known neurogenic chromatin modulators at 3 hpd. Despite the enrichment of genes encoding several neurogenic transcription factors such as Eomes, Pou3f1, and Foxo3 (Fig. 2A; Supplementary S4A–S4C), a handful of genes encoding neurogenic chromatin modulators including Six3, Neurod1, Hdac11, Pou3f1, and Hes1 were downregulated in Chd5−/− cells at this time point (Fig. 2A), suggesting premature differentiation with transcriptional defects in Chd5−/− cells. We hypothesized the transcription factor Six3 to be an important downstream target of Chd5 in this process, as Six3 is essential for proper forebrain and eye development; Six3−/− mice lack most of the head structures anterior to the midbrain, including the eyes and nose, and die at birth.[27] Furthermore, 1%-2% of the Chd5+/− and Chd5−/− female mice are born with eyes lacking the neuroretina (Supplementary Fig. S4D–S4E), similar to the loss of eye phenotype seen in Six3−/− mice. To test whether Chd5 directly regulates the expression of Six3, we transduced Chd5−/− NSCs with a construct expressing an inducible version of Chd5 cDNA. Importantly, overexpression of Chd5 in differentiating cells at 3 hpd rescued the downregulation of Six3 at both the transcript (Fig. 2B) and protein level (Fig. 2C; Supplementary Fig. S4F–S4G). These findings indicate that Chd5 is sufficient for positively regulating the expression of Six3 in NSCs at an early stage of differentiation.
Figure 2.
Chd5 regulates the expression of Wnt5a via proneural transcription factor Six3. (A) Heatmap showing scaled expression values of neurogenic chromatin modulators at 3 hpd (n = 3); FDR<0.05. (B) Relative mRNA expression of Six3 in WT and Chd5−/− samples with overexpression of empty vector (EV) or Chd5 at 3 hpd. ***<.001; ****<.0001. ANOVA followed by a two-tailed Student’s t-test. (C) Representative Western blots of WT and Chd5−/− samples with overexpression of EV or Chd5 at 3 hpd, assessed for the expression of Chd5-FLAG, Chd5, Six3, and β-actin. See also Supplementary Fig. S4F–5FG. (D) Normalized TPM counts of Wnt5a in cells at 0, 3, and 48 hpd as determined by RNA-seq. n = 2 for 0 hpd and n = 3 for 3 and 48 hpd samples. (For 0 hpd-data reanalyzed from NCBI GEO database[18], accession GSE80583). (E) Representative Western blots of WT and Chd5−/− samples at 3 hpd, assessed for the expression of pCamKII, Gap43 and β-actin. (F) Genome browser tracks for the Wnt5a promoter region using data from CUT&RUN experiments in WT cells with an antibody against Six3-FLAG and IgG, showing binding of Six3 at a locus 2-3 kb upstream of Wnt5a transcription start site. (G) Relative mRNA expression of Wnt5a in WT and Chd5−/− cells with overexpression of EV or Six3 at 3 hpd. *<.05; **<.01. ANOVA followed by a two-tailed Student’s t-test (n =4). (H) Relative mRNA expression of Wnt5a in WT and Chd5−/− samples with overexpression of EV or Six3 at 48 hpd. *<.05; **<.01. ANOVA followed by a two-tailed Student’s t-test (n = 4). (I) Representative Western blots of WT and Chd5−/− samples with overexpression of EV or Six3 at 3 and 48 hpd, assessed for Six3, Six3-FLAG, Wnt5a, and β-actin. (# indicates overexpressed Six3-FLAG). See also Supplementary Fig. S5C–S5F. (J) Schematic diagram showing the relationship between Chd5, Six3, and Wnt5a. (K) Representative Western blots of WT and Chd5−/− cortical lysates assessed for Chd5, Wnt5a, pCamKII, Gap43, and β-actin.
Six3 Represses Wnt5a in Differentiating Neural Stem Cells
During embryonic neurogenesis, Six3 promotes the differentiation of NSCs into neurons. The function of Six3 is essential in NSCs, as conditional depletion of Six3 in NSCs causes severe developmental defects, whereas conditional depletion in newborn neurons has a more subtle effect.[28,29] Earlier in development, Six3 represses the expression of Wnt1 and Wnt8b [27,30], and activates the expression of Shh and Foxg1.[31,32] Since the known factors downstream of Six3 are indispensable for brain development, we hypothesized that Six3 might be functioning as a transcriptional regulator in the NSCs of the CNS as well. To identify factors that were potentially repressed by Six3, we checked the expression levels of the known downstream factors of Six3 as well as the other Wnt ligands in addition to Wnt1 and Wnt8b at an early differentiation stage. This allowed us to discover that Wnt5a was upregulated in Chd5−/− cells at 0 and 3 hpd (Fig. 2D; Supplementary Fig. S5A), which indicated that it was a potential downstream target of Six3 in NSCs during early neuronal differentiation.
Wnt5a has been reported to function through both β-catenin-dependent and independent pathways.[19,20,33] To determine the downstream pathway affected by upregulation of Wnt5a in this context, we probed for the expression of β-catenin in the nuclear lysates of WT and Chd5−/− cells, as it is an indicator of canonical Wnt/β-catenin pathway. We did not detect any changes in nuclear β-catenin in Chd5−/− cells compared to WT cells (Supplementary Fig. S5B). However, pCamKII and Gap43, known downstream targets of Wnt5a in the non-canonical Wnt/calcium pathway,[19] were upregulated in Chd5−/− cells 3 hpd (Fig. 2E). Furthermore, GSEA analysis using the calcium signaling pathway gene set showed a positive enrichment in Chd5−/− cells at 3 hpd (see Supplementary Fig. S2D), further confirming transcriptional upregulation of genes encoding components of the Wnt/calcium pathway.
To test whether Six3 directly regulated the expression of Wnt5a, we utilized CUT&RUN using an antibody against FLAG in WT cells with Six3-FLAG overexpression at 3 hpd. Through this approach, we detected binding of Six3 at a locus 2-3kb upstream of the Wnt5a transcriptional start site (TSS) (Fig. 2F). Furthermore, overexpression of Six3 in Chd5−/− NSCs rescued Wnt5a expression at both the transcript (Fig. 2G) and the protein level (Fig. 2I; Supplementary Fig. S5C–S5D) at 3 hpd. These experiments established Six3 as a repressor of Wnt5a in NSCs following differentiation for 3 h.
The role of Wnt5a has been well-defined in neurons as a regulator of axon guidance and postsynaptic dendrite formation.[19–22] Even though Wnt5a was upregulated in Chd5−/− cells at 0 and 3 hpd, it was downregulated at 48 hpd (see Fig. 2D), a time when endogenous Six3 was reduced in WT cells (Fig. 2I), and the function of Wnt5a is crucial for axonal and dendritic growth. Furthermore, GSEA analysis of RNA-seq data at 48 hpd showed negative enrichment of calcium signaling pathway in Chd5−/− cells (see Supplementary Fig. S3D), suggesting transcriptional downregulation of the Wnt/calcium pathway genes. However, with overexpression of Six3 and initial repression of Wnt5a in Chd5−/− cells at 3 hpd (see Fig. 2G–2I), the downregulation of Wnt5a at 48 hpd was rescued at the transcript (Fig. 2H) and protein level (Fig. 2I; Supplementary Fig. S5E–S5F). Hence, we can conclude that Chd5 positively regulates Six3, which is a repressor of Wnt5a (Fig. 2J). Furthermore, these findings indicate that transcriptional deregulation during early differentiation influences events later on that precludes fate specification and maturation.
Chd5 is dispensable for producing neurons in vivo, but expression of Wnt/calcium signaling factors in the brain is compromised.
Chd5−/− primary NSCs generate fewer neurons compared to the WT NSCs in culture.[18] However, Chd5−/− adult mice did not have significant changes in brain size, weight, or generation of neurons and astrocytes (Supplementary Fig. S6A–S6F). Through the RNA-sequencing of differentiated cells in culture, we concluded that Chd5−/− cells have defects in transcriptional activation of genes that regulate neuron maturation programs, including upregulation of Wnt5a. Therefore, we assessed whether the adult brain tissues—which largely consist of differentiated cell types—have compromised expression of genes encoding Wnt5a and/or its downstream Wnt/calcium pathway factors. We performed Western blot analysis using whole brain lysates of P28 mice and found that lysates from Chd5−/− animals had lower levels of Wnt5a as well as its downstream factors pCamKII and Gap43 (Fig. 2K). These findings demonstrate that the molecular defects seen in differentiated cells in culture are recapitulated in vivo, even if the differentiation defect is compensated.
Overexpression of Six3 Rescues Differentiation Defects Caused by Chd5 Deficiency
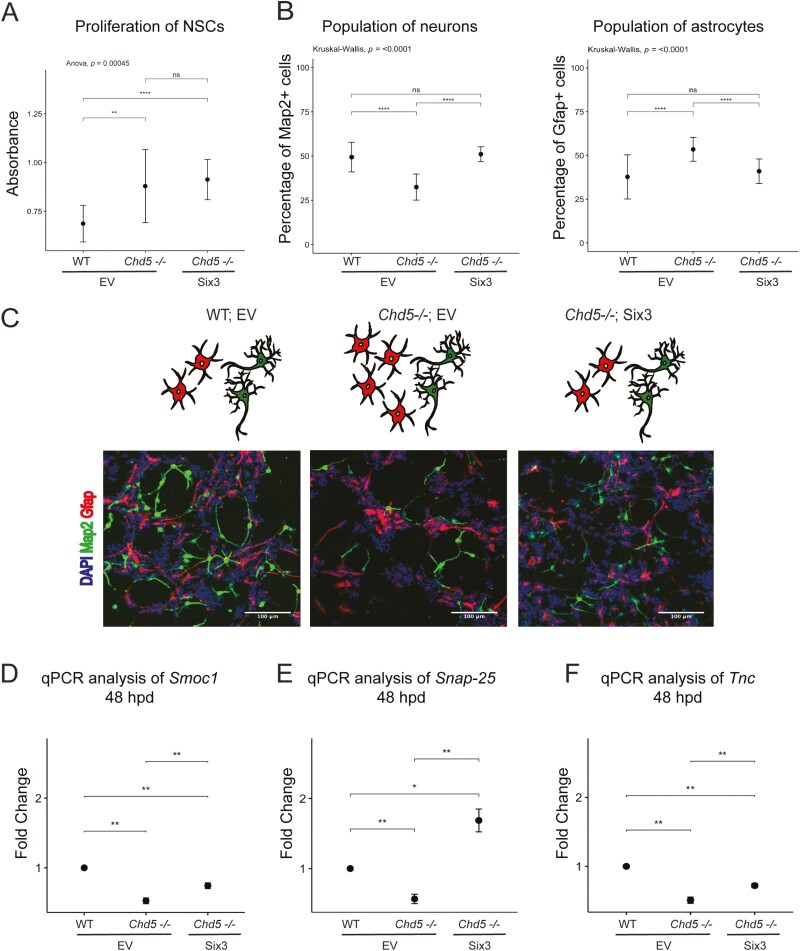
We have shown that Six3 is an important downstream target of Chd5 (see Fig. 2A–2J). We therefore tested whether Six3 can rescue the cellular defects caused by Chd5 deficiency. The 2 major cellular phenotypes seen in Chd5−/− cells are proliferation and differentiation defects.[18] To investigate whether Six3 rescues the proliferation defect, we treated WT and Chd5−/− cells transduced with empty vector control (EV) or Six3 with the thymidine analog, BrdU, for 24 h, and assayed for BrdU incorporation. Overexpression of Six3 did not rescue the proliferation defect caused by Chd5 deficiency (Fig. 3A). However, when we differentiated these cells for 5 days, Chd5−/− cells with Six3 overexpression produced more neurons and fewer astrocytes than Chd5−/− cells transduced with EV (Fig. 3B–3C). Hence, Six3 is a downstream factor of Chd5 that effectively rescues differentiation, but not proliferative defects, caused by Chd5 deficiency.
Figure 3.
Overexpression of Six3 rescues differentiation defects caused by Chd5 deficiency. (A) Absorbance measured from WT and Chd5−/− NSCs with overexpression of empty vector (EV) or Six3 after BrdU incorporation for 24 h and detection by BrdU cell proliferation assay kit. **>.01; ****>.0001. ANOVA followed by a two-tailed Student’s t-test. (B) Quantification of Map2+ neurons and Gfap+ astrocytes generated by WT and Chd5−/− cells with overexpression of EV or Six3 at 5 days post-differentiation (n = 28). ****<.0001; ns = not significant. Kruskal-Wallis followed by a two-tailed Student’s t-test. (C) Representative immunofluorescent images of WT and Chd5−/− cells with overexpression of EV or Six3 at 5 days post-differentiation and analyzed for Map2 (green), Gfap (red), and DAPI nuclear signal (blue), accompanied by a schematic on the top. Scale bar = 100 μm. Relative mRNA expression of Smoc1 (D), Snap-25 (E) and Tnc (F) in WT and Chd5−/− cells with overexpression of EV or Six3 at 48 hpd. *<.05; **<.01. ANOVA followed by a two-tailed Student’s t-test (n = 4).
Furthermore, we wanted to test whether transcriptional defects caused by Chd5 deficiency in differentiated cells (aside from Wnt5a), were rescued by Six3 expression. We examined the expression of three calcium responsive factors that were downregulated in Chd5−/− samples: secreted modular calcium-binding protein-1 (Smoc1), synaptosome associated protein-25 (Snap-25), and troponin C (Tnc).[34–36] Overexpression of Six3 rescued the downregulation of these 3 factors at 48 hpd (Fig. 3D–3F), further highlighting Six3 as an important neurogenic transcription factor downstream of Chd5 that transcriptionally represses Wnt5a during early neuronal differentiation, and can alleviate defects caused by Chd5 deficiency to respond to differentiation cues, thus reestablishing appropriate differentiation and maturation programs.
Discussion
Cell intrinsic and extrinsic factors facilitate deterministic transcriptional programs that steer NSCs toward proliferation versus differentiation and maturation into various cell types. ATP-dependent chromatin remodelers along with a number of interacting partners, including transcription factors and histone modifiers, initiate and/or maintain required epigenetic and transcriptional changes while the cells are undergoing fate specification. Defects in these steps can obstruct the achievement of the transcriptional states required for proper functioning of the differentiated cell types. Here, we define the role of Chd5 and its downstream target Six3 during neurogenesis. Through molecular analyses, we identify Six3 as an important transcription factor downstream of Chd5 that regulates neuronal differentiation by keeping Wnt5a in a repressed state in NSCs at an early differentiation time point. The role of Wnt5a has been well-defined in neurons,[20] but its upstream regulation is not well understood. Here we show that early repression of Wnt5a in progenitors is necessary for cells to be able to enhance its expression post-differentiation when its function is required. This study supports the notion that increased transcriptional noise in progenitors abrogates their ability to respond to differentiation cues and maintain a coherent differentiation trajectory.
Chd proteins have been reported to regulate multiple cellular functions during stem cell differentiation into various lineages.[12] Similarly, Chd5 regulates proliferation of NSCs and neuronal differentiation.[18] Here we show that overexpression of Six3 in Chd5−/− NSCs rescues differentiation, but not the proliferation defects caused by Chd5 loss. Six3 is a positive regulator of NSC proliferation; [37] however, the role of Chd5 in this cellular process seems to be independent of Six3, as even with lower levels of Six3, Chd5−/− cells proliferate more. Therefore, Six3 is a transcription factor downstream of Chd5 that promotes neuronal differentiation. We also showed that genes encoding other proneural transcription factors like Neurod1, Hes1, and Pou3f4 are also downregulated, whereas Eomes, Pou3f1, and Foxo3 are aberrantly upregulated in Chd5−/− cells. Hence, we envision Chd5 as regulating other downstream pathways through additional factors, similar to Chd2 regulating genes encoding the transcription factors REST, Neurod1, and Neurog2 at different stages of neurogenesis,[38,39] and more studies are needed to elucidate the full range of functions of Chd5 during brain development.
Through this study, we provide evidence to show that defects in early events during the neuronal differentiation process can have profound consequences for proper fate specification and maturation into fully functional neurons. Developmental defects cause NDDs; unsurprisingly, mutations in CHD5 have also been seen in patients with NDDs.[15] To find causal links between mutations in genes encoding chromatin remodelers such as CHD5 and dysfunctions seen in patients, we need to understand the role of these proteins during normal development. Functional studies in cell culture models and mice can give us a better insight into how the loss of functional CHD5 causes defects in neuron differentiation and maturation, ultimately leading to clinical manifestations seen in patients.
Conclusion
Our data indicate that Six3 is a proneural transcription factor downstream of Chd5 that represses neurogenic factor Wnt5a at early differentiation stages; this is essential for the upregulation of Wnt5a post-differentiation, when its function is required. Moreover, overexpression of Six3 in Chd5−/− NSCs rescues transcriptional and differentiation defects caused by Chd5 deficiency. In summary, Chd5 represses premature upregulation of neurogenic factor Wnt5a through Six3 in early neuronal differentiation, so that the cells are amenable to respond to the differentiation cues and turn on maturation programs.
Supplementary Material
Acknowledgments
We are grateful to Osama El Demerdash at Cold Spring Harbor Laboratory (CSHL) for helping with CUT&RUN data analysis and Matt Fisher in the Mills laboratory for helpful comments on the manuscript.
Contributor Information
Padmina Shrestha, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA; Department of Molecular and Cell Biology, Stony Brook University, Stony Brook, NY, USA.
Anbalagan Jaganathan, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA.
Dhananjay Huilgol, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA; Department of Neurobiology, Duke University Medical Center, Durham, NC, USA.
Carlos Ballon, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA.
Yon Hwangbo, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA.
Alea A Mills, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA.
Funding
We thank the CSHL Cancer Center Shared Resources, including Animal and Tissue Imaging, Next Generation Sequencing, Microscopy and Antibody, which are supported by the CSHL Cancer Center Support Grant. This project was also supported through the Darlene Carbone Brain Tumor Foundation, the Bradley Zankel Foundation, the Edward Davis Foundation, and the Cold Spring Harbor Laboratory-Northwell Health Affiliation. Research in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA190997 and Award Number P30CA045508 and the Office of the Director, National Institutes of Health under Award Number R21OD018332. D.H. was supported by the Human Frontier Science Program long-term fellowship LT000075/2014-L and NARSAD Young Investigator Grant no. 26327. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
The authors declare no potential conflicts of interest.
Author Contributions
P.S.: performed and designed experiments, data analysis, manuscript writing—original draft, reviewing and editing. A.J.: performed and designed experiments. D.H.: designed experiments, manuscript writing—review and editing. C.B., Y.H.: performed experiments. A.A.M.: designed experiments, funding acquisition, and manuscript writing—original draft, reviewing, and editing.
Data Availability
The RNA-seq and CUT&RUN data were deposited to Gene Expression Omnibus (GEO) and can be found with GEO accession numbers GSE208168 and GSE208169, respectively.
References
- 1. Kriegstein, A. R., Subramanian, L., Obernier, K., and Alvarez-Buylla, A. (2020). Neural stem cells among glia. In Rubenstein J, Rakic P, Chen B, Kwan KY, eds. Patterning and Cell Type Specification in the Developing CNS and PNS (pp. 775–806). Elsevier. 10.1016/b978-0-12-814405-3.00031-x [DOI] [Google Scholar]
- 2. Qian X, Shen Q, Goderie SK, et al. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28(1):69-80. 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 3. Geoffroy CG, Critchley JA, Castro DS, et al. Engineering of dominant active basic helix-loop-helix proteins that are resistant to negative regulation by postnatal central nervous system antineurogenic cues. Stem Cells. 2009;27(4):847-856. 10.1002/stem.17. [DOI] [PubMed] [Google Scholar]
- 4. Hirabayashi Y, Itoh Y, Tabata H, et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131(12):2791-2801. 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 5. Muroyama Y, Kondoh H, Takada S.. Wnt proteins promote neuronal differentiation in neural stem cell culture. Biochem Biophys Res Commun. 2004;313(4):915-921. 10.1016/j.bbrc.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 6. Caballero IM, Manuel MN, Molinek M, et al. Cell-autonomous repression of Shh by transcription factor Pax6 regulates diencephalic patterning by controlling the central diencephalic organizer. Cell Rep. 2014;8(5):1405-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ideno H, Imaizumi K, Shimada H, et al. Human PSCs determine the competency of cerebral organoid differentiation via FGF signaling and epigenetic mechanisms. iScience. 2022;25(10):105140. 10.1016/j.isci.2022.105140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sokpor G, Castro-Hernandez R, Rosenbusch J, Staiger JF, Tuoc T.. ATP-dependent chromatin remodeling during cortical neurogenesis. Front Neurosci. 2018;12:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgold T, Barber M, Kloet S, et al. The nucleosome remodelling and deacetylation complex suppresses transcriptional noise during lineage commitment. EMBO J. 2019;38(12). 10.15252/embj.2018100788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaji K, Caballero IM, MacLeod R, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8(3):285-292. 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 11. Reynolds N, Latos P, Hynes-Allen A, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10(5):583-594. 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodman JV, Bonni A.. Regulation of neuronal connectivity in the mammalian brain by chromatin remodeling. Curr Opin Neurobiol. 2019;59:59-68. 10.1016/j.conb.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cardoso AR, Lopes-Marques M, Oliveira M, et al. Genetic variability of the functional domains of chromodomains helicase DNA-binding (CHD) proteins. Genes. 2021;12(11):1827. 10.3390/genes12111827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bagchi A, Papazoglu C, Wu Y, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128(3):459-475. 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 15. Parenti I, Lehalle D, Nava C, et al. ; Undiagnosed Diseases Network, Friedman, J. Missense and truncating variants in CHD5 in a dominant neurodevelopmental disorder with intellectual disability, behavioral disturbances, and epilepsy. Hum Genet. 2021;140(7):1109-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Egan CM, Nyman U, Skotte J, et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell. 2013;26(3):223-236. [DOI] [PubMed] [Google Scholar]
- 17. Nitarska J, Smith JG, Sherlock WT, et al. A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 2016;17(6):1683-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hwang D-W, Jaganathan A, Shrestha P, et al. Chromatin-mediated translational control is essential for neural cell fate specification. Life Sci Alliance. 2018;1(4):e201700016. 10.26508/lsa.201700016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arredondo SB, Guerrero FG, Herrera-Soto A, et al. Wnt5a promotes differentiation and development of adult-born neurons in the hippocampus by noncanonical Wnt signaling. Stem Cells. 2020;38(3):422-436. [DOI] [PubMed] [Google Scholar]
- 20. Horigane S-I, Ageta-Ishihara N, Kamijo S, et al. Facilitation of axon outgrowth via a Wnt5a-CaMKK-CaMKIα pathway during neuronal polarization. Mol Brain. 2016;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramos-Fernández E, Arrázola MS, Oliva CA, et al. Wnt5a promotes hippocampal postsynaptic development and GluN2B-induced expression via the eIF2α HRI kinase. Sci Rep. 2021;11(1):7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Varela-Nallar L, Alfaro IE, Se rrano FG, Parodi J, Inestrosa NC.. Wingless-type family member 5A (Wnt-5a) stimulates synaptic differentiation and function of glutamatergic synapses. Proc Natl Acad Sci USA. 2010;107(49):21164-21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deleyrolle LP, Reynolds BA.. Isolation, expansion, and differentiation of adult Mammalian neural stem and progenitor cells using the neurosphere assay. Methods Mol Biol. 2009;549:91-101. [DOI] [PubMed] [Google Scholar]
- 24. Marshall GP 2nd, Ross HH, Suslov O, et al. Production of neurospheres from CNS tissue. Methods Mol Biol. 2008;438:135-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gallagher, S., Winston, S. E., Fuller, S. A., and Hurrell, J. G. (2001). Immunoblotting and immunodetection. In Coligan John E.et al., eds. Current Protocols in Immunology, Chapter 8, Unit 8.10. [Google Scholar]
- 26. Skene PJ, Henikoff S.. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife. 2017;6:6. 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lagutin OV, Zhu CC, Kobayashi D, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17(3):368-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song X, Chen H, Shang Z, et al. Homeobox Gene Six3 is required for the differentiation of D2-type medium spiny neurons. Neurosci Bull. 2021;37(7):985-998. 10.1007/s12264-021-00698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu Z, Liang Q, Song X, et al. SP8 and SP9 coordinately promote D2-type medium spiny neuron production by activating Six3 expression. Development. 2018;145(14) 10.1242/dev.165456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu W, Lagutin O, Swindell E, Jamrich M, Oliver G.. Neuroretina specification in mouse embryos requires Six3-mediated suppression of Wnt8b in the anterior neural plate. J Clin Invest. 2010;120(10):3568-3577. 10.1172/jci43219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Geng X, Speirs C, Lagutin O, et al. Haploinsufficiency of Six3 fails to activate Sonic hedgehog expression in the ventral forebrain and causes holoprosencephaly. Dev Cell. 2008;15(2):236-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Geng X, Acosta S, Lagutin O, Gil HJ, Oliver G.. Six3 dosage mediates the pathogenesis of holoprosencephaly. Development. 2016;143(23):4462-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li T, Chan RWS, Lee C-L, et al. WNT5A interacts with FZD5 and LRP5 to regulate proliferation and self-renewal of endometrial mesenchymal stem-like sells. Front Cell Dev Biol. 2022;8:837827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Genchev GZ, Kobayashi M, Kobayashi T, Lu H.. Molecular dynamics provides new insights into the mechanism of calcium signal transduction and interdomain interactions in cardiac troponin. FEBS Open Biol. 2021;11(7):1841-1853. 10.1002/2211-5463.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toft-Bertelsen TL, Ziomkiewicz I, Houy S, Pinheiro PS, Sørensen JB.. Regulation of Ca2+ channels by SNAP-25 via recruitment of syntaxin-1 from plasma membrane clusters. Mol Biol Cell. 2016;27(21):3329-3341. 10.1091/mbc.e16-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vannahme C, Smyth N, Miosge N, et al. Characterization of SMOC-1, a novel modular calcium-binding protein in basement membranes. J Biol Chem. 2002;277(41):37977-37986. 10.1074/jbc.m203830200. [DOI] [PubMed] [Google Scholar]
- 37. Del Bene F, Tessmar-Raible K, Wittbrodt J.. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427(6976):745-749. [DOI] [PubMed] [Google Scholar]
- 38. Kim YJ, Khoshkhoo S, Frankowski JC, et al. Chd2 is necessary for neural circuit development and long-term memory. Neuron. 2018;100(5):1180-1193.e6. 10.1016/j.neuron.2018.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen T, Ji F, Yuan Z, Jiao J.. CHD2 is required for embryonic neurogenesis in the developing cerebral cortex. Stem Cells. 2015;33(6):1794-1806. 10.1002/stem.2001. [DOI] [PubMed] [Google Scholar]
- 40. Yao Z, van Velthoven CTJ, Nguyen TN, et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell. 2021;184(12):3222-3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and CUT&RUN data were deposited to Gene Expression Omnibus (GEO) and can be found with GEO accession numbers GSE208168 and GSE208169, respectively.