Abstract
It is well established that air travel plays a key role in the global spread of many enteric and respiratory diseases, including COVID-19. Even with travel restrictions (e.g. mask wearing, negative COVID-19 test prior to departure), SARS-CoV-2 may be transmitted by asymptomatic or pre-symptomatic individuals carrying the virus. Due to the limitation of current clinical surveillance approaches, complementary methods need to be developed to allow estimation of the frequency of SARS-CoV-2 entry across international borders. Wastewater-based epidemiology (WBE) represents one such approach, allowing the unbiased sampling of SARS-CoV-2 carriage by passenger cohorts entering via airports. In this study, we monitored sewage in samples from terminals (n = 150) and aircraft (n = 32) at three major international airports in the UK for 1–3 weeks in March 2022. As the raw samples were more turbid than typical municipal wastewater, we used beef extract treatment followed by polyethylene glycol (PEG) precipitation to concentrate viruses, followed by reverse transcription quantitative PCR (RT-qPCR) for the detection of SARS-CoV-2 and a faecal indicator virus, crAssphage. All samples taken from sewers at the arrival terminals of Heathrow and Bristol airports, and 85% of samples taken from sites at Edinburgh airport, were positive for SARS-CoV-2. This suggests a high COVID-19 prevalence among passengers and/or airport staff members. Samples derived from aircraft also showed 93% SARS-CoV-2 positivity. No difference in viral prevalence was found before and after COVID-19 travel restrictions were lifted. Our results suggest that WBE is a useful tool for monitoring the global transfer rate of human pathogens and other disease-causing agents across international borders and should form part of wider international efforts to monitor and contain the spread of future disease outbreaks.
1. Introduction
Since the start of the COVID-19 pandemic in March 2020, SARS-CoV-2 has been responsible for over 0.5 billion confirmed cases and over six million deaths worldwide [1]. Approximately 40–45% are asymptomatic or associated with mild symptoms, hence, the number of cases based on clinical surveillance is typically underestimated [2–4]. Although SARS-CoV-2 is a respiratory pathogen, ca. 43% of symptomatic patients shed the virus in their faeces [5] and the virus has also been detected in the faeces of asymptomatic and mildly symptomatic infected individuals [6, 7]. Due to the high prevalence of the virus in faeces, the RNA genome of SARS-CoV-2 can be detected and quantified in domestic wastewater. Hence, by using wastewater-based epidemiology (WBE), the temporal dynamics of viral RNA concentrations in wastewater can be measured and, subsequently, related to case numbers [8]. It has also been shown that WBE can be used as an early warning system and as a predictive approach for the monitoring and mitigation of the COVID-19 pandemic [8–11]. As WBE can provide unbiased information at the community level, it has been implemented in many countries as a complementary monitoring tool for COVID-19 and other viral diseases [12–15]. Furthermore, WBE has been successfully utilised to support monitoring of highly transient populations in near-source settings at the building scale, such as university campuses [16–18], suggestive of its utility to monitor smaller, localised, and dynamic populations.
The usefulness of WBE for international border control has also been investigated, focusing on airplane wastewater surveillance, suggesting that SARS-CoV-2 can be detected, quantified, and sequenced in a static setting distinct from classical piped flows (i.e., sewer networks) [19–21]. There has been only a single study, to date, reporting SARS-CoV-2 RNA levels in sewage from a wastewater treatment plant associated with a major airport [9]. However, there is still no comprehensive, spatio-temporal study of the value of WBE for airport-associated public health assessment.
As data quality predominantly depends on the efficiency of sample processing, several studies have investigated the usefulness of different wastewater concentration methods for SARS-CoV-2 detection [22–27]. To date, filtration, ultrafiltration, and precipitation methods have been widely used for COVID-19 WBE [28–31]. However, the viral recoveries also depend on the volume of the samples and the physico-chemical properties of the wastewater [32, 33]. As the composition of wastewater near-source and at specific locations with water-preserving waste systems (e.g., aircrafts, ferries, mobile toilets) is often more concentrated than the sewage collected at wastewater treatment plants, the currently used WBE methods may need to be modified to optimise performance.
The aim of this study was to investigate the value of WBE for COVID-19 monitoring and public health protection, at three airports in the United Kingdom. Samples were collected from sewers close to the airport terminals, from vacuum trucks collecting wastewater from incoming aircraft, and at a wastewater treatment plant (WWTP) in the vicinity of the airport. We also trialled different wastewater concentration methods tailored for highly turbid samples with the best method coupled with RT-qPCR for viral RNA quantification.
2. Methods and materials
2.1 Sample collection and spiking
Wastewater sampling was conducted by external partners (2030 Labs, Veolia and Aqua Enviro, UK) in liaison with airport staff. Sampling was authorised by Heathrow Airport Ltd, Bristol Airport Ltd and Edinburgh Airport Ltd.
2.1.1 Set 1—Spiked samples for method validation
For method validation, 24 1-litre grab wastewater samples were collected at Edinburgh airport on the 7th and 8th March 2022. From the 24 samples, six 4-litre composites were created by mixing. Four of the composite mixtures were spiked with different concentrations of heat-inactivated SARS-CoV-2 Wuhan and Alpha variants (provided by Prof Andrew Weightman, Cardiff University) to reach final concentrations of approximately 104–105 genome copies (gc)/ml (Table 1). After thorough mixing, approximately 220 ml aliquots were prepared and processed in triplicates as described below.
Table 1. Samples (taken at Edinburgh airport wastewater treatment plant inlet point) spiked with the Wuhan strain and the Alpha variant for SARS-CoV-2 for the study (gc: Genome copies).
| Sample code | Virus spiking type and approx. concentration | Number of replicates |
|---|---|---|
| EDI-neat1 | None | 3 |
| EDI-neat2 | None | 3 |
| EDI-Wuhan_low | Wuhan strain: 104 gc/ml | 3 |
| EDI-Wuhan_high | Wuhan strain: 105 gc/ml | 3 |
| EDI-Wuhan_high-Alpha_low | Wuhan strain: 105 gc/ml | 3 |
| Alpha variant: 104 gc/ml | ||
| EDI-Wuhan_low-Alpha_high | Wuhan strain: 104 gc/ml | 3 |
| Alpha variant: 105 gc/ml |
2.1.2 Set 2—Surveillance samples
The virus concentration method using beef extract elution and polyethylene glycol (PEG) precipitation (see Section 2.2 for details) that recovered most of the spiked viruses was applied to the processing of the further samples taken at Edinburgh, Heathrow and Bristol airports between 8th and 31st March 2022 (n = 180; Table 2). At Edinburgh airport, samples were taken from manholes to capture the outflow of the international terminal (JR1-3; approx. location 55°56’50.7"N 3°21’45.1"W), from a pumping station (P1; approx. location 55°56’39.7"N 3°22’23.3"W) where vacuum trucks used to collect wastewater from aircraft deposit their contents, and at the WWTP (approx. location 55°56’36.6"N 3°23’55.8"W) where all sewage from the airport is discharged. At Heathrow airport, samples were taken from a manhole at the Central Terminal Area (CTA; approx. location 51°28’22.2"N 0°27’03.4"W) capturing terminals T1, T2, and T3. Ten additional samples were also taken directly from the vacuum trucks, which collected aircraft wastewater directly from ten individual aircraft during the day of sampling. At Bristol airport, samples were taken from manholes to sewers collecting wastewater predominantly serving the arrival terminal (MH2; approx. location 51°23’10.6"N 2°42’53.0"W) and from the entire airport (MH3; approx. location 51°23’15.4"N 2°42’20.5"W). Additional samples were taken from a deposit sites where vacuum trucks collecting aircraft wastewater unload their content using an autosampler programmed to take grab samples during each wastewater discharge event (MH1; approx. location 51°23’09.5"N 2°42’13.4"W). These samples contained a mixture of wastewater originating from 4–17 planes (S1 Table). At each location, 24-hour composite samples were taken using autosamplers, except when samples were taken from vacuum trucks at Heathrow airport. In that case grab samples were taken using a clean wastewater sampling device.
Table 2. Sampling sites, sample types, numbers, and physico-chemical properties for wastewater collected at three UK airports.
| Sampling site | Sampling period | n | Sample type | pH | Turbidity NTU | Conductivity μS/cm | Ammonium mg N/l | Orthophosphate mg P/l |
|---|---|---|---|---|---|---|---|---|
| EDI-JR1 | 8–23 March 2022 | 44 | 2-hour composite | 7.79 (0.07) | 7.28 (2.02) | 1038 (196) | 27.7 (1.83) | 5.5 (1.25) |
| EDI-JR2 | 8–23 March 2022 | 16 | 24-hour composite | 7.44 (0.12) | 85.7 (9.1) | 1576 (206) | 71.6 (6.54) | 11.0 (5.25) |
| EDI-JR3 | 8–19 March 2022 | 8 | 24-hour composite | 7.64 (0.09) | 92.2 (30.9) | 1330 (81) | 81.9 (13.49) | 8.9 (3.11) |
| EDI-P1 | 8–23 March 2022 | 15 | 12-hour composite | 7.54 (0.11) | 54.5 (10.6) | 1547 (103) | 67.2 (8.79) | 5.9 (1.71) |
| EDI-WWTP | 8–24 March 2022 | 13 | 24-hour composite | 7.32 (0.02) | 34.3 (16.1) | 844 (22) | 20.7 (4.03) | 6.3 (2.60) |
| LHR-CTA wet well | 8–24 March 2022 | 15 | 24-hour composite | 7.59 (0.08) | 202 (40.2) | 3169 (212) | 129.2 (6.91) | 24.0 (5.71) |
| LHR-Vacuum truck | 16 March 2022 | 10 | Grab | 8.84 (0.13) | 1205 (989) | 88.8 (11.98) | 109.7 (14.22) | |
| BRS-MH2 | 21–28 March 2022 | 24 | 21-hour composite | 8.77 (0.05) | 366 (103) | 2498 (295) | 120.9 (5.31) | 8.3 (1.16) |
| BRS-MH3 | 21–29 March 2022 | 28 | 21-hour composite | 8.06 (0.08) | 497 (157) | 2026 (111) | 106.5 (5.27) | 11.6 (1.30) |
| BRS-MH1 Vacuum truck | 25–28 March 2022 | 7 | Grab | 8.78 (0.29) | 1410 (354) | 12718 (3060) | 83.7 (13.90) | 93.7 (23.30) |
Where applicable, numbers represent mean values with the standard error shown in brackets. EDI: Edinburgh airport; LHR: London Heathrow airport; BRS: Bristol airport; NTU: nephelometric turbidity unit. Due to sample availability, some samples were not tested for physico-chemical properties.
The sample pH, turbidity, electrical conductivity, ammonium, and orthophosphate ion concentrations were measured as described previously [8]. Formaldehyde was measured in the samples derived from aircrafts using a Formaldehyde Assay Kit (Abcam, UK) following the manufacturer’s instructions. Total chorine was measured in the aircraft wastewater samples taken at Heathrow and Bristol samples after centrifugation (10,000 x g, 10 min) using a total chlorine meter (Hanna Instruments, USA) according to the manufacturer’s instructions.
2.2 Sample process for virus concentration
We trialled five sample concentration methods on the samples spiked with SARS-CoV-2 (Table 1) and used the best performing method on the samples taken for surveillance (Table 2).
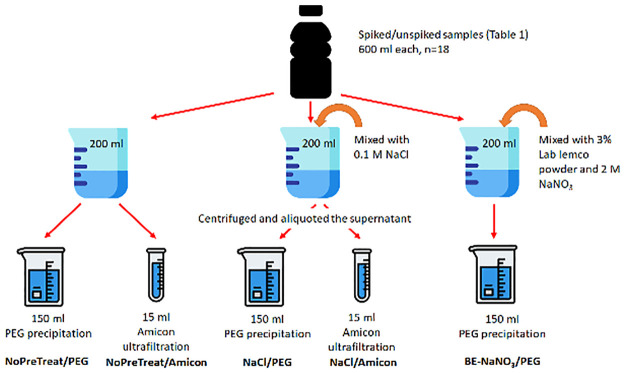
The samples spiked with SARS-CoV-2 (Table 1) were divided into three 200-ml aliquot sets and were spiked with approximately 105 gc phi6 bacteriophage as a process control [32]. Process positive controls (water spiked with phi6) and process negative controls (water only) were also concentrated along with each batch of samples to investigate recovery efficiency and cross-contamination. We used two pre-treatments (sodium chloride and beef extract-sodium nitrate) and two types of concentration (polyethylene glycol (PEG) precipitation and ultrafiltration) on each sample (Fig 1).
Fig 1. Sample pre-treatment and concentration methods used on the wastewater samples.
PEG: polyethylene glycol.
One set of the 200-ml aliquots was not pre-treated, being only concentrated using PEG precipitation and ultrafiltration, as described elsewhere [33]. The samples were centrifugated at 10,000 x g at 4°C for 10 min. Then, 15 ml of the supernatant was transferred to a 10 kDa Amicon Ultra-15 centrifugal filters (Merck Life Science UK Ltd, UK). The samples were centrifuged at 5,000 x g for 30–60 min to reach a final volume of 200–500 μl, resulting in NoPreTreat/Amicon samples. The filtrates were discarded. For NoPreTreat/PEG samples, another 150 ml of the primary supernatant was also separated, and the pH was adjusted to 7.0–7.5 and mixed with PEG8000 and NaCl to reach final concentrations of 10% and 2%, respectively, and incubated at 4°C for 16 hours. The mixture was centrifuged at 10,000 x g at 4°C for 30 min. The resulting pellet was then subjected to RNA/DNA extraction.
The second set of samples were mixed with NaCl to reach a final concentration of 0.1 M. The mixtures were incubated at 50 rev min-1 at room temperature for 30 min. Then, the samples were centrifuged to clarify solid matter, from which 150 ml of the supernatant was PEG-precipitated (NaCl/PEG) and 15 ml was ultrafiltered (NaCl/Amicon), as described above.
The third set of samples were mixed with Lab Lemco beef extract (Oxoid, USA) and sodium nitrate to reach the final concentration of 3% and 2 M, respectively [33]. After the pH of the mixtures were adjusted to 3.5–5.5, the solutions were incubated at 50 rev min-1 at room temperature for 30 min. Subsequently, the samples were centrifuged and PEG-precipitated, as described above (resulting in BE-NaNO3/PEG samples). As this method performed the best (see Section 3.1 for details), this approach was used on the surveillance samples (Table 2).
2.3 Viral RNA/DNA extraction
The viral nucleic acids were extracted from the PEG pellets and the ultrafiltered concentrate using the NucliSens extraction kit (BioMerieux, France) on a Kingfisher 96 Flex system (Thermo Scientific, USA) as described previously [32, 34]. The final volume of the eluent was 100 μl. To assess extraction performance and cross-contamination, extraction positive (deionised water spiked with phi6) and negative controls (distilled water) were used.
2.4 Viral RNA/DNA quantification
We quantified SARS-CoV-2 and phi6 RNA using a duplex RT-qPCR and crAssphage using a qPCR assay on a QuantStudio Flex 6 real-time PCR machine (Applied Biosystems Inc., USA) as previously described [32, 33]. Each assay contained a dilution series of standards of known concentrations [33] and non-template controls, which were negative in each reaction. Assay details and performance are summarised in Table 3.
Table 3. RT-qPCR and qPCR assay details and performance for the viral targets.
| Oligo reference | Assay | Standard curve | |||
|---|---|---|---|---|---|
| Slope | R2 | Efficiency % | |||
| SARS-CoV-2 (N1) | [35] | TaqMan Viral 1-step RT-qPCR master mix (Applied Biosystems Inc., USA) | -3.51 –-3.21 | 0.993–0.999 | 92.6–104.9 |
| Phi6 | [36] | -3.55 –-3.11 | 0.969–0.999 | 91.4–109.9 | |
| CrAssphage | [37] | QuantiNova Probe qPCR mix (Qiagen, Germany) | -3.41 –-3.18 | 0.994–0.998 | 96.3–106.3 |
2.5 Data analysis
The preliminary qPCR data analysis and quality control was performed using the QuantStudio Flex 6 real-time PCR software v1.7.1 (Applied Biosystems Inc., USA). The viral concentrations were expressed as gc/μl RNA/DNA extract. The viral concentrations (gc/ml) in the concentrated samples were calculated as:
| (1) |
For the surveillance samples, concentrations were interpreted as gc/l wastewater. Virus recoveries for the phi6 process control virus were calculated as:
| (2) |
Subsequent data analysis and statistical tests were carried out in R version 4.1.2 [38], utilising packages “readxl”, “tidyr”, “dplyr”, “tidyverse”, and “ggplot2”. We tested for significant differences in viral concentration (gc/ml) across methods using a Kruskal-Wallis rank sum test and Pairwise Wilcoxon Tests. Spearman correlation was used to assess correlation between chemical and viral data.
Metadata can be found in S2 Table.
3. Results
3.1 Wastewater concentration method selection
Of the five methods tested, BE-NaNO3/PEG precipitation method showed the highest virus recovery in comparison to the other methods, both for the SARS-CoV-2 N1 gene fragment and for the phi6 process control virus (Table 4).
Table 4. Summary statistics for SARS-CoV-2 (N1 gene fragment) and phi6 showing higher viral recovery using BE-NaNO3 pre-treatment.
| Virus | Method | Mean concentration (gc/ml) | SD | Median concentration (gc/ml) | IQR |
|---|---|---|---|---|---|
| SARS-CoV-2 | BE-NaNO3/PEG | 1858 | 2629 | 943 | 1375 |
| NaCl/PEG | 1227 | 2094 | 347 | 1450 | |
| NaCl/Amicon | 655 | 877 | 248 | 760 | |
| NoPreTreat/PEG | 433 | 442 | 192 | 838 | |
| NoPreTreat/Amicon | 858 | 1208 | 283 | 959 | |
| Phi6 | BE-NaNO3/PEG | 4193 | 1706 | 4094 | 2891 |
| NaCl/PEG | 58 | 96 | 13 | 52 | |
| NaCl/Amicon | 15 | 28 | 4 | 5 | |
| NoPreTreat/PEG | 8 | 6 | 8 | 12 | |
| NoPreTreat/Amicon | 28 | 32 | 13 | 38 | |
| CrAssphage | BE-NaNO3/PEG | 16707 | 12875 | 15202 | 19972 |
| NaCl/PEG | 12698 | 9775 | 13844 | 16730 | |
| NaCl/Amicon | 5280 | 857 | 5453 | 1241 | |
| NoPreTreat/PEG | 5237 | 7275 | 1134 | 4695 | |
| NoPreTreat/Amicon | 3124 | 3124 | 3031 | 1076 |
gc: genome copies; SD: standard deviation; IQR: interquartile range.
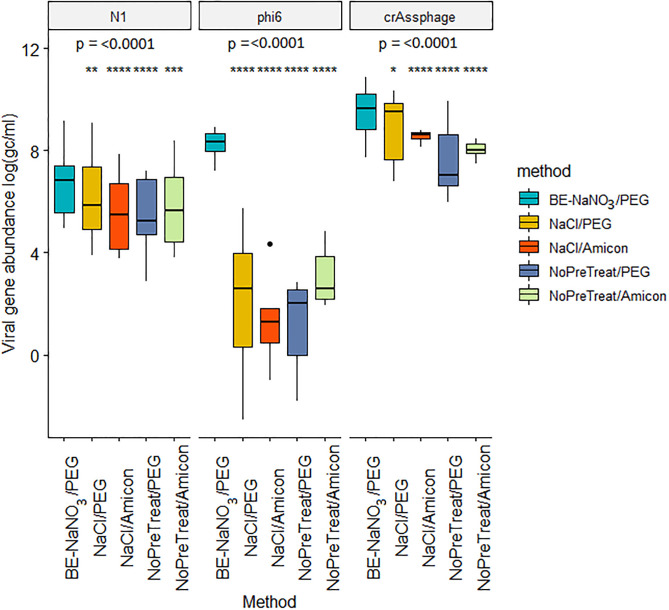
A Shapiro-Wilk normality test showed that the distribution of the data departed significantly from normality for SARS-CoV-2 (W = 0.76, p<0.01), Phi6 (W = 0.57, p<0.01) and crAssphage (W = 0.76, p<0.01). The BE-NaNO3 method was significantly better than the other four methods for SARS-CoV-2, phi6 recovery and crAssphage (Fig 2) using a Kruskal Wallis rank sum test followed by Pairwise Wilcoxon Tests. The surveillance samples were therefore processed using a BE-NaNO3 pre-treatment and PEG precipitation.
Fig 2. Boxplots showing the viral gene abundance recovered for N1, phi6 and crAssphage with different processing methods of spiked wastewater.
Global p-value is shown for each target, and significance codes (≤0.0001 ‘****’, ≤0.001 ‘***’, ≤0.01 ‘**’, ≤0.05 ‘*’) indicate statistically significant difference in results of BE-NaNO3/PEG methods compared to each of the remaining methods.
3.2 Surveillance of SARS-CoV-2 in wastewater at airports
The wastewater samples were spiked with phi6 process control virus prior to concentration to assess viral recovery efficiency. Overall, the control virus was recovered in most samples, however, no virus recovery was noted in the samples derived from vacuum trucks at Heathrow airport (Table 5). The recovery rates were also low for samples taken from vacuum trucks at the Bristol site (33%) and at the Heathrow CTA site (50%). In many cases, crAssphage and SARS-CoV-2 were detected in samples where no Phi6 recovery probably due to low spiking concentrations.
Table 5. SARS-CoV-2 and crAssphage detection rates and concentrations along with Phi6 process control recoveries for airport wastewater surveillance.
| Sampling site | SARS-CoV-2 (N1) | crAssphage | Phi6 | |||
|---|---|---|---|---|---|---|
| Positivity rate | Concentration gc/l | Positivity rate | Concentration gc/l | Positivity rate | Recovery % | |
| EDI-JR1 | 75% | 3.1x103 (9.7x102) | 100% | 5.3x106 (4.7x106) | 100% | 47.6 (4.3) |
| EDI-JR2 | 100% | 8.9x104 (2.1x104) | 100% | 4.9x107 (3.6x107) | 100% | 19.7 (4.6) |
| EDI-JR3 | 100% | 3.9x105(2.7x105) | 100% | 2.6x107 (1.5x107) | 100% | 41.7 (18.7) |
| EDI-P1 | 100% | 6.7x104 (1.3x104) | 100% | 1.8x107 (7.5x106) | 64% | 53.3 (27.7) |
| EDI-WWTP | 92% | 2.4x105 (1.2x105) | 100% | 2.9x107 (1.6x107) | 82% | 13.1 (3.2) |
| LHR-CTA wet well | 100% | 8.2x104 (1.5x104) | 100% | 3.1x107 (5.8x106) | 50% | 27.0 (12.1) |
| LHR-VT | 80% | 1.6x106 (8.9x105) | 40% | 5.6x107 (1.8x107) | 0% | - |
| BRS-MH2 | 100% | 1.8x105 (4.4x104) | 100% | 2.3x107 (7.3x106) | 75% | 57.8 (8.0) |
| BRS-MH3 | 100% | 4.7x105 (1.6x105) | 100% | 5.3x107 (2.0x107) | 79% | 23.4 (6.1) |
| BRS-MH1 VT | 100% | 4.0x106 (2.3x106) | 100% | 7.4x107 (5.0x107) | 33% | 44.8 (34.7) |
Where applicable, numbers represent mean values with the standard error shown in brackets. gc: genome copies. Due to sample availability, some samples were not tested for crAssphage. VT–Vacuum Truck.
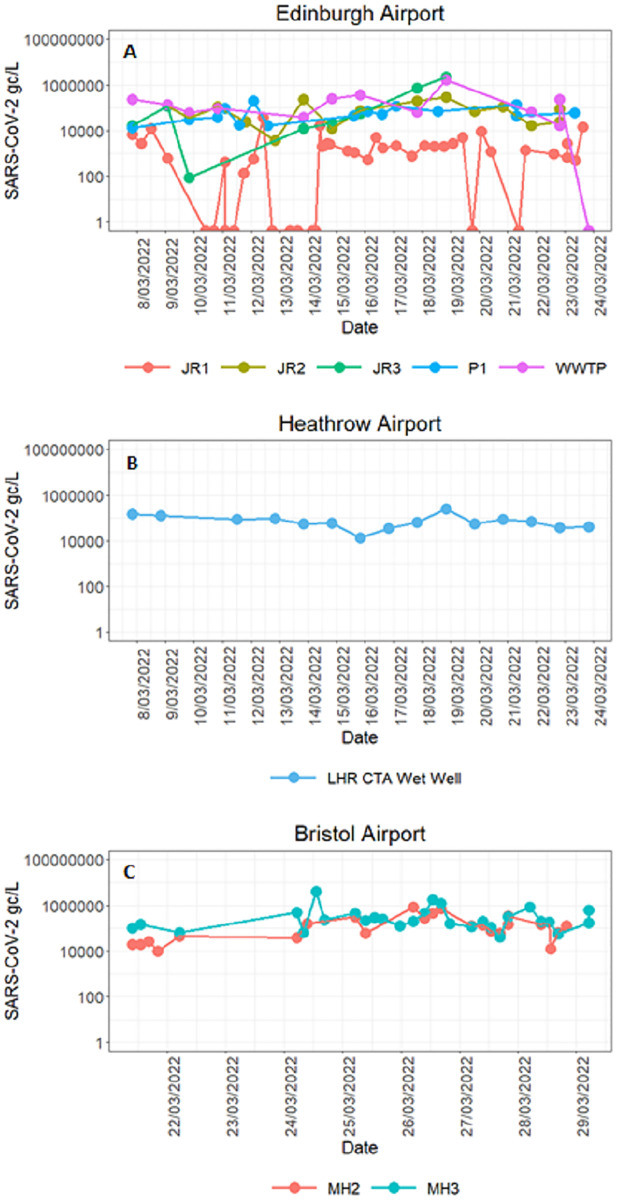
The majority of the wastewater samples taken at the three airports were positive for SARS-CoV-2 RNA and crAssphage DNA. The lowest SARS-CoV-2 concentration and detection rates were observed in the JR1 Edinburgh airport samples (Table 5). The remaining samples taken at Edinburgh were all positive, except for one sample taken at the WWTP, with recorded concentrations of between 1x102 and 2x106 gc/l. All samples taken at Edinburgh airport contained crAssphage at high concentrations (Table 5). The sample pH and orthophosphate levels showed little variation among the sampling sites, however, considerably higher turbidity, electrical conductivity and ammonium levels were noted at JR2, JR3, and P1 sites compared to JR1 and WWTP samples (Table 2). No significant trends in SARS-CoV-2 levels over time were observed at any of the sampling sites (Fig 3).
Fig 3. SARS-CoV-2 RNA concentrations in genome copies (gc)/l at (A) Edinburgh, (B) Heathrow and (C) Bristol airports.
At Heathrow and Bristol airports, all sewage samples derived from the terminals were positive for SARS-CoV-2 and crAssphage, with concentrations in the range of 1x104 − 4x106 gc/l and 2x107 − 6x107 gc/l, respectively (Table 5; Fig 3). At these sites the ammonium and turbidity levels in the samples were higher than those observed in samples collected at the Edinburgh sites. At Bristol sites, the sample pH levels were also higher than the pH in samples collected at the other two locations (Table 5). No correlation between viral concentrations and chemical data was found.
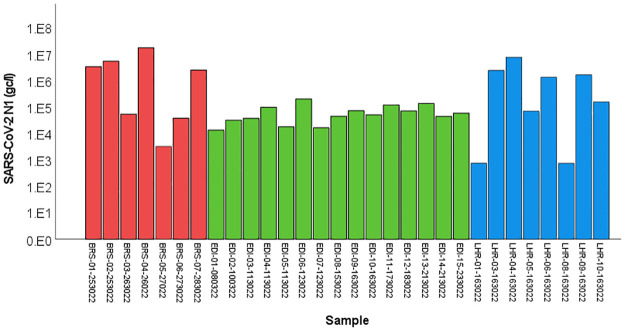
The content of vacuum trucks carrying sewage derived from aircraft were also tested for SARS-CoV-2. All samples taken at Edinburgh and Bristol sites were positive for SARS-CoV-2 and all but two samples taken at Heathrow returned positive (Fig 4). CrAssphage was detected in all samples taken from vacuum trucks at Edinburgh and Bristol airports and 40% of the Heathrow vacuum truck samples were also positive for that virus. No correlation between the crAssphage and SARS-CoV-2 detection rates or concentrations were identified. Sample pH, turbidity, orthophosphate levels and, in some cases, electrical conductivity were also notably higher than those levels in the rest of the samples (Table 5). In order to assess the presence of biocides often used for aircraft wastewater treatment, we tested the samples for the presence of formaldehyde and chlorine. The concentration of formaldehyde in the samples was negligible (<0.5mg/l) and chlorine was not detected either. Due to the viscosity of the samples taken from the vacuum truck deposit site at Heathrow airport, turbidity could not be measured.
Fig 4. SARS-CoV-2 RNA concentration in samples taken from vacuum trucks at Bristol (BRS–red bars), at Edinburgh (EDI–green bars) and at Heathrow (LHR–blue bars) airports.
4. Discussion
International air travel has had a significant impact on the rapid spread of COVID-19 and greatly contributed to the development of the pandemic. It is now established that travellers arriving from mainland Europe to the UK in early 2020 were responsible for introducing ca. 1300 SARS-CoV-2 lineages, resulting in the first wave of the pandemic [39]. Even with significant non-pharmaceutical interventions being in place (i.e., social distancing, face covering, negative COVID test results prior to travel), in-flight transmission continued to occur with an attack rate (i.e. the rate of infected individuals in a population at risk) of 10–17% [40]. Due to the successful roll-out of vaccination programmes worldwide, COVID-19 restrictions have been lifted in many airports and during flights. While vaccination reduces the number of COVID-19-related hospitalisations and deaths, it has limited capacity to reduce the spread of infections and to contain outbreaks [41]. With continued transmission, which occurs more rapidly in confined spaces [42], such as aircraft, the introduction of novel lineages via air travel is a significant risk. Besides the clinical surveillance of passengers, research on alternative approaches to reduce transmission in confined spaces, such as updating air circulation systems onboard flights, is a priority.
In this study, we assessed the usefulness of WBE for the quantitative assessment of SARS-CoV-2 in wastewater arrival locations at three international airports in the UK. The samples were collected from i) sewers connected to the arrival halls in the airport terminals, ii) vacuum trucks and deposit sites where vacuum trucks collecting aircraft wastewater unload their contents, and iii) a wastewater treatment plant receiving wastewater serving the airport. We noted considerable differences in the solids content and other physico-chemical properties of wastewater taken from the sewers and aircraft in comparison to municipal wastewater (Table 2). Hence, we subsequently trialled different approaches to enhance viral nucleic acid recovery and provide consistent results from highly turbid samples.
Most virus concentration methods applied to wastewater samples begin with the exclusion of solid matter via filtration or centrifugation. However, previous research suggests that up to 25% of SARS-CoV-2 RNA may be retained in the solid matrix of municipal wastewater [22, 23] and this ratio may be even higher in highly turbid samples. Therefore, in this study we trialled pre-treatment methods to detach viruses and viral nucleic acids from solids and found that the application of beef extract significantly increases recovery compared to the application of salt or no treatment (Fig 2). Beef extract has been shown to enhance detachment of viruses and nucleic acids from solid matter in wastewater and environmental samples, while at the same time binding viral particles, which assists their precipitation [33, 43–45]. However, when centrifugation is used to clarify the sample after beef extract treatment, the solution is not suitable for subsequent ultrafiltration due to the remaining proteins clogging the filter, further increasing the processing time.
The application of NaCl also enhanced viral recovery compared to samples where no pre-treatment was applied, potentially through the detachment of viral particles from solid particles. It can be explained by an additional competition created by Na+ and Cl- ions for the polar binding sites on the surface of solid particles, which usually comprises organic matter. NaCl treatment did not require pH adjustment and the resulting samples were suitable for ultrafiltration and, hence, it may be applied for WBE when sample processing is a time-limiting step.
After the samples (with or without pre-treatment) were clarified using centrifugation, the resulting supernatants were concentrated using PEG precipitation or ultrafiltration. When no pre-treatment was applied, the ultrafiltration-based concentration performed similar or better than PEG precipitation, as shown previously [30, 33]. When salt pre-treatment was used, the PEG precipitation gave slightly higher viral recoveries than ultrafiltration (Fig 2). The best performing method was the beef extract-based pre-treatment coupled with PEG precipitation, which enabled the consistent virus concentration in 150 ml samples and, hence, this method was used for subsequent SARS-CoV-2 monitoring.
Using the BE-NaNO3 concentration method, we successfully recovered SARS-CoV-2 RNA and the DNA of the faecal indicator virus, crAssphage. However, we noted that the process control virus (phi6) used to assess viral recovery was less frequently detected in the samples than SARS-CoV-2 and crAssphage, especially in the samples derived from vacuum trucks. Previous studies also noted lower recoveries for phi6 than for coronaviruses in concentrated wastewater [46, 47], suggesting that phi6 bacteriophage may not be a useful indicator for SARS-CoV-2 recovery in all types of wastewater samples. This may be due to the decreased genetic stability of phi6 in comparison to coronaviruses or crAssphage when exposed to complex wastewater matrices. Nonetheless, the phi6 virus may be used as a process control for less challenging sample matrices, such as municipal wastewater [16, 32, 48].
Our results suggest that the highest SARS-CoV-2 concentration and detection rates may be achieved when samples collected close to source are analysed. In wastewater samples taken from sewers close to the airport terminals, for almost all cases SARS-CoV-2 was detected at high concentrations, whereas samples further away from the source and at the receiving wastewater treatment plant demonstrated lower detection rates. This may be due to viral decay and/or accumulation in biofilm in the sewers [49, 50].
All but two samples derived from aircraft were also positive for SARS-CoV-2, and all those collected from airplanes landing at Bristol airport were SARS-CoV-2 positive. However, as those samples contained wastewater from several aircraft, the origin of the viruses could not be determined. Similarly, at Edinburgh airport, where all aircraft-derived samples were positive for SARS-CoV-2, it was not possible to determine the flight origin as the samples were taken from a deposit site, where cross-contamination is inevitable. Whole genome sequencing may also be useful to determine the origin of novel strains [51], but these approaches may be hard to implement for difficult matrices, such as aircraft wastewater. One of the two samples from aircraft landing at Heathrow airport that tested negative for SARS-CoV-2 were also negative for crAssphage, suggesting that the wastewater may have contained disinfectants and other cleaning agents [52], indicated by unusual colour and pH, which may degrade viruses and viral RNA. Hence, it is possible that the wastewater contained SARS-CoV-2 before such treatment but is unrecoverable after chemical treatment. More detailed studies are needed to assess viral degradation and recovery under such conditions [53].
The high SARS-CoV-2 detection frequencies in wastewater from terminal sewers and aircraft suggest that passengers arriving at the airports and/or staff members working at the terminals had ongoing COVID-19 infections. The COVID-19 restrictions were lifted in England on the 18th March 2022, and no differences in the concentrations of SARS-CoV-2 in wastewater before and after that date were noted. This finding may suggest that the travel restrictions, such as negative COVID-19 tests, social distancing and mask wearing, were not suitable to filter asymptomatic and pre-symptomatic individual. Previous studies have found the effectiveness of these travel restrictions depends on early intervention, enforcement and compliance, and the sensitivity of screening tests [54, 55].
Overall, our results suggest that WBE may be used as a simple monitoring tool for SARS-CoV-2 and other viral diseases at airports and aircraft, to identify outbreak hotspots internationally and to observe trends in the infection prevalence [19, 20], however, the approach has limitations, which must be considered. Cross-contamination may occur as one vacuum truck is used for emptying sewage from several aircraft with no cleaning in between planes. In order to avoid that, sampling from aircraft may be a viable option, however, considering the number of flights arriving at international airports daily, that may not be feasible due to logistics issues and high costs. Furthermore, sampling of aircraft may be biased due to in-flight toilet usage and behaviour trends. A recent study assessing toilet use on short- and long-haul flights, discovered that aircraft wastewater probably captures 8–14% of SARS-CoV-2 cases [56]. The same study estimated that approximately 62% of the long-haul flights would have wastewater containing SARS-CoV-2, which is a slightly lower prevalence that observed here (80–100%). The lower estimate in the former study is most likely due to underreporting of defecation habits when undertaking social behaviour surveys, uncertainties in the proportion of individuals that shed SARS-CoV-2 in faeces, and the lack of consideration for capture of other bodily fluids in aircraft toilet wastewater (e.g. saliva) [56].
Even with highly effective extraction methods, wastewater samples derived from aircraft may contain solid matter and additives which reduce the chances of successful virus detection [53]. Monitoring sewage from airport terminals is less challenging, but also less informative on the origin of the pathogen due to the high number of people contributing to a sample. In some cases this wastewater may capture both inbound and outbound passengers as well as office staff working in the same terminal. Nonetheless, the regular sampling of airport and aircraft wastewater can be useful as targeted monitoring system for emerging diseases and other agents (e.g. anti-microbial resistance genes) that have not yet become endemic in the country. Using qPCR or digital PCR-based quantitative analysis of novel viral agents would enable rapid assessment and also help shortlisting samples for further analysis, e.g. whole genome sequencing and metagenomics, enabling deeper understanding on the pathogens circulating globally.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
We thank Daphne Beniston and Andrew Singer (Accelerated Capability Environment, Homeland Security Group, UK) for their help contribution organizing the project. We also thank Prof. Andrew Weightman (Cardiff University, UK) for providing the inactivated SARS-CoV-2 virus stocks. We thank 20/30 Labs (UK), Veolia (UK) and Aqua Enviro (UK) for conducting sampling and sample transport and the management and field staff at all the airports for their logistical support.
Data Availability
Data is available in supplementary material.
Funding Statement
KF, RW, NAS, JMSG, MJW, DLJ: The work was supported by the Environmental Monitoring for Health Protection Programme using funds provided by the UK Department of Health and Social Care. The funder was involved in sampling design and organised sampling through external partners. DLJ: The Centre for Environmental Biotechnology Project was funded though the European Regional Development Fund (ERDF) by Welsh Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. WHO Coronavirus (COVID-19) Dashboard. 2020 [cited 22 Jun 2022]. https://covid19.who.int/
- 2.Byambasuren O, Cardona M, Bell K, Clark J, McLaws ML, Glasziou P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: Systematic review and meta-analysis. Journal of the Association of Medical Microbiology and Infectious Disease Canada. 2020;5: 223–234. doi: 10.3138/jammi-2020-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alene M, Yismaw L, Assemie MA, Ketema DB, Mengist B, Kassie B, et al. Magnitude of asymptomatic COVID-19 cases throughout the course of infection: A systematic review and meta-analysis. PLoS One. 2021;16: e0249090. doi: 10.1371/journal.pone.0249090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oran DP, Topol EJ. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann Intern Med. 2020;173: 362–368. doi: 10.7326/M20-3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Cen M, Hu M, Du L, Hu W, Kim JJ, et al. Prevalence and Persistent Shedding of Fecal SARS-CoV-2 RNA in Patients With COVID-19 Infection: A Systematic Review and Meta-analysis. Clin Transl Gastroenterol. 2021;12: E00343. doi: 10.14309/ctg.0000000000000343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.kyung Park S, Lee CW, Park D il, Woo HY, Cheong HS, Shin HC, et al. Detection of SARS-CoV-2 in Fecal Samples From Patients With Asymptomatic and Mild COVID-19 in Korea. Clinical Gastroenterology and Hepatology. 2021;19: 1387–1394.e2. doi: 10.1016/j.cgh.2020.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su L, Ma X, Yu H, Zhang Z, Bian P, Han Y, et al. The different clinical characteristics of corona virus disease cases between children and their families in China—the character of children with COVID-19. Emerg Microbes Infect. 2020;9: 707–713. doi: 10.1080/22221751.2020.1744483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillary LS, Farkas K, Maher KH, Lucaci A, Thorpe J, Distaso MA, et al. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021; 117214. doi: 10.1016/j.watres.2021.117214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett. 2020. 7: 511–516. doi: 10.1021/acs.estlett.0c00357 [DOI] [PubMed] [Google Scholar]
- 10.Ahmed W, Angel N, Edson J, Bibby K, Brien JWO, Choi PM, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of the Total Environment. 2020; 138764. doi: 10.1016/j.scitotenv.2020.138764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ai Y, Davis A, Jones D, Lemeshow S, Tu H, He F, et al. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Science of The Total Environment. 2021;801: 149757. doi: 10.1016/j.scitotenv.2021.149757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xagoraraki I. Can we predict viral outbreaks using wastewater surveillance? Journal of Environmental Engineering. 2020;146. doi: 10.1061/(ASCE)EE.1943-7870.0001831 [DOI] [Google Scholar]
- 13.Feng L, Zhang W, Li X. Monitoring of regional drug abuse through wastewater-based epidemiology—A critical review. Sci China Earth Sci. 2018;61: 239–255. doi: 10.1007/s11430-017-9129-x [DOI] [Google Scholar]
- 14.Choi PM, Tscharke BJ, Donner E, O’Brien JW, Grant SC, Kaserzon SL, et al. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends in Analytical Chemistry. 2018;105: 453–469. [Google Scholar]
- 15.Bivins A, North D, Ahmad A, Ahmed W, Alm E, Been F, et al. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight against COVID-19. Environ Sci Technol. 2020;54: 7754–7757. doi: 10.1021/acs.est.0c02388 [DOI] [PubMed] [Google Scholar]
- 16.Scott LC, Aubee A, Babahaji L, Vigil K, Tims S, Aw TG. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ Res. 2021;200: 111374. doi: 10.1016/j.envres.2021.111374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Liu P, Zhang H, Ibaraki M, VanTassell J, Geith K, et al. Early warning of a COVID-19 surge on a university campus based on wastewater surveillance for SARS-CoV-2 at residence halls. Science of The Total Environment. 2022;821: 153291. doi: 10.1016/j.scitotenv.2022.153291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karthikeyan S, Nguyen A, McDonald D, Zong Y, Ronquillo N, Ren J, et al. Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. mSystems. 2021;6: e00793–21. doi: 10.1128/mSystems.00793-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albastaki A, Naji M, Lootah R, Almeheiri R, Almulla H, Almarri I, et al. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: The use of wastewater based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Science of The Total Environment. 2021;760: 143350. doi: 10.1016/j.scitotenv.2020.143350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed W, Bivins A, Simpson SL, Bertsch PM, Ehret J, Hosegood I, et al. Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia. Environ Int. 2022;158: 106938. doi: 10.1016/j.envint.2021.106938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed W, Bivins A, Smith WJM, Metcalfe S, Stephens M, Jennison A v., et al. Detection of the Omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater. Science of The Total Environment. 2022;820: 153171. doi: 10.1016/j.scitotenv.2022.153171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forés E, Bofill-Mas S, Itarte M, Martínez-Puchol S, Hundesa A, Calvo M, et al. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Science of The Total Environment. 2021;768: 144786. doi: 10.1016/j.scitotenv.2020.144786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed W, Bivins A, Simpson SL, Smith WJM, Metcalfe S, McMinn B, et al. Comparative analysis of rapid concentration methods for the recovery of SARS-CoV-2 and quantification of human enteric viruses and a sewage-associated marker gene in untreated wastewater. Science of The Total Environment. 2021;799: 149386. doi: 10.1016/j.scitotenv.2021.149386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Mazurowski L, Dewan A, Carine M, Haak L, Guarin TC, et al. Longitudinal monitoring of SARS-CoV-2 in wastewater using viral genetic markers and the estimation of unconfirmed COVID-19 cases. Science of The Total Environment. 2022;817: 152958. doi: 10.1016/j.scitotenv.2022.152958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonçalves J, Koritnik T, Mioč V, Trkov M, Bolješič M, Berginc N, et al. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Science of The Total Environment. 2021;755: 143226. doi: 10.1016/j.scitotenv.2020.143226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juel MAI, Stark N, Nicolosi B, Lontai J, Lambirth K, Schlueter J, et al. Performance evaluation of virus concentration methods for implementing SARS-CoV-2 wastewater based epidemiology emphasizing quick data turnaround. Science of The Total Environment. 2021;801: 149656. doi: 10.1016/j.scitotenv.2021.149656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez R, Curtis K, Bivins A, Bibby K, Weir MH, Yetka K, et al. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186: 116296. doi: 10.1016/j.watres.2020.116296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Y, Ellenberg RM, Graham KE, Wigginton KR. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ Sci Technol. 2016;50: 5077–5085. doi: 10.1021/acs.est.6b00876 [DOI] [PubMed] [Google Scholar]
- 29.Corpuz MVA, Buonerba A, Vigliotta G, Zarra T, Ballesteros F, Campiglia P, et al. Viruses in wastewater: occurrence, abundance and detection methods. Science of The Total Environment. 2020;745: 140910. doi: 10.1016/j.scitotenv.2020.140910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mailepessov D, Arivalan S, Kong M, Griffiths J, Low SL, Chen H, et al. Development of an efficient wastewater testing protocol for high-throughput country-wide SARS-CoV-2 monitoring. Science of The Total Environment. 2022; 154024. doi: 10.1016/j.scitotenv.2022.154024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed W, Bertsch PM, Bivins A, Bibby K, Farkas K, Gathercole A, et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Science of The Total Environment. 2020;739: 139960. doi: 10.1016/j.scitotenv.2020.139960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kevill JL, Pellett C, Farkas K, Brown MR, Bassano I, Denise H, et al. A comparison of precipitation and filtration-based SARS-CoV-2 recovery methods and the influence of temperature, turbidity, and surfactant load in urban wastewater. Science of The Total Environment. 2022;808: 151916. doi: 10.1016/j.scitotenv.2021.151916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farkas K, Pellett C, Alex-Sanders N, Bridgman MTP, Corbishley A, Grimsley JMS, et al. Comparative Assessment of Filtration- and Precipitation-Based Methods for the Concentration of SARS-CoV-2 and Other Viruses from Wastewater. Faucher SP, editor. Microbiol Spectr. 2022;10. doi: 10.1128/spectrum.01102-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farkas K, Hillary LS, Thorpe J, Walker DI, Lowther JA, McDonald JE, et al. Concentration and quantification of SARS-CoV-2 RNA in wastewater using polyethylene glycol-based concentration and qRT-PCR. Methods Protoc. 2021;4: 1–9. doi: 10.3390/MPS4010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CDC. 2019-Novel Coronavirus (2019-nCoV) Real-time rRT-PCR Panel Primers and Probes. Atlanta, Georgia: Centers for Disease Control and Prevention; 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- 36.Gendron L, Verreault D, Veillette M, Moineau S, Duchaine C. Evaluation of filters for the sampling and quantification of RNA phage aerosols. Aerosol Science and Technology. 2010;44: 893–901. doi: 10.1080/02786826.2010.501351 [DOI] [Google Scholar]
- 37.Stachler E, Kelty C, Sivaganesan M, Li X, Bibby K, Shanks OC. Quantitative crAssphage PCR assays for human fecal pollution measurement. Environ Sci Technol. 2017;51: 9146–9154. doi: 10.1021/acs.est.7b02703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2021. http://www.r-project.org
- 39.du Plessis L, McCrone JT, Zarebski AE, Hill V, Ruis C, Gutierrez B, et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science (1979). 2021;371: 708–712. doi: 10.1126/science.abf2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy N, Boland M, Bambury N, Fitzgerald M, Comerford L, Dever N, et al. A large national outbreak of COVID-19 linked to air travel, Ireland, summer 2020. Eurosurveillance. 2020;25: 2001624. doi: 10.2807/1560-7917.ES.2020.25.42.2001624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore S, Hill EM, Tildesley MJ, Dyson L, Keeling MJ. Vaccination and non-pharmaceutical interventions for COVID-19: a mathematical modelling study. Lancet Infect Dis. 2021;21: 793–802. doi: 10.1016/S1473-3099(21)00143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Z, Lan Y. Modeling COVID-19 infection in a confined space. Nonlinear Dyn. 2020;101: 1643–1651. doi: 10.1007/s11071-020-05802-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farkas K, Cooper DM, McDonald JE, Malham SK, de Rougemont A, Jones DL, et al. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Science of the Total Environment. 2018;634: 1174–1183. doi: 10.1016/j.scitotenv.2018.04.038 [DOI] [PubMed] [Google Scholar]
- 44.Lewis GD, Metcalf TG. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl Environ Microbiol. 1988;54: 1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machado-Moreira B, Monteiro S, Santos R, Martinez-Murcia A, Rajkovic A, Smigic N, et al. Impact of beef extract used for sample concentration on the detection of Escherichia coli DNA in water samples via qPCR. J Microbiol Methods. 2020;168: 105786. doi: 10.1016/j.mimet.2019.105786 [DOI] [PubMed] [Google Scholar]
- 46.Torii S, Oishi W, Zhu Y, Thakali O, Malla B, Yu Z, et al. Comparison of five polyethylene glycol precipitation procedures for the RT-qPCR based recovery of murine hepatitis virus, bacteriophage phi6, and pepper mild mottle virus as a surrogate for SARS-CoV-2 from wastewater. Science of The Total Environment. 2022;807: 150722. doi: 10.1016/j.scitotenv.2021.150722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alamin M, Tsuji S, Hata A, Hara-Yamamura H, Honda R. Selection of surrogate viruses for process control in detection of SARS-CoV-2 in wastewater. Science of The Total Environment. 2022;823: 153737. doi: 10.1016/j.scitotenv.2022.153737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherchan SP, Shahin S, Ward LM, Tandukar S, Aw TG, Schmitz B, et al. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Science of The Total Environment. 2020;743: 140621. doi: 10.1016/j.scitotenv.2020.140621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales Medina WR, D’Elia S, Fahrenfeld NL. Accumulation of SARS-CoV-2 RNA in Sewer Biofilms. ACS Environmental Science and Technology Water. 2021. [cited 8 Jul 2022]. doi: 10.1021/ACSESTWATER.1C00345 [DOI] [Google Scholar]
- 50.Petrovich ML, ben Maamar S, Hartmann EM, Murphy BT, Poretsky RS, Wells GF. Viral composition and context in metagenomes from biofilm and suspended growth municipal wastewater treatment plants. Microb Biotechnol. 2019;12: 1324–1336. doi: 10.1111/1751-7915.13464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurano MT, Ramaswami S, Zappile P, Dimartino D, Boytard L, Ribeiro-dos-Santos AM, et al. Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York City region. Genome Res. 2020;30: 1781–1788. doi: 10.1101/gr.266676.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Yang J, Zhao N, Sheng L, Zhao Y, Tang Z, et al. Preliminary evaluation of evaluation of the efficiency of aircraft liquid waste treatment using resolvable sanitizing liquid: a case study in Changchun. Environ Monit Assess. 2011;183: 95–101. doi: 10.1007/s10661-011-1909-x [DOI] [PubMed] [Google Scholar]
- 53.Ahmed W, Bertsch PM, Angel N, Bibby K, Bivins A, Dierens L, et al. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J Travel Med. 2020;27: 1–11. doi: 10.1093/jtm/taaa116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bou-Karroum L, Khabsa J, Jabbour M, Hilal N, Haidar Z, Abi Khalil P, et al. Public health effects of travel-related policies on the COVID-19 pandemic: A mixed-methods systematic review. Journal of Infection. 2021;83: 413–423. doi: 10.1016/j.jinf.2021.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ayouni I, Maatoug J, Dhouib W, Zammit N, Fredj S ben, Ghammam R, et al. Effective public health measures to mitigate the spread of COVID-19: a systematic review. BMC Public Health. 2021;21: 1015. doi: 10.1186/s12889-021-11111-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones DL, Rhymes JM, Wade MJ, Kevill JL, Malham SK, Grimsley JMS, et al. Suitability of aircraft wastewater for pathogen detection and public health surveillance. Science of The Total Environment. 2023;856: 159162. doi: 10.1016/j.scitotenv.2022.159162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
Data is available in supplementary material.