Abstract
A saturated health care system with a lack of evidence-based antiviral medicine and ignorance of antimicrobial stewardship during pandemics has prompted clinicians to prescribe a broad-spectrum antibiotic more often. A prospective, cross-sectional study of COVID-infected patients was conducted to gain insight into antibiotic prescribing practices and their impact on antimicrobial resistance. The antibiotic susceptibility test was performed using the disc diffusion method. 318 patients met the study’s inclusion criteria, with a mean age of 46 years and 55% (175) of them being males. Antibiotics were prescribed for 93.72% (209) of mild cases, 92.45% (49) of moderate cases, 96.15% (25) of severe cases, and 100% (16) of critical cases of COVID-19. A total of 95 samples were sent in for culture and antibiotic sensitivity testing, with 58.95% (56) confirming growth. The majority of the growth was found to contain E. coli (14). In 54.9% of cases, antibiotics with less than 50% sensitivity to curing bacterial infection were detected. In the study, we found that antibiotics were being used unnecessarily in excessive quantities and that more than half of the antibiotics were less sensitive to isolated bacteria.
Introduction
Antibiotics are being utilized unnecessarily in the treatment of COVID-19. Excessive and needless antibiotic use raises the risk of antimicrobial resistance (AMR) in the future by promoting the emergence of multidrug-resistant (MDR) bacteria [1]. The World Health Organization (WHO) recently revised its guidelines against the use of empirical antibiotics in COVID-19 [2], which has sparked concerns about AMR. Unless there is a clinical sign of a bacterial infection, recent guidance discourages antibiotic therapy or prophylaxis for individuals with mild COVID-19 symptoms or those with suspected or confirmed moderate COVID-19 illness [3]. Antibiotic resistance patterns must be transparently monitored and reported to combat bacterial resistance [4].
COVID-19 resembles bacterial pneumonia in conjunction with clinical presentation, including fever, cough, and lung infiltrates [5]. It could be a reason to treat ill patients with empirical antibiotics as a preventive measure. On the other hand, a saturated health care system with a lack of evidence-based antiviral medicine and ignorance of antimicrobial stewardship during pandemics has prompted clinicians to prescribe broad-spectrum antibiotics more often [2, 6]. Zhou F et al. observed that antibiotics were given to 95% of the patients, antivirals were given to 21%, and 15% of them had suffered from secondary bacterial infection [7]. Goyal P et al. observed that only 5.6% of patients experienced bacteraemia [8].
Here, the study intends to get insight into antibiotic prescribing practices in COVID-19 treatment and its impact on antibiotic resistance.
The method and methodology
Study design
A prospective, cross-sectional study was conducted in the COVID ward of Adichunchanagiri Hospital & Research Centre (AH & RC) after obtaining approval from the Intuitional Ethical Committee (IEC/AH & RC/AC/002/2021).
Study participants
The study included all patients diagnosed using the diagnostic method reverse transcription- polymerase chain reaction (RT-PCR) and admitted to the COVID wards between August 1st and November 30th, 2021, regardless of gender. The study did not include those diagnosed with other diagnostic methods than RT-PCR.
Study procedures and data collection
All COVID-19 patients hospitalized in AH & RC’s COVID wards were assessed for inclusion criteria, and those who met them were considered for the study. Although the study was part of a daily medication evaluation by a clinical pharmacist for the patient’s safety, informed consent was obtained from patients who were capable of giving it or from the caretaker for severe and critical patients.
The data collection form was designed and validated, contains various sections which include demographic details (age, gender, comorbidities, and Coronavirus prevention measures), the severity of the disease (mild, moderate, severe, and critical) [9], anti-microbial medications prescribed (antiviral, antibiotic, and antiprotozoal), and an antibiotic susceptibility test (AST) report. Relevant information was gleaned from the patient’s case report. During data collection, patients and their caretakers were briefed about coronavirus prevention strategies and asked if they were following them appropriately. The sample processing for AST was not enforced on the treating physician; instead, it was left to their discretion.
Antibiotic susceptibility test
AST was performed by the clinical microbiologist at the central laboratory of AH&RC as a part of routine diagnostics. In brief, samples were inoculated into MacConkey Agar, Blood Agar, and Chocolate Agar and incubated for 24 hours before AST. The disc-diffusion method is used on Muller-Hinton agar for AST. The Clinical and Laboratory Standards Institute (CLSI) guidelines 2020 were used to interpret the result of the AST test [10].
Data analysis
All of the collected data was imported into MS Excel, and statistical analysis was performed employing the Statistical Package for the Social Sciences (SPSS) version 26 program. The frequency and percentage of all nominal variables such as age, gender, coronavirus prevention measures, the severity of the disease, bacterial isolation, antibiotic use, and antibiotic resistance were measured. The Chi-Square test was used to find out the relationship between the different variables. A significant association was defined as one with a P-value less than 0.05.
Result
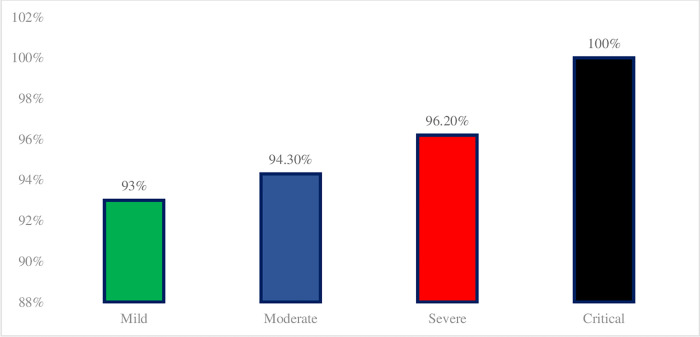
During the study’s duration, 356 Covid-infected individuals were admitted to the hospital, with 38 of them failing to satisfy the study’s inclusion criteria, bringing the number of subjects to 318. There were 55% (175) males and 45% (143) females. The average age was 45.33 ± 17.33 with an interquartile range (IQR) of 26.25 (Q3 = 57.25 and Q1 = 31). The majority of patients were above the age of 60, representing 19.8% (63) of the total, while the minority of patients were under the age of 20, representing only 4.7% (15) of the total. At least one comorbidity was present in 25.5% (81) of the patients. During the study period, 5.7% (18) of the patients lost their lives. Overall, 87.7% (279) of patients said they used hand sanitizer, 79.9% (254) said they cleaned their hands with soap and water, 58.5% (186) said they wore a mask, and 51.3% (163) said they maintained a safe distance. COVID-19 symptoms were classified as mild in 70.13% (223) of the patients, moderate in 16.67% (53) of the patients, severe in 8.2% (26) of the patients, and critical in 5.0% (16) of the patients (Table 1).
Table 1. Distribution of patient’s characteristics, antimicrobial prescribed, and the severity of the disease.
| Patient’s characteristic | Antibiotic Prescribed | Total number | Percentage (n = 318) | ||||
|---|---|---|---|---|---|---|---|
| Yes | No | P-value | |||||
| Gender (Total participant = 318) | Male | 165 | 10 | 175 | 55% | 0.163 | |
| Female | 132 | 11 | 143 | 45% | |||
| Severity | Mild | 207 (93%) | 14 | 223 | 70.13% | 0.644 | |
| Moderate | 50 (94.3%) | 3 | 53 | 16.67% | |||
| Severe | 25 (96.15%) | 1 | 26 | 8.2% | |||
| Critical | 16 (100%) | 0 | 16 | 5% | |||
| Comorbidity | 80 | 25.2% | |||||
| Death | 18 | 5.7% | |||||
| Agreed to use Hand Sanitizer | 279 | 87.7% | |||||
| Agreed to Wash Hands | 254 | 79.9% | |||||
| Agreed to Using Mask | 186 | 58.5% | |||||
| Agreed to maintain safe distance | 163 | 51.3% | |||||
| Antibiotic prescription | Total | 299 | 94% | ||||
| Single | 201 | 63.2% | |||||
| Two | 78 | 24.5% | |||||
| Three | 18 | 5.7% | |||||
| Four | 2 | 0.6% | |||||
| Antiprotozoal prescription | 252 | 79.3% | |||||
| Antiviral prescription | 245 | 77% | |||||
| Sent for Culture | 95 | 29.87% | |||||
| Growth | 56 (58.95%) | 17.6% | |||||
Antibiotics were prescribed for 93.7% (298) of the prescriptions, antiprotozoal (HCQS & Ivermectin) were prescribed for 79.3% (252), and antiviral (Oseltamivir, Favipiravir, Remdesivir, and Ritonavir/Lopinavir) were prescribed for 77% (245). Antibiotics were prescribed for 93% (207) of mild cases, 94.3% (50) of moderate cases, 96.15% (25) of severe cases, and 100% (16) of critical cases of COVID-19 (Table 1 & Fig 1). Azithromycin was prescribed at 73.6% (234), followed by piperacillin/tazobactam at 14.5% (46), ceftriaxone 13.5% (43), meropenem 6.9% (22), doxycycline 6.6% (21), amoxicillin/clavulanic acid 3.5% (11), ofloxacin 3.1% (10), amikacin 2.8% (9), metronidazole 2.5% (8), colistin 1.3% (4), ciprofloxacin 0.9% (3), cefotaxime 0.9% (3), cefixime 0.6% (2), linezolid 0.6% (2), and rifaximin 0.6% (2). 63.2% (201) were prescribed single antibiotics, 24.5% (78) with two antibiotics in a prescription, 5.7% (18) with triple antibiotics in a prescription, and 0.6% (2) with quad antibiotics in a prescription (Tables 1 & 2). Combinations of azithromycin and ceftriaxone were predominant, followed by azithromycin and piperacillin/tazobactam, azithromycin and meropenem, and azithromycin and amikacin.
Fig 1. Distribution of antibiotics prescribed for the treatment of COVID-19 based on severity.
Table 2. Distribution of antibiotics prescribed.
| Antibiotic | Total number | Percentage |
|---|---|---|
| Azithromycin | 235 | 73.9% |
| Piperacillin/tazobactam | 46 | 14.5% |
| Ceftriaxone | 43 | 13.5% |
| Doxycycline | 21 | 6.6% |
| Amikacin | 9 | 2.8% |
| Metronidazole | 8 | 2.5% |
| Meropenem | 22 | 6.9% |
| Ciprofloxacin | 3 | 0.9% |
| Cefixime | 2 | 0.6% |
| Cefotaxime | 3 | 0.9% |
| Amoxicillin/clavulanic | 11 | 3.5% |
| Ofloxacin | 10 | 3.1% |
| Rifaximin | 2 | 0.6% |
| Linezolid | 2 | 0.6% |
| Colistin | 4 | 1.3% |
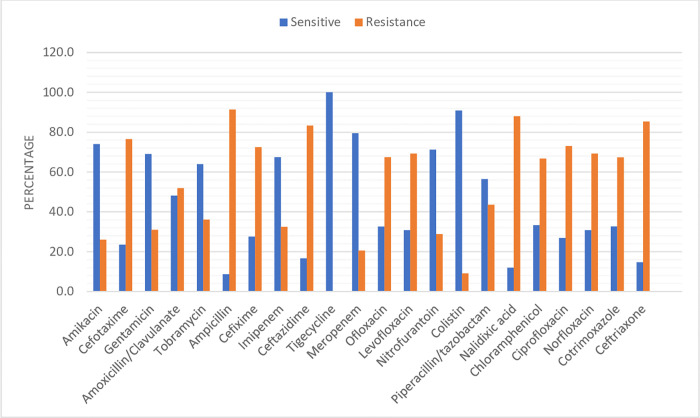
95 (29.9%) specimens were sent for AST testing out of 318 patients, with 16.59% (37) being mild, 66% (35) being moderate, 80.8% (21) being severe, and 12.5% (2) being critical. 58.9% (56) of them confirmed growth. E. Coli was isolated in the majority of the samples, 25% (14) followed by Klebsiella Pneumonia 17.9% (10), Pseudomonas spp. 14.3% (8), Enterobacter spp. 10.7% (6), Enterococcus 10.7% (6), Staphylococcus [MR CONS] 8.9% (5), non-fermenting gram-negative bacteria [NFGNB] 7.1% (4), Providencia Stuartii 1.8% (1), Proteus Mirabillis 1.8% (1), and Citrobacter Freundii 1.8% (1) (Table 3). Regardless to the isolated bacteria, the overall AST result was 100% sensitive to tigecycline, 90.9% to colistin, 79.9% to meropenem, 74% to amikacin, 71.2% to nitrofurantoin, 69% to gentamicin, 67.5% to imipenem, 63.9% to tobramycin, 56.4% to piperacillin/tazobactam, 48.1% to amoxicillin/clavulanate, 33.3% to chloramphenicol, 32.7% to cotrimoxazole, 32.6% to ofloxacin, 30.8% to norfloxacin, 30.8% to levofloxacin, 27.5% to cefixime, 26.9% to ciprofloxacin, 23.5% to cefotaxime, 16.7% to ceftazidime, 14.6% to ceftriaxone, 12% to nalidixic acid, and 8.7% to ampicillin (Table 4 & Fig 2).
Table 3. Distribution of isolated bacteria.
| Organisms Detected | Frequency | Percentage (n = 56) |
|---|---|---|
| CITROBACTER FREUNDII | 1 | 1.8% |
| E. COLI | 14 | 25% |
| ENTEROBACTER SPP | 6 | 10.7% |
| KLEBSIELLA PNEUMONIA | 10 | 17.9% |
| NFGNB | 4 | 7.1% |
| PROTEUS MIRABILLIS | 1 | 1.8% |
| PROVIDENCIA STUARTII | 1 | 1.8% |
| PSEUDOMONAS SPP | 8 | 14.3% |
| STAPHYLOCOCCUS [MR CONS] | 5 | 8.9% |
| ENTEROCOCCUS | 6 | 10.7% |
| TOTAL | 56 | 100% |
Table 4. Distribution of antibiotics sensitive and resistant irrespective of organism detected.
| Name | Total Isolated Organism (n = 56) | Excluding Not Tested | |||
|---|---|---|---|---|---|
| Sensitive | Resistance | Not tested | Sensitive | Resistance | |
| Amikacin | 66.1% (37) | 23.2% (13) | 10.7% (6) | 74% | 26% |
| Cefotaxime | 21.4% (12) | 69.6% (39) | 8.9% (5) | 23.5% | 76.5% |
| Gentamicin | 51.8% (29) | 23.2% (13) | 25% (14) | 69% | 31.0% |
| Amoxicillin/Clavulanate | 46.4% (26) | 50% (28) | 3.6% (2) | 48.1% | 51.9% |
| Tobramycin | 41.1% (23) | 23.2% (13) | 35.7% (20) | 63.9% | 36.1% |
| Ampicillin | 7.1% (4) | 75% (42) | 17.9% (10) | 8.7% | 91.3% |
| Cefixime | 19.6% (11) | 51% (29) | 28.6% (16) | 27.5% | 72.5% |
| Imipenem | 48.2% (27) | 23.2% (13) | 28.6% (16) | 67.5% | 32.5% |
| Ceftazidime | 12.5% (7) | 62.5% (35) | 25% (14) | 16.7% | 83.3% |
| Tigecycline | 16.1% (9) | 0 | 83.9% (47) | 100% | 0 |
| Meropenem | 55.4% (31) | 14.3% (8) | 30.4% (17) | 79.5% | 20.5% |
| Ofloxacin | 25% (14) | 51.8% (29) | 23.2% (13) | 32.6% | 67.4% |
| Levofloxacin | 28.6% (16) | 64.3% (36) | 7.1% (4) | 30.8% | 69.2% |
| Nitrofurantoin | 66.1% (37) | 26.8% (15) | 4.1% (4) | 71.2% | 22.8% |
| Colistin | 17.9% (10) | 1.8% (1) | 80.4% (45) | 90.9% | 9.1% |
| Piperacillin/tazobactam | 39.3% (22) | 30.4% (17) | 30.4% (17) | 56.4% | 43.6% |
| Nalidixic acid | 10.7% (6) | 78.6% (44) | 10.7% (6) | 12% | 88% |
| Chloramphenicol | 3.6% (2) | 7.1% (4) | 89.3% (50) | 33.3% | 66.7% |
| Ciprofloxacin | 25% (14) | 67.9% (38) | 7.1% (4) | 26.9% | 73.1% |
| Norfloxacin | 28.6% (16) | 64.3% (36) | 7.1% (4) | 30.8% | 69.2% |
| Cotrimoxazole | 28.6% (16) | 58.9% (33) | 12.5% (7) | 32.7% | 67.3% |
| Ceftriaxone | 10.7% (6) | 62.5% (35) | 26.8% (15) | 14.6% | 85.4% |
Fig 2. The distribution of antibiotic sensitivity and resistance was based on the results of the AST test.
There were no statistically significant differences in the antibiotics prescribed between the severity of COVID-19 and the p-value obtained was 0.644. Similarly, the antibiotics prescribed had a p-value of 0.163, indicating that no statistically significant differences existed between the genders. However, statistically significant differences in the severity of diseases were observed in the sample collection for the antibiotic susceptibility test (AST), with a p-value of 0.000.
Discussion
With this study, we intended to get insight into antibiotic prescribing practices in COVID-19 treatment and its impact on antibiotic resistance. In the study, we observed that the antibiotic prescribed was unaffected by the severity of COVID-19. At least 93.7% of the patients were prescribed at least one antibiotic, and azithromycin alone accounted for 73.6%. Similar findings were reported by WHO and Getahun H et al. [3, 11]. Antibacterial medications were preferred over antiviral drugs, which is similar to the results of Zhou F et al [7]. However, the number of antivirals used was seen to have increased. Antibiotics were prescribed in 93% of mild cases and 94.3% of moderate cases, despite COVID-19’s revised guidelines prohibiting the use of empirical antibiotics. It could be influenced by the findings of studies aimed at enhancing treatment outcomes by combining antibiotics with antiviral or antiprotozoal drugs when no cure is available [12–15].
Although antibiotics were started before the specimens were collected for AST, the growth was present in 58.9% of the total specimens sent for AST. This shows that antibiotics are being used inappropriately and the process of specimen collection was contrary to the Treatment Guidelines for Antimicrobial Use in Common Syndromes [16]. We found that E. coli, K. Pneumoniae, and Pseudomonas spp. were the most commonly isolated bacteria, while Marina G et al. [17] reported that Pseudomonas aeruginosa and Enterobacterales were the most prevalent in their study. The isolation of organisms is being influenced by the geographical region. 54.5% (12) of the antibiotics used for AST showed more than 50% resistance to the isolated pathogens. Antibiotics such as ampicillin, nalidixic acid, ceftriaxone, ceftazidime, cefotaxime, cefixime, and ciprofloxacin, which are commonly prescribed, were resistant in more than 70% of AST results. The inappropriate and overuse of antibiotics without an indication could be a cause of the issue. It’s also possible that the current pandemic’s focus on minimizing the immediate impact on individuals has obscured the longer-term threat of AMR [6].
Hand sanitizer was used by 92.43% of patients, indicating that it is an important component of COVID-19 transmission prevention awareness campaigns. Ochwoto M et al. reported that half of the hand sanitizers tested have efficacy levels that are lower than the WHO formulation in their study [18]. The pandemic’s desperate circumstances prevented quality assurance of the supplied hand sanitizer. Ineffective hand sanitizers encourage the growth of drug-resistant bacteria, increasing the risk of antibiotic cross-resistance [19].
The findings of the investigation highlight the terrible situation of antibiotics. Its inappropriate and overused nature can be more fatal if a pathogen develops multi-drug resistance. To save antibiotics in the future, no compromises should be made in the implementation of antimicrobial stewardship programs. AMR is caused by a variety of factors, including antibiotic overuse and misuse, as well as exposure to low-quality biocidal agents.
Our study’s limitation is that it was a single-center study with a small sample size, and we assessed antibiotic resistance independently of the isolated bacterium. The fact that AH&RC is located in a rural part of the Mandya district may have impacted its decision to ignore coronavirus protective measures such as safe distance and mask use. Further multi-center studies with a large sample size are required to demonstrate the impact of the COVID pandemic on AMR.
Conclusion
In the study, we identified that antibiotics were being used unnecessarily in excessive amounts and that more than half of the antibiotics tested were less sensitive to curing bacterial illness, regardless of the isolated bacterium. The current pandemic’s focus on reducing the immediate impact on humans may have hidden the longer-term threat of AMR, overuse of antibiotics, and exposure to inadequate biocidal agents, which might lead to an increase in AMR. To corroborate the findings, a multi-center study with larger sample size is necessary.
Acknowledgments
We thank Dr. B. Ramesh (Dean & Principal, Sri Adichunchanagiri College of Pharmacy), Dr. M. G. Shivaramu (Principal, Adichunchanagiri Institute of Medical Sciences), and Dr. B. G. Sagar (Superintendent, Adichunchanagiri Hospital and Research Center) for providing a favourable environment to conduct the study.
Data Availability
Data supporting the findings of this study are available with in the article and supplementary materials.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Rawson TM, Ming D, Ahmad R, Moore LSP, Holmes AH. Antimicrobial use, drug-resistant infections and COVID-19. Nat Rev Microbiol [Internet]. 2020;18(8):409–10. doi: 10.1038/s41579-020-0395-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abelenda-Alonso G, Padullés A, Rombauts A, Gudiol C, Pujol M, Alvarez-Pouso C, et al. Antibiotic prescription during the COVID-19 pandemic: a biphasic pattern. Infection Control & Hospital Epidemiology. 2020. Nov;41(11):1371–2. doi: 10.1017/ice.2020.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical management of COVID-19 Interim Guidance—May 2020. Geneva: World Health Organization; 2020. Available from: https://www.who.int/publications-detail/clinical-management-of-covid-19
- 4.Yam ELY. COVID-19 will further exacerbate global antimicrobial resistance. Int Soc Travel Med. 2020;27((6)). doi: 10.1093/jtm/taaa098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beović B, Doušak M, Ferreira-Coimbra J, Nadrah K, Rubulotta F, Belliato M, et al. Antibiotic use in patients with COVID-19: a ‘snapshot’ Infectious Diseases International Research Initiative (ID-IRI) survey. Journal of antimicrobial chemotherapy. 2020. Nov;75(11):3386–90. doi: 10.1093/jac/dkaa326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawson TM, Moore LS, Castro-Sanchez E, Charani E, Davies F, Satta G, et al. COVID-19 and the potential long-term impact on antimicrobial resistance. Journal of antimicrobial chemotherapy. 2020. Jul 1;75(7):1681–4. doi: 10.1093/jac/dkaa194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The lancet. 2020. Mar 28;395(10229):1054–62. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, et al. Clinical characteristics of Covid-19 in New York city. New England Journal of Medicine. 2020. Jun 11;382(24):2372–4. doi: 10.1056/NEJMc2010419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021. Apr 1;70(4):698–706. doi: 10.1136/gutjnl-2020-323020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical Laboratory Standards Institute; 2020. Available from: https://clsi.org/standards/products/microbiology/documents/m100/ [Google Scholar]
- 11.Getahun H, Smith I, Trivedi K, Paulin S, Balkhy HH. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull World Health Organ. 2020;98(7):19–20. doi: 10.2471/BLT.20.268573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautret P, Lagier JC, Parola P, Meddeb L, Mailhe M, Doudier B, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. International journal of antimicrobial agents. 2020. Jul 1;56(1):105949. doi: 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Mahmud R, Rahman MM, Alam I, Ahmed KGU, Kabir AKMH, Sayeed SKJB, et al. Ivermectin in combination with doxycycline for treating COVID-19 symptoms: a randomized trial. J Int Med Res. 2021;49(5). doi: 10.1177/03000605211013550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-kuraishy HM, Al-Gareeb AI, Qusty N, Cruz-Martins N, El-Saber Batiha G. Sequential doxycycline and colchicine combination therapy in Covid-19: The salutary effects. Pulm Pharmacol Ther [Internet]. 2021. Apr;67(January):102008. doi: 10.1016/j.pupt.2021.102008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arshad S, Kilgore P, Chaudhry ZS, Jacobsen G, Wang DD, Huitsing K, et al. Treatment with hydroxychloroquine, azithromycin, and combination in patients hospitalized with COVID-19. International journal of infectious diseases. 2020. Aug 1;97:396–403. doi: 10.1016/j.ijid.2020.06.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Indian Council of Medical Research. Treatment Guidelines for Antimicrobial Use in Common Syndromes. Indian Counc Med Res Dep Heal Res New Delhi, India [Internet]. 2017;1–106. Available from: http://www.icmr.nic.in/guidelines/treatment guidelines for antimicrobial.pdf
- 17.Gysin M, Acevedo CT, Haldimann K, Bodendoerfer E, Imkamp F, Bulut K, et al. Antimicrobial susceptibility patterns of respiratory Gram-negative bacterial isolates from COVID-19 patients in Switzerland. Annals of clinical microbiology and antimicrobials. 2021. Dec;20(1):1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochwoto M, Muita L, Talaam K, Wanjala C, Ogeto F, Wachira F, et al. Anti-bacterial efficacy of alcoholic hand rubs in the Kenyan market, 2015. Antimicrobial Resistance & Infection Control. 2017. Dec;6(1):1–6. Available from: doi: 10.1186/s13756-017-0174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caselli E. Hygiene: microbial strategies to reduce pathogens and drug resistance in clinical settings. Microbial Biotechnology. 2017. Sep;10(5):1079–83. doi: 10.1111/1751-7915.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings of this study are available with in the article and supplementary materials.