Abstract
Changes in plant auxin levels can be perceived and converted into cellular responses by auxin signal transduction. AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) proteins are auxin transcriptional inhibitors that play important roles in regulating auxin signal transduction. The stability of Aux/IAA proteins is important for transcription initiation and downstream auxin-related gene expression. Here, we report that the Aux/IAA protein IAA17 interacts with the small ubiquitin-related modifier (SUMO) E3 ligase METHYL METHANESULFONATE-SENSITIVE 21 (AtMMS21) in Arabidopsis (Arabidopsis thaliana). AtMMS21 regulated the SUMOylation of IAA17 at the K41 site. Notably, root length was suppressed in plants overexpressing IAA17, whereas the roots of K41-mutated IAA17 transgenic plants were not significantly different from wild-type roots. Biochemical data indicated that K41-mutated IAA17 or IAA17 in the AtMMS21 knockout mutant was more likely to be degraded compared with nonmutated IAA17 in wild-type plants. In conclusion, our data revealed a role for SUMOylation in the maintenance of IAA17 protein stability, which contributes to improving our understanding of the mechanisms of auxin signaling.
Introduction
Auxin is an important hormone involved in plant development and environmental adaptation. Plants perceive the changes taking place in auxin levels and then convert the signal into plant cell responses through the auxin signal transduction pathway (Yu et al., 2022). In the auxin signal transduction pathway, the interaction of AUXIN/INDOLE-3-ACETIC ACIDs (Aux/IAAs) with AUXIN RESPONSE FACTORs (ARFs) can repress ARF activity in normal conditions (Salehin et al., 2015). Once the auxin receptor receives the auxin signal, Aux/IAAs are degraded by the 26S proteasome, which derepresses ARFs and initiates an auxin-responsive gene expression cascade (Salehin et al., 2015). Auxin signal transduction contributes to plant response to environmental changes and adjusts growth and development (Lv et al., 2021; Zhang et al., 2022). Therefore, Aux/IAAs play a crucial role in the auxin signal transduction pathway.
There are 29 members of the Aux/IAA proteins family in Arabidopsis (Arabidopsis thaliana) (Paponov et al., 2008), which usually contains 4 conserved Domains I-IV (Li et al., 2009). Among them, Domain I can be interacted by a TOPLESS corepressor to bring about repression function (Causier et al., 2012); III and IV are protein–protein interaction domains where ARFs and Aux/IAA proteins interact with each other (Hagen, 2015); these domains enable Aux/IAA to inhibit the transcriptional activity of the ARF protein. Domain II is the key domain that determines the stability of the Aux/IAA protein. It contains a conserved “degron” motif, which is the interaction site between Aux/IAA and the auxin receptor protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1) (Calderon-Villalobos et al., 2010). Once the auxin signal is received by the auxin receptor TIR1, the interaction between TIR1 protein and Aux/IAA protein is enhanced (Dharmasiri et al., 2003). The receptor protein TIR1 is also the core component of the SCF ubiquitin ligase complex. SCFTIR1 can mediate the Aux/IAA protein into the ubiquitination pathway for degradation (Tan et al., 2007). Consequently, a high concentration of auxin induces the degradation of Aux/IAA. The stability of Aux/IAAs is an important factor in the auxin signal transduction pathway.
Due to the difference in the sequences of Domain II, the half-lives of 29 Aux/IAAs and the extent to which their half-lives reduce in response to auxin vary greatly (Leyser, 2018).
Mutation in Domain II greatly enhances the half-life of IAA17, which increases the stability of the protein and further affects root stem cell niches and root growth (Rouse et al., 1998; Ouellet et al., 2001; Tian et al., 2014). Besides, the rate of Aux/IAA degradation can be changed through competitive interactions with other proteins. RGA-LIKE PROTEIN 3 (RGL3) and TIR1 competitively bind to IAA17 in the presence of auxin, suggesting that RGL3 plays a role in IAA17 stabilization (Shi et al., 2017). Moreover, photoactivated photoreceptors cryptochrome 1 (CRY1) and phytochrome B (phyB) can directly interact with Aux/IAA to regulate the hypocotyl elongation by stabilizing the Aux/IAA proteins (Xu et al., 2018). The protein stability of Aux/IAA can also be regulated by posttranslational modification. In Arabidopsis, SCFTIR1-mediated ubiquitination marks IAA3 and IAA16 proteins for degradation (Maraschin Fdos et al., 2009). In Rice, degradation of OsIAA9 and OsIAA26 is mediated by ubiquitination (Chen et al., 2018).
SUMOylation, which involves the transfer of a polypeptide small ubiquitin-like modifier (SUMO) onto protein substrates, is a type of important posttranslational modification that is widely found in plants. One of the major functions of SUMOylation is regulating protein stability (Verma et al., 2018). Previous research has revealed that the T-DNA insertion mutant mms21-1 of SUMO E3 ligase METHYL METHANESULFONATE-SENSITIVE 21 (AtMMS21) exhibits root development defects due to a misregulation of auxin-related transcription factors (Huang et al., 2009; Ishida et al., 2009; Xu et al., 2013). Mutation of AtMMS21 affects the gene expression of the downstream auxin signaling pathway (Ishida et al., 2009; Xu et al., 2013), suggesting that AtMMS21-mediated SUMOylation may be involved in the regulation of the auxin signaling pathway. It has been established that SUMOylation is related to the regulation of the auxin signal transduction pathway. SUMOylation modifications in TIR1 and ARF7 have been discovered and have roles in receptor stability maintenance and repressor complex recruitment (Kirsten, 2017; Orosa-Puente et al., 2018). However, there is currently no evidence that Aux/IAA can be SUMOylated in plants, which may be due to the fact that large members of the Aux/IAA family are full of functional redundancy (Kirsten, 2017). The mechanism by which SUMOylation regulates the auxin signal transduction pathway remains unclear.
In the present study, the SUMOylation of IAA17, a member of the Arabidopsis Aux/IAA proteins, is identified and validated. Furthermore, we characterize the SUMOylation-mediated stability of IAA17, which is involved in root growth regulation, which contributes to improving our understanding of auxin signal transduction mechanisms. In all, we discover a key mechanism by which SUMOylation is involved in regulating Aux/IAA protein degradation.
Results
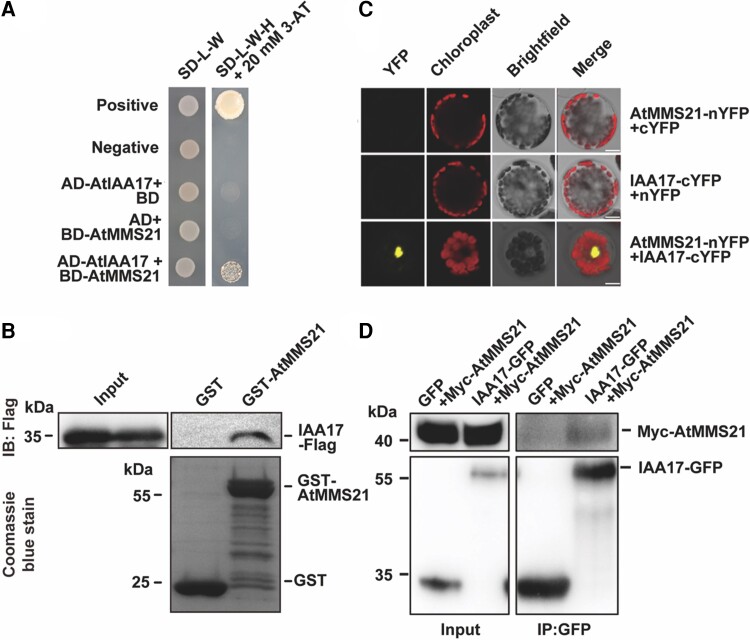
IAA17 interacts with AtMMS21 in vitro and in vivo
Previous reports show that AtMMS21 regulates the gene expression of the downstream auxin signaling pathway (Xu et al., 2013). Here, we are interested in revealing whether the SUMO E3 ligase AtMMS21 targets key components in the auxin signal transduction pathway. Yeast 2-hybrid screen was employed to identify AtMMS21-interacting proteins involved in auxin signal transduction. As key components in the auxin signal transduction pathway, Aux/IAA family proteins were examined to reveal their interaction with AtMMS21. The results showed that the Aux/IAA protein member IAA17 interacted with AtMMS21 in yeast cells (Figure 1A; Supplemental Figure 1). To further confirm the physical interaction between IAA17 and AtMMS21, an in vitro pull-down assay was implemented. Flag-tagged IAA17 was pulled down by GST-AtMMS21 compared with control samples, implying a potential association between IAA17 and AtMMS21 (Figure 1B). To further explore their interaction in plant cells, the bimolecular fluorescence complementation assay (BiFC) was performed. Both nYFP-fused AtMMS21 and cYFP-fused IAA17 were co-expressed in Arabidopsis protoplasts, and confocal microscopy revealed that IAA17 and AtMMS21 interacted in the nucleus (Figure 1C). Furthermore, their interaction was validated by a co-immunoprecipitation assay using Myc-tagged AtMMS21 and GFP-tagged IAA17 in transiently expressed protoplasts. The result revealed that IAA17-GFP was specifically immunoprecipitated with Myc-AtMMS21 (Figure 1D). Taken together, these results support the conclusion that IAA17 interacts with AtMMS21.
Figure 1.
AtMMS21 interacts with IAA17. A, The interaction between AtMMS21 (fused with the DNA-binding domain [BD]) and IAA17 (fused with the activation domain [Ad]) was determined by using a yeast 2-hybrid assay. Twenty millimolar 3-AT (3-amino-1,2,4-triazole) were used to select 2-hybrid interactions. pBD-53 + pAD-T were co-transformed to yeast cells as positive control, and pBD-Lam + pAD-T were co-transformed as negative control. B, The interaction between AtMMS21 and IAA17 was detected using an in vitro pull-down assay. GST-AtMMS21 and IAA17-Flag were expressed in Escherichia coli. The IAA17-Flag protein associated with the immobilized GST-AtMMS21 was detected by immunoblotting analysis. The levels of GST and GST-AtMMS21 were visualized using Coomassie Blue staining. C, The interaction between AtMMS21 and IAA17 was detected by using a BiFC assay. AtMMS21 fused with nYFP and IAA17 fused with cYFP were co-expressed in Arabidopsis protoplast. Bars, 10 μm. YFP, the autofluorescence from chloroplasts and bright-field signals were detected and merged. D, The interaction between AtMMS21 and IAA17 in an in vivo co-immunoprecipitation assay. 35S:Myc-AtMMS21 and 35S:IAA17-GFP were co-transformed in protoplasts. Total protein extracts were immunoprecipitated with anti-GFP beads. The proteins from lysates (left) and immunoprecipitated proteins (right) were detected using an anti-GFP or anti-Myc antibody.
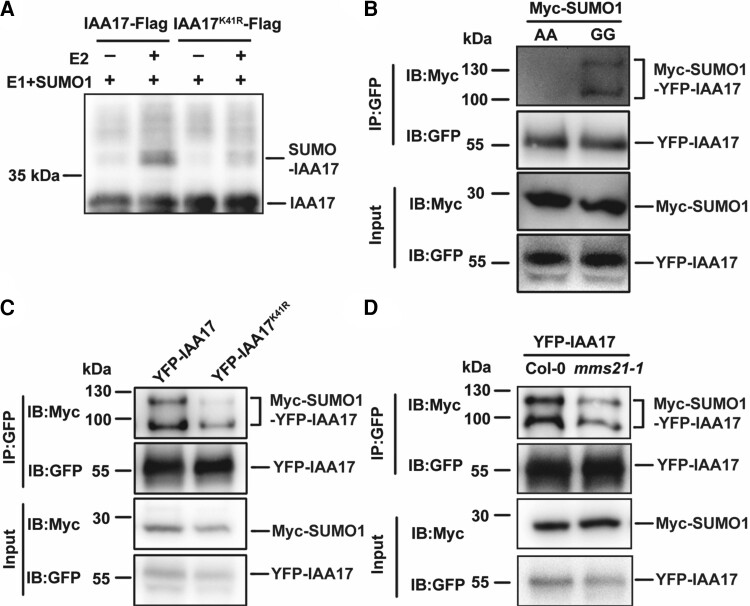
AtMMS21 regulates the SUMOylation of IAA17 at lysine 41
The interaction of IAA17 with AtMMS21 suggests that IAA17 may be a substrate for SUMOylation since AtMMS21 is a SUMO E3 ligase (Duan et al., 2009). To detect whether IAA17 is covalently modified by the SUMO protein, the SUMOylation reaction was performed in a reconstituted Escherichia coli system with Arabidopsis SUMOylation machinery proteins (Okada et al., 2009). In the presence of the SUMO E1 and E2 enzyme, a higher molecular weight IAA17-Flag signal was detected in the sample containing SUMO1 by immunoblotting, indicating that IAA17 is covalently modified by SUMO1 (Figure 2A). According to the SUMOylation site prediction of GPS-SUMO 2.0 (Zhao et al., 2014), the lysine at position 41 of IAA17 has the potential for SUMOylation. Thus, we mutated Lys-41 of IAA17 and detected the SUMOylation in E. coli compared with wild-type IAA17. Compared with IAA17-Flag, SUMO1-modified IAA17K41R-Flag decreased substantially or almost disappeared (Figure 2A). To further analyze the SUMOylation of IAA17 in plant cells, we performed transient SUMOylation experiments in Arabidopsis protoplasts. Firstly, we transiently co-expressed YFP-IAA17 with Myc-SUMO1GG (wild-type) or Myc-SUMO1AA (a conjugation-deficient mutant) in Arabidopsis protoplasts. We then immunoprecipitated YFP-IAA17 with an anti-GFP antibody and detected the immunoprecipitated proteins with an anti-Myc antibody. Higher molecular weight SUMOylated YFP-IAA17 bands (SUMO1-IAA17) were detected when YFP-IAA17 was co-expressed with Myc-SUMO1GG but not with Myc-SUMO1AA. This indicates that IAA17 is SUMOylated and is a substrate of SUMO1 in plant cells. To further verify the SUMO modification site of IAA17 in plant cells, the wild-type or Lys-41 mutation version of IAA17 (IAA17K41R) was co-expressed with Myc-tagged SUMO1GG in Arabidopsis protoplasts (Figure 2B). The anti-Myc antibody detected the SUMOylated IAA17 bands in Arabidopsis protoplasts cells co-expressing YFP-IAA17 and Myc-SUMO1GG (Figure 2C). However, the SUMOylated IAA17 bands were greatly reduced in cells co-expressing YFP-IAA17K41R and Myc-SUMO1GG (Figure 2C). This result reveals that Lys-41 of IAA17 is the main site of SUMO1 modification. We previously demonstrated that IAA17 interacted with the SUMO E3 ligase AtMMS21, and then we determined whether SUMO modification of IAA17 was mediated by AtMMS21. We transiently co-expressed YFP-IAA17 with Myc-SUMO1GG in Arabidopsis protoplasts of Col-0 and mms21-1. The result shows that the SUMOylated IAA17 bands are greatly reduced in the mms21-1 background compared with Col-0, suggesting that SUMOylation of IAA17 is mediated by AtMMS21 (Figure 2D).
Figure 2.
IAA17 is SUMOylated at its K41 residue and mediated by AtMMS21. A, The SUMO conjugation of IAA17 was detected by using a reconstituted SUMOylation system in Escherichia coli. Flag-tagged IAA17 or the mutant with Lys-41 was expressed in Escherichia coli. In the presence of E1 and SUMO1 with or without E2, the unconjugated and SUMO-conjugated IAA17-Flag were detected by using an anti-Flag antibody. B, In vivo SUMOylation of IAA17 was detected in Arabidopsis protoplast. YFP-IAA17 and Myc-SUMO1GG or Myc-SUMO1AA were co-transformed in Arabidopsis protoplast. Total proteins were immunoprecipitated with an anti-GFP antibody, and the immunoprecipitated proteins were detected with anti-GFP and anti-Myc antibodies. C, K41 is a major SUMOylation site of IAA17 in vivo. YFP-IAA17 or YFP-IAA17K41R was co-expressed with Myc-SUMO1 in Arabidopsis protoplast. Total proteins were immunoprecipitated with an anti-GFP antibody, and the immunoprecipitated proteins were detected with anti-GFP and anti-Myc antibodies. D, SUMOylation of IAA17 is mediated by AtMMS21. YFP-IAA17 and Myc-SUMO1 were co-transformed in Col-0 and mms21-1 protoplast. Total proteins were immunoprecipitated with an anti-GFP antibody, and the immunoprecipitated proteins were detected with anti-GFP and anti-Myc antibodies.
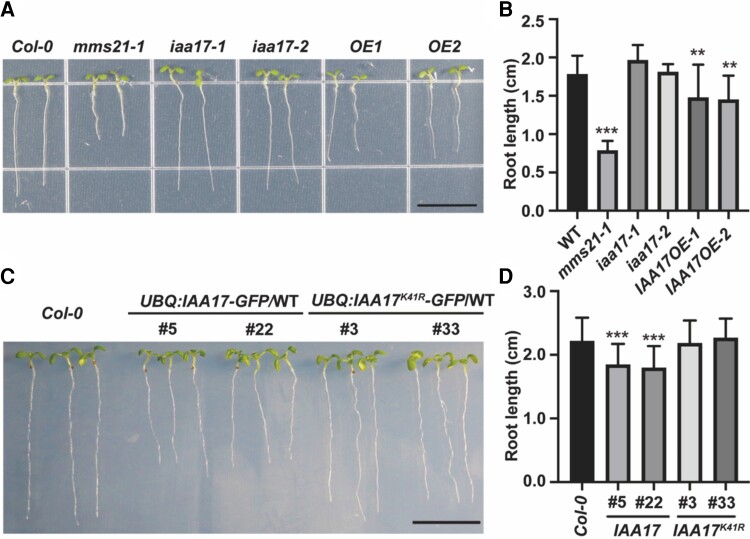
SUMOylation of IAA17 is necessary for its function in root development
Previous reports indicate that the deletion of AtMMS21 results in a short root phenotype in Arabidopsis (Huang et al., 2009); the stable IAA17 mutant axr3 also shows the inhibition of root development (Rouse et al., 1998). Due to the interaction of AtMMS21 and IAA17, they may co-regulate root development. We compared the root development of Col-0, mms21-1, IAA17 T-DNA insertion mutants iaa17-1 and iaa17-2, and IAA17 overexpression lines IAA17-OE1 and IAA17-OE2 (Figure 3, A and B). The T-DNA insertion mutants iaa17-1 and iaa17-2 did not differ significantly from Col-0 in root development at 7 days after germination (DAG) (Figure 3, A and B). However, IAA17 overexpression lines OE1 and OE2 showed a reduced root length compared with Col-0 (Figure 3, A and B). Also, mms21-1 exhibited a more severe impaired root growth and development than IAA17 overexpression lines. We crossed the mms21-1 mutant with IAA17 overexpression lines and compared its root length with that of mms21-1. The result showed that the root lengths of the hybrid plants and mms21-1 were not significantly different (Supplemental Figure 2). This supports the notion that AtMMS21 and IAA17 act on the same signaling pathway required for root development and that AtMMS21 may be an upstream regulator of IAA17.
Figure 3.
SUMOylation of IAA17 is involved in root development. A, The phenotypes of root developmental defect in mms21-1 and IAA17 overexpression lines (OE). The photograph was taken 7 days after germination. Bar, 1 cm. B, The measurement of the root lengths of Col-0, mms21-1, OE1, OE2, and T-DNA insertion mutants iaa17-1 and iaa17-2. The data are means ± Sd from at least 15 seedlings in 3 biological independent experiments. **P < 0.01, ***P < 0.001, Student's t test. C, The phenotypes of the root development of Col-0, UBQ:IAA17-GFP/WT, and UBQ:IAA17K41R-GFP/WT. The photograph was taken 7 days after germination. Bar, 1 cm. D, The measurement of the root lengths of Col-0, UBQ:IAA17-GFP/WT, and UBQ:IAA17K41R-GFP/WT. The data are means ± Sd from at least 15 seedlings in 3 biological independent experiments. ***P < 0.001, Student's t test.
To investigate the mechanisms by which AtMMS21 and IAA17 affect root growth, we used the GFP-tagged IAA17 to explore the physiological function of SUMOylated IAA17 in Arabidopsis. GFP fused to the N or C terminus of the IAA17 protein does not affect the nuclear localization and function of IAA17 (Supplemental Figure 3; Moss et al., 2015). We selected transgenic lines with similar constitutive transcript levels of wild-type and Lys-41 mutant versions of IAA17-GFP for a further comparison of root phenotypes. The results showed that the wild-type version of the IAA17-GFP transgenic line exhibited a shortened root length compared with the Lys-41 mutant and Col-0 (Figure 3, C and D; Supplemental Figure 4). However, there is no significant difference between Lys-41 mutant IAA17-GFP and Col-0, suggesting that only SUMOylated IAA17 serves function properly in root growth and development.
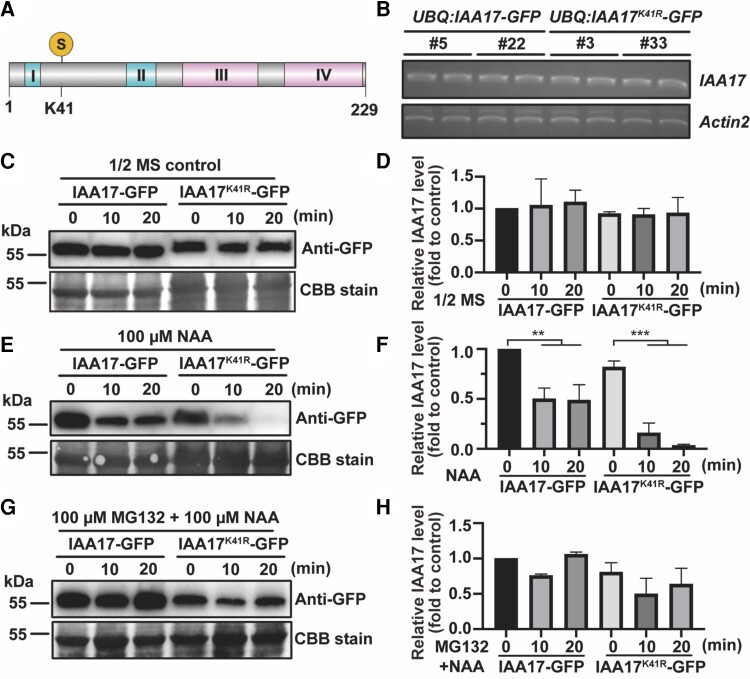
Stability of the IAA17 protein is dependent on AtMMS21-mediated SUMOylation
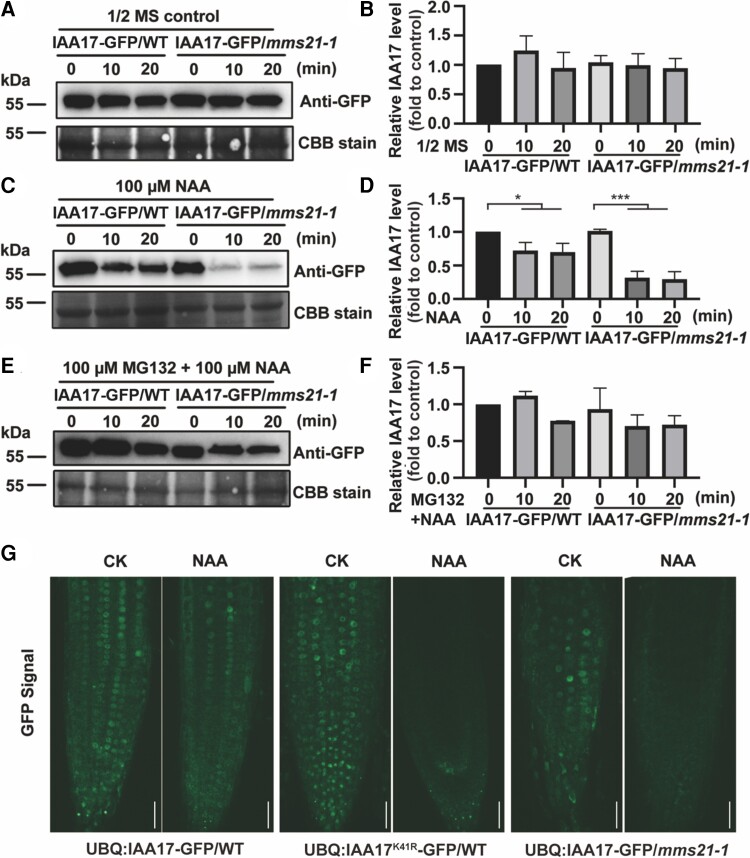
The IAA17 gain-of-function mutant axr3 encodes a stable IAA17 that inhibits root growth and development (Rouse et al., 1998). Our data show that only the wild-type IAA17, but not Lys-41 mutant IAA17, inhibits root growth and development. Therefore, we hypothesized that the SUMOylation of IAA17 is related to the stability of the IAA17 protein. Similar to other Aux/IAA proteins, IAA17 has 4 conserved domains, and Lys-41 residue is located outside the conserved domains (Figure 4A). We selected transgenic lines with similar constitutive transcript levels of wild-type and Lys-41 mutant versions of IAA17-GFP for further determination of protein levels (Figure 4B; Supplemental Figure 5). 1-Naphthylacetic acid (NAA) treatment induced the degradation of IAA17 (Supplemental Figure 6). The protein stabilities of the wild-type and Lys-41 mutant versions of IAA17-GFP were compared under NAA treatment conditions. Transgenic Arabidopsis UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT were treated by using a 1/2 Murashige and Skoog (MS) medium with or without 100 μM NAA. Immunoblotting analysis showed that the wild-type and Lys-41 mutant versions of IAA17-GFP remained stable under 1/2 MS normal conditions (Figure 4, C and D). Interestingly, the IAA17K41R-GFP signal decreased rapidly after NAA treatment, whereas the wild-type IAA17-GFP showed a higher stability (Figure 4, E and F). To further verify that SUMOylation-mediated stabilization of IAA17 is indeed related to the proteasomal degradation pathway, transgenic plants were treated with 1/2 MS containing 100 µM NAA and 100 µM MG132. The GFP signal of UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT showed no degradation, indicating that the SUMOylation of IAA17 is resistant to the 26S proteasomal degradation pathway (Figure 4, G and H).
Figure 4.
SUMOylation of IAA17 inhibits IAA17 degradation. A, Schematic diagram of an IAA17 protein structure. The relative positions of Domains I, II, III, and IV in the IAA17 protein structure are indicated. S, the position of the SUMOylation site, K41. B, Gene expression of IAA17 in UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT. The relative expression of IAA17 in UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT was detected by RT-PCR. Actin2 was used as a loading control. C, IAA17 protein levels in UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT. UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT were treated with 1/2 MS for 0, 10, and 20 min. The protein levels of IAA17-GFP in the seedlings were detected by immunoblotting analysis using a GFP antibody. The loading controls from Coomassie Blue staining are shown at the bottom. D, Quantification of an IAA17-GFP blot in (C). The quantification of IAA17-GFP in 0 min is set to 1. The data are means ± Sd from 2 biological independent experiments. E, IAA17 protein levels in UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT under NAA treatment. UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT were treated with 1/2 MS containing 100 µM NAA for 0, 10, and 20 min. The protein levels of IAA17-GFP in the seedlings were detected by immunoblotting analysis using a GFP antibody. The loading controls from Coomassie Blue staining are shown at the bottom. F, Quantification of the IAA17-GFP blot in (E). The quantification of IAA17-GFP in 0 min is set to 1. The data are means ± Sd from 2 biological independent experiments. *P < 0.05, ***P < 0.001, Student's t test. G, IAA17 protein levels in UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT under NAA and MG132 treatment. UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT were treated with 1/2 MS containing 100 µM NAA and 100 µM MG132 for 0, 10, and 20 min. The protein levels of IAA17-GFP in the seedlings were detected by immunoblotting analysis using a GFP antibody. The loading controls from Coomassie Blue staining are shown at the bottom. H, Quantification of the IAA17-GFP blot in (G). The quantification of IAA17-GFP in 0 min is set to 1. The data are means ± Sd from 2 biological independent experiments. NAA, 1-Naphthylacetic acid.
To further confirm that the stability of IAA17 is regulated by AtMMS21-mediated SUMOylation, we compared the stability of the IAA17 protein in Col-0 and mms21-1 backgrounds. Transgenic Arabidopsis UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1 were treated by 1/2 MS with or without 100 μM NAA. Compared with the control group (Figure 5, A and B), the IAA17-GFP signal in the mms21-1 background rapidly decreased after NAA treatment (Figure 5, C and D). However, the IAA17 protein in Col-0 showed a higher stability than that in the mms21-1 background (Figure 5, C and D). To prove that the rapid degradation of IAA17 in mms21-1 is also via the 26S proteasome, UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1 were treated with 1/2 MS containing 100 µM NAA and 100 µM MG132. Immunoblotting analysis showed that MG132 inhibited IAA17 degradation in mms21-1, revealing that AtMMS21-medatied SUMOylation of IAA17 regulates protein stability by affecting the 26S proteasomal degradation pathway (Figure 5, E and F).
Figure 5.
AtMMS21 mediates the SUMOylation of IAA17 regulating IAA17 degradation. A, IAA17 protein levels in UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1. UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1 were treated with 1/2 MS for 0, 10, and 20 min. The protein levels of IAA17-GFP in the seedlings were detected by immunoblotting analysis using a GFP antibody. The loading controls from Coomassie Blue staining are shown at the bottom. B, Quantification of the IAA17-GFP blot in (A). The quantification of IAA17-GFP in 0 min is set to 1. The data are means ± Sd from 2 biological independent experiments. C, IAA17 protein levels in UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1 under NAA treatment. UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1 were treated with 1/2 MS containing 100 µM NAA for 0, 10, and 20 min. The protein levels of IAA17-GFP in the seedlings were detected by immunoblotting analysis using a GFP antibody. The loading controls from Coomassie Blue staining are shown at the bottom. D, Quantification of the IAA17-GFP blot in (C). The quantification of IAA17-GFP in 0 min is set to 1. The data are means ± Sd from 2 biological independent experiments. *P < 0.05, ***P < 0.001, Student's t test. E, IAA17 protein levels in UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1 under NAA and MG132 treatment. UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1 were treated with 1/2 MS containing 100 µM NAA and 100 µM MG132 for 0, 10, and 20 min. The protein levels of IAA17-GFP in the seedlings were detected by immunoblotting analysis using a GFP antibody. The loading controls from Coomassie Blue staining are shown at the bottom. F, Quantification of the IAA17-GFP blot in (E). The quantification of IAA17-GFP in 0 min is set to 1. The data are means ± Sd from 2 biological independent experiments. G, IAA17-GFP signals in the roots of UBQ:IAA17-GFP/WT, UBQ:IAA17K41R-GFP/WT, and UBQ:IAA17-GFP/mms21-1 seedlings under NAA treatment. Five-day-old seedlings were treated with 100 µM NAA for 5 min. and the GFP signals were observed and collected via microscopy with the same settings. Bars, 50 μm. NAA, 1-Naphthylacetic acid.
To further investigate the degradation of the IAA17 protein under NAA treatment conditions, the GFP intensity in the roots of UBQ:IAA17-GFP/WT, UBQ:IAA17K41R-GFP/WT, and UBQ:IAA17-GFP/mms21-1 were compared (Figure 5G). The GFP intensity decreased rapidly under NAA treatment in UBQ:IAA17K41R-GFP/WT and UBQ:IAA17-GFP/mms21-1. However, it was more stable in UBQ:IAA17-GFP/WT under NAA treatment. Taken together, the AtMMS21-mediated SUMOylation of IAA17 increases the stability of the IAA17 protein.
SUMOylation of IAA17 inhibits the ubiquitination of IAA17
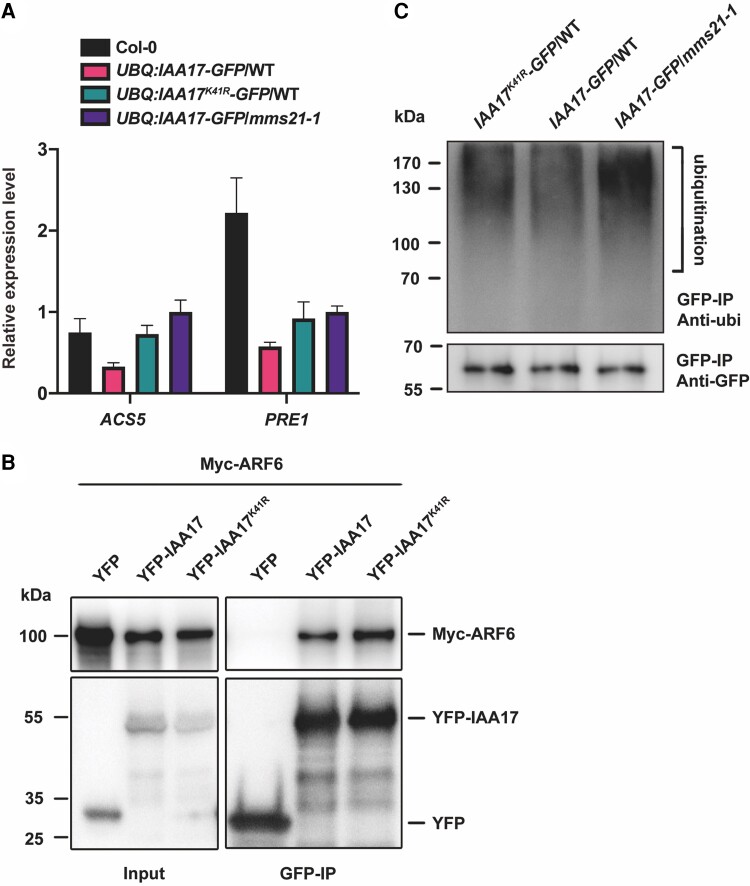
Previous studies have shown that Aux/IAAs interact with ARFs to repress ARF activity (Salehin et al., 2015), and IAA17 interacts with ARF6 (Mao et al., 2020). To further verify whether the SUMOylation-mediated degradation of IAA17 affects downstream gene expression, the gene expressions of ARF6-binding targets ACS5 and PRE1 were detected by RT-qPCR (Oh et al., 2014). The results showed that the transcriptional levels of ACS5 and PRE1 were repressed in wild-type IAA17-GFP overexpression plants but not in IAA17-GFP overexpression plants mutated at the SUMOylation site (Figure 6A). IAA17 overexpression also failed to suppress the gene expression of ARF6-binding targets in AtMMS21 knockout mutants (Figure 6A). This result is consistent with the conclusion that SUMOylation of IAA17 is critical for its molecular function in regulating IAA17 degradation.
Figure 6.
IAA17 SUMOylation regulates the expression of downstream genes by affecting ubiquitination. A, The transcript levels of ACS5 and PRE1 in UBQ:IAA17-GFP/WT, UBQ:IAA17K41R-GFP/WT, and UBQ:IAA17-GFP/mms21-1 transgenic plants. RNA was prepared from 7-day-old seedlings. Actin2 was used as an internal control. The error bars indicate the ± Sd of 3 independent assays. B, The interaction of IAA17 and IAA17K41R with ARF6. The effect of SUMOylation on the interaction between IAA17 and ARF6 was detected via immunoprecipitation. C, The ubiquitination levels of IAA17 in UBQ:IAA17-GFP/WT, UBQ:IAA17K41R-GFP/WT, and UBQ:IAA17-GFP/mms21-1 transgenic plants were detected by immunoprecipitation. The total proteins were extracted from 7-day-old seedlings of UBQ:IAA17-GFP/WT, UBQ:IAA17K41R-GFP/WT, and UBQ:IAA17-GFP/mms21-1. The protein extracts were incubated with GFP beads. After washing, the precipitated signals were measured via immunoblotting analysis with a GFP or ubiquitin antibody.
To further investigate how the SUMOylation of IAA17 regulates the auxin signaling pathway, the interaction between IAA17 and downstream ARF6 was tested by co-immunoprecipitation. Immunoblot analysis showed that IAA17 mutated at the SUMOylation site exhibited the same interaction with ARF6 compared with wild-type IAA17 (Figure 6B), indicating that SUMOylation does not affect the interaction with ARF6.
Degradation of the Aux/IAA protein depends on the ubiquitination pathway (Tan et al., 2007). To further investigate whether the SUMOylation of IAA17 regulates the stability of the IAA17 protein by affecting the ubiquitination pathway, we performed an immunoprecipitation experiment in UBQ:IAA17-GFP/WT, UBQ:IAA17K41R-GFP/WT, and UBQ:IAA17-GFP/mms21-1 plants using anti-GFP beads. Immunoblot analysis showed that IAA17 SUMOylation site mutation or AtMMS21 knockout enhanced IAA17 ubiquitination (Figure 6C), indicating that SUMOylation blocks IAA17 ubiquitination.
SUMOylation of IAA17 reduces under auxin treatment
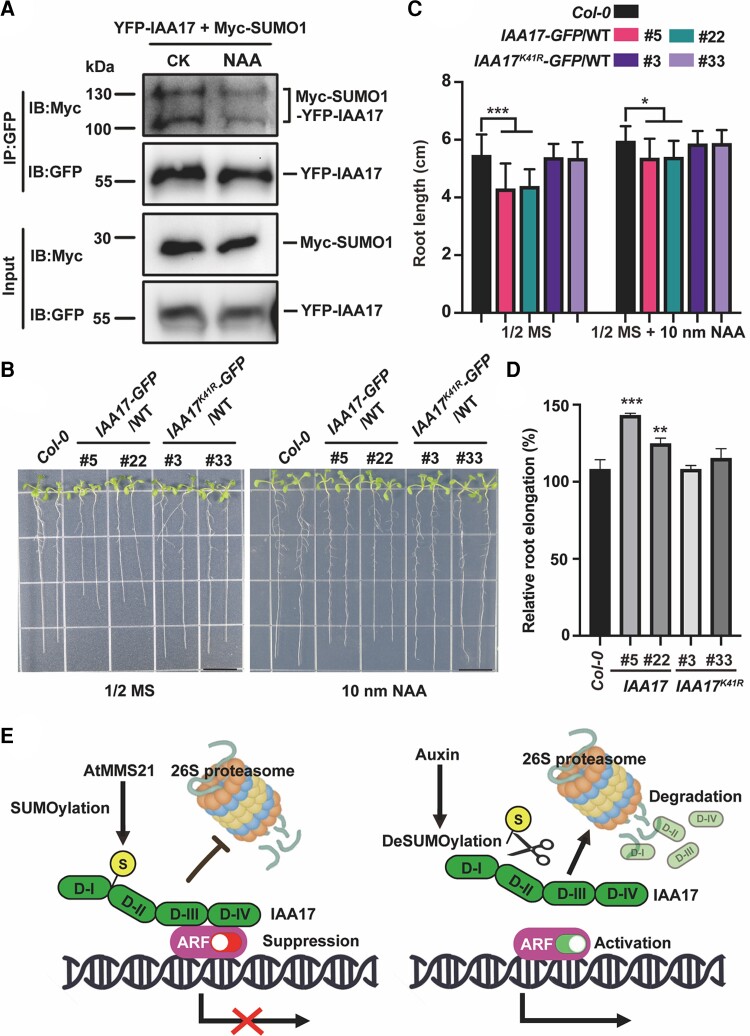
Due to the fact that auxin induces the degradation of IAA17 (Dreher et al., 2006), what follows is whether auxin is involved in the degradation of IAA17 by affecting the SUMOylation level of the IAA17 protein. To further investigate the link between IAA17 SUMOylation and auxin pathway signaling, we compared the SUMOylation level of IAA17 under control and NAA treatment conditions (Figure 7A). YFP-IAA17 and Myc-SUMO1GG were co-expressed in Arabidopsis protoplasts followed by treatment with 100 µM NAA. The SUMOylated YFP-IAA17 bands (IAA17-SUMO1) under normal and NAA treatment conditions were compared. The immunoblotting results showed that the SUMOylation of IAA17 reduced under NAA treatment, suggesting that high auxin levels might lead to a de-SUMOylation of IAA17.
Figure 7.
Auxin reduces the SUMOylation level of IAA17. A, In vivo SUMOylation of IAA17 was detected in Arabidopsis protoplast treated with or without 100 µM NAA. YFP-IAA17 and Myc-SUMO1 were co-transformed in Arabidopsis protoplast. The transformed Arabidopsis protoplasts were treated with 100 µM NAA for 30 min. Total proteins were immunoprecipitated with an anti-GFP antibody, and the immunoprecipitated proteins were detected with anti-GFP and anti-Myc antibodies. B, The root length of 14-day-old seedlings grown at 1/2 MS or 1/2 MS supplemented 10 nM NAA. The photo was taken 10 days after germination. Bars, 1 cm. C, The measurement of the root length of 10-day-old seedlings grown at 1/2 MS or 1/2 MS supplemented 10 nM NAA. The data are means ± Sd from at least 15 seedlings in 3 biological independent experiments. *P < 0.05, ***P < 0.001, Student's t test. NAA, 1-Naphthylacetic acid. D, relative root elongation rate comparison with control and NAA treatment. The ratio of the root length under NAA treatment and control conditions is calculated as the relative root elongation rate. The error bars indicate the Sd of 3 biological repeats. **P < 0.01, ***P < 0.001, and Student's t test. E, Proposed model for AtMMS21-mediated SUMOylation of IAA17 involved in the auxin signaling pathway. This model is based on the information provided in this study and the references cited in the discussion.
To further verify whether high auxin levels reduce the SUMOylation of IAA17 in plants, the roots of transgenic plants under NAA treatment were compared (Figure 7B). Under normal conditions, the root length of UBQ:IAA17-GFP/WT reduced more than that of the Col-0 and Lys-41 mutant versions of IAA17-GFP (Figure 7C). However, the root length difference between UBQ:IAA17-GFP/WT and UBQ:IAA17K41R-GFP/WT reduced under NAA treatment (Figure 7C). The root elongation rate also indicated that the dysplastic root growth phenotype of UBQ:IAA17-GFP/WT was partially restored under NAA treatment (Figure 7C and D). Taken together, our results suggest that auxin reduces the SUMOylation of IAA17 and that IAA17 with a reduced SUMOylation level is more likely to be degraded.
Discussion
The Aux/IAA family repressor proteins are an integral part of the auxin signaling machinery. In the presence of auxin, Aux/IAAs are degraded by the action of the ubiquitin E3 ligase SCFTIR1/AFB, resulting in a derepression of ARF transcription (Salehin et al., 2015; Weijers and Wagner, 2016). Therefore, the stability of Aux/IAA is crucial for its function as transcriptional repressors in auxin signaling. Although the mechanisms of Aux/IAA degradation are well known, the factors that regulate Aux/IAA stability remain unclear. In this study, we validated the interaction of IAA17 and AtMMS21 and identified AtMMS21-mediated SUMOylation of IAA17. We further characterized the regulatory mechanism by which SUMOylation enhances the stability of the IAA17 protein.
We identified the SUMOylation of IAA17 in vivo and in vitro and found that there were at least 2 SUMO-modified bands of IAA17 as shown by the immunoblotting analysis (Figure 2B), suggesting that IAA17 is a multiple SUMO1-modified protein in Arabidopsis. The Lys-41 mutation of IAA17 causes an attenuation of 2 SUMO modification bands (Figure 2C). However, K41-mutated IAA17 still has weak SUMO modifications, suggesting that IAA17 may have other SUMO1 modification sites in addition to the main SUMOylation site of Lys-41. Immunoblotting analysis revealed that the SUMO modification bands attenuated rather than vanished in the mms21-1 mutant (Figure 2D), indicating that AtMMS21 knockout results in a decreased SUMOylation of the IAA17 protein. Although AtMMS21 mediates the SUMOylation of IAA17, we cannot exclude the possibility that the SUMO-conjugating enzyme E2 or other SUMO ligases may co-regulate the SUMOylation of IAA17 with AtMMS21 in plants.
SUMOylation contributes to regulating protein stability, and SUMO may indulge in an interplay with ubiquitin to inhibit ubiquitin-dependent degradation (Zhang et al., 2017; Wang et al., 2018, 2020). Similarly, we found that IAA17 stability was regulated by SUMOylation by blocking IAA17 ubiquitination (Figures 4–6C). Interestingly, we found that the SUMOylation site, K41, was located near Domain II, which is important for IAA17 degradation. Domain II contains a conserved “degron” motif, which is the interaction site between IAA17 and the auxin receptor protein TIR1 (Tan et al., 2007). TIR1 also acts as a core component of the SCF ubiquitin ligase complex, leading to IAA17 ubiquitination and proteasomal degradation (Tan et al., 2007). Previous studies have shown that mutation in Domain II of IAA17 disrupts the interaction with TIR1 and increases the stability of IAA17 (Gray et al., 2001; Ouellet et al., 2001). The mutation of K41 affects the stability of IAA17, even though the SUMOylation site, K41, is not in the sequence of Domain II. In addition, the protein stability of IAA17-GFP also reduces in mms21-1 mutant plants. Therefore, SUMO attachment may disrupt the interaction between IAA17 and TIR1, thereby suppressing the ubiquitin-dependent degradation of IAA17. In future work, it will be interesting to determine whether the interaction between IAA17 and TIR1 is SUMO-dependent.
Previous studies have shown that SUMOylation plays a critical role in regulating the auxin signal transduction pathway (Orosa-Puente et al., 2018). ARF7 is rapidly SUMOylated after auxin treatment, and subsequently SUMOylated ARF7 interacts with IAA3 to regulate the initiation of lateral roots (Orosa-Puente et al., 2018). However, the SUMOylation status of IAA3, IAA18, and IAA28 was investigated, and it was found that none of these Aux/IAA proteins was SUMOylated in planta (Kirsten, 2017). Although Aux/IAAs at the center of an auxin signaling network have been extensively studied, 29 Aux/IAAs with distinct auxin response characteristics exist in Arabidopsis. It is plausible that different Aux/IAA members have a specific regulatory mechanism in auxin signaling. Previous studies showed auxin increased the degradation of IAA17 (Gray et al., 2001, 2003). We found that the SUMOylation of IAA17 reduced under auxin treatment. IAA17 with reduced SUMOylation is more likely to be degraded. We hypothesized that when plants receive the auxin signal, the SUMO modifier on IAA17 is removed by the SUMO protease, thereby reducing the stability of IAA17. However, which protease cleaves SUMO from the IAA17 protein remains unknown.
In conclusion, our studies reveal that AtMMS21-mediated SUMOylation of IAA17 stabilizes the IAA17 protein, regulating auxin signaling transduction and root development. Therefore, we propose a model in which the SUMOylation of IAA17 regulates auxin signaling (Figure 7E). In normal conditions, IAA17 is SUMOylated by the SUMO E3 ligases AtMMS21. SUMO attachment disrupts the degradation of IAA17. When plants receive the auxin signal, SUMOylation of IAA17 decreases, leading to a 26S proteasomal degradation of IAA17 and a derepression of transcription by ARF. Subsequently, auxin-responsive genes associated with root development are expressed, which helps plants respond to auxin signals.
Materials and methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) seeds of Columbia-0 (Col-0, wild-type) and mms21-1 (CS848340) were isolated as described previously (Huang et al., 2009). The seeds of knockout mutants iaa17-1 (SALK_065697) and iaa17-2 (SALK_011820) and transgenic IAA17-OE1 and IAA17-OE2 were provided by the Shi Laboratory (Shi et al., 2015). The IAA17-OE/mms21-1 mutants were generated by genetic crossing. For the observation of IAA17 in stable transgenic plants, the full-length coding region of IAA17 was amplified and cloned into the pCAMBIA1300-GFP vector to express IAA17-GFP under the control of a UBQ10 promoter. Transgenic plants were generated by Agrobacterium tumefaciens-mediated transformation via the floral dip method (Clough and Bent, 1998) and selected on Murashige and Skoog (MS) medium plates in the presence of antibiotics. Homozygous lines were used in the experiments.
Seeds were surface-sterilized using 75% ethanol for 2 min followed by 5 min in 2.5% sodium hypochlorite solution, rinsed 5 times with sterile water, plated on MS plates with 1.5% sucrose and 0.8% agar, and then stratified at 4°C in darkness for 3 days. Plants were grown under long-day conditions (16 h of light/8 h of dark) at 22°C.
Yeast 2-hybrid assay
Yeast 2-hybrid assays were performed according to the manufacturer's instructions for the Matchmaker GAL4-based Two-Hybrid System 3 (Clontech). The coding sequence (CDS) of IAA17 was cloned into the pGBKT7 vector. The CDS of AtMMS21 was cloned into the pGADT7 vector. Protein interactions were tested by using a stringent method of selection (synthetic-defined minimal yeast media/–Leu/–Trp/–His) with 5, 10, 15, or 20 mM 3-amino-1,2,4-triazole. pBD-53 + pAD-T were co-transformed into yeast cells as positive controls, and pBD-Lam + pAD-T were co-transformed as negative controls. The primers used in this study are listed in Supplemental Data set 1.
In vitro pull-down assay
The CDS of AtMMS21 was cloned into pGEX4T-1 and the CDS of IAA17 fused with a Flag tag was cloned into pCDFDuet-1. The proteins were then separately expressed in BL21(DE3). The pull-down assay was conducted following the same protocol described previously (Jiang et al., 2019).
Bimolecular fluorescent complementation assay
To analyze the interaction of IAA17 and AtMMS21, a BiFC (Bimolecular Fluorescent Complimentary) assay was performed. The CDS of AtMMS21 was cloned into the pSAT6-nEYFP vector, and the CDS of IAA17 was cloned into the pSAT6-cEYFP vector. Protoplast transformation was performed as described previously (Yoo et al., 2007). After transformation, the YFP signals were examined by confocal microscopy (Leica LSM800). Excitation/emission wavelengths 514 nm/530 to 600 nm were set in this experiment.
Co-immunoprecipitation assays
To analyze the interaction of IAA17 and AtMMS21, co-immunoprecipitation (CO-IP) assays were performed as described previously (Wang et al., 2020). IAA17-GFP and Myc-AtMMS21 were transiently co-expressed in Arabidopsis protoplasts. Total proteins were prepared in the CO-IP extraction buffer containing 150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 1 mM MgCl2, 20% [v/v] glycerol, 0.2% [v/v] Nonidet P-40, and 1× Protease Inhibitor Cocktail (Roche, #04693159001). After centrifugation at 13,000×g for 15 min, the supernatant was collected and incubated with anti-GFP affinity beads at 4°C for 2 h. After incubation, the affinity beads were collected and rinsed 5 times with a washing buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 20% [v/v] glycerol, and 0.02% [v/v] Nonidet P-40). The immunoprecipitated proteins were detected with anti-GFP and anti-Myc antibodies.
SUMOylation assay
The in vitro SUMOylation assay in E. coli was performed as previously described (Okada et al., 2009). IAA17-Flag was cloned into pCDFDuet-1 and then transformed for expression in the bacteria carrying pET28-SAE1a-His6-AtSAE2 (E1) with pACYCDuet-1-SUMO1GG or pACYCDuet-1-SCE1-SUMO1GG. Cells were harvested and used for immunoblotting analysis with an anti-Flag antibody (TransGen Biotech). The in vivo SUMOylation assay was performed as previously described with modifications (Niu et al., 2019). In brief, YFP-IAA17 and Myc-SUMO1 were each cloned into the pBluescript vector. YFP-IAA17 and Myc-SUMO1GG or SUMO1AA were transiently co-expressed in Arabidopsis protoplasts. Cells were harvested and extracted in an extract buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 20% [v/v] glycerol, 0.2% [v/v] Nonidet P-40, 20 mM N-ethylmaleimide, and 1× Protease Inhibitor Cocktail (Roche, #04693159001). The protein extracts were incubated in anti-GFP affinity beads at 4°C for 2 h. The immunoprecipitated proteins were detected with anti-GFP and anti-Myc antibodies.
Gene expression and protein level analyses
RNA was extracted using the Plant RNAprep Pure Kit (Magen) following the manufacturer's instructions. RNA was reverse-transcribed using a PrimeScript RT reagent kit (Takara), and the resulting cDNA was used in RT-qPCR using SYBR Premix Ex Taq (Takara) in a Bio-Rad CFX 96 system (C1000 thermal cycler) and detected by using Bio-Rad CFX Manager software.
Total proteins were extracted by adding a protein sample buffer (250 mM Tris–HCl pH = 6.8, 40% [v/v] glycerol, 8% [v/v] SDS, 5% [v/v] β-mercaptoethanol, 0.04% [v/v] bromphenol blue) to ground materials and boiling for 5 min. The extracts were then subjected to SDS–PAGE. The levels of GFP-tagged IAA17 were detected through immunoblotting analysis using an anti-GFP antibody (TransGen Biotech).
Statistical analysis
The data in this article are means ± Sd from 3 independent experiments. Significance analysis was performed using Student's t test via GraphPad Prism 8: *P < 0.05; **P < 0.01; and ***P < 0.001.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AtMMS21 (At3G15150), IAA17 (AT1G04250), SUMO1 (At4G26840), ACS5 (AT5G65800), PRE1 (AT5G39860), and UBQ10 (At4G05320).
Supplementary Material
Acknowledgments
We thank Prof. Haitao Shi (Hainan University) for kindly providing iaa17-1, iaa17-2, IAA17-OE1, and IAA17-OE2 seeds used in this study.
Contributor Information
Cheng Zhang, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Yi Yang, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Zhibo Yu, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Jun Wang, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Ruihua Huang, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Qiuna Zhan, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Shangze Li, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Jianbin Lai, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Shengchun Zhang, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Chengwei Yang, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, PR China.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Interaction of IAA17 with AtMMS21 in yeast 2-hybrid assays.
Supplemental Figure S2 . Root length of mms21-1, IAA17-OE1/mms21-1, and IAA17-OE2/mms21-1.
Supplemental Figure S3 . Subcellular localization of YFP-IAA17 and IAA17-GFP.
Supplemental Figure S4 . Root lengths of Col-0, UBQ:IAA17-GFP/WT, and UBQ:IAA17K41R-GFP/WT.
Supplemental Figure S5 . Relative expression of IAA17 in UBQ:IAA17-GFP/WT and UBQ:IAA17-GFP/mms21-1.
Supplemental Figure S6 . NAA induces the degradation of IAA17 proteins.
Supplemental Data Set 1 . Primers used in this study.
Funding
This work was supported by the Natural Science Foundation of Guangdong (2018B030308002), the Major Program of Guangdong Basic and Applied Research (2019B030302006), the National Natural Science Foundation of China (31870301, 32000151), the China Postdoctoral Science Foundation (2020M672675), and the Program for Changjiang Scholars.
References
- Calderon-Villalobos LI, Tan X, Zheng N, Estelle M (2010) Auxin perception–structural insights. Cold Spring Harb Perspect Biol 2(7): a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Lloyd J, Stevens L, Davies B (2012) TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal Behav 7(3): 325–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ma B, Zhou Y, He SJ, Tang SY, Lu X, Xie Q, Chen SY, Zhang JS (2018) E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc Natl Acad Sci U S A 115(17): 4513–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6): 735–743 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M (2003) Auxin action in a cell-free system. Curr Biol 13(16): 1418–1422 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18(3): 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H (2009) Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell 35(5): 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414(6861): 271–276 [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15(6): 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G (2015) Auxin signal transduction. Essays Biochem 58: 1–12 [DOI] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, et al. (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60(4): 666–678 [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21(8): 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Mao N, Hu H, Tang J, Han D, Liu S, Wu Q, Liu Y, Peng C, Lai J, et al. (2019) A SWI/SNF subunit regulates chromosomal dissociation of structural maintenance complex 5 during DNA repair in plant cells. Proc Natl Acad Sci U S A 116(30): 15288–15296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsten WC (2017) The role of SUMOylation in the auxin response pathway. Thesis (Doctoral). Durham University, Durham theses
- Leyser O (2018) Auxin signaling. Plant Physiol 176(1): 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Cheng Y, Murphy A, Hagen G, Guilfoyle TJ (2009) Constitutive repression and activation of auxin signaling in Arabidopsis. Plant Physiol 149(3): 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B, Wei K, Hu K, Tian T, Zhang F, Yu Z, Zhang D, Su Y, Sang Y, Zhang X, et al. (2021) MPK14-mediated Auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol Plant 14(2): 285–297 [DOI] [PubMed] [Google Scholar]
- Mao Z, He S, Xu F, Wei X, Jiang L, Liu Y, Wang W, Li T, Xu P, Du S, et al. (2020) Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225(2): 848–865 [DOI] [PubMed] [Google Scholar]
- Maraschin Fdos S, Memelink J, Offringa R (2009) Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59(1): 100–109 [DOI] [PubMed] [Google Scholar]
- Moss BL, Mao H, Guseman JM, Hinds TR, Hellmuth A, Kovenock M, Noorassa A, Lanctot A, Villalobos LI, Zheng N, et al. (2015) Rate motifs tune Auxin/Indole-3-acetic acid degradation dynamics. Plant Physiol 169(1): 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D, Lin XL, Kong X, Qu GP, Cai B, Lee J, Jin JB (2019) SIZ1-Mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol Plant 12(2): 215–228 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3:e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama J, Tanaka K (2009) Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol 50(6): 1049–1061 [DOI] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. (2018) Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362(6421): 1407–1410 [DOI] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13(4): 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1(2): 321–337 [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279(5355): 1371–1373 [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based Auxin perception: mechanism and role in plant growth and development. Plant Cell 27(1): 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Liu W, Wei Y, Ye T (2017) Integration of auxin/indole-3-acetic acid 17 and RGA-LIKE3 confers salt stress resistance through stabilization by nitric oxide in Arabidopsis. J Exp Bot 68(5): 1239–1249 [DOI] [PubMed] [Google Scholar]
- Shi H, Reiter RJ, Tan DX, Chan Z (2015) INDOLE-3-ACETIC ACID INDUCIBLE 17 positively modulates natural leaf senescence through melatonin-mediated pathway in Arabidopsis. J Pineal Res 58(1): 26–33 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446(7136): 640–645 [DOI] [PubMed] [Google Scholar]
- Tian H, Wabnik K, Niu T, Li H, Yu Q, Pollmann S, Vanneste S, Govaerts W, Rolcík J, Geisler M, et al. (2014) WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol Plant 7(2): 277–289 [DOI] [PubMed] [Google Scholar]
- Verma V, Croley F, Sadanandom A (2018) Fifty shades of SUMO: its role in immunity and at the fulcrum of the growth-defence balance. Mol Plant Pathol 19(6): 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liu Y, Shi Y, Han D, Wu Y, Ye W, Yang H, Li G, Cui F, Wan S, et al. (2020) SUMOylation stabilizes the transcription factor DREB2A to improve plant thermotolerance. Plant Physiol 183(1): 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Qu GP, Kong X, Yan Y, Li J, Jin JB (2018) Arabidopsis small ubiquitin-related modifier protease ASP1 positively regulates abscisic acid signaling during early seedling development. J Integr Plant Biol 60(10): 924–937 [DOI] [PubMed] [Google Scholar]
- Weijers D, Wagner D (2016) Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67(1): 539–574 [DOI] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L, et al. (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant 11(4): 523–541 [DOI] [PubMed] [Google Scholar]
- Xu P, Yuan D, Liu M, Li C, Liu Y, Zhang S, Yao N, Yang C (2013) AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol 161(4): 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7): 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu Z, Zhang F, Friml J, Ding Z (2022) Auxin signaling: research advances over the past 30 years. J Integr Plant Biol 64(2): 371–392 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lai J, Wang F, Yang S, He Z, Jiang J, Li Q, Wu Q, Liu Y, Yu M, et al. (2017) A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development. Plant Physiol 173(3): 1574–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Li C, Qu X, Liu J, Yu Z, Wang J, Zhu J, Yu Y, Ding Z (2022) A feedback regulation between ARF7-mediated auxin signaling and auxin homeostasis involving MES17 affects plant gravitropism. J Integr Plant Biol 64(7): 1339–1351 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J (2014) GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42(W1): W325–W330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.