Abstract
A key function of SUMOylation is the coordinated modification of numerous proteins to optimize plant growth and resistance to environmental stress. Plant cuticular wax is deposited on the surface of primary plant organs to form a barrier that provides protection against changes in terrestrial environments. Many recent studies have examined cuticular wax biosynthetic pathways and regulation. However, whether SUMOylation is involved in the regulation of cuticle wax deposition at the posttranslational level remains unclear. Here, we demonstrate that a small ubiquitin-like modifier (SUMO) E3 ligase, SAP AND MIZ1 DOMAIN CONTAINING LIGASE1 (MdSIZ1), regulates wax accumulation and cuticle permeability in apple (Malus domestica Borkh), SUMO E2 CONJUGATING ENZYME 1(MdSCE1) physically interacts with MdMYB30, a transcription factor involved in the regulation of cuticle wax accumulation. MdSIZ1 mediates the SUMOylation and accumulation of MdMYB30 by inhibiting its degradation through the 26S proteasome pathway. Furthermore, MdMYB30 directly binds to the β-KETOACYL-COA SYNTHASE 1 (MdKCS1) promoter to activate its expression and promote wax biosynthesis. These findings indicate that the MdSIZ1–MdMYB30–MdKCS1 module positively regulates cuticular wax biosynthesis in apples. Overall, the findings of our study provide insights into the regulation pathways involved in cuticular wax biosynthesis.
MdSIZ1 mediates the SUMOylation and accumulation of MdMYB30 protein, which activates expression of MdKCS1 to promote cuticular wax biosynthesis in apple.
Introduction
The evolution of the cuticle in higher organisms facilitated the transition to life on land, as it limits transpiration water loss and prevents desiccation and pathogen attack (Balabanidou et al., 2018; Berhin et al., 2022). Cuticular wax is impregnated with a cutin matrix, and epicuticular wax accumulates on the surface; depending on their content and composition, wax films and different crystal structures can be formed (Jetter and Riederer, 2016; Elejalde-Palmett et al., 2021). Plants have evolved hydrophobic cuticular wax on the surface of various organs, including fruits, leaves, stems, and flowers (Lara et al., 2015; Kong et al., 2020a). Cuticular wax serves as a water-permeable barrier to prevent water loss, as well as a barrier that provides protection against biotic (pathogens) and abiotic (UV radiation) stress; it also improves the storability of fruit and vegetable products, extends their shelf life, and enhances their appearance quality (fruit skin texture and glossiness) (Ziv et al., 2018; Lara et al., 2019; Chen et al., 2021; García-Coronado et al., 2022; Yang et al., 2022).
The biosynthesis of cuticular wax varies among species, organs, developmental stages, and environmental conditions (Fich et al., 2016; Bhanot et al., 2021). Substantial advances have been made in our understanding of cuticular wax biosynthesis. Cuticular wax is synthesized in epidermal cells starting with C16 and C18 fatty acids in the plastids; they are then elongated to C24–C34 very-long-chain fatty acids (VLCFA) in the endoplasmic reticulum (ER). β-ketoacyl-coenzyme A (CoA) synthases (KCSs) play vital roles in the biosynthesis of C26 and longer VLCFAs, and they are involved in the first and rate-limiting step in fatty acid elongation (Chen et al., 2020). KCS1 encodes β-KETOACYL-COA SYNTHASE 1, which is involved in VLCFA synthesis (Joubès et al., 2008). Alkanes, secondary alcohols, aldehydes, and ketones are then formed via an alkane-forming pathway, and primary alcohols and wax esters are formed by an alcohol-forming pathway (Kan et al., 2022; Lu et al., 2022). Finally, wax is biosynthesized in the apoplast, and wax is deposited in the outermost layer of the cell wall; therefore, cuticular wax needs to be exported from the ER to outside the cell wall by transporters (Elejalde-Palmett et al., 2021; Lee et al., 2021).
The regulation of cuticular wax biosynthesis in Arabidopsis (Arabidopsis thaliana) has been widely investigated. MYB30 modulates the hypersensitive response (HR) via VLCFAs by targeting the FATTY ACYL-ACP THIOESTERASES B(FATB) for activation (Raffaele et al., 2008). MdMYB30 is a crucial element in the formation of the plant cuticle and confers apple (Malus domestica), fruits with pathogen tolerance (Zhang et al., 2019). SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9) forms a complex with DECREASE WAX BIOSYNTHESIS (DEWAX) that prevents SPL9 from binding to the promoter of ECERIFERUM1 (CER1) in Arabidopsis; this finding indicates that plants optimize wax synthesis during daily cycles (Li et al., 2019). ECERIFERUM7 (CER7) was shown to regulate the levels of small RNA species that control ECERIFERUM3 (CER3) expression by gene silencing at the post-transcriptional level using cer7 suppressor mutants (Lam et al., 2015). Cuticular wax biosynthesis is regulated not only at the transcriptional and post-transcriptional levels but also at the posttranslational level. Leucine-rich repeat receptor-like protein kinases, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASEs (SERKs) phosphorylated receptor-like protein kinases GASSHO1 (GSO1) and GASSHO2 (GSO2) respond to TWISTED SEED1 (TWS1) and regulate embryonic cuticle integrity (Zhang et al., 2022). The RING-type E3 ligase DHS targets the ubiquitination substrate ROC4 (an HD-ZIP transcription factor) to fine-tune wax biosynthesis and the drought stress response in rice (Oryza sativa) (Wang et al., 2018). The Kelch repeat F-box protein SMALL AND GLOSSY LEAVES1 (SAGL1) mediates the protein stability of CER3 by the 26S proteasome in response to changes in humidity (Kim et al., 2019). However, whether cuticular wax formation is regulated by SIZ1-mediated SUMOylation has not been investigated.
SUMOs (small ubiquitin-related modifiers) are reversible posttranslational modifications (PTMs) that can be attached to protein substrates in a process called SUMOylation, which alters the localization, function, activity, and/or stability of substrate proteins (Li et al., 2021, 2022; Fang et al., 2022; Sahin et al., 2022). The process of attaching SUMO peptides by deSUMOylating proteases (SENPs in mammals; ubiquitin-like proteases (ULPs) in yeast) first involves exposing a C-terminal di-glycine motif via SUMO E1-activating enzymes (SAE1 in Arabidopsis). This is followed by conjugation of the target protein via SUMO E2 conjugases (SCE1 in Arabidopsis; also called UBC9 in some animals and fungi). SUMO E3 ligases (SIZ1, MMS21/HPY2) then mediate the attachment of SUMO peptides to E1 and E2 target substrates (Castaño-Miquel et al., 2017; Kong et al., 2020b; Roy and Sadanandom, 2021).
In apple, the SUMO E3 ligase MdSIZ1 is involved in multiple biological processes, including anthocyanin accumulation, iron homeostasis, and the formation of lateral roots (Zhou et al., 2017, 2019; Zhang et al., 2021; Jiang et al., 2022); however, whether MdSIZ1-mediated SUMOylation regulates cuticular wax biosynthesis remains unclear. In this study, we investigated the positive regulatory mechanisms underlying the PTM of MdSIZ1 in regulating cuticular wax biosynthesis. MdSIZ1-overexpressing apple leaves exhibit increased wax deposition and reduced cuticle permeability. Furthermore, the apple E2 conjugase MdSCE1 physically interacts with MdMYB30. MdSIZ1 mediates the SUMOylation and stabilization of MdMYB30, which directly activates the transcription of MdKCS1 to promote cuticular wax accumulation in the apple. These observations indicate that the MdSIZ1-dependent posttranslational regulatory mechanism contributes to the positive regulation of cuticular wax biosynthesis in apple. The MdSIZ1–MdMYB30–MdKCS1 module might be a key regulator in apple cuticular wax biosynthesis.
Results
MdSIZ1 positively regulates SUMO conjugates and wax biosynthesis in apple
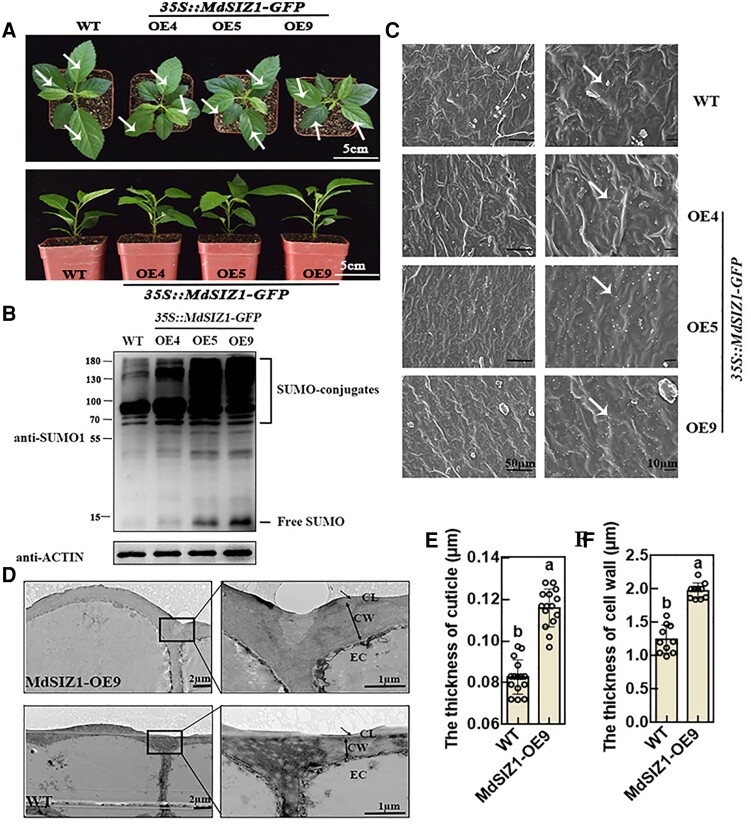
To investigate the function of MdSIZ1 in wax biosynthesis in apples, we generated MdSIZ1-OE plants (overexpressing GFP-fused MdSIZ1) through Agrobacterium tumefaciens strains EHA105-mediated genetic transformation. Three MdSIZ1 transgenic lines were obtained, and these were named MdSIZ1-OE4, MdSIZ1-OE5, and MdSIZ1-OE9, respectively (Figure 1A; Supplemental Figure 1). We speculated that MdSIZ1 might be associated with protein SUMOylation and examined levels of protein SUMOylation in MdSIZ1-OE plants. The results revealed that levels of protein SUMOylation, including SUMO conjugates and free SUMO, were higher in MdSIZ1-OE plants than in WT plants (Figure 1B). Scanning electron microscopy (SEM) was then used to examine epicuticular wax crystals. Granular wax crystals were observed on both WT and MdSIZ1-OE apple leaves; the density of wax crystals was higher on MdSIZ1-OE leaves than on WT leaves (Figure 1C). Transmission electron microscopy (TEM) analysis of the leaf epidermal cells of WT and MdSIZ1-OE9 plants revealed that the thickness of the cuticle and epidermal cell wall was approximately 1.5- and 1.6-fold higher in MdSIZ1-OE9 apple leaves than in WT leaves, respectively (Figure 1, D–F).
Figure 1.
SUMO conjugates and cuticular wax crystals of WT and MdSIZ1-OE apple plants. A, Comparison of leaf phenotypes. Two-month-old plants are shown. Scale bar, 5 cm. Vertical view: top images; side view: bottom images. White arrows indicate the leaves used for glossiness measurements in Supplemental Figure 3. B, SUMO conjugates in WT and MdSIZ1-OE apple plants. Immunoblot analysis was performed using anti-SUMO1 and anti-actin antibodies; anti-actin antibody was used to ensure equivalent protein loading. C, SEM images of cuticular wax crystals of apple leaves. Scale bar, 50/10 µm. White arrows indicate the typical shapes of epicuticular wax crystals (ellipse). D, TEM images show the cuticular layer and epidermal cell structure of apple leaves. CL, cuticular layer; CW, cell wall; EC, epidermal cell. Scale bar, 2/1 µm. Single arrows indicate the cuticle layer measured via ImageJ. The thickness of the cuticle (E) and cell wall (F) of WT and MdSIZ1-OE9 leaves. Averages from three biological replicates and their SDs are shown. Error bars represent standard deviation (Sd; n = 15/10). Different letters indicate significant differences (analysis of variance [ANOVA], Student's t test; P < 0.05).
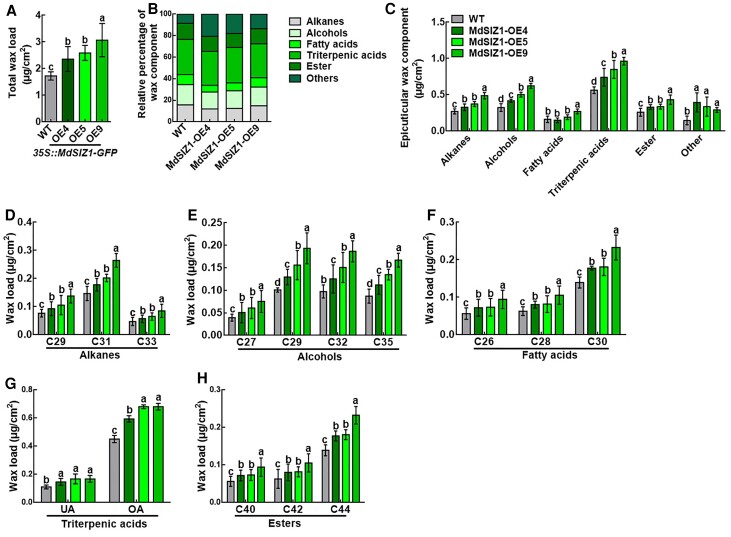
Next, the leaf cuticular wax composition of WT and MdSIZ1-OE plants was analyzed by gas chromatography–mass spectrometry (GC–MS). Total wax loads were 1.36, 1.49, and 1.77 times higher in MdSIZ1-OE4, MdSIZ1-OE5, and MdSIZ1-OE9 leaves compared with WT leaves, respectively (Figure 2A; Supplemental Data 1). We also analyzed the composition of cuticular wax, including alkanes, alcohols, fatty acids, triterpenic acids, and esters (Figure 2, B and C). Alkanes (mainly C29, C31, and C33) were 21.0%, 37.5%, and 79.9% higher (Figure 2, C and D), alcohols (mainly C27, C29, C32, and C35) were 28.68%, 54.78%, and 91.83% higher (Figure 2, C and E), total fatty acids (mainly C26, C28, and C30) were 1.3%, 18.08%, and 66.32% higher (Figure 2, C and F), triterpenic acids (mainly UA and OA) were 31.6%, 50.9%, and 71.1% higher (Figure 2, C and G), and total esters (mainly C40, C42, and C44) were 27.56%, 29.9%, and 67.0% higher (Figure 2, C and H) in MdSIZ1-OE4/5/9 leaves compared with WT leaves, respectively. MdSIZ1 was ectopically expressed in Arabidopsis (EE13, EE19, and EE33) following Zhou et al. (2019) to evaluate the function of MdSIZ1 in Arabidopsis. The wax loading and the levels of protein SUMOylation of MdSIZ1 transgenic Arabidopsis (EE13, EE19, and EE33), WT (Col), and the siz1-2 mutant were examined. We first analyzed the SUMO conjugate profile in a total protein immunoblot; an increase in SUMO conjugates was observed in 35S::MdSIZ1-Myc ectopically expressed in Arabidopsis (#13, #19, and #33), and the SUMO conjustes were slightly weaker in siz1-2 plants than in Col plants (Supplemental Figure 2A). SEM analysis revealed that the wax crystals on the leaf surface of MdSIZ1-EE were denser, and the cuticular wax crystals were sparser on the surface of siz1-2 mutant leaves than on the surface of Col leaves (Supplemental Figure 2B). The wax crystals on the surface of inflorescence stems were similar to those on the surface of the leaves (Supplemental Figure 2C). Total wax loads were 1.15, 1.22, and 1.23 times higher in MdSIZ1-EE13, MdSIZ1-EE19, and MdSIZ1-EE33 Arabidopsis leaves compared with WT leaves, respectively, and the total wax load was 1.19 times lower in siz1-2 mutant leaves compared with WT leaves (Supplemental Figure 2D). Wax deposition on Arabidopsis inflorescence stems was similar to that on the leaves (Supplemental Figure 2E). These results indicate that MdSIZ1-mediated SUMOylation positively regulates wax biosynthesis in apple.
Figure 2.
Cuticular wax composition in WT and MdSIZ1-OE apple leaves. A, Total wax load, (B) Relative percentage of wax component, (C) Composition of cuticular waxes, (D) Total alkane load, (E) Total alcohol load, (F) Total fatty acid load, (G) Total triterpenic acid load, UA: ursolic acid; OA: oleanolic acid, and (H) Total ester load. Wax coverage is expressed as μg cm−2 leaf surface area. Each wax constituent is designated by the carbon chain length and is labeled by chemical class along the x-axis. Error bars indicate standard deviation (Sd; n = 3). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05).
MdSIZ1 alters leaf cuticular membrane permeability
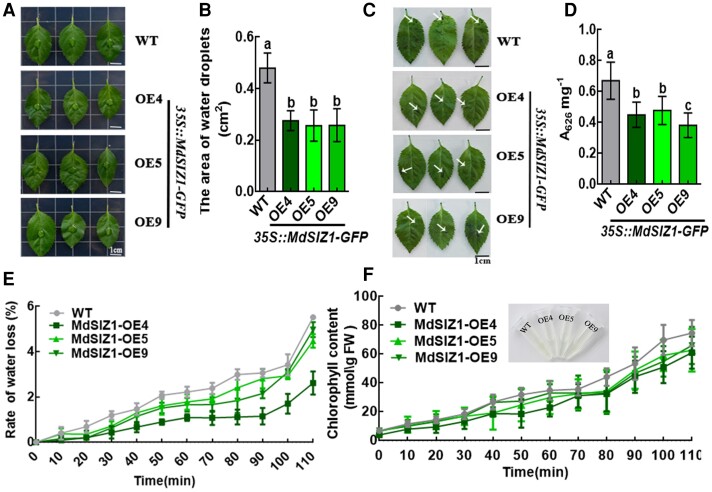
Plants often exhibit higher glossiness with less cuticular wax accumulation. However, we did not observe any difference in glossiness between WT and MdSIZ1-OE apple leaves according to L*, a*, and b* values (Supplemental Figure 3). Because cuticular wax is closely associated with cuticular membrane properties, we performed several assays to characterize cuticular membrane properties in transgenic apple leaves. First, we applied the same volume of water droplets to apple leaves and found that the contact area was smaller in MdSIZ1-OE leaves than in WT control leaves, indicating that the hydrophobicity of MdSIZ1-OE leaves was higher (Figure 3, A and B). Second, the barrier properties of the leaf cuticle were enhanced in MdSIZ1-OE lines according to Toluidine blue (TB) staining (Figure 3, C and D). Third, we analyzed the water loss rates of detached leaves and discovered that MdSIZ1-OE plants lost water much more slowly than WT plants (Figure 3E). Lastly, we conducted a chlorophyll leaching experiment in which leaves from both MdSIZ1-OE and WT plants were submerged in 80% ethanol for different lengths of time, and the chlorophyll concentration in the solution was determined. The rate of chlorophyll extraction was much slower from MdSIZ1-OE leaves than from WT leaves (Figure 3F); the lower elution of chlorophyll from MdSIZ1-OE leaves indicates a decrease in cuticle permeability.
Figure 3.
MdSIZ1 reduces leaf cuticle permeability. A, Water droplets attached to the apple leaves. Leaves sampled from the same location of apple seedlings. B, The contact area between the water droplets and the leaf surface. Error bars indicate standard deviation (Sd; n = 10). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05). C, Toluidine blue (TB) staining (0.5% [v/v] solution) of apple leaves. White arrows indicate the cuticle stained blue with TB. Scale bar, 1 cm. D, TB uptake was quantified in MdSIZ1 transgenic and WT plants in (C) using a spectrophotometer with three biological replicates. Error bars indicate standard deviation (Sd; n = 3). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05). E, Water loss rate of apple leaves. The decrease in weight every 10 min at room temperature corresponds to the decrease in moisture. Error bars indicate standard deviation (Sd; n = 3) (F) Chlorophyll leaching assays of apple leaves. Chlorophyll μmol/mg FW (fresh weight, FW). Error bars indicate standard deviation (Sd; n = 3).
MdSIZ1 transgenic Arabidopsis seedlings (EE13, EE19, and EE33) were used to further explore the function of MdSIZ1. TB leaching (Supplemental Figure 4, A–C and E) and water loss (Supplemental Figure 4D) were lower in MdSIZ1-OE Arabidopsis compared with WT Arabidopsis. The leaves of the siz1-2 mutant were not stained by TB because of its pleiotropic growth defects, including its smaller leaves and greater number of siliques (Supplemental Figure 4A). High TB leaching was observed in siz1-2 mutant seedlings two weeks after germination (Supplemental Figure 4E). Overall, MdSIZ1 might alter leaf cuticle permeability by affecting wax accumulation.
MdSCE1 physically interacts with MdMYB30 as a substrate for MdSIZ1
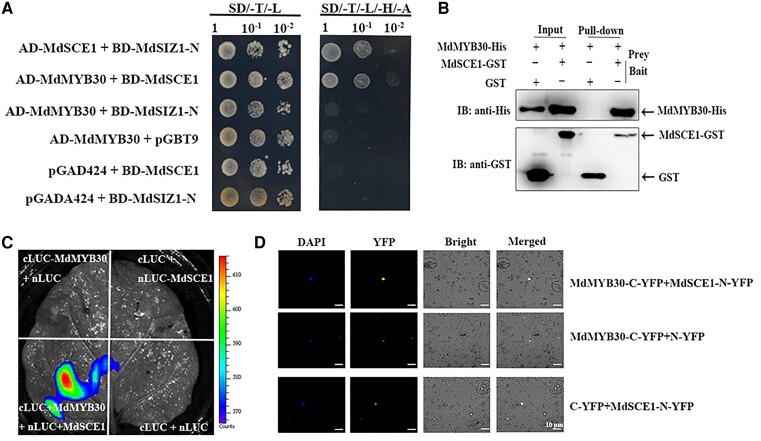
The SUMO E3 ligase MdSIZ1 can mediate the conjugation of SUMO to target proteins (Zhou et al., 2019), and the E2 conjugating enzyme interacts directly with target proteins. E3 ligase then catalyzes the conjugation of SUMO to the attachment site on the substrate in the process of SUMOylation (Niu et al., 2019). To identify the target of MdSIZ1 in wax biosynthesis, the E2 conjugating enzyme MdSCE1 was used as bait to screen its interacting proteins by yeast-two-hybrid (Y2H) assays. MdMYB30, a well-established MYB transcription factor that plays a key role in regulating wax biosynthesis in apple was found to interact with MdSCE1. Subsequently, Y2H assays revealed that MdSCE1, but not MdSIZ1 (MdSIZ1-N), physically interacted with MdMYB30 (Figure 4A). A pull-down assay was performed to confirm the interaction between MdSCE1 and MdMYB30. His-MdMYB30 and GST-MdSCE1 proteins were expressed and purified (Input). The His-MdMYB30 proteins were enriched by GST-MdSCE1, but not by GST alone, indicating that the two proteins interact in vitro (Figure 4B). We then fused MdMYB30 to the C-terminal half of LUC (cLUC-MdMYB30) and MdSCE1 to the N-terminal half of LUC (nLUC-MdSCE1) and transiently introduced both fusion proteins into Nicotiana benthamiana leaf epidermal cells. N. benthamiana leaves that co-expressed cLUC-MdMYB30 and nLUC-MdSCE1 produced a strong LUC signal. By contrast, LUC activity was barely detectable when leaves were infiltrated with other combinations. The results showed that MdSCE1 interacted with MdMYB30 (Figure 4C). Bimolecular fluorescence complementation (BiFC) assays were conducted by co-transfecting different combinations of genes fused to N-YFP or C-YFP into N. benthamiana leaves, which revealed a direct interaction between MdSCE1 and MdMYB30. Cells co-expressing MdMYB30-C-YFP and MdSCE1-N-YFP generated a yellow fluorescence signal in the nucleus, and the measurable YFP signal was undetectable when either fusion protein was expressed alone in N. benthamiana cells (Figure 4D). Overall, our data suggest that MdSCE1 physically interacts with MdMYB30 both in vitro and in vivo.
Figure 4.
MdSCE1 specifically interacts with MdMYB30 in vitro and in vivo. A, Yeast two-hybrid assay showing the specific interaction of MdSCE1 with MdMYB30, but not with MdSIZ1. MdSCE1 and MdSIZ1-N were fused to the GAL4-binding domain (BD) in pGBT9, whereas MdMYB30 and MdSCE1 were fused to the GAL4 activation domain (Ad) in pGAD424. Transformed yeast cells were grown on SD/-Trp/-Leu (SD/-T-L) (left). The direct protein interactions were assayed on a SD/-Trp/-Leu/-His/-Ade (SD/-T/-L/-H/-A) (right) plate. The pGBT9 and pGAD424 vectors were used as negative controls. The interaction between MdSCE1 and MdSIZ1 was used as a positive control. B, An in vitro pull-down assay revealed an interaction between MdMYB30 and MdSCE1. His-MdMYB30 protein was incubated with immobilized GST or GST-MdSCE1 protein, and immunoprecipitated fractions were detected by anti-His or anti-GST antibody. GST-MdSCE1 protein was incubated with immobilized His-MdMYB30 or GST protein, and immunoprecipitated fractions were detected by anti-GST antibody. C, Luciferase complementation imaging assay demonstrating that MdMYB30 interacts with MdSCE1 in vivo. The leaves of N. benthamiana were infiltrated with agrobacteria as indicated. Constructs were combined at a 1:1 ratio. The empty nLUC or cLUC vector was used as a negative control. D, Bimolecular fluorescence complementation (BiFC) analysis of the interaction between MdMYB30 and MdSCE1. Empty nYFP or cYFP vector was used as a negative control.
MdSIZ1 facilitates SUMOylation and increases MdMYB30 stability
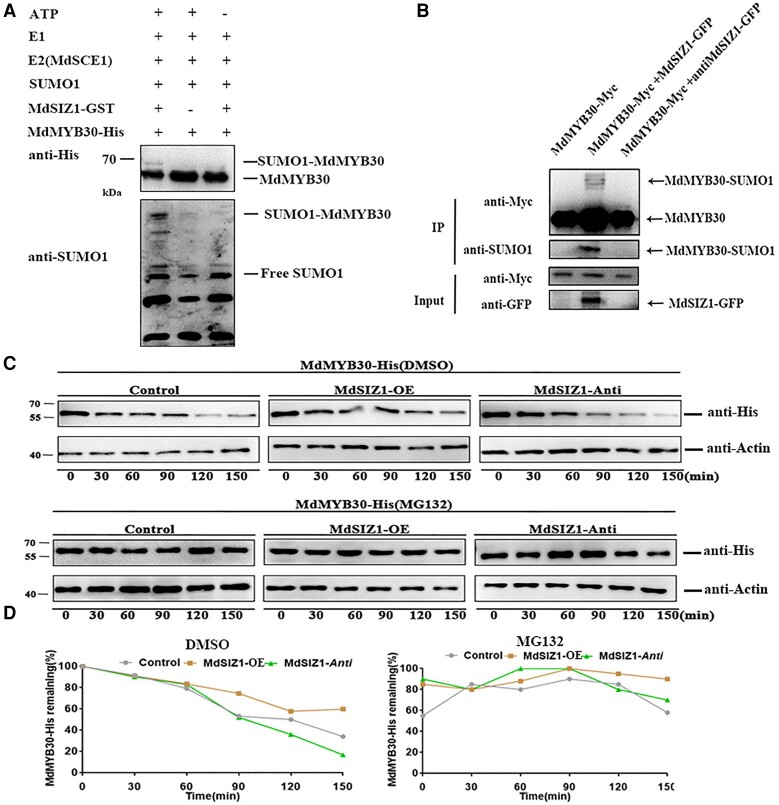
The altered wax deposition of the MdSIZ1 transgenic apple leaves and the strong physical interaction of MdSCE1 with MdMYB30 prompted us to investigate whether MdMYB30 was SUMOylated by MdSIZ1. To examine whether MdMYB30 proteins are substrates of MdSIZ1, in vitro SUMOylation assays were conducted. MdMYB30-His and MdSIZ1-GST protein were incubated with the SUMO1, E1, and E2 (MdSCE1) proteins, and SUMO-conjugated MdMYB30 was undetectable when either anti-His or anti-SUMO1 antibodies were applied in the absence of MdSIZ1 or ATP; however, SUMO-conjugated MdMYB30 was detectable when all components were present (Figure 5A). This indicated that MdMYB30 was SUMOylated by MdSIZ1 protein in vitro. Next, we obtained MdMYB30-Myc, MdMYB30-Myc + MdSIZ1-GFP, and MdMYB30-Myc + anti-MdSIZ1-GFP transgenic apple calli (Supplemental Figure 5). SUMO-conjugated MdMYB30 was detected in MdMYB30-Myc + MdSIZ1-GFP but not in MdMYB30-Myc or MdMYB30-Myc + anti-MdSIZ1-GFP apple calli (Figure 5B). This indicated that MdSIZ1 SUMOylates MdMYB30 in vitro and in vivo.
Figure 5.
MdMYB30 protein was SUMOylated and stabilized by MdSIZ1 in apple. A, MdSIZ1 SUMOylated MdMYB30 protein in vitro. (+) denotes presence, and (−) denotes absence. B, MdSIZ1-SUMOylated MdMYB30 protein in vivo. Input: the whole-cell lysates before treatment. IP, soluble pellet fraction. Arrowhead indicates that SUMOylated or non-SUMOylated MdMYB30 was detected via immunoblot analysis with anti-Myc, anti-SUMO1, and anti-GFP antibodies. C, MdMYB30 protein degradation assay in vitro. His-MdMYB30 protein abundance was detected using an anti-His antibody. ACTIN was used as an internal control. DMSO was used as a negative control for MG132. D, The degradation rate of His-MdMYB30 in vitro measured with Bio-Rad QUANTITY ONE software. The ratio of protein abundance to ACTIN at the initial time 0 was taken as 100%.
SUMO conjugation affects transcription factor function through activation, repression, or protein stabilization processes (Miura et al., 2007). We examined whether MdSIZ1-mediated SUMOylation affects the stability of MdMYB30. The total proteins extracted from Control (WT), MdSIZ1-OE, and anti-MdSIZ1 transgenic apple calli were incubated with His-MdMYB30 protein. The abundance of His-MdMYB30 was detected by immunoblotting using an anti-His antibody. The results showed that the His-MdMYB30 protein abundance was more stable in the protein extracts of MdSIZ1-OE transgenic apple calli, whereas MdMYB30 protein degraded faster in anti-MdSIZ1 transgenic apple calli than in WT calli. In addition, the degradation of MdMYB30 in anti-MdSIZ1 calli was inhibited by treatment of MG132, indicating that MdSIZ1-mediated SUMOylation increased the protein stability of MdMYB30 and blocked its degradation via the 26S proteasome pathway (Figure 5, C and D).
MdSIZ1 promotes wax accumulation in a MdMYB30-dependent manner
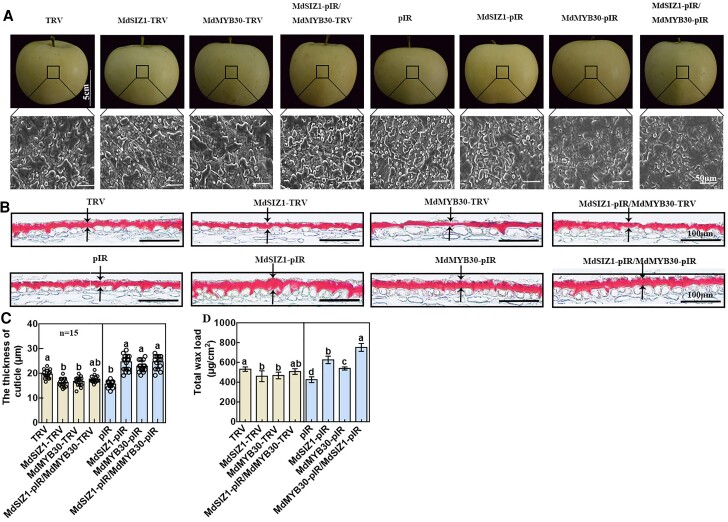
To demonstrate whether the SUMOylation of MdMYB30 by MdSIZ1 regulates cuticular wax biosynthesis in apple, a viral vector-based transformation method in apple fruit peel was performed. The constructs MdSIZ1-TRV, MdMYB30-TRV, and MdSIZ1-pIR/MdMYB30-TRV were used to activate or inhibit the expression of MdSIZ1 or MdMYB30, and the overexpression vectors MdSIZ1-pIR, MdMYB30-pIR, and MdSIZ1-pIR/MdMYB30-pIR were obtained; the empty vectors pIR and TRV were used as controls. RT-qPCR analyses verified that the genes fused to the viral constructs were efficiently expressed in apple peel (Supplemental Figure 6). The density of the wax crystals (Figure 6A), cuticle thickness (Figure 6, B and C), and total wax load (Figure 6D) were decreased in MdSIZ1-TRV and MdMYB30-TRV, and the phenotype of MdSIZ1-pIR/MdMYB30-TRV was similar to that of MdMYB30-TRV. Furthermore, MdSIZ1-pIR, MdMYB30-pIR, and MdSIZ1-pIR/MdMYB30-pIR apple fruit were characterized by a greater number of wax crystals (Figure 6A), cuticle thickness (Figure 6, B and C), and total wax load (Figure 6D) than the pIR vector; the total wax load of MdSIZ1-pIR/MdMYB30-pIR was much higher than that of MdSIZ1-pIR or MdMYB30-pIR. We also examined the wax crystals that were removed by chloroform and fruit cuticle before injection. SEM images showed that no wax crystals were deposited after cuticle treatment with chloroform, and the pattern of wax crystal accumulation on the cuticle before injection was similar to that observed in the TRV (or pIR) control (Supplemental Figure 7). These results suggested that MdSIZ1 positively regulates cuticle wax biosynthesis in an MdMYB30-dependent manner.
Figure 6.
Wax deposition in virus-induced transgenic apple fruits. A, SEM images of apple epicuticular wax crystals in apple fruits. Scale bar, 5 cm/50 μm. B, Light microscopy showing the cuticle in apple fruits indicated by Oil Red Ostaining. Scale bar, 100 μm. Black arrows indicate the cuticle sections measured via ImageJ. C, The cutinization of the epidermal cell wall of apple fruits indicated by Oil Red O staining measured via ImageJ. Error bars indicate standard deviation (Sd; n = 15). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05). D, The total wax load of apple fruits. Error bars indicate standard deviation (Sd; n = 3). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05).
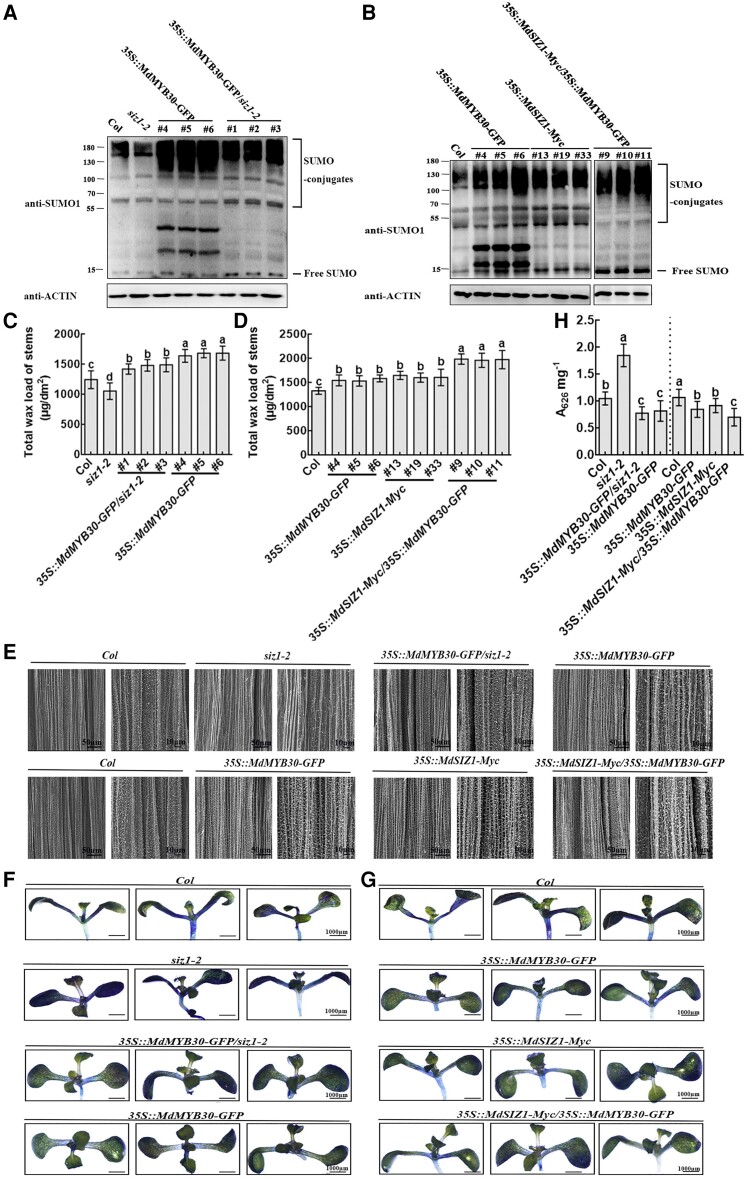
In addition, we obtained 35S::MdMYB30-GFP/siz1-2 (#1, #2, #3) and 35S::MdSIZ1-Myc/35S::MdMYB30-GFP (#9, #10, #11) transgenic Arabidopsis (Supplemental Figure 8) and analyzed their SUMO conjugate profile, wax deposition, and cuticle permeability. The overexpression of MdSIZ1 is linked to increases in SUMO conjugates, which demonstrates an association between sumoylation and wax accumulation (Figures 1B and 2). To further determine how sumoylation status determines wax loading, we analyzed the SUMO conjugate profile in a total protein immunoblot of transgenic Arabidopsis in Supplemental Figure 8 and compared it with that of Col. The decrease in SUMO conjugates in siz1-2 mutant was restored to near the level observed in 35S::MdMYB30-GFP by ectopic expression of MdMYB30 in the siz1-2 mutant (Figure 7A). SUMO conjugate levels were higher in 35S::MdMYB30-GFP and 35S::MdSIZ1-Myc than in Col and lower in 35S::MdMYB30-GFP and 35S::MdSIZ1-Myc than in 35S::MdSIZ1-Myc/35S:: MdMYB30-GFP (Figure 7B). As expected, when MdMYB30 was ectopically expressed in the siz1-2 mutant, the stem wax content of 35S::MdMYB30-GFP/siz1-2 was restored to near to the wax level of 35S::MdMYB30-GFP (Figure 7C); the density of wax crystals revealed by SEM (Figure 7E) and cuticle permeability indicated by TB staining (Figure 7, F and H) were consistent with the observed wax content and SUMO conjugate levels. Furthermore, the wax loading and density of wax crystals were increased and cuticle permeability was decreased in 35S::MdSIZ1-Myc/35S::MdMYB30-GFP transgenic Arabidopsis compared with 35S::MdSIZ1-Myc or 35S::MdMYB30-GFP Arabidopsis (Figure 7, D, E, G, and H). These results indicated that AtSIZ1 upstream of MdMYB30 and modulates wax accumulation by SUMOylating MdMYB30.
Figure 7.
SIZ1 acts upstream of MdMYB30 to regulate wax accumulation. A-B, SUMO conjugates in transgenic Arabidopsis lines. Immunoblot analysis was performed using anti-SUMO1 and anti-actin antibodies; anti-actin antibody was used to ensure equivalent protein loading. C-D, Total wax load, (E) SEM image, and (F-G) TB staining (0.5% [v/v] solution); the images were digitally extracted for comparison. H, TB uptake was quantified in the transgenic Arabidopsis lines in (F) and (G) using a spectrophotometer with three biological replicates. Error bars indicate standard deviation (Sd; n = 3). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05).
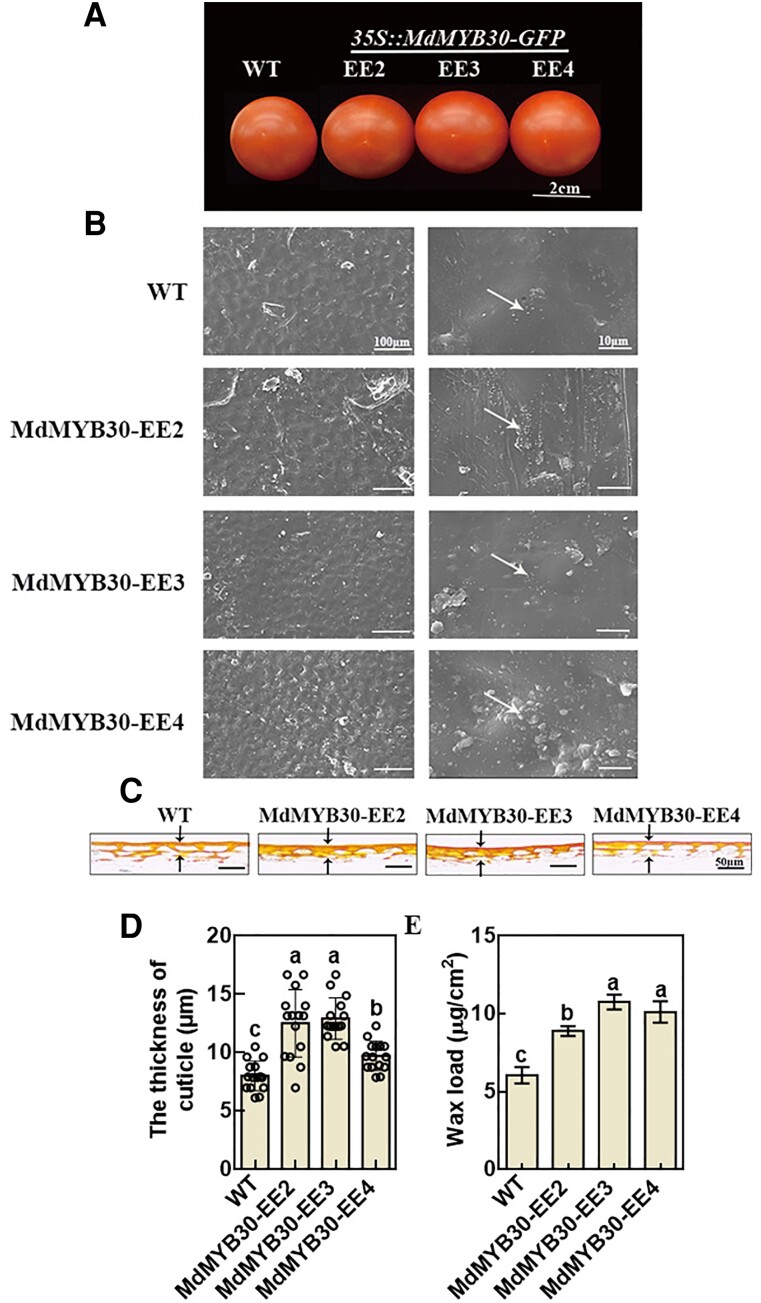
The role of MdMYB30 in wax accumulation was examined in tomato fruits. We ectopically expressed MdMYB30 in tomato (Figure 8A; Supplemental Figure 9) and analyzed the density of wax crystals and cuticle thickness by SEM and Oil Red O staining, respectively. Wax crystals were denser (Figure 8B) and wax loads (Figure 8E) and the thickness of the cuticle at the red ripe developmental stages were higher in MdMYB30-EE plants compared with WT plants according to Oil Red O staining of cross-sections of the pericarp (Figure 8, C and D). In sum, tomato fruit ectopically expressing MdMYB30 shows increased cuticular wax deposition.
Figure 8.
MdMYB30 increased cuticle accumulation in tomato fruits. A, Phenotypes of WT and MdMYB30-EE tomato fruits. Scale bar, 2 cm. B, SEM images of cuticular wax crystals on tomato fruits. Bars, 100/10 µm. White arrows indicate the typical shapes of epicuticular wax crystals (ellipse). C, The cuticle of the epidermal cell wall of tomato fruits by Oil Red O staining. D, The cuticle thickness of tomato fruits by Oil Red O staining. Error bars represent standard deviation (Sd; n = 15). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05). E, Total wax load of tomato fruits in the red ripe stage. Error bars indicate standard deviation (Sd; n = 3). Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05).
MdKCS1 is a direct target of the MdSIZ1–MdMYB30 cascade in the regulation of wax accumulation
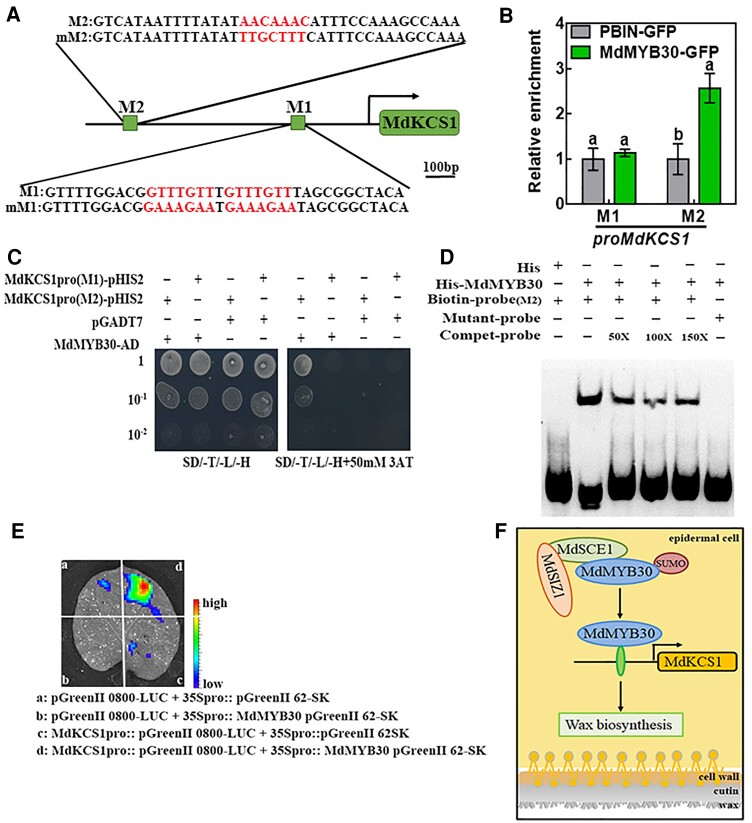
According to the above results, MdMYB30 controls wax biosynthesis by acting downstream of SIZ1. We explored the role of the MdSIZ1–MdMYB30 cascade in this pathway. In Arabidopsis, KCS1 has an important function in cuticular development, and MYB30 regulates the expression of KCS1 (Todd et al., 1999; Joubès et al., 2008; Marino et al., 2013). We analyzed the promoter sequences of wax-related genes, including MdKCS1, MdLACS1, MdCYP86A4, and MdKCS19; some conserved MBS motifs (CANNTG) and the AC1 motif (M2: AACAAAC), which have been previously identified as MdMYB30-binding sites (Li et al., 2009; Kishi-Kaboshi et al., 2018), were detected (Figure 9A; Supplemental Figure 10A; Supplemental Table 2). To investigate whether these motifs in wax-related genes promoter are direct targets of MdMYB30, we conducted chromatin immunoprecipitation (ChIP)-qPCR using MdMYB30-GFP and pBIN transgenic calli by immunoprecipitation with anti-GFP antibody. We found that the AC1 (M2) fragment in the MdKCS1 promoter was more enriched in MdMYB30-GFP calli than in pBIN empty vector, indicating that MdMYB30 binds to M2, but not to M1 in the MdKCS1 promoter or other MBS motifs in the promoter of other wax-related (Figure 9B; Supplemental Figure 10B). Next, we co-expressed MdMYB30-Ad + MdKCS1(M1)/MdKCS1(M2)-pHIS2 and pGADT7 + MdKCS1(M1)/MdKCS1(M2)-pHIS2 in yeast cells to perform Y1H assays, and the results of the Y1H assays were consistent with the results of ChIP-qPCR (Figure 9C). We subsequently performed an electrophoretic mobility shift assay (EMSA) with biotin probes and the protein of MdMYB30 fused to His-tag (His-MdMYB30) or His protein. A mobility shift was observed in the presence of His-MdMYB30 but not His. The binding band was weakened with the addition of the competitive probe, and this was not observed with the addition of the mutant probe (Figure 9D). These results suggested that MdKCS1 is a direct target of MdMYB30.
Figure 9.
MdMYB30 directly binds to the MdKCS1 promoter. A, A schematic diagram depicting the M1 and M2 motifs in the MdKCS1 promoter. Red letters indicate binding motifs and their corresponding mutation sequences. Scale bar, 100 bp. B, ChIP-qPCR analysis for testing the binding of MdMYB30 to the M1 and M2 motifs in the promoter sequence of MdKCS1. Enrichment of PBIN-GFP was used as a reference and set to 1. Data are mean ± Sd of three independent replicates. Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05). C, Yeast one-hybrid assay to assess the binding of MdMYB30 to MdKCS1 promoter DNA. The yeast strains were grown on SD/-Trp/-Leu/-His/and SD/-Leu/-Trp/-His/ + 50 mM 3-AT media for 3 d. Empty pGADT7 vector was used as a negative control. The experiments were repeated three times, and similar results were obtained across all replicates. D, EMSA for testing the binding of MdMYB30 to the putative M2 motif in the MdKCS1 promoters. Competitive probe (Compet-probe) refers to the unlabeled probe. 50×, 100×, and 150× represent the multiples of the competitor probe. The AACAAAC (M2) motifs were mutated to TTGCTTT in M2. E, Transient expression assays showing that MdMYB30 activated the expression of MdKCS1pro::LUC. Representative images of Nicotiana benthamiana leaves 72 h after infiltration. F, Schematic diagram of the working model for the roles of MdSIZ1 and MdMYB30. MdSIZ1 positively regulates the level of MdMYB30 by promoting its SUMOylation. MdMYB30 directly binds to and activates the expression of MdKCS1, which positively regulates wax biosynthesis.
To determine whether MdMYB30 can repress or activate the expression of MdKCS1. We performed a dual-luciferase reporter assay in N. benthamiana leaves. The 35S::MdMYB30 pGreenII 62-SK plasmid vector was able to drive the expression of the reporter gene MdKCS1pro::pGreenII 0800-LUC, and luminescence signals were substantially weaker and almost undetectable when other groups were present, indicating that MdMYB30 activated the expression of MdKCS1 in vivo (Figure 9E). The expression of MdKCS1 was significantly increased in MdMYB30-overexpressing plants and decreased in plants in which the expression of MdMYB30 was inhibited (Supplemental Figure 6). In conclusion, these findings indicated that MdSIZ1 positively regulates wax biosynthesis by SUMOylating and stabilizing MdMYB30 protein, which directly regulates the expression of the wax-related structural gene MdKCS1 (Figure 9F).
Discussion
Wax deposition affects plant cuticle properties and the quality of apple fruit
Cuticular wax affects the drought resistance, nonstomatal water loss, and cuticle membrane permeability of plants (Nawrath, 2006). For example, in Arabidopsis, MYB96 promotes the biosynthesis of cuticular wax under drought conditions (Seo et al., 2011). OsROC4 positively regulates wax loads and the drought stress response by decreasing nonstomatal water loss (Wang et al., 2018). Serk1/2/3 cotyledons show cuticle defects, as indicated by the rapid passage and permeation of TB through the cuticle, whereas the cuticle of WT plants remains intact and unstained (Zhang et al., 2022). Differences in the cuticle level are often associated with changes in cuticle permeability, and this in turn affects the quality of the apple fruit. These phenotypic changes observed in our study might from increases in the wax level in MdSIZ1-OE plants. The cuticle permeability of MdSIZ1-OE leaves was reduced compared with WT leaves based on the low permeation of TB through the cuticle of the transgenic leaves (Figure 3, C and D); the water loss and rate of chlorophyll extraction were reduced in MdSIZ1-OE leaves compared with WT leaves (Figure 3, E and F), which are closely associated with shelf life, post-harvest storage, disease resistance, and mechanical damage. The thickness of the cuticle is related to the gloss characteristics of plant (Petit et al., 2014). The MdSIZ1-mediated formation of cuticular wax might be important for determining the appearance quality of apple fruit; however, the glossiness of the leaves was the same in MdSIZ1-OE lines and WT plants (Figure 1A; Supplemental Figure 3). In future studies, the glossiness of the surface of stable transgenic apple fruits should be examined. However, fruit brightness is independent of variation in wax load according to an analysis of tomato mutants with altered cuticle composition and architecture (Petit et al., 2014). Therefore, the factors associated with glossiness are complex; they might not only depend on wax levels but also on cuticular wax composition or alteration of the epidermal cells and overall surface structure.
In this study, we showed that the SUMO E3 ligase MdSIZ1 plays a role in cuticular wax deposition by analyzing the phenotypes of apple seedlings, Arabidopsis, and tomato fruits, including the accumulation of epicuticular crystals (Figure 1), cuticular wax load (Figures 2 and 8), cuticular membrane permeability (Figure 3), and leaf brightness (Supplemental Figure 3); all results were consistent. Our findings revealed a direct relationship between cuticular wax and cuticle permeability; however, the components of wax load that are responsible for changes in cuticle permeability remain unclear.
SUMOylation plays a critical role in regulating cuticle biosynthesis
PTMs are involved in the regulation of cellular signaling pathways (Roy and Sadanandom, 2021). SUMOylation is a key PTM in plants that plays an important role in plant growth, development, and responses to biotic and abiotic stress (Conti et al., 2014; Sahin et al., 2022). Both SUMO conjugates and free SUMO were more abundant in MdSIZ1-OE plants than in WT plants. MdSIZ1 plays a role in wax biosynthesis in apple by mediating the SUMOylation of MdMYB30 protein (Figures 1B and 7, A and B; Supplemental Figure 2A); this revealed a positive association between the level of wax loading and the intensity of protein SUMOylation.
The list of critical cellular processes affected by SUMOylation is growing. Unlike the ubiquitin system (where hundreds of E3s act as substrate adapters), the SUMO system has only two E3s in Arabidopsis, SIZ1 and MMS21; the molecular mechanisms underlying this PTM thus remain unclear. Zhang et al. isolated MdSIZ1 in apple and identified it as a SUMO E3 ligase that is capable of responding to ABA stress (Zhang et al., 2016). Like ubiquitin, SUMO is conjugated to its substrates through an E1-E2-E3 cascade (Saitoh et al., 1997; Santner and Estelle, 2010). Unlike ubiquitin, E3 ligase directly recognizes and catalyzes the ubiquitination of its substrates; in the SUMO system, E2 (SCE1) binds to the substrate, and E3 catalyzes the SUMOylation of the targets (Shu and Yang, 2017; Niu et al., 2019). Therefore, either E2 (SCE1) or E3 (SIZ1) can interact with the target protein at different residues during the process of SUMOylation. Unfortunately, we did not find direct interactions between MdMYB30 and MdSIZ1 using the Y2H system, but SUMO E2 conjugating enzyme (MdSCE1) interacted directly with the MdMYB30 protein (Figure 4A); therefore, we conducted further studies of MdSCE1. We speculate that MdSIZ1 might enhance the conjugating enzyme activity of MdSCE1 to form a SUMO–MdSCE1–MdSIZ1–MdMYB30 complex, thereby facilitating SUMO modification and thus wax deposition. DREB2A interacts with the E2-targeted protein SCE1 and is SUMOylated at its K163 residue to improve plant thermotolerance (Wang et al., 2020). Arabidopsis SUMO E3 ligase interacts with the E3-targeted protein SIZ1, which negatively regulates abscisic acid signaling by interacting with and SUMOylating ABSCISIC ACID INSENSITIVE 5 (ABI5) at the K391 SUMO conjugation residue (Miura et al., 2009). Apple MdSIZ1 interacts with MdbHLH104, a basic helix–loop–helix transcription factor, and the SUMOylation at residues K139 and K153 is critical for its function in iron acquisition and homeostasis (Zhou et al., 2019). Here, we detected a direct interaction between MdSCE1 and MdMYB30, but not MdSIZ1 (Figure 4).
SUMO can be attached to protein substrates by SUMOylation, which can affect protein function, activity, localization, and/or stability (Wilkinson and Henley, 2010; Augustine and Vierstra, 2018). Consistent with this idea, our results showed that the increased cuticular wax accumulation stemmed from the stability of the MdMYB30 protein SUMOylated by MdSIZ1 (Figures 5 and 7, A and B). MG132 is a 26S proteasome inhibitor, treatment with MG132 in the cell-free assay indicated that MdSIZ1 might stabilize the MdMYB30 protein by inhibiting its degradation via the 26S proteasome (Figure 5, C and D).
MdMYB30 functions as a target protein of MdSIZ1 through SUMOylation in the regulation of apple cuticular wax accumulation
The conjugation of SUMO to target proteins is particularly important for the regulation of plant development and responses to abiotic stresses. We have previously studied the function of MdSIZ1 in anthocyanin biosynthesis, Fe homeostasis, and root formation in apples (Zhou et al., 2017, 2019; Zhang et al., 2021). Here, a function of MdSIZ1 in cuticular wax biosynthesis by the SUMOylation of MdMYB30 was identified in apple. We demonstrated that MdSIZ1 and its putative SUMOylation target MdMYB30 are critical regulators of wax biosynthesis, which complements the functional network of MdSIZ1 in apple. MdSIZ1 mediates the SUMO modification of MdMYB30 protein to promote its accumulation. MdMYB30 directly binds to the promoter sequence of MdKCS1 to activate its transcription, which positively regulates cuticular wax biosynthesis (Figure 9F).
Previous studies have found that SIZ1 can respond to a variety of environmental signals (Miura et al., 2005; Park et al., 2011). siz1-2 T-DNA insertion alleles that cause freezing and chilling sensitivities were complemented genetically by expressing SIZ1, and SIZ1-dependent SUMOylation of INDUCER OF CBF EXPRESSION 1(ICE1) was moderately induced by cold, indicating that SIZ1 controls low-temperature adaptation in plants (Miura et al., 2007). The stability of DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) is maintained by SUMOylation under high temperature, which enhances the regulatory mechanisms underlying the heat stress response in plants (Wang et al., 2020). SIZ1 regulates Arabidopsis growth, and this SUMO E3 ligase plays a role in the response to drought stress (Catala et al., 2007). Our results indicate that the MdSIZ1–MdMYB30–MdKCS1 pathway affected cuticular wax biosynthesis. Our findings also provide insights into the regulatory mechanism by which cuticular wax deposition was affected by environmental conditions, such as low temperature and drought.
Materials and methods
Plant material and growth conditions
Murashige & Skoog medium with 0.1 mmol L−1 indole-3-acetic acid (IAA) and 0.5 mmol L−1 6-benzylaminopurine (6-BA) with a pH of 5.9, 3% (w/v) sucrose, and 0.8% (w/v) agar was used to subculture tissue-cultured “Gl3” apple (Malus domestica Borkh) plants under a 14-h:10-h, light:dark photoperiod at 24°C and 60% relative humidity (RH) for 25 d.
The Columbia ecotype (Col-0), the siz1-2 mutant (Salk_065397), and the MdSIZ1-OE13/19/33 Arabidopsis (Arabidopsis thaliana) line from Zhou et al. (2019) and Zhang et al. (2021) were used. These seeds were sterilized with 2.6% NaClO for 10 min and then washed in distilled water seven times. All A. thaliana seeds were germinated on MS medium with a pH of 5.9, 3% (w/v) sucrose, and 0.8% (w/v) agar.
Bagged apple fruits (Malus domestica cv. “Red Delicious”) were harvested from mature trees in an orchard located in Tai’an (Shandong, China) at 120 d after full bloom. Nicotiana benthamiana was cultivated in a growth chamber at 24°C and 65% RH under a 16-h/8-h light/dark cycle for three weeks for the transient expression assay.
“Orin” apple (Malus domestica Borkh.) calli were used in this study. MS medium (pH 6.0, 3% (w/v) sucrose, and 0.8% (w/v) agar) contained 0.5 mmol 1−1 benzylaminopurine (6-BA) and 0.5 mmol l−1 2,4-dichlorophenoxyacetic acid (2,4-D) for calli growth at 25°C in the dark. Subculturing was conducted every 2 weeks to maintain activity for genetic transformation and other assays. Wild-type tomato (Ailsa Craig) was used to obtain transgenic tomato fruits; 35S::MdMYB30-GFP transgenic tomatoes (T2) were used in subsequent studies.
RNA extraction and reverse transcription quantitative PCR analyses
RNA extraction and reverse transcription quantitative PCR (RT-qPCR) analyses of all plant material were performed by the method described in Zhou et al. (2017). All primers are shown in Supplemental Table 1. MdActin rRNA was used as an internal control.
Skin color and gloss measurements
The L*a*b* system was used to detect the light reflected using a color difference meter (ThreeNH, Shenzhen, China). The L* value represents color brightness (values from 100 to 0 indicate colors from white to black). The a* value represents changes from red to green and varies from +60 to −60. The b* value represents changes from yellow to blue and varies from +60 to −60. Measurements were taken from each apple leaf at the same location on the plant at least 15 times, and the results were averaged.
Water loss assay
Leaves of apple seedlings and Arabidopsis from the same location were collected, left at room temperature, and weighed at different time points. Time-course calculations of water loss were made to determine the percentage of the initial fresh weight at each time point.
Chlorophyll leaching assays
Leaves were cut into segments (∼2 cm) and immersed in 30 mL of 80%(v/v) ethanol at room temperature and 220 rpm in the dark. Aliquots of 1 mL were removed for chlorophyll quantification at the indicated time points. The chlorophyll concentration was quantified using a Thermo BIOMATE3 spectrophotometer at wavelengths of 664 and 647 nm. The chlorophyll content was determined using the following formula: chlorophyll content (mmol g−1) = (19.53 * A647 + 7.93 * A664)/FW (Fresh weight).
Toluidine blue staining
Toluidine blue solution (0.05%, w/v) was used to stain the leaves of apple seedlings, and Arabidopsis seedlings were immersed in this solution for 10 min. They were then washed with water repeatedly until the water was no longer visibly blue. Cotyledons (1 g) were stained, washed, and then immersed in 1 ml of 80% (v/v) ethanol for 2 h with continuous shaking. The resulting liquid was used to measure the absorbance value at 626 nm using a spectrophotometer.
Cuticle staining
Pericarp pieces of transgenic apple fruits and tomato fruits were collected. The pericarp equator was cut into 2-mm cubes for fixation in FAA buffer (water: formaldehyde: glacial acetic acid: ethanol 1:2:10:7). Pericarp pieces were used to cut 6-μm sections for staining with Oil red O. Sections were observed via light microscopy. For cuticle thickness measurements, sections were stained with Oil red O, and the thickness was determined for each biological replicate; an average of 10–15 measurements were taken per replicate. Oil red O stain dyes the apple cuticle red. The thickness of the cuticle was measured along anticlinal pegs, which are indicated by the arrows in ImageJ (https://imagej.en.softonic.com/).
Scanning electron microscopy and transmission electron microscopy
SEM and TEM were used to observe epicuticular wax and cutin crystallization patterns. Apple peels were collected from apple and tomato fruits. Samples were vacuum-dried at −80°C in a 110-Pa vacuum for 24 h (FDU–1110, 50/60 Hz, 1.7 kVA) and viewed by cryo-SEM as described by Lü et al. (2009).
The deposition of cuticle was observed by TEM as described in Chen et al. (2003). Briefly, 1.2-mm discs of apple leaves were fixed in 2.5% glutaraldehyde solution for 24 h at 4°C. They were rinsed three times for 5 min in phosphate buffer and post-fixed in a fresh mixture of osmium tetroxide 1% (EMS) with 1.5% potassium ferrocyanide in phosphate buffer for 2 h at room temperature. The samples were then washed two times in distilled water and dehydrated in acetone solution. Ultrathin sections 60 nm thick were cut using a Leica Ultracut (Leica Mikrosysteme GmbH) and placed on a copper slot grid (2 × 1 mm, EMS) coated with a polystyrene film (Sigma Aldrich). Micrographs were taken using a transmission electron microscope (JEOL, Japan).
Cuticular wax analysis
The wax levels of leaves of WT and MdSIZ1 transgenic apple seedings and transiently transformed fruits were measured. Apple fruits were immersed in chloroform for 1 min twice with 20 μg of n-tetracosane as an internal standard. The chloroform was then evaporated under gaseous N2, and the residue was derivatized with 150 μL of bis-N, N-(trimethylsilyl) trifluoroacetamide (BSTFA, Sigma, St. Louis, MO, USA) for 60 min at 70°C. The reactants were evaporated again and dissolved in chromatography chloroform solvent. Samples were injected out of chloroform in a gas chromatograph coupled to a mass spectrometer (ionization voltage: 10–200 eV; resolution: R > 2 M (RWHM); scanning speed: 10,000 u/s). The temperature program was as follows: 2 min at 50°C, increase from 50°C to 140°C at 20°C min−1, increase at 2°C min−1 to 320°C, and held at 320°C for 10 min. Peaks were quantified on the basis of their FID ion current and library NIST17.1.
Yeast one-hybrid and yeast two-hybrid assay
The full-length open reading frames of MdMYB30 and MdSCE1 cloned into the pGADT7 vector were fused in-frame with the GAL4 activation domain. The promoters of MdKCS1 (2-kb upstream regions from the start codon) were amplified and cloned into the pHIS2 vector (Clontech) and linearized at the SmaI or SacII sites. The resulting constructs were transformed into Y187 yeast strains using the PEG6000-induced method (Clontech, Dalian, China). The yeast strains were screened using the selective medium SD/-Leu/-Trp/-His/+ 3-AT (3-amino-1,2,4-triazole, 50 mM). The correct clones revealed that MdMYB30 was able to bind to their promoter.
Y2H assays were conducted using the lithium acetate method described by Zhang et al. (2021). MdSIZ1-N (1–427 aa) was the domain excluding autonomous activation. The full-length MdSCE1 and MdSIZ1-N sequences (1–427 aa) were inserted into the pGBT9 vector. The co-expressed yeast cells were diluted 1, 10−1, and 10−2 with water and inoculated in SD/-T/-L and SD/-T/-L/-H/-A synthetic defined (SD) media at 28°C for 2 d.
Pull-down assays
The full-length cDNA fragment of MdSCE1 was inserted in-frame into the PGEX–4T–1 vector for GST-tag fusion (GST, glutathione S-transferase). The coding sequences of MdMYB30 were amplified and inserted into the pET-32a-c (+) vector for fusion with His-tag. The resulting vectors were transformed into E. coli strain (BL21) to express the protein.
For the in vitro pull-down assay, the His-MdMYB30 protein and GST-MdSCE1 or GST were incubated together with the pull-down buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 Mm DTT) with continuous rocking at 4°C for 2–3 h. The beads were washed five times with wash buffer (20 mM Tris, pH 7.4, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100); the pull-downed protein was separated by 10% SDS–PAGE, and the anti-GST (Abmart M20007, 1/1000) or anti-His (Abmart M30111, 1/1000) antibodies were added to detect protein by immunoblot analysis.
Bimolecular fluorescence complementation assays
The full-length cDNA fragment of MdSCE1 was fused to the N-terminal half of yellow fluorescent protein (N-YFP-MdSCE1), and the MdMYB30 sequence was fused to the C-terminal half of YFP (MdMYB30-C-YFP). N-YFP-MdSCE1 and MdMYB30-C-YFP were transiently co-expressed in Nicotiana benthamiana leaves. N-YFP and C-YFP were used as negative controls. Infected N. benthamiana was cultivated for 16 h in the dark and then subjected to a 16-h:8-h light:dark photoperiod at 25°C. After 48 h, the fluorescence signal was detected with a laser confocal microscope (Zeiss LSM 880 Meta, Jena, Germany) with the excitation and emission wavelengths set to 488 nm/505–550 nm for YFP signals. DAPI was used to label transformed cell nuclear.
SUMOylation assays
For in vitro SUMOylation assays, proteins were incubated in 20 µL of reaction buffer (50 mM Tris–HCl, 100 mM NaCl, 15% glycerol, 5 mM ATP, 10 mM MgCl2, pH 7.8) with or without 1 µg of SUMO1 protein (Abcam, Cambridge, UK), E1 (0.5 µg of human SAE1 full and 0.5 µg of SAE2/UBA2 peptide), E2 (2 µg of MdSCE1 customized by Abmart), and 8 µg of GST-MdSIZ1 or His-MdMYB30 protein for 3 h at 37°C. Next, 10% SDS-polyacrylamide gel immunoblot analysis was performed with anti-His and anti-SUMO1 antibodies (Abcam). For in vivo SUMOylation assays, three 3-week-old transgenic apple calli were extracted with IP lysis/wash buffer, and soluble extracts were IPed with anti-Flag antibody using a Pierce Crosslink IP kit following the manufacturer's instructions. Lastly, 8% SDS-polyacrylamide gel immunoblot analysis was performed with anti-Myc (Abmart), anti-GFP(Abmart), and anti-SUMO1(Abcam) antibodies. For the SUMO conjugate assay, total proteins of plant tissue were extracted with 50 mM Tris–HCl (pH 8), 150 mM NaCl, 0.2% Triton X-100, 1 mM PMSF, and 5 mM EDTA. Next, 10% SDS-polyacrylamide gel immunoblot analysis was performed with anti-SUMO1 and anti-actin antibodies (Abcam). Anti-actin antibody was used to ensure equivalent protein loading.
Protein degradation assays
In vitro protein degradation assays were performed as described by Zhang et al. (2021). Total protein extract from WT, MdSIZ1, and anti-MdSIZ1 transgenic apple calli and His-MdMYB30 protein treated with dimethyl sulfoxide (DMSO) or 50 µM of Z-Leu-Leu-Leu- CHO aldehyde (MG132, 26S proteasome inhibitor) for 8 h were incubated with degradation buffer (25 mM Tris–HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM phenylmethylsulfonylfluoride (PMSF), 5 mM DTT, and 10 mM ATP, pH 7.5) and sampled at 0, 30, 60, 90, 120, and 150 min. An anti-His antibody was used to measure protein abundance. Anti-ACTIN was used as an internal control, and DMSO was used as a negative control for MG132. The degradation rate of protein in vitro or in vivo was measured with Bio-Rad QUANTITY ONE software. The ratio of protein abundance to anti-ACTIN at the initial time 0 was taken as 100%.
Chromatin immunoprecipitation–qPCR assay
Chromatin Immunoprecipitation–qPCR (ChIP–qPCR) assays were performed as described previously (Saleh et al., 2008). Briefly, two-week-old PBIN-GFP and MdbMYB30-GFP transgenic apple calli were crosslinked in fixation buffer (10 mM Tris–HCl pH 8.0, 0.4 M sucrose, 0.05% Triton X–100, 1% formaldehyde) and vacuumed for 10 min; 1 M glycine solution was added to quench the reaction. The calli were ground into fine powder and broken into 200–800-bp DNA fragments with an ultrasonic crushing apparatus (JY92-IIN, SCIENTZ, Ningbo, China). After centrifugation, the supernatant was diluted to 1 ml. Anti-GFP antibody (Sigma) was used for immunoprecipitation with MdMYB30 protein–DNA fragments and incubated overnight at 4°C. Next, 20 mL of protein A and protein G beads mixture (1:1 in volume) was added to each tube and incubated at 4°C for 1 h. After incubation, beads were washed five times with different wash buffers. The eluates were de-crosslinked by adding 20 µL of 5 M NaCl, and the DNA was purified. Finally, the GFP-specific enrichment of the fragments from MdKCS1 promoters was analyzed by qPCR, and three technical replicates were conducted for each sample. qPCR was performed using a Step One Real-time Detection System and SYBR Green PCR Master Mix (CWBIO, Beijing, China). Primers used are listed in Supplemental Table 1.
Electrophoretic mobility shift assay
The coding sequences of MdMYB30 were amplified and inserted into the pET-32a-c(+) vector for fusion with His-tag. MdMYB30-His and His were expressed in Escherichia coli strain (BL21), subsequently induced with 3 mM IPTG (isopropyl-b-D-thiogalactopyranoside) for 6 h at 37°C, and purified using glutathione sepharose beads (Thermo Scientific, San Jose, CA, USA). The 30-bp length oligonucleotides containing the AACAAAC motif of the promoters of MdKCS1 were synthesized and labeled with a biotin chemiluminescent EMSA kit (Beyotime, Shanghai, China) following the manufacturer's protocol. The AACAAAC motif mutated to TTGCTTT was used as the mutated probe. Probe primer sequences are shown in Supplemental Table 1.
The recombined protein of MdMYB30-His was incubated in a 10-mL system (2 µL of 5× binding buffer, 1 µL of biotin-labeled double-stranded binding consensus oligonucleotides, and 7 µL of MdMYB30-His protein) at 24°C for 30 min. For competition assays, unlabeled competitors were added to the reaction at 50 and 100-fold molar excess.
Luciferase complementation imaging assays and dual-luciferase assay
The coding regions of MdSCE1 and MdMYB30 were inserted into pCAMBIA-nLUC and pCAMBIA-cLUC to generate the MdMYB30-cLUC and MdSCE1-nLUC constructs, respectively. The empty vectors were used as a negative control. 35S::MdMYB30-62SK was created by cloning its coding sequences into a pGreen II 62–SK vector via T4 ligase (Clontech, Dalian, China). The 2-kb promoter including the AACAAAC motif regions of MdKCS1 was inserted into the pGreenII 0800-LUC vector to obtain MdKCS1pro::pGreenII 0800–LUC. Agrobacterium tumefaciens strain GV3101 containing different combinations of the above constructs was cultured overnight at 28°C with constant shaking at 220 rpm. The Agrobacterium cells were collected by centrifugation at 5,000 rpm for 10 min, and the pellet was suspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) at a final concentration of OD600 = 0.6. Resuspensed Agrobacterium was then mixed in different combinations and incubated at 24°C without shaking for 3 h prior to infiltration. After infiltration for 48 h, the N. benthamiana leaves were sprayed with 100 mM luciferin and placed in the dark for 2 min; the luciferase signals were captured using an IVIS Spectrum imaging system (Xenogen, Alameda, CA, USA).
Viral vector constructs and apple fruit injection assays
The full-length cDNA sequences of MdSIZ1 and MdMYB30 were cloned into the pIR vectors to obtain overexpression vector constructs. To construct antisense expression vectors, the 300-bp fragments of MdSIZ1 and MdMYB30 were cloned into tobacco rattle virus (TRV); the overexpression and antisense recombinant vectors were named MdSIZ1-pIR, MdMYB30-pIR and MdSIZ1-TRV, MdMYB30-TRV, respectively.
Fruit injection assays were conducted following the previously described procedure in Li et al. (2012). The plasmid of the overexpression constructs and the auxiliary vector IL60-1 were diluted with injection buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) to 10 μg mL−1. Fresh-bagged “Red Delicious” apple (M. domestica) fruits were injected with mixed plasmids using a needleless syringe. Agrobacterium cells (GV3101) was transformed with antisense recombinant vector and resuspended in infiltration buffer (10 mM MES (pH 5.6), 10 mM MgCl2, 150 mM acetosyringone) at OD600 = 0.6. Extraction of the surface wax of apple fruit was performed after placing fruits in the dark for 7 d. Three apples and three technical replicates were conducted for each test, and fruits injected with either pIR or TRV were used as controls.
Statistical analyses
The numbers of samples and replicates are indicated in the figure legends. Statistically significant differences between control and experimental groups were determined by one-way ANOVA (Student's t test; P < 0.05).
Accession numbers
Sequence data from this article can be found in the Genome Database for Rosaceae (GDR) (https://www.rosaceae.org/) data libraries under accession numbers. MdSIZ1 (MD11G1131700), MdSCE1 (MD15G1286800), MdMYB30 (MD09G1051000), MdKCS1 (MD01G1087400). The siz1 mutant Arabidopsis (Arabidopsis thaliana) seeds were obtained from TAIR mutant library (https://www.arabidopsis.org/). siz1-2 (Salk_065397).
Supplementary Material
Acknowledgments
We would like to thank Prof. Takaya Moriguchi of the National Institute of Fruit Tree Science, Japan, for “Orin” apple calli. We would also like to thank EditorBar Language Editing Company for providing linguistic assistance during the preparation of this manuscript. We also thank Prof. Zhi-Hong Zhang from Shenyang Agricultural University, China, for providing “GL-3” apple seedlings, and Prof. Ilan Sela from Hebrew University, Israel, for providing TRV and pIR binary vectors.
Contributor Information
Ya-Li Zhang, State Key Laboratory of Crop Biology, Shandong Collaborative Innovation Center of Fruit & Vegetable Quality and Efficient Production, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An 271018, Shandong, China.
Yi Tian, National Agricultural Engineering Center for North Mountain Region of the Ministry of Science and Technology, Mountainous Area Research Institute of Hebei Province, Hebei Agricultural University, Baoding, Hebei 071001, China.
Yao-Yang Man, State Key Laboratory of Crop Biology, Shandong Collaborative Innovation Center of Fruit & Vegetable Quality and Efficient Production, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An 271018, Shandong, China.
Chun-Ling Zhang, State Key Laboratory of Crop Biology, Shandong Collaborative Innovation Center of Fruit & Vegetable Quality and Efficient Production, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An 271018, Shandong, China.
Yi Wang, State Key Laboratory of Crop Biology, Shandong Collaborative Innovation Center of Fruit & Vegetable Quality and Efficient Production, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An 271018, Shandong, China.
Chun-Xiang You, State Key Laboratory of Crop Biology, Shandong Collaborative Innovation Center of Fruit & Vegetable Quality and Efficient Production, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An 271018, Shandong, China.
Yuan-Yuan Li, State Key Laboratory of Crop Biology, Shandong Collaborative Innovation Center of Fruit & Vegetable Quality and Efficient Production, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An 271018, Shandong, China.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Identification of MdSIZ1 overexpressed apple seedlings.
Supplemental Figure S2 . The accumulation of SUMO conjugates and wax loading of WT, MdSIZ1-EE 13/19/33, and siz1-2 mutant Arabidopsis.
Supplemental Figure S3 . Analysis of the glossiness of WT and MdSIZ1-OE apple leaves.
Supplemental Figure S4 . The cuticle permeability of WT, MdSIZ1-EE 13/19/33, and siz1-2 mutant Arabidopsis.
Supplemental Figure S5 . Relative expression level of MdMYB30 and MdSIZ1 in the MdMYB30-Myc/MdSIZ1-GFP and MdMYB30-Myc/anti-MdSIZ1-GFP transgenic apple calli.
Supplemental Figure S6 . Relative expression of MdSIZ1, MdMYB30, and MdKCS1 in virus-induced transformed apple fruits.
Supplemental Figure S7 . SEM images of apple epicuticular wax crystals.
Supplemental Figure S8 . Identification of 35S::MdSIZ1-Myc/35S::MdMYB30-GFP and 35S::MdMYB30/siz1-2 transgenic Arabidopsis seedlings at the DNA level.
Supplemental Figure S9 . Identification of MdMYB30 ectopically expressing tomato fruits.
Supplemental Figure S10 . The interaction of MdMYB30 with the promoters of wax-related genes.
Supplemental Table S1 . Primers for cDNA synthesis, amplification, and sequencing.
Supplemental Table S2 . The promoter sequence of MdKCS1.
Supplemental Dataset S1 . Cuticular wax composition of apple leaves. Wax coverage is expressed as μg cm−2 leaves surface area.
Funding
This work was supported by the National Natural Science Foundation of China (32072539) and Natural Science Foundation of Shandong Province (ZR2022JQ14).
References
- Augustine RC, Vierstra RD (2018) SUMOylation: re-wiring the plant nucleus during stress and development. Curr Opin Plant Biol 45(Pt A): 143–154 [DOI] [PubMed] [Google Scholar]
- Balabanidou V, Grigoraki L, Vontas J (2018) Insect cuticle: a critical determinant of insecticide resistance. Curr Opin Insect 27(2214): 68–74 [DOI] [PubMed] [Google Scholar]
- Berhin A, Nawrath C, Hachez C (2022) Subtle interplay between trichome development and cuticle formation in plants. New Phytol 233(5): 2036–2046 [DOI] [PubMed] [Google Scholar]
- Bhanot V, Fadanavis SV, Panwar J (2021) Revisiting the architecture, biosynthesis and functional aspects of the plant cuticle: there is more scope. Environ Exp Bot 183(0098): 104364 [Google Scholar]
- Castaño-Miquel L, Mas A, Teixeira I, Seguí J, Perearnau A, Thampi BN, Schapire AL, Rodrigo N, La Verde G, Manrique S, et al. (2017) SUMOylation inhibition mediated by disruption of SUMO E1-E2 interactions confers plant susceptibility to necrotrophic fungal pathogens. Mol Plant 10(5): 709–720 [DOI] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua N-H (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19(9): 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin S-M, Boroff V-L, Liu X, Jenks M-A (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15(5): 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hu WJ, Mishra N, Wei J, Lu HL, Hou YQ, Qiu XY, Yu SF, Wang CL, Zhang H, et al. (2020) AKR2A interacts with KCS1 to improve VLCFAs contents and chilling tolerance of Arabidopsis thaliana. Plant J 103(4): 1575–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sun Z, Wu K, Zhang Q, Song Y, Wang T, Fu D, Cao J, Luo Y, Qu G (2021) Dynamic changes in wax and cutin compounds and the relationship with water loss in ‘Red Fuji ‘and ‘Golden Delicious’ apples during shelf life. Int J Food Sci Technol 56(12): 6335–6344 [Google Scholar]
- Conti L, Nelis S, Zhang C, Woodcock A, Swarup R, Galbiati M, Tonelli C, Napier R, Hedden P, Bennett M, et al. (2014) Small ubiquitin-like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev Cell 28(1): 102–110 [DOI] [PubMed] [Google Scholar]
- Elejalde-Palmett C, San Segundo IM, Garroum I, Charrier L, De Bellis D, Mucciolo A, Guerault A, Liu J, Zeisler-Diehl V, Aharoni A, et al. (2021) ABCG Transporters export cutin precursors for the formation of the plant cuticle. Curr Biol 31(10): 2111–2123 [DOI] [PubMed] [Google Scholar]
- Fang S, Hou X, Liang X (2022) SIZ1-mediated SUMOylation responds to multiple abiotic stresses in plants. Environ Exp Bot 201(0098): 104977 [Google Scholar]
- Fich EA, Segerson NA, Rose JKC (2016) The plant polyester cutin: biosynthesis, structure, and biological roles. Annu Rev Plant Biol 67(1): 207–233 [DOI] [PubMed] [Google Scholar]
- García-Coronado H, Tafolla-Arellano JC, Hernández-Oñate MÁ, Burgara-Estrella AJ, Robles-Parra JM, Tiznado-Hernández ME (2022) Molecular biology, composition and physiological functions of cuticle lipids in fleshy fruits. Plants 11(9): 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter R, Riederer M (2016) Localization of the transpiration barrier in the epi-and intracuticular waxes of eight plant species: water transport resistances are associated with fatty acyl rather than alicyclic components. Plant Physiol 170(2): 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Zhou L-J, Gao H-N, Wang X-F, Li Z-W, Li Y-Y (2022) The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol 189(4):2044–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67(5): 547–566 [DOI] [PubMed] [Google Scholar]
- Kan Y, Mu X-R, Zhang H, Gao J, Shan J-X, Ye W-W, Lin H-X (2022) TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat Plants 8(1): 53–67 [DOI] [PubMed] [Google Scholar]
- Kim H, Yu S-I, Jung SH, Lee B-H, Suh MC (2019) The F-box protein SAGL1 and ECERIFERUM3 regulate cuticular wax biosynthesis in response to changes in humidity in Arabidopsis. Plant Cell 31(9): 2223–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M, Seo S, Takahashi A, Hirochika H (2018) The MAMP-responsive MYB transcription factors MYB30, MYB55 and MYB110 activate the HCAA synthesis pathway and enhance immunity in rice. Plant Cell Physiol 59(5): 903–915 [DOI] [PubMed] [Google Scholar]
- Kong X, Hong Y, Hsu Y-F, Huang H, Liu X, Song Z, Zhu J-K (2020b) SIZ1-mediated SUMOylation of ROS1 enhances its stability and positively regulates active DNA demethylation in Arabidopsis. Mol Plant 13(12): 1816–1824 [DOI] [PubMed] [Google Scholar]
- Kong L, Liu Y, Zhi P, Wang X, Xu B, Gong Z, Chang C (2020a) Origins and evolution of cuticle biosynthetic machinery in land plants. Plant Physiol 184(4): 1998–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Zhao L, Eveleigh N, Yu Y, Chen X, Kunst L (2015) The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol 167(2): 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara I, Belge B, Goulao LF (2015) A focus on the biosynthesis and composition of cuticle in fruits. J Agric Food Chem 63(16): 4005–4019 [DOI] [PubMed] [Google Scholar]
- Lara I, Heredia A, Domínguez E (2019) Shelf life potential and the fruit cuticle: the unexpected player. Front Plant Sci 10(2019): 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-J, Kim KY, Zhang J, Yamaoka Y, Gao P, Kim H, Hwang J-U, Suh MC, Kang B, Lee Y (2021) Arabidopsis seedling establishment under waterlogging requires ABCG5-mediated formation of a dense cuticle layer. New Phytol 229(1): 156–172 [DOI] [PubMed] [Google Scholar]
- Li R-J, Li L-M, Liu X-L, Kim J-C, Jenks MA, Lü S (2019) Diurnal regulation of plant epidermal wax synthesis through antagonistic roles of the transcription factors SPL9 and DEWAX. Plant Cell 31(11): 2711–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Ma J, Liu H, Wang X, Li J, Li Z, Li M, Sui S, Liu D (2021) Overexpression of CpSIZ1, a SIZ/PIAS-type SUMO E3 ligase from wintersweet (Chimonanthus praecox), delays flowering, accelerates leaf senescence and enhances cold tolerance in Arabidopsis. Plant Mol Biol Rep 39(2): 301–316 [Google Scholar]
- Li Y-Y, Mao K, Zhao C, Zhao X-Y, Zhang H-L, Shu H-R, Hao Y-J (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160(2): 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58(2): 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou S, Liu Z, Lu L, Dang H, Li H, Chu B, Chen P, Ma Z, Zhao S, et al. (2022) Fine-tuning of SUMOylation modulates drought tolerance of apple. Plant Biotechnol J 20(5): 903–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H-C, Lam S-H, Zhang D, Hsiao Y-Y, Li B-J, Niu S-C, Li C-Y, Lan S, Tsai W-C, Liu Z-J (2022) R2R3-MYB genes coordinate conical cell development and cuticular wax biosynthesis in Phalaenopsis aphrodite. Plant Physiol 188(1): 318–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma D-K, Parsons E-P, Rowland O, Jenks M-A (2009) Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J 59(4): 553–564 [DOI] [PubMed] [Google Scholar]
- Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, Pouzet C, Jauneau A, Roby D, Rivas S (2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat Commun 4(1): 1–9 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun D-J, Hasegawa PM (2007) SIZ1-mediated Sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19(4): 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci U S A 106(13): 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci U S A 102(21): 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C (2006) Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol 9(3): 281–287 [DOI] [PubMed] [Google Scholar]
- Niu D, Lin X-L, Kong X, Qu G-P, Cai B, Lee J, Jing JB (2019) SIZ-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol Plant 12(2): 215–228 [DOI] [PubMed] [Google Scholar]
- Park BS, Song JT, Seo HS (2011) Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun 2(1): 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J, Bres C, Just D, Garcia V, Mauxion J-P, Marion D, Bakan B, Joubès J, Domergue F, Rothan C (2014) Analyses of tomato fruit brightness mutants uncover both cutin-deficient and cutin-abundant mutants and a new hypomorphic allele of GDSL lipase. Plant Physiol 164(2): 888–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20(3): 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Sadanandom A (2021) SUMO mediated regulation of transcription factors as a mechanism for transducing environmental cues into cellular signaling in plants. Cell Mol Life Sci 78(6): 2641–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, de Thé H, Lallemand-Breitenbach V (2022) Sumoylation in physiology, pathology and therapy. Cells 11(5): 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Pu RT, Dasso M (1997) SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci 22(10): 374–376 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, Al-Abdallat A, Xia Y-N, Lu G-Q, Ladunga I, et al. (2008) The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20(3): 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M (2010) The ubiquitin-proteasome system regulates plant hormone signaling. Plant J 61(6): 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park M-J, Go YS, Park C-M (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23(3): 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Yang W (2017) E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol 58(9): 1461–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17(2): 119–130 [DOI] [PubMed] [Google Scholar]
- Wang F, Liu Y, Shi Y, Han D, Wu Y, Ye W, Yang H, Li G, Cui F, Wan S, et al. (2020) SUMOylation stabilizes the transcription factor DREB2A to improve plant thermotolerance. Plant Physiol 183(1): 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tian X, Zhao Q, Liu Z, Li X, Ren Y, Tang J, Fang J, Xu Q, Bu Q (2018) The E3 ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice. Plant Cell 30(1): 228–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428(2): 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Yang X, Wang L, Zheng B, Cai Y, Ogutu CO, Zhao L, Peng Q, Liao L, Zhao Y, et al. (2022) Two R2R3-MYB genes cooperatively control trichome development and cuticular wax biosynthesis in Prunus persica. New Phytol 234(1): 179–196 [DOI] [PubMed] [Google Scholar]
- Zhang R-F, Guo Y, Li Y-Y, Zhou L-J, Hao Y-J, You C-X (2016) Functional identification of MdSIZ1 as a SUMO E3 ligase in apple. J Plant Physiol 198(0176): 69–80 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li X, Wang W, Li H, Cui Y, Zhu Y, Kui H, Yi J, Li J, Gou X (2022) SERKs regulate embryonic cuticle integrity through the TWS1-GSO1/2 signaling pathway in Arabidopsis. New Phytol 233(1): 313–328 [DOI] [PubMed] [Google Scholar]
- Zhang C-L, Wang G-L, Zhang Y-L, Hu X, Zhou L-J, You C-X, Li Y-Y, Hao Y-J (2021) Apple SUMO E3 ligase MdSIZ1 facilitates SUMOylation of MdARF8 to regulate lateral root formation. New Phytol 229(4): 2206–2222 [DOI] [PubMed] [Google Scholar]
- Zhang Y-L, Zhang C-L, Wang G-L, Wang Y-X, Qi C-H, Zhao Q, You C-X, Li Y-Y, Hao Y-J (2019) The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol 19(1): 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L-J, Li Y-Y, Zhang R-F, Zhang C-L, Xie X-B, Zhao C, Hao Y-J (2017) The SUMO E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low temperature conditions in apple. Plant Cell Environ 40(10): 2068–2080 [DOI] [PubMed] [Google Scholar]
- Zhou L-J, Zhang C-L, Zhang R-F, Wang G-L, Li Y-Y, Hao Y-J (2019) The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis. Plant Physiol 179(1): 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv C, Zhao Z, Gao YG, Xia Y (2018) Multifunctional roles of plant cuticle during plant-pathogen interactions. Front Plant Sci 9(2018): 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.