Abstract
The increased use of antimicrobial compounds such as copper into nanoparticles changes how living cells interact with these novel materials. The increased use of antimicrobial nanomaterials combats infectious disease and food spoilage. Fungal infections are particularly difficult to treat because of the few druggable targets, and Saccharomyces cerevisiae provides an insightful model organism to test these new materials. However, because of the novel characteristics of these materials, it is unclear how these materials interact with living cells and if resistance to copper-based nanomaterials could occur. Copper nanoparticles built on carboxymethylcellulose microfibril strands with copper (CMC-Cu) are a promising nanomaterial when imported into yeast cells and induce cell death. The α-arrestins are cargo adaptors that select which molecules are imported into eukaryotic cells. We screened α-arrestins mutants and identified Aly2, Rim8, and Rog3 α-arrestins, which are necessary for the internalization of CMC-Cu nanoparticles. Internal reactive oxygen species in these mutants were lower and corresponded to the increased viability in the presence of CMC-Cu. Using lattice light-sheet microscopy on live cells, we determined that CMC-Cu were imported into yeast within 30 min of exposure. Initially, the cytoplasmic pH decreased but returned to basal level 90 min later. However, there was heterogeneity in response to CMC-Cu exposure, which could be due to the heterogeneity of the particles or differences in the metabolic states within the population. When yeast were exposed to sublethal concentrations of CMC-Cu no resistance occurred. Internalization of CMC-Cu increases the potency of these antimicrobial nanomaterials and is likely key to preventing fungi from evolving resistance.
Keywords: copper nanoparticles, endocytosis, α-arrestins, S. cerevisiae, lattice light-sheet microscopy, reactive oxygen species
Graphical Abstract
Graphical Abstract.
Positively charged copper nanoparticles built on carboxymethylcellulose particles associate with the negatively charged cell wall of S. cerevisiae. CMC-Cu entry is mediated through numerous α-arrestins. Once assimilated in cells, the CMC-Cus increase internal ROS levels likely through Fenton reactions.
Introduction
Cellulose is a vastly abundant renewable resource. It is a fibrous substance composed of high molecular weight homopolymers of β-1,4-linked anhydro-D-glucose units that are insoluble in water.1 Utilization of cellulosic raw material protects copper nanoparticles from self-aggregation and fast oxidation,2–4 a common problem with metallic nanoparticles.5 The high number of Na-carboxyl groups facilitates copper's reduction on the cellulose derivative carboxymethyl cellulose (CMC) and makes it an appealing organic support structure to construct copper nanoparticles.4,6 These copper nanoparticles are synthesized in situ on carboxymethylcellulose microfibril strands (CMC-Cu) and exhibit mostly Cu+ and Cu0 forms averaging 15 nm in diameter.4 There are several technique variations in the synthesis of copper nanoparticles including thermal reduction, chemical reduction, sono-chemical reduction, metal vapor synthesis, vacuum vapor deposition, radiation methods, microemulsion techniques, and laser ablation. Variations in the copper composition and cellulose biopolymer have been successfully synthesized resulting in changes to their interaction with microorganisms.7–10 These are distinct from fully metallic nanoparticles that are relatively stable and are found transported from roots to stems11 and into organs.12 The toxicity of nanoparticles of fully metallic nanoparticles would be different from hybrid particles built on different substrates. Cellulose is biodegradable while metallic nanoparticles’ fate would vary depending on their characteristics leading to aggregation, interaction with natural organic matter, or sedimentation. While CMC-Cu nanoparticles are similar in size as metallic copper nanoparticles they are approximately 3.8 times less toxic than metallic copper nanoparticles.13,14
Copper nanoparticles are a promising technology that may inhibit pervasive drug-resistant pathogens, nosocomial pathogens, and food spoilage microorganisms that pose a serious threat to human and livestock health.15 Copper is essential to organisms at trace levels; however, at higher concentrations it becomes toxic. Human cells have been shown to have a low sensitivity to transition metal,16 while microorganisms maintain a high sensitivity.17 The response induced by copper in microorganisms has been widely studied. The expression of genes in response to copper is tightly regulated to maintain a fine balance of this essential, yet damaging metal in all organisms.18–20 Copper's regulation also includes the compartmentalization of copper enzymes, the use of metallochaperones to deliver copper to targets, and the localization and turnover of copper transporters.21–24 Copper is used sparingly for protein functions, usually, as an electron donor/acceptor, in which it switches redox states between Cu I and Cu II, or as an electron carrier.25
The eukaryotic organism Saccharomyces cerevisiae specifically imports vital copper to the Golgi apparatus for incorporation into cuproenzymes and distribution in the mitochondria.26,27 Two metal cell surface reductases, Fre1 and Fre2, facilitate copper reduction from Cu II to usable Cu I.28,29 In copper-deficient environments, Mac1 induces the transcription of CTR1 and CTR3, which encode high-affinity copper transporters that mediate cuprous (Cu I) uptake.30–33 However, yeast strains derived from S288c such as the one used in this study contain a transposon that blocks the expression of CTR3.34 Ctr1 is internalized for vacuole degradation through Rsp5-dependent endocytosis when copper concentrations are above 50 μM.35 Rsp5, an E3 ubiquitin ligase ubiquitinates cargo and interacts through its WW domains with the (PPxY) motifs on the arrestins, the endocytic adaptors, which serve as adaptors between Rsp5 and membrane protein targeted for degradation.36–38 Once inside the vacuole, the Ctr1/Cu complex is degraded.35 There are differences in previous studies on whether Ctr1 is trafficked to the vacuole for degradation, which are likely due to differences in the expression of the Ctr1-GFP construct in the different studies.35,39 Copper can then be chaperoned by Atx1, Cox17, and Ccs122 to areas of the cell as needed for a multiplicity of functions including mitochondrial oxidative phosphorylation and protection against oxidative stress.35 In high concentrations of copper, the transcription factor Ace1/Cup2 induces expression of the CUP1 and CRS5, which encode metallothioneins, and SOD1 encodes cytosolic copper-zinc superoxide dismutase 6.40–42
Endocytosis internalizes cargo from the plasma membrane and has a critical role in nutrient uptake, damaged protein turnover, membrane composition, and the response to extracellular signals. Two protein families, arrestins and G-protein-coupled receptor (GPCR) kinases (GRKs) mediate the downregulation of receptor signaling. Originally, arrestins were discovered for their role during downregulation of clathrin-mediated endocytosis where GRKs recognize and phosphorylate GPCRs to which arrestins bind at the plasma membrane. Yeast contain a family of arrestins that have been suggested as a general model for transporter regulation.43 Yeast α-arrestins target specific plasma membrane proteins for endocytosis by recruiting endocytic promotion factors such as adaptins and clathrin while excluding heterotrimeric G proteins.44,45 A less characterized pathway, clathrin-independent endocytosis, has been shown to utilize specific yeast arrestins to promote endocytosis of different cargos.46 Clathrin-independent endocytosis requires Rsp5, other components of the endocytosis system, and their regulators, but not Rsp5 binding to α-arrestins. The α-arrestins are critical for response to starvation, redox homeostasis regulation, innate immune response, and tumor suppression.47 The mechanisms for understanding cargo selection are not well understood, but evidence has shown that α-arrestins are important in both clathrin-mediated and clathrin-independent endocytosis by different mechanisms.46
The major features of lattice light sheet microscopy (LLSM), which make it superior to confocal or two-photon microscopy in the context of this paper are its superior photon efficiency (resistance to photobleaching) due to the highly parallel widefield imaging and lack of photobleaching of out of focus planes that confocal microscopy cannot avoid.48,49 Further, superior z-resolution of LLSM (estimated 5× higher) due to the crossed longitudinal and transverse point spread functions of the excitation and detection optics provide a greatly improved and nearly isotropic 3D point spread function,50 which is routinely deconvolved during image reconstruction.51 The LLSM has higher detection efficiency due to the lack of pinholes and the higher numerical aperture of the collection optics used, combined with the superior efficiency of scientific complementary metal oxide semiconductor (sCMOS) detectors compared to photo multiplier tubes (PMT).52 Similar statements apply to the comparison of two-photon microscopy, which has the advantage of greater depth resolution in tissues, but similarly still obtains lower collection efficiency due to lower numerical aperture and suffers from photobleaching and photodegradation due to the serial data collection employed.53,54 These features of LLSM make volumetric measurements possible over longer times at lower powers, optimizing the so-called “photon budget”; thus the results obtained are more representative due to the lack of photobleaching across the field of view or with depth that would occur in a confocal or two-photon microscope, both of which require serial acquisition. Scanning electron microscopy and transmission electron microscopy, both being nonoptical, cannot distinguish optical properties and of course, have the additional disadvantages of the former being a surface-only method and both former and latter usually requiring significant sample preparation and modification, thus neither could perform the large-scale population assay in an aqueous environment at near-physiological conditions we have obtained with the LLSM in this work.
The hybrid cellulosic-copper nanoparticle materials are effectively incorporated into both a thermoplastic resin and polyvinyl alcohol with the intent to increase the utility of low-value wood products. The feasibility of using these antimicrobial films in the packaging and healthcare industry needs to be assessed through systematic analysis. Characterizing nanoparticle toxicity and transport of these cellulose copper nanoparticles provides critical insights into advancing the field of nanotoxicity. However, the exact mechanism by which copper nanoparticles induce toxicity has not been determined, representing a major knowledge gap. In this work, we studied the uptake and cellular response of select wild-type and mutant yeast strains to cellulose-enveloped copper nanoparticles at the single-cell level using lattice light-sheet fluorescence microscopy, a novel three-dimensional dynamical volumetric fluorescence imaging method. We hypothesized that these cellulose copper nanoparticles are attracted to the negative charge of the cell wall. Interaction with the cellular lipids and copper transporters triggers endocytosis, which leads to copper nanoparticle degradation in the vacuole and subsequent toxicity to the yeast cell. A compromised vacuole may lead to cellular damage by the release of vacuolar degradation enzymes and/or copious cupric (Cu2+) reactive oxygen species (ROS). In this work, we employed lattice light-sheet microscopy to image yeast cellular response. Four-dimensional imagery of live yeast incubated in the presence of CMC-Cu at varying concentrations was recorded over time with the LLSM, the integrated intensity per cell was analysed, presented here to assess the role of arrestins in endocytosing CMC-Cu, and their subsequent cellular toxicity. This variation of light-sheet microscopy uses orthogonal excitation and detection optics to minimize out-of-focus excitation by illuminating only a thin layer of fluorophores during imaging. The custom-built instrument uses so-called “nondiffracting” Bessel beam illumination in an optical lattice configuration. This achieves superior optical sectioning with high fluorescence collection efficiency.55,56 Several lines of evidence presented here support the hypothesis that CMC-Cu is internalized. The toxicity of CMC-Cu was greater than an equivalent amount of soluble Cu, interpreted in terms of enhanced uptake due to the cellulose overcoat. We assessed the role of select α-arrestins, in response to CMC-Cu. The α-arrestins single mutants were evaluated and roles in cell viability were classified in different categories based on how they affect copper and CMC-Cu response compared to the wild-type strain. At least five different α-arrestins were required for the acquisition of CMC-Cu and mutants increased resistance to CMC-Cu compared to wild-type yeast. Individual knockouts of ALY1, ALY2, SPO23, ROD1, and RIM8 α-arrestins reduced toxicity while loss of BUL1 increased sensitivity. The aly2 and rim8 mutants reduced the assimilation of CMC-Cu and subsequent ROS production. CMC-Cu induced diffuse ROS in the cytoplasm and vacuoles. Using LLSM, we discovered that CMC-Cu reduced dynamic cellular movement which correlates to damage to membranes. Here we have analysed ROS production by CMC-Cu providing the groundwork to couple other molecules to CMC for intercellular transport.
Methods
Yeast strains and growth conditions
All strains were from the BY4741 (his3Δ, ura3Δ, leu2Δ, met15Δ) yeast knockout collection except for art10, which was in the B4742 (his3Δ, ura3Δ, leu2Δ, lys2Δ) background. Strains were grown on yeast minimal (YM) media supplemented with the necessary nutrients to cover existing auxotrophies in yeast. Log-phase yeast were exposed to 400 μM copper sulfate or 157 μM CMC-Cu for 90 min at 30°C, then washed with water and plated onto yeast extract peptone dextrose (YPD) plates. The amounts of soluble copper and CMC-Cu were previously determined empirically by the amount necessary to cause the same reduction of growth and internal concentration of copper in a copper-sensitive strain.13 Three biological replicates for each mutant were carried out. Each trial had one to two mutants with BY4741 as the control. Eighteen biological replicates of BY4741 were averaged and compared with the mutants’ viability. A Student's t-test was conducted with two tails and type 3 in Excel. Yeast were diluted to 0.1 OD and diluted 100 000 in water and 200 μL were plated. Colony-forming units were counted after 2 days of growth at 30°C. Colony-forming units were normalized to BY4741 (wild-type) grown in YM (untreated). In serial dilution growth assays, yeast were grown overnight, diluted to 0.1 OD, and then diluted 10-fold. Approximately 5 μL was spotted on YM plates (yeast minimal media with dextrose supplemented with histidine, uracil, leucine, methionine, and lysine) with and without 1 mM soluble copper. Plates were incubated at 30°C for 3 days and then photographed.
Staining of nanoparticles
CMC-Cu were stained using fluorescein isothiocyanate (FITC) (ThermoFisher-A1363) or AF568 Hydrazine (ThermoFisher-A104371). A total of 2.5 μM FITC or 1 μg/mL AF568 was added into a CMC-Cu solution and then vortexed before incubating for overnight at 4°C in a rotisserie. Log-phase yeast cells were centrifuged at 3000 rpm for 5 min, the supernatant was removed, and the yeast was diluted with 1 mL of phosphate buffered saline (PBS). Yeast cells were diluted into an OD600 of 0.1 and incubated with dye only (FITC or AF568) and stained CMC-Cu with FITC or AF568 for 90 min. Cells were excited at 488 nm for FITC and 560 nm for AF568 nm with an exposure time of 50 ms and scanned over a volume consisting of 101 slices, with intervals of 500 nm between planes. The scale is 5 μm.
LLSM
Yeast cells were immobilized on coated coverslips using 0.25 mg/mL concanavalin A for 5 min and washed with sterilized water twice. A total of 20 μL of yeast suspension was dropped onto a 5 mm diameter coverslip (place a coverslip on top of parafilm to create a hydrophobic environment outside the coverslip). Cells were scanned over a volume consisting of 101 slices, with intervals of 500 nm between planes. To measure endocytosis, cells were exposed for 50 ms, and excited at 50 mW of 488 nm and 560 nm for yeast cell or CMC-Cu stained with FITC or AF568, respectively. In LLSM the detection objective image plane is tilted to the axis of sample movement, as a result, it is required to deskew the images in postprocessing, this can be done in image J,57 by using the TransformJ—Affine command (αxz = (Δz/Δxy)*sin 58.2o) and subsequently scaling the image with the TransfomJ—Scale command (Z = (Δz/Δxy)*cos 58.2o). Image intensity is measured using Image J by removing the background and used maximum intensity projection (MIP) projection to draw ROI and use sum intensity projection to calculate the intensity. The design and component specifications for the instrument, patented as the “Bessel Beam Microscope” by Eric Betzig of Howard Hughes Medical Institute (HHMI), were made available to South Dakota School of Mines and Technology through a license agreement with HHMI.58 The resulting imaging system provides optical sectioning properties superior to commercially available light sheet microscopes.56
Reactive oxygen species (ROS) intensity was measured using Image J. The first step was to draw a region of interests (ROIs) using MIP image at channel 488 nm. Yeast cells were segmented using standard thresholding techniques (Image → Threshold) then ROIs were created using analyse particles (Analyse → Analyse particles). The ROS was detected for images at channel 642 nm, background intensity on the images was removed (Process → Math → Subtract), the Z slices were summed (sum intensity projection image), and ROIs were applied to the sum image at channel 642 nm to calculate the ROS intensity (Analyse measure).
Volumetric image analysis
MIPs were rendered from the 4D data using ImageJ. Cell centroid tracking and membrane velocity analysis were performed using in-house analysis routines in Chimera X.
ROS detection using CellROx deep red (ThermoFisher, #C10422)
CellROx is a cell-permiable dye with absorption/emission maxima of ∼644/665 nm. Control (untreated) and treated yeast sample with GFP conjugate CMC-Cu were prepared and incubated for 30 min with CMC-Cu then added 10 μM CellROx and continued incubation for another 60 min at room temperature using the rotisserie. After incubation cells were washed with PBS three times. Cells were plated in a 5 mm coverslip that was coated with ConA overnight and incubated for 5–10 min before imaging. Cells were imaged at 488 nm to detect the CMC-Cu uptake and 640 nm to detect the ROS.
Confocal microscopy
Slide preparation: Glass microscope slides were prepared for confocal microscopy by coating in concanavalin A. The concanavalin A solution of 12.5 mg/L was prepared using MilliQ (MQ) water on a magnetic stir plate for 4 h before vacuum filter sterilizing with a 220 nm filter membrane. Glass slides were cleaned with 100% ethanol, lint-free cloth, and ethanol-cleaned gloves. Clean slides were dipped five times in concanavalin A solution before submerging for 2 min and then allowing to air dry in a fume hood for 1 h. Dry slides were dipped five times in MQ water before a final rinse in a separate MQ aliquot and sequential drying for 1 h.
Sample preparation: FM464 was used to visualize live cell vesicle endocytosis to the vacuole by transferring mid-log phase BY4741 to 1.5 Eppendorf tubes. After pelleting cells at room temperature, the cells were resuspended in 50 μL YPD + 1.6 μL of FM464 at 10 mM in DMSO (dimethyl sulfoxide). Cells were incubated in the YPD + dye for 1 h at 30°C in a water bath. The cells were washed in YPD and incubated in 5 ml for 90 min. Dyed cells were then washed in MQ water before resuspending in 25 μL YM media for transfer to concanavalin A-coated slides. A Nikon A1R/NSIM-E microscope with the 60× objective was used to capture images. The images were analysed using Nikon General Analysis to obtain mean intensity measurements for each yeast cell.
Statistical analysis: In this analysis, violin plots were generated using GraphPad Prism version 8 which shows the frequency distribution of data and plot lines at the median and quartiles. (Unpaired t) test and one-way ANOVA (Tukey test) were used to measure the statistical analysis of two groups and more than two groups of data, respectively. P-value higher than 0.05 means the value is not significant (ns), and P-value less than or equal to ≤0.05, ≤0.01, ≤0.001, and ≤0.0001 mean significant difference that corresponds to *, **, ***, and ****, respectively.
Results
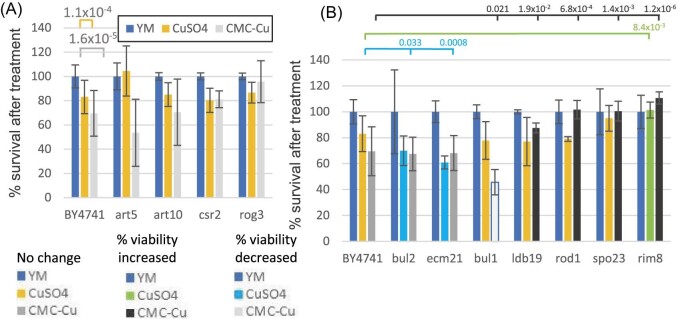
To determine which α-arrestins are responsible for regulating the import of CMC-Cu into the cell, we evaluated the viability of yeast with each of the 11 α-arrestin knockouts. Previously, we had empirically demonstrated that CMC-Cu are more efficient at transporting more copper into yeast than the equivalent amount of soluble copper. When yeast were exposed to 157 μM of CMC-Cu, viability and internal concentrations of copper were equal to yeast that were exposed to 400 μM of CuSO4.13 When exposed to soluble copper, the loss of aly1 reduced viability to 60%, and the aly2 mutant had 50% viability compared to 85% of wild-type.13 When exposed to CMC-Cu, both aly1 and aly2 mutants had no reduction of viability in contrast to wild-type cells, which decreased to 70%.13 The response to soluble copper and CMC-Cu of other mutants were separated into categories. The first class had no statistical significant difference in reduction in cellular viability to wild-type cells after 90 min exposure to soluble copper or CMC-Cu and included art5, art10, csr2, ldb19, and rog3 (Fig. 1A). The second class contained mutants that only had an altered response to soluble copper (Fig. 1B). In this class, the bul1 and ecm1 mutants further reduced viability to 70% and 60%, respectively, but the response to CMC-Cu did not change from wild-type cells. In the third class, the bul1 mutant's viability in CMC -Cu exposure decreased to 46% and viability in soluble copper did not change. In the fourth class, rod1 and spo23 were completely resistant to CMC-Cu (101% and 100% viability, respectively) and sensitivity to soluble copper did not change. The last group only contained rim8 mutant, which was completely resistant to both soluble copper and CMC-Cu at these concentrations. The rim8 mutant has numerous targets that are distinct from aly1 and aly2 mutants that rescued viability from CMC-Cu exposure but made the cells more sensitive to soluble copper and represent a fifth class of α-arrestins phenotypes.
Fig. 1.
Viability of α-arrestin mutants treated with soluble copper or CMC-Cu. Mutants were grown to log-phase and then exposed to 400 μM soluble copper (orange) or 157 μM CMC-Cu (gray) for 90 min. Colony-forming units were normalized to BY4741 (wild-type) untreated in YM media (blue) and graphed as a percentage of viability. Mutants were then classified as to whether the response was different from wild-type. A. Mutants that had the same response as BY4741 to CuSO4 and CMC-Cu. B. Mutants that had changes in their copper or CMC-Cu viability. The direction of change was noted with colors increased copper viability (green), decreased copper viability (aqua), increased CMC-Cu viability (black), and decreased CMC-Cu viability (gray). The P-value of the change is noted above.
The α-arrestins are involved in remodeling plasma membranes by targeting a wide range of proteins including amino acid permeases.43 Structurally related, after the whole genome-wide duplication, some α-arrestins retained their paralog. Aly1 and Aly2 are 42% identical while Rod1 and Rog3 are 46% identical at the protein level (Table 1). The rod1 knockout was CMC-Cu resistant while the rog3 knockout was not statistically significantly resistant (P-value = 0.1); however, the emc21 knockout was more copper sensitive while its paralog csr2 was not. Aly1, Aly2, Rog3, Rod1, Ecm21, and Csr2 have the most sequence similarity shared between them. Ecm21 and its paralog Csr2 are 45% identical/66% similar and these mutants do not affect CMC-Cu viability. The bul1 mutation lowered the resistance to CMC-Cu while the mutant of its paralog bul2 conferred increased sensitivity to CMC-Cu and these paralogs shared 53% sequence identity. The growth of serially diluted yeast cultures on solid media measures the response to chronic exposure to the presence of a chemical. Because we wanted to measure the increase in resistance to soluble copper, 2.5 times more copper was used in growth assays because more copper is needed in the soluble form than CMC-Cu to affect growth.13 The growth of α-arrestin mutants was measured in the presence of 1 mM soluble copper (Supplementary Fig. S1). Yeast were diluted 10-fold and allowed to grow on solid media. The ldb19, rim8, bul1, and ecm21 mutants were copper sensitive while art10 and spo23 mutants were more resistant to copper. Of note, at this high level of copper, the cells turned a rust color due to the met15 marker present in the BY4741 background. The art10 mutant in the BY4742 background has a wild-type allele of MET15 and is a mutant for lys2, which provides evidence for the lack of color in their colonies. The mutant viability with soluble copper did not appear to correlate with their chronic response to copper. However, the BY4741 strain contains 14 copies of CUP1 that encodes the major copper metallothionein and confers copper resistance compared to other yeast strains59 but the copper concentration in the media used here is limiting for growth. Adding 50 μM of additional copper sulfate improves the growth of BY4741.13 Serial growth assays are most suited to measuring large differences in growth on the order of 2- to 1000-fold differences over days.
Table 1.
Summary of α-arrestin mutant viability to 90 min exposure to soluble copper or CMC-Cu compared to wild-type
| Name | Alias | Soluble copper | CMC-Cu | Paralog | Percentage identity/similarity/gaps | Source |
|---|---|---|---|---|---|---|
| ALY1 | ART6 | Down | Up | ALY2 | 42%/58%/13% | Rong-Mullins et al.13 |
| ALY2 | ART3 | Down | Up | ALY1 | 42%/58%/13% | Rong-Mullins et al.13 |
| ART5 | ART5 | Up | None | |||
| ART10 | None | None | ||||
| CSR2 | ART8 | None | None | ECM21 | 45%/66%/11% | |
| ROG3 | ART7 | None | None | ROD1 | 46% 63%/11% | |
| BUL2 | Down | None | BUL1 | 53%/69%/5% | ||
| ECM21 | ART2 | Down | None | CSR2 | 45%/66%/11% | |
| BUL1 | None | Down | BUL2 | 53%/69%/5% | ||
| LDB19 | ART1 | None | Up | |||
| ROD1 | ART4 | None | Up | ROG3 | 46% 63%/11% | |
| SPO23 | None | Up | ||||
| RIM8 | ART9 | Up | Up |
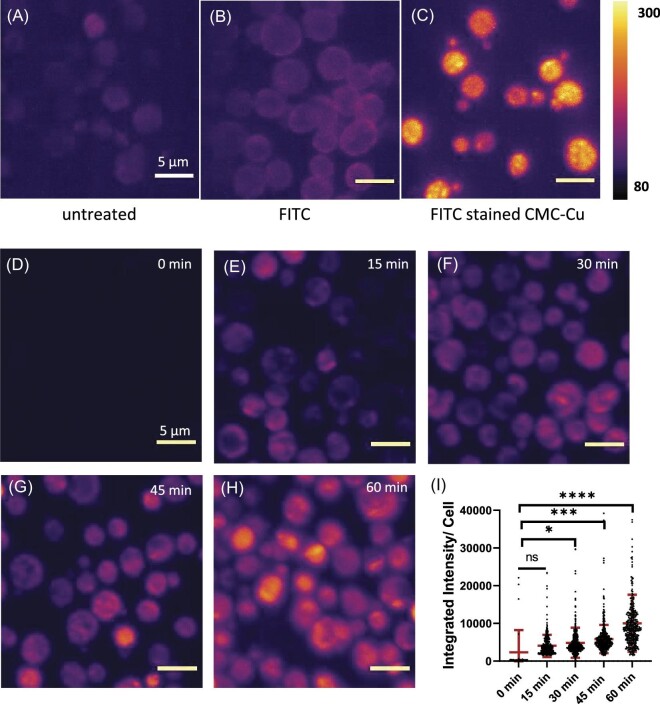
To determine if the addition of copper changed the internalization of nanoparticles, CMC-Cu were stained with FITC and incubated with BY4741. Previously, CMC-Cu internalization was measured by prestaining the nanoparticle with FITC then the internalization was determined by flow cytometry and confocal microscopy.13 Unstained yeast were visible because at 488 nm there was weak autofluorescence (Fig. 2A) and yeast incubated with FITC alone showed increased staining of the cellular periphery (Fig. 2B). Only when yeast incubated with FITC-stained CMC-Cu did internal staining appear (Fig. 2C). To assess if the CMC itself can be internalized, both CMC and CMC-Cu were stained with AF568 hydrazine instead of FITC because of the weak autofluorescence. Cells measured at 568 nm do not have autofluorescence (Fig. 2D). Yeast internalized more CMC alone (Supplementary Fig. S2A) than CMC-Cu (Supplementary Fig. S2B). The integrated intensity of CMC-Cu was 0.65 counts/s for CMC and 0.42 counts/s CMC-Cu (Supplementary Fig. S2C). Next, we determined the kinetics of CMC-Cu internalization over 60 min. Within 30 min of CMC-Cu exposure, there was statistically significant CMC-Cus internalization increasing over the next hour (Fig. 2D–I). The average fluorescence was measured for 400 yeast. Over the time course, the staining begins in the cytoplasm with the stained nanoparticles excluded from what appeared to be a vacuole. By 30 min some cells start to have stained CMC-Cu throughout the cell, which increases over time. By 45 min, the staining intensity increased so by 60 min yeast were brighter and there are cells with areas of the cell brighter than other parts.
Fig. 2.
Maximum intensity projection of yeast A. BY4741 without treatment B. BY4741 were incubated with 2.5 μM FITC C. BY4741 were incubated 2.5 μM FITC stained CMC-Cu. D. BY4741 treated with CMC E. BY4741 treated with CMC-Cu, both CMC and CMC-Cu were stained by AF568 Hydrazine overnight, further BY4741 cells were incubated with CMC and CMC-Cu for 90 min. F. Violin plot of integrated intensity per cell for CMC (E) vs. CMC-Cu (D)treated yeast, using unpaired t-test with P-value **** ≤0.0001. AF568 stained CMC-Cu were incubated with BY4741 for G. 0 min, H. 15 min, I. 30 min, J. 45 min, and K. 60 min. L. The plot for integrated intensity per cell from 0 to 60 min. In this analysis, one-way ANOVA was used for the analysis with P-value higher than 0.05 means the value is not significant (ns), P-value less than or equal to ≤0.05, ≤0.01, ≤0.001, and ≤0.0001 mean significant difference which corresponds to *, **, ***, and ****, respectively.
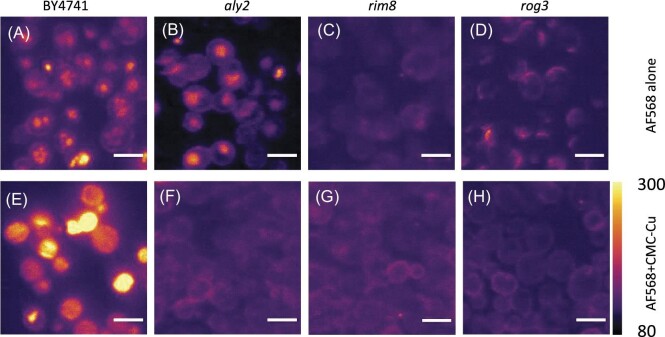
We selected three α-arrestin mutants to determine their role in endocytosis and CMC-Cu transport. The viability of aly2 mutant is sensitive to soluble copper and resistant to CMC-Cu.13 The rim8 mutant demonstrated increased viability when exposed to soluble copper and CMC-Cu. The rog3 mutant had higher resistance to soluble copper and CMC-Cu exposure but it was not statistically significantly different from wild-type viability and growth did not change under chronic copper exposure. We measured the average fluorescence for these strains incubated with AF568-stained CMC-Cu (Fig. 3). BY4741 and the aly2 mutant internalized the dye in what appeared to be vacuoles (Fig. 3A and B). The rim8 and rog3 mutants did not have increased internal staining (Fig. 3C and D). Yeast were incubated with CMC-Cu prestained with AF568 and the internalization was measured in BY4741 and the α-arrestin mutants. Incubation with prestained CMC-Cu caused bright emission from BY4741 (Fig. 3E) while the CMC-Cu-resistant mutants did not show CMC-Cu fluorescence (Fig. 3F–H). These results suggest that the mutants reduced endocytosis of CMC-Cu and therefore, the CMC-Cu nanoparticles are not internalized.
Fig. 3.
Maximum intensity projection of yeast treated with AF568 alone and stained CMC-Cu A. BY4741 with AF568 alone. B. aly2 mutant with AF568 alone. C. rim8 mutant with AF568 alone. D. rog3 mutant with AF568 alone. E. BY4741 with AF568 stained CMC-Cu. F. aly2 mutant with AF568 stained CMC-Cu. G. rim8 mutant with AF568 stained CMC-Cu. H. rog3 mutant with AF568 stained CMC-Cu.
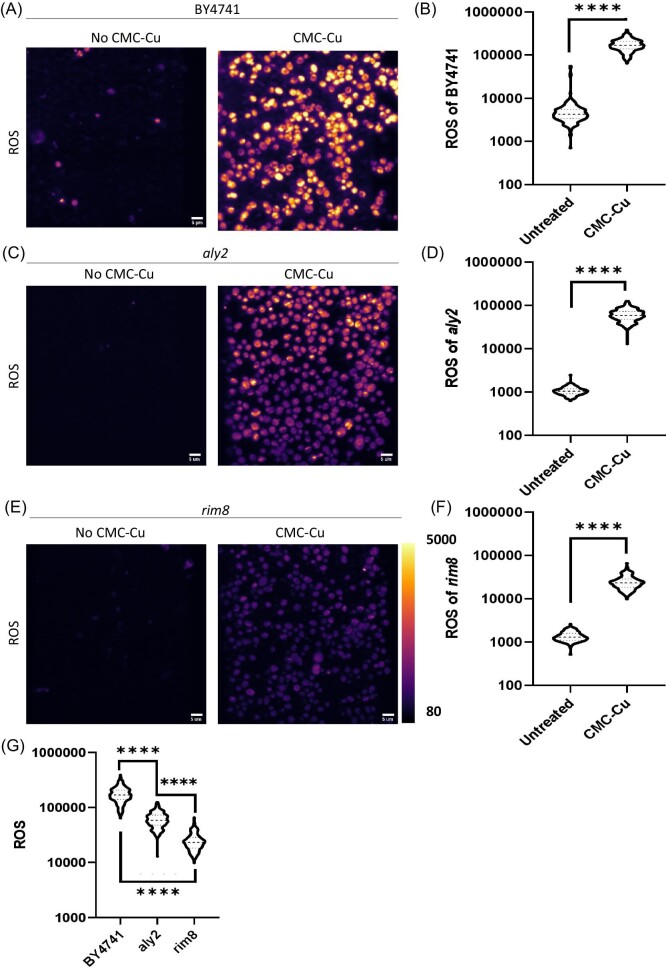
Treatment of yeast with antioxidants reduces the toxicity of CMC-Cu.13 Like soluble copper, CMC-Cu generates ROS and primarily damages lipids.60 To measure the internal levels of ROS, yeast were stained with CellROx (Fig. 4A). With no treatment, there was a low level of ROS consistent with endogenous ROS was generated primarily due to protons leaking from the mitochondria that increase as cells age (reviewed in Zhao et al.61). Autofluorescence was used to visualize cells with α-arrestin mutants that were treated with CMC or CMC-Cu. The ROS levels in aly2 mutant were lower than in wild-type yeast (Fig. 4C) as well as in rim8 mutant (Fig. 4E). Directly comparing ROS between mutants shows that rim8 mutants had less ROS than the aly2 mutant and wild-type yeast (Fig. 4G).
Fig. 4.
ROS of untreated vs. CMC-Cu treated of BY4741, aly2, and rim8 and intensity plot of ROS for untreated vs. CMC-Cu. All cells (BY4741, aly2, rim8) were incubated with CMC-Cu for 30 min and CellROx to measure ROS production. A. BY4741 untreated and CMC-Cu treated, B. Quantification of ROS in BY4741 CMC-Cu treated cells, C. aly2 mutant untreated and CMC-Cu treated, D. Quantification of ROS in aly2 mutant CMC-Cu treated cells, E. rim8 mutant untreated and CMC-Cu treated, F. Quantification of ROS in rim8 mutant CMC-Cu treated cells, G. ROS generated by CMC-Cu exposure of BY4741, aly2, and rim8 presented in the same graph. In this analysis, (Unpaired t) test was used for the analysis of Fig. B, D, and F, and one-way ANNOVA was used for the analysis of Fig. G with P-value higher than 0.05 means the value is not significant (ns), P-value less than or equal to ≤0.05, ≤0.01, ≤0.001, and ≤0.0001 mean significant difference which corresponds to *, **, ***, and ****, respectively.
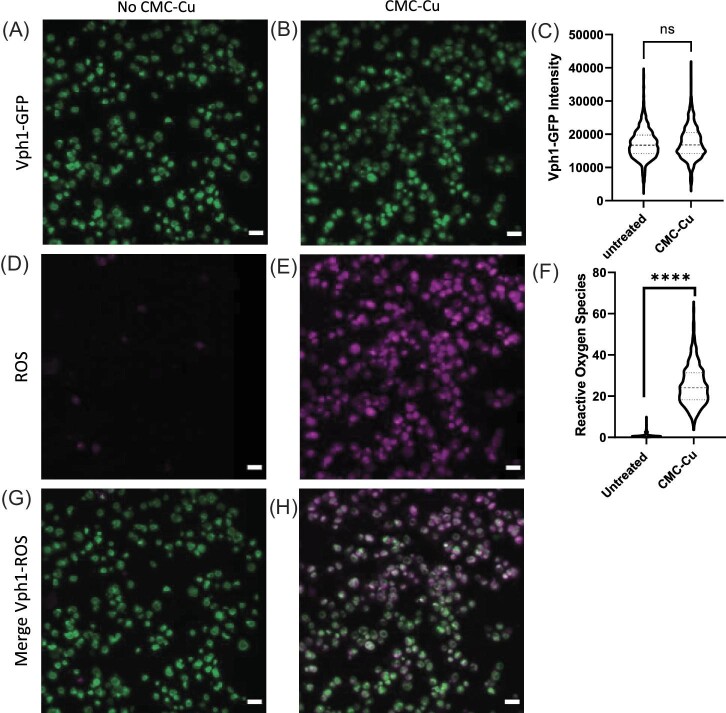
Because internalization by α-arrestins brings the cargo to the vacuole and vacuoles are acidified, vacuolar localization of CMC-Cu could generate ROS in the vacuole. We used Vph1-GFP in BY4741 to label the membrane of the vacuole (Supplementary Fig. S3). Vph1 is a transmembrane protein in the vacuole and inserts into the membrane.62 The GFP tag in Vph1 faces the cytoplasm and GFP is a pH-sensitive fluorescent protein.63 Vph1-GFP colocalizes with the vacuolar dye FM4-64 (Supplementary Fig. S3). We quantified the colocation of ROS production with CMC-Cu exposure (Fig. 5). After 90 min of CMC-Cu exposure (Fig. 5A and B), Vph1-GFP levels were unchanged (Fig. 5C). Untreated cells had nearly no detectable ROS (Fig. 5D). ROS production was diffused in the cytoplasm after 90 min of CMC-Cu exposure (Fig. 5E) and over 20-fold higher (Fig. 5F). Then, we measured the amount of colocalization between Vph1 and ROS and there was little overlap (Fig. 5G and H). To better visualize any possible overall localization, we rendered 3D images (Supplementary Fig. S4) and slice by slice images (Supplementary Fig. S5) diffuse overlap between the vacuole and ROS. By sorting through the slices, we noted that some cells had prominent ROS signal surrounding the vacuole, while mostly being diffuse in the cytoplasm (Supplementary Fig. S5). To measure the dynamics of Vph1-GFP, cells were mounted on the LLSM imaging chamber and scanned (Supplementary Fig. S4B). Vacuoles are dynamic organelles and in untreated cells, they moved around inside the cell during the 26 min movie. The cells were then exposed to CMC-Cu and then rescanned. After CMC-Cu treatment, all loss Vph1-GFP was lost in frame 30 about 4 min after scanning (Supplementary Fig. S4B). All previous experiments were conducted after 90 min of exposure and 90 min the Vph1-GFP levels were the same as untreated. However, after 4 min of exposure Vph1-GFP levels diminished only to recover sometime between 26 and 90 min. We concluded that the CMC-Cu quenched the GFP signaling possibly by the acidification of the cytoplasm induced by CMC-Cu but then the pH of the cytoplasm returned to normal by 90 min.
Fig. 5.
Colocalization of vacuole and ROS production when yeast were treated with CMC-Cu. Cells were incubated with CMC-Cu for 30 min and added with 10 μM CellROx deep red and continuously incubated for another 60 min. Cells were imaged at 488 nm to image CMC-Cu and at 640 nm to detect reactive oxygen species. A. Vph1-GFP in green when untreated. B. Vph1-GFP in green in CMC-Cu treated yeast (90 min) C. Quantification of Vph1 intensity for untreated vs. CMC-Cu from panel A and B. D. ROS with CellROx staining in purple when untreated, E. ROS staining in purple when treated. F. Quantitation of ROS from panel C and D. G. Merged of ROS and Vph1-GFP without treatment, H. Merged of ROS and Vph1-GFP when exposed to CMC-Cu. Unpaired t-test P-value less than or equal to ≤0.0001 mean significant difference that corresponds to ****.
To overcome the likely pH-dependent quenching of GFP to localize ROS production relative to the vacuole during short exposures, we analysed the effect of CMC-Cu on the yeast vacuole using FM4-64 staining. BY4741 and aly2 mutants were stained with FM4-64 for 30 min then imaged after 75 min to stain the vacuole instead of the membrane (Fig. 6). First cells were imaged without any treatment for 100 frames (4.56 s/frame with a total scanning time of 6.3 min) as a control sample/no treatment. Then cells at different regions of interest (ROI) were chosen and CMC-Cu was added to the chamber. Cells were imaged in three movies of 25 min intervals (4.56 s/frame 200 frames) at a different region of interest to prevent photobleaching. The cells appeared very active before treatment and less dynamic after CMC-Cu treatment. We hypothesize that cellular metabolism is reduced upon generation of ROS after exposure to CMC-Cu and subsequent internalization and breakdown of the CMC-Cu nanoparticles. To test this hypothesis we quantified the motion of the cells themselves and the dynamical deformations of the cell membranes by tracking the cell centroids and the membrane motion/velocity of each cell using the four-dimensional image analysis package Chimera X. We calculated the centroid positions of each cell and tracked these in time to quantify locomotion of each cell. We then computed the change in membrane velocity across the membrane of each cell. The measurements are limited by the choice of the threshold value and the signal-to-noise ratio of the data. No discernable difference in centroid tracking was observed before and after exposure to CMC-Cu (average displacement in the range of 2.5–3 pixels or 275–330 nm), leading us to conclude that any apparent locomotion of the cells is independent of exposure to CMC-Cu and is possibly an artifact of the sample scanning during the LLSM imaging. Similarly, our analysis of the membrane deformation showed localized membrane deformations in a punctate or “pocked” arrangement in subcellular detail (deformation ranging from 0 to 275 nm), shown in false color images (Supplementary Fig. S7), but the frequency and amplitude of these deformations did not appear to depend on exposure to CMC-Cu. However, the calculation is highly dependent on the choice of the threshold value, which in turn is limited by the signal-to-noise ratio, as it affects the ability to form contiguous isosurfaces upon which the calculation is dependent. We posit that the motion of the vacuoles in the interior of the cell is affected by exposure to CMC-Cu, but due to low signal to noise, we are unable to track this motion for the obtained data sets. Future experiments with a better signal-to-noise ratio may reveal more quantifiable data that could quantify this motion and more robustly address our hypothesis. The level of detail and the combination of spatial and temporal resolution obtained with the LLSM here uniquely affords these observations. Over time, the intensity of Vph1 was also reduced/photobleached. There was diffuse staining within the cytoplasm and in the vacuole, supporting the colocalization of Vph1 with ROS.
Fig. 6.
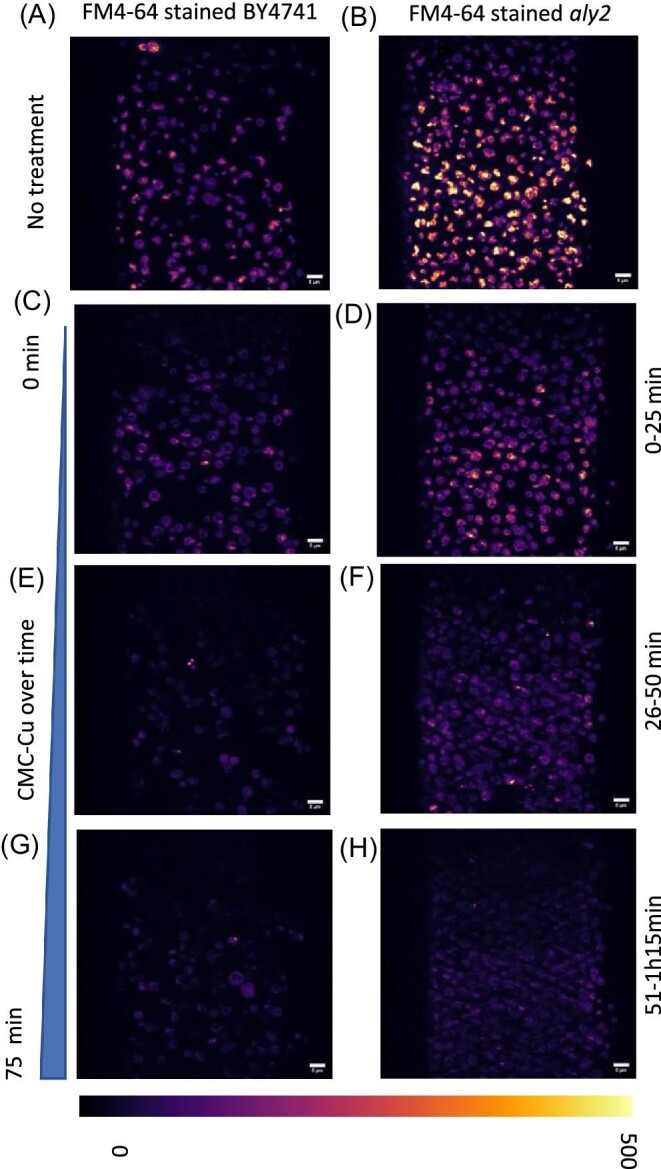
Visualization of vacuolar dynamics with CMC-Cu exposure. Yeast were stained with FM4-64 and exposed to CMC-Cu. Cells were stained with FM4-64 for 30 min then imaged for 75 min track. Cells were imaged before CMC-Cu treatment as a control sample/no treatment then CMC-Cu were added to the imaging chamber. Cells were imaged in three movies of 25 min (4.56 s/frame 200 frames) intervals at a different region of interest to prevent photobleaching. A. BY4741 untreated, B. aly2 untreated, C. BY4741 exposed to CMC-Cu for 0–25 min, D. aly2 exposed to CMC-Cu for 0–25 min, E. BY4741 exposed to CMC-Cu for 26–50 min, F. aly2 exposed to CMC-Cu for 26–50 min, G. BY4741 exposed to CMC-Cu for 51–75 min, H. aly2 exposed to CMC-Cu for 51–7.5 min.
While the loss of one α-arrestin rescued a reduction in viability from acute CMC-Cu exposure, the increased ROS levels were still detected in a single α-arrestin mutant. We attempted to evolve CMC-Cu-resistant yeast by serial passaging BY4741 in the presence of CMC-Cu. However, in over 15 passages only slight resistance to CMC-Cu was noted in one of the six independent passages (data not shown). Genomic sequencing of one strain found mutations in RRG8, BPT1, and YBL081W in the CMC-Cu passaged yeast and not in the untreated yeast. RRG8 is required for mitochondrial respiration.64 Loss of YBL081w decreases plasma membrane electron transport.65 BPT1 encodes a vacuolar ABC transmembrane transporter involved in heavy metal detoxification.66,67 Knockouts of these genes were individually tested assuming that most mutations are deactivating mutations. While rrg8 knockouts had increased CMC-Cu resistance, ybl081w and bpt1 knockouts had decreased resistance (Supplementary Fig. S7). Notably, no mutations in the α-arrestins were detected.
Discussion
The biodegradability and efficient production of antimicrobial materials are important to combat infectious diseases. How living organisms interact with these materials provides insight into transport mechanisms. Mutations in these pathways could lead to antimicrobial resistance. We showed that BY4741 did not internalize FITC or AF568H dyes alone, but CMC and CMC-Cu-stained nanoparticles are internalized by BY4741. S. cerevisiae like other eukaryotes have α-arrestins that provide specificity to target the internalization and trafficking of cargos on and in the cell membrane. Spo23, Rod1, Rog3, Ldb1, Rim8, Aly1, and Aly2 are key adaptors for the import of CMC-Cu. This leads us to conclude that yeast are indeed importing the CMC-Cu nanoparticles and ROS production occurs primarily in the cytoplasm. The CMC-Cu reduced the pH of the cytoplasm, quenching GFP while inducing ROS during the initial exposure leading to peroxidation of lipids.
At least seven α-arrestins serve as adaptors for CMC-Cu import. There are four pairs of α-arrestins paralogs retained from the whole genome duplication in S. cerevisiae. As genomes evolve, one duplicated gene can be lost. However, in a subset of paralogs, one copy is retained and the paralogs can divide the functions of the ancestral protein between the paralogs known as subfunctionalization, or neofunctionalization that results in a new function in one paralog while the other retains the ancestral functions.68,69 Each α-arrestin targets multiple overlapping proteins but arrestins are known for the target that defines their characterization. Aly1 and Aly2 are responsible for the endocytosis of Dip5 during aspartic and glutamic acid excess.70 Rog3 downregulates Ste2 to desensitize to α-factor when cells fail to mate71 and is upregulated in an atx1 knockout when exposed to soluble copper.72 Emc21 and Bul1 downregulate amino acid transporters.73,74 Bul1 and Bul2 serve as adapters for Ctr1 during copper exposure35; however, the mutants did not have the viability to acute exposure to soluble copper but bul1 showed increased sensitivity to chronic exposure and acute exposure to CMC-Cu. Ldb19 also regulated Ste2.71 Unlike other mutants, bul1 mutants were more sensitive to CMC-Cu. Loss of Rim8 confers both resistance to cadmium, nickel, zinc, and sensitivity to arsenic to numerous metals including cadmium.75 While some α-arrestin have known roles in metal metabolism, others have roles in regulating nutrient transporters in response to changes in the availability of nutrients. There was no single α-arrestin target that is known to be common among the seven α-arrestin that have a role in CMC-Cu import.
The α-arrestins serve as adapters for Rps5, the Ub ligase that recognizes the PPXY motif ([L/V/P]xY) in over 240 plasma membrane proteins.76 Plasma membrane proteins that do not have a PPXY motif use α-arrestins which have PY motifs as adaptors.45 The functions of α-arrestins are regulated by other proteins such as Rho1 and the GDP/GTP exchange protein Rom1.46 The α-arrestins have nonredundant cargos and directly interact with the cytoplasmic portions of plasma membrane proteins. We propose that Aly1, Aly2, Rod1, Rog3, Rim8, and Spo23 trigger endocytosis of CMC particles as these mutants were CMC-Cu resistant and aly2, rim8, and rod1 mutants had fewer CMC internalized. Both CMC and CMC-Cu were internalized so the positive charge from the copper is not likely contributing to endocytosis. It is possible that CMC directly interact with an α-arrestin target(s); however, when screening the literature we could not find one target that was common in the mutants that increased CMC-Cu resistance. α-arrestin induce conformational shifts of its target through electrostatic interaction exposing acidic patches (reviewed in Kahlhofer et al.77). CMC nanoparticles may interact with multiple targets through their positive charge that are subject to α-arrestin-dependent internalization or induce internalization by interacting with the membrane directly.
Once inside the cell, at least some of the CMC-Cu are trafficked to the surface of the vacuole. Intracellular trafficking moves vesicles to and away from the membrane. The vacuole divides and grows correlated to the rate of growth. These subcellular structures lost their dynamic movement when exposed to CMC-Cu. Imaging live cells as they were exposed to CMC-Cu showed that cells became less dynamic, but due to signal-to-noise issues, we did not quantitate it. CMC-Cu damage lipids,60 which could reduce the movement of the membranes. Other studies have shown that when cells are stressed by starvation, internal organelle movement becomes less dynamic based on pH.78 While substantial CMC-Cu resistance could not be evolved in the lab, a few mutants of note were detected, and individual knockouts of these genes did change the growth in the presence of CMCs. BPT1 encodes a protein that affects metal metabolism in the vacuole and RRG8 encodes a protein important for mitochondrial function. During normal growth, the mitochondria are a major source of ROS as electrons occasionally leak from oxidative phosphorylation. As cells age or are damaged, the electron potential decreases and is a potent trigger for apoptosis. While mitochondria generate energy through ATPs produced by the tricarboxylic acid (TCA) cycle, it comes at a cost. The intermediates of the TCA cycle also serve as precursors for amino acids and other metabolites necessary for growth. Cells with lower electron potential can also change how drugs are metabolized. For example, 4-nitroquinoline-1-oxide is reduced more in cells with more electron potential generating more ROS, and thus these cells are more sensitive to the drug than yeast that have mitochondria biased toward amino acid production.79,80 The ROS from CMC-Cu damages lipids primarily at the mitochondria,60 which may be the mechanism triggering cell death. Other types of copper nanoparticles may interact with different pathways and have different mechanisms to induce endocytosis.8
Conclusion
Genetic variation in response to soluble copper correlated with CMC-Cu response13 and no variation has been noted specifically for nanoparticles. Genetic variation within a species contributes to variation in phenotype but some genetic variation has a more prominent impact than others. Master variators are polymorphic proteins in a species that have large effects on phenotypes, and small differences in transcription factors can have more pronounced effects.79 RoundUp-resistant yeast were isolated after as little as one passage and after six passages and RoundUp-resistant yeast had hundreds of mutations that contributed to the resistance.81 Similar results were found when yeast were exposed to MCHM (4-methylcyclohexanemethanol), a synthetic hydrotrope.82,83 Given that after multiple attempts no substantial increased resistance to CMC-Cu could be found, cellulosic copper nanoparticles are a promising antimicrobial material.
Supplementary Material
Acknowledgements
Marjin Ford shared the Vph1-GFP labeled strain. Gloria Oporto provided CMC-Cu. Victoria Bovard, Nicholas Miller, Shayla Koerner, Zach Lynn, Cassie Plishka, and Ashley Noah attempted to evolve CMC-Cu-resistant yeast in Genomics Capstone class funded by the WVU Department of Biology. The authors acknowledge funding from the State of South Dakota through the Board of Regents Collaborative grant IMAGEN: Biomaterials in South Dakota. The lattice light sheet microscope was developed under a license agreement with Howard Hughes Medical Institute, Janeila Farms. The authors acknowledge the assistance of Dr Brandon Scott in the experiments and computational analysis of the Lattice Light Sheet images and Michael DiBacco for critical reading of the manuscript.
Contributor Information
Ni Putu Dewi Nurmalasari, Department of Nanoscience & Biomedical Engineering, South Dakota School of Mines and Technology, Rapid City, SD, USA.
Matthew J Winans, Department of Biology, West Virginia University, Morgantown, WV, USA.
Katelyn Perroz, Department of Biology, West Virginia University, Morgantown, WV, USA.
Victoria R Bovard, Department of Biology, West Virginia University, Morgantown, WV, USA.
Robert Anderson, Department of Nanoscience & Biomedical Engineering, South Dakota School of Mines and Technology, Rapid City, SD, USA.
Steve Smith, Department of Nanoscience & Biomedical Engineering, South Dakota School of Mines and Technology, Rapid City, SD, USA.
Jennifer E G Gallagher, Department of Biology, West Virginia University, Morgantown, WV, USA.
Funding
M.J.W. was supported by the National Science Foundation's Integrative Graduate Education and Research Traineeship: Research and Education in Nanotoxicology at West Virginia University (IGERT: REN@WVU) award number 1144676. K.P. was supported by National Science Foundation Research Experience for Undergraduates funding (DMR1559880). We want to acknowledge the WVU Genomics Core Facility, Morgantown WV for the support provided to help make this publication possible and National Institutes of Health #U54 GM104942 grant to the West Virginia Clinical and Translational Science Institute provided financial support to the West Virginia University Genomics Facility, which in turn provides financial support to the Core Facility.
Authors contributions
N.P.D.N. and M.J.W. conducted the LLSM experiments. N.P.D.N. also carried out the statistical analysis of data. M.J.W. contributed to the conception of the experiments, verified the dyes and grew the cells. K.P. carried out the CFU experiments. V.R.B. carried out the In-Lab evolutions. R.A. assisted in the LLSM experiments. S.S. directed the work at SDSMT. J.E.G.G. directed the work and wrote the paper with assistance from M.J.W. and S.S.
Conflicts of interest
The authors declare no conflicts of interest.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Habibi Y., Lucia L. A., Rojas O. J., Cellulose nanocrystals: chemistry, self-assembly, and applications, Chem. Rev., 2010, 110 (6), 3479–3500. 10.1021/cr900339w [DOI] [PubMed] [Google Scholar]
- 2. Padalkar S., Capadona J. R., Rowan S. J., Weder C., Won Y. -. H., Stanciu L. A., Moon R. J., Natural biopolymers: novel templates for the synthesis of nanostructures, Langmuir, 2010, 26 (11), 8497–8502. 10.1021/la904439p [DOI] [PubMed] [Google Scholar]
- 3. Shi Z., Zang S., Jiang F., Huang L., Lu D., Ma Y., Yang G., In Situ nano-assembly of bacterial cellulose-polyaniline composites, RSC Adv., 2012, 2 (3), 1040–1046. 10.1039/c1ra00719j [DOI] [Google Scholar]
- 4. Zhong T., Oporto G. S., Armstrong J., Jaczynski J., Tesfai A. T., Antimicrobial properties of the hybrid copper nanoparticles-carboxymethyl cellulose, Wood Fiber Sci, 2013, 45, 215–222. [Google Scholar]
- 5. Cioffi N., Ditaranto N., Torsi L., Sabbatini L., Approaches to synthesis and characterization of spherical and anisotropic copper nanomaterials, Nanotechnol. Life Sci., 2010, 1, 3–69. [Google Scholar]
- 6. Owens C. W., Oporto G. S., Söderberg B. C. G., Lambson K. E., Lignocellulosic micro- and nanomaterials as copper frames for the evaluation of the Copper(I)-catalyzed azide-alkyne cycloaddition, J. Nanomater., 2017, 2017, 1–6. 10.1155/2017/9461615 [DOI] [Google Scholar]
- 7. Hassanzadeh M., Sabo R., Rudie A., Reiner R., Gleisner R., Oporto G. S.., Nanofibrillated cellulose from appalachian hardwoods logging residues as template for antimicrobial copper, J. Nanomater., 2017, 2017, 1–14. 10.1155/2017/2102987 [DOI] [Google Scholar]
- 8. Winans M. J., Gallagher J. E. G., Jaczynski J., Oporto G., Pick your poison: benzalkonium chloride and copper enable nanocellulose derivatives to form antimicrobial properties against a spectrum of microorganisms, bioRxiv, 2019, 783076. 10.1101/783076 [DOI] [Google Scholar]
- 9. Zhong T., Oporto G. S., Jaczynski J., Jiang C., Nanofibrillated cellulose and copper nanoparticles embedded in polyvinyl alcohol films for antimicrobial applications, Biomed. Res. Int. Hindawi., 2015, 2015, 456834. 10.1155/2015/456834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang C., Oporto G. S., Zhong T., Jaczynski J., TEMPO nanofibrillated cellulose as template for controlled release of antimicrobial copper from pva films, Cellulose, 2015, 23, 1–10. 10.1007/s10570-015-0834-5 [DOI] [Google Scholar]
- 11. Hong J., Rico C. M., Zhao L., Adeleye A. S., Keller A. A., Peralta-Videa J. R., Gardea-Torresdey J. L., Toxic effects of copper-based nanoparticles or compounds to lettuce (Lactuca sativa) and alfalfa (Medicago sativa), Environ. Sci. Process Impacts. NIH Public Access, 2015, 17 (1), 177. 10.1039/C4EM00551A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee I.-.C., Ko J.-.W., Park S.-.H., Lim J.-.O., Shin I.-.S., Moon C., Kim S.-.H., Her J.-.D., Kim J.-.C., Comparative toxicity and biodistribution of copper nanoparticles and cupric ions in rats, Int. J. Nanomed., 2016, 11, 2883. 10.2147/IJN.S106346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rong-Mullins X., Winans M. J., Lee J. B., Lonergan Z. R., Pilolli V. A., Weatherly L. M., Carmenzind T. W., Jiang L., Cumming J. R., Oporto G. S., Gallagher J. E. G., Proteomic and genetic analysis of the response of s. cerevisiae to soluble copper leads to improvement of the antimicrobial function of cellulosic copper nanoparticles, Metallomics, 2017, 9 (9), 1304–1315. 10.1039/C7MT00147A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ameh T., Sayes C. M., The potential exposure and hazards of copper nanoparticles: a review, Environ. Toxicol. Pharmacol., 2019, 71, 103220. 10.1016/J.ETAP.2019.103220 [DOI] [PubMed] [Google Scholar]
- 15. Ingle A. P., Duran N., Rai M., Bioactivity, mechanism of action, and cytotoxicity of copper-based nanoparticles: a review, Appl. Microbiol. Biotechnol, 2014, 98 (3), 1001–1009. 10.1007/s00253-013-5422-8. [DOI] [PubMed] [Google Scholar]
- 16. Hostynek J. J., Maibach H. I., Copper hypersensitivity: dermatologic aspects, Dermatol. Ther., 2004, 17 (4), 328–333. 10.1111/j.1396-0296.2004.04035.x [DOI] [PubMed] [Google Scholar]
- 17. Borkow G., Gabbay J., Putting copper into action: copper-impregnated products with potent biocidal activities, FASEB J, 2004, 18 (14), 1728–1730. 10.1096/fj.04-2029fje [DOI] [PubMed] [Google Scholar]
- 18. Gerstein A. C., Ono J., Lo D. S., Campbell M. L., Kuzmin A., Otto S. P., Too much of a good thing: the unique and repeated paths toward copper adaptation, Genetics, 2015, 199 (2), 555–571. 10.1534/genetics.114.171124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sparks D. L., Schreurs B. G., Trace amounts of copper in water induce ß-Amyloid plaques and learning deficits in a rabbit model of alzheimer's disease, Proc. Natl. Acad. Sci., 2003, 100 (19), 11065–11069. 10.1073/pnas.1832769100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiao G., Fan Q., Wang X., Zhou B.., Huntington disease arises from a combinatory toxicity of polyglutamine and copper binding, Proc. Natl. Acad. Sci., 2013, 110 (37), 14995–15000. 10.1073/pnas.1308535110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rademacher C., Masepohl B., Copper-responsive gene regulation in bacteria, Microbiol (United Kingdom), 2012;158 (10), 2451–2464. 10.1099/mic.0.058487-0 [DOI] [PubMed] [Google Scholar]
- 22. Robinson N. J., Winge D. R., Copper metallochaperones, Annu. Rev. Biochem., 2010, 79, 537–562. 10.1146/annurev-biochem-030409-143539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rutherford J. C., Bird A. J., Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells, Eukaryot. Cell, 2004, 3 (1), 1–13. 10.1128/EC.3.1.1-13.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y., Hodgkinson V., Zhu S., Weisman G. A., Petris M. J., Advances in the understanding of mammalian copper transporters, Adv. Nutr., 2011, 2, 129–137. 10.3945/an.110.000273.Figure [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Solomon E. I., Heppner D. E., Johnston E. M., Ginsbach J. W., Cirera J., Qayyum M., Kieber-Emmons M. T., Kjaergaard C. H., Hadt R. G., Tian L., Copper active sites in biology, Chem. Rev., 2014, 114 (7), 3659–3853. 10.1021/cr400327t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nevitt T., Öhrvik H., Thiele D. J., Charting the travels of copper in eukaryotes from yeast to mammals, Biochim. Biophys. Acta (BBA)-Molecular Cell Res., Elsevier; 2012, 1823 (9), 1580–1593. 10.1016/j.bbamcr.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turski M. L., Brady D. C., Kim H. J., Kim B.-.E., Nose Y., Counter C. M., Winge D. R., Thiele D. J., A novel role for copper in ras/mitogen-activated protein kinase signaling, Mol. Cell. Biol., 2012, 32 (7), 1284–1295. 10.1128/mcb.05722-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Georgatsou E., Alexandraki D., Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae, Mol. Cell. Biol., 1994, 14 (5), 3065–3073. 10.1128/mcb.14.5.3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Georgatsou E., Mavrogiannis L. A., Fragiadakis G. S., Alexandraki D., The yeast fre1p/fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated mac1p activator, J. Biol. Chem., 1997, 272 (21), 13786–13792. 10.1074/jbc.272.21.13786 [DOI] [PubMed] [Google Scholar]
- 30. Jungmann J., Reins H. A., Lee J., Romeo A., Hassett R., Kosman D., Jentsch S., MAC1, a nuclear regulatory protein related to cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast, EMBO J., 1993, 12 (13), 5051–5056. 10.1002/j.1460-2075.1993.tb06198.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dancis A., Haile D., Yuan D. S., Klausner R. D., The Saccharomyces cerevisiae copper transport protein (Ctr1p), J. Biol. Chem., 1994, 269 (41), 25660–25667. 10.1016/S0021-9258(18)47300-0 [DOI] [PubMed] [Google Scholar]
- 32. Hassett R., Kosman D. J., Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae, J. Biol. Chem. 1995, 270 (1), 128–134. 10.1074/jbc.270.1.128 [DOI] [PubMed] [Google Scholar]
- 33. Labbé S., Zhu Z., Thiele D. J., Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway, J. Biol. Chem., 1997, 272 (25), 15951–15958. 10.1074/jbc.272.25.15951 [DOI] [PubMed] [Google Scholar]
- 34. Knight S. A. B., Labbé S., Kwon L. F., Kosman D. J., Thiele D. J., A widespread transposable element masks expression of a yeast copper transport gene, Genes Dev., 1996, 10 (15), 1917–1929. 10.1101/gad.10.15.1917 [DOI] [PubMed] [Google Scholar]
- 35. Liu J., Sitaram A., Burd C. G., Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase, Traffic, 2007, 8 (10), 1375–1384. 10.1111/j.1600-0854.2007.00616.x [DOI] [PubMed] [Google Scholar]
- 36. Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P., A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins, Lett. Nat., 2002, 416 (6879), 451–455. 10.1038/416451a [DOI] [PubMed] [Google Scholar]
- 37. Reider A., Wendland B., Endocytic adaptors - social networking at the plasma membrane, J. Cell. Sci., 2011, 124 (10), 1613–1622. 10.1242/jcs.073395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shih S. C., Katzmann D. J., Schnell J. D., Sutanto M., Emr S. D., Hicke L., Epsins and Vps27p/hrs contain ubiquitin-binding domains that function in receptor endocytosis, Nat. Cell. Biol., 2002, 4 (5), 389–393. 10.1038/ncb790 [DOI] [PubMed] [Google Scholar]
- 39. Wu X., Sinani D., Kim H., Lee J., Copper transport activity of yeast ctr1 is down-regulated via its c terminus in response to excess copper, J. Biol. Chem., American Society for Biochemistry and Molecular Biology; 2009, 284 (7), 4112. 10.1074/JBC.M807909200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thiele D. J., ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene, Mol. Cell. Biol., 1988, 8 (7), 2745–2752. 10.1128/mcb.8.7.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gralla E. B., Thiele D. J., Silar P., Valentine J. S., ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene, Proc. Natl. Acad. Sci. USA, 1991, 88 (19), 8558–8562. 10.1073/pnas.88.19.8558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Culotta V. C., Howard W. R., Liu X. F., CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae, J. Biol. Chem., Department of Environmental Health Sciences, Johns Hopkins University School of Hygiene and Public Health, Baltimore, Maryland 21205.; 1994, 269 (41), 25295–25302. 10.1016/S0021-9258(18)47246-8 [DOI] [PubMed] [Google Scholar]
- 43. Nikko E., Pelham H. R. B., Arrestin-mediated endocytosis of yeast plasma membrane transporters, Traffic, 2009, 10 (12), 1856–1867. 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore C. A. C., Milano S. K., Benovic J. L., Regulation of receptor trafficking by grks and arrestins, Annu. Rev. Physiol., 2007, 69 (1), 451–482. 10.1146/annurev.physiol.69.022405.154712 [DOI] [PubMed] [Google Scholar]
- 45. Lin C. H., Macgurn J. A., Chu T., Stefan C. J., Emr S. D.., Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface, Cell, 2008, 135 (4), 714–725. 10.1016/j.cell.2008.09.025 [DOI] [PubMed] [Google Scholar]
- 46. Prosser D. C., Pannunzio A. E., Brodsky J. L., Thorner J., Wendland B., O'donnell A. F., Alpha-arrestins participate in cargo selection for both clathrin-independent and clathrin-mediated endocytosis, J. Cell. Sci., 2015, 128, 4220–4234. 10.1242/jcs.175372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zbieralski K., Wawrzycka D., α-Arrestins and their functions: from yeast to human health, Int. J. Mol. Sci., 2022, 23 (9), 4988. Multidisciplinary Digital Publishing Institute; 2022;23: 4988. 10.3390/IJMS23094988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen B.-.C., Legant W. R., Wang K., Shao L., Milkie D. E., Davidson M. W., Janetopoulos C., Wu X. S., Hammer J. A., Liu Z., English B. P., Mimori-Kiyosue Y., Romero D. P., Ritter A. T., Lippincott-Schwartz J., Fritz-Laylin L., Mullins R. D., Mitchell D. M., Bembenek J. N., Reymann A.-.C., Böhme R., Grill S. W., Wang J. T., Seydoux G., Tulu U. S., Kiehart D. P., Betzig E., Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution, Sci, 2014, 346 (6208), 346. 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinn S. E., Huang L., Kerkvliet J. G., Swanson J. A., Smith S., Hoppe A. D., Anderson R. B., Thiex N. W., Scott B. L., The structural dynamics of macropinosome formation and pi3-kinase-mediated sealing revealed by lattice light sheet microscopy, Nat Commun., 2021, 12 (1), 10.1038/S41467-021-25187-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Teranikar T., Lim J., Ijaseun T., Lee J., Development of planar illumination strategies for solving mysteries in the sub-cellular realm, Int. J. Mol. Sci., 2022, 23 (3) 23. 10.3390/IJMS23031643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmied C., Stamataki E., Tomancak P., Open-Source solutions for spimage processing, Methods Cell. Biol., 2014, 123, 505–529. 10.1016/B978-0-12-420138-5.00027-6 [DOI] [PubMed] [Google Scholar]
- 52. Nishihara T., Matsumura M., Imoto T., Okumura K., Sakano Y., Yorikado Y. et al. , An experimental cmos photon detector with 0.5e-rms temporal noise and 15 μm pitch active sensor pixels, 2017 IEEE International Electron Devices Meeting (IEDM). 2017. pp. 16.1.1–16.1.4. 10.1109/IEDM.2017.8268400 [DOI]
- 53. Liu P., Zhao R., Li H., Zhu T., Li Y., Wang H., Zhang X.-.D., Near-infrared-II deep tissue fluorescence microscopy and application, Nano. Res., 2023, 16 (1), 692–714. 10.1007/s12274-022-4836-y [DOI] [Google Scholar]
- 54. Ajuwon T. E. T., Smith S., Spectrally resolved ratiometric multiphoton fluorescence and second harmonic imaging of noninvasive vascular scaffolds in porcine arterial walls, In SPIE Photonics West BIOS: Multiphoton Microscopy in the Biomedical Sciences XXIII, San Francisco. 2023. pp. 12384–12353. [Google Scholar]
- 55. Planchon T. A., Gao L., Milkie D. E., Davidson M. W., Galbraith J. A., Galbraith C. G., Betzig E., Rapid three-dimensional isotropic imaging of living cells using bessel beam plane illumination, Nat. Methods, 2011, 8 (5), 417–423. 10.1038/nmeth.1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gao L., Shao L., Chen B.-.C., Betzig E., 3D live fluorescence imaging of cellular dynamics using bessel beam plane illumination microscopy, Nat Protoc, 2014, 9 (5), 1083–1101. 10.1038/nprot.2014.087 [DOI] [PubMed] [Google Scholar]
- 57. Meijering E. H. W., Niessen W. J., Viergever M. A., Quantitative evaluation of convolution-based methods for medical image interpolation, 2001, 5, 111–126. [DOI] [PubMed] [Google Scholar]
- 58. Joanne M.; Theurich HHMI CAO . Research License agreement between HHMI and SDSMT for patent applications 13/160,492 and 13/844,405. 2014. [Google Scholar]
- 59. Zhao Y., Strope P. K., Kozmin S. G., Mccusker J. H., Dietrich F. S., Kokoska R. J., Petes T. D., Structures of naturally evolved cup1 tandem arrays in yeast indicate that these arrays are generated by unequal nonhomologous recombination, G3 (Bethesda). Department of Molecular Genetics and Microbiology and University Program in Genetics and Genomics, Duke University Medical Center, Durham, North Carolina 27710.; Department of Molecular Genetics and Microbiology and University Program in Genetic(TRUNCATED: Zhao et al; 2014;4 (11), 2259–2269. 10.1534/g3.114.012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Winans M. J., Gallagher J. E. G., Metallomic and lipidomic analysis of S. cerevisiae response to cellulosic copper nanoparticles uncover drivers of toxicity, Metallomics, Royal Society of Chemistry; 2020, 12 (5), 799–812. 10.1039/d0mt00018c [DOI] [PubMed] [Google Scholar]
- 61. Zhao R.-.Z., Jiang S., Zhang L., Yu Z. ‑. B.., Mitochondrial electron transport chain, ros generation and uncoupling (review) [Internet], Int. J. Mol. Med., Spandidos Publications; 2019, pp. 3–15. 10.3892/ijmm.2019.4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Varlakhanova N. V., Alvarez F. J. D., Brady T. M., Tornabene B. A., Hosford C. J., Chappie J. S. et al. , Structures of the fungal dynamin-related protein vps1 reveal a unique, open helical architecture, J. Cell. Biol., 2018, 217, 3608–3624. 10.1083/JCB.201712021/VIDEO-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. dos Santos N. V., Saponi C. F., Ryan T. M., Primo F. L., Greaves T. L., Pereira J. F. B., Reversible and irreversible fluorescence activity of the enhanced green fluorescent protein in pH: insights for the development of pH-biosensors, Int. J. Biol. Macromol., 2020, 164, 3474–3484. 10.1016/J.IJBIOMAC.2020.08.224 [DOI] [PubMed] [Google Scholar]
- 64. Merz S., Westermann B., Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae, Genome Biol., 2009, 10 (9), R95. 10.1186/GB-2009-10-9-R95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Herst P. M., Perrone G. G., Dawes I. W., Bircham P. W., Berridge M. V., Plasma membrane electron transport in Saccharomyces cerevisiae depends on the presence of mitochondrial respiratory subunits, FEMS Yeast Res, Oxford Academic; 2008, 8 (6), 897–905. 10.1111/J.1567-1364.2008.00418.X [DOI] [PubMed] [Google Scholar]
- 66. Klein M., Mamnun Y. M., Eggmann T., Schüller C., Wolfger H., Martinoia E., Kuchler K., The ATP-binding cassette (ABC) transporter bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast, FEBS Lett., 2002, 520 (1-3), 63–67. 10.1016/S0014-5793(02)02767-9 [DOI] [PubMed] [Google Scholar]
- 67. Sousa C. A., Hanselaer S., Soares E. V., ABCC subfamily vacuolar transporters are involved in Pb (Lead) detoxification in Saccharomyces cerevisiae, Appl. Biochem. Biotechnol., 2015;175 (1), 65–74. 10.1007/s12010-014-1252-0 [DOI] [PubMed] [Google Scholar]
- 68. Wolfe K. H., Shields D. C., Molecular evidence for an ancient duplication of the entire yeast genome, Nature, 1997, 387 (6634), 708–713. 10.1038/42711 [DOI] [PubMed] [Google Scholar]
- 69. Escalera-Fanjul X., Quezada H., Riego-Ruiz L., González A., Whole-genome duplication and yeast's fruitful way of life [Internet], Trends Genet., 2019; 35 (1), 42–54. 10.1016/j.tig.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 70. Hatakeyama R., Kamiya M., Takahara T., Maeda T., Endocytosis of the aspartic acid/glutamic acid transporter dip5 is triggered by substrate-dependent recruitment of the rsp5 ubiquitin ligase via the arrestin-like protein aly2, Mol. Cell. Biol., 2010, 30 (24), 5598–5607. 10.1128/MCB.00464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Alvaro C. G., O'Donnell A. F., Prosser D. C., Augustine A. A., Goldman A., Brodsky J. L., Cyert M. S., Wendland B., Thorner J., Specific α-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the g-protein-coupled receptor ste2, Mol. Cell. Biol., 2014, 34 (14), 2660–2681. 10.1128/MCB.00230-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cankorur-Cetinkaya A., Eraslan S., Kirdar B., Transcriptomic response of yeast cells to atx1 deletion under different copper levels, BMC Genom., 2016, 17, 1–14. 10.1186/S12864-016-2771-6/FIGURES/3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O'donnell A. F., The running of the buls: control of permease trafficking by α-arrestins bul1 and bul2, Mol. Cell. Biol., 2012, 32 (22), 4506–4509. 10.1128/MCB.01176-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Villers J., Savocco J., Szopinska A., Degand H., Nootens S., Morsomme P., Study of the plasma membrane proteome dynamics reveals novel targets of the nitrogen regulation in yeast, Mol. Cell. Proteomics, 2017, 16 (9), 1652–1668. 10.1074/mcp.M116.064923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ruotolo R., Marchini G., Ottonello S., Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast, Genome Biol., 2008, 9 (4), R67. 10.1186/gb-2008-9-4-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Helliwell S. B., Losko S., Kaiser C. A., Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease, J. Cell. Biol., 2001, 153 (4), 649–662. 10.1083/JCB.153.4.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kahlhofer J., Leon S., Teis D., Schmidt O., The α-arrestin family of ubiquitin ligase adaptors links metabolism with selective endocytosis, Biol. Cell., 2021, 113 (4), 183–219. 10.1111/BOC.202000137 [DOI] [PubMed] [Google Scholar]
- 78. Munder M. C., Midtvedt D., Franzmann T., NüSke E., Otto O., Herbig M., Ulbricht E., Müller P., Taubenberger A., Maharana S., Malinovska L., Richter D., Guck J., Zaburdaev V., Alberti S., A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy, Elife, 2016, 5. 10.7554/ELIFE.09347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gallagher J. E. G., Zheng W., Rong X., Miranda N., Lin Z., Dunn B., Zhao H., Snyder M. P., Divergence in a master variator generates distinct phenotypes and transcriptional responses, Genes Dev., 2014, 28 (4), 409–421. 10.1101/gad.228940.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rong-Mullins X., Ayers M. C., Summers M., Gallagher J. E. G., Transcriptional profiling of Saccharomyces cerevisiae reveals the impact of variation of a single transcription factor on differential gene expression in 4nqo, fermentable, and nonfermentable carbon sources, G3 Genes Genome. Genet. 2018, 8 (2), 607–619. 10.1534/g3.117.300138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ravishankar A., Pupo A., Gallagher J. E. G., Resistance mechanisms of Saccharomyces cerevisiae to commercial formulations of glyphosate involve dna damage repair, the cell cycle, and the cell wall structure, G3 Genes Genome. Genet., 2020, 10 (6), g3.401183.2020. 10.1534/g3.120.401183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ayers M. C., Sherman Z. N., Gallagher J. E. G., Oxidative stress responses and nutrient starvation in mchm treated Saccharomyces cerevisiae, Genome. Genet., 2020, 10 (12), 4665–4678. 10.1534/g3.120.401661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pupo A., Ayers M. C., Sherman Z. N., Vance R. J., Cumming J. R., Gallagher J. E. G., MCHM Acts as a hydrotrope, altering the balance of metals in yeast, Biol. Trace Elem. Res., 2020, 195 (1), 260–271. 10.1007/s12011-019-01850-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.