ABSTRACT
Mitochondrial homeostasis requires a dynamic balance of fission and fusion. The actin cytoskeleton promotes fission, and we found that the mitochondrially localized myosin, myosin 19 (Myo19), is integral to this process. Myo19 knockdown induced mitochondrial elongation, whereas Myo19 overexpression induced fragmentation. This mitochondrial fragmentation was blocked by a Myo19 mutation predicted to inhibit ATPase activity and strong actin binding but not by mutations predicted to affect the working stroke of the motor that preserve ATPase activity. Super-resolution imaging indicated a dispersed localization of Myo19 on mitochondria, which we found to be dependent on metaxins. These observations suggest that Myo19 acts as a dynamic actin-binding tether that facilitates mitochondrial fragmentation. Myo19-driven fragmentation was blocked by depletion of either the CAAX splice variant of the endoplasmic reticulum (ER)-anchored formin INF2 or the mitochondrially localized F-actin nucleator Spire1C (a splice variant of Spire1), which together polymerize actin at sites of mitochondria–ER contact for fission. These observations imply that Myo19 promotes fission by stabilizing mitochondria–ER contacts; we used a split-luciferase system to demonstrate a reduction in these contacts following Myo19 depletion. Our data support a model in which Myo19 tethers mitochondria to ER-associated actin to promote mitochondrial fission.
Keywords: Myo19, Mitochondria, Fission, Endoplasmic reticulum, Actin
Summary: Mitochondrial homeostasis requires regulated fission, which frequently occurs at mitochondria–ER contacts. Mitochondrially associated Myo19 binds ER-associated F-actin, tethering the organelles to promote fission.
INTRODUCTION
Mitochondria are highly dynamic organelles that are remodeled by ongoing fusion and fission. Fission plays a role in mitochondrial biogenesis and enables damaged parts of mitochondria to be cordoned off for mitophagy, whereas fusion facilitates mitochondrial content mixing, which can lead to complementation (Youle and van der Bliek, 2012). The proper balance of these processes is key to mitochondrial, and thus cellular, health, whereas dysregulation of the balance between fission and fusion is implicated in diseases of energy-demanding tissues, such as the heart and the brain (Archer, 2013; Ong et al., 2010; Wang et al., 2011). For this reason, deciphering the molecular mechanisms underpinning mitochondrial fission and fusion has been the subject of intense research.
The final step of mitochondrial fission centers on the activity of dynamin-related protein 1 (Drp1, also known as Dnm1l). This protein associates with the mitochondrial outer membrane and oligomerizes to form a ring around the organelle. Oligomerized Drp1 hydrolyzes GTP and undergoes a conformational change that forces scission of mitochondrial membranes (Labrousse et al., 1999; Losón et al., 2013). Drp1 activity is tightly regulated, with phosphorylation at serine 616 leading to increased mitochondrial fission (Kashatus et al., 2015; Taguchi et al., 2007). The recruitment of Drp1 is also regulated. The receptor Fis1 recruits Drp1 to the endzones of mitochondria for fission aimed at segregating damaged mitochondrial tips for recycling, whereas the receptor Mff recruits Drp1 to the midzone of mitochondria for fission in periods of cellular growth and proliferation (Kleele et al., 2021).
In Mff-regulated midzone mitochondrial fission, the actin cytoskeleton is required upstream of Drp1 recruitment. Specifically, the mitochondrial outer membrane-embedded protein Spire1C (a splice variant of Spire1) and the CAAX splice variant of the endoplasmic reticulum (ER) membrane-anchored protein inverted formin 2 (INF2-CAAX) collaborate to generate filamentous (F)-actin at sites of mitochondria–ER (mito–ER) contact, stabilizing these sites for fission (Chakrabarti et al., 2018; Manor et al., 2015). Perhaps separately, downstream of actin polymerization, but upstream of Drp1-mediated membrane scission, non-muscle myosin II plays a role in membrane constriction (Korobova et al., 2014). Non-muscle myosin II generates forces that deform the surrounding actin filament network, which in turn applies pressure on mitochondria (Yang and Svitkina, 2019). These findings clearly implicate actin in mitochondrial fission but do not fully define the mechanisms involved, inviting further investigation.
We note that the mitochondrially localized myosin – myosin 19 (Myo19) – has not been investigated as a fission regulator. The N-terminus of Myo19 is a conserved motor domain that binds to F-actin in an ATP-dependent manner with a high duty ratio (Adikes et al., 2013; Lu et al., 2014; Quintero et al., 2009; Ušaj and Henn, 2017). The sequence of the C-terminal tail domain is unique to this myosin class and targets the protein to the outer mitochondrial membrane (OMM), both by direct insertion and via binding to the proteins Miro1 (Rhot1) and Miro2 (Rhot2), which also directly insert into the OMM (Hawthorne et al., 2016; López–Doménech et al., 2018; Shneyer et al., 2016). Other OMM-binding partners of Myo19 remain to be investigated, including metaxins 1, 2 and 3 (MTX1–3). Two proteomics studies identified MTX3 as a binding partner for Myo19 (Bocanegra et al., 2020; Oeding et al., 2018). More recently, the related proteins MTX1 and MTX2 were shown to bind Miro1 and Miro2 and recruit trafficking kinesin (TRAK) proteins, which compete with Myo19 for access to mitochondria (Zhao et al., 2021). Thus, there is evidence to suggest that metaxins associate with Myo19.
Several cellular functions have been ascribed to Myo19. In interphase, Myo19 can promote mitochondrial motility, notably in filopodia along bundled actin filaments (Quintero et al., 2009; Shneyer et al., 2016, 2017). During mitosis, Myo19 tethers mitochondria to cytoplasmic actin cables, in turn organizing the ER and ensuring that daughter cells inherit equivalent mitochondrial mass (Moore et al., 2021; Rohn et al., 2014).
We recognized that Myo19 is a good candidate for a facilitator of mitochondrial fission. This is because this motor interacts with mitochondria and F-actin, and F-actin is implicated in mitochondrial fission. Furthermore, a class of fission events occur at mito–ER contacts, where Myo19 might localize through interactions with two of its adaptors, Miro1 and Miro2, which are present there (Friedman et al., 2011; Modi et al., 2019). Supporting this possibility, Myo19 was identified as a hit in a proteomic study of mito–ER contact sites (Hung et al., 2017). Furthermore, Myo19 might be functionally relevant at mito–ER contact sites, as Myo19 depletion leads to disorganization of the ER in mitosis (Moore et al., 2021).
Here, we describe Myo19 as a promoter of mitochondrial fission. Cellular depletion of Myo19 led to dramatic hyperfusion of the mitochondrial network, whereas Myo19 overexpression drove fission. We introduced point mutations to (1) inhibit the progression of the myosin through its ATPase cycle while locking the motor in a state that does not bind strongly to actin, or (2) inhibit the working stroke of Myo19 without affecting its ATPase activity and actin binding. We found that an active motor is required to induce mitochondrial fragmentation, but a full working stroke is not. This result is consistent with a model in which Myo19 is able to dynamically tether mitochondria to actin filaments in an ATP-dependent manner. Supporting the tether hypothesis, super-resolution microscopy to determine the nanoscale distribution of Myo19 indicated that the motor is sparsely distributed across mitochondria and primarily monomeric. We probed the mechanism underlying this distribution and found that metaxins contribute to the mitochondrial recruitment of Myo19, in parallel with a known role for Miro proteins. We found that Myo19-dependent mitochondrial fission is dependent on ER-associated actin filaments nucleated by INF2-CAAX and Spire1C, as depletion of either INF2-CAAX or Spire1C prevented Myo19-induced mitochondrial fission. This led us to query the role of Myo19 at mito–ER contacts, where we used a split-luciferase reporter to show that Myo19 depletion led to a loss of these sites. Taken together, these data support a model in which ATP-dependent Myo19 tethering of mitochondria to ER-associated actin filaments stabilizes mito–ER contact sites, thus facilitating mitochondrial fission.
RESULTS
Myo19 fragments mitochondria via F-actin tethering
To test whether Myo19 influences mitochondrial morphology, we used siRNA to knock down the motor protein in HeLa cells, achieving 96.6% depletion (Fig. 1A). We visualized mitochondria using the fluorescent dye MitoTracker Deep Red FM and confocal microscopy. Myo19-depleted cells, which also transiently expressed GFP to guide random sampling, featured a mitochondrial network that was more interconnected than seen in control cells treated with a non-targeting oligonucleotide (Fig. 1B; Fig. S1A). In a blinded qualitative analysis, we categorized the mitochondrial network of each cell as either ‘hyperfused’, ‘tubular’, intermediate or fragmented, with hyperfused defined as essentially a continuous reticulum and fragmented defined as many individual organelles with aspect ratios of ∼1 (Fig. 1C; Fig. S1B). The percentage of cells with hyperfused mitochondria was consistently higher for the Myo19 knockdown condition (Fig. 1D), with 52±1% (mean±s.e.m.) of Myo19-depleted cells characterized by a hyperfused mitochondrial network in comparison to only 12±6% of control cells. In a complementary quantitative analysis, we used machine learning to segment mitochondria, allowing us to determine the average size of these organelles per cell. In agreement with our initial qualitative analysis, we found that the area of a mitochondrion per cell was significantly higher in cells lacking Myo19 compared to that of controls (Fig. 1D): 2.3±0.1 μm2 and 1.3±0.1 μm2 for Myo19-depleted and control cells, respectively. Although we noted a significant change in mitochondrial size, this was not simply a result of perinuclear clustering of mitochondria, as we found this clustering did not occur with our Myo19 siRNA (Fig. S1C). Moreover, we also produced cells with more interconnected mitochondria by treating with an alternative Myo19 siRNA (Fig. S1D). These data indicate that Myo19 promotes fragmentation of mitochondria.
Fig. 1.
Myo19 fragments mitochondria via tethering to F-actin. (A) Western blot for Myo19 in cells treated with the control oligonucleotide or Myo19 siRNA, accompanied by quantitation. (B) Images of MitoTracker Deep Red FM signal in control or Myo19-depleted cells. (C) Cartoon demonstrating hyperfused, reticular, intermediate and fragmented mitochondrial networks. (D) Qualitative (left) and quantitative (right) analyses of mitochondrial morphology in cells with or without Myo19. For qualitative analysis, two-way ANOVA and Dunnett's multiple comparisons test was used. For quantitative analysis, a two-tailed unpaired Student's t-test was used. (E) Cartoon demonstrating Myo19 domain architecture and location of the mutations used. (F) Images of MitoTracker Deep Red FM and GFP signal in cells expressing GFP alone, WT GFP–Myo19 or GFP–Myo19 G417A. (G) For these groups, analysis of mitochondrial morphology was conducted qualitatively (left; two-way ANOVA and Dunnett's multiple comparisons test used) and quantitatively (right; one-way ANOVA and Dunnett's multiple comparison test). (H) Images of cells expressing either WT GFP–Myo19 or GFP–Myo19 Δ2IQ showing MitoTracker Deep Red FM and GFP signals. (I) For these groups, the analysis of mitochondrial morphology was conducted qualitatively. Two-way ANOVA and Dunnett's multiple comparisons test were used. (J) Images of cells expressing either WT GFP–Myo19 or GFP–Myo19 R659L. (K) Qualitative analysis for J, performed as in I. Three independent experiments were performed; n=20–30 cells for each experiment. In graphs, the bars represent the means. In the images, cell boundaries are outlined. All scale bars: 10 μm. ns, not significant; *P<0.05; **P<0.005; ***P<0.0005.
We next examined the effects of Myo19 overexpression. We found more fragmented mitochondrial networks in GFP–Myo19-expressing cells compared to cells expressing GFP alone (Fig. S2A). Overexpression of C-terminally tagged Myo19–GFP also produced fragmented mitochondria (Fig. S2B). To test whether our observations from HeLa cells could be replicated in another cell type, we carried out parallel studies in C2C12 mouse myoblasts. We chose this cell line because muscle is a tissue that is particularly reliant on mitochondria due to high energy demand. In C2C12 cells expressing Halo–MsMyo19, mitochondria were smaller compared to those of controls (Fig. S2C). Thus, Myo19 promotes mitochondrial fragmentation in both HeLa and C2C12 cells.
We next interrogated what properties of Myo19 are required to promote mitochondrial fragmentation. First, we tested whether Myo19-induced mitochondrial fragmentation requires binding to F-actin by expressing GFP–Myo19 G417A, a construct with a mutation predicted to reduce F-actin-binding affinity (Fig. 1E). The analogous mutation in Dictyostelium Myosin II reduces F-actin binding, and this myosin has strong sequence homology to human Myo19 in the relevant domain (Adikes et al., 2013; Sasaki et al., 1998). Specifically, the G417A mutation in the Switch II region of the motor domain is predicted to impair the ability of Myo19 to hydrolyze ATP once it is bound, thus locking the myosin in a state with weak affinity to F-actin. We overexpressed GFP, WT GFP–Myo19, or GFP–Myo19 G417A in HeLa cells, visualizing mitochondria with the dye MitoTracker Deep Red FM. Cells expressing high levels of Myo19 were imaged and, after verifying similar Myo19 expression levels between groups, we determined the percentage of cells with mitochondrial networks characterized as hyperfused or tubular, intermediate, or fragmented (Fig. 1F,G; Fig. S2D). A significantly higher percentage of cells featured fragmented mitochondria in the WT GFP–Myo19 condition compared to cells expressing GFP alone [30.0±8.7% (mean±s.e.m.) versus 1.3±1.3%], whereas the control condition was indistinguishable from cells expressing the GFP–Myo19 G417A mutant (1.3±1.3% versus 5.3±2.9%). Next, we segmented mitochondria using machine learning and measured the average size of a mitochondrion per cell. We were able to train our machine-learning program to find that cells expressing WT GFP–Myo19 had smaller mitochondria (1.91±0.10 μm2) compared to those expressing GFP alone (2.37±0.06 μm2), whereas cells expressing GFP–Myo19 G417A had larger mitochondria (2.92±0.11 μm2) compared to those of controls. Thus, we conclude that Myo19 must bind F-actin to promote fragmentation of the mitochondrial network.
Myosins not only bind F-actin but can also generate force to do work, inducing either cytoskeletal filament sliding or cargo motility. To test whether the working stroke of Myo19 was required to induce mitochondrial fragmentation, we generated a construct with a shortened lever arm that lacks the two C-terminal IQ motifs (Δ2IQ). The effect of this mutation on the mechanochemical cycle of Myo19 has been partially characterized and, based on these observations as well as studies on the effects of the analogous mutation in other myosins, we predict that the deletion of the two IQ motifs should still allow Myo19 to dynamically bind and unbind F-actin in an ATP-dependent manner, but produce a motor with a reduced working stroke (Fig. 1E) (Lu et al., 2014; Uyeda et al., 1996). Using the same experimental paradigm as described above, we found that this mutation did not affect the degree of mitochondrial fragmentation, which was the same upon overexpression of either WT GFP–Myo19 or GFP–Myo19 Δ2IQ (Fig. 1H,I; Fig. S2D). The percentage of cells featuring fragmented mitochondria was 29±4% (mean±s.e.m.) for the WT GFP–Myo19 condition and 24±4% for the GFP–Myo19 Δ2IQ condition. These data suggest that Myo19 functions as a dynamic actin-binding tether to induce mitochondrial fragmentation, as the process depends on the actin-interacting motor domain but not the lever arm that is required for the myosin to perform work. We further tested this tethering model in an analogous experiment using GFP–Myo19 R659L, a mutation predicted – based on studies with another myosin, though not yet shown for Myo19 – to uncouple the motor domain and lever arm, maintaining F-actin-binding ability and ATPase activity but inhibiting the working stroke (Snoberger et al., 2021) (Fig. 1E). As with GFP–Myo19 Δ2IQ, GFP–Myo19 R659L fragmented mitochondria to the same extent as wild-type (WT) Myo19 (34.5±7.1% of cells fragmented for WT GFP–Myo19 and 38.9±5.8% of cells fragmented for GFP–Myo19 R659L) (Fig. 1J,K; Fig. S2D). Taken together, our data support the hypothesis that Myo19 does not function as a transport motor to fragment the mitochondrial network, rather, it might function as an ATP-dependent F-actin tether.
Sub-organellar distribution of Myo19
To further examine the tethering hypothesis, we investigated the nanoscale distribution of Myo19 on mitochondria. First, we performed immunofluorescence on fixed HeLa cells using a wide range of anti-Myo19 antibody concentrations and then imaged the samples with stochastic optical reconstruction microscopy (STORM) (Fig. 2A). Epifluorescence images of TOMM20 were used to segment mitochondria; we used the resulting mask to perform an automated analysis for each anti-Myo19 antibody concentration, calculating the nearest neighbor distance of localization clusters as well as the number of localizations per cluster (Fig. 2B). Over a range of primary antibody concentrations spanning more than three orders of magnitude the number of localizations per cluster did not change. In contrast, the nearest neighbor distance between clusters decreased with increasing antibody, saturating at about 130 nm. These data suggest that Myo19 is predominantly organized in a dispersed rather than grouped pattern on mitochondria, with each motor separated by ∼130 nm.
Fig. 2.
Sub-organellar distribution of Myo19. (A) Images of endogenous Myo19 and TOMM20 acquired on an Oxford Nanoimager (ONI). The TOMM20 image was taken using conventional epifluorescence. The accompanying Myo19 image was acquired with STORM – shown is an ONI-rendered localization map. A magnified view of the boxed regions is shown, in which the TOMM20 signal is outlined. Box sizing/placement and TOMM20 outline was determined with manual visual registration. Scale bar: 0.5 μm. (B) For the mitochondrial Myo19 STORM signals, the numbers of localizations per cluster (left) and nearest neighbor distances of localization clusters (right) were plotted as a function of the relative amount of the anti-Myo19 antibody. Boxes show the 25–75th percentiles, and bars represent the 10–90th percentiles. Data are from two independent experiments; n=4 cells total per condition, where one and two cells were excluded from the ‘512’ and ‘256’ groups, respectively, due to drift. (C) For antibody dilution ‘1’, the number of localizations per Myo19 cluster were plotted as a histogram (red curve) and fitted to a log-normal distribution (blue curve) as the single Myo19 calibration function. The parameters µ and σ are shown in the box. PDF, probability density function. (D) Percentages of Myo19 clusters that contain one or two heavy chains were obtained by fitting the frequency distribution of the numbers of localizations of the experimental data to a combination of weighted single- and double-Myo19 calibration functions. (E) Example traces of Myo19 spots that demonstrated one-step and two-step bleaching. A.U., arbitrary units. (F) Percentage of localization clusters from STORM (left) representing a single Myo19 (1 M19) and two Myo19s (2 M19). Bars represent averages and dots represent findings from ‘128’ and ‘2048’ antibody concentrations. Percentage of total Myo19 (M19) bleaching events characterized as one-step and two-step (right). Bars represent averages and each dot represents the mean from three videos of the same chamber. Data are from three independent experiments, where for each experiment, n=686–1287 spots.
To more accurately define the nanoscale distribution of Myo19 on mitochondria, we further analyzed our STORM data using an approach that allows us to estimate the percentage of molecules that are monomeric or are part of higher-order assemblies (Cella Zanacchi et al., 2019; Ehmann et al., 2014; Gyparaki et al., 2021; Zanacchi et al., 2017). The number of protein targets within super-resolution clusters can be deduced using a calibration function that corresponds to the localization cluster coming from a single protein. To establish a single Myo19 calibration function, we used images from samples stained with our lowest concentration of antibody, as in these images, clusters of localizations were likely to represent individual motors, given the patterns observed in Fig. 2B. The frequency distribution of number of localizations per Myo19 cluster was fitted to a log-normal distribution to extract the parameters μ (mean) and σ (variance) (Fig. 2C). Next, we fitted the frequency distribution of cluster localizations from images of samples stained with saturating amounts of antibodies to the convolution of the weighted calibration function to estimate the percentages of single- and double-heavy-chain populations. Considering the saturating antibody concentrations that were good fits (dilutions 2048 and 128 from Fig. 2B), we found that Myo19 primarily existed as a single motor (87–88%) on mitochondria (Fig. 2D). These fits predicted a low percentage of dimers (12–13%). Some saturating antibody concentrations (dilutions 1024 to 256 from Fig. 2B) only properly fit a model that included three-molecule assemblies. Notably, though, these cases still predicted that Myo19 is mostly monomeric (Fig. S3A). This sparse, monomeric organization suggests that small teams of Myo19 motors do not act to transport mitochondria in the unorganized sub-cortical actin network, recently described by electron microscopy (Yang and Svitkina, 2019). The distribution we observe does, however, allow Myo19 to induce mitochondrial fragmentation via tethering to F-actin.
We next tested the hypothesis that Myo19 is largely monomeric with an orthogonal approach. First, we overexpressed Myo19 labeled with the fluorescent Halo ligand (Halo–Myo19) in Cos7 cells and lysed the cells in detergent to solubilize membranes. The resulting clarified lysate contained soluble fluorescent Myo19 motors suitable for single-molecule analysis by total internal reflection fluorescence (TIRF) microscopy (Fenton et al., 2021). After non-specifically adhering labeled Myo19 to a coverslip, we photo-bleached the samples while imaging using TIRF microscopy and recorded the fluorescence profiles of each spot. Most spots bleached in a single step, whereas some bleached in two steps (Fig. 2E). We determined the percentage of spots that fit each category and found that on average, 71.8% of spots bleached in one step, whereas 24.9% of spots bleached in two steps (Fig. 2F). A small percentage bleached in three steps. Although this analysis of soluble proteins might selectively enrich a subpopulation of cellular Myo19, these data in conjunction with the analysis of super-resolution data describing Myo19 bound to intact mitochondria both support the hypothesis that Myo19 is largely monomeric and that the distribution of Myo19 on mitochondria facilitates linkage to dispersedly organized actin filament networks to facilitate fragmentation.
Metaxins are essential for Myo19 recruitment to mitochondria
The distinct sub-organellar distribution of Myo19 prompted us to ask how this motor is patterned on the mitochondria. Although there is an established role for Miro proteins in the localization of Myo19 to mitochondria (López–Doménech et al., 2018), we wondered whether additional factors might be involved. In particular, we focused on a possible role for metaxins. Proteomics studies indicated that MTX3 interacts with the Myo19 tail domain and more recent work showed that MTX1 and MTX2 associate with TRAK proteins, which Myo19 can compete with for access to mitochondria (Bocanegra et al., 2020; Oeding et al., 2018; Zhao et al., 2021). However, a possible role for metaxins in recruiting and stabilizing Myo19 on mitochondria has not yet been examined.
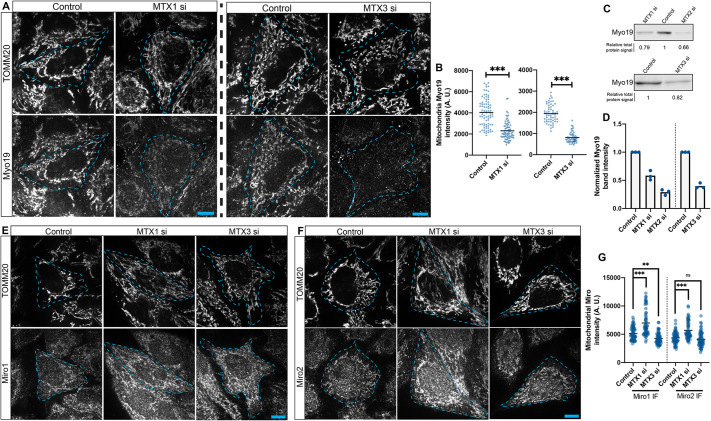
We treated HeLa cells with siRNAs against metaxins and validated their efficiency in the knockdown of MTX1, MTX2 and MTX3 by western blotting, achieving 87%, 79% and 89% knockdown, respectively (Fig. S4A). Next, we tested the effects of knockdown of MTX1, MTX2 or MTX3 individually on the localization of Myo19 to mitochondria, using antibodies to Myo19 and the mitochondrial marker TOMM20 (Fig. 3A). We imaged the samples with confocal microscopy and then segmented mitochondria by intensity thresholding the TOMM20 staining to measure mean mitochondrial Myo19 signal per cell (Fig. 3B). MTX2-depleted cells could not be imaged owing to high levels of cell death, as assessed by phase-contrast microscopy (Fig. S4B). For cells depleted of MTX1 or MTX3, mitochondrial Myo19 levels were significantly lower compared to controls (the means were 57% and 42% of control levels, respectively). In a complementary experiment, we collected lysates of cells treated with a control oligonucleotide or siRNAs against either MTX1, MTX2 or MTX3 for SDS-PAGE and immunoblotted for Myo19 (Fig. 3C). Myo19 was found to be reduced in all three knockdown conditions, showing 58±5% (mean±s.e.m.), 29±3%, and 40±3% of endogenous levels, respectively (Fig. 3D). We tested whether this result was due to decreased mitochondrial content by using images of knockdown cells to record the percentage of the cellular area occupied by mitochondria, as defined by the TOMM20 signal. In fact, we found that the mitochondrial content of cells was actually slightly increased after MTX1 knockdown and unchanged with MTX3 depletion (Fig. S4C). Taken together, these data support the Myo19 localization data by suggesting that metaxins stabilize Myo19 on mitochondria, as cytosolic Myo19 is readily degraded by the proteosome (López–Doménech et al., 2018).
Fig. 3.
Metaxins are essential for Myo19 recruitment to mitochondria. (A) Immunofluorescence images of TOMM20 and Myo19 signals in cells treated with either a control oligonucleotide, or MTX1 or MTX3 siRNAs. (B) For these groups, quantitation of mitochondrial Myo19 intensity is shown. A two-tailed unpaired Student's t-test was used to determine significance. (C,D) Western blots (C) for Myo19 in cells treated with either a control oligonucleotide, or MTX1, MTX2 siRNA or MTX3 siRNAs, and quantitation of the bands (D). (E,F) Immunofluorescence images of either control, MTX1-depleted or MTX3-depleted cells, in which TOMM20 and Miro1 (E) or Miro2 (F) signals are shown. (G) Quantitation of mitochondrial Miro1 and Miro2 intensity in control cells, cells lacking MTX1 and cells lacking MTX3. One-way ANOVA and Dunnett's multiple comparisons test was used. Three independent experiments were performed; n=20–30 cells for each experiment. In graphs, the bars represent the means. In the images, cell boundaries are outlined. All scale bars: 10 μm. A.U., arbitrary units; IF, immunofluorescence. ns, not significant; **P<0.005; ***P<0.0005.
The proteins Miro1 and Miro2 are both known to recruit Myo19 to mitochondria, and metaxins directly bind Miro proteins by analytical gel filtration, so we wanted to check whether metaxins indirectly stabilize Myo19 on mitochondria by maintaining Miro protein levels on the organelle (Zhao et al., 2021). We depleted either MTX1 or MTX3 as before, and probed for Miro1 and Miro2 by immunofluorescence (Fig. 3E,F). The mitochondrial signal of either of the Miro proteins was not meaningfully decreased by knockdown of MTX1 or MTX3 (Fig. 3G). Specifically, after MTX1 and MTX3 depletion, cells showed, on average, 136% and 83% of endogenous Miro1 levels, respectively, and 130% and 94% of endogenous Miro2 levels, respectively. Increased levels of Miro proteins would not be expected to negatively affect Myo19 levels, given that Miro proteins recruit Myo19 to mitochondria. Thus, these observations suggest that metaxins regulate Myo19 recruitment or stability on mitochondria directly, rather than indirectly by affecting the levels of Miro proteins localized to this organelle.
As metaxins either recruit or stabilize Myo19 on mitochondria, and Myo19 seems to promote mitochondrial fragmentation, we tested the hypothesis that mitochondrial morphology is regulated by metaxins. Specifically, we tested whether knockdown of MTX3, which most potently reduced Myo19 levels, led to more interconnected mitochondria. We first treated HeLa cells with either a non-targeting oligonucleotide or an siRNA directed to MTX3, then transfected both experimental groups with GFP to allow random sampling on the microscope, and finally we imaged the cells after labeling mitochondria with MitoTracker Deep Red FM. In a blinded analysis, we characterized the mitochondrial network of each cell as either hyperfused, tubular, intermediate or fragmented (Fig. 1C). Surprisingly, we found that mitochondrial morphology was not significantly altered by depletion of MTX3 (Fig. S4D). Potentially, MTX3 knockdown might not have been sufficient to reduce the mitochondrial levels of Myo19 below a critical threshold required to affect mitochondrial fission. Alternatively, metaxin knockdown might simultaneously affect the recruitment of other proteins, such as microtubule-based motors (Zhao et al., 2021), and this could oppose Myo19 depletion-induced mitochondrial hyperfusion.
F-actin neither organizes Myo19 nor is remodeled by this motor
We next tested whether the actin-binding motor domain of Myo19 contributes to its sub-organellar distribution on mitochondria. We depolymerized actin with the drug latrunculin B, then used conventional epifluorescence microscopy followed by STORM on cells in which Myo19 staining revealed mitochondria to assess the nanoscale distribution of endogenous mitochondrial Myo19 (Fig. S4E). Again, we determined the nearest neighbor distance of localization clusters (Fig. S4F). The median was 118.7 nm, similar to the 126.6 nm recorded for cells with intact F-actin (Fig. 2C). Thus, disruption of cellular F-actin did not markedly influence the sub-organellar distribution of Myo19.
We also wondered whether Myo19 overexpression was sufficient to alter the density of F-actin around mitochondria. To test this possibility, we overexpressed either GFP alone or GFP–Myo19 and analyzed cells by confocal microscopy, labeling the mitochondrial matrix with an anti-Hsp60 antibody and F-actin with phalloidin. We did not observe increased phalloidin staining proximal to the Hsp60 signal in cells expressing GFP–Myo19 (Fig. S5A,B). Swapping the anti-Hsp60 antibody for an antibody against the mitochondrial outer membrane marker TOMM20, we were able to measure average peri-mitochondrial F-actin intensity in many cells and found that this metric was not significantly different between cells expressing GFP alone and GFP–Myo19-expressing cells (Fig. S5C). Finally, we visualized F-actin live by expressing LifeAct and, again, saw no accumulation of F-actin on mitochondria in cells overexpressing GFP–Myo19 (Fig. S5D). Taken together, these data indicate that Myo19 overexpression is not sufficient to recruit additional F-actin to mitochondria and thus induce fragmentation.
Myo19 binds Spire1C- and INF2-CAAX-generated F-actin to fragment mitochondria
We then reasoned that Myo19 must interact with F-actin already in its surroundings to promote mitochondrial fragmentation and set out to identify this F-actin population. Spire1C is the only actin filament nucleator identified to specifically target mitochondria. Moreover, this F-actin nucleator has been shown to facilitate mitochondrial fission (Manor et al., 2015). Therefore, we tested the dependence of Myo19-induced mitochondrial fragmentation on Spire1C expression by treating cells with either a non-targeting oligonucleotide or an siRNA to Spire1C previously shown to effectively silence this protein (Moore et al., 2021). For both these conditions, we concurrently overexpressed either GFP or GFP–Myo19 and visualized mitochondria with the dye MitoTracker Deep Red FM (Fig. 4A). We imaged cells expressing high levels of Myo19 and, after verifying similar levels of Myo19 expression between groups, we determined the percentage of cells with mitochondrial networks characterized as hyperfused or tubular, intermediate, or fragmented in a blinded analysis (Fig. 1C). Only those cells transfected with both the non-targeting oligonucleotide and GFP–Myo19 exhibited a significant increase in fragmented mitochondria: 31.7±0.9% (mean±s.e.m.), compared to 7.0±5.1% for the control oligonucleotide/GFP condition, 2.7±2.7% for the Spire1C siRNA/GFP condition, and 6.3±1.3% for the Spire1C/GFP–Myo19 condition (Fig. 4B; Fig. S5E). Notably, GFP–Myo19 overexpression failed to fragment mitochondria in a Spire1C knockdown background. These results indicate that Myo19 specifically engages Spire1C-generated actin filaments to fragment mitochondria.
Fig. 4.
Myo19 binds Spire1C- and INF2-CAAX-generated F-actin to fragment mitochondria. (A,C) Images of MitoTracker Deep Red FM and GFP signals in cells treated with control oligonucleotide (A,C), Spire1C siRNA (A) or INF2-CAAX siRNA (C) and concurrently expressing either GFP alone or GFP–Myo19. (B,D) For these groups, qualitative analyses of mitochondrial morphology are shown. Three independent experiments were performed; n=20–30 cells for each experiment. In graphs, the bars represent the means and two-way ANOVA with Dunnett's multiple comparisons test used. In the images, cell boundaries are outlined. Scale bars: 10 μm. *P<0.05; ***P<0.0005.
To promote mitochondrial fission, Spire1C interacts with the ER-anchored splice isoform of the formin INF2, INF2-CAAX, to generate F-actin, so we next tested whether INF2-CAAX is also required for Myo19-induced mitochondrial fragmentation with the same experimental paradigm as described above. Using a previously validated INF2-CAAX siRNA, we found that Myo19-induced mitochondrial fragmentation was significantly inhibited in cells depleted of this protein (Moore et al., 2021). Mitochondrial fragmentation was common (53.0±16.1%) in cells treated with the non-targeting oligonucleotide while overexpressing GFP–Myo19, whereas fragmentation was observed in only 10.5±6.0% of cells overexpressing GFP–Myo19 that were depleted of INF2-CAAX (Fig. 4C,D; Fig. S5E). These data support previous studies indicating critical roles for Spire1C and INF2-CAAX in mitochondrial fission (Korobova et al., 2013; Manor et al., 2015), and extend this model by implicating Myo19 in this pathway. We propose that Myo19 binds actin filaments generated locally by mitochondrially localized Spire1C and ER-localized INF2-CAAX.
Myo19 facilitates mitochondrial fission
We next sought to determine whether Myo19 produces a more fragmented mitochondrial network by stimulating mitochondrial fission as opposed to inhibiting fusion. To test this possibility, we used siRNA to deplete Myo19 from HeLa cells and conducted immunofluorescence for the essential fission mediator Drp1 and the mitochondrial marker TOMM20, visualizing the samples with confocal microscopy (Fig. 5A). We used TOMM20 immunofluorescence to segment mitochondria by intensity thresholding and found that Myo19 knockdown led to increased Drp1 levels on this organelle, measured as average fluorescence intensity (122% of control levels) and density of puncta: 0.39±0.02 puncta/μm2 (mean±s.e.m.) for the controls and 0.54±0.02 puncta/μm2 for the Myo19 siRNA condition (Fig. 5B). Next, we collected cell lysates after Myo19 siRNA treatment and performed western blotting to quantify total Drp1 and phospho-S616 Drp1 levels. When phosphorylated at serine 616, Drp1 more readily promotes mitochondrial fission (Kashatus et al., 2015; Taguchi et al., 2007). We also blotted for Mfn2, an essential mediator of mitochondrial fusion (Santel and Fuller, 2001). We found that Mfn2 levels were unaltered with Myo19 knockdown (107±9% of control levels), whereas total Drp1 levels were increased (138±15% of control levels) and phospho-S616 Drp1 levels were decreased (56±11% of control levels) (Fig. 5C). Thus, Myo19 depletion altered the regulation of Drp1 and not Mfn2, consistent with a role for Myo19 in mitochondrial fission and not fusion.
Fig. 5.
Myo19 facilitates mitochondrial fission. (A) Immunofluorescence images of TOMM20 and Drp1 signals in cells treated with either a control oligonucleotide or Myo19 siRNA. (B) For these groups, quantitation of mitochondrial Drp1 intensity and number of Drp1 puncta per μm2 mitochondria. A two-tailed unpaired Student's t-test was used. A.U., arbitrary units. (C) Western blots (top) for total Drp1, phospho-S616 Drp1 and Mfn2 in cells with and without Myo19, with accompanying quantification (bottom). (D) Images of MitoTracker Deep Red FM, GFP and SNAP signals in cells expressing SNAP alone or SNAP–Drp1 K38A and concurrently expressing either GFP alone or GFP–Myo19. (E) For these groups, qualitative analysis of mitochondrial morphology is shown. Two-way ANOVA with Dunnett's multiple comparisons test was used. (F) Western blots (left) for Mff and Fis1 in cells treated with control oligonucleotide or Myo19 siRNA, and accompanying quantification (right). ≥3 independent experiments were performed; n=20–30 cells for each experiment. In graphs, the bars represent the means. In the images, cell boundaries are outlined. Scale bars: 10 μm. ***P<0.0005.
To directly test whether Myo19 facilitates mitochondrial fission, we asked whether mitochondrial fragmentation induced by Myo19 overexpression depended on Drp1. We overexpressed either SNAP alone or SNAP–Drp1 K38A; K38A is a well-characterized mutation that blocks GTP hydrolysis and, thus, the protein acts as a dominant-negative inhibitor of fission (Smirnova et al., 1998). Concurrently, we also overexpressed either GFP alone or GFP–Myo19. We stained mitochondria with the dye MitoTracker Deep Red FM and visualized cells expressing high levels of Myo19 with confocal microscopy. After verifying relatively equal Myo19 expression between groups, we categorized the mitochondrial network in each cell as hyperfused or tubular, intermediate, or fragmented in a blinded manner (Fig. 1C, Fig. 5D; Fig. S6A). Only when SNAP control and GFP–Myo19 were co-expressed did we observe substantial fragmentation of the mitochondrial network, with 29.7±1.8% (mean±s.e.m.) of cells exhibiting fragmented mitochondria, compared to 8.3±4.4% when SNAP alone and GFP alone were co-expressed, 3.3±1.7% when SNAP–Drp1 K38A and GFP were co-expressed, and 7.8±2.1% when SNAP–Drp1 K38A and GFP–Myo19 were co-expressed (Fig. 5E). These data provide direct support for the model that Myo19 facilitates mitochondrial fission.
We also examined how levels of different Drp1 receptors responded to Myo19 depletion. For cells treated with either a control oligonucleotide or a Myo19 siRNA, we collected lysates and subjected them to SDS-PAGE and immunoblotting with antibodies against the Drp1 receptors Fis1 and Mff. Fis1 recruits Drp1 for mitochondrial fission that occurs outside of mito–ER contacts, whereas Mff recruits Drp1 for mito–ER contact fission (Kleele et al., 2021). We found that only Mff levels were altered with Myo19 depletion: 152±25% (mean±s.e.m.) of control levels (Fig. 5F). Fis1 levels remained unchanged. The observation that Mff levels were altered but Fis1 levels were not, in conjunction with the dependence we noted on Spire1C and INF2-CAAX activity, suggested that Myo19 plays a role in mito–ER contact mitochondrial fission specifically.
Myo19 promotes mitochondrial fission by bringing mitochondria and the ER in close apposition
We next asked whether Myo19 tethering induces fission by stabilizing mito–ER contacts, as ER-anchored INF2-CAAX F-actin brings the ER and mitochondria in close apposition during fission (Chakrabarti et al., 2018). The bridge between these two organelles could be formed by association of INF2-CAAX-generated F-actin and Myo19. To test this hypothesis, we first treated HeLa cells with either a control oligonucleotide or an siRNA to Myo19, using AcGFP–Sec61β to label the ER and MitoTracker Deep Red FM to label the mitochondrial network. Although there was no obvious loss of overlap in the Myo19 knockdown condition, this approach lacked sufficient resolution to unambiguously identify contacts, owing to the extensive nature of the cellular ER (Fig. 6A).
Fig. 6.
Myo19 promotes mitochondrial fission by bringing mitochondria and the ER in close apposition. (A) Images of Mitotracker Deep Red FM and AcGFP–Sec61β for cells treated with either a control oligonucleotide or Myo19 siRNA. Scale bar: 10 μm. Cell boundaries are outlined. One biological replicate was conducted. (B) Images of staining for TOMM20 and Myo19 in mito–ER contact reporter cells. Blue arrowheads indicate Myo19/split-GFP colocalization and orange arrowheads indicate slight overlap of the Myo19 signal with split-GFP puncta, or where it is adjacent with split-GFP puncta. Quantification of all overlap is shown, with the bar representing the mean and dots representing individual cells. One biological replicate was conducted. Scale bar: 2 μm. (C) Cartoon of the split-luciferase system (top). In the presence and absence of Myo19, quantification (bottom) of luminescence derived from the proximity of two halves of luciferase targeted to mitochondria and ER is shown. A two-tailed paired t-test was used (five independent experiments). A.U., arbitrary units. *P<0.05. (D) Cartoon of model, depicting metaxin-mediated Myo19 association with mitochondria. Cartoon also shows Myo19 as a link to INF2-CAAX-generated F-actin and thus the ER for mitochondrial fission.
We also asked whether Myo19 is present at mito–ER contacts, turning to a recently described HeLa cell line stably expressing a doxycycline-inducible fluorescent reporter of mito–ER contacts (Tashiro et al., 2020). These cells constitutively express a mitochondrial outer membrane marker tagged with a non-fluorescent GFP fragment; upon treatment with doxycycline, these cells express an ER membrane marker tagged with the complementary GFP fragment needed for fluorescence. After adding doxycycline, we fixed the cells and conducted immunofluorescence using an anti-Myo19 antibody and an antibody against the mitochondrial marker TOMM20 (Fig. 6B). Analysis of the resulting images revealed that the majority of mito–ER contact puncta either co-localized with, or were directly adjacent to, the Myo19 signal, in line with the idea that Myo19 is indeed present at mito–ER contacts.
Finally, we tested our hypothesis that Myo19 acts to stabilize mito–ER contacts by employing a tool free from the resolution constraints of light microscopy. We used a luminescence reporter of mito–ER contacts previously demonstrated to respond to perturbations of these sites (Cho et al., 2017). We treated cells with either a control oligonucleotide or a Myo19 siRNA and then co-transfected the cells with plasmids coding for complementary halves of luciferase – one half targeted to the ER membrane and the other targeted to the mitochondrial outer membrane (Fig. 6C). In this system, when mitochondria and the ER are in close proximity, the complementary luciferase fragments associate, leading to the generation of luminescence in the presence of substrate. Thus, greater luminescence indicates a greater degree of mito–ER contact. Within most experimental replicates, Myo19 depletion caused reduced luminescence: 55±16% (mean±s.e.m.) of control. These data suggest that Myo19 stabilizes mito–ER contacts, and this function of Myo19 might allow this unconventional myosin to promote mitochondrial fission.
DISCUSSION
Mitochondrial fission and fusion dynamics are key to maintaining the health of the mitochondrial network within a cell. For this reason, deciphering the underlying mechanisms of these processes has been the subject of intense research. Here, we present evidence that Myo19 is targeted to mitochondria by metaxins, where this myosin tethers mitochondria to ER-associated actin to enhance ER–mitochondria contacts and to promote mitochondrial fission (Fig. 6D).
First, we found that Myo19 promotes mitochondrial fragmentation. Although previous studies examining the cellular effects of Myo19 depletion generally did not report a phenotype of hyperfused mitochondria, we found that a robust knockdown of the endogenous protein might be required to reveal this phenotype. We were able to achieve 97% depletion of endogenous Myo19, compared to ∼70% depletion of mRNA in at least one other study (Rohn et al., 2014). Of note, a study published during the course of this work catalogued the cellular effects of Myo19 knockout and found mitochondria to be more interconnected in prometaphase (Majstrowicz et al., 2021); here, we extend this observation to interphase cells. Our observation that mitochondrial hyperfusion is induced by Myo19 depletion is complemented by our data indicating that Myo19 overexpression is sufficient to induce mitochondrial fragmentation. The extent of fragmentation observed might scale with levels of overexpression; thus, we ensured that we compared phenotypes in cells expressing similar levels of either WT or mutated Myo19.
Myo19-induced mitochondrial fragmentation is dependent on the ability of the motor to bind F-actin, as a point mutant predicted to bind more weakly to F-actin was insufficient to induce mitochondrial fragmentation. Surprisingly, however, expression of either a Myo19 mutant with a shortened lever arm or a mutant that is predicted to uncouple the lever arm from the motor domain did not alter the extent of mitochondrial fragmentation observed, compared to that seen upon expression of WT Myo19. Based on structural and biophysical studies of other myosins, we predict that these Myo19 mutants should dynamically bind and unbind F-actin in an ATP-dependent manner, like the WT motor, but should exhibit a reduced working stroke. Together, these findings suggest that Myo19 functions as an ATP-dependent tether rather than as a transport motor in mitochondrial fragmentation. This observation is consistent with the identified roles for Myo19 in mitosis and mitocytosis (Jiao et al., 2021; Moore et al., 2021). In mitosis, Myo19 tethers mitochondria to cytoplasmic actin cables, facilitating the equitable distribution of mitochondria between daughter cells (Moore et al., 2021; Rohn et al., 2014). In mitocytosis, Myo19 tethers mitochondria to cortical actin so that they can be trafficked into migrasomes, which form at the lagging edge of cells. Of note, tethering roles have also been assigned to other myosins; for example, some myosin I motors are force-dependent actin anchors (McIntosh and Ostap, 2016). It will be interesting to probe the force dependence of Myo19 in future experiments. Moreover, in neurons, myosin V retains synaptic vesicles at the plasma membrane but does not play a role in transporting them or facilitating exocytosis (Maschi et al., 2018).
The nanoscale distribution of Myo19 further supports a role for this myosin in tethering mitochondria to actin filaments for mitochondrial fragmentation. We collected STORM images over a wide range of antibody dilutions and found that Myo19 assemblies are spaced ∼130 nm apart from one another on average. Although the nearest neighbor distance between clusters increased with decreasing concentrations of antibody, we found that the average number of localizations per cluster remained the same. This observation suggests that Myo19 is primarily monomeric when bound to mitochondria. If Myo19 were a dimer, or assembled into teams, this relationship would be different – as antibody concentration decreased, both metrics would fall, with average number of localizations per cluster doing so first. The notion that Myo19 is mainly monomeric and is distributed at a spacing of ∼130 nm on mitochondria is also supported by our analysis of our STORM images and our photobleaching experiments. One limitation of this latter experiment is that we might have selected for a specific population of Myo19 by collecting soluble protein. Still, in cell lysates generated from Myo19-expressing cells, we found most spots bleached in a single step, consistent with Myo19 mainly existing as a monomer.
The dispersed arrangement of Myo19 molecules is inconsistent with a model in which these motors transport mitochondria along the unorganized sub-cortical F-actin network that surrounds these organelles (Yang and Svitkina, 2019), as kinetics experiments have revealed that only Myo19 teams can mediate processive motility along single actin filaments (Ušaj and Henn, 2017). Thus, based on the sub-organellar distribution of Myo19 and the results of our experiments with mutants of the IQ motifs and the lever arm hinge, we propose that this myosin functions primarily to tether mitochondria to F-actin for mitochondrial fragmentation. Still, we acknowledge that our data leave open the possibility that there are domains on mitochondria where Myo19 is mostly monomeric but is more closely packed, and that these domains could represent hubs for processive motility, although we did not see clear evidence of heterogenous distribution in our analysis. Moreover, the dispersed mitochondrial distribution of Myo19 still enables this motor to transport this organelle in environments in which F-actin is more highly ordered, such as filopodia where F-actin is unipolar and bundled. In such an environment, despite a relatively large distance between organelle-bound Myo19 motors, they could still work on the same track and thus function as a team for processive motility of a mitochondrion. Indeed, others have observed Myo19-mediated mitochondrial motility in filopodia (Sato et al., 2022; Shneyer et al., 2016).
To better understand how the sub-organellar distribution of Myo19 is achieved, we probed for regulators that might function in addition to Miro1 and Miro2. We initially focused on metaxins because these mitochondrially associated proteins cooperate with Miro proteins in localizing TRAK proteins to mitochondria, and TRAK proteins can compete with Myo19 for association with mitochondria (Oeding et al., 2018; Zhao et al., 2021). Furthermore, proteomics studies have suggested an association between MTX3 and the tail of Myo19, but still unexplored was the functional significance of this putative interaction (Bocanegra et al., 2020; Oeding et al., 2018). Addressing this gap in knowledge, we found that depletion of either MTX1 or MTX3 was sufficient to significantly reduce the recruitment of Myo19 to mitochondria, without disrupting the localization of Miro1 and Miro2. We consider there to be two main possibilities for how metaxins recruit Myo19 to mitochondria. First, these proteins could independently bind to Myo19. More likely, however, is the possibility that metaxins could associate with Myo19 in a Miro-dependent manner. In this model, metaxins either cooperate with Miro proteins to form a Myo19-binding site on mitochondria or, alternatively, metaxins stabilize a Myo19-binding-competent conformation of Miro, without directly contacting Myo19. Work published during the completion of this study favors the former, as the authors showed a direct interaction between Myo19 and MTX3 with purified proteins (Shi et al., 2022). Regardless of the mode by which metaxins stabilize Myo19 on mitochondria, we unexpectedly found that depletion of MTX3 did not significantly alter mitochondrial morphology. As our results predict that hyperfused mitochondria would be promoted by decreased Myo19 recruitment to mitochondria, we suggest that MTX3 knockdown might also reduce the mitochondrial association of other proteins and that a combination of phenotypes accounts for the lack of an observed alteration in mitochondrial morphology. Indeed, the depletion of metaxins has been shown to alter microtubule-based trafficking of mitochondria, presumably by reducing mitochondrial levels of plus- and minus-end-directed motors (Zhao et al., 2021). Moreover, metaxins have been implicated in the import of β-barrel proteins into the mitochondrial outer membrane (Höhr et al., 2015).
Myo19 might act as an F-actin tether for mitochondrial fragmentation, but what specific population of actin filaments is involved? Our data indicate that Myo19 does not recruit F-actin to mitochondria, although we cannot rule out the possibility that our F-actin probes lacked the appropriate sensitivity to visualize small scale changes in F-actin enrichment. Instead, we asked whether Myo19 interacts with local F-actin populations to fragment mitochondria. There is a population of F-actin that assembles on mitochondria as a wave and might independently drive fission (Moore et al., 2016); however, depletion of Myo19 had no effect on the size of the actin wave (Moore et al., 2021) and, in initial experiments, this intervention did not clearly prevent wave-mediated mitochondrial fragmentation.
Next, we considered the F-actin nucleator Spire1C, which is localized to mitochondria and collaborates with the ER-resident formin INF2-CAAX to facilitate mitochondrial fission. We hypothesized that Myo19 interacts specifically with Spire1C/INF2-CAAX-generated actin filaments in order to induce mitochondrial fission. Our data support this hypothesis. INF2-CAAX-generated F-actin is bundled by fascin for mitochondrial fission, and F-actin bundles have been directly observed at mitochondrial fission sites with platinum-replica electron microscopy (Lin et al., 2019; Yang and Svitkina, 2019). Filament bundling might serve to locally increase the number of binding sites for Myo19, lowering the probability that the motor diffuses away as it progresses through its mechanochemical cycle.
The observation that Myo19 overexpression fragments mitochondria in a Spire1C- and INF2-CAAX-dependent manner suggests that this motor specifically promotes mitochondrial fission, instead of inhibiting fusion, and our data investigating the requirement for Drp1 activity support this interpretation. Mechanistically, the most likely possibility is that Myo19-dependent tethering of mitochondria to ER-associated actin filaments induces mitochondrial fission by stabilizing mito–ER contacts, which are the sites of many fission events. We tested this possibility by first verifying the presence of Myo19 at mito–ER contacts as defined by split-GFP fluorescence. Then, we used a split-luciferase system and found that Myo19 depletion reduces mito–ER contacts. These data support our hypothesis that Myo19 stabilizes mito–ER contacts specifically for mitochondrial fission.
Mitochondria are the major cellular source of ATP and myosins dynamically associate with actin upon ATP binding, hydrolysis and product release – thus, we wonder whether Myo19 constitutes a sensor of oxidative phosphorylation, as others have hypothesized (Adikes et al., 2013). Given our findings, the motor could respond by increasing or decreasing the degree of mito–ER contacts. Indeed, mito–ER contacts are enhanced and mitochondrial fission is upregulated in hypoxic conditions in which ATP production is diminished and, thus, Myo19–actin binding should be promoted (Chai et al., 2021). How Myo19-mediated linkage of mitochondria to the ER promotes mitochondrial fission is unclear. There is evidence to support a model in which Ca2+ transfer between the two organelles is critical to fission (Chakrabarti et al., 2018). Other studies suggest a mechanical model, in which ER linkages might render mitochondria stable in space, so they might be stretched and thinned to a breaking point through the action of microtubule-based motors (Helle et al., 2017; Mahecic et al., 2021). Further study of the questions outlined here is warranted, given the critical role mitochondrial fission plays in maintaining cellular homeostasis and in the pathogenesis of many debilitating disease states.
MATERIALS AND METHODS
Reagents
The following primary antibodies were used for western blots and immunofluorescence: anti-Myo19 (Abcam, ab174286), anti-TOMM20 (Santa Cruz Biotechnology, sc-17764), anti-Miro1 (Abcam, ab211363), anti-Miro2 (Proteintech, 11237-1-AP), anti-MTX1 (Proteintech, 15529-1-AP), anti-MTX2 (Santa Cruz Biotechnology, sc-514231), anti-MTX3 (Abcam, ab222048), anti-Hsp60 (Atlas Antibodies, HPA001523), anti-Drp1 (Santa Cruz Biotechnology, sc-101270, immunofluorescence), anti-Drp1 (Abcam, ab56788, western blotting), anti-phospho-S616 Drp1 (Cell Signaling Technology, 3455), anti-Mfn2 (Santa Cruz Biotechnology, sc-515647), anti-Fis1 (Santa Cruz Biotechnology, sc-376447) and anti-Mff (Santa Cruz Biotechnology, sc-398617). We used the following secondary antibodies: Alexa Fluor 405 anti-mouse IgG (Invitrogen, A31553), Alexa Fluor 546 anti-rabbit IgG (Invitrogen, A10040), Alexa Fluor 647 anti-rabbit IgG (Invitrogen, A31573), Alexa Fluor 488 anti-mouse IgG (Invitrogen, A11029), Alexa Fluor 488 anti-rabbit IgG (Invitrogen, A11034), anti-mouse IgG (Jackson ImmunoResearch, 115-005-003) custom co-labeled with Alexa Fluor 405 and Alexa Fluor 488, anti-rabbit IRDye 680RD (LI-COR, 926-68073), anti-mouse IRDye 800CW (LI-COR, 626-32212), and anti-rabbit IRDye 800CW (LI-COR, 926-32213).
The dyes used were: MitoTracker Deep Red FM (Thermo Fisher Scientific, M22426), HaloTag TMR ligand (Promega, G825A), JF549-SNAP (provided by Luke Lavis, Janelia Farms), JF646-Halo (Promega) and Alexa Fluor 647 phalloidin (Thermo Fisher Scientific, A22287).
The Myo19 plasmids used in this study include those encoding human GFP–Myo19 and Myo19–GFP from O.A.Q.-C. (Quintero et al., 2009). We generated additional plasmids encoding the following Myo19 constructs by mutagenesis and subcloning: GFP–Myo19 G417A, GFP–Myo19 Δ2IQ, GFP–Myo19 R659L and Halo–MsMyo19 (subcloned from pcR2.1; MsMyo19; supplied by O.A.Q.-C.). Additional plasmids used were: pHaloTag vector (Promega), pSNAPf (New England Biolabs), SNAP–Drp1 K38A (Drp1 K38A sequence from Addgene 45161, subcloned into pSNAPf), AcGFP–Sec61β (Addgene, 15108), ER split-luciferase construct cDNA3-CRluc92-ER (Cho et al., 2017), mitochondrial split-luciferase construct pcDNA-Mit-NRluc91 (both gifts from Jeffrey Golden, Cedars-Sinai Medical Center) (Cho et al., 2017) and LifeAct-mScarlet-N1(Addgene, 85054).
The siRNAs used were: Control (ON-TARGETplus, Horizon, D-001810-01-20), Myo19 (Santa Cruz, sc-93640), Myo19 (ON-TARGETplus, Horizon, 5′-CAAAGGAGCCAGUGAGGACUU-3′), MTX1 (Santa Cruz Biotechnology, sc-88250) MTX2 (Santa Cruz Biotechnology, sc-95035), MTX3 (Santa Cruz Biotechnology, sc-92062), Spire1C (Moore et al., 2021) and INF2-CAAX (Moore et al., 2021).
Cell culture and transfections
HeLa-M cells (Andrew Peden, Cambridge Institute for Medical Research, Cambridge, UK; authenticated by STR profiling and tested negative for mycoplasma), C2C12 mouse myoblasts [American Type Culture Collection (ATCC), Manassas, VA, USA; CRL-1772], and Cos7 cells (ATCC, CRL-1651) were grown using maintenance medium – Dulbecco's modified Eagle medium (Corning, 10-017-CV) supplemented with 10% fetal bovine serum (HyClone SH30071.03) and 1% GlutaMAX (Gibco, 35050061). Mito–ER contact reporter HeLa cells (Tashiro et al., 2020) were grown in the same medium but with 300 µg/ml hygromycin (Corning, 30-240-CR) and 500 µg/ml G418 (Univ. of Penn. Cell Center). Cells were maintained in an incubator at 37°C, 5% CO2 and passaged using trypsin. C2C12 cells were maintained at <75% confluency to prevent differentiation. For imaging experiments, cells were plated on #1.5 glass-bottomed dishes (Mattek, P35G-1.5-20-C).
Plasmid transfections were performed with FuGENE 6 transfection reagent (Promega, E269A). Myo19 constructs or control vectors were transfected 48 h prior to imaging (except for Fig. S5D, where transfection of all components was performed 24 h before), whereas for the split-luciferase assay, plasmids were transfected 60 h prior to luminescence analysis. If cells were pre-treated with siRNA, the medium was replaced prior to plasmid transfection.
siRNA transfections were performed at a final concentration of 40 nM oligonucleotide using Lipofectamine RNAiMAX (Invitrogen, 13778-150). siRNAs to Myo19, Spire1C, INF2-CAAX and metaxins were added 72 h prior to imaging or cell harvesting.
Western blotting
Cells were pelleted, washed with PBS, and RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS and protease inhibitors) was added for lysis. For the split-luciferase assay, RIPA buffer was added to PBS-washed adhered cells in the 96-well plate. After a 30-min incubation on ice with agitation, lysates were clarified by a 10 min, 4°C spin at 17,000 g. A bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) was performed to measure total protein concentration. SDS-PAGE gels were loaded with equal amounts of protein for each experiment, then transferred onto PVDF membranes (Millipore Immobilon FL 0.45 μm). Membranes were blocked and stained with REVERT Total Protein Dye (LI-COR, 926-11011), then incubated overnight at 4°C with antibodies at the following concentrations: anti-Myo19 (1:2000), anti-MTX1 (1:600), anti-MTX2 (1:500), anti-MTX3 (1:1000), anti-Drp1 (1:500), anti-phospho-S616-Drp1 (1:1000), anti-Mfn2, anti-Mff (1:500) or anti-Fis1 (1:50). Membranes were then incubated with fluorescent secondary antibodies (LI-COR, 1:20,000) for 1 h at room temperature and imaged using an Odyssey CLx imaging system. Blots were analyzed with Image Studio. Antibody signals were normalized to total protein stain signals. See Fig. S7 for uncropped blots.
Photobleaching experiment
Cos7 cells expressing the Halo-tagged mouse Myo19 were labeled with 2.5 µM Janelia Fluor 646-Halo ligand (Promega) for 15 min, washed twice with Ca2+- and Mg2+-free Dulbecco PBS (dPBS; GIBCO), returned to the culture medium and left in the incubator for 30 min. Cells were then washed twice with dPBS, collected in dPBS and pelleted at 5000 g for 5 min. The cell pellet was then lysed in 40 mM HEPES, 1 mM EDTA, 120 mM NaCl, 0.1% Triton X-100 and 1 mM magnesium ATP (pH 7.4) supplemented with protease inhibitors (1 mM PMSF, 0.01 mg/ml Nα-p-tosyl-L-arginine methyl ester, 0.01 mg/ml leupeptin, 0.001 mg/ml pepstatin A and 1 mM dithiothreitol). The lysate was left on ice for 10 min and clarified at 17,000 g for 10 min at 4°C.
Flow chambers were prepared using plasma-cleaned #1.5 glass coverslips (Thermo Fisher Scientific) coated with 0.5% colloidin (Electron Microscopy Sciences) attached to glass slides (Corning) using double-sided tape and vacuum grease (Dow Corning). Cell lysates were diluted in BRB80 buffer (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.80), flowed into the chamber, and incubated at room temperature to allow non-specific surface binding. The dilution and incubation times were adjusted for each video to ensure that there were enough puncta visible on the surface while maintaining distance between each puncta (dilutions ranged from 1:100 to 1:1000 and incubation times ranged from 10 to 30 s). The chamber was washed with BRB80 and sealed with vacuum grease. In each video, there were 600–700 bleaching events.
Split-luciferase assay
For the split luciferase assay, cells were seeded into a 96 well plate and, following transfection with siRNAs and plasmids, cells were incubated with EnduRen Live Cell Substrate (Promega, E6481), as prescribed by the manufacturer. Luminescence was recorded using a plate reader (BioTek, CYTATION 5) and duplicate values were averaged. Luminescence values were normalized within each biological replicate based on protein concentration, as measured with a BCA assay, and then further normalized based on reporter expression levels, as assessed by western blotting.
Immunofluorescence
Cells were fixed at 37°C with 4% paraformaldehyde for 10 min. For mito–ER contact reporter cells, 200 ng/ml doxycycline was added prior to fixation. After three PBS washes, cells were permeabilized for 5 min with 0.2% Triton X-100. Cells were blocked in buffer containing 0.5% bovine serum albumin (BSA), 2.5% normal goat serum and 2.5% normal donkey serum at 37°C for 30 min, washed with PBS, then incubated overnight in primary antibody diluted in 0.5% BSA at 4°C. The dilutions for primary antibodies were as follows: anti-Myo19 (for STORM, 1:10–1:20,480, saturating for confocal microscopy), anti-TOMM20 (1:40), anti-Miro1 (1:100), anti-Miro2 (1:300), anti-Hsp60 (1:100) and anti-Drp1 against total Drp1 (1:50). The next day, cells were washed three times with PBS and incubated with secondary antibody and phalloidin diluted to 1:300 and 1:40–1:50 in 0.5% BSA, respectively, for a 1 h room-temperature incubation. A custom anti-mouse secondary antibody was used at a 1:100 dilution for STORM experiments. For confocal microscopy experiments, cells were mounted in Slow Fade Diamond Mounting Media (Thermo Fisher Scientific, S36972) for imaging.
Confocal and TIRF microscopy
All confocal microscopy was conducted with an UltraView VoX Perkin Elmer spinning-disk system on a Nikon Eclipse Ti Microscope using an Apochromat 100×1.49 numerical aperture oil immersion objective (Nikon). Imaging was performed with an EM-charge-coupled device camera (C9100; Hamamatsu Photonics) or a CMOS camera (Orca-Fusion, Hamamatsu Photonics). For quantitated experiments, microscope acquisition parameters were held constant and, within each biological replicate, the same number of cells were imaged per group (n=20–30, except the peri-mitochondrial actin experiment, for which n=50). Prior to imaging live cells, MitoTracker Deep Red FM was added to the maintenance medium at 1:1000–1:2000 for 10 min. After one wash, the cells were put in imaging medium, composed of 10% fetal bovine serum and 1% GlutaMAX in Leibovitz's medium (Gibco, 11415-064), and set for ∼1 h at 37°C in the microscope chamber. In assessing mitochondrial morphology after Myo19 or MTX3 depletion, the cells imaged were the ones that first came into the field of view when scanning in the GFP-alone channel. For quantitated experiments involving overexpression of Myo19, plates were scanned in the Myo19 channel and the cells imaged were the ones that were in a visually determined range of expression level that was high relative to all transfected cells. For the R659L experiment, WT GFP–Myo19-expressing cells were imaged first and mutant cells were imaged subsequently to have a dataset in which the two conditions expressed equally. Equal Myo19 expression between groups was ensured (see the Image analysis section in the Materials and Methods). Control, i.e. GFP/SNAP-alone-expressing, cells in these experiments were imaged at random. When imaging fixed cells in quantitated experiments (except Fig. S5C, see above), plates were scanned in the TOMM20 channel and cells were randomly chosen for imaging.
For the photobleaching experiment, the same imaging set up was utilized but for TIRF microscopy. Illuminating the samples to excite the labels on Myo19, three 10 min time-lapse videos were captured per chamber. Videos were captured at 1 fps.
STORM
Fixed and immunostained cells were transferred to β-mercaptoethylamine hydrochloride (MEA) buffer (7.62 mg/ml MEA, 4.95% glucose and a glucose oxidase/catalase oxygen-scavenging system, all in PBS). Prior to the expiration of 2.5 h, cells were imaged by STORM using an Oxford Nanoimager S system (ONI) with a 1.4 numerical aperture 100× oil immersion objective (Olympus) and a Hamamatsu Flash 4 V3 sCMOS camera. For the antibody titration experiment, the plate was scanned in the TOMM20 channel, and we picked cells to image randomly (two per biological replicate), whereas for the latrunculin experiment, we ensured that the mitochondrial network could be visualized in the Myo19 channel before choosing cells to image (three technical replicates). For each cell imaged, a conventional epifluorescence image in both the TOMM20/custom Alexa Fluor 488-labeled secondary antibody channel and the Myo19/commercial Alexa Fluor 647-labeled secondary antibody channel was acquired. Following this, the latter channel was imaged with STORM, where a 15 ms exposure for 25,000 frames was used, given that at this frame, blinking had decayed substantially. Images in which drift was apparent were excluded (one and two from the ‘512’ and ‘256’ groups, respectively, in anti-Myo19 antibody titration experiment).
Image analysis
To analyze mitochondrial morphology, for overexpression experiments, the average Myo19 signal across each cell was found in Fiji, using cytosolic Myo19 signals and gaps in the MitoTracker signal to identify cell boundaries. Only cells with Myo19 signals within one standard deviation of the mean across groups were further analyzed. Qualitative analysis of mitochondrial morphology was carried out as follows: the mitochondrial network of cells was characterized as being either hyperfused, reticular, intermediate or fragmented by an investigator blinded to the experimental condition. To quantitatively analyze mitochondrial morphology, a subset of images was used to train the machine learning program Ilastik (https://www.ilastik.org/) to identify mitochondria, then Ilastik was used to segment mitochondria and the resulting binary masks were used to determine the mean mitochondrial area per cell. Three iterations, each time with additional training, were required to produce the reported trend for the overexpression experiment, as in the first two iterations we noticed that low-signal mitochondria and clustered mitochondria were not being segmented properly. Myo19-overexpressing cells with unusual signs of stress, such as blebbing or multiple and/or fractured nuclei were further excluded; sampling three out of six experiments, this corresponded to less than 3.9% of total cells examined.
In order to analyze the percentage of mitochondrial area in the perinuclear space, we segmented mitochondria in individual cells using auto-intensity thresholding in Fiji and measured the total area. Then, for each cell, we used a combination of the GFP fill channel and the MitoTracker channel to manually outline the nucleus. Specifically, nuclei were identified as the large circular mitochondria-free area within cells, and in the GFP channel as the large intracellular circular area of differing intensity. We then used the ‘enlarge’ function to expand this region of interest (ROI) by 4 μm, a ‘perinuclear’ cutoff used by others studying Myo19 (Oeding et al., 2018). We measured the area of mitochondria within this expanded ROI. Then, we divided this perinuclear mitochondrial area by the initial total mitochondrial area to obtain the reported percentage metric. Cells were excluded from analysis if their nucleus could not be faithfully outlined with our markers.
Mitochondrial Drp1 intensity and puncta density analyses were carried out as follows: mitochondria were segmented in Fiji based on intensity thresholding of the TOMM20 signal and the resulting masks were used to determine the average intensity or the density of puncta in the Drp1 channel. To determine puncta density, Drp1 signal was intensity thresholded, the ‘analyze particles’ function was run with a lower limit of 0.05 μm2, and the number of particles detected was divided by the area of mitochondria. In the experiments concerning metaxins, mitochondrial Myo19 and Miro protein intensity was determined by intensity thresholding the TOMM20 signal within the mask measuring the relevant signal, and subtracting the background signal defined as the mean fluorescence in a region unoccupied by cells. The same pipeline was applied when measuring peri-mitochondrial actin.
For analysis of the percentage of cellular area occupied by mitochondria after knockdown of metaxins, the cells were outlined using non-specific staining from the anti-Miro2 antibody as a guide using Fiji. Mitochondria were segmented using intensity thresholding of the anti-TOMM20 antibody signal and their area was measured. We divided this mitochondrial area by the total cellular area to get the reported percentage metric.
To analyze the percentage of mito–ER contacts with overlapping Myo19 in Fiji, for each cell, we used intensity thresholding to segment mito–ER contact puncta and Myo19 puncta. Only Myo19 puncta within the mitochondrial area (as defined by segmenting the TOMM20 signal) were included in further analysis. We used the multi-measure tool to determine which of the mito–ER contact puncta overlapped at all with Myo19 puncta.
In analyzing STORM data, a subset of conventional epifluorescence TOMM20 images were used to train the machine learning program Ilastik. Using the trained program, mitochondria were segmented for all images. For Myo19 localizations within mitochondria, MATLAB was used to perform Voronoi tessellation analysis similarly to Levet et al. (2015). A Voronoi cluster was defined as a collection of Voronoi polygons with areas smaller than a manually chosen threshold. The x- and y-coordinates of localizations were used to generate Voronoi polygons, and the shoelace algorithm was applied in determining their areas. Localizations with areas below the threshold and sharing a Voronoi polygon edge were grouped together into different clusters and, for each cluster, the nearest neighbor distance and number of localizations were recorded. For the anti-Myo19 antibody titration experiment, clusters that had density (number of localizations per area) beyond two standard deviations from the mean were first excluded as they typically represent imaging artifacts.
To calculate the number of Myo19 heavy chains per cluster, we used the Myo19 STORM images obtained by staining with anti-Myo19 antibody at a 1:20,480 dilution as the single Myo19 heavy-chain calibration. The number of localizations per cluster was plotted as a histogram distribution and fitted to a lognormal function as below:
where PDF is the probability density function, the x represents the number of localizations, and the mean (µ) and variance (σ) were obtained from the fit. The double Myo19 heavy-chain calibration function was obtained by a linear convolution of the single Myo19 heavy-chain calibration function using a custom-written MATLAB code (Gyparaki et al., 2021). The frequency distribution of the number of localizations per Myo19 cluster from images stained by saturating amount of antibody were fitted to the combination of calibration functions with weights w1 and w2 corresponding to the proportions of one or two Myo19 heavy chains.
Concerning the photobleaching experiment, for each separate puncta in a video, a circular region of interest was drawn in ImageJ and the intensity over time of that region was measured. The ‘Make Band’ function was used to generate a 2-pixel-wide band from the circular ROI and this resulting background intensity was measured for the duration of the video. The intensity of the puncta for each frame was background adjusted. These background-corrected data were then shifted such that the minimum value was zero before normalizing between zero and one. The normalized data were smoothed using a Savitzky–Golay filter with a 41-frame window using a parabolic fitting. The first derivative of the smoothed curve was calculated, and this was smoothed using the same Savitzky–Golay filter. A bleach step was counted if the smoothed first derivative had a value less than or equal to −0.005 and the data point 25 s later on the smooth normalized curve had a difference in intensity value less than or equal to −0.2. The percentage of single or double bleach steps was recorded.
Supplementary Material
Acknowledgements
We thank Mariko Tokito for technical assistance in cloning. We are also grateful for aid from Melina Gyparaki and Arian Arab with STORM experiments.
Footnotes
Author contributions
Conceptualization: S.M.C., O.A.Q.-C., E.M.O., M.L., E.L.F.H.; Methodology: S.M.C., C.P.T., Q.T., M.L.; Software: M.L.; Validation: S.M.C.; Formal analysis: S.M.C., C.P.T., Q.T., E.E.B., M.L.; Investigation: S.M.C., C.P.T., E.E.B., J.A.R.; Resources: E.L.F.H.; Data curation: S.M.C.; Writing - original draft: S.M.C., C.P.T., Q.T., M.L., E.L.F.H.; Writing - review & editing: S.M.C., C.P.T., Q.T., O.A.Q.-C., E.M.O., M.L., E.L.F.H.; Visualization: S.M.C., C.P.T.; Supervision: E.M.O., M.L., E.L.F.H.; Project administration: E.L.F.H.; Funding acquisition: O.A.Q.-C., E.L.F.H.
Funding
This work was funded by support from the National Institutes of Health (R35 GM126950 to E.L.F.H.; RM1 GM136511 to E.L.F.H., E.M.O. and M.L.; and R01 GM133842 to M.L.), Sigma Xi [Grants in Aid of Research (GIAR) to S.M.C.] and the School of Arts and Sciences, University of Richmond (Faculty Research Fellowship to O.A.Q.-C.; and Summer Undergraduate Research Fellowship to J.A.R.). Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.260612.reviewer-comments.pdf
References
- Adikes, R. C., Unrath, W. C., Yengo, C. M. and Quintero, O. A. (2013). Biochemical and bioinformatic analysis of the myosin-XIX motor domain: analysis of the MYO19 motor domain. Cytoskeleton 70, 281-295. 10.1002/cm.21110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer, S. L. (2013). Mitochondrial dynamics — mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 369, 2236-2251. 10.1056/NEJMra1215233 [DOI] [PubMed] [Google Scholar]
- Bocanegra, J. L., Fujita, B. M., Melton, N. R., Cowan, J. M., Schinski, E. L., Tamir, T. Y., Major, M. B. and Quintero, O. A. (2020). The MyMOMA domain of MYO19 encodes for distinct Miro-dependent and Miro-independent mechanisms of interaction with mitochondrial membranes. Cytoskeleton 77, 149-166. 10.1002/cm.21560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella Zanacchi, F., Manzo, C., Magrassi, R., Derr, N. D. and Lakadamyali, M. (2019). Quantifying protein copy number in super resolution using an imaging-invariant calibration. Biophys. J. 116, 2195-2203. 10.1016/j.bpj.2019.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, P., Cheng, Y., Hou, C., Yin, L., Zhang, D., Hu, Y., Chen, Q., Zheng, P., Teng, J. and Chen, J. (2021). USP19 promotes hypoxia-induced mitochondrial division via FUNDC1 at ER-mitochondria contact sites. J. Cell Biol. 220, e202010006. 10.1083/jcb.202010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti, R., Ji, W.-K., Stan, R. V., De Juan Sanz, J., Ryan, T. A. and Higgs, H. N. (2018). INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol. 217, 251-268. 10.1083/jcb.201709111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, I.-T., Adelmant, G., Lim, Y., Marto, J. A., Cho, G. and Golden, J. A. (2017). Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum–mitochondrial contacts. J. Biol. Chem. 292, 16382-16392. 10.1074/jbc.M117.795286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann, N., Van De Linde, S., Alon, A., Ljaschenko, D., Keung, X. Z., Holm, T., Rings, A., Diantonio, A., Hallermann, S., Ashery, U.et al. (2014). Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 5, 4650. 10.1038/ncomms5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton, A. R., Jongens, T. A. and Holzbaur, E. L. F. (2021). Mitochondrial adaptor TRAK2 activates and functionally links opposing kinesin and dynein motors. Nat. Commun. 12, 4578. 10.1038/s41467-021-24862-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J. R., Lackner, L. L., West, M., Dibenedetto, J. R., Nunnari, J. and Voeltz, G. K. (2011). ER tubules mark sites of mitochondrial division. Science 334, 358-362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyparaki, M. T., Arab, A., Sorokina, E. M., Santiago-Ruiz, A. N., Bohrer, C. H., Xiao, J. and Lakadamyali, M. (2021). Tau forms oligomeric complexes on microtubules that are distinct from tau aggregates. Proc. Natl. Acad. Sci. USA 118, e2021461118. 10.1073/pnas.2021461118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne, J. L., Mehta, P. R., Singh, P. P., Wong, N. Q. and Quintero, O. A. (2016). Positively charged residues within the MYO19 MyMOMA domain are essential for proper localization of MYO19 to the mitochondrial outer membrane: basic residues mediate MYO19 binding to mitochondria. Cytoskeleton 73, 286-299. 10.1002/cm.21305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle, S. C. J., Feng, Q., Aebersold, M. J., Hirt, L., Grüter, R. R., Vahid, A., Sirianni, A., Mostowy, S., Snedeker, J. G., Šarić, A.et al. (2017). Mechanical force induces mitochondrial fission. Elife 6, e30292. 10.7554/eLife.30292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhr, A. I. C., Straub, S. P., Warscheid, B., Becker, T. and Wiedemann, N. (2015). Assembly of β-barrel proteins in the mitochondrial outer membrane. Biochim. Biophys. Acta 1853, 74-88. 10.1016/j.bbamcr.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Hung, V., Lam, S. S., Udeshi, N. D., Svinkina, T., Guzman, G., Mootha, V. K., Carr, S. A. and Ting, A. Y. (2017). Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6, e24463. 10.7554/eLife.24463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, H., Jiang, D., Hu, X., Du, W., Ji, L., Yang, Y., Li, X., Sho, T., Wang, X., Li, Y.et al. (2021). Mitocytosis, a migrasome-mediated mitochondrial quality-control process. Cell 184, 2896-2910.e13. 10.1016/j.cell.2021.04.027 [DOI] [PubMed] [Google Scholar]
- Kashatus, J. A., Nascimento, A., Myers, L. J., Sher, A., Byrne, F. L., Hoehn, K. L., Counter, C. M. and Kashatus, D. F. (2015). Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537-551. 10.1016/j.molcel.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleele, T., Rey, T., Winter, J., Zaganelli, S., Mahecic, D., Perreten Lambert, H., Ruberto, F. P., Nemir, M., Wai, T., Pedrazzini, T.et al. (2021). Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature 593, 435-439. 10.1038/s41586-021-03510-6 [DOI] [PubMed] [Google Scholar]
- Korobova, F., Ramabhadran, V. and Higgs, H. N. (2013). An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464-467. 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova, F., Gauvin, T. J. and Higgs, H. N. (2014). A role for myosin II in mammalian mitochondrial fission. Curr. Biol. 24, 409-414. 10.1016/j.cub.2013.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse, A. M., Zappaterra, M. D., Rube, D. A. and Van Der Bliek, A. M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815-826. 10.1016/s1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Levet, F., Hosy, E., Kechkar, A., Butler, C., Beghin, A., Choquet, D. and Sibarita, J. B. (2015). SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat Methods 12, 1065-1071. 10.1038/nmeth.3579 [DOI] [PubMed] [Google Scholar]
- Lin, S., Huang, C., Gunda, V., Sun, J., Chellappan, S. P., Li, Z., Izumi, V., Fang, B., Koomen, J., Singh, P. K.et al. (2019). Fascin controls metastatic colonization and mitochondrial oxidative phosphorylation by remodeling mitochondrial actin filaments. Cell Rep. 28, 2824-2836.e8. 10.1016/j.celrep.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López–Doménech, G., Covill–Cooke, C., Ivankovic, D., Halff, E. F., Sheehan, D. F., Norkett, R., Birsa, N. and Kittler, J. T. (2018). Miro proteins coordinate microtubule– and actin–dependent mitochondrial transport and distribution. EMBO J. 37, 321-336. 10.15252/embj.201696380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón, O. C., Song, Z., Chen, H. and Chan, D. C. (2013). Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659-667. 10.1091/mbc.e12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z., Ma, X.-N., Zhang, H.-M., Ji, H.-H., Ding, H., Zhang, J., Luo, D., Sun, Y. and Li, X. (2014). Mouse myosin-19 is a plus-end-directed, high-duty ratio molecular motor. J. Biol. Chem. 289, 18535-18548. 10.1074/jbc.M114.569087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahecic, D., Carlini, L., Kleele, T., Colom, A., Goujon, A., Matile, S., Roux, A. and Manley, S. (2021). Mitochondrial membrane tension governs fission. Cell Rep. 35, 108947. 10.1016/j.celrep.2021.108947 [DOI] [PubMed] [Google Scholar]
- Majstrowicz, K., Honnert, U., Nikolaus, P., Schwarz, V., Oeding, S. J., Hemkemeyer, S. A. and Bähler, M. (2021). Coordination of mitochondrial and cellular dynamics by the actin-based motor Myo19. J. Cell Sci. 134, jcs255844. 10.1242/jcs.255844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor, U., Bartholomew, S., Golani, G., Christenson, E., Kozlov, M., Higgs, H., Spudich, J. and Lippincott-Schwartz, J. (2015). A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife 4, e08828. 10.7554/eLife.08828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschi, D., Gramlich, M. W. and Klyachko, V. A. (2018). Myosin V functions as a vesicle tether at the plasma membrane to control neurotransmitter release in central synapses. Elife 7, e39440. 10.7554/eLife.39440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcintosh, B. B. and Ostap, E. M. (2016). Myosin-I molecular motors at a glance. J. Cell Sci. 129, 2689-2695. 10.1242/jcs.186403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi, S., López-Doménech, G., Halff, E. F., Covill-Cooke, C., Ivankovic, D., Melandri, D., Arancibia-Cárcamo, I. L., Burden, J. J., Lowe, A. R. and Kittler, J. T. (2019). Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat. Commun. 10, 4399. 10.1038/s41467-019-12382-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A. S., Wong, Y. C., Simpson, C. L. and Holzbaur, E. L. F. (2016). Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 7, 12886. 10.1038/ncomms12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A. S., Coscia, S. M., Simpson, C. L., Ortega, F. E., Wait, E. C., Heddleston, J. M., Nirschl, J. J., Obara, C. J., Guedes-Dias, P., Boecker, C. A.et al. (2021). Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 591, 659-664. 10.1038/s41586-021-03309-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeding, S. J., Majstrowicz, K., Hu, X.-P., Schwarz, V., Freitag, A., Honnert, U., Nikolaus, P. and Bähler, M. (2018). Identification of Miro1 and Miro2 as mitochondrial receptors for myosin XIX. J. Cell Sci. 131, jcs219469. 10.1242/jcs.219469 [DOI] [PubMed] [Google Scholar]
- Ong, S.-B., Subrayan, S., Lim, S. Y., Yellon, D. M., Davidson, S. M. and Hausenloy, D. J. (2010). Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 121, 2012-2022. 10.1161/CIRCULATIONAHA.109.906610 [DOI] [PubMed] [Google Scholar]
- Quintero, O. A., Divito, M. M., Adikes, R. C., Kortan, M. B., Case, L. B., Lier, A. J., Panaretos, N. S., Slater, S. Q., Rengarajan, M., Feliu, M.et al. (2009). Human Myo19 is a novel myosin that associates with mitochondria. Curr. Biol. 19, 2008-2013. 10.1016/j.cub.2009.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn, J. L., Patel, J. V., Neumann, B., Bulkescher, J., Mchedlishvili, N., Mcmullan, R. C., Quintero, O. A., Ellenberg, J. and Baum, B. (2014). Myo19 ensures symmetric partitioning of mitochondria and coupling of mitochondrial segregation to cell division. Curr. Biol. 24, 2598-2605. 10.1016/j.cub.2014.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel, A. and Fuller, M. T. (2001). Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867-874. 10.1242/jcs.114.5.867 [DOI] [PubMed] [Google Scholar]
- Sasaki, N., Shimada, T. and Sutoh, K. (1998). Mutational analysis of the switch II loop of Dictyostelium Myosin II. J. Biol. Chem. 273, 20334-20340. 10.1074/jbc.273.32.20334 [DOI] [PubMed] [Google Scholar]
- Sato, O., Sakai, T., Choo, Y.-Y., Ikebe, R., Watanabe, T. M. and Ikebe, M. (2022). Mitochondria-associated myosin 19 processively transports mitochondria on actin tracks in living cells. J. Biol. Chem. 298, 101883. 10.1016/j.jbc.2022.101883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, P., Ren, X., Meng, J., Kang, C., Wu, Y., Rong, Y., Zhao, S., Jiang, Z., Liang, L., He, W.et al. (2022). Mechanical instability generated by Myosin 19 contributes to mitochondria cristae architecture and OXPHOS. Nat. Commun. 13, 2673. 10.1038/s41467-022-30431-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneyer, B. I., Ušaj, M. and Henn, A. (2016). Myo19 is an outer mitochondrial membrane motor and effector of starvation-induced filopodia. J. Cell Sci. 129, 543-556. 10.1242/jcs.175349 [DOI] [PubMed] [Google Scholar]
- Shneyer, B. I., Ušaj, M., Wiesel-Motiuk, N., Regev, R. and Henn, A. (2017). ROS induced distribution of mitochondria to filopodia by Myo19 depends on a class specific tryptophan in the motor domain. Sci. Rep. 7, 11577. 10.1038/s41598-017-11002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Shurland, D. L., Ryazantsev, S. N. and van der Bliek, A. M. (1998). A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351-358. 10.1083/jcb.143.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoberger, A., Barua, B., Atherton, J. L., Shuman, H., Forgacs, E., Goldman, Y. E., Winkelmann, D. A. and Ostap, E. M. (2021). Myosin with hypertrophic cardiac mutation R712L has a decreased working stroke which is rescued by omecamtiv mecarbil. Elife 10, e63691. 10.7554/eLife.63691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, N., Ishihara, N., Jofuku, A., Oka, T. and Mihara, K. (2007). Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521-11529. 10.1074/jbc.M607279200 [DOI] [PubMed] [Google Scholar]
- Tashiro, S., Kakimoto, Y., Shinmyo, M., Fujimoto, S. and Tamura, Y. (2020). Improved split-GFP systems for visualizing organelle contact sites in yeast and human cells. Front. Cell Dev. Biol. 8, 571388. 10.3389/fcell.2020.571388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ušaj, M. and Henn, A. (2017). Kinetic adaptation of human Myo19 for active mitochondrial transport to growing filopodia tips. Sci. Rep. 7, 11596. 10.1038/s41598-017-11984-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda, T. Q., Abramson, P. D. and Spudich, J. A. (1996). The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. USA 93, 4459-4464. 10.1073/pnas.93.9.4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Song, P., Du, L., Tian, W., Yue, W., Liu, M., Li, D., Wang, B., Zhu, Y., Cao, C.et al. (2011). Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in parkinson disease. J. Biol. Chem. 286, 11649-11658. 10.1074/jbc.M110.144238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. and Svitkina, T. M. (2019). Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat. Cell Biol. 21, 603-613. 10.1038/s41556-019-0313-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle, R. J. and Van Der Bliek, A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062-1065. 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanacchi, F. C., Manzo, C., Alvarez, A. S., Derr, N. D., Garcia-Parajo, M. F. and Lakadamyali, M. (2017). A DNA origami platform for quantifying protein copy number in super-resolution. Nat. Methods 14, 789-792. 10.1038/nmeth.4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Song, E., Wang, W., Hsieh, C.-H., Wang, X., Feng, W., Wang, X. and Shen, K. (2021). Metaxins are core components of mitochondrial transport adaptor complexes. Nat. Commun. 12, 83. 10.1038/s41467-020-20346-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.