Abstract
Cognitive disorders are prevalent in people with HIV (PWH) despite antiretroviral therapy. Given the heterogeneity of cognitive disorders in PWH in the current era and evidence that these disorders have different etiologies and risk factors, scientific rationale is growing for using data-driven models to identify biologically defined subtypes (biotypes) of these disorders. Here, we discuss the state of science using machine learning to understand cognitive phenotypes in PWH and their associated comorbidities, biological mechanisms, and risk factors. We also discuss methods, example applications, challenges, and what will be required from the field to successfully incorporate machine learning in research on cognitive disorders in PWH. These topics were discussed at the National Institute of Mental Health meeting on “Biotypes of CNS Complications in People Living with HIV” held in October 2021. These ongoing research initiatives seek to explain the heterogeneity of cognitive phenotypes in PWH and their associated biological mechanisms to facilitate clinical management and tailored interventions.
Keywords: HIV, cognitive impairment, HIV-associated neurocognitive disorders, machine learning, deep learning
Despite the success of current antiretroviral therapies (ART) in achieving viral suppression and improving longevity of people with human immunodeficiency virus (PWH), the mechanisms underlying human immunodeficiency virus (HIV)-associated neurocognitive disorders (HAND) remain poorly understood and effective adjunctive therapies are still lacking [1]. While the prevalence of HIV-associated dementia has declined with viral suppression on ART, milder forms of HAND remain prevalent [1, 2]. The reported rates of HAND vary widely between studies of PWH who are virally suppressed, with estimates typically ranging from 20% to 50% [1–7]. Given the heterogeneity of cognitive disorders in PWH in the current ART era and evidence that these disorders have different etiologies and risk factors, scientific rationale is growing for using data-driven approaches to identify biologically defined subtypes (biotypes). Progress in research on blood and cerebrospinal fluid (CSF) biomarkers of HAND and wider availability of neuroimaging has further intensified interest in using data-driven approaches to understand biotypes of cognitive disorders in PWH.
The current research classification for cognitive disorders in PWH is the HAND criteria (often referred to as the Frascati criteria) published in 2007 [8]. These criteria rely on neurocognitive testing and assessment of functional ability to perform activities of daily living. While the HAND diagnostic scheme has significantly contributed to the conceptual framework for central nervous system (CNS) complications of HIV, methodologies vary across studies. Furthermore, the HAND diagnostic scheme utilizes a symptom-based approach to meet thresholds for diagnosis and determination of HAND severity (asymptomatic neurocognitive impairment, mild neurocognitive disorder, and HIV-associated dementia). Criteria vary across studies, particularly regarding the ascertainment of functional impairment. Moreover, potential biases in test construction and normative comparisons needed to establish HAND diagnoses have the potential to overpathologize cognitive impairment among individuals from racially/ethnically diverse populations [9]. Not surprisingly, studies have reported modest or inconsistent associations between HAND diagnostic categories and neuroimaging or blood/CSF biomarkers in PWH receiving ART [2].
Given the complex and highly dimensional nature of data available in the current era of NeuroHIV research, sophisticated approaches are needed to define cognitive subtypes and their associated comorbidities, biological mechanisms, and risk factors, and to develop diagnostic tools and personalized treatments [2]. One such approach is to employ data-driven models (machine learning) to discover, characterize, and explain distinct CNS biotypes among PWH, with the goal of discovering features to support clinical interventions. Data-driven models are also needed for initiatives using the Research Domain Criteria (RDoC) framework developed by the National Institute of Mental Health (NIMH) to integrate findings from measures of cognitive, affective, and psychosocial processes that cut across traditional diagnostic categories [10]. Applications of machine learning to understand cognitive disorders and mental health in PWH have enormous potential to advance the understanding of these disorders, but it requires availability of high-quality datasets, rich metadata, and combined expertise from stakeholders across multiple disciplines.
To review the state of the science and how to address gaps in knowledge, the NIMH held a virtual meeting on “Biotypes of CNS Complications in People Living with HIV” (21–22 October 2021). A working group was convened to lead a session on machine learning to understand cognitive phenotypes in PWH. Here, we summarize talks and discussions led by the machine learning working group on the following topics: (1) use of machine learning to advance the RDoC paradigm; (2) machine learning approaches for cognitive phenotyping in PWH; (3) what is required to make machine learning successful, including common data elements, high-quality datasets, and handling of confounds; (4) integration and harmonization of data across studies; and (5) validation of machine learning models. We also discuss how machine learning can lead to discoveries that can improve clinical management and development of personalized therapies.
THE RESEARCH DOMAIN CRITERIA INITIATIVE
The RDoC framework aims to cut across predefined diagnostic classifications to identify and characterize the mechanisms that underlie individual variability in complex mental health conditions, including cognitive impairment [10]. The goal of the initiative is to identify novel therapeutic targets to inform the development and implementation of tailored prevention and intervention strategies. Central to the RDoC approach is the integration of highly dimensional data across multiple units of analysis, from genes to circuits to behaviors. Traditional analytic methods are not ideally suited to accomplish this task. Previous work in HIV-uninfected individuals with mental health conditions (eg, psychosis, depression) have successfully overcome the inherent limitations of traditional analytics via use of machine learning methods that identify biotypes and potential underlying mechanisms using data-driven clustering and classification algorithms.
Because RDoC is an integrated transdiagnostic system for understanding neurobehavioral dysfunction, it provides more flexibility in analysis and interpretation than the HAND classification scheme. Machine learning studies that integrate laboratory biomarkers, multimodal neuroimaging, cognitive performance, and psychosocial factors/comorbidities may enable new insights into the diversity of cognitive phenotypes among PWH and reveal shared features between CNS disorders in PWH and other neurological and psychiatric disorders [11]. The combination of machine learning with methods for causal modeling represents a potentially powerful approach to traversing levels of analysis (eg, from genomic to proteomic to metabolomic), and generating hypotheses about mechanisms underlying HAND. The RDoC framework does not specify relationships among the units of analysis, so investigators will need to develop prior hypotheses about predicted associations to avoid problems with model misspecification and overfitting [12]. While machine learning is often described as an “agnostic” approach, prior work underscores the importance of incorporating a hypothesis-driven approach to machine learning applications in the behavioral sciences.
MACHINE LEARNING METHODS FOR CLINICAL PHENOTYPING
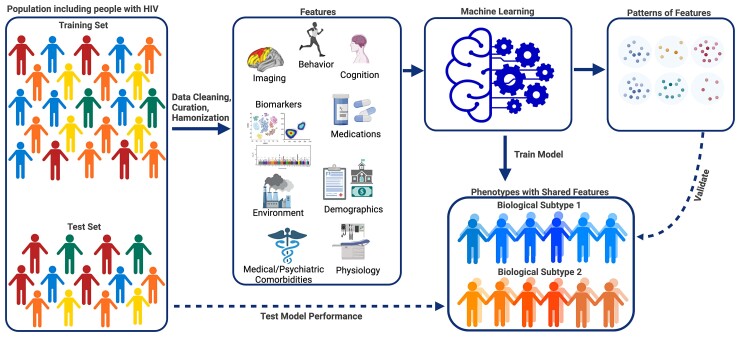
A graphical representation of machine learning to study cognitive phenotypes in PWH is shown in Figure 1. Machine learning approaches leverage large complex multimodal data to identify novel patterns that cannot be ascertained using traditional statistical approaches. Machine learning algorithms vary by design, assumptions, and limitations, so proper selection of a method depends on the question being addressed and data available, including the types (categorical and continuous) of input features (predictors) used for analysis (Table 1 and Table 2) [13, 14]. In this section, we summarize 3 overarching categories of machine learning: supervised learning, unsupervised learning, and deep learning (which can be trained using supervised or unsupervised approaches).
Figure 1.
Graphical representation of machine learning to understand cognitive phenotypes in people with HIV (PWH). Machine learning is a relatively new approach for identifying cognitive phenotypes in PWH. Availability of multimodal data provides an opportunity to study physiological, molecular, behavioral, environmental, and external factors that may contribute to an individual’s phenotype. To understand cognitive phenotypes in PWH, machine learning can analyze diverse data types, including neuroimaging, neurocognitive test performance, behavioral, and physiological data, using supervised or unsupervised approaches. A machine learning pipeline typically starts with a large representative dataset that has undergone data cleaning, curation, and harmonization. Features are selected/extracted from the initial dataset and used as inputs to train models to make classifications or predictions on a subject level. Models are then tested and validated on independent datasets. Deep learning techniques can model complex relationships between inputs and outputs and accept larger datasets and a wide variety of data inputs. As datasets become larger and more complex, machine learning methods will enable the identification of biological subtypes (biotypes) of cognitive disorders in PWH.
Table 1.
Machine Learning Approaches for Clinical Phenotyping
| Type of Machine Learning | Algorithms | Description |
|---|---|---|
| Supervised | Machine learning methods that train predictive models on labeled data sets. | |
| Categorical classification | Logistic regression, decision trees, support vector machine, k-nearest neighbors | Prediction of categorical outcomes using linear or nonlinear combinations of input data. |
| Continuous prediction | Linear regression, ridge/LASSO models, support vector machine, Gaussian process regression | Prediction of continuous outcomes. Input data can be continuous or categorical. |
| Unsupervised | Machine learning methods that identify patterns in unlabeled data sets. | |
| Clustering | k -means, fuzzy c-means, expectation maximization, DBScan, Gaussian mixture model, hierarchical methods (divisive, agglomerative) | Machine learning approaches that group unlabeled data based on similar patterns of features to identify latent classes and predict class membership of new data. |
| Dimensionality reduction | Principal component analysis, factor analysis, linear discriminant analysis, backward feature elimination, random forests | Methods to transform data sets with many features into lower-dimensional forms by selecting important features or combining features to capture variance in the data set while preserving data relationships as much as possible. |
| Deep learning | Multilayer perceptron, convolutional neural networks, recurrent neural networks, autoencoders | Machine learning methods that use training on artificial neural networks for representation learning; utilizes layers of nodes and edges resembling a simplified biological layered neural network to learn patterns or associations in large data sets by generating predictions from the input data and comparing them with ground truth annotations. The activation of a node or “neuron” depends on a weighted combination of inputs from the previous layer. |
Table 2.
Example Applications of Machine Learning for Cognitive Phenotyping in People with HIV
| Study Goals | Dataset | Data Type | Methods | Reference |
|---|---|---|---|---|
| Identify similarities in cognitive profiles among PWH and determine features associated with cognitive profiles | Cognitive testing and associated sociodemographic and clinical data |
Multisite clinical and cognitive testing
(N = 1646) |
Self-organizing maps and clustering (unsupervised deep learning); random forest (supervised) | Dastgheyb et al [15] |
| Determine predictors of cognitive impairment subtypes in PWH using sociodemographic, clinical, and cognitive test data | Cognitive testing and associated sociodemographic and clinical data | Multisite clinical and cognitive testing dataset (N = 370) | Univariate and multiple logistic regression, random forest models (supervised) | Tu et al [16] |
| Predict neurocognitive trajectories in children with perinatal HIV using demographics, clinical blood markers, and mental health indices | Cognitive testing and associated sociodemographic and clinical data | Multisite clinical and cognitive testing dataset (N = 285) | Gradient boosted multivariate regression; feature selection with SciKit and PDPBox (supervised) | Paul et al [17] |
| Determine relationships between sleep health and cognitive function based on HIV serostatus and investigate interpretation based on analytical approaches | Cognitive testing, questionnaires, actigraphy data | Multisite actigraphy, pulse oximetry, and cognitive testing dataset (N = 463) | Partial least-squares regression, multidimensional construct, and random forest (supervised); latent class analysis (unsupervised) | De Francesco et al. [18] |
| Classify HIV infection based on structural MRI data and associated regional volumetric data and determine which regions are implicated in HIV infection | MRI and associated diagnoses | Single-site MRI dataset (N = 310) | Multiple kernel learning; chained and single-step regularization and support vector machine (supervised) | Adeli et al [19] |
| Predict diagnosis and cognitive measures in individuals with alcohol use disorder and HIV using structural MRI | MRI and associated diagnoses | Single-site MRI dataset (N = 549) | Customized sparse logistic regression with joint feature-sample selection compared with joint feature-sample selection with sparse feature selection and support vector machine (supervised) | Adeli et al [20] |
| Predict HIV-associated cognitive impairment using clinical and MRI-derived features including grey matter volumes and white matter integrity | MRI, clinical features, and associated diagnoses | Single-site MRI and cognitive testing merged datasets (N = 101) | Support vector machine; feature selection with LASSO regression (supervised) | Xu et al [21] |
| Classify HIV and cognitive impairment status using minimally processed structural MRI | T1-weighted MRIs and associated diagnoses | Merged MRI datasets (N = 1449) | Convolutional neural network with domain-specific predictors (supervised deep learning) | Zhang and Zhao et al [22] |
| Determine resting state networks that differentiate between groups based on HIV serostatus and cognitive status | MRI and associated diagnoses | Merged MRI datasets (N = 1806) | Relief feature selection and convolutional neural network (supervised deep learning) | Luckett et al [23] |
| Classify frail status based on neuroimaging features (volumetric data, arterial spin labeling, resting state functional MRI) | MRI and associated diagnoses | Single-site MRI dataset (N = 105) | Gradient-boosted multivariate regression; feature selection with SciKit and PDPBox (supervised) | Paul et al [24] |
Abbreviations: HIV, human immunodeficiency virus; MRI, magnetic resonance imaging; PWH, people with HIV.
Supervised algorithms can be separated into classification algorithms for prediction of categorical outcomes (eg, cognitively impaired vs unimpaired) and regression algorithms for prediction of continuous outcomes (eg, neurocognitive scores). Training of algorithms consists of learning how to process the input features so the method can reproduce the outputs (outcome). Classification algorithms for categorical outcomes include decision trees, random forest, support vector machines (SVM), logistic regression, naive Bayesian models, and k-nearest neighbor, while algorithms for continuous outcomes include linear and polynomial regressions. Novel supervised learning methods are also being developed to understand cognitive disorders in PWH [17, 19, 20]. For example, Adeli et al developed a novel supervised method incorporating a step that uses sparse classification technology to identify brain regions impacted by HIV that yielded better classification accuracy when applied to magnetic resonance imaging (MRI) datasets than SVM or other traditional approaches [19]. In another study, these investigators utilized another supervised method to discover altered patterns in brain MRI in people with alcohol use disorder, HIV, or their comorbidity, and to identify diagnostic scores that predicted individual membership in these groups [20].
Unsupervised algorithms are useful when questions lack a known output, that is, where the assignment of samples to groups is unknown. Unsupervised learning methods include clustering and dimensionality reduction (Table 1). Clustering methods include k-means, hierarchical clustering, and latent class analysis, while dimensionality reduction methods include principal component analysis, factor analysis, linear discriminant analyses, and Uniform Manifold Approximation and Projection (UMAP). Common outputs include a grouping or summary of the input features, for example predictors of cognitive profiles (eg, demographic features, biomarkers, clinical phenotypes, mental health factors, neuroimaging parameters). Examples of unsupervised approaches to discover cognitive phenotypes in PWH are discussed in more detail in the next section [15, 25].
Deep learning can handle a larger number of features compared to the approaches mentioned above. Deep learning can extract features informative for diagnosis directly from raw or minimally processed input data using neural networks for representation learning [14]. Example applications in the field of HIV include rapid diagnostic testing [26], predicting HIV infection among social networks [27], and engagement in care [28]. Deep learning performs well for analysis of imaging data because neural networks can handle complex unstructured data; however, a drawback is computational complexity and potential uncertainty about the features that drive classification accuracy (the “black box dilemma”) [15]. Nevertheless, studies have successfully implemented deep learning to investigate neuroimaging signatures of cognitive impairment and frailty in PWH [29], classify individuals by HIV or cognitive status within specific age bins [23], or distinguish HAND from mild cognitive impairment due to non-HIV conditions [22]. Other high-dimensional datasets have also been used for deep learning to understand cognitive phenotypes in PWH (Table 2) and applications of these methods for discovery of clinical phenotypes will continue to rise.
Any of these algorithms mentioned above can be coupled with techniques for clustering and dimensionality reduction (Table 1). Machine learning pipelines in RDoC and NeuroHIV research often begin with clustering followed by classification/prediction methods to discover features and mechanisms that explain the clusters [11, 12, 15, 20, 30]. One potential problem with clustering methods is discovering clusters that are not meaningful. Challenges encountered in dimensionality reduction can include spurious associations due to confounding factors or noise in the data. Alternative methods such as factor mixture modeling may help to identify latent classes and latent variables, but typically require large sample sizes [31].
CLUSTERING TO IDENTIFY COGNITIVE PHENOTYPES IN PWH
Unsupervised clustering algorithms are widely used across diverse fields of medicine to identify clinical phenotypes [13, 14]. Here, we discuss examples using unsupervised clustering to identify cognitive subtypes in PWH based on similar features (eg, symptoms, clinical manifestations, cognitive impairment). Supervised methods can then be applied to find sociodemographic, clinical, and neuroimaging/biomarker features predictive of cluster membership [15, 17, 30].
An example is a study of 388 PWH with chronic infection and at least 3 months of ART use who participated in neurocognitive testing and multimodal MRI. The objective was to identify data-driven cognitive phenotypes and underlying features that correspond to subgroup classification. Paul et al [25] first applied hierarchical density-based clustering (HDBScan) with a UMAP variant to identify 4 cognitive phenotypes that differed in the degree of impairment on tests sensitive to frontal-subcortical disruption (ie, psychomotor speed, learning and recall, executive function). Gradient boosted multivariate regression, a form of ensemble machine learning, was then employed to identify combinations of features that could distinguish individual membership according to impaired versus normal cognitive performance. HIV disease markers, resting-state functional MRI and volumetrics, substance use, and psychosocial variables were included as potential input features. Interestingly, the analysis revealed that alterations in brain regions involved in addiction, substance use, and psychosocial factors (rather than HIV disease metrics) classified individuals into the data-driven cognitive subgroups [25].
Another example is a study by Dastgheyb et al that analyzed baseline neuropsychological data from the Women’s Interagency HIV Study (WIHS), a multicenter, longitudinal study of women with and without HIV [15]. An unsupervised deep learning algorithm called self-organizing maps (SOM, kohonen package in R) [32, 33] was used to reduce dimensionality and identify cognitive patterns among 1646 women (929 virally suppressed women with HIV, 717 women without HIV). SOMs reduce data complexity by taking information with multiple attributes (eg, neurocognitive test scores with timed values or number of correct answers) and generating a 2-dimensional output. The algorithm uses a competitive deep learning process to determine which nodes (“neurons”) respond (“activate”) to a set of data inputs and adjust weights of each node accordingly. The resulting network can be visualized in a structured topographic map. The authors then used clustering algorithms (mclust R package) [34] to identify 5 cognitive profiles in the cohort including impairments in (1) cognitive sequencing, (2) speed, (3) learning and recognition, (4) learning and memory, and (5) combination of learning, processing speed, attention, and executive functioning.
DISCOVERING PREDICTORS OF COGNITIVE PHENOTYPES IN PWH
Supervised machine learning is widely used to discover predictors of disease and has been successfully applied to neuroimaging and cognitive data. Dastgheyb et al used a supervised random forest classifier to identify factors associated with cognitive cluster membership in the WIHS study described above [15]. Random forest is an ensemble classification method that is sensitive to training data; thus, minor changes in training sets can result in different tree structures. To ensure predictive validity, bootstrap aggregations (10-fold resampling, repeated 5 times) was performed on a subset of the data (“training” data from 70% of the cohort) to ensure algorithms (“trees”) were trained on different subsets of data to make robust predictions. To identify potential predictors, variable (“feature”) importance was identified, and top variables associated with each profile were validated in the testing dataset (remaining 30% of the cohort). Among virally suppressed women with HIV, severity of depressive symptoms was a distinguishing feature in 4 out of 5 cluster-defined cognitive profiles, while stress-related self-reports were associated with 2 profiles (sequencing and learning/memory), reinforcing the concept that mental health factors can associate with some cognitive profiles and remain a potentially modifiable target for improving cognition in PWH. A similar approach was used to reveal sex differences in patterns and predictors of cognitive phenotypes in a study of data from the University of California, San Diego’s HIV Neurobehavioral Research Program [35].
DISCOVERY OF MENTAL HEALTH DETERMINANTS OF HIV DISEASE SEVERITY AFTER ART INITIATION
Paul et al combined group-based multitrajectory analysis, traditional statistics, and ensemble machine learning to investigate causal pathways of persistent CD4/CD8 T-cell ratio trajectories modeled before and after ART initiation in acute HIV infection [36]. This approach allowed for simultaneous modeling of CD4+ and CD8+ T-cell trajectories over 144 weeks to determine the relative contributions of perturbations in each T-cell population as an independent or synergistic determinant of persistent inversion in the ratio despite suppressive ART. In a study of over 400 individuals who initiated ART within 30 days of infection and maintained viral control for 144 weeks, 49% failed to achieve CD4/CD8 T-cell ratio normalization after prolonged use of suppressive ART. Latent trajectory analysis identified 2 specific immune risk phenotypes for chronic inversion of the ratio, one with incomplete CD4+ T-cell recovery after treatment onset and another with high CD8+ T-cell count before and after treatment. Treatment after Fiebig stage II (approximately 7 days after initial infection) was a robust predictor of chronic CD4/CD8 ratio inversion. Machine learning analysis further revealed that mental health factors during early stages of infection strongly predicted inversion due to incomplete CD4+ T-cell recovery. These results emphasize the contribution and potential synergy between mental health factors and HIV pathogenesis at time of treatment onset as factors contributing to long-term immune dysregulation despite optimal treatment of HIV.
DATA HARMONIZATION ACROSS COHORTS
Data harmonization is a critical step for machine learning using common measures across multiple cohorts. The amount of data on CNS complications in PWH is growing rapidly. Available resources for machine learning include data from cross-sectional studies and well-characterized longitudinal cohorts, including multisite clinical trials (eg, AIDS Clinical Trials Group studies) and large observational studies (eg, Multicenter AIDS Cohort Study [MACS]/WIHS Combined Cohort Study, Pharmacokinetic and Clinical Observations in People Over Fifty [POPPY], Comorbidity in Relation to AIDS [COBRA]). Data from such studies, however, are typically “siloed” and integration is often limited by scientific and methodological barriers, including differences in study design, use of different instruments to measure the same construct, and divergent populations (eg, virally suppressed vs viremic). Improved access to suppressive ART poses an additional barrier for comparison of older and newer studies. Ethical and legal barriers related to conducting human subjects research create further challenges by impeding data sharing [13].
Despite these hurdles, pooling data across studies offers important advantages: (1) larger samples provide increased power to identify the contribution of multiple factors that individually lack predictive strength; (2) larger samples improve the performance and reliability of machine learning models; (3) harmonized data can reduce biases introduced by methodological choices, promoting cross-study equivalence; and (4) incorporation of diverse characteristics in larger samples can facilitate novel discovery, for instance by increasing power for detection of effect modifiers. Pooling data across cohorts to increase demographic diversity also tends to improve generalizability, albeit at a cost of individual prediction.
Data harmonization is a multistep process requiring delineation of research objectives, documentation of methods, and selection of core variables (data schema) to improve consistency and enable pooling and statistical cocalibration. The lack of common data elements in NeuroHIV research, however, limits data integration and harmonization. Therefore, it is crucial to plan prospectively for data harmonization by establishing infrastructures to ensure consistent data collection, establishing common data elements, and preparing metadata to enhance efficient data use. Quality of the data must be assessed prior to dissemination, and FAIR (findable, accessible, interoperable, reusable) standards should be met [37]. Dissemination and reproducibility will be further enhanced by establishing minimum reporting standards [38].
Methodological issues involved in retrospective data harmonization for studies on CNS complications in PWH are illustrated by studies that pooled data from the CNS HIV Antiretroviral Therapy Effects Research (CHARTER) and National NeuroAIDS Tissue Consortium (NNTC) cohorts [39, 40]. Collectively, these cohorts include approximately 4000 PWH. Extensive data harmonization across CHARTER and NNTC has been performed through a combination of manual and computational techniques to maintain uniformity [41]. Methods to assess cognitive function and behavior have significant overlap between CHARTER and NNTC, but measures vary widely when compared to other large cohort studies (eg, MACS/WIHS, POPPY, COBRA, Neurocognitive Assessment in the Metabolic and Aging Cohort [NAMACO]). Using factor analysis or transforming neurocognitive test data into derived summary variables (eg, global deficit scores) or applying other psychometric methods that produce a robust summary score [42] can help to minimize such differences [41, 43], albeit at the expense of specificity of the individual tests. These approaches will facilitate data pooling across studies using different neuropsychological tests to reveal novel phenotypes and predictors of disease.
Machine learning using pooled data across cohorts requires addressing additional challenges, including differences in study design, cohort characteristics, data processing, and handling of missing data. Quality control standards can also vary across studies, leading to issues in interpretation, validity, and generalizability. Latent confounds can differ between cohorts; for example, aging and medically complex populations may experience high rates of some confounds that are uncommon in younger cohorts. Recruitment bias is another confounder that can influence findings; for example, nonrandom enrollment of participants willing to perform test batteries and provide biosamples may select individuals less likely to be employed [44]. Information bias is exemplified by a focus on factors presumed in advance to be important, ie, “searching under a streetlight.” This phenomenon also has the potential to artificially inflate health disparities in underrepresented populations when sociocultural factors are not considered. Unbalanced study designs can affect classification problems. When unequal representation of groups is unavoidable, choice of model performance metrics becomes crucial. In such cases, the F1 score (weighted sum of precision and sensitivity) can be more informative than area-under-the-curve (AUC).
While these challenges are considerable, they can be addressed through machine learning models that account for them, use of common data elements, and data harmonization [22]. In addition, shared standards for data acquisition, processing, interpretation, and reporting will increase the validity and reproducibility of findings derived from pooled samples [37, 38]. Although machine learning approaches can alleviate some problems related to differences across datasets, the need for careful data inspection and curation by researchers with domain expertise remains. Bringing together multidisciplinary teams including data scientists, statisticians, clinicians, and stakeholders to discuss study goals, data quality, data processing, and validation can help to mitigate these challenges and facilitate more robust machine learning analyses and more accurate, clinically relevant prediction models [13].
VALIDATION OF MACHINE LEARNING MODELS FOR PHENOTYPING IN PWH
As machine learning becomes more commonly used for identifying cognitive phenotypes in PWH, it is important to establish best practices based upon frameworks in published guidelines [38, 45, 46]. A set of recommended best practices for machine learning to understand cognitive phenotypes in PWH is shown in Table 3. Early development of rigorous study design, selection of appropriate models, and use of representative training sets will be critical for improving challenging issues such as collinearity, missingness, biases, and overfitting. The need for interpretable models must be balanced with approaches that increase predictive power, as some highly predictive models may not be interpretable or actionable (“black box” models). While internal and external validation to ensure reproducibility, transparency, and rigor remain the idealized objectives, methods have not yet been standardized for studies on cognitive phenotyping (eg, documenting and publishing code, use of large or representative training sets, reducing biases while curating data). Techniques such as cross-validation to assess model performance on independent test datasets are suggested as guidance for judging high-quality studies. Given that there are multiple types of cross-validation procedures including k-fold, repeated random subsampling, and leave-one-out, a fundamental understanding of available methods and their pros and cons is critical.
Table 3.
Recommended Best Practices for Machine Learning to Understand Cognitive Phenotypes in People with HIV
| Well-curated high-quality data |
| Cohort size should take into account study goals and quality, dimensionality, and completeness of dataset (ie, no “one size fits all”) |
| Larger studies can be complemented by more focused studies with smaller cohorts |
| Make decisions on best methods to handle missing data |
| Choose models and metrics that take into account the balance of classes |
| Internal/external validation to demonstrate model stability and reproducibility |
| Representative training sets from diverse cohorts (including people with common comorbidities) to increase generalizability |
| Accurately report and account for biases and confounders |
| Measures that can be used across international settings |
LIMITATIONS AND CHALLENGES
It is important to acknowledge limitations and potential pitfalls when using machine learning to identify, characterize, and predict cognitive phenotypes in PWH. Machine learning applications remain challenging in the absence of established accuracy and reporting standards [13]. The complexity of many machine learning methods can hinder the core objectives of reproducibility and transparency. Care must be taken to balance accuracy with audience accessibility and ease of interpretability, given that clinical medicine requires both high classification accuracy and interpretability.
Another limitation is the size and scope of datasets needed to train reliable machine learning models, a problem that is exacerbated by intersite differences but minimized by data harmonization. Issues of data quality must also be considered, including presence of noisy, skewed, missing, or flawed measures. The axiom of “garbage in–garbage out” describes the tendency for data-quality issues to produce unreliable conclusions or inflate prediction accuracy [47]. Robust internal and external validation are therefore important to assess model performance.
Although not all machine learning methods assume normality, it is important to examine data distributions, including identification of missingness and outliers with potential to disproportionately affect models and conclusions. Missing values are a common source of error, especially when missingness is not at random, and researchers must consider the best approach for handling missing data. It is also important to accurately model potential confounders to prevent models from reaching spurious conclusions [48].
The risk of overfitting to training data must be considered in the development and interpretation of machine learning approaches. Overfitted models effectively “memorize” particularities of the training set rather than learning true relationships among study variables, and thus inflate performance on training data but fail to generalize. A method to avoid overfitting is early stopping, where training is monitored and halted when performance on holdout data reaches a plateau.
Given phenotypic heterogeneity and complexity among PWH, large studies with thousands of cases are likely to be needed to discover clinically meaningful cognitive phenotypes. Larger studies can be complemented by more focused work using smaller cohorts to gain deeper understanding of underlying mechanisms and distinct biotypes through analysis of blood/CSF and neuroimaging biomarkers.
CONCLUSIONS
These discussions highlight opportunities and challenges of data harmonization, multimodal data integration, and machine learning to identify cognitive phenotypes in PWH by elucidating relationships within diverse clinical and biomarker data. Machine learning presents new opportunities to better characterize CNS complications in PWH outside of conventional classifications, for example by identifying cognitive subtypes beyond HAND categories and elucidating their relationships to comorbidities and mental health. Data-driven approaches may help researchers discover complex relationships among blood/CSF biomarkers, neuroimaging, and neurobehavioral profiles in PWH. For such methods to succeed, computational expertise applied to large, diverse datasets is necessary, but not sufficient. These resources must be supplemented with clinical and analytical expertise enhanced by best practices in data acquisition, processing, and harmonization. Pitfalls and sources of error must be anticipated and mitigated, including missing data, confounders, and recruitment or information bias. Machine learning approaches offer enormous potential to enhance the clinical relevance of research on CNS complications of HIV, but clinical applications will require advances in model usability and interpretability. To enhance clinical utility, rigorous methods and standards should be adopted to promote reproducibility, validity, and generalizability. These principles will increase the likelihood of success for machine learning applications to improve future clinical care and develop personalized therapies for PWH.
Contributor Information
Shibani S Mukerji, Massachusetts General Hospital, Boston, Massachusetts, USA; Dana-Farber Cancer Institute, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Kalen J Petersen, Washington University in Saint Louis, Saint Louis, Missouri, USA.
Kilian M Pohl, Stanford University, Stanford, California, USA; SRI International, Menlo Park, California, USA.
Raha M Dastgheyb, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Howard S Fox, University of Nebraska Medical Center, Omaha, Nebraska, USA.
Robert M Bilder, University of California, Los Angeles, California, USA.
Marie-Josée Brouillette, McGill University, Montreal, Quebec, Canada.
Alden L Gross, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA.
Lori A J Scott-Sheldon, Division of AIDS Research, National Institute of Mental Health, National Institutes of Health, Bethesda, Maryland, USA.
Robert H Paul, Missouri Institute of Mental Health, University of Missouri, Saint Louis, Missouri, USA.
Dana Gabuzda, Dana-Farber Cancer Institute, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Notes
Disclaimer. The views expressed in this manuscript do not necessarily represent the views of the National Institutes of Health, the Department of Health and Human Services, or the United States Government.
Financial support. This work was supported by the National Institutes of Health (grant numbers 5R01MH110259 and 1R56MH115853 to D. G.; 5K23MH115812 to S. S. M.; 1F32MH129151 to K. J. P.; 5R01MH113406 and 5U01AA017347 to K. M. P.; 5R03MH123290 and P30AI094189 to R. M. D; 5U24MH100925 and 5P30MH062261 to H. S. F.; 5R01MH118514 and 5R01MH114152 to R. M. B.; and 5R01MH113560, 5R01MH113406, 1R01MH128868, and 5R0MH118031 to R. H. P.).
Supplement sponsorship. This article appears as part of the supplement “State of the Science of Central Nervous System Complications in People With HIV,” sponsored by the National Institutes of Health, National Institute of Mental Health.
References
- 1. Winston A, Spudich S. Cognitive disorders in people living with HIV. Lancet HIV 2020; 7:e504-13. [DOI] [PubMed] [Google Scholar]
- 2. Nightingale S, Dreyer AJ, Saylor D, Gisslen M, Winston A, Joska JA. Moving on from HAND: why we need new criteria for cognitive impairment in persons living with human immunodeficiency virus and a proposed way forward. Clin Infect Dis 2021; 73:1113–8. [DOI] [PubMed] [Google Scholar]
- 3. De Francesco D, Underwood J, Post FA, et al. . Defining cognitive impairment in people-living-with-HIV: the POPPY study. BMC Infect Dis 2016; 16:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molsberry SA, Cheng Y, Kingsley L, et al. . Neuropsychological phenotypes among men with and without HIV disease in the multicenter AIDS cohort study. AIDS 2018; 32:1679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morgello S, Gensler G, Sherman S, et al. . Frailty in medically complex individuals with chronic HIV. AIDS 2019; 33:1603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schouten J, Su T, Wit FW, et al. . Determinants of reduced cognitive performance in HIV-1-infected middle-aged men on combination antiretroviral therapy. AIDS 2016; 30:1027–38. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y, Liu M, Lu Q, et al. . Global prevalence and burden of HIV-associated neurocognitive disorder: a meta-analysis. Neurology 2020; 95:e2610-21. [DOI] [PubMed] [Google Scholar]
- 8. Antinori A, Arendt G, Becker J, et al. . Updated research nosology for HIV-associated neurocognitive disorders. Neurology 2007; 69:1789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paul R, Tsuei T, Cho K, et al. . Ensemble machine learning classification of daily living abilities among older people with HIV. EClinicalMedicine 2021; 35:100845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Insel T, Cuthbert B, Garvey M, et al. . Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry 2010; 167:748–51. [DOI] [PubMed] [Google Scholar]
- 11. Galatzer-Levy IR, Ruggles K, Chen Z. Data science in the research domain criteria era: relevance of machine learning to the study of stress pathology, recovery, and resilience. Chronic Stress (Thousand Oaks) 2018; 2: 2470547017747553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bilder RM. Wrangling the matrix: lessons from the RDoC working memory domain. In: Parnas J, Kendler KS, Zachar P, eds. Levels of analysis in psychopathology: cross-disciplinary perspectives. Cambridge: Cambridge University Press, 2020:59–77. [Google Scholar]
- 13. Goecks J, Jalili V, Heiser LM, Gray JW. How machine learning will transform biomedicine. Cell 2020; 181:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greener JG, Kandathil SM, Moffat L, Jones DT. A guide to machine learning for biologists. Nat Rev Mol Cell Biol 2022; 23:40–55. [DOI] [PubMed] [Google Scholar]
- 15. Dastgheyb RM, Buchholz AS, Fitzgerald KC, et al. . Patterns and predictors of cognitive function among virally suppressed women with HIV. Front Neurol 2021; 12:604984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tu W, Chen PA, Koenig N, et al. . Machine learning models reveal neurocognitive impairment type and prevalence are associated with distinct variables in HIV/AIDS. J Neurovirol 2020; 26:41–51. [DOI] [PubMed] [Google Scholar]
- 17. Paul RH, Cho KS, Belden AC, et al. . Machine-learning classification of neurocognitive performance in children with perinatal HIV initiating de novo antiretroviral therapy. AIDS 2020; 34:737–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Francesco D, Sabin CA, Winston A, et al. . Sleep health and cognitive function among people with and without HIV: the use of different machine learning approaches. Sleep 2021; 44:zsab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adeli E, Kwon D, Zhao Q, et al. . Chained regularization for identifying brain patterns specific to HIV infection. Neuroimage 2018; 183:425–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adeli E, Zahr NM, Pfefferbaum A, Sullivan EV, Pohl KM. Novel machine learning identifies brain patterns distinguishing diagnostic membership of human immunodeficiency virus, alcoholism, and their comorbidity of individuals. Biol Psychiatry Cogn Neurosci Neuroimaging 2019; 4:589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu Y, Lin Y, Bell RP, et al. . Machine learning prediction of neurocognitive impairment among people with HIV using clinical and multimodal magnetic resonance imaging data. J Neurovirol 2021; 27:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J, Zhao Q, Adeli E, et al. . Multi-label, multi-domain learning identifies compounding effects of HIV and cognitive impairment. Med Image Anal 2022; 75:102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luckett PH, Paul RH, Hannon K, et al. . Modeling the effects of HIV and aging on resting-state networks using machine learning. J Acquir Immune Defic Syndr 2021; 88:414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paul RH, Cho KS, Luckett P, et al. . Machine learning analysis reveals novel neuroimaging and clinical signatures of frailty in HIV. J Acquir Immune Defic Syndr 2020; 84:414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paul RH, Cho K, Belden A, et al. . Cognitive phenotypes of HIV defined using a novel data-driven approach. J Neuroimmune Pharmacol. 2022; 17: 515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Turbe V, Herbst C, Mngomezulu T, et al. . Deep learning of HIV field-based rapid tests. Nat Med 2021; 27:1165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiang Y, Fujimoto K, Li F, et al. . Identifying influential neighbors in social networks and venue affiliations among young MSM: a data science approach to predict HIV infection. AIDS 2021; 35:S65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olatosi B, Sun X, Chen S, et al. . Application of machine-learning techniques in classification of HIV medical care status for people living with HIV in South Carolina. AIDS 2021; 35:S19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luckett P, Paul RH, Navid J, et al. . Deep learning analysis of cerebral blood flow to identify cognitive impairment and frailty in persons living with HIV. J Acquir Immune Defic Syndr 2019; 82:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paul RH, Shikuma CM, Chau NVV, et al. . Neurocognitive trajectories after 72 weeks of first-line anti-retroviral therapy in Vietnamese adults with HIV-HCV co-infection. Front Neurol 2021; 12:602263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lubke GH, Muthén B. Investigating population heterogeneity with factor mixture models. Psychol Methods 2005; 10:21–39. [DOI] [PubMed] [Google Scholar]
- 32. Wehrens R, Kruisselbrink J. Flexible self-organizing maps in kohonen 3.0. J Stat Softw 2018; 87:1–18. [Google Scholar]
- 33. Wehrens R, Buydens LMC. Self- and super-organizing maps in R: the kohonen package. J Stat Softw 2007; 21:1–19. [Google Scholar]
- 34. Scrucca L, Fop M, Murphy TB, Raftery AE. Mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R J 2016; 8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 35. Rubin LH, Sundermann EE, Dastgheyb R, et al. . Sex differences in the patterns and predictors of cognitive function in HIV. Front Neurol 2020; 11:551921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paul R, Cho K, Bolzenius J, et al. . Individual differences in CD4/CD8 T-cell ratio trajectories and associated risk profiles modeled from acute HIV infection. Psychosom Med 2022; in press. [DOI] [PMC free article] [PubMed]
- 37. Reiser L, Harper L, Freeling M, Han B, Luan S. FAIR: a call to make published data more findable, accessible, interoperable, and reusable. Mol Plant 2018; 11:1105–8. [DOI] [PubMed] [Google Scholar]
- 38. Hernandez-Boussard T, Bozkurt S, Ioannidis JPA, Shah NH. MINIMAR (MINimum information for medical AI reporting): developing reporting standards for artificial intelligence in health care. J Am Med Inform Assoc 2020; 27:2011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heaton RK, Clifford DB, FranklinDR, Jr, et al. . HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER study. Neurology 2010; 75:2087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morgello S, Gelman BB, Kozlowski PB, et al. . The National NeuroAIDS Tissue Consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol 2001; 27:326–35. [DOI] [PubMed] [Google Scholar]
- 41. Heithoff AJ, Totusek SA, Le D, et al. . The integrated National NeuroAIDS Tissue Consortium database: a rich platform for neuroHIV research. Database (Oxford) 2019; 2019:bay134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sanford R, Fernandez Cruz AL, Scott SC, et al. . Regionally specific brain volumetric and cortical thickness changes in HIV-infected patients in the HAART era. J Acquir Immune Defic Syndr 2017; 74:563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dawes S, Suarez P, Casey CY, et al. . Variable patterns of neuropsychological performance in HIV-1 infection. J Clin Exp Neuropsychol 2008; 30:613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mayo NE, Brouillette MJ, Fellows LK. Estimates of prevalence of cognitive impairment from research studies can be affected by selection bias. J Acquir Immune Defic Syndr 2018; 78:e7–8. [DOI] [PubMed] [Google Scholar]
- 45. Rivera S C, Liu X, Chan AW, et al. . Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat Med 2020; 26:1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu X, Cruz Rivera S, Moher D, et al. . Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med 2020; 26:1364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kilkenny MF, Robinson KM. Data quality: “garbage in–garbage out”. Health Inf Manag 2018; 47:103–5. [DOI] [PubMed] [Google Scholar]
- 48. Zhao Q, Adeli E, Pohl KM. Training confounder-free deep learning models for medical applications. Nat Commun 2020; 11:6010. [DOI] [PMC free article] [PubMed] [Google Scholar]