Abstract
Background: Dates palm (Phoenix dactylifera L.) fruits are among the most widely used fruits in the Middle East and African nations. Numerous researchers confirmed the presence of phytochemicals in P. dactylifera L. fruit and its by-products with broad-ranging biological activities.
Objectives: In the present work, phytochemical and biological assessments of two different cultivars of date fruit (Shishi M1 and Majdool M2 grown in the Ha’il region of Saudi Arabia) have been carried out.
Methods: Date fruits were extracted and analyzed by gas chromatography-mass spectrometry (GS-MS),liquid chromatography-mass spectrometry (LC-MS) and Fourier-transform infrared spectroscopy (FT-IR)techniques. The lyophilized methanolic extracts were analyzed for their in-vitro antiproliferative andcytotoxicity against colon cancer (HCT116) cell line. To identify the possible constituents responsible for the bioactivity, in-silico molecular docking and molecular dynamics (MD) simulation studies were carried out.
Results: Both cultivars exhibited in-vitro anticancer activity (IC50 = 591.3 μg/mL and 449.9 μg/mL for M1 and M2, respectively) against colon cancer HCT-116 cells. The computational analysis results indicated procyanidin B2 and luteolin-7-O-rutinoside as the active constituents.
Conclusion: Based on these results, we conclude that these cultivars could be a valuable source for developing health promoter phytochemicals, leading to the development of the Ha’il region, Saudi Arabia.
Keywords: date fruits, extraction, GC-MS, Ha’il region, molecular docking, molecular dynamics, P. dactylifera L.
Introduction
Natural products (NPs) play an instrumental role in drug design and remain an inspiration for discovering new drug candidates (Newman and Cragg, 2016). Being the largest source of new pharmacophores, 60%–70% of drugs used today are based directly or indirectly on NPs (Atanasov et al., 2021). Indeed, enormous diversity in their chemical structure, broad-ranging bioactivity, low toxicity, and ability to bind with different proteins (targets) gives natural compounds an edge over synthetic ones (Ali et al., 2010). In the quest for new drug candidates, research is being carried out on the extraction, isolation, and identification of bioactive compounds found in plants, animals and microbes (Altemimi et al., 2017). Among a large pool of NPs, the dates palm (Phoenix dactylifera L.), a member of the Asteraceae family, has garnered an immense interest (Al-Alawi et al., 2017; Maqsood et al., 2020). Date fruits of the date palm tree (P. dactylifera L.) is one of the most consumed fruits worldwide, especially in the Middle East and Asian countries (Complexity, 2022). Apart from their high nutritional and commercial values (Mia et al., 2020), date fruits and their by-products have also attracted researchers due to their potential health benefits (Maqsood et al., 2020). Their antibacterial, antifungal, antiviral, antidiabetic, anticancer, anti-inflammatory, antioxidant, antiangiogenic and other protective effects along with negligible side effects, are particularly interesting (Vayalil, 2002; Maqsood et al., 2020). It has been demonstrated that date fruits are rich in carbohydrates, protein, fibres, minerals, vitamins, phenolic acids, flavonoids, and other phytochemicals responsible for bioactivities. The chemical composition depends on various factors, including the type of cultivar, geographical location, irrigation method, ripening stage, processing time, extracting solvents, etc. (Borochov-Neori et al., 2013) Based on this knowledge, various groups investigated the chemical composition of date fruits and seeds native to different regions (Vayalil, 2012; Al-Alawi et al., 2017; Mia et al., 2020; Echegaray et al., 2021; Ibrahim et al., 2021). The group led by Aviram (Borochov-Neori et al., 2013) has conducted studies on the chemical and biological analysis of several varieties of date fruits (Maqsood et al., 2020). They, along with others, confirmed that the phytoconstituents and bioactivity of the date fruits are the function of the parameters mentioned above. For example, a pilot study showed that Medjool or Hallawi varieties of date fruits vary in phenolics, catechins and quercetin derivative content and antioxidant effect (Rock et al., 2009). The same group reported anti-atherogenic properties of acetone extracts of Hallawi in addition to the eight other variants (Borochov-Neori et al., 2013). As per the group, phenolic compounds exerted anti-atherogenic properties via low-density lipoprotein (LDL) oxidation and serum-mediated cholesterol efflux. On the other hand, alcoholic extract of the Tunisian variety was found to inhibit α-glucosidase and α-amylase enzymes with low IC50 values (El Abed et al., 2017). Zhang et al. (2013) performed an extensive chemical and biochemical profiling of Ajwa date fruits. They identified several new compounds such as bis (2-ethylhexyl) terephthalate and bis (2-ethylheptyl) phthalate) in addition to glycoside, terpenoids, triglyceride, phthalates, etc. They noted that the aqueous and organic extract exerts dose-dependent antioxidant and anti-inflammatory effects.
In Saudi Arabia, more than 400 varieties of date fruit are cultivated, which vary in appearance, nutrition, and nutritional value (Zhang et al., 2017). Several researchers reported that the varieties such as Barni, Khalas, and Ajwa show unique biological activities (Eid et al., 2013; Assirey, 2015; Hamad et al., 2015). Recently, Amir and co-workers (Alghamdi et al., 2018) studied the nutritional value of several varieties of date fruits found in Ha’il province; however, the biological activity of the date fruits remains unclear. Prompted by this, we carried out extraction, characterization, in-vitro antiproliferative and cytotoxicity assay of two varieties of date fruits (Shishi M1 and Majdool M2) grown in the Ha’il region of Saudi Arabia). Then, to further identify the possible potential constituent present, ligand-based virtual screening was performed. This is followed by molecular dynamics simulation studies to identify the stability of promising compounds with possible receptor.
Materials and methods
General
All solvents used for isolation and purification were of ACS reagent grade (Sigma-Aldrich Chemical Co., St. Louis, MO, United States). Lyophilization was carried out on BenchTop Manifold Freeze Dryer (MILLROCK, United States) for 24 h at a condenser temperature of −45°C equipped with Edward pump. Attenuated-total-reflectance IR spectra were recorded on pure samples on diamond using a Shimadzu IRSpirit-T spectrometer.
Sample collection, extraction, and sample preparation
Two different varieties of date fruits (Tamr stage, Supplementary Table S1) grown in Ha’il province were collected from the local market. Authors (AKDA and KMA) and local farmers authenticated the samples, and a voucher specimen was deposited. The samples were stored and kept in a −20°C freezer. First, three pieces of date fruits from each variant (Supplementary Table S1) were pitted to remove seeds and cut into small pieces. Then, cold extraction was performed by shaking and mixing fruit materials in methanol (MeOH) overnight at room temperature, followed by filtration. The residue was further extracted twice with MeOH for 1 h. Finally, the extracts were combined and concentrated using rotatory evaporation at room temperature. The resulting viscous honey-like liquid was lyophilized to afford light yellow water-soluble powder and stored at −20°C till further analysis.
Chromatographic studies
Date fruit extracts were analyzed by LC-MS system using a reverse phase C18 column (Accucore, 150 × 4.6, 2.6 μm). The LC-MS system comprised a Waters Alliance 2695 HPLC pump, an autosampler, a vacuum degasser, and a column compartment attached to a XEVO-TQD detector with electrospray ionization (ESI). The following gradient of solvents were used: acetonitrile (mobile phase A) and 5 mM acetic acid (mobile phase B); ratio of A to B, 0–1 min, 5:95; 1–10 min, 5:95 to 30:70; 10–16 min, 30:70 to 60:40; 16–24 min, 60:40 to 80:20; 24–32 min, 80:20; 32–40 min, 5:95. In all cases, the columns were reequilibrated between injections with the equivalent of 5 mL of the mobile phase. During the full scan by MS/MS, mass acquisition was set from 150 to 2000 Da. This method utilized ESI-LC/MS/MS operating in MRM mode. The ESI settings were the following capillary voltage, 3.5 kV; cone voltage, 40 V; the flow of desolvation gas (Argon gas), 650 L/h; flow of cone gas, 30 L/h. Gas Chromatography-Mass Spectrometry (GC-MS) analysis was carried out using Agilent 8890/5977B Series (Agilent 5977B EI/CI MSD) spectrometer. The segments were recognized by examination of their delay times and mass spectra with those of the NIST 11 mass spectral database.
Cell culture and maintenance
The human colon cancer cell HCT116 was acquired from American Type Culture Collection (ATCC). McCoy’s 5 A media supplemented with 10% v/v Fetal Bovine Serum (FBS), and 1% antibiotic-antimycotic solution (1 mL contains 10,000 U Penicillin, 10 mg Streptomycin and 25 µg Amphotericin B) was used to grow and maintain HCT116 cells. A humidified environment constituted the standard conditions for cell culture at 37°C with 5% CO2.
Cell viability assay
To determine the cytotoxicity of M1 and M2 extracts on colon cancer HCT-116 cell line, MTT assay was used. In a 96-well plate, the cells (5 × 103 cells/well) were cultured for 24 h. The cells were treated with M1 and M2 at varied concentrations (100, 500, 1,000, and 5,000 μg/mL) for 24 h, respectively. Each well received 10 µL of MTT solution (5 mg/mL) and was subjected to further 3 h incubation at 37°C. In order to dissolve the purple formazan crystals, 100 µL of dimethyl sulfoxide (DMSO) was added to each well. A microplate reader measured absorbance at 570 nm (Bio-Rad, United States). The cell viability was expressed as a percentage (%) over the untreated control. For calculating the IC50 value, GraphPad Prism Professional software was used.
Morphological analysis
The effects of M1 and M2 extract treatment on the morphology of HCT-116 cells were investigated using a phase contrast microscope. Briefly, HCT-116 cells (5 × 103) were cultivated in a 96-well plate before M1 and M2 (100, 500, 1,000, and 5,000 μg/mL) treatment. The alteration in morphology of M1 and M2-treated cells was then examined using a phase contrast microscope (Labomed, United States).
Trypan blue exclusion assay
Trypan blue dye exclusion assay was performed further to confirm the M1 and M2-mediated cytotoxicity in HCT-116 cells. A hemocytometer and a microscope were used to count the cells (5 × 104) after they had been co-cultured with and without M1 and M2 (100, 500, 1,000, and 5,000 μg/mL), respectively for 24 h. The proportion of dead cells in each treatment set from studies done in triplicates was used to express the results.
Lactate dehydrogenase release assay
In accordance with the manufacturer’s instructions, the lactate dehydrogenase (LDH) release assay kit was used to measure the level of cellular cytotoxicity. First, M1 and M2 were applied to the HCT-116 cells using a 96-well plate at different doses (100, 500, 1,000, and 5,000 μg/mL) for 24 h. LDH release kit was then used to detect released LDH in both the M1 and M2—treated HCT-116 cells in the incubation medium.
Computational details
Computational calculations were carried out on a Dell workstation (Galax GeForce GTX 1660 Ti) equipped with 8-core processors, 64 GB Ram, and NVIDIA graphics card.
Receptor and ligands preparation
The co-crystal structure of Bcl-2 complex (PDB ID: 5JSN) was selected for virtual screening and molecular dynamics studies. In the crystallographic structure of this complex, there is a gap at position 33–86, which was fixed by homology modelling using the Swiss modeller tool. Based on literature reports (Mia et al., 2020), a total of ninety-four (n = 94) phytoconstituents of different chemical classes (Supplementary Table S2) were selected. Chemical structures (.sdf format) of the compounds were retrieved from the NCBI PubChem database (Wang et al., 2009). The downloaded files were converted to .pdb format using the Open Babel software. The ligand files were prepared using AutoDock Tools 1.5.7 (the Scripps Research Institute, La Jolla, CA, United States) software and finally written as .pdbqt file format for docking studies (Ahamad et al., 2021a).
Active site prediction
The Bcl-2 protein (PDB ID: 5JSN) was given as input to identify the active site, which gives significant insight into recognizing surface structural pockets, shape and volume of every pocket, internal cavities of protein and surface areas. Next, the active site and the interactive residues were selected using PDBsum and CASTp online tools (Laskowski et al., 2018; Tian et al., 2018). The ligands were prepared using AutoDock Tools (ADT), and saved in pdbqt format (Trott and Olson, 2010).
Protein preparation and grid generation
The 3D structure of Bcl-2 was prepared using the ADT protein preparation wizard. The polar and missing hydrogen atoms were added, while water molecules and hetero-atoms were deleted (Forli et al., 2016). Energy minimization was performed with a default constraint of 0.3 Å root mean square (RMS) and charges were assigned. After protein preparation, clean structure was saved as pdbqt file. Grid box (84 Å × 82 Å × 84 Å) was generated around the centroid of compounds with assigned X, Y, and Z axis.
Virtual screening and binding affinity calculation
To identify the potential compounds found in P. dactylifera L., a dataset of ninety-four compounds was utilized for the virtual screening. The pdbqt files were provided as input and screened against Bcl-2 (Duffy and Avery, 2012). Top two compounds (ranked based on the binding energy scores and the docking poses) were selected for further studies (Trott and Olson, 2010; Forli et al., 2016). The compounds with favourable binding poses were identified with the help of the lowest free energy (ΔG), defined using the equation as follows,
Where (∆Gcomplex), (∆Greceptor), and (∆Gligand) are the average values of Gibbs free energy for the complex, receptor, and ligand, respectively. The stability of the docked complex between the receptor-ligand exhibits more negative scores, revealing the high potency of the inhibitor. All the other docking parameters were kept default, and the docked complexes final visualization was performed using PyMOL tool (DeLano, 2002). The active pocket of Bcl-2 and docked pose of the top-ranked compounds were compared to find interactive orientations.
Molecular dynamics (MD) simulation
MD simulations were performed for the best-docked complexes with maximum binding affinity scores using GROningen MAchine for Chemical Simulations (GROMACS) version 5.18.3. Package (Abraham et al., 2015). The topology of Bcl-2 was generated using GROMOS9643a1 force field (Van Der Spoel et al., 2005). Due to the lack of suitable force field parameters for a drug-like molecule in the GROMACS software, the PRODRG server was used for the generation of molecular topologies and coordinate files (Schüttelkopf and Van Aalten, 2004). All the systems were solvated using a simple point charge model (SPC/E) in a cubic box. To neutralize the system 0.15 M counter ions (Na+ and Cl−) were added. The energy minimization of all the neutralized systems was performed using the steepest descent and conjugate gradients (50,000 steps for each). The constant number of particles, volume, and temperature (NVT) ensemble and constant number of particles, pressure, and temperature (NPT) ensemble were run for system equilibration (Ahamad et al., 2021b). Steepest descent followed by conjugate gradient algorithms was utilized on enzyme-ligand complexes. The NVT ensemble was employed at a constant temperature of 300 K and a constant pressure of 1bar. The SHAKE algorithm was used to confine the H atoms at their equilibrium distances and periodic boundary conditions. Moreover, the Particle Mesh Ewald (PME) method defines long-range electrostatic forces (Abraham et al., 2015). The cut-offs for van der Waals and columbic interactions were set as 1.0 nm. LINC algorithm was used to constrain the bonds and angles. Using the NPT ensemble, production runs were performed for 500 ns, with time integration. The energy, velocity, and trajectory were updated at the time interval of 10 ps The analysis is performed by using Cα-atom deviations of the protein calculated using root mean square deviations (RMSD). The relative fluctuations of each amino acid were defined with root mean square fluctuations (RMSF). To measure the compactness of a given molecule radius of gyrations (Rg) is implemented, and the solvent accessible surface area (SASA) was employed to know the electrostatic contributions of molecular solvation (Ahamad et al., 2018; Ahamad et al., 2019).
Results
Extraction and characterization of Shishi (M1) and Majdool (M2) fruits
Aqueous, organic or mixture solvents can extract a natural product, depending upon the analyte of interest (Nematallah et al., 2018). In the past, it has been demonstrated that when date fruits are extracted with an organic solvent, it yields bioactive compounds able to inhibit colon, liver and cervical cancerous cell lines in vitro (Mansour et al., 2011; Ravi, 2017). Especially, using a polar solvent such as methanol allows the extraction of various components (MHM et al., 2015). It was found that the alcoholic extract of the date fruits effectively inhibits α-glucosidase and α-amylase enzymes with low IC50 values in both in-vitro and in-vivo (El Abed et al., 2017). In 2016, Khan et al. (2016) demonstrated that the methanolic extract of Ajwa Date (Saudi origin) inhibits breast cancer (MCF-7) cell lines via cell cycle arrest and apoptosis. Motivated by this, we selected methanol as the extracting solvent in this study too. Cold extraction of finely cut M1 (36.11 gm) and M2 (46.22 gm) followed by concentration and lyophilization yielded M1 and M2 as a light-yellow water-soluble powder (9.1 and 15 gm of M1 and M2, respectively). To identify the extract components, chromatographic (LC-MS & GC-MS) and spectroscopic (FT-IR) techniques (Supplementary Figures S1, S2) were employed.
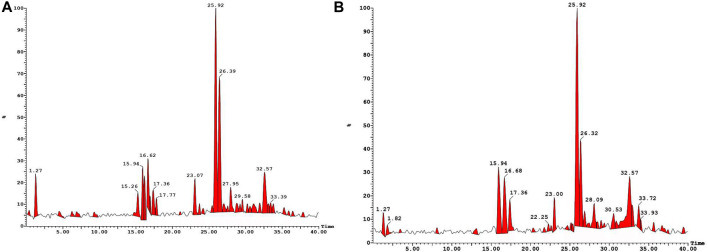
Lyophilized products were analyzed by LC-MS (negative mode) using gradient mobile phase as it allows easy detection of phenolic acids & flavonoids as they contain acidic hydroxy group. It has been reported that the methanolic extract contains phenolic acids, flavonoid diglucosides, monoglucosides, acylated monoglucosides, free aglycones, lipids and others when analyzed under similar conditions (Farag et al., 2014). Table 1 collects the identities and molecular/fragment ions of some major components present in M1 and M2 as identified by comparing LC-MS (negative mode, Figure 1) results with the literature. Both varieties exhibit similar chromatograms, with M1 having relatively more fraction than M2.
TABLE 1.
LC-MS (negative mode) results of M1 and M2.
| S. No | M-H | Mol. Form. | Identification | Ref. |
|---|---|---|---|---|
| 1. | 180.1 | C6H12O | β-D-Glucopyranose | Najm et al. (2021) |
| 2. | 341.1 | C15H18O9 | Caffeic acid hexoside | Farag et al. (2014) |
| 3. | 179.1 | C16H15O8 | O-Caffeoyl shikimic acid | Otify et al. (2019) |
| 4. | 322.0 | C12H19O10 | Anhydro dihexose | Otify et al. (2019) |
| 5. | 425.3 | C17H29O12 | Acyl sucrose | Najm et al. (2021) |
| 6. | 463.1 | C21H20O12 | Isoquercetin | Farag et al. (2014) |
| 7. | 476.0 | C12H21O12 | Isorhamnetin-3-O-glucoside | Najm et al. (2021) |
| 8. | 476.0 | C23H43NO7P | Sphingolipid conjugate I | Otify et al. (2019) |
| 9. | 311.3 | C18H31O4 | Dihydroxy linolenic acid | Farag et al. (2014) |
| 10. | 277.4 | C18H29O2 | Linolenic acid | Farag et al. (2014) |
| 11. | 255.6 | C16H31O2 | Palmitic acid | Farag et al. (2014) |
| 12. | 279.4 | C18H31O2 | Linoleic acid | Farag et al. (2014) |
| 13. | 283.2 | C18H36O2 | Stearic acid | Eid et al. (2013) |
| 14. | 195.0 | C10H10O4 | ferulic acid | Nematallah et al. (2018) |
| 15. | 326.3 | C18H32O5 | Trihydroxy octadecadienoic acid | Nematallah et al. (2018) |
| 16. | 311.4 | C18H32O4 | Dihydroxy octadecadienoic acid | Nematallah et al. (2018) |
| 17. | 594.9 | C27H30O15 | Luteolin-7-O-rutinoside | Nematallah et al. (2018) |
FIGURE 1.
LC-MS (negative mode) traces of methanolic extracts of M1 (A) and M2 (B).
GC-MS analysis further indicated the presence of several phytochemical belongings of different classes. For example, quinic acid, oleic acid, trans-13-octadecenoic acid, stearic acid, O-caffeoyl shikimic acid, luteolin, trihydroxy-octadecenoic acid, stearic acid linoleic acid, 6-hydroxy 7 methoxy coumarin, 4-hydroxy 6-methylcoumarin and amino acids were tentatively identified (Supplementary Table S3). These components and other metabolites have been well-identified in different varieties of date fruits (Farag et al., 2016; Abdul-Hamid et al., 2019; Perveen and Bokahri, 2020; Souda et al., 2020; Ibrahim et al., 2021).
The FTIR spectrum of the methanolic extracts (Supplementary Figure S1) is also consistent with previous literature (Alam et al., 2022). It has been reported that IR spectrum of dates extracts exhibits multiple peaks responsible for functionalities present in lipid (2,960–2,850 cm−1), amide (3,299–3,399 cm−1 and 1,591–1,529 cm−1 for amine and 1,619–1,691 for carbonyl) and carbohydrates (900–1,200 cm−1). As it is clear, the IR spectra of M1 and M2 are identical. The spectrum shows a stretching vibrations band at 3,280 cm−1 attributed to -OH group, bands at 2,888 and 2,930 cm−1 attributed to Csp3-H stretching vibration, aromatic C=C stretching vibrations at 1,622 cm−1 and C–O deformation vibrations of aliphatic alcohols at 1,009 cm−1 (Alam et al., 2022).
Biological studies
Antiproliferative and cytotoxic effect
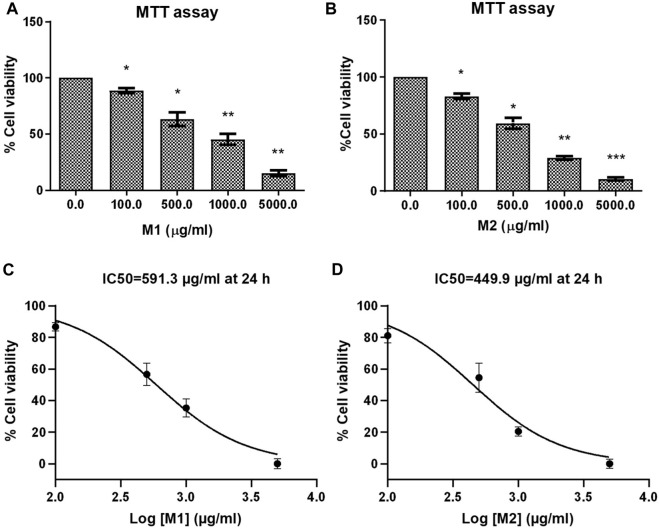
Using the MTT test, the antiproliferative and cytotoxic effects of the M1 and M2 date extract were assessed against colon cancer HCT116 cells for 24 h (Figures 2A, B). The M1 and M2 extracts exhibited strong and dose-dependent cytotoxic potential in HCT116 cells. The % cell viability of M1-and M2-treated HCT116 cells were found to be 88.80% ± 1.33%, 63.26% ± 3.47%, 45.24% ± 2.80%, and 15.28 % ± 1.53%; and 83.07% ± 2.37%, 59.35% ± 4.72%, 28.90% ± 1.49% and 10.63 % ± 1.47% at a dose of 100, 500, 1,000, and 5,000 μg/mL, respectively. IC50 values were determined to be 591.3 μg/mL and 449.9 μg/mL for M1 and M2, respectively, revealing the inhibitory potential (Figures 2C, D). Our findings thus demonstrated that both the M1 and M2 inhibit colon cancer cell proliferation in a dose-dependent manner.
FIGURE 2.
Effect of date extracts M1 and M2 on HCT-116 cells. (A, B) Percent (%) cell viability of HCT-116 cells treated with different doses of M1 and M2 (100–5,000 μg/mL) for 24 h. The results shown are the mean ± SEM of three independent experiments performed in triplicate (ns > 0.01, *p < 0.01, **p < 0.001, and ***p < 0.0001 represent significant differences compared with control). (C, D) Graph showing IC50 of M1 and M2 against HCT-116 colon cancer cell at 24 h.
Morphological alterations
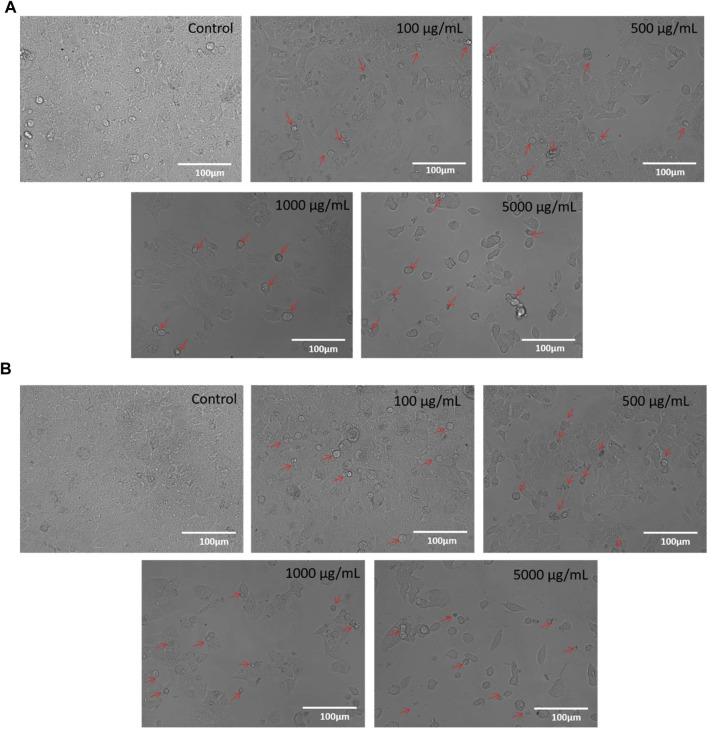
Under a phase contrast microscope, the images of control and M1 & M2-treated HCT-116 cells revealed discernible morphological alterations. The control cells showed increased cell growth and intact cell shape. However, in a dose-dependent manner (100, 500, 1,000, and 5,000 μg/mL), significant morphological modifications were observed in the M1 and M2-treated HCT-116 cells (Figures 3A, B). Moreover, M1 and M2-treated HCT-116 cells showed increased detachment and cytoplasmic shrinkage, which led to a rise in the number of floating cells. The findings thus support the hypothesis that treatment with M1 and M2 causes cytotoxicity in HCT-116 colon cancer cells.
FIGURE 3.
(A): Phase-contrast images of HCT-116 cells treated with either vehicle or different doses of M1 (100–5,000 μg/mL) for 24 h. The photomicrographs shown are the representatives of three independent experiments. (B): Phase-contrast images of HCT-116 cells treated with either vehicle or different doses of M2 (100–5,000 μg/mL) for 24 h. The photomicrographs shown are the representatives of three independent experiments.
M1 and M2 causes cell death in HCT-116 cells
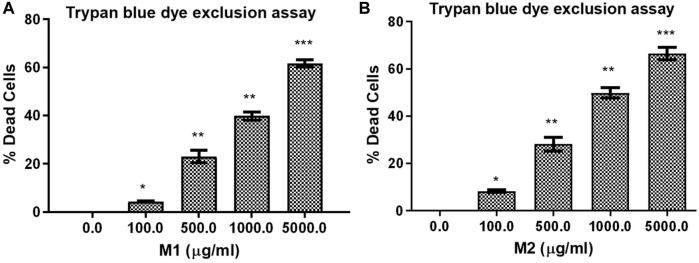
Trypan blue dye exclusion assay was used to assess how M1 and M2-treated HCT-116 cells lost viability. Figures 4A, B illustrates the considerable increase in cell mortality in HCT-116 cells after exposure to M1 and M2 at various doses (100, 500, 1,000, and 5,000 μg/mL) for 24 h. This result supported the cytotoxic action of M1 and M2 on colon cancer cells.
FIGURE 4.
Trypan blue dye exclusion assay. Percent (%) dead cells in HCT-116 cells treated with different doses of (A) M1 and (B) M2 (100–5,000 μg/mL) for 24 h. The results shown are the mean ± SEM of three independent experiments performed in triplicate (ns > 0.01, *p < 0.01, **p < 0.001, and ***p < 0.0001 represent significant difference compared with control).
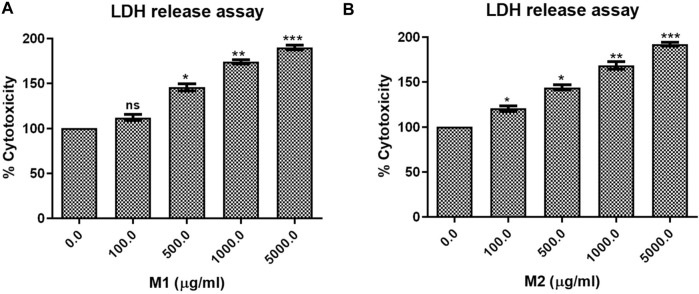
Release of cellular LDH in HCT-116 cells
LDH release assay displayed that treatment with both the M1 and M2 in HCT-116 cells mediated significant release of LDH, which showed the degree of cellular membrane damage post-treatment. Higher M1 and M2 concentrations were found to be significantly more cytotoxic, as evidenced by increased cytotoxicity in HCT-116 cells (Figures 5A, B). The percentage cytotoxicity in M1-treated HCT-116 cells, after 24 h of treatment, was found to be 112.08% ± 3.42%, 145.78% ± 3.88%, 174.07% ± 2.25%, and 190.03% ± 2.64% at 100, 500, 1,000, and 5,000 μg/mL dose, respectively. Similarly, after 24 h of treatment with M2, HCT-116 cells exhibited percent cytotoxicity of 120.53% ± 3.13%, 144.07% ± 3.00%, 168.46% ± 4.29%, 192.15% ± 1.98% at 100, 500, 1,000, and 5,000 μg/mL dose, respectively. Thus, our results suggest that both the M1 and M2 were able to decrease the viability and proliferation in colon cancer cells.
FIGURE 5.
LDH release assay. Percent cytotoxicity in HCT-116 cells treated with different doses of (A) M1 and (B) M2 (100–5,000 μg/mL) for 24 h. The results shown are the mean ± SEM of three independent experiments performed in triplicate (ns > 0.01, *p < 0.01, **p < 0.001, and ***p < 0.0001 represent significant differences compared to control).
In Silico studies
Virtual screening
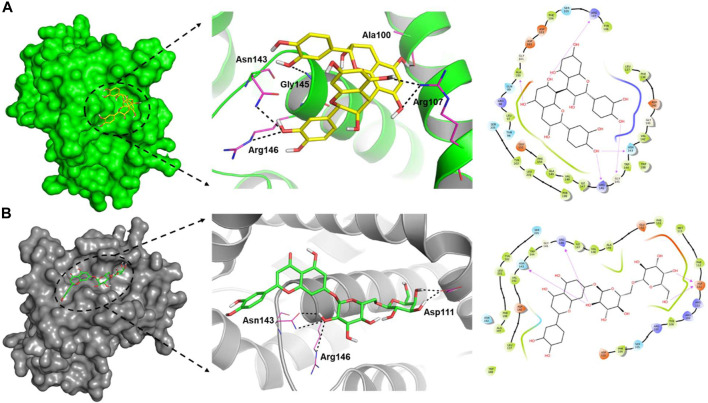
It has been demonstrated that the Phoenix dactylifera L. extract exhibits anticancer activity by modulating Bcl-2-family proteins (Khan et al., 2021). To identify the critical component(s) responsible for the anticancer activity, we conducted exhaustive in silico studies. To this end, ligand-based virtual screening was performed using ninety-four compounds found in P. dactylifera against the receptor (PDB: 5JSN). It has been reported that the small molecule may interact with the various receptors of Bcl-2 protein via multiple non-covalent interactions (Khosravi et al., 2022; Taghizadeh et al., 2022). Among others, Lys22, Arg26, Asp102, Ser105, Arg106, Arg109, Phe112, Val156, Val159, Asp163, Glu160, and Glu209 which participates in H-bonds and steric interactions (Khosravi et al., 2022). Based on the free binding energies and docking poses, virtual screening of the ligands resulted in procyanidin B2 and luteolin-7-O-rutinoside as the most potent candidates (Table 2). As depicted in Figure 6A, procyanidin B2 interacted with various amino acid residues via H-bonding (Ala100, Arg107, Asn143, Gly145, and Arg146) and other non-covalent interactions (such as hydrophobic and Van der Waal’s) with a total binding energy of −9.3 kcal/mol. On the other hand, luteolin-7-O-rutinoside formed H-bond with Asp111, Asn143 and Arg146 amino acids and yielded a binding energy of −9.1 kcal/mol (Figure 6B). Overall results revealed that the proposed two compounds have an edge over the Bcl-2 complexes attributable to more potent binding abilities.
TABLE 2.
Ligands name, 3D structures, SMILE format and the virtual screening outputs.
| S. No. | Ligand | 3D structure | SMILE format | Binding score (kcal/mol) | H-bond residues |
|---|---|---|---|---|---|
| 1 | Procyanidin B2 |
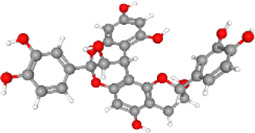
|
C1C(C(OC2=C1C(=CC3 =C2C4C(C(O3)(OC5= CC(=CC(=C45)O)O)C6=CC(=C(C=C6)O)O)O)O)C7=CC(=C(C=C7)O)O)O | −9.3 | Ala100, Arg107, Asn143, Gly145, and Arg146 |
| 2 | Luteolin-7-O-rutinoside |
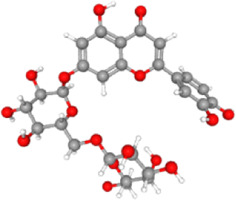
|
C1=CC(=C(C=C1C2=CC(=O)C3=C(C=C(C=C3O2)OC4C(C(C(C(O4)COC5C(C(C(C(O5)CO)O)O)O)O)O)O)O)O)O | −9.1 | Asp111, Asn143 and Arg146 |
FIGURE 6.
Surface view of the ligand-binding pocket (left), binding poses and interacting fragments (centre) and the H-bonds, hydrophobic interactions, van der Waal’s interactions around 4.0 Ǻ of the binding cavity (right) of procyanidin B2 (A) and luteolin-7-O-rutinoside (B) with Bcl-2 (PDB ID: 5JSN).
Molecular dynamics (MD) simulations
To understand the complex stability and interaction profile of the most promising hit compounds inside the active site of Bcl-2, MD simulations of Bcl-2-native, procyanidin B2 and luteolin-7-O-rutinoside complexes were performed on a 500 nanosecond (ns) scale. In addition, structural parameters, including RMSD, RMSF, SASA, and Rg were evaluated as a function of time and discussed in the following sub-sections.
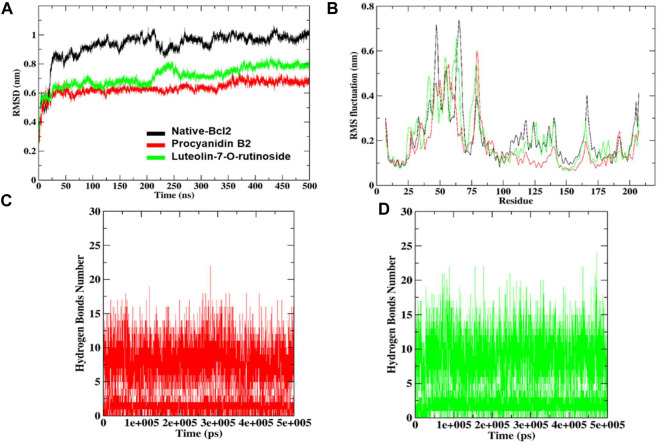
RMS-deviation and RMS-fluctuations
The docked complexes were subjected to RMSD analysis to assess the residual flexibility of the Bcl-2 receptor. It was noted that the native protein exhibits higher RMSD fluctuation and reaches equilibrium between 0.8 nm and 1.0 nm. However, in the presence of procyanidin B2, it reached an equilibrium at 0.6 nm and showed steady RMSD (average RMSD value 0.92 nm, Table 3), which remained stable over the 500 ns MD simulation ( Figure 7A ). Similarly, luteolin-7-O-rutinoside and Bcl-2 complexes showed stable equilibrium at 0.6 nm–0.7 nm. Furthermore, they displayed minimal fluctuation over the 500 ns MD simulation. The average RMSD of Luteolin-7-O-rutinoside and Bcl-2 complexes was 0.71 nm. Overall, both procyanidin B2 and luteolin-7-O-rutinoside complexes exhibit stable RMSD values and have a stable binding with Bcl-2 under the given simulation conditions. This also indicates that the studied compounds reached stable and reliable dynamic equilibriums, which bolstered the docking results.
TABLE 3.
The average RMSD, Rg, and SASA of the native and ligand-protein complexes.
| System | RMSD (nm) | Rg (nm) | SASA (nm)2 |
|---|---|---|---|
| Native | 0.92 | 1.56 | 111.73 |
| Procyanidin B2 | 0.63 | 1.65 | 112.77 |
| Luteolin-7-O-rutinoside | 0.71 | 1.57 | 115.39 |
FIGURE 7.
RMSD and RMSF analysis of the complexes of native protein Bcl-2, complexes of shortlisted ligands (A), combined RMS fluctuations (B). The number of hydrogen bonds formed by procyanidin B2 (C) and luteolin-7-O-rutinoside (D).
Furthermore, RMSF analysis was implemented to identify the flexible and rigid regions of the complexes and to measure the average atomic flexibility of the Cα-atoms of native Bcl-2 and docked complexes. In the case of native Bcl-2, amino acids residues such as Pro46 ∼0.53 nm, Gly47 ∼ 0.71 nm, Ile48 ∼ 0.64 nm, Arg63 ∼ 0.58 nm, Asp64 ∼ 0.65 nm, Pro65 ∼ 0.73 nm and Val66 ∼ 0.67 nm showed higher fluctuations (Figure 7B). However, fluctuation at 104–112, 162–163 and 201–207 amino acids residue also was found to be higher while other amino acids remain stable. For example, in a complex with procyanidin B2, RMS fluctuations were found in the region Gly79 ∼ 0.60 nm and Ala80 ∼0.57 nm, which is acceptable as these amino acids did not participate in the binding. Similarly, the complex with luteolin-7-O-rutinoside showed RMS-fluctuations at Gly54 ∼0.57 nm, Ala61 ∼0.55 nm and Arg63 ∼0.66 nm values. Overall, the RMSF displayed the highest degree of flexibility, exhibiting stable active site residues interaction compared to the native protein.
Hydrogen bond monitoring
To underpin the stability of the ligand-protein complex, the number of H-bond was monitored by analyzing the MD trajectories (Figures 7C, D). As can be seen, both compounds procyanidin B2 and luteolin-7-O-rutinoside formed 17 and 22 hydrogen bonds, respectively, which increased/remained the same during the 500 ns MD simulation.
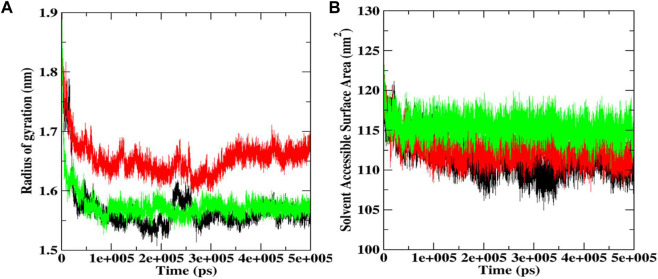
Radius of gyration (Rg) and solvent accessible surface area (SASA)
Rg helps determine protein folding and unfold upon ligand binding, thus giving an idea about the stability of the complex during the simulation. A higher Rg indicates a less compact structure, while a lower Rg means more compactness (Sharma et al., 2022). We found that the average Rg values for the native Bcl-2 protein (1.56 nm) and luteolin-7-O-rutinoside complex (1.57 nm) were almost similar, indicating that the protein will likely maintain a relatively steady value and is stably folded (Figure 8A; Table 2). However, in the case of the procyanidin B2 complex, the average Rg value was 1.65 nm, indicating unfolded structure.
FIGURE 8.
Rg (A) and SASA plot (B) during 500 ns MD simulations docked complexes of native Bcl-2, and complexes with procyanidin B2 and luteolin-7-O-rutinoside.
SASA was also conducted to ascertain the interactions between the protein-ligand complex and solvent during the 500 ns MD simulation (Figure 8B; Table 2). It was noted that the average SASA value for the complexes (112.77 and 115.39 nM2) of procyanidin B2 and luteolin-7-O-rutinoside, respectively, was better than the native Bcl-2 protein (111.73 nM2).
Discussion
It has been long understood that the phytochemicals found in Phoenix dactylifera L. target and inhibit several important biochemical pathways contributing to disease development (Farag et al., 2014; Lamia and Mukti, 2021). The ethnopharmacological significance of P. dactylifera L., such as antioxidant, anti-inflammatory anticancer, antimicrobial, etc., is now well established (El Abed et al., 2018). The amount of phytoconstituents, and thus the bioactivity, depends on several factors, including the part of the plant (fruits, seeds, etc.), stage, geographical location, and others. For example, it has been demonstrated that date fruit seeds extract shows anticancer activity against pancreatic (Habib et al., 2014), colorectal (Rezaei et al., 2015), liver (Al-Sheddi, 2019), lung (Al-Sheddi, 2019), and breast (Al-Sheddi, 2019) and other (Al-Zubaidy et al., 2016; Hilary et al., 2021; Habib et al., 2022; Khan et al., 2022) cancer cell lines. On the other hand, Siddiqui et al. (2019) reported that the pulp extract of the Ajwa variety exhibit antiproliferative activity against human liver cancer cells (HepG2, IC50 = 20.03 and 16.78 mg/mL at 24 and 48 h periods, respectively). Moreover, Khan et al. (2021) demonstrated the apoptosis-inducing potential of Ajwa date pulp extract against human triple-negative breast cancer cells (MDA-MB-231, IC50 = 17.45 and 16.67 mg/mL at 24 and 48 h, respectively). Khattak et al. (2020) have reported the antiproliferative property of Emirati date fruits extract on human triple-negative breast cancer cell line MDA-MB-231. Besides, the antioxidant and apoptotic potentials of the whole fruit (flesh and pit extracts) is also known (Shahbaz et al., 2022). In addition to the above-mentioned factors, the polarity of extracting solvents also plays an important role; therefore both aqueous and organic solvent systems have been investigated in the past. In a study, it was found that the aqueous extract of a number of date varieties (Saudi Arabian origin) was less bioactive than the methanolic counterparts (Zhang et al., 2017). In a remarkable study, Khan et al. (2016) noted that the methanolic extract of Ajwa date fruits exhibit strong anticancer effect on human breast adenocarcinoma (MCF7) (Khan et al., 2016). Besides, other researchers also noted the antitumor activity of methanolic extracts (Mansour et al., 2011; Thouri et al., 2019). Therefore, in the present study, we selected methanol as the solvent to extract date fruits of Shishi (M1) and Majdool (M2) cultivars grown in Ha’il region of Saudi Arabia.
As mentioned, (vide-infra), the methanolic extract concentrates were subjected to lyophilization and the resulting water-soluble products were used for further studies without any further purification. LC-MS (negative mode) and GC-MS analyses of the extract revealed the presence of various phytochemicals in both varieties. We noted that the chromatograms of M1 have more peaks than M2; therefore, the former has relatively more constituents. Among the main constituents identified were flavonoids, sphingolipids, and fatty acids classes of phytochemicals. Several researchers already report the presence of these constituents in a wide variety of date fruits (see references in the result section). At the same time, we firmly believe the presence of other components escaped the detection. MTT assay of the extracts against colon cancer cells (HCT-116) revealed a dose-dependent inhibitory nature of the compounds (IC50 = 591.3 μg/mL and 449.9 μg/mL for M1 and M2 at 24, respectively).
It has been demonstrated that the P. dactylifera L. extract exhibits anticancer activity by modulating Bcl-2-family proteins which is also expressed in the HCT-116 cell line (El-Far et al., 2021). Considering this, attempts have been made to identify the principal agent(s) present in the extract using computational approaches, including virtual screening and MD studies. Ligand-based virtual screening identified procyanidin B2 and luteolin-7-O-rutinoside as the most probable candidates since they could bind with Bcl-2 protein efficiently through various amino acids. MD simulation study further strengthens this observation. Considering the earlier reported values and inhibition mechanism on other cell lines, we believe that the anticancer potential of both Shishi and Majdool date extracts against colon cancer cells is interesting and requires further biochemical investigation.
Conclusion
In conclusion, the anticancer activity of methanolic extract of two varieties of dates fruits (Shishi M1 and Majdool M2) grown in the Ha’il region of Saudi Arabia has been compared. The results of GC-MS and Ft-IR studies indicated the presence of various components in the M1 and M2 extracts, which are responsible for dose-dependent cytotoxicity against colon cancer cells (HCT116 cells) through morphological modifications, including cellular membrane damage. The IC50 value was 591.3 μg/mL and 449.9 μg/mL for M1 and M2, respectively. Furthermore, Trypan blue dye exclusion assay further supported the cytotoxic action of M1 and M2 on colon cancer cells. Extensive virtual screening combined with MD simulations studies at 500 ns revealed that procyanidin B2 and luteolin-7-O-rutinoside could be possible agents for the bioactivities. Overall, our data strongly suggest that the consumption of date fruits might prove helpful against colon cancer. Moreover, we also believe that these varieties of date fruits could be utilized as a source of bioactive phytochemicals, leading to the development of Ha’il, Saudi Arabia.
Acknowledgments
This research has been funded by Scientific Research Deanship at University of Ha’il—Saudi Arabia through project number “RD-21 085”.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2023.1138057/full#supplementary-material
References
- Abdul-Hamid N. A., Abas F., Ismail I. S., Tham C. L., Maulidiani M., Mediani A., et al. (2019). Metabolites and biological activities of phoenix dactylifera L. pulp and seeds: A comparative MS and nmr based metabolomics approach. Phytochem. Lett. 31, 20–32. 10.1016/j.phytol.2019.03.004 [DOI] [Google Scholar]
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Ahamad S., Hassan M. I., Dwivedi N. (2018). Designing of phenol-based β− carbonic anhydrase1 inhibitors through QSAR, molecular docking, and MD simulation approach. 3 Biotech. 8, 256. 10.1007/s13205-018-1278-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahamad S., Islam A., Ahmad F., Dwivedi N., Hassan M. I. (2019). 2/3D-QSAR, molecular docking and MD simulation studies of FtsZ protein targeting benzimidazoles derivatives. Comput. Biol. Chem. 78, 398–413. 10.1016/j.compbiolchem.2018.12.017 [DOI] [PubMed] [Google Scholar]
- Ahamad S., Kanipakam H., Birla S., Ali M. S., Gupta D. (2021a). Screening Malaria-box compounds to identify potential inhibitors against SARS-CoV-2 Mpro, using molecular docking and dynamics simulation studies. Eur. J. Pharmacol. 890, 173664. 10.1016/j.ejphar.2020.173664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahamad S., Kanipakam H., Kumar V., Gupta D. (2021b). A molecular journey to check the conformational dynamics of tau tubulin kinase 2 mutations associated with Alzheimer's disease. RSC Adv. 11, 1320–1331. 10.1039/d0ra07659g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Alawi R. A., Al-Mashiqri J. H., Al-Nadabi J. S., Al-Shihi B. I., Baqi Y. (2017). Date palm tree (phoenix dactylifera L.): Natural products and therapeutic options. Front. plant Sci. 8, 845. 10.3389/fpls.2017.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sheddi E. (2019). Anticancer potential of seed extract and pure compound from Phoenix dactylifera on human cancer cell lines. Pharmacogn. Mag. 15, 494–499. 10.4103/pm.pm_623_18 [DOI] [Google Scholar]
- Al-Zubaidy N., Al-Zubaidy A., Sahib H. (2016). The anti-proliferative activity of phoenix dactylifera seed extract on MCF-7 breast cancer cell line. Int. J. Pharm. Sci. Rev. Res. 41, 358–362. [Google Scholar]
- Alam M. Z., Ramachandran T., Antony A., Hamed F., Ayyash M., Kamal-Eldin A. (2022). Melanin is a plenteous bioactive phenolic compound in date fruits (Phoenix dactylifera L.). Sci. Rep. 12, 6614–6712. 10.1038/s41598-022-10546-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghamdi A. A., Awadelkarem A. M., Hossain A., Ibrahim N. A., Fawzi M., Ashraf S. A. (2018). Nutritional assessment of different date fruits (Phoenix dactylifera L.) varieties cultivated in Hail province Saudi Arabia. Biosci. Biotechnol. Res. Commun. 11, 263. 10.21786/bbrc/11.2/11 [DOI] [Google Scholar]
- Ali I., Rahisuddin S. K., Haque A., El-Azzouny A. (2010). Natural products: Human friendly anticancer medications. Egypt Pharma J. 9, 133–179. [Google Scholar]
- Altemimi A., Lakhssassi N., Baharlouei A., Watson D. G., Lightfoot D. A. (2017). Phytochemicals: Extraction, isolation, and identification of bioactive compounds from plant extracts. Plants 6, 42. 10.3390/plants6040042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assirey E. R. (2015). Nutritional composition of fruit of 10 date palm (Phoenix dactylifera L.) cultivars grown in Saudi Arabia. J. Taibah Univ. Sci. 9, 75–79. 10.1016/j.jtusci.2014.07.002 [DOI] [Google Scholar]
- Atanasov A. G., Zotchev S. B., Dirsch V. M., Supuran C. T. (2021). Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 20, 200–216. 10.1038/s41573-020-00114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borochov-Neori H., Judeinstein S., Greenberg A., Volkova N., Rosenblat M., Aviram M. (2013). Date (Phoenix dactylifera L.) fruit soluble phenolics composition and anti-atherogenic properties in nine Israeli varieties. J. Agric. food Chem. 61, 4278–4286. 10.1021/jf400782v [DOI] [PubMed] [Google Scholar]
- Complexity O.-T. O. O. E. (2022). Dates, fresh or dried. Available at https://oec.world/en/profile/hs/dates-fresh-or-dried#:∼:text=Trade%20in%20Dates%2C%20fresh%20or .
- Delano W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsl. protein Crystallogr. 40, 82–92. [Google Scholar]
- Duffy S., Avery V. M. (2012). Development and optimization of a novel 384-well anti-malarial imaging assay validated for high-throughput screening. Am. J. Trop. Med. Hyg. 86, 84–92. 10.4269/ajtmh.2012.11-0302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echegaray N., Gullón B., Pateiro M., Amarowicz R., Misihairabgwi J. M., Lorenzo J. M. (2021). Date fruit and its by-products as promising source of bioactive components: A review. Food Rev. Int. 2021, 1–22. 10.1080/87559129.2021.1934003 [DOI] [Google Scholar]
- Eid N. M., Al-Awadi B., Vauzour D., Oruna-Concha M. J., Spencer J. P. (2013). Effect of cultivar type and ripening on the polyphenol content of date palm fruit. J. Agric. food Chem. 61, 2453–2460. 10.1021/jf303951e [DOI] [PubMed] [Google Scholar]
- El Abed H., Chakroun M., Abdelkafi-Koubaa Z., Drira N., Marrakchi N., Mejdoub H., et al. (2018). Antioxidant, anti-inflammatory, and antitumoral effects of aqueous ethanolic extract from Phoenix dactylifera L. parthenocarpic dates. BioMed Res. Int. 2018, 1542602. 10.1155/2018/1542602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Abed H., Chakroun M., Fendri I., Makni M., Bouaziz M., Drira N., et al. (2017). Extraction optimization and in vitro and in vivo anti-postprandial hyperglycemia effects of inhibitor from Phoenix dactylifera L. parthenocarpic fruit. Biomed. Pharmacother. 88, 835–843. 10.1016/j.biopha.2017.01.129 [DOI] [PubMed] [Google Scholar]
- El-Far A. H., Ragab R. F., Mousa S. A. (2021). Date palm bioactive compounds: Nutraceuticals, functional nutrients, and pharmaceuticals," in the date palm genome, (Germany: Springer; ) [Google Scholar]
- Farag M. A., Handoussa H., Fekry M. I., Wessjohann L. A. (2016). Metabolite profiling in 18 Saudi date palm fruit cultivars and their antioxidant potential via UPLC-qTOF-MS and multivariate data analyses. Food & Funct. 7, 1077–1086. 10.1039/c5fo01570g [DOI] [PubMed] [Google Scholar]
- Farag M. A., Mohsen M., Heinke R., Wessjohann L. A. (2014). Metabolomic fingerprints of 21 date palm fruit varieties from Egypt using UPLC/PDA/ESI–qTOF-MS and GC–MS analyzed by chemometrics. Food Res. Int. 64, 218–226. 10.1016/j.foodres.2014.06.021 [DOI] [PubMed] [Google Scholar]
- Forli S., Huey R., Pique M. E., Sanner M. F., Goodsell D. S., Olson A. J. (2016). Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 905–919. 10.1038/nprot.2016.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib H. M., El-Fakharany E. M., Souka U. D., Elsebaee F. M., El-Ziney M. G., Ibrahim W. H. (2022). Polyphenol-rich date palm fruit seed (phoenix dactylifera L.) extract inhibits labile iron, enzyme, and cancer cell activities, and DNA and protein damage. Nutrients 14, 3536. 10.3390/nu14173536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib H., Platat C., Almaqbali F., Ibrahim W. (2014). Date seed (Phoenix dactylifera) extract reduces the proliferation of pancreatic cancer cells, DNA damage and superoxide-dependent iron release from ferritin in vitro (829.20). FASEB J. 28, 829. 10.1096/fasebj.28.1_supplement.829.20 [DOI] [Google Scholar]
- Hamad I., Abdelgawad H., Al Jaouni S., Zinta G., Asard H., Hassan S., et al. (2015). Metabolic analysis of various date palm fruit (Phoenix dactylifera L.) cultivars from Saudi Arabia to assess their nutritional quality. Molecules 20, 13620–13641. 10.3390/molecules200813620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilary S., Kizhakkayil J., Souka U., Al-Meqbaali F., Ibrahim W., Platat C. (2021). In-vitro investigation of polyphenol-rich date (Phoenix dactylifera L.) seed extract bioactivity. Front. Nutr. 8, 667514. 10.3389/fnut.2021.667514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S. A., Ayad A. A., Williams L. L., Ayivi R. D., Gyawali R., Krastanov A., et al. (2021). Date fruit: A review of the chemical and nutritional compounds, functional effects and food application in nutrition bars for athletes. Int. J. Food Sci. Technol. 56, 1503–1513. 10.1111/ijfs.14783 [DOI] [Google Scholar]
- Khan F., Ahmed F., Pushparaj P. N., Abuzenadah A., Kumosani T., Barbour E., et al. (2016). Ajwa date (Phoenix dactylifera L.) extract inhibits human breast adenocarcinoma (MCF7) cells in vitro by inducing apoptosis and cell cycle arrest. PloS one 11 11, e0158963. 10.1371/journal.pone.0158963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A., Siddiqui S., Ahmad I., Singh R., Mishra D. P., Srivastava A. N., et al. (2021). Phytochemicals from Ajwa dates pulp extract induce apoptosis in human triple-negative breast cancer by inhibiting AKT/mTOR pathway and modulating Bcl-2 family proteins. Sci. Rep. 11, 10322–10414. 10.1038/s41598-021-89420-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A., Singh R., Siddiqui S., Ahmad I., Ahmad R., Upadhyay S., et al. (2022). Anticancer potential of phoenix dactylifera L. Seed extract in human cancer cells and pro-apoptotic effects mediated through caspase-3 dependent pathway in human breast cancer MDA-MB-231 cells: An in vitro and in silico investigation. BMC complementary Med. Ther. 22, 68–19. 10.1186/s12906-022-03533-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak M. N. K., Shanableh A., Hussain M. I., Khan A. A., Abdulwahab M., Radeef W., et al. (2020). Anticancer activities of selected Emirati Date (Phoenix dactylifera L.) varieties pits in human triple negative breast cancer MDA-MB-231 cells. Saudi J. Biol. Sci. 27, 3390–3396. 10.1016/j.sjbs.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A., Sharifi I., Tavakkoli H., Molaakbari E., Bahraminegad S., Salarkia E., et al. (2022). Cytotoxicity of amphotericin B and AmBisome: In silico and in vivo evaluation employing the chick embryo model. Front. Pharmacol. 13, 1. 10.3389/fphar.2022.860598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia F. S., Mukti R. F. (2021). Bangladeshi wild date palm fruits (Phoenix sylvestris): Promising source of anti-cancer agents for hepatocellular carcinoma treatment. Int. J. Appl. Sci. Biotechnol. 9, 32–37. 10.3126/ijasbt.v9i1.36110 [DOI] [Google Scholar]
- Laskowski R. A., Jabłońska J., Pravda L., Vařeková R. S., Thornton J. M. (2018). PDBsum: Structural summaries of PDB entries. Protein Sci. 27, 129–134. 10.1002/pro.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour R. B., Lassoued S., Dammak I., Elgaied A., Besbes S., Attia H., et al. (2011). Cytotoxicity evaluation and antioxidant activity of date seed oil from ‘‘Deglet-nour Tunisian cultivar”(Phoenix dactylifera L.). Nat. Prod. 7, 16–20. [Google Scholar]
- Maqsood S., Adiamo O., Ahmad M., Mudgil P. (2020). Bioactive compounds from date fruit and seed as potential nutraceutical and functional food ingredients. Food Chem. 308, 125522. 10.1016/j.foodchem.2019.125522 [DOI] [PubMed] [Google Scholar]
- Mhm A. E.-A., El-Mesalamy A., Yassin F., Khalil S. (2015). Identification phenolic and biological activities of methanolic extract of date palm pollen (Phoenix dactylifera). J. Microb. Biochem. Technol. 7, 047–050. 10.4172/1948-5948.1000180 [DOI] [Google Scholar]
- Mia M. a.-T., Mosaib M. G., Khalil M. I., Islam M. A., Gan S. H. (2020). Potentials and safety of date palm fruit against diabetes: A critical review. Foods 9, 1557. 10.3390/foods9111557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm O. A., Addnan F. H., Mohd-Manzor N. F., Elkadi M. A., Abdullah W. O., Ismail A., et al. (2021). Identification of phytochemicals of phoenix dactylifera L. Cv Ajwa with UHPLC-ESI-QTOF-MS/MS. Int. J. Fruit Sci. 21, 848, 10.1080/15538362.2021.1939227 [DOI] [Google Scholar]
- Nematallah K. A., Ayoub N. A., Abdelsattar E., Meselhy M. R., Elmazar M. M., El-Khatib A. H., et al. (2018). Polyphenols LC-MS2 profile of Ajwa date fruit (Phoenix dactylifera L.) and their microemulsion: Potential impact on hepatic fibrosis. J. Funct. foods 49, 401. 10.1016/j.jff.2018.08.032 [DOI] [Google Scholar]
- Newman D. J., Cragg G. M. (2016). Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661. 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- Otify A. M., El-Sayed A. M., Michel C. G., Farag M. A. (2019). Metabolites profiling of date palm (phoenix dactylifera L.) commercial by-products (pits and pollen) in relation to its antioxidant effect: A multiplex approach of MS and nmr metabolomics. Metabolomics 15, 119–217. 10.1007/s11306-019-1581-7 [DOI] [PubMed] [Google Scholar]
- Perveen K., Bokahri N. A. (2020). Comparative analysis of chemical, mineral and in-vitro antibacterial activity of different varieties of date fruits from Saudi Arabia. Saudi J. Biol. Sci. 27, 1886–1891. 10.1016/j.sjbs.2019.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi L. (2017). Bioactivity of Phoenix dactylifera seed and its phytochemical analysis. Int. J. Green Pharm. (IJGP) 11. 10.22377/ijgp.v11i02.1037 [DOI] [Google Scholar]
- Rezaei M., Khodaei F., Hooshmand N. (2015). Date seed extract diminished apoptosis event in human colorectal carcinoma cell line. MOJ Toxicol. 1, 00017. 10.15406/mojt.2015.01.00017 [DOI] [Google Scholar]
- Rock W., Rosenblat M., Borochov-Neori H., Volkova N., Judeinstein S., Elias M., et al. (2009). Effects of date (phoenix dactylifera L., Medjool or Hallawi variety) consumption by healthy subjects on serum glucose and lipid levels and on serum oxidative status: A pilot study. J. Agric. food Chem. 57, 8010–8017. 10.1021/jf901559a [DOI] [PubMed] [Google Scholar]
- Schüttelkopf A. W., Van Aalten D. M. (2004). Prodrg: A tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. Sect. D. Biol. Crystallogr. 60, 1355–1363. 10.1107/s0907444904011679 [DOI] [PubMed] [Google Scholar]
- Shahbaz K., Asif J. A., Liszen T., Nurul A. A., Alam M. K. (2022). Cytotoxic and antioxidant effects of phoenix dactylifera L. (Ajwa date extract) on oral squamous cell carcinoma cell line. BioMed Res. Int. 2022, 1–16. 10.1155/2022/5792830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Joshi T., Mathpal S., Joshi T., Pundir H., Chandra S., et al. (2022). Identification of natural inhibitors against Mpro of SARS-CoV-2 by molecular docking, molecular dynamics simulation, and MM/PBSA methods. J. Biomol. Struct. Dyn. 40, 2757–2768. 10.1080/07391102.2020.1842806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui S., Ahmad R., Khan M. A., Upadhyay S., Husain I., Srivastava A. N. (2019). Cytostatic and anti-tumor potential of Ajwa date pulp against human hepatocellular carcinoma HepG2 cells. Sci. Rep. 9, 245. 10.1038/s41598-018-36475-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souda B., Rami R., Jalloul B., Mohamed D. (2020). Roasted date palm seeds (phoenix dactylifera) as an alternative coffee: Chemical composition and bioactive properties. Biomass Convers. Biorefinery 12, 3771. 10.1007/s13399-020-00896-7 [DOI] [Google Scholar]
- Taghizadeh M. S., Niazi A., Moghadam A., Afsharifar A. (2022). Experimental, molecular docking and molecular dynamic studies of natural products targeting overexpressed receptors in breast cancer. Plos one 17 17, e0267961. 10.1371/journal.pone.0267961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouri A., La Barbera L., Canuti L., Vegliante R., Jelled A., Flamini G., et al. (2019). Antiproliferative and apoptosis-inducing effect of common Tunisian date seed (var. Korkobbi and Arechti) phytochemical-rich methanolic extract. Environ. Sci. Pollut. Res. 26, 36264–36273. 10.1007/s11356-019-06606-9 [DOI] [PubMed] [Google Scholar]
- Tian W., Chen C., Lei X., Zhao J., Liang J. (2018). CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic acids Res. 46, W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A. J. (2010). AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005). Gromacs: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Vayalil P. K. (2002). Antioxidant and antimutagenic properties of aqueous extract of date fruit (Phoenix dactylifera L. Arecaceae). J. Agric. Food Chem. 50, 610–617. 10.1021/jf010716t [DOI] [PubMed] [Google Scholar]
- Vayalil P. K. (2012). Date fruits (phoenix dactylifera linn): An emerging medicinal food. Crit. Rev. food Sci. Nutr. 52, 249–271. 10.1080/10408398.2010.499824 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiao J., Suzek T. O., Zhang J., Wang J., Bryant S. H. (2009). PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic acids Res. 37, W623–W633. 10.1093/nar/gkp456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-R., Aldosari S. A., Vidyasagar P. S., Nair K. M., Nair M. G. (2013). Antioxidant and anti-inflammatory assays confirm bioactive compounds in Ajwa date fruit. J. Agric. food Chem. 61, 5834–5840. 10.1021/jf401371v [DOI] [PubMed] [Google Scholar]
- Zhang C.-R., Aldosari S. A., Vidyasagar P. S., Shukla P., Nair M. G. (2017). Health-benefits of date fruits produced in Saudi Arabia based on in vitro antioxidant, anti-inflammatory and human tumor cell proliferation inhibitory assays. J. Saudi Soc. Agric. Sci. 16, 287–293. 10.1016/j.jssas.2015.09.004 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.