Abstract
HDAC3 is one of the main targets of histone deacetylase (HDAC) inhibitors in clinical development as cancer therapies, yet the in vivo role of HDAC3 in solid tumors is unknown. We identified a critical role for HDAC3 in Kras-mutant lung cancer. Using genetically engineered mouse models (GEMMs), we found that HDAC3 is required for lung tumor growth in vivo. HDAC3 was found to direct and enhance the transcription effects of the lung cancer lineage transcription factor NKX2-1 to mediate expression of a common set of target genes. We identified FGFR1 as a critical previously unidentified target of HDAC3. Leveraging this, we identified that an HDAC3-dependent transcriptional cassette becomes hyperactivated as Kras/LKB1-mutant cells develop resistance to the MEK inhibitor trametinib, and this can be reversed by treatment with the HDAC1/HDAC3 inhibitor entinostat. We found that the combination of entinostat plus trametinib treatment elicits therapeutic benefit in the Kras/LKB1 GEMM.
HDAC3 is essential for Kras, Lkb1-dependent lung tumor growth and therapeutic resistance in genetic engineered mouse models.
INTRODUCTION
Targeted therapies have begun to prove themselves as successful treatments against cancer types harboring specific, defined vulnerabilities. However, only a small subset of tumor types have targeted therapies currently available, as such agents only exist for a limited number of oncogenic drivers. Moreover, tumors characterized by loss of tumor suppressor genes provide no clear targets against which to develop inhibitors. Transcriptional dependencies of tumors have emerged as definable and therapeutically tractable liabilities that can be oncogene-agnostic (1). Much recent effort has focused on targeting epigenetic regulators (e.g., Brd4) as a means to globally affect transcription in such tumors (2–6). One case in point is histone deacetylase (HDAC) inhibitors, which were originally developed to antagonize the reduced global histone acetylation observed in many tumor types (7, 8). Several HDAC inhibitors are now Food and Drug Administration (FDA) approved to treat hematopoietic malignancies (9), although efficacy of HDAC inhibitors in solid tumors has been disappointingly limited. Recent efforts to identify effective approaches to HDAC inhibitor combination therapy have gained traction in specific tumor types (10–14). However, current FDA-approved inhibitors target multiple HDACs, and better therapeutic potential may be realized with more selective inhibitors aimed at one or two HDACs. Despite the fact that HDAC inhibitors are already in the clinic, little analysis of the disruption of the four class I HDACs has been performed in genetically engineered tumor models in mice that might help narrow down which are most important in different tumor contexts in vivo. Recent studies indicate that the HDAC inhibitor entinostat, which is selective to HDAC1 and HDAC3, exhibits potent effects in boosting therapeutic response in specific contexts (15–18). However, even with increased understanding of their therapeutic potential, the molecular mechanisms that mediate tumor growth control by individual HDACs in vivo remain poorly understood.
There are four members of the class I HDAC family: HDAC1, HDAC2, HDAC3, and HDAC8. HDAC3 is unique among them in requiring the nuclear receptor co-repressor (NCoR) complex for its enzymatic activity (19), forming a core complex of nuclear receptor corepressor 1/2 (NCoR1/SMRT), transducin beta like 1 x-linked (TBL1X), TBL1X/Y related 1 (TBL1XR1), G protein pathway suppressor 2 (GPS2), and HDAC3. HDAC3 has been shown to deacetylate histone and nonhistone proteins and can function, in part, through deacetylase-independent mechanisms (19). Tissue-specific deletion of HDAC3 in metabolic tissues in mice has identified notable biological functions and deregulation of distinct nonoverlapping transcriptional programs unique to each corresponding tissue (19). Collectively, these studies reveal that HDAC3 function is not uniformly through global control of histone acetylation but is nuanced and directed in a tissue-specific fashion. For example, HDAC3 deletion in brown adipose tissue causes mice to become hypothermic and succumb to acute cold exposure (20), but HDAC3 deletion in the liver induces hypertrophy and metabolic alterations (21–23), and the genes controlled by HDAC3 in each tissue are distinct and relate to tissue-specific functions. Despite clinical advancement of inhibitors of class I HDACs as therapeutics, any potential role of HDAC3 in tumorigenesis remains largely unknown, as its in vivo function and mechanism of action has predominantly been examined in metabolic tissues.
The liver kinase B1 (LKB1/STK11) tumor suppressor is mutated in ~20% of lung adenocarcinoma (LUAD), often concurrently with Kristen rat sarcoma viral oncogene (Kras) mutation (24–27). LKB1 is a serine/threonine kinase that directly activates a family of 14 downstream kinases in the adenosine monophosphate (AMP)–activated protein kinase (AMPK) family (28). Recent studies dissecting which of these kinases are most critical to LKB1 tumor suppressor function in the lung revealed that AMPK may not be as critical as originally hypothesized (29) and that two AMPK-related kinases, salt inducible kinase 1 (SIK1) and SIK3, are the most important for suppressing growth of lung tumors (30, 31). Notably, only two sets of substrates of the SIK1/3 kinases are well established: the cyclic AMP response element–binding protein (CREB)–regulated transcription coactivator family of CREB coactivators and the class IIa family of HDACs (32). Class IIa HDACs, which lack catalytic activity themselves, are hypothesized to function as transcriptional co-regulators of HDAC3 (33, 34). Our genetic analysis suggested that class IIa HDACs may be key targets of LKB1 tumor suppression, but there are three redundant class IIa HDACs expressed in non–small cell lung cancer (NSCLC) cells, all of which may rely on HDAC3. Therefore, we first explored a role for HDAC3 in lung cancer tumorigenesis using Kras-mutant genetically engineered mouse models (GEMMs).
RESULTS
HDAC3 is essential for lung tumorigenesis in vivo
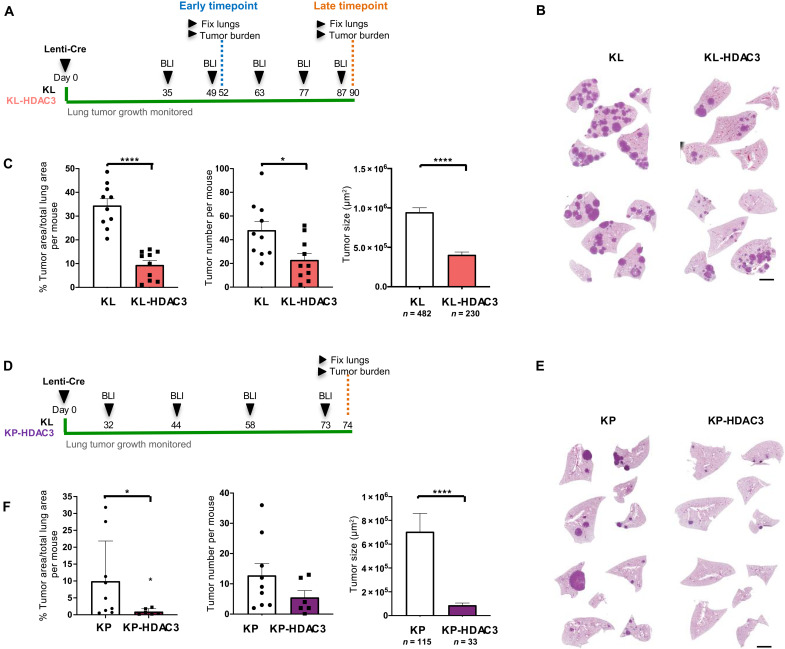
To assess the role of HDAC3 in solid tumors in vivo, we used two mouse models engineered to recapitulate the most common subtypes of Kras-mutant NSCLC: mutant Kras combined with LKB1 loss, KrasLSL-G12D/+ STK11−/− (KL), and mutant Kras combined with p53 loss, KrasLSL-G12D/+ p53−/− (KP). We first examined mice harboring KrasLSL-G12D/+, STK11L/L, and ROSA26LSL-luciferase, with or without conditional HDAC3L/L (KL-HDAC3). In these mice, intratracheal administration of lentivirus expressing Cre recombinase (Lenti-Cre) simultaneously activates KrasG12D and deletes LKB1 (STK11) to initiate tumorigenesis in the lung epithelium and, for those bearing HDAC3L/L, coincidentally deletes HDAC3. Simultaneously, Cre recombinase induces expression of firefly luciferase in infected cells, allowing for noninvasive longitudinal bioluminescence imaging (BLI) of NSCLC tumor development in the whole animal, as we have reported previously (29, 31, 35, 36). Tumor growth was markedly reduced in KL-HDAC3 mice compared to KL littermate controls at both early and late time points, exhibiting significantly less tumor area, tumor number, and smaller tumor size (Fig. 1, A to C, and fig. S1, A to C). Thus, we conclude that HDAC3 supports tumor initiation and tumor growth in the KL model of NSCLC. Using a similar experimental design, we generated mice harboring KrasLSL-G12D/+, p53L/L, ROSA26LSL-luciferase, HDAC3L/L (KP-HDAC3) to test the role of HDAC3 in the KP model of NSCLC. Tumor growth was markedly reduced in KP-HDAC3 mice compared to KP littermate controls, with significantly less tumor area and smaller tumor size and a trend toward smaller tumor number (Fig. 1, D to F, and fig. S1D). We conclude that HDAC3 is of critical importance for growth of NSCLC tumors driven by both KL and KP genotypes.
Fig. 1. HDAC3 is essential for lung tumorigenesis in vivo in KL and KP GEMM models of NSCLC.
(A) Schematic of experimental design in KrasG12D/+, LKB1L/L (KL), and KL-HDAC3L/L (KL-HDAC3) mouse models administered lentivirus expressing Cre recombinase (Lenti-Cre). (B) Representative hematoxylin and eosin (H&E)–stained sections from the late time point. Scale bar, 1000 μm. (C) Quantitation from H&E-stained sections from the late time point cohort: Tumor area as a percentage of total lung area per mouse (n = 10), tumor number per mouse (n = 10), and average tumor size (n = 482 or 230 as indicated). (D) Schematic of experimental design in KrasG12D/+, p53L/L (KP), and KP-HDAC3L/L (KP-HDAC3) mouse models administered Lenti-Cre. (E) Representative H&E-stained sections. Scale bar, 1000 μm. (F) Quantitation from H&E-stained sections: tumor area as a percentage of total lung area per mouse (n = 9 or 6 as indicated), tumor number per mouse (n = 9 or 6 as indicated), and average tumor size (n = 115 or 33 as indicated). Values are expressed as means ± SEM. *P < 0.05 and ****P < 0.0001, determined by two-tailed Mann-Whitney test.
HDAC3 genome occupancy in primary tumors predicts cooperation with NK2 homeobox 1
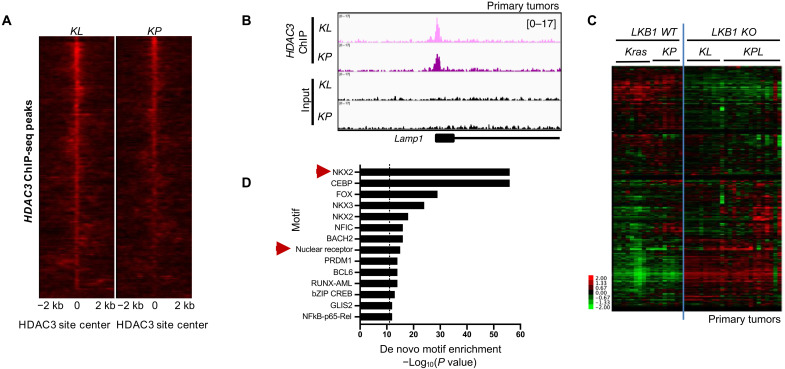
Transcriptional programs regulated by HDAC3 are markedly tissue context specific, in a manner often dictated by the identity of the cooperating transcription factors (19). Therefore, to understand the molecular mechanism mediating the function of HDAC3 in NSCLC, we set out to identify the transcription factors with which HDAC3 cooperates in this cellular context. To this end, we performed HDAC3 chromatin immunoprecipitation sequencing (ChIP-seq) on KL and KP primary tumors to identify genome-wide, endogenously bound HDAC3 target loci in vivo (Fig. 2, A and B, and fig. S2A). A total of 1522 peaks were bound by HDAC3 in both KL and KP tumors (Fig. 2A), corresponding to 753 nonredundant genes with at least one HDAC3 binding site within ±25 kb of the transcription start site (TSS).
Fig. 2. HDAC3 genome occupancy in primary tumors.
(A) A total of 1522 HDAC3 ChIP-seq peaks common to KL and KP primary tumors. (B) Example of HDAC3 ChIP-seq peaks at genomic regions bound by HDAC3 in both KL and KP primary tumors. (C) Heatmap of RNA-seq data showing FPKM (fragments per kilobase of transcript per million) read counts from primary tumors from LKB1 wild type (Kras and KP) and LKB1 knockout (KO) (KL and KPL) models for the 753 nonredundant genes associated with at least one HDAC3 ChIP-seq peak within 25 kb of the TSS. Kras, KrasLSL-G12D/+; KL, KrasLSL-G12D/+ Stk11−/−; KP, KrasLSL-G12D/+ p53−/−; KPL, KrasLSL-G12D/+ Stk11−/− p53−/−. (D) Homer de novo motif enrichment analysis of the HDAC3-bound peaks in (A). All significantly enriched motifs are listed.
We next plotted the expression of these 753 direct HDAC3 target genes across RNA sequencing (RNA-seq) data from individual primary lung tumors dissected from four different Kras-mutant GEMMs of NSCLC: the Kras, KP, KL, and KPL (Fig. 2C) models. This analysis revealed an unexpected LKB1-dependent gene expression pattern, where gene expression was distinctly different in LKB1 wild type (Kras and KP) versus LKB1 knockout (KO) (KL and KPL) tumor cells. Thirty-nine percent of HDAC3 direct target genes were differentially expressed between Kras and KL tumors (fig. S2B). LKB1-mutant tumors exhibit unique gene expression signatures clinically (37, 38), but the involvement of HDAC3 in LKB1-specific gene expression is unknown. The data thus far suggest that LKB1 loss affects the expression of HDAC3 target genes, suggesting an important role for HDAC3 in these tumors.
To this end, we next sought to identify the transcription factors with which HDAC3 cooperates in the lung tumor context. HDAC3 enzymatic activity requires interaction with NCoRs (39), and consistently, de novo motif enrichment analysis of the HDAC3 ChIP-seq binding sites (Fig. 2A) revealed the classical motif recognized by a number of nuclear receptors (Fig. 2D). However, the most enriched de novo motif was that of the transcription factor NK2 homeobox 1/thyroid transcription factor 1 (NKX2-1/TTF-1; Fig. 2D). This suggested an unexpected functional overlap between HDAC3 and NKX2-1. This finding was particularly relevant to the NSCLC context, as NKX2-1 is highly expressed in up to 85% and amplified in up to 15% of human LUAD cases, to the extent that it is used as a clinical biomarker of the disease (26, 40–43).
Functionally, NKX2-1 is considered a lineage-survival oncogene, and it has an established role enforcing a lineage-specific differentiation program in lung and LUAD (41, 44–46). Lineage-specific oncogenic transcription factors are appreciated addictions of cancer but often lack entry points for therapeutic intervention (47), as is the case for NKX2-1 in the lung cancer context. NKX2-1 is an appreciated but undruggable transcriptional addiction of LUAD (47), and identifying druggable regulators of NKX2-1 function is of great interest. Moreover, HDAC3 has recently been implicated as a regulator of lineage specification in normal T cells and glial cells (48, 49). Together, the interesting implication is that because NKX2-1 is the lineage transcription factor in LUAD, perhaps HDAC3 is a druggable regulator of this particular lineage factor addiction.
HDAC3 cooperates with NKX2-1 to regulate the expression of a common set of target genes
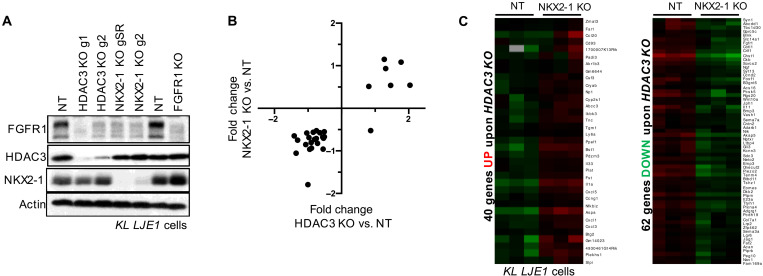
To explore this, we set out to assess whether HDAC3 and NKX2-1 coordinately control a common set of target genes in NSCLC cells. First, we created an immortalized cell line, KL LJE1 cells, from an explanted KL primary lung tumor, as described in Methods. Using CRISPR-Cas9, we then genomically disrupted HDAC3 or NKX2-1 in KL LJE1 cells. KL LJE1 cells were infected with lentivirus expressing Cas9 and small guide RNA (sgRNA) directed against a nontargeting sequence (NT), HDAC3, or NKX2-1. Two independent sgRNAs directed against HDAC3 or NKX2-1 were used. Subsequent puromycin selection generated a pooled population of NT, HDAC3 KO, or NKX2-1 KO cells, and immunoblotting verified deletion (Fig. 3A).
Fig. 3. HDAC3 cooperates with NKX2-1 to regulate the expression of a common set of target genes.
(A) Western blot analysis of HDAC3, NKX2-1, or FGFR1 KO by CRISPR-Cas9 in polyclonal lysates from KL LJE1 cells. (B) Plot of fold change upon HDAC3 KO compared to NKX2-1 KO for the genes significantly deregulated (adjusted P < 0.05; fold, ±0.5) upon loss of both factors in KL LJE1 cells. (C) Heatmap of RNA-seq data showing FPKM read counts for genes commonly up-regulated (left) or down-regulated (right) upon both HDAC3 KO and NKX2-1 KO in KL cells, as defined from red box regions on heatmap in fig. S3D.
We profiled these cell lines by RNA-seq. Comparison between RNA-seq datasets identified a common set of gene deregulation upon HDAC3 KO and NKX2-1 KO and that each of these KOs affect gene expression changes with the same directionality in KL NSCLC cells (Fig. 3B). Comparison with published data from Krasmut tumors deleted for NKX2-1 (44) verified that, for target genes co-regulated by both factors, HDAC3 predominantly promotes the gene expression program driven by NKX2-1 (fig. S3A) and loss of either protein results in reduced expression of common target genes. We next queried which fraction of the NKX2-1 transcriptional program is regulated by HDAC3. Of the genes most deregulated upon NKX2-1 KO, 83% were also modulated upon HDAC3 KO (fig. S3B), including the established NKX2-1 target Tnc (50). This suggests that the most NKX2-1–dependent genes are nearly all under the control of HDAC3. To query the extent to which HDAC3 is involved in the regulation of NKX2-1 target genes across a broader set of genes, we extended this analysis to the 68 genes deregulated upon NKX2-1 KO by the stronger sgRNA (gSR) and found that 72% of NKX2-1 target genes were also modulated upon HDAC3 KO in these cells. This suggests that nearly three quarters of the NKX2-1 transcriptional program is co-regulated by HDAC3. To identify the direct targets of endogenous HDAC3 in these cells, we performed HDAC3 ChIP-seq (fig. S3C) on NT and HDAC3 KO KL LJE1 cells. Overlay with the RNA-seq data revealed that 31% of the genes regulated by both NKX2-1 and HDAC3 were associated with at least one HDAC3 ChIP-seq peak, indicative of direct regulation by HDAC3. Notably, HDAC3 was not observed to be binding at or near the NKX2-1 genomic locus in HDAC3 ChIP-seq experiments.
We next assessed what fraction of the total HDAC3 transcriptional response in these cells is regulated by NKX2-1. For genes differentially expressed upon HDAC3 KO, we plotted gene expression from NT and NKX2-1 KO cells (Fig. 3C and fig. S3D). Of the 171 genes up-regulated upon HDAC3 KO, 21% were also up-regulated upon NKX2-1 KO, but only 3% were down-regulated upon NKX2-1 KO. Of the 165 genes down-regulated upon HDAC3 KO, 38% were also down-regulated upon NKX2-1 KO, and none were up-regulated upon NKX2-1 KO. We conclude that NKX2-1 regulates the expression of ~30% of the HDAC3-dependent genes in KL NSCLC cells. Notably, genes both activated and repressed by NKX2-1/HDAC3 are direct targets of HDAC3, suggesting that HDAC3 is not solely acting as a canonical repressor (19) on the NKX2-1–regulated genes in KL cells.
We observed that HDAC3 and NKX2-1 predominantly promote the expression of a set of common target genes in KL cells. One co-regulated target of both NKX2-1 and HDAC3 (Fig. 3C) is Fgfr1, which we selected for further validation. Fibroblast growth factor (FGF) receptor 1 (FGFR1) is one of four receptor tyrosine kinases that make up the FGFR protein family. FGFRs, receptors for FGFs, have been widely implicated in promoting tumor growth, and multiple small-molecule inhibitors of FGFRs are in various stages of development as cancer therapies (51, 52). Our RNA-seq analysis identified that Fgfr1 mRNA was down-regulated upon both NKX2-1 KO and HDAC3 KO. Western blotting revealed that both HDAC3 and NKX2-1 are required for maintenance of FGFR1 protein expression in KL cells (Fig. 3A). We found that HDAC3, NKX2-1, and FGFR1 all support tumor cell growth in KL LJE1 cells (fig. S3E).
Considering the LKB1-dependent expression pattern of HDAC3 target genes in primary lung tumors (Fig. 2C), we also tested the impact of HDAC3 and NKX2-1 in an LKB1–wild-type cell line derived from a KP GEMM tumor, KP T3 cells (31). Unlike in KL cells, neither HDAC3 KO nor NKX2-1 KO affected FGFR1 protein expression in KP T3 cells (fig. S3F). We also found that NKX2-1 did not support KP T3 cell growth (fig. S3G) as it did in KL LJE1 cells (fig. S3E). Next, we used RNA-seq to identify the transcriptional targets common to both HDAC3 and NKX2-1 in this KP cell system. Performing the same analysis on RNA-seq data from KP T3 cells (fig. S3H) as we had performed on KL LJE1 cells (fig. S3D) revealed remarkably little overlap between the target genes co-regulated by HDAC3 and NKX2-1 in KL LJE1 cells and KP T3 cells (fig. S3I). Last, RNA-seq data from primary GEMM lung tumors (n ≥ 8) revealed that Fgfr1 mRNA was expressed at higher levels in tumors without LKB1 compared to tumors with LKB1 (fig. S3J), indicating that Fgfr1 expression differences between tumor cells with or without LKB1 is observed across multiple independently arising tumors.
HDAC3 and NKX2-1 co-regulate target genes that are aberrantly engaged upon trametinib resistance
Notably, FGFR1 has been shown to mediate resistance to the FDA-approved mitogen-activated protein kinase (MAPK) kinase (MEK) inhibitor, trametinib, that acts downstream of Kras to suppress signaling through the MAPK cascade (53). However, therapies directed against Kras effectors activate compensatory pathways that limit their efficacy as single agents, and many current efforts are directed toward elucidating combination therapy approaches that would potentiate clinical benefit from existing Kras effector inhibitors. Using a short hairpin RNA screen, Manchado et al. (53) identified FGFR1 as a mediator of trametinib resistance. In the KP lung tumor GEMM where trametinib treatment alone is largely ineffective because of rapidly acquired resistance, they found that cotreatment of trametinib with the FGFR1 inhibitor ponatinib induced tumor regression in this aggressive in vivo model of NSCLC (53).
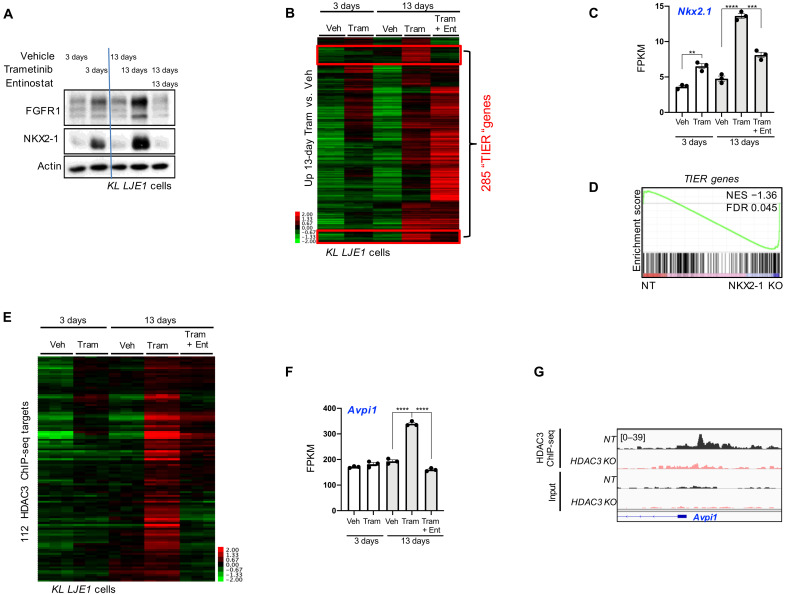
Because HDAC3 is required for FGFR1 protein expression (Fig. 3A), we hypothesized that HDAC inhibition may be an alternative therapeutic approach for blocking trametinib resistance–induced FGFR1 hyperactivation. To test this, we performed short-term (3-day) and long-term (13-day) treatments with trametinib. We selected entinostat (MS-275) as our HDAC inhibitor of choice for molecular studies, as it is one of the most selective clinically tolerated HDAC inhibitors, preferentially inhibiting HDAC1 and HDAC3. FGFR1 protein was strongly induced upon 13 days of trametinib in a manner that could be reversed by cotreatment with entinostat (Fig. 4A). Moreover, NKX2-1 protein itself was regulated in a similar fashion (Fig. 4A): long-term trametinib-induced, entinostat-reversed (“TIER”). This suggested that up-regulation of NKX2-1 activity is an HDAC-dependent component of the trametinib resistance response, and FGFR1 behaves as a readout of this molecular event in KL cells. We confirmed that neither FGFR1 protein nor mRNA levels were affected by 1 μM entinostat treatment alone (fig. S4, A and B). Blunted FGFR1 induction from trametinib and entinostat cotreatment corresponded with a notable reduction in extracellular signal–regulated kinase (ERK) activity, as assessed by phosphorylation at Thr202/Tyr204 (fig. S4A). Using the A549 cell line, we also found that FGFR1 behaved as a TIER gene in human KRAS, LKB1-mutant lung cancer cells (fig. S4C). Comparing NT versus HDAC3 KO cells revealed that HDAC3 was required for the trametinib-enhanced FGFR1 expression (fig. S4D). Both HDAC3 and NKX2-1 were required for maximal cellular resistance to trametinib (fig. S4E).
Fig. 4. HDAC3 and NKX2-1 common target genes are aberrantly engaged upon trametinib resistance.
(A) Western blot analysis of protein lysates from KL LJE1 cells treated with vehicle, 10 nM trametinib, or 1 μM entinostat for 3 or 13 days. (B) Heatmap of RNA-seq data showing FPKM read counts across all treatment conditions for the 2141 genes significantly up-regulated (adjusted P < 0.05; fold, >±0.5) upon 13 day of trametinib compared to 13 days of vehicle in KL LJE1 cells. Veh, vehicle; Tram, trametinib; Ent, entinostat. Red boxes identify TIER genes. (C) Nkx2-1 mRNA levels (FPKM) across all treatment conditions (n = 3) from RNA-seq data in (B). (D) Gene set enrichment analysis (GSEA) of the 285 TIER genes queried across RNA-seq data from NKX2-1 KO versus NT KL LJE1 cells. (E) Heatmap of RNA-seq data showing FPKM read counts across all treatment conditions for the 112 TIER genes that are HDAC3 ChIP-seq target genes. (F) Avpi1 mRNA levels across all treatment conditions from RNA-seq data from cells in (B) (n = 3). (G) HDAC3 ChIP-seq data in NT and HDAC3 KO KL LJE1 cells at the Avpi1 genomic locus. Values are expressed as means ± SEM. **P < 0.01, ***P < 0.001, and ****P < 0.0001, determined by two-tailed Student’s t test.
Because genes associated with direct HDAC3 genome binding had displayed an LKB1-dependent gene expression pattern in primary tumors (Fig. 2C), we next queried whether FGFR1 expression behaved in an LKB1-dependent manner. We reintroduced LKB1 into KL LJE1 cells and queried the impact on FGFR1 response to treatment, which revealed that FGFR1 only behaved as a TIER gene in the absence of LKB1 (fig. S4, F and G). Consistently, FGFR1 expression did not follow the TIER pattern in KP cells (fig. S4H). Together, the implication is that HDAC inhibitors that target HDAC3 such as entinostat may block the induction of a transcriptional program that becomes hyperactivated as Kras, LKB1-mutant lung cancer cells develop resistance to trametinib.
Because NKX2-1 is a transcription factor with multiple target genes, we next explored whether FGFR1 is part of a larger set of HDAC3-dependent NKX2-1 target genes that become up-regulated upon trametinib resistance. To this end, we profiled KL LJE1 cells treated as in Fig. 4A by RNA-seq. To identify genes exhibiting the TIER gene expression pattern, we first defined the genes up-regulated upon 13 days of trametinib compared to vehicle (2141 genes) and then plotted their gene expression across all five treatment conditions (Fig. 4B). This analysis identified that 285 of 2141 (13%) genes displayed the TIER expression pattern. One of these 285 TIER genes was Nkx2-1 itself (Fig. 4C), consistent with the immunoblot results in Fig. 4A. To identify whether a broader set of NKX2-1–dependent genes behaved similarly, we queried the 285 TIER genes against RNA-seq data from NKX2-1 KO cells using gene set enrichment analysis (GSEA). The TIER gene set was negatively enriched in cells deleted for NKX2-1 (Fig. 4D), indicating that a group of TIER genes are NKX2-1–dependent in their basal gene expression pattern as well. We plotted TIER gene expression across RNA-seq data from NT and NKX2-1 KO cells to identify these genes (fig. S4I). Thus, induction of a cassette of NKX2-1–regulated genes is a component of the trametinib resistance transcriptional program that can be reversed by HDAC inhibition. We hypothesized that entinostat blunts the induction of NKX2-1 target gene expression through inhibition of HDAC3 and, thus, predicted that a set of HDAC3 direct target genes (fig. S3C) would be members of the TIER gene cassette. We found that 112 of the 285 (39%) TIER genes were associated with at least one HDAC3 ChIP-seq binding site (Fig. 4E). Moreover, many of these 112 HDAC3-bound TIER genes display LKB1-dependent gene expression patterns in primary tumors (fig. S4J). Together, identification of the TIER genes revealed a set of direct HDAC3 target genes hyperactivated in KL cells upon trametinib resistance. Together, we have discovered that trametinib resistance accentuates the necessity for the HDAC3/NKX2-1 pathway in NSCLC.
Avpi1 provides an example of how a TIER gene behaves across the experimental conditions queried (Fig. 4, F and G, and fig. S4, K and L). Avpi1 is a TIER gene (Fig. 4F) and is directly bound by HDAC3 (Fig. 4G), and its expression is reduced upon deletion of HDAC3 in KL NSCLC cells (fig. S4K). Thus, Avpi1 expression in lung tumor cells is directly regulated by HDAC3, induced by trametinib resistance, and reduced by HDAC inhibitor treatment in KL cells. Querying The Cancer Genome Atlas (TCGA) human Lung Adenocarcinoma dataset we found that, in tumors harboring KRAS amplification or mutation at G12, 71% of the tumors with high AVPI1 expression harbored STK11 mutation, corresponding with the genetics of the KL cell lines used in our study. High AVPI1 expression within this patient cohort correlated with substantially shorter overall survival (8.48 versus 88.07 median months overall; fig. S4L). One potential implication is that, particularly for this subset of patients destined for poor outcome whose tumors harbor high expression of HDAC3 target genes, HDAC3 inhibition strategies may be worthwhile exploring.
Trametinib plus entinostat treatment elicits therapeutic benefit in the KL GEMM model
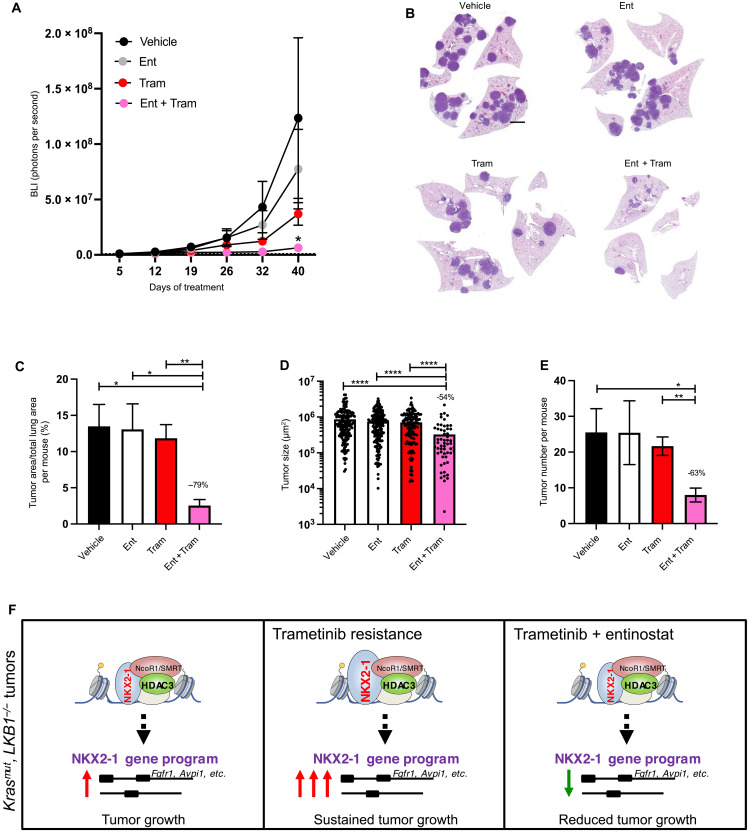
Identification of the TIER genes revealed that trametinib resistance accentuates the necessity for the HDAC3 pathway in a manner that can be reversed by cotreatment with the HDAC inhibitor entinostat in KL NSCLC cells. Thus, to assess therapeutic efficacy of the combination treatment in vivo, we next sought to treat KL mice with entinostat, trametinib, or the combination of entinostat plus trametinib (“Ent + Tram”; fig. S5A). Lung tumorigenesis was initiated in KL mice (day 0), and tumor growth was monitored with BLI. Thirty-four days after Cre recombinase (Lenti-Cre) administration, mice were randomized into treatment groups and treatment was initiated. Drug was administered by oral gavage throughout a 42-day treatment course, and material collected at end point was used for quantitation of tumor burden. BLI indicated that after 42 days of treatment, the Ent + Tram group exhibited the lowest tumor burden of all treatment groups (Fig. 5A). Hematoxylin and eosin (H&E)–stained lung sections from the treatment mice at end point illustrated that Ent + Tram mice harbored distinctly less tumor burden than all other treatment groups (Fig. 5B). Quantitation of tumor burden from the H&E-stained sections from all mice in the study confirmed that neither entinostat nor trametinib alone affected tumor burden compared to vehicle control, but the Ent + Tram drug combination elicited significantly reduced tumor burden compared to all other treatment groups (Fig. 5C). The Ent + Tram group contained smaller and fewer tumors than other treatment groups (Fig. 5, D and E). These data identified that entinostat and trametinib, which are both clinically viable drugs that do not elicit efficacy as single agent treatments for lung cancer, impart therapeutic efficacy in the KL GEMM model when administered simultaneously.
Fig. 5. Trametinib plus entinostat combination treatment elicits therapeutic efficacy in KL NSCLC GEMM in vivo.
(A) Average longitudinal BLI data. (B) Representative H&E-stained sections at experimental end point. Scale bar, 1000 μm. (C to E) Quantitation from H&E-stained sections: (C) tumor area as a percentage of total lung area per mouse, (D) average tumor size, and (E) tumor number per mouse. (F) Model of HDAC3 cooperation with NKX2-1 to support KL tumor growth basally and in the context of trametinib resistance. Values are expressed as means ± SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001, determined by t test with Welch’s correction.
DISCUSSION
While HDAC inhibitors are FDA-approved in hematopoietic malignancies, their efficacy in solid tumors has been waning, which has been conjectured to be due to limiting toxicity from current agents inhibiting multiple HDAC family members, as well as lack of insights into the optimal clinical contexts where HDAC inhibitors may synergize with other currently approved or in-development therapeutics (54). In spite of years of clinical study, very little genetic analysis of individual class I HDACs in GEMMs has been performed despite extensive data that HDAC1/2 form completely distinct protein complexes with completely distinct downstream targets from HDAC3 or HDAC8. HDAC3 is infrequently directly mutated, nor is its expression frequently altered in cancer, which has contributed to HDAC3 function in cancer being overlooked despite its appreciated importance in normal tissue homeostasis. Notably, however, recent studies have identified HDAC3 as playing a specific role in lymphomas (55–57), rhabdomyosarcoma (58), melanomas (59), and pancreatic cancer (60) settings. Genetic deletion of HDAC3 in normal tissues in vivo has not characterized HDAC3 as a ubiquitous regulator of cell growth or proliferation, instead identifying a diverse range of tissue-specific functions (19). In liver, HDAC3 loss was associated with severe metabolic derangements after 5 months, and subsequently, hepatocellular carcinoma formation was observed 10 months after deletion (61), indicating that HDAC3 is not only not required for growth of all tumors as we observe here in the lung but also, in liver loss of HDAC3, leads to more, not less, tumor burden. However, mutation of the bona fide but poorly characterized HDAC3 protein complex member TBL1XR1 has been elegantly proven to drive immunoblastic lymphoma in vivo by aberrantly redirecting HDAC3 complex function (55). CREBBP, which is frequently mutated in B cell lymphoma, was also reported to drive in vivo tumorigenesis via HDAC3 (56, 57). These recent studies suggest that tumor formation and/or maintenance via misregulation of HDAC3 activity may occur more often than is currently appreciated via multiple upstream mechanisms. In contexts where it supports tumor formation, HDAC3 is a promising molecular target partly because it is therapeutically targetable and it lacks a functionally redundant homolog (unlike HDAC1-HDAC2). Our interest in HDAC3 stemmed from our dissection of key targets of the LKB1 tumor suppressor in Kras-mutant NSCLC, which led us to the hyperactivation of the class IIa HDACs and their common binding partner HDAC3 as potential key mediators of the consequences of LKB1 deficiency (31).
Globally, we found that a large fraction of HDAC3 target genes in NSCLC tumors in vivo are also dependent on the lineage-specific transcription factor NKX2-1. This fits with the many studies of HDAC3 function in various tissues in mice that reveal that HDAC3 controls tissue-specific gene expression across a broad set of tissues examined (muscle, adipose, liver, neurons, and immune cells) (19). NKX2-1 is a member of the NKX subclass of homeobox proteins, which is a large class of transcription factors that share a related DNA binding domain, the homeodomain. Much of the understanding of the NKX subclass of homeodomain proteins, which contain a tyrosine at position 54 not present in other homeodomains, originated from developmental biology studies that identified notable mutant phenotypes. For example, studies of tinman, the Drosophila homolog of murine Nkx2-5, identified that it is a critical regulator of mesodermal cell fate required for cardiac development (62). Cloning of vertebrate Nkx2-1 (TTF-1, Titf1, and T/ebp; vnd in Drosophila) facilitated the identification of its selective expression and activity in thyroid, lung, and restricted areas of the brain (63, 64), and the timing of its expression at the onset of organ development immediately implicated NKX2-1 as a lineage determining factor (64, 65). Subsequent studies confirmed the requirement for NKX2-1 in proper lung and thyroid development, and NKX2-1 dysfunction is associated with human disease in these tissues (41, 66). In LUAD, NKX2-1 is considered a lineage-specific oncogene when overexpressed (in ~80% of cases) or amplified (in up to 15% of cases), and notably, NKX2-1 is the most amplified gene in human LUAD (26, 40–43). However, NKX2-1 expression has also been reported to associate with favorable prognosis in early-stage LUAD (67), and in the Kras and KP lung cancer models, NKX2-1 suppresses tumor growth, enforces a lineage-specific differentiation program (44), and restrains metastatic potential (46). In contrast, in an EGFR-driven NSCLC GEMM model, NKX2-1 inactivation suppressed lung tumorigenesis (68), highlighting the duality of NKX2-1 with respect to both tumor-promoting and tumor-suppressing functions. The molecular determinants that drive these divergent functions remain to be fully identified. In early-stage LUAD, oncogenic KRAS was shown to lead to loss of lineage identity in alveolar epithelial progenitor (AT2) cells associated with reduced NKX2-1 transcriptional output (69). NKX2-1 expression can be affected by multiple upstream transcription factors, and its transcriptional activity can be modulated in a context-dependent manner by posttranslational modification and/or cooperation with additional transcription factors and cofactors (41), much of which remains to be comparatively elucidated between NSCLC subtypes. Our study identifies selective cooperation between NKX2-1 and HDAC3 on a specific set of target genes in LKB1-mutant cells. We do not observe mucinous dedifferentiation upon HDAC3 deletion in KL or KP tumors in vivo, suggesting that HDAC3 inactivation is not equivalent to complete NKX2-1 deletion in vivo (44) but that HDAC3 functions instead as a co-regulator of NKX2-1. We hypothesize that having NKX2-1 present to bind DNA (even in the context of reduced NKX2-1 function) differs from its complete absence after genetic deletion in that complete absence of NKX2-1 may allow other transcription factors to bind DNA in the latter case that may otherwise be occupied by NKX2-1 in the former situation. Our data implicates HDAC3 as a druggable regulator of NKX2-1 in KL lung tumors.
Beyond basal gene expression regulation, we found that the activity of the HDAC3/NKX2-1 complex is induced in KL NSCLC cells as a resistance mechanism to MEK inhibitors, an effect reversed by HDAC3 inhibition. Multiple existing targeted therapies induce clinically well-documented acquired resistance, and efforts to identify molecular resistance mechanisms have become major lines of investigation. Our finding that the LUAD lineage transcription factor NKX2-1 is hyperactivated by trametinib resistance suggests a connection between resistance mechanisms and lineage identity. It is interesting to hypothesize that one possible cellular option for responding to therapy could be to modulate the lineage identity program. In a BRAF-driven NSCLC GEMM model, NKX2-1 was found to modulate response to BRAF/MEK-inhibitor targeted therapy (70). HDAC3 has also been connected to lineage identity and, independently, targeted therapy response. In normal cells, HDAC3 has been reported to regulate lineage specification in T cell and glial cell contexts (48, 49). In rhabdomyosarcoma, a CRISPR screen identified the NCoR/HDAC3 complex as a major suppressor of differentiation (58). It is possible that HDAC3 is a regulator of lineage fate in normal cells more broadly, and an outstanding question is how broadly this mechanism is leveraged to support tumor formation and growth.
Our data have revealed a specific therapeutic context, trametinib resistance, where HDAC3 inhibition may have utility in NSCLC. Four independent groups identified HDAC inhibition as a promising approach for overcoming trametinib resistance in vivo in melanoma and pancreatic cancer (59, 60, 71, 72). In two of the studies, HDAC3 specifically was identified as the critical target eliciting therapeutic response (59, 60). Together, recent evidence suggests that cooperation between trametinib and HDAC inhibition may be emerging as a general principal across different tumor types. Our work extends this phenomenon to NSCLC and brings a previously unidentified mechanism to the fore. HDAC3 inhibition was found to overcome Bcl-2-like protein 11 (BIM) polymorphism–driven endothelial growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) resistance in EGFR-mutant lung cancer (73), suggesting that HDAC3 may also mediate NSCLC targeted therapy resistance beyond trametinib. Together, our findings suggest a model where HDAC3 cooperates with the lineage factor NKX2-1 in LUAD cells (Fig. 5F), and these findings motivate further exploration of the role of HDAC3 in epithelial tumors and resistance to targeted therapies.
METHODS
Cell culture and cell lines
All cell lines were incubated at 37°C and were maintained in an atmosphere containing 5% CO2. Cells were tested for Mycoplasma (Lonza) using the manufacturer’s conditions and were deemed negative. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal bovine serum (FBS; Gibco) and were continuously maintained under antibiotic selection for stable cell lines. Proliferation assays were performed by plating 2 × 103 cells per well of a six-well plate, and cells were counted 5 days after plating. Trametinib was used at 10 nM, and entinostat was used at 1 μM or as indicated. Treatments were for 3 or 13 days. Medium was changed and fresh drug was added every 2 days.
Generating primary tumor cell lines
Cell lines from KL primary tumors are not readily available because of the fact that, unlike KP tumor cells that lack p53, explanted KL primary tumor cells do not grow in culture, presumed to be from p53 activation-dependent growth arrest. To circumvent this issue, we immortalized explanted KL tumor cells before onset of growth arrest. We plucked individual tumors from KL mice, and after dissociation and collagenase treatment, isolated cells were immortalized with SV40 T antigen and subsequently purified by epithelial cellular adhesion molecule (Epcam+) cell sorting to generate the epithelial lung tumor cell line KL LJE1. Specifically, to generate the KL LJE1 cell line, individual primary tumors were dissected from the lungs of KL mice, mechanically dissociated, and then digested for 45 min in digestion medium [10% FBS, penicillin-streptomycin, Collagenase/Dispase (1 mg/ml; Roche) in DMEM] at 37°C. Cells were strained through 70-um nylon cell strainer, spun at 2000 rpm for 5 min, resuspended in 1 ml of complete medium plus 5 μl of Fungizone (Lifetech), and plated in a 24 well dish. Twenty-four hours later, cells were infected by adding 1 ml of T antigen–expressing lentivirus to each well, and twenty-four hours later, viral medium was removed and replaced with complete medium with Fungizone. Cells were cultured in Fungizone for 4 weeks and then Epcam+ sorted.
CRISPR-Cas9 studies
sgRNAs targeting mouse HDAC3 were selected using the optimized CRISPR design tool (http://crispr.mit.edu). The gSR gRNA sequence targeting NKX2-1 was obtained from the work of Sanchez-Rivera et al. (74), and the other gRNA targeting NKX2-1, g2, was designed with the GPP sgRNA designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). gRNAs targeting FGFR1 were designed with the Benchling program (www.benchling.com/crispr/). Guides with high targeting scores and low probability of off-target effects were chosen. At least three independent sgRNA sequences were tested for each gene. Oligonucleotides for sgRNAs were synthesized by IDT, annealed in vitro, and subcloned into Bsm BI–digested lentiCRISPRv.2-puro (Addgene, 52961). Validation of guide specificity was assessed by Western blot. Assays were carried out within six passages of thawing early passage frozen cell stocks. Oligonucleotide sequences are listed in table S1.
Lentiviral production and titering
Lentiviruses made from pLentiCRISPRv.2 were produced by cotransfection of the lentiviral backbone constructs and packaging plasmids pSPAX2 (Addgene, 12260) and pMD2.G (Addgene, 12259). Lipofectamine 2000 (Thermo Fisher Scientific) was used as a transfection reagent at a ratio of 3:1 Lipofectamine/DNA. Viral supernatant was collected from 293 cells 48 hours after transfection, 0.45 μm–filtered, supplemented with polybrene, and applied to destination cells for 24 hours. Destination cells were allowed to recover from infection 24 hours before being subjected to selection with 2 μg/ml. Resulting stably transduced lines were frozen down immediately after selection. Large-scale viral preps of Lenti Pgk-Cre (a gift from T/ Jacks) were made by the University of Iowa Viral Vector Core. For titering, lentiviral preps for mouse experiments (Pgk-Cre) were functionally titered by transduction of a reporter line (293-LSL-GFP), which turns on expression of GFP (green fluorescent protein) upon Cre-mediated recombination and allows quantitation of functional titers derived from the percent of GFP-positive cells.
Mouse studies
All procedures using animals were approved by the Salk Institute Institutional Animal Care and Use Committee. All mice were maintained on the FVB/n background. Kras (KrasLSLG12D/+; R26LSL;luc/luc), KL (KrasLSLG12D/+;Lkb1fl/fl;R26LSL;luc/luc), KP (KrasLSLG12D/+;p53fl/fl; R26LSL;luc/luc), and KPL (KrasLSLG12D/+;Lkb1fl/fl;p53fl/fl; R26LSL;luc/luc) mice in FVB/n have been previously described (29, 36). The Hdac3fl/fl conditional floxed mouse has also been described (23). In this study, Hdac3fl/fl was crossed into the FVB/n K background before crossing into the KL or KP genotypes to generate KL-HDAC3fl/fl and KP-HDAC3fl/fl experimental mice. All experiments used a mixture of female and male mice. Lentivirus expressing Cre recombinase (4 × 105 plaque-forming units per mouse) was delivered by intratracheal intubation to each mouse to initiate lung tumorigenesis, according by the protocol of DuPage (75). Experimental end point was defined across experiments as the time point at which the experimental cohorts of KL or KP mice reached BLI tumor burden of 108 mean photon flux or earlier as indicated. At end point, all mice in that experiment were collected at that point. All animals at experimental end point were included for analysis of lung tumor burden and tumor size analysis. No animals were excluded from longitudinal BLI measurements and graphs.
Bioluminescence imaging
BLI was performed biweekly using IVIS Spectrum (Caliper Life Sciences) using Living Image software (PerkinElmer). Mice were injected intraperitoneally with d-luciferin (150 mg/kg; Caliper Life Sciences), anesthetized with isoflurane, and imaged both ventrally and dorsally 10 min after luciferin injection. The total lung photon flux for each animal is calculated by the combination of ventral and dorsal photon flux calculated within a region of interest encompassing the thorax.
In vivo entinostat and trametinib treatment
Mice were intratracheally intubated with lentivirus expressing Cre recombinase to initiate tumorigenesis and imaged weekly starting 4 weeks after Cre. Treatment was initiated at day 34 after Cre (Fig. 5). In vivo treatment doses were selected on the basis of publications (76–79). Entinostat was diluted to 1 mg/ml in vehicle (0.5% methyl cellulose in water), vortexed, sonicated for 10 min, and administered at 10 mg/kg. Trametinib was diluted to 2 mg/ml in corn oil, vortexed, and sonicated for 20 min, and this stock was stored up to 3 days at 4°C. On the day of dosing, trametinib stock was diluted 1:10 in corn oil to 0.2 mg/ml and administered at 1 mg/kg. Drug vials were kept on a rack on a heat pad during dosing to maintain fluidity of the corn oil. Drug(s) and/or vehicle(s) were administered by sequential oral gavage starting at ~9 a.m. daily throughout the duration of the treatment, each mouse being gavaged twice on each dosing day (once to deliver entinostat or vehicle and once to deliver trametinib or vehicle). On weekly imaging days, mice were given a drug holiday to mitigate any potential toxicity and reduce stress to the animals. Because mice were gavaged twice daily for an extended duration, an additional drug holiday was built into the dosing schedule starting after 3 weeks of treatment: Mice were administered drug using the following schedule: 4 days on, 1 day off, 1 day on, and 1 day off (imaging day). Body weight of mice was recorded every 2 days and did not indicate drug-induced toxicity in any treatment group throughout the treatment experiments. The experiment was terminated when the vehicle treatment group reached disease end point due to high lung tumor burden. Primary tumors were collected from the mouse with the highest tumor burden in each treatment group: Mice were dosed with drug and euthanized 2 hours later, at which point tumors were plucked from lungs and immediately flash-frozen. To collect lungs at treatment end point, mice were euthanized 2 hours after final dose of drug (~9 a.m.), at which point lungs were inflated and formalin-fixed.
Immunohistochemistry and image analysis
Lungs from mice were collected at each experimental end point as noted in the figures, fixed in formalin for 18 to 22 hours, transferred to 70% ethanol, and paraffin-embedded (FFPE) at the Tissue Technology Shared Resources at University of California, San Diego. Sections (5 μm) from FFPE tissues were prepared and stained with H&E. For immunohistochemistry, slides were deparaffinized and rehydrated, and antigen retrieval was performed in citrate buffer for 13 min at high heat (~95°C). Endogenous peroxidase activity was quenched with 10-min hydrogen peroxide in methanol. Using the ImmPRESS HRP Ig (Peroxidase) Polymer Detection Kits (Vector Laboratories), slides were blocked and incubated overnight with primary antibody diluted in blocking buffer, and secondary antibody steps were carried out according to the manufacturer’s instructions. Staining was visualized with ImmPACT DAB peroxidase substrate (Vector Laboratories, SK-4105), further counterstained with hematoxylin, dehydrated through ethanol and xylenes, and mounted with Cytoseal 60 (Thermo Scientific). H&E-stained and immunostained slides were scanned using a PerkinElmer slide scanner (Pannoramic MIDI Digital Side Scanner) for further downstream analysis using the Panoramic Viewer software, inForm v2.1 image analysis software (Cambridge Research and Instrumentation), or QuPath software (80).
Lung tumor burden
Total lung tumor burden was quantitated from H&E sections using inForm v2.1 image analysis software (Cambridge Research and Instrumentation) in a nonbiased manner. Briefly, the Trainable Tissue Segmentation method was trained to identify tumor, normal lung, vessel, and space. This program was then applied to all H&E images, and each of the resulting mapped images was then screened to verify that accurate tissue segmentation had occurred. The quantitation data from this analysis was then used to calculate the percentage of tumor area as normalized to total lung area (tumor area + normal lung area).
Tumor size quantitation
Quantitation of each individual tumor was measured from H&E sections using morphometric analysis in Pannoramic viewer software (PerkinElmer), which calculates the size of each identified tumor by area in squared micrometers. The area of all tumors found in the five lobes of each mouse was exported and compiled to plot the number of tumors per mouse and the average size of every tumor in the cohort.
mRNA preparation and mRNA sequencing
mRNA was collected from cells harvested within two passages after thaw. mRNA was isolated using the Quick-RNA Miniprep Kit (Zymo Research), including deoxyribonuclease treatment. RNA integrity numbers were determined using the Agilent TapeStation before library preparation. mRNA-seq libraries were prepared using the TruSeq RNA library preparation kit (version 2), according to the manufacturer’s instructions (Illumina). Libraries were quantified, pooled, and sequenced by single-end 50 base pairs using the Illumina HiSeq 2500 platform at the Salk Next-Generation Sequencing Core. Raw sequencing data were demultiplexed and converted into FASTQ files using CASAVA (version 1.8.2).
Bioinformatic analysis of RNA-seq data
Sequenced reads were quality-tested using the online FASTQC tool (www.bioinformatics.babraham.ac.uk/projects/fastqc) and aligned to the mouse mm10 genome using the STAR aligner version 2.4.0 k (81). Raw gene expression was quantified across all annotated exons using HOMER (82), and differential gene expression was carried out using the getDiffExpression.pl command. Differentially expressed genes were defined as having a false discovery rate (FDR) of <0.05 and a log2 fold change of >0.5.
GSEA was carried out with the GenePattern interface (https://genepattern.broadinstitute.org) using preranked lists generated from FDR values. Queried datasets used were gene lists from genes differentially expressed upon tamoxifen-driven NKX2-1 KO in Kras tumors (44). Heatmaps were generated by clustering using the Cluster 3.0 program (log2 transform data, center genes, and hierarchical clustering with average linkage) (83) and then visualized with Java TreeView version 1.1.6r4 (84).
ChIP-sequencing
Primary tumors
Individually dissected, flash-frozen primary tumors were combined from three different mice into one pool of 130 mg of primary tumors per replicate per genotype. Equivalent masses of tumors were used from each of the three mice to ensure equal representation. Two independent pools of tumors per genotype were processed separately to generate two biological replicate pool of cross-linked, sonicated chromatin for ChIP. Four independent ChIPs were performed on each pool of sonicated chromatin and then pooled together to generate one replicate for ChIP-seq. To cross-link, tumors were Dounce-homogenized in cross-linking buffer [1% formaldehyde in phosphate-buffered saline (PBS)], incubated with end-over-end rotation for 15 min at room temperature, and then quenched with 2.5 M glycine for 5 min. Samples were spun at 600g for 5 min, washed with cold PBS, and resuspended in ChIP buffer [radioimmunoprecipitation assay (RIPA)] (see the “Immunoprecipitation” section for recipe) with protease inhibitors. Samples were sonicated in a Covaris LE220 for 8 min (duty factor, 2; 105 W; and 200 cycles per burst) and spun down, and the supernatant was saved. For each ChIP, 100 μl of lysate was combined with 900 μl of ChIP buffer, while 50 μl was used for input. Hdac3 ab7030 antibody (10 μg) and H3K27ac ab4729 antibody (2 μg) were used for each ChIP. Lysate was incubated overnight with antibody. Washed and preblocked Dynabeads Protein A (20 μl) were incubated for 2 hours, rotating with each sample at 4°C. Washes were performed with 5-min incubations of each buffer while rotating at 4°C. Samples were washed thrice with cold ChIP buffer, once with room temperature ChIP buffer, and once with room temperature Tris-EDTA (TE) (pH 8) and then spun down. Elution of ChIP and input samples was done by incubating samples with elution buffer [50 mM tris-HCl (pH 7.5), 10 mM EDTA, and 1% SDS] overnight at 65°C. Beads were pelleted and discarded, and 200 μl of eluate was combined with 194 μl of low-EDTA TE and 100 μg of proteinase K and incubated for 2 hours at 37°C. Ribonuclease A (RNase A) (8 μl) was added, and samples were incubated for 30 min at 37°C. The MinElute PCR Purification Kit (QIAGEN, 28006) was used to isolate DNA, which was eluted in 15 μl of elution buffer (EB) at 55°C. Four ChIPs were combined into one sample for ChIP-sequencing.
KL LJE1 cells
ChIP-seq was carried out on DSG [di(N-succinimidyl) glutarate] + formaldehyde cross-linked, sonicated nuclear extracts. Cells were washed in PBS and then cross-linked by 30 min of incubation in 2 mM DSG (Thermo Fisher Scientific, NC0054325). Aspirate and incubate for 15 min with 1% formaldehyde before 5 min of quench with 125 mM glycine. Cells were washed in cold PBS, scraped, spun down, and washed again in PBS before nuclei isolation. Nuclei were isolated by resuspension in CiA NP-Rinse 1 (50 mM Hepes, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100), incubated for 10 min at 4°C with end-over-end rotation, and then centrifuged at 1200g for 5 min at 4°C. Samples were then resuspended in CiA NP-Rinse 2 [10 mM tris (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, and 200 mM NaCl], incubated for 10 min at 4°C with end-over-end rotation, and centrifuged at 1200g for 5 min at 4°C. Tubes were washed twice with Covaris Shearing Buffer [0.1% SDS, 1 mM EDTA (pH 8), and 10 mM tris-HCl (pH 8)] to remove salt and centrifuged at 1200g at 4°C for 3 min. Samples were diluted to a concentration of 2.5 × 106 cells per 130 μl in ChIP buffer [RIPA; 50 mM tris-HCl (pH 7.5), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% NaDOC (sodium deoxycholate), and 0.1% SDS] with protease inhibitors and sonicated in a Covaris LE220 for 8 min (duty factor, 2; 105 W; and 200 cycles per burst). Sonicated material was spun down, and supernatant was used for ChIP. Lysate from 5 million cells was diluted in ChIP buffer to a final volume of 1 ml. Fifty microliters was used for input. Hdac3 CST-85057 antibody (10 μg) was used for each ChIP. Lysate was incubated overnight with antibody. Washed and preblocked Dynabeads Protein A (20 μl) were incubated for 2 hours, rotating with each sample at 4°C. Washes were performed with 5-min incubations of each buffer while rotating at 4°C. Samples were washed thrice with cold ChIP buffer, once with room temperature ChIP buffer, and once with room temperature TE (pH 8) and then spun down. Elution of ChIP and input samples was done by incubating samples with elution buffer [50 mM tris-HCl (pH 7.5), 10 mM EDTA, and 1% SDS] overnight at 65°C. Beads were pelleted and discarded, and 200 μl of eluate was combined with 194 μl of low-EDTA TE and 100 μg of proteinase K and incubated for 2 hours at 37°C. RNase A (8 μl) was added, and samples were incubated for 30 min at 37°C. MinElute PCR Purification Kit (QIAGEN, 28006) was used to isolate DNA, which was eluted in 15 μl of EB at 55°C.
Bioinformatic analysis of ChIP-seq data
Sequenced reads were aligned to the mouse mm10 genome using the STAR aligner version 2.4.0k (81). HOMER (82) was used for data processing. For KL LJE1 cell line ChIP-seq data, peaks were called using the getDifferentialPeaksReplicates.pl command using HDAC3 ChIP-seq data from NT cells as target (-t), HDAC3 ChIP-seq data from HDAC3 KO cells as background (-b), and input sequencing data from NT cells as input (-i), with -style factor and -F 3. For primary tumor ChIP-seq data, peaks were called for each replicate individually using the findPeaks command with parameters -style factor -F 3 using HDAC3 ChIP-seq as target and input sequencing data as input (-i). Peaks were merged using the mergePeaks command to generate a consolidated file containing all HDAC3 ChIP-seq peaks identified in KL and KP tumors. The getDifferentialPeaks command with -F 3 -same was used to identify peaks bound in both KL and KP tumors. The annotatePeaks.pl command with the -ghist -hist 25 option was used to visualize binding at each peak independently across samples, and Java TreeView was used to visualize the output. The annotatePeaks.pl command with -hist 25 was used to plot average reads across all peaks relative to peak center for each replicate separately. BedGraph files were also generated and visualized with Integrative Genomics Viewer version 2.5.1.
Western blots
Protein lysates in CST buffer [20 mM tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, 50 mM NaF, 1 mM EDTA, 1 mM EGTA, 2.5 mM pyrophosphate, 2 mM β-glycerol-phosphate, 1 mM orthovanadate, and 0.01 mM calyculin A] with protease inhibitors were equilibrated for protein levels using a BCA protein assay kit (Pierce), resolved on 8% SDS–polyacrylamide gel electrophoresis gels, and transferred to membranes. Membranes were blocked in milk, incubated overnight at 4°C in diluted primary antibody, washed with TBS-T, incubated for 1 hour in secondary antibody diluted in in TBS-T plus milk, washed in TBS-T, and developed using SuperSignal ECL. Secondary antibodies were anti-rabbit (Millipore, AP132P) and anti-mouse (Millipore, AP124P).
Immunoprecipitation
Immunoprecipitation was carried out on dithiobis(succinimidyl propionate) (DSP)–cross-linked, sonicated nuclear lysates. Cells were washed in PBS and then cross-linked by 30-min incubation in 1 mM DSP (Thermo Scientific, 22585), followed by 5 min of quench with 2.5 M glycine. Cells were washed in PBS, scraped, spun down, and washed again in PBS before nuclei isolation. Nuclei were isolated by resuspension in CiA NP-Rinse 1 (50 mM Hepes, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100), incubated for 10 min at 4°C with end-over-end rotation, and then centrifuged at 1200g for 5 min at 4°C. Samples were then resuspended in CiA NP-Rinse 2 [10 mM tris (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, and 200 mM NaCl], incubated for 10 min at 4°C with end-over-end rotation, and centrifuged at 1200g for 5 min at 4°C. Tubes were washed twice with Covaris Shearing Buffer [0.1% SDS, 1 mM EDTA (pH 8), and 10 mM tris-HCl (pH 8)] to remove salt, and centrifuged at 1200g at 4°C for 3 min. Samples were diluted to a concentration of 2.5 × 106 cells per 130 μl in ChIP buffer (RIPA) [50 mM tris-HCl (pH 7.5), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% NaDOC, and 0.1% SDS] with protease inhibitors and sonicated in a Covaris LE220 for 8 min (duty factor, 2; 105 W; and 200 cycles per burst). Sonicated material was spun down, and supernatant was used for immunoprecipitation: Material (400 μl) was incubated with 3 μl of NKX2-1 antibody (Abcam, ab76013) per immunoprecipitation overnight with rotation at 4°C. Prewashed Protein A Dynabeads (20 μl) were added per tube and incubated for 4 hours at 4°C with rotation. Samples were washed five times with CST buffer (see the “Western blots” section) before adding 25 μl of 6× loading dye and 50 μl of CST per tube and eluting by boiling for 5 min. Input and immunoprecipitation samples were subsequently assessed by Western blot.
TCGA analysis of Firehose LUAD dataset
The results shown are in whole based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/. TCGA datasets were queried using cBioPortal (www.cbioportal.org) (85, 86). Methods for data generation, normalization, and bioinformatics analyses were previously described in the TCGA LUAD publication (Cancer Genome Atlas Research 2014). mRNA data used for this analysis was RNA Seq V2 RSEM with z score thresholds of 1.8.
Homer motif enrichment analysis
Homer motif enrichment analysis can be found at http://homer.ucsd.edu/homer/motif/.
Antibodies and reagents
Western blotting
Antibodies from Cell Signaling Technology (Danvers, MA, USA) were diluted 1:1000 (Hdac3, CST-85057; Fgfr1, CST-9740; and ERK, CST-4695) or 1:2000 (phospho-ERK Thr202/Tyr204, CST-4370). Nkx2-1 was from Abcam (ab76013) and was diluted 1:1500. Anti-actin (#A5441) from Sigma-Aldrich was diluted 1:10,000.
Chromatin immunoprecipitation
Hdac3 from Abcam (ab7030) was used on primary tumors, and Hdac3 from Cell Signaling Technology (CST-85057) was used on KL LJE1 cells.
Immunoprecipitation
Nkx2-1 raised in rabbit from (Abcam, ab76013) was used to immunoprecipitate, and HDAC3 raised in mouse (CST-3949) was used to detect coimmunoprecipitated HDAC3.
Drugs
Entinostat (MS-275) was obtained from Selleck Chemicals (S1053). Trametinib was obtained from LC Laboratories (T-8123).
Statistical analyses
Statistical analyses are described in each figure and were all performed using GraphPad Prism 9. Results are expressed as means ± SEM unless otherwise indicated.
Acknowledgments
We thank D. Hargreaves and J. Remsberg for support with ChIP-seq assay development, G. Liang for advice on in vivo entinostat dosing, and S. Lowe for advice on in vivo trametinib dosing. We also thank J. Van Nostrand for constructive criticism of the manuscript.
Funding: This study was supported by grants to R.J.S. from the NIH (R35CA220538 and P01CA120964) and The Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002. L.J.E. was supported by a postdoctoral fellowship from the American Cancer Society (PF-15-037-01-DMC), and research reported in this publication was supported by the National Cancer Institute (NCI) of the NIH under award number K22CA251636. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. S.N.B was supported by NCI training grant 5T32CA009370 to the Salk Institute Cancer Center and NCI 5F32CA206400. This work was supported by the NGS and the Razavi Newman Integrative Genomics and Bioinformatics Core Facilities of the Salk Institute with funding from the NIH-NCI CCSG: P30 014195, the Chapman Foundation, and the Helmsley Charitable Trust. Tissue Technology Shared Resource is supported by an NCI Cancer Center Support Grant (CCSG P30CA23100).
Author contributions: L.J.E designed all experiments, performed all experiments except as noted here, and wrote the paper. S.D.C., S.N.B, and J.T.B. assisted with cloning and KO cell line generation and validation. S.N.B. and J.T.B. assisted with in vivo drug dosing studies. S.D.C. assisted with qRT-PCR. D.S.R. assisted with animal husbandry. T.J.R. assisted with genotyping, BLI imaging, and in vivo drug dosing. E.T. assisted with in vivo drug dosing. C.K.M. performed cell culture experiments, Western blot analysis, and qRT-PCR for fig. S4 (A to H). I.G. generated cell lines used for fig. S4 (D and E). L.J.E. and R.J.S. conceived the project, and R.J.S. supervised the project and wrote and edited the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or Supplementary Materials. RNA-seq and ChIP-seq data have been deposited to the Gene Expression Omnibus (GEO) data repository with accession number GSE164759. Previously published sequencing data that were reanalyzed here is available under the GEO accession code GSE36473. Source data are provided for all experiments. Correspondence and requests for materials should be addressed to R.J.S. or L.J.E.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Other Supplementary Material for this : manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.J. E. Bradner, D. Hnisz, R. A. Young, Transcriptional addiction in cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.T. De Raedt, E. Beert, E. Pasmant, A. Luscan, H. Brems, N. Ortonne, K. Helin, J. L. Hornick, V. Mautner, H. Kehrer-Sawatzki, W. Clapp, J. Bradner, M. Vidaud, M. Upadhyaya, E. Legius, K. Cichowski, PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514, 247–251 (2014). [DOI] [PubMed] [Google Scholar]
- 3.J. Shi, C. R. Vakoc, The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.N. Tasdemir, A. Banito, J. S. Roe, D. Alonso-Curbelo, M. Camiolo, D. F. Tschaharganeh, C. H. Huang, O. Aksoy, J. E. Bolden, C. C. Chen, M. Fennell, V. Thapar, A. Chicas, C. R. Vakoc, S. W. Lowe, BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 6, 612–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.T. Shimamura, Z. Chen, M. Soucheray, J. Carretero, E. Kikuchi, J. H. Tchaicha, Y. Gao, K. A. Cheng, T. J. Cohoon, J. Qi, E. Akbay, A. C. Kimmelman, A. L. Kung, J. E. Bradner, K. K. Wong, Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin. Cancer Res. 19, 6183–6192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S. L. Guerra, O. Maertens, R. Kuzmickas, T. De Raedt, R. O. Adeyemi, C. J. Guild, S. Guillemette, A. J. Redig, E. S. Chambers, M. Xu, H. Tiv, S. Santagata, P. A. Jänne, S. J. Elledge, K. Cichowski, A deregulated HOX gene axis confers an epigenetic vulnerability in KRAS-mutant lung cancers. Cancer Cell 37, 705–719.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. W. Johnstone, J. D. Licht, Histone deacetylase inhibitors in cancer therapy. Cancer Cell 4, 13–18 (2003). [DOI] [PubMed] [Google Scholar]
- 8.A. C. West, R. W. Johnstone, New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124, 30–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.E. Ceccacci, S. Minucci, Inhibition of histone deacetylases in cancer therapy: Lessons from leukaemia. Br. J. Cancer 114, 605–611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Z. Lu, J. Zou, S. Li, M. J. Topper, Y. Tao, H. Zhang, X. Jiao, W. Xie, X. Kong, M. Vaz, H. Li, Y. Cai, L. Xia, P. Huang, K. Rodgers, B. Lee, J. B. Riemer, C. P. Day, R. W. C. Yen, Y. Cui, Y. Wang, Y. Wang, W. Zhang, H. Easwaran, A. Hulbert, K. B. Kim, R. A. Juergens, S. C. Yang, R. J. Battafarano, E. L. Bush, S. R. Broderick, S. M. Cattaneo, J. R. Brahmer, C. M. Rudin, J. Wrangle, Y. Mei, Y. J. Kim, B. Zhang, K. K. H. Wang, P. M. Forde, J. B. Margolick, B. D. Nelkin, C. A. Zahnow, D. M. Pardoll, F. Housseau, S. B. Baylin, L. Shen, M. V. Brock, Epigenetic therapy inhibits metastases by disrupting premetastatic niches. Nature 579, 284–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.M. J. Topper, M. Vaz, K. B. Chiappinelli, C. E. De Stefano Shields, N. Niknafs, R.-W. C. Yen, A. Wenzel, J. Hicks, M. Ballew, M. Stone, P. T. Tran, C. A. Zahnow, M. D. Hellmann, V. Anagnostou, P. L. Strissel, R. Strick, V. E. Velculescu, S. B. Baylin, Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284–1300.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D. O. Adeegbe, Y. Liu, P. H. Lizotte, Y. Kamihara, A. R. Aref, C. Almonte, R. Dries, Y. Li, S. Liu, X. Wang, T. Warner-Hatten, J. Castrillon, G. C. Yuan, N. Poudel-Neupane, H. Zhang, J. L. Guerriero, S. Han, M. M. Awad, D. A. Barbie, J. Ritz, S. S. Jones, P. S. Hammerman, J. Bradner, S. N. Quayle, K.-K. Wong, Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov. 7, 852–867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.C. F. Malone, C. Emerson, R. Ingraham, W. Barbosa, S. Guerra, H. Yoon, L. L. Liu, F. Michor, M. Haigis, K. F. Macleod, O. Maertens, K. Cichowski, mTOR and HDAC inhibitors converge on the TXNIP/thioredoxin pathway to cause catastrophic oxidative stress and regression of RAS-driven tumors. Cancer Discov. 7, 1450–1463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. Morel, D. Jeffery, S. Aspeslagh, G. Almouzni, S. Postel-Vinay, Combining epigenetic drugs with other therapies for solid tumours—Past lessons and future promise. Nat. Rev. Clin. Oncol. 17, 91–107 (2020). [DOI] [PubMed] [Google Scholar]
- 15.L. Ny, H. Jespersen, J. Karlsson, S. Alsén, S. Filges, C. All-Eriksson, B. Andersson, A. Carneiro, H. Helgadottir, M. Levin, I. Ljuslinder, R. Olofsson Bagge, V. R. Sah, U. Stierner, A. Ståhlberg, G. Ullenhag, L. M. Nilsson, J. A. Nilsson, The PEMDAC phase 2 study of pembrolizumab and entinostat in patients with metastatic uveal melanoma. Nat. Commun. 12, 5155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. S. Truong, M. Zhou, B. Krishnan, T. Utsumi, U. Manocha, K. G. Stewart, W. Beck, T. L. Rose, M. I. Milowsky, X. He, C. C. Smith, L. M. Bixby, C. M. Perou, S. E. Wobker, S. T. Bailey, B. G. Vincent, W. Y. Kim, Entinostat induces antitumor immune responses through immune editing of tumor neoantigens. J. Clin. Invest. 131, 138560 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.B. J. Christmas, C. I. Rafie, A. C. Hopkins, B. A. Scott, H. S. Ma, K. A. Cruz, S. Woolman, T. D. Armstrong, R. M. Connolly, N. A. Azad, E. M. Jaffee, E. T. Roussos Torres, Entinostat converts immune-resistant breast and pancreatic cancers into checkpoint-responsive tumors by reprogramming tumor-infiltrating MDSCs. Cancer Immunol. Res. 6, 1561–1577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. Orillion, A. Hashimoto, N. Damayanti, L. Shen, R. Adelaiye-Ogala, S. Arisa, S. Chintala, P. Ordentlich, C. Kao, B. Elzey, D. Gabrilovich, R. Pili, Entinostat neutralizes myeloid-derived suppressor cells and enhances the antitumor effect of PD-1 inhibition in murine models of lung and renal cell carcinoma. Clin. Cancer Res. 23, 5187–5201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. J. Emmett, M. A. Lazar, Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell Biol. 20, 102–115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. J. Emmett, H. W. Lim, J. Jager, H. J. Richter, M. Adlanmerini, L. C. Peed, E. R. Briggs, D. J. Steger, T. Ma, C. A. Sims, J. A. Baur, L. Pei, K. J. Won, P. Seale, Z. Gerhart-Hines, M. A. Lazar, Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 546, 544–548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Z. Sun, R. A. Miller, R. T. Patel, J. Chen, R. Dhir, H. Wang, D. Zhang, M. J. Graham, T. G. Unterman, G. I. Shulman, C. Sztalryd, M. J. Bennett, R. S. Ahima, M. J. Birnbaum, M. A. Lazar, Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 18, 934–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Z. Sun, D. Feng, B. Fang, S. E. Mullican, S. H. You, H. W. Lim, L. J. Everett, C. S. Nabel, Y. Li, V. Selvakumaran, K. J. Won, M. A. Lazar, Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell 52, 769–782 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. K. Knutson, B. J. Chyla, J. M. Amann, S. Bhaskara, S. S. Huppert, S. W. Hiebert, Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 27, 1017–1028 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.L. Ding, G. Getz, D. A. Wheeler, E. R. Mardis, M. D. McLellan, K. Cibulskis, C. Sougnez, H. Greulich, D. M. Muzny, M. B. Morgan, L. Fulton, R. S. Fulton, Q. Zhang, M. C. Wendl, M. S. Lawrence, D. E. Larson, K. Chen, D. J. Dooling, A. Sabo, A. C. Hawes, H. Shen, S. N. Jhangiani, L. R. Lewis, O. Hall, Y. Zhu, T. Mathew, Y. Ren, J. Yao, S. E. Scherer, K. Clerc, G. A. Metcalf, B. Ng, A. Milosavljevic, M. L. Gonzalez-Garay, J. R. Osborne, R. Meyer, X. Shi, Y. Tang, D. C. Koboldt, L. Lin, R. Abbott, T. L. Miner, C. Pohl, G. Fewell, C. Haipek, H. Schmidt, B. H. Dunford-Shore, A. Kraja, S. D. Crosby, C. S. Sawyer, T. Vickery, S. Sander, J. Robinson, W. Winckler, J. Baldwin, L. R. Chirieac, A. Dutt, T. Fennell, M. Hanna, B. E. Johnson, R. C. Onofrio, R. K. Thomas, G. Tonon, B. A. Weir, X. Zhao, L. Ziaugra, M. C. Zody, T. Giordano, M. B. Orringer, J. A. Roth, M. R. Spitz, I. I. Wistuba, B. Ozenberger, P. J. Good, A. C. Chang, D. G. Beer, M. A. Watson, M. Ladanyi, S. Broderick, A. Yoshizawa, W. D. Travis, W. Pao, M. A. Province, G. M. Weinstock, H. E. Varmus, S. B. Gabriel, E. S. Lander, R. A. Gibbs, M. Meyerson, R. K. Wilson, Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. Sanchez-Cespedes, P. Parrella, M. Esteller, S. Nomoto, B. Trink, J. M. Engles, W. H. Westra, J. G. Herman, D. Sidransky, Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 62, 3659–3662 (2002). [PubMed] [Google Scholar]
- 26.J. D. Campbell, A. Alexandrov, J. Kim, J. Wala, A. H. Berger, C. S. Pedamallu, S. A. Shukla, G. Guo, A. N. Brooks, B. A. Murray, M. Imielinski, X. Hu, S. Ling, R. Akbani, M. Rosenberg, C. Cibulskis, A. Ramachandran, E. A. Collisson, D. J. Kwiatkowski, M. S. Lawrence, J. N. Weinstein, R. G. W. Verhaak, C. J. Wu, P. S. Hammerman, A. D. Cherniack, G. Getz; Cancer Genome Atlas Research Network, M. N. Artyomov, R. Schreiber, R. Govindan, M. Meyerson, Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48, 607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.F. Skoulidis, J. V. Heymach, Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 19, 495–509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. M. Lizcano, O. Göransson, R. Toth, M. Deak, N. A. Morrice, J. Boudeau, S. A. Hawley, L. Udd, T. P. Mäkelä, D. G. Hardie, D. R. Alessi, LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.L. J. Eichner, S. N. Brun, S. Herzig, N. P. Young, S. D. Curtis, D. B. Shackelford, M. N. Shokhirev, M. Leblanc, L. I. Vera, A. Hutchins, D. S. Ross, R. J. Shaw, R. U. Svensson, Genetic analysis reveals AMPK is required to support tumor growth in murine kras-dependent lung cancer models. Cell Metab. 29, 285–302.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. W. Murray, J. J. Brady, M. K. Tsai, C. Li, I. P. Winters, R. Tang, L. Andrejka, R. K. Ma, C. A. Kunder, P. Chu, M. M. Winslow, An LKB1-SIK axis suppresses lung tumor growth and controls differentiation. Cancer Discov. 9, 1590–1605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.P. E. Hollstein, L. J. Eichner, S. N. Brun, A. Kamireddy, R. U. Svensson, L. I. Vera, D. S. Ross, T. J. Rymoff, A. Hutchins, H. M. Galvez, A. E. Williams, M. N. Shokhirev, R. A. Screaton, R. Berdeaux, R. J. Shaw, The AMPK-related kinases SIK1 and SIK3 mediate key tumor-suppressive effects of LKB1 in NSCLC. Cancer Discov. 9, 1606–1627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.N. J. Darling, P. Cohen, Nuts and bolts of the salt-inducible kinases (SIKs). Biochem. J. 478, 1377–1397 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Martin, R. Kettmann, F. Dequiedt, Class IIa histone deacetylases: Regulating the regulators. Oncogene 26, 5450–5467 (2007). [DOI] [PubMed] [Google Scholar]
- 34.M. M. Mihaylova, R. J. Shaw, Metabolic reprogramming by class I and II histone deacetylases. Trends Endocrinol. Metab. 24, 48–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R. U. Svensson, S. J. Parker, L. J. Eichner, M. J. Kolar, M. Wallace, S. N. Brun, P. S. Lombardo, J. L. van Nostrand, A. Hutchins, L. Vera, L. Gerken, J. Greenwood, S. Bhat, G. Harriman, W. F. Westlin, H. J. Harwood Jr., A. Saghatelian, R. Kapeller, C. M. Metallo, R. J. Shaw, Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D. B. Shackelford, E. Abt, L. Gerken, D. S. Vasquez, A. Seki, M. Leblanc, L. Wei, M. C. Fishbein, J. Czernin, P. S. Mischel, R. J. Shaw, LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 23, 143–158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. M. Kaufman, J. M. Amann, K. Park, R. R. Arasada, H. Li, Y. Shyr, D. P. Carbone, LKB1 loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. J. Thorac. Oncol. 9, 794–804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.L. Chen, B. E. Engel, E. A. Welsh, S. J. Yoder, S. G. Brantley, D. T. Chen, A. A. Beg, C. Cao, F. J. Kaye, E. B. Haura, M. B. Schabath, W. D. Cress, A sensitive nanostring-based assay to score STK11 (LKB1) pathway disruption in lung adenocarcinoma. J. Thorac. Oncol. 11, 838–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. G. Guenther, O. Barak, M. A. Lazar, The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21, 6091–6101 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. Tanaka, K. Yanagisawa, K. Shinjo, A. Taguchi, K. Maeno, S. Tomida, Y. Shimada, H. Osada, T. Kosaka, H. Matsubara, T. Mitsudomi, Y. Sekido, M. Tanimoto, Y. Yatabe, T. Takahashi, Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 67, 6007–6011 (2007). [DOI] [PubMed] [Google Scholar]
- 41.T. Yamaguchi, Y. Hosono, K. Yanagisawa, T. Takahashi, NKX2-1/TTF-1: An enigmatic oncogene that functions as a double-edged sword for cancer cell survival and progression. Cancer Cell 23, 718–723 (2013). [DOI] [PubMed] [Google Scholar]
- 42.H. Watanabe, J. M. Francis, M. S. Woo, B. Etemad, W. Lin, D. F. Fries, S. Peng, E. L. Snyder, P. R. Tata, F. Izzo, A. C. Schinzel, J. Cho, P. S. Hammerman, R. G. Verhaak, W. C. Hahn, J. Rajagopal, T. Jacks, M. Meyerson, Integrated cistromic and expression analysis of amplified NKX2-1 in lung adenocarcinoma identifies LMO3 as a functional transcriptional target. Genes Dev. 27, 197–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.K. A. Kwei, Y. H. Kim, L. Girard, J. Kao, M. Pacyna-Gengelbach, K. Salari, J. Lee, Y. L. Choi, M. Sato, P. Wang, T. Hernandez-Boussard, A. F. Gazdar, I. Petersen, J. D. Minna, J. R. Pollack, Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 27, 3635–3640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. L. Snyder, H. Watanabe, M. Magendantz, S. Hoersch, T. A. Chen, D. G. Wang, D. Crowley, C. A. Whittaker, M. Meyerson, S. Kimura, T. Jacks, Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol. Cell 50, 185–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.G. Mollaoglu, A. Jones, S. J. Wait, A. Mukhopadhyay, S. Jeong, R. Arya, S. A. Camolotto, T. L. Mosbruger, C. J. Stubben, C. J. Conley, A. Bhutkar, J. M. Vahrenkamp, K. C. Berrett, M. H. Cessna, T. E. Lane, B. L. Witt, M. E. Salama, J. Gertz, K. B. Jones, E. L. Snyder, T. G. Oliver, The lineage-defining transcription factors SOX2 and NKX2-1 determine lung cancer cell fate and shape the tumor immune microenvironment. Immunity 49, 764–779.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. M. Winslow, T. L. Dayton, R. G. W. Verhaak, C. Kim-Kiselak, E. L. Snyder, D. M. Feldser, D. D. Hubbard, M. J. DuPage, C. A. Whittaker, S. Hoersch, S. Yoon, D. Crowley, R. T. Bronson, D. Y. Chiang, M. Meyerson, T. Jacks, Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.L. A. Garraway, W. R. Sellers, Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer 6, 593–602 (2006). [DOI] [PubMed] [Google Scholar]
- 48.R. L. Philips, J. H. Lee, K. Gaonkar, P. Chanana, J. Y. Chung, S. R. Romero Arocha, A. Schwab, T. Ordog, V. S. Shapiro, HDAC3 restrains CD8-lineage genes to maintain a bi-potential state in CD4+CD8+ thymocytes for CD4-lineage commitment. eLife 8, e43821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L. Zhang, X. He, L. Liu, M. Jiang, C. Zhao, H. Wang, D. He, T. Zheng, X. Zhou, A. Hassan, Z. Ma, M. Xin, Z. Sun, M. A. Lazar, S. A. Goldman, E. N. Olson, Q. R. Lu, Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev. Cell 36, 316–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.V. Gocheva, A. Naba, A. Bhutkar, T. Guardia, K. M. Miller, C. M. Li, T. L. Dayton, F. J. Sanchez-Rivera, C. Kim-Kiselak, N. Jailkhani, M. M. Winslow, A. del Rosario, R. O. Hynes, T. Jacks, Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc. Natl. Acad. Sci. U.S.A. 114, E5625–E5634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. Dai, Z. Zhou, Z. Chen, G. Xu, Y. Chen, Fibroblast growth factor receptors (FGFRs): Structures and small molecule inhibitors. Cell 8, 614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.I. S. Babina, N. C. Turner, Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 17, 318–332 (2017). [DOI] [PubMed] [Google Scholar]
- 53.E. Manchado, S. Weissmueller, J. P. Morris, C. C. Chen, R. Wullenkord, A. Lujambio, E. de Stanchina, J. T. Poirier, J. F. Gainor, R. B. Corcoran, J. A. Engelman, C. M. Rudin, N. Rosen, S. W. Lowe, A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534, 647–651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.K. J. Falkenberg, R. W. Johnstone, Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 13, 673–691 (2014). [DOI] [PubMed] [Google Scholar]
- 55.L. Venturutti, M. Teater, A. Zhai, A. Chadburn, L. Babiker, D. Kim, W. Béguelin, T. C. Lee, Y. Kim, C. R. Chin, W. T. Yewdell, B. Raught, J. M. Phillip, Y. Jiang, L. M. Staudt, M. R. Green, J. Chaudhuri, O. Elemento, P. Farinha, A. P. Weng, M. D. Nissen, C. Steidl, R. D. Morin, D. W. Scott, G. G. Privé, A. M. Melnick, TBL1XR1 mutations drive extranodal lymphoma by inducing a pro-tumorigenic memory fate. Cell 182, 297–316.e27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Jiang, A. Ortega-Molina, H. Geng, H. Y. Ying, K. Hatzi, S. Parsa, D. McNally, L. Wang, A. S. Doane, X. Agirre, M. Teater, C. Meydan, Z. Li, D. Poloway, S. Wang, D. Ennishi, D. W. Scott, K. R. Stengel, J. E. Kranz, E. Holson, S. Sharma, J. W. Young, C. S. Chu, R. G. Roeder, R. Shaknovich, S. W. Hiebert, R. D. Gascoyne, W. Tam, O. Elemento, H. G. Wendel, A. M. Melnick, CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 7, 38–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.P. Mondello, S. Tadros, M. Teater, L. Fontan, A. Y. Chang, N. Jain, H. Yang, S. Singh, H. Y. Ying, C. S. Chu, M. C. J. Ma, E. Toska, S. Alig, M. Durant, E. de Stanchina, S. Ghosh, A. Mottok, L. Nastoupil, S. S. Neelapu, O. Weigert, G. Inghirami, J. Baselga, A. Younes, C. Yee, A. Dogan, D. A. Scheinberg, R. G. Roeder, A. M. Melnick, M. R. Green, Selective inhibition of HDAC3 targets synthetic vulnerabilities and activates immune surveillance in lymphoma. Cancer Discov. 10, 440–459 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. P. Phelps, J. N. Bailey, T. Vleeshouwer-Neumann, E. Y. Chen, CRISPR screen identifies the NCOR/HDAC3 complex as a major suppressor of differentiation in rhabdomyosarcoma. Proc. Natl. Acad. Sci. U.S.A. 113, 15090–15095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O. Maertens, R. Kuzmickas, H. E. Manchester, C. E. Emerson, A. G. Gavin, C. J. Guild, T. C. Wong, T. de Raedt, C. Bowman-Colin, E. Hatchi, L. A. Garraway, K. T. Flaherty, S. Pathania, S. J. Elledge, K. Cichowski, MAPK pathway suppression unmasks latent DNA repair defects and confers a chemical synthetic vulnerability in BRAF-, NRAS-, and NF1-mutant melanomas. Cancer Discov. 9, 526–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Z. Wang, S. Hausmann, R. Lyu, T.-M. Li, S. M. Lofgren, N. M. Flores, M. E. Fuentes, M. Caporicci, Z. Yang, M. J. Meiners, M. A. Cheek, S. A. Howard, L. Zhang, J. E. Elias, M. P. Kim, A. Maitra, H. Wang, M. C. Bassik, M.-C. Keogh, J. Sage, O. Gozani, P. K. Mazur, SETD5-coordinated chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. Cancer Cell 37, 834–849.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.S. Bhaskara, S. K. Knutson, G. Jiang, M. B. Chandrasekharan, A. J. Wilson, S. Zheng, A. Yenamandra, K. Locke, J. L. Yuan, A. R. Bonine-Summers, C. E. Wells, J. F. Kaiser, M. K. Washington, Z. Zhao, F. F. Wagner, Z. W. Sun, F. Xia, E. B. Holson, D. Khabele, S. W. Hiebert, Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 18, 436–447 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R. P. Harvey, NK-2 homeobox genes and heart development. Dev. Biol. 178, 203–216 (1996). [DOI] [PubMed] [Google Scholar]
- 63.S. Guazzi, M. Price, M. de Felice, G. Damante, M. G. Mattei, R. di Lauro, Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 9, 3631–3639 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D. Lazzaro, M. Price, M. de Felice, R. Di Lauro, The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development 113, 1093–1104 (1991). [DOI] [PubMed] [Google Scholar]
- 65.M. N. Stanfel, K. A. Moses, R. J. Schwartz, W. E. Zimmer, Regulation of organ development by the NKX-homeodomain factors: An NKX code. Cell. Mol. Biol. (Noisy-le-grand) (suppl. 51), OL785–OL799 (2005). [PubMed] [Google Scholar]
- 66.L. P. Fernandez, A. Lopez-Marquez, P. Santisteban, Thyroid transcription factors in development, differentiation and disease. Nat. Rev. Endocrinol. 11, 29–42 (2015). [DOI] [PubMed] [Google Scholar]
- 67.V. K. Anagnostou, K. N. Syrigos, G. Bepler, R. J. Homer, D. L. Rimm, Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J. Clin. Oncol. 27, 271–278 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Y. Maeda, T. Tsuchiya, H. Hao, D. H. Tompkins, Y. Xu, M. L. Mucenski, L. du, A. R. Keiser, T. Fukazawa, Y. Naomoto, T. Nagayasu, J. A. Whitsett, KrasG12D and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J. Clin. Invest. 122, 4388–4400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. F. M. Dost, A. L. Moye, M. Vedaie, L. M. Tran, E. Fung, D. Heinze, C. Villacorta-Martin, J. Huang, R. Hekman, J. H. Kwan, B. C. Blum, S. M. Louie, S. P. Rowbotham, J. S. de Aja, M. E. Piper, P. J. Bhetariya, R. T. Bronson, A. Emili, G. Mostoslavsky, G. A. Fishbein, W. D. Wallace, K. Krysan, S. M. Dubinett, J. Yanagawa, D. N. Kotton, C. F. Kim, Organoids model transcriptional hallmarks of oncogenic KRAS activation in lung epithelial progenitor cells. Cell. Stem. Cell. 27, 663–678 e668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R. Zewdu, E. M. Mehrabad, K. Ingram, P. Fang, K. L. Gillis, S. A. Camolotto, G. Orstad, A. Jones, M. C. Mendoza, B. T. Spike, E. L. Snyder, An NKX2-1/ERK/WNT feedback loop modulates gastric identity and response to targeted therapy in lung adenocarcinoma. eLife 10, e66788 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.L. Wang, R. L. de Oliveira, S. Huijberts, E. Bosdriesz, N. Pencheva, D. Brunen, A. Bosma, J.-Y. Song, J. Zevenhoven, G. T. L.-d. Vries, H. Horlings, B. Nuijen, J. H. Beijnen, J. H. M. Schellens, R. Bernards, An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell 173, 1413–1425.e14 (2018). [DOI] [PubMed] [Google Scholar]
- 72.F. Faiao-Flores, M. F. Emmons, M. A. Durante, F. Kinose, B. Saha, B. Fang, J. M. Koomen, S. P. Chellappan, S. S. Maria-Engler, U. Rix, J. D. Licht, J. W. Harbour, K. S. M. Smalley, HDAC inhibition enhances the in vivo efficacy of MEK inhibitor therapy in uveal melanoma. Clin. Cancer Res. 25, 5686–5701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. Tanimoto, S. Takeuchi, S. Arai, K. Fukuda, T. Yamada, X. Roca, S. T. Ong, S. Yano, Histone deacetylase 3 inhibition overcomes BIM deletion polymorphism-mediated osimertinib resistance in egfr-mutant lung cancer. Clin. Cancer Res. 23, 3139–3149 (2017). [DOI] [PubMed] [Google Scholar]
- 74.F. J. Sanchez-Rivera, T. Papagiannakopoulos, R. Romero, T. Tammela, M. R. Bauer, A. Bhutkar, N. S. Joshi, L. Subbaraj, R. T. Bronson, W. Xue, T. Jacks, Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.M. DuPage, A. L. Dooley, T. Jacks, Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.J. Abraham, Y. Nuñez-Álvarez, S. Hettmer, E. Carrió, H. I. H. Chen, K. Nishijo, E. T. Huang, S. I. Prajapati, R. L. Walker, S. Davis, J. Rebeles, H. Wiebush, A. T. McCleish, S. T. Hampton, C. R. R. Bjornson, A. S. Brack, A. J. Wagers, T. A. Rando, M. R. Capecchi, F. C. Marini, B. R. Ehler, L. A. Zarzabal, M. W. Goros, J. E. Michalek, P. S. Meltzer, D. M. Langenau, R. D. LeGallo, A. Mansoor, Y. Chen, M. Suelves, B. P. Rubin, C. Keller, Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes Dev. 28, 1578–1591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.L. Shen, M. Ciesielski, S. Ramakrishnan, K. M. Miles, L. Ellis, P. Sotomayor, P. Shrikant, R. Fenstermaker, R. Pili, Class I histone deacetylase inhibitor entinostat suppresses regulatory T cells and enhances immunotherapies in renal and prostate cancer models. PLOS ONE 7, e30815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.R. T. Kurmasheva, A. Bandyopadhyay, E. Favours, V. D. Pozo, S. Ghilu, D. A. Phelps, S. W. Erickson, C. J. Peer, W. D. Figg, M. A. Smith, P. J. Houghton, Evaluation of entinostat alone and in combination with standard-of-care cytotoxic agents against rhabdomyosarcoma xenograft models. Pediatr. Blood Cancer 66, e27820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. Ruscetti, J. Leibold, M. J. Bott, M. Fennell, A. Kulick, N. R. Salgado, C. C. Chen, Y. J. Ho, F. J. Sanchez-Rivera, J. Feucht, T. Baslan, S. Tian, H. A. Chen, P. B. Romesser, J. T. Poirier, C. M. Rudin, E. de Stanchina, E. Manchado, C. J. Sherr, S. W. Lowe, NK cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 362, 1416–1422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.P. Bankhead, M. B. Loughrey, J. A. Fernández, Y. Dombrowski, D. G. McArt, P. D. Dunne, S. McQuaid, R. T. Gray, L. J. Murray, H. G. Coleman, J. A. James, M. Salto-Tellez, P. W. Hamilton, QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.S. Heinz, C. Benner, N. Spann, E. Bertolino, Y. C. Lin, P. Laslo, J. X. Cheng, C. Murre, H. Singh, C. K. Glass, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.M. J. L. de Hoon, S. Imoto, J. Nolan, S. Miyano, Open source clustering software. Bioinformatics 20, 1453–1454 (2004). [DOI] [PubMed] [Google Scholar]
- 84.A. J. Saldanha, Java Treeview—Extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- 85.E. Cerami, J. Gao, U. Dogrusoz, B. E. Gross, S. O. Sumer, B. A. Aksoy, A. Jacobsen, C. J. Byrne, M. L. Heuer, E. Larsson, Y. Antipin, B. Reva, A. P. Goldberg, C. Sander, N. Schultz, The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.J. Gao, B. A. Aksoy, U. Dogrusoz, G. Dresdner, B. Gross, S. O. Sumer, Y. Sun, A. Jacobsen, R. Sinha, E. Larsson, E. Cerami, C. Sander, N. Schultz, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Table S1