SUMMARY
The PI3K inhibitor alpelisib is clinically approved for the treatment of metastatic ER+ breast cancers harboring hotspot mutations in PIK3CA, which encodes a subunit of PI3K. Prospective clinical trial results demonstrated benefit from alpelisib for the treatment of advanced ER+ breast cancers harboring PIK3CA mutations in the hotspots of exons 7, 9, and 20. However, 20% of PIK3CA mutations occur in non-hotspot regions. A recent article demonstrated that patients with cancers bearing non-hotspot PIK3CA mutations also derived benefit from alpelisib, which will inform clinical decision-making moving forward.
In this issue of Clinical Cancer Research, Rugo and colleagues evaluated the efficacy of alpelisib for advanced ER+ breast cancer with PIK3CA mutations as detected using comprehensive genomic profiling (CGP) (1). PIK3CA encodes the p110α catalytic subunit of phosphatidylinositol 3-kinase (PI3K), and PIK3CA mutations are commonly found in ER+ breast and other cancers (2). Given the incidence of PIK3CA mutations and the understanding of its role in promoting cancer cell growth and survival, efforts were undertaken to develop targeted therapeutics that inhibit PI3K.
Early clinical attempts to inhibit p110α tested agents with little isoform selectivity (e.g., buparlisib, pictilisib), covering p110α, p110β, p110γ, and p110δ. These agents were poorly tolerated, which led to their discontinued development (3) and the pursuit of isoform-selective agents. Alpelisib is the first-in-class p110α-selective PI3K inhibitor approved for the treatment of advanced ER+ breast cancer in combination with endocrine therapy. The SOLAR-1 Phase III trial demonstrated increased efficacy from the combination of alpelisib with endocrine therapy vs. placebo with endocrine therapy alone in treating patients with advanced ER+ breast cancer bearing PI3KCA hotspot mutations.
While missense PIK3CA mutations occur in ~35% of advanced ER+ breast cancers, ~80% of such mutations are localized to hotspot regions in exons 7, 9, and 20 (1). These hotspot mutations are well-characterized as enzymatically activating and oncogenic (4), and were evaluated in the SOLAR-1 trial that led to the FDA approval of alpelisib. However, ~20% of PIK3CA mutations occur in non-hotspot regions that are less characterized (Figure 1), leaving clinicians with little information with which to inform the decision of whether to prescribe alpelisib for non-hotspot-bearing patients. Rugo et al. conducted a retrospective analysis of 10,750 patients with CGP of all protein-coding regions of PIK3CA. This revealed increased progression-free survival (PFS) with the combination of alpelisib/fulvestrant vs. placebo/fulvestrant (4.0 vs. 2.5 months) among non-hotspot-bearing patients. However, patients with any PIK3CA mutation had even longer PFS with the drug combination (5.9 months), indicating that hotspot PIK3CA-mutant tumors may more responsive to alpelisib than non-hotspot-mutant tumors. Another outcome of this study validates the efficacy of alpelisib + fulvestrant in patients previously treated with a CDK4/6 inhibitor, which has become a preferred first-line treatment for ER+ breast cancer. These findings are in agreement with those of the BYLieve trial demonstrating alpelisib efficacy in a post-CDK4/6 inhibitor setting (5).
Fig. 1-. Distribution of non-hotspot PIK3CA mutations in patients with advanced ER+ breast cancer.
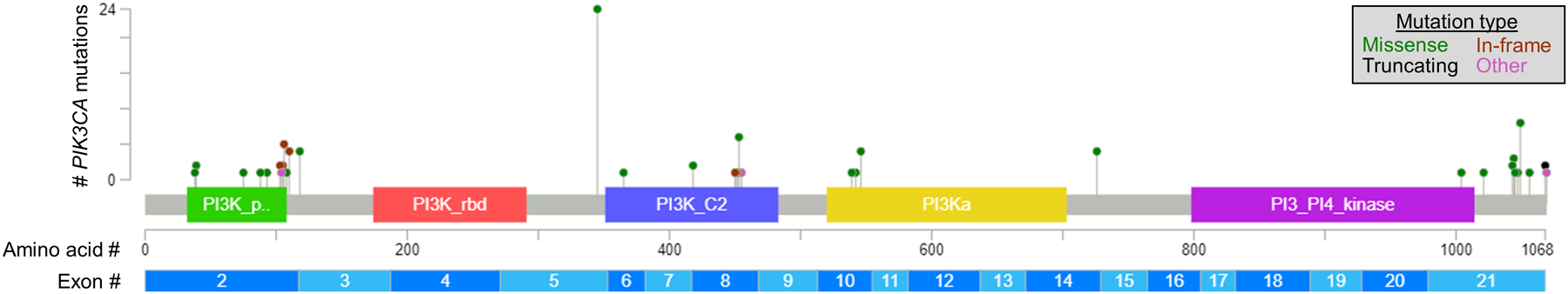
Among 77 patients found to have non-hotspot mutations in PIK3CA without co-occurring hotspot mutations, 93 non-hotspot mutations were detected among 17 plasma and 60 tumor specimens. Data were extracted from Fig. 5 in ref. (1).
At present, there are 3 FDA-approved companion diagnostic tests for PIK3CA mutations. The therascreen PIK3CA RGQ PCR Kit (QIAGEN GmbH) is a real-time PCR assay to qualitatively detect 11 defined mutations within the hotspot regions of exons 7, 9, and 20. The SOLAR-1 trial used a PCR-based profiling assay before transitioning to the therascreen kit. While PCR-based assays only detect a limited number of pre-specified mutations, CGP can comprehensively identify mutations (and copy number alterations and fusions) in a large panel of genes; however, CGP is more expensive and requires more intensive sample processing and data analysis. The FoundationOne CDx and Liquid CDx tests (Foundation Medicine) are CGP assays that cover the protein-coding regions of hundreds of genes including PIK3CA using DNA/RNA acquired from tumor tissue or plasma, respectively. While FoundationOne tests report all PIK3CA mutations detected in a specimen, only the same 11 hotspot mutations are flagged as having an “FDA-approved therapeutic option” (i.e., alpelisib). Thus, FoundationOne tests are the only FDA-approved assays that detect non-hotspot PIK3CA mutations. However, many cancer centers have established in-house CGP pipelines analogous to the FoundationOne tests, enabling routine and complete profiling of PIK3CA. Since the study by Rugo et al. warrants reconsideration of using hotspot vs. any missense mutations in PIK3CA as an indicator for alpelisib, a diagnostic test that provides complete coverage of the PIK3CA coding region must become widely available, and this may be best achieved using CGP.
In the field of precision oncology, open questions remain regarding the reliability of using (A) plasma DNA to reflect genetic profiles of tumors, and (B) genetic profiles of archived tumor specimens (vs. newly acquired biopsies) to inform treatment decisions. Rugo et al. used CGP of patient-matched but asynchronously collected tumor and plasma specimens. Based on the PIK3CA status of tumor specimens, 75% of plasma specimens showed the same mutations; the 25% discordance was attributable in part to lower allelic frequencies of mutations in some tumors (1). A meta-analysis revealed that longer time periods between sampling of tumor and plasma contribute to discordance in PIK3CA status in breast cancer patients (6), suggesting that genetic profiling of plasma DNA may be more reflective of current tumor PIK3CA status. However, the frequencies of missense PIK3CA mutations in primary and metastatic ER+/HER2- breast tumors are similar [51% vs. 48%] (7–9). Furthermore, genetic profiling of patient-matched primary and metastatic tumors from the AURORA trial showed that PIK3CA mutations were nearly completely concordant (10), suggesting that such mutations are early events in cancer development. These data collectively suggest that primary tumor tissue is generally suitable for PIK3CA mutational profiling in patients with metastatic ER+ breast cancer, and plasma DNA profiling should be considered as a secondary option if tumor DNA profiling is not feasible.
Funding acknowledgment:
The authors are supported by the National Cancer Institute (R01CA200994, R01CA262232, and R01CA267691).
Footnotes
Conflict of interest statement: All authors declare that they have no conflict of interest to disclose regarding the information presented in this article.
REFERENCES CITED:
- 1.Rugo HS, Raskina K, Schrock AB, Madison RW, Graf RP, Sokol ES, et al. Biology and targetability of the extended spectrum of PIK3CA mutations (PIK3CAm) detected in breast carcinoma. Clinical Cancer Research. 2022;CCR-22-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–5. [DOI] [PubMed] [Google Scholar]
- 3.Armaghani AJ, Han HS. Alpelisib in the Treatment of Breast Cancer: A Short Review on the Emerging Clinical Data. Breast Cancer (Dove Med Press). 2020;12:251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, DeLong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. Elsevier; 2005;7:561–73. [DOI] [PubMed] [Google Scholar]
- 5.Rugo HS, Lerebours F, Ciruelos E, Drullinsky P, Ruiz-Borrego M, Neven P, et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): one cohort of a phase 2, multicentre, open-label, non-comparative study. The Lancet Oncology. 2021;22:489–98. [DOI] [PubMed] [Google Scholar]
- 6.Galvano A, Castellana L, Gristina V, La Mantia M, Insalaco L, Barraco N, et al. The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis. Ther Adv Med Oncol. 2022;14:17588359221110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Jiang B, Guo J, Shao H, Del Priore IS, Chang Q, et al. INK4 Tumor Suppressor Proteins Mediate Resistance to CDK4/6 Kinase Inhibitors. Cancer Discov. 2022;12:356–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aftimos P, Oliveira M, Irrthum A, Fumagalli D, Sotiriou C, Gal-Yam EN, et al. Genomic and Transcriptomic Analyses of Breast Cancer Primaries and Matched Metastases in AURORA, the Breast International Group (BIG) Molecular Screening Initiative. Cancer Discov. 2021;11:2796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]


