Abstract
3D printing has become one of the primary fabrication strategies used in biomedical research. Recent efforts have focused on the 3D printing of hydrogels to create structures that better replicate the mechanical properties of biological tissues. These pose a unique challenge, as soft materials are difficult to pattern in three dimensions with high fidelity. Currently, a small number of biologically derived polymers that form hydrogels are frequently reused for 3D printing applications. Thus, there exists a need for novel hydrogels with desirable biological properties that can be used as 3D printable inks. In this work, we establish the printability of Multidomain Peptides (MDPs), a class of self-assembling peptides that form a nanofibrous hydrogel at low concentrations. MDPs with different charge functionalities are optimized as distinct inks and are used to create complex three-dimensional structures, including multi-MDP prints. Additionally, printed MDP constructs are used to demonstrate charge-dependent differences in cellular behavior in vitro. This work presents the first time that self-assembling peptides have been used to print layered structures with overhangs and internal porosity. Overall, MDPs are a promising new class of 3D printable inks that are uniquely peptide-based and rely solely on supramolecular mechanisms for assembly.
Keywords: 3D printing, self-assembling peptides, hydrogels, supramolecular chemistry, biomaterials
Graphical Abstract
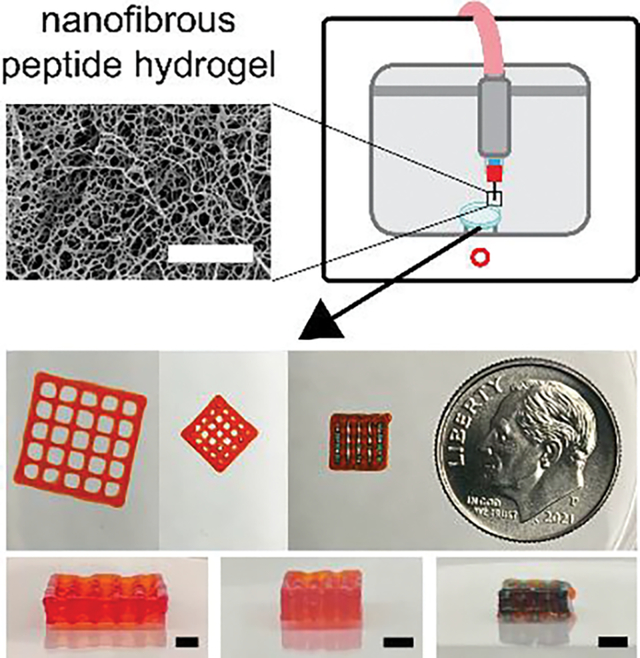
In this work, Multidomain Peptides, a class of self-assembling peptides, are used to 3D print complex, three dimensional structures. Multiple MDPs are demonstrated as printable and oppositely charged MDPs are printed together into layered, porous constructs. MDPs are found to support high cell viability regardless of charge, while charge is shown to influence cellular behavior and morphology.
1. Introduction
Three-dimensional (3D) printing of hydrogels allows researchers to replicate the geometrical complexity that naturally exists in biological structures using soft, tissue-like materials. Advancements in both extrusion[1] and light-based[2] 3D printing have led to the creation of structures that mimic the heart and lungs. Although multiple 3D printing methods exist, extrusion-based printing has been most widely adopted due to its simplistic working principle and ease of use.[3,4] Inks used in extrusion 3D printing mainly consist of chemically modified versions of gelatin,[5–12] alginate,[13–16] hyaluronic acid,[17–20] collagen,[1,21,22] decellularized extracellular matrix,[23–25] or combinations of these.[26–35] The traditional approach for developing 3D printable inks has been to chemically modify naturally-derived hydrogel materials to make them more printable, while attempting to maintain their favorable biological properties. Recently, two generalizable strategies have been proposed to allow for the printing of a wide range of hydrogel materials, which both include a stabilization method to allow for short term print fidelity, followed by a crosslinking method to impart long-term stability.[36,37] Despite these advances, naturally derived materials suffer from batch to batch variability, frequently require chemical modification to have sufficient mechanical properties, and have predefined bioactivity.[38]
A more favorable approach for the creation of new 3D printable inks is to design synthetic materials with specified mechanical, biological, and chemical properties. One class of materials that offers this design freedom is self-assembling peptides (SAPs).[39–42] SAPs are peptides that assemble into nanostructures due to the orchestration of well-designed supramolecular forces. These nanostructures then interact in such a way to generate a macroscopic hydrogel. SAPs are easy to synthesize, chemically well-defined and purifiable, and offer design flexibility to achieve a wide range of material properties. In addition, their synthesis is inherently modular, and the properties of SAPs can be simply altered by changing their primary sequence or through the incorporation of bioactive peptide sequences. Because SAPs consist solely of amino acid building blocks, their chemistry and degradation products are biologically friendly, which is a common drawback of hydrogels made from other polymers. Thus, SAPs offer the flexibility of synthetic hydrogel materials, while also maintaining the biocompatibility of naturally derived ones.
SAPs are an attractive soft material for extrusion 3D printing because they commonly have shear thinning and rapidly self-healing properties due to their noncovalent assembly mechanism.[4,43] Despite this, there has been limited 3D printing work involving SAPs. The Hauser lab was the first to demonstrate the 3D printing of SAPs, using tetrapeptides and PBS in a coaxial printing system to create centimeter sized constructs and encapsulate cells.[44,45] Although this work presented multiple breakthroughs, limitations included the use of noncanonical amino acids and limited print fidelity and complexity. The other major work showing the extrusion 3D printing of SAPs, from the Stupp lab, demonstrated the shear alignment of nanofibers using a peptide amphiphile ink.[46] The authors invented a fabrication method that included extrusion printing followed by spraying a salt gelling agent at each layer. Although this novel process was demonstrated to create anisotropic, supramolecular hydrogels, further development of the fabrication process is needed to allow for the creation of overhanging structures or other complex 3D designs, which were not demonstrated. There have been a few other attempts at printing SAP inks, but these works either used SAPs as part of a composite ink or lacked thorough 3D printing characterization.[47–49]
In this work, we present the 3D printing of Multidomain Peptides (MDPs). MDPs are a class of SAP that are designed to form a nanofibrous hydrogel at physiological pH and ionic strength.[50] MDPs have the general structure of alternating hydrophobic and hydrophilic amino acids flanked by charged residues (Figure 1a). Shielding of the charged amino acids by counterions leads to the creation of a “hydrophobic sandwich,” β-sheet fibrilization, and at appropriate concentration, rapid gelation into a nanofibrous hydrogel.[50–53] MDPs have been shown to promote vascularization, innervation, and high levels of cell infiltration when implanted in vivo,[54] and have been employed for nerve regeneration,[55,56] cancer treatment,[57–59] and vaccine delivery.[60] MDPs of different charge and chemical functionality have been found to provoke varying immune responses in vivo, so the MDP used for a given application will vary based on desired use.[53] To expand the capabilities of MDPs, we present here the 3D printing assessment, optimization, and in vitro characterization of anionic and cationic MDPs (Figure 1b).
Figure 1.
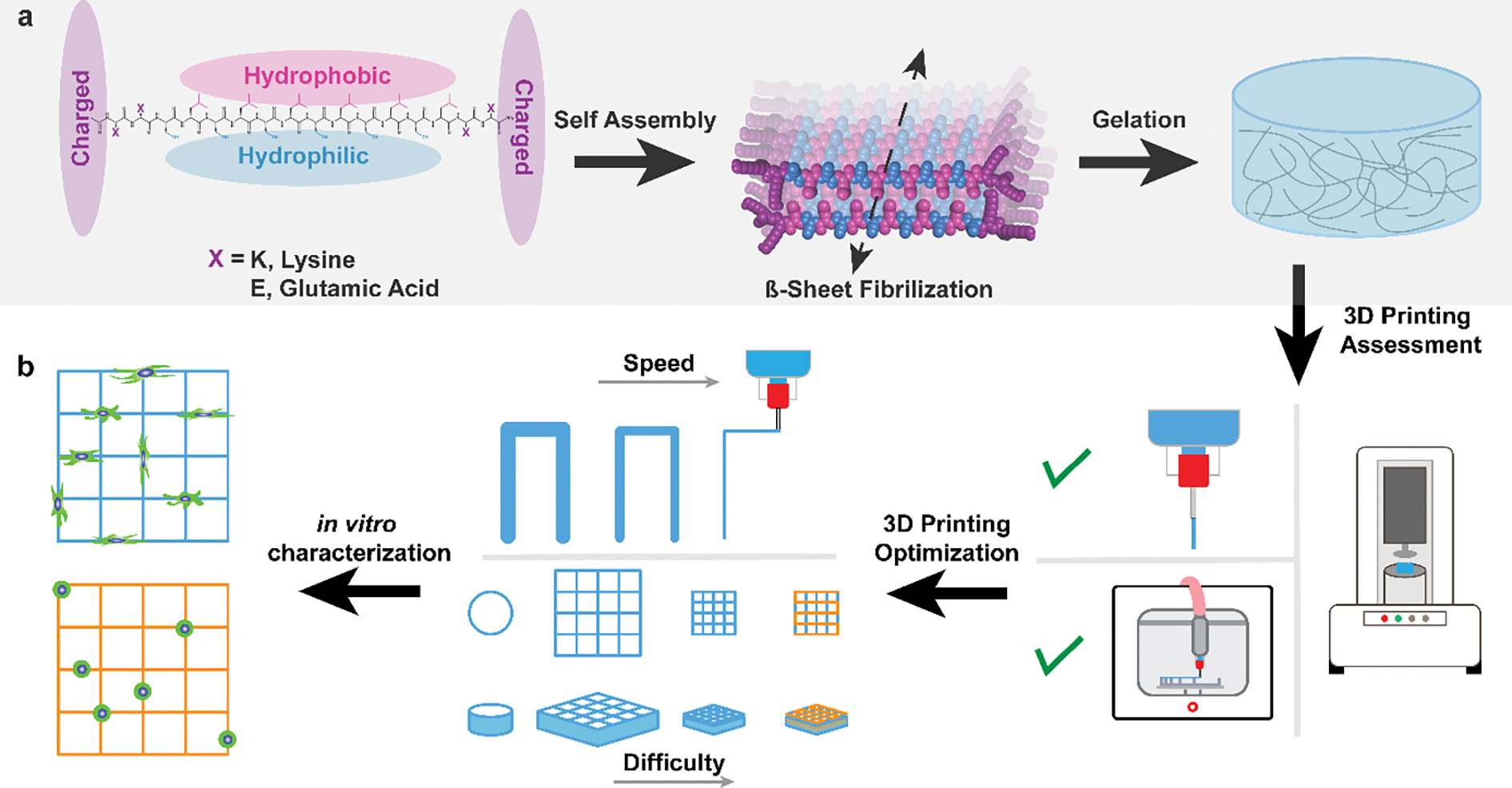
MDP assembly and 3D printing schematic. a) Structure of MDPs with one hydrophilic face (blue), one hydrophobic face (pink), and charged domains at either side (purple). Under physiological conditions MDPs self-assemble and undergo β-sheet fibrilization. This results in the formation of MDP nanofibers and gelation. b) The process undertaken in this study, which includes the assessment of MDPs as a 3D printable ink candidate, 3D printing optimization with multiple MDP inks, and the printing of constructs with increasing difficulty (including multimaterial printing). Finally, MDP inks with opposite charge were used to create 3D structures and observe differing in vitro characteristics.
2. Results and Discussion
2.1. Multidomain Peptide Characterization
The cationic MDP used in this work, named K2, has the sequence K2(SL)6K2 whereas the anionic MDP, named E2, has the sequence E2(SL)6E2 (Figure S1, Supporting Information).[51–53] Both K2 and E2 are N-terminally acetylated and C-terminally amidated. They were synthesized via Solid Phase Peptide Synthesis and confirmed with matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (Figure S2, Supporting Information). The peptides were dissolved in Hank’s Balanced Salt Solution (HBSS) at 1 wt% to analyze the secondary structure and macroscopic assembly. HBSS was used as the solvent throughout the study to create a hydrogel that contains the necessary osmotic pressure, salt content, glucose concentration, and pH to optimize cellular survival in vitro. These peptides have previously been characterized at lower concentrations of peptide and ionic strength, so re-characterization was performed to ensure that changing these variables, which could be necessary for 3D printing, did not alter the secondary structure of the MDPs. Circular dichroism (CD) on K2 and E2 showed a characteristic β-sheet secondary structure, with a maximum around 198 nm and minimum around 218 nm (Figure 2a). K2 forms a slightly stronger β-sheet as indicated by the higher maximum and lower minimum, which can be attributed to differences between the amino group on Lysine and the carboxyl group on Glutamic Acid. Attenuated Total Reflectance Fourier Transform Infrared (ATR-FTIR) Spectroscopy on K2 and E2 reveal a peak at around 1620 cm−1 and 1695 cm−1, which correspond to an amide I peak and the formation of an antiparallel β-sheet (Figure 2b).
Figure 2.
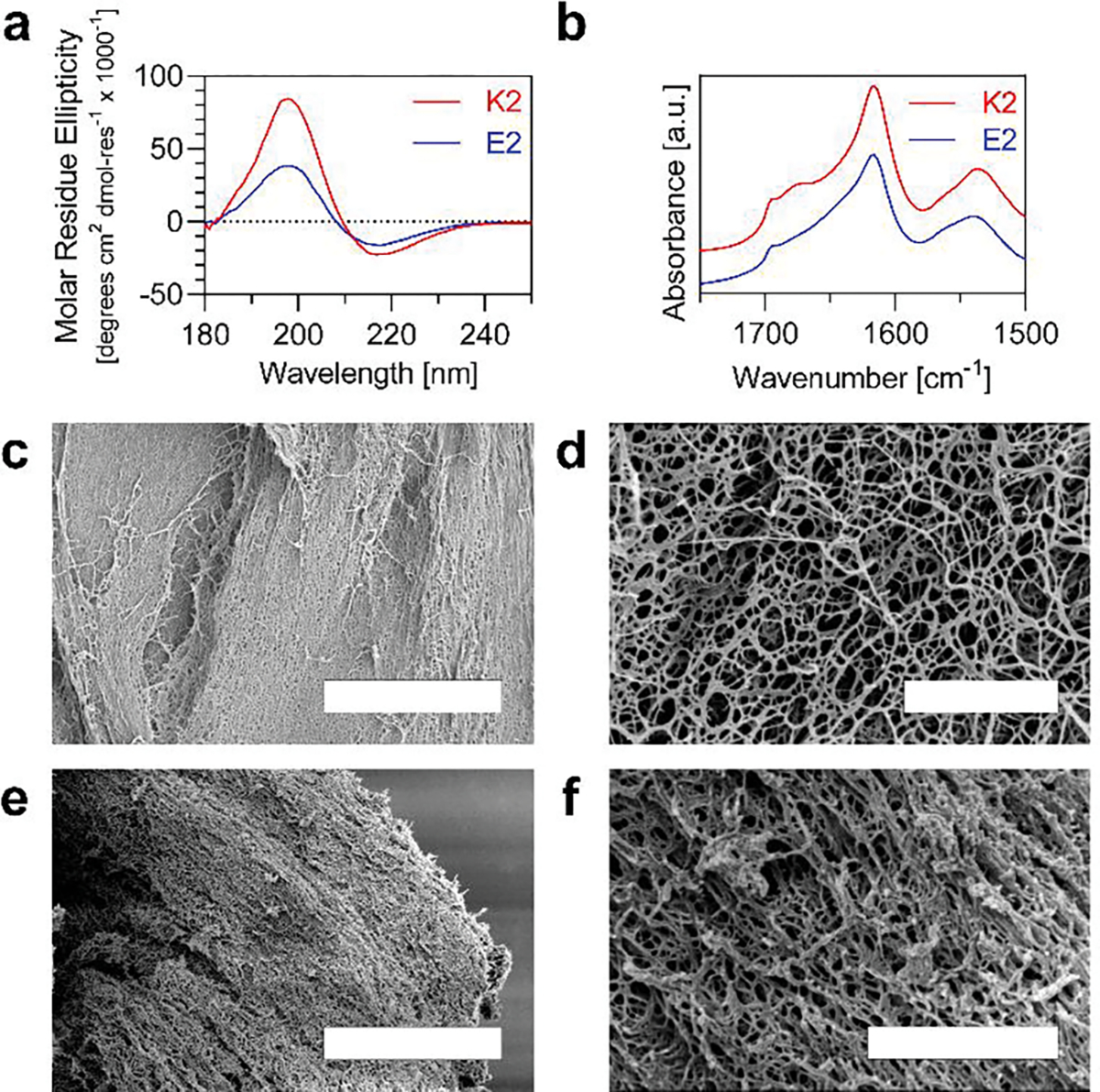
Characterization of MDP secondary Structure and nanofiber network. a) Circular Dichroism of K2 and E2 from 180 – 250 nm. b) Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy of K2 and E2 from 1500 – 1750 cm−1. c-f) Scanning Electron Microscopy of K2 (c,d) and E2 (e,f) nanofilaments that formed a hydrogel. (Scale bars = 5 μm in c, e and 1 μm in d, f).
Scanning electron microscopy was used to observe the fibrous network that forms in both K2 and E2 hydrogels (Figure 2c – f). At low magnification, an expansive, dense network of fibers are seen in both MDPs (Figure 2c, e), whereas at higher magnification, MDP nanofibers are visualized creating a web-like structure (Figure 2d, f).
Rheological testing was performed on both K2 and E2 to assess their potential use as extrudable inks. 3D printing has been shown using inks with a range of storage moduli, so a frequency sweep from 0.1 to 100 rad/s at 1% strain was performed on 1 – 4 wt% K2 and E2 to understand how increasing peptide concentration affects the resulting gel storage modulus (G′) (Figure 3a, e). 4 wt% was chosen as the upper peptide concentration limit because dissolving MDPs at higher concentrations was difficult to accomplish. At 1 rad/s, 1 – 4% K2 has G′ values of 396, 1400, 3000, and 4580 Pa, respectively. In contrast, 1 – 4% E2 has G′ values of 172, 283, 583, and 1040 Pa, respectively. Thus, K2 gels have storage moduli that are on average 4.2X greater than E2 gels at equal concentrations, although both MDPs demonstrate around a 2X increase in storage modulus for each additional wt% (2.4X for K2 and 1.8X for E2).
Figure 3.
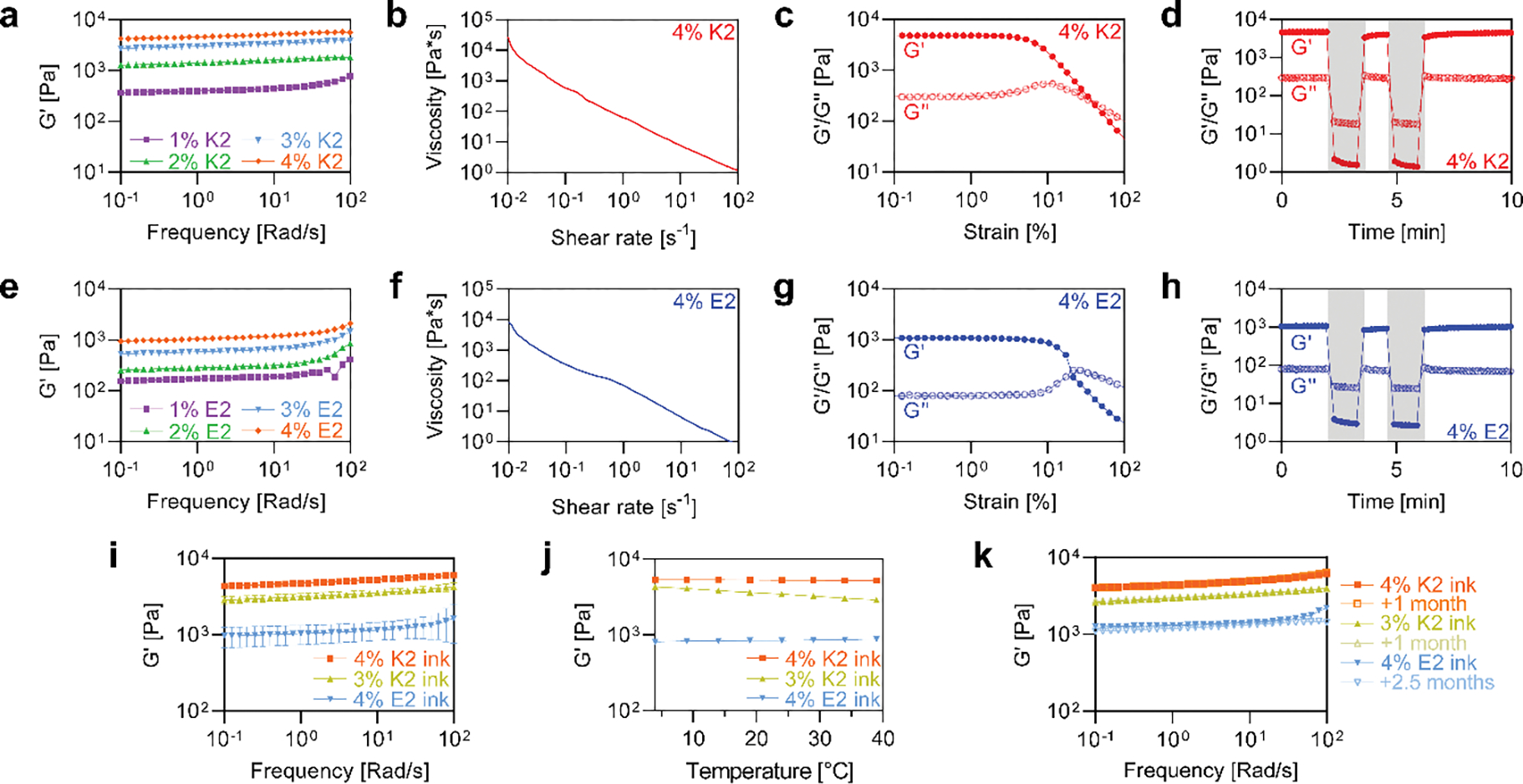
Rheology of MDP gels and MDP inks. Frequency sweeps from 0.1 – 100 rad/s on 1 – 4 wt% a) K2 and e) E2. Shear sweeps from 0.01 – 100 s−1 on 4 wt% b) K2 and f) E2. Strain sweeps from 0.1 – 100% on 4 wt% c) K2 and g) E2. Oscillatory high and low strains on 4 wt% d) K2 and h) E2. White regions represent 1% strains and grey regions represent 500% strain. i) Frequency sweeps from 0.1 – 100 rad/s on MDP inks (n=3). j) Temperature sweeps from 4 – 37°C on MDP inks. k) Frequency sweeps from 0.1 – 100 rad/s on MDP inks after storage at 4°C for >1 month.
4 wt% K2 and E2, which had the highest G′ values, were subjected to additional rheological testing to ensure they were shear thinning, shear yielding, and rapidly self-healing, which are essential properties for a 3D printable ink.[61,62] A shear sweep from 0.01 – 100 s−1 was performed on 4 wt% K2 and E2 (Figure 3b, f). At shear rates of 0.1, 1, and 10 s−1, K2 has viscosity values of 577, 63, and 7.7 Pa*s. At the same shear rates, E2 has viscosity values of 336, 70, and 6.4 Pa*s. Importantly, both 4 wt% MDPs had a negative slope, which means that they are both shear thinning.
A strain sweep from 0.1 – 100% strain at 1 rad/s was also performed on both 4 wt% MDPs (Figure 3c, g). Throughout the linear viscoelastic region, the ratio between the storage and loss modulus for both K2 and E2 were very similar (16 and 14 at 1% strain, respectively). In contrast, the storage modulus of K2 (4810 Pa) was 4.5X that of E2 (1080 Pa) at 1% strain, which corroborates the results of the frequency sweeps. Both K2 and E2 exhibit shear yielding behavior at high strains, although the storage/loss modulus crossover occurs at around 40% strain for K2, compared to 20% strain for E2.
To assess the self-recovery of K2 and E2, a series of low (1%) and high (500%) strains were applied to each gel as the storage and loss moduli were measured (Figure 3d, h). Both MDPs exhibited an inversion of the moduli during high strain, indicating liquid-like behavior, followed by rapid recovery during low strain. K2 and E2 both exhibited a recovery of storage modulus to 86% of the pre-strain value within 1 minute of low shear conditions. This rapid recovery of the storage modulus following liquification is necessary, but not sufficient, for fiber formation during 3D printing.
2.2. Multidomain Peptide Ink Printing Optimization
MDP inks were created by dissolving peptides in dye supplemented HBSS. Before making large batches of ink (>1 mL), preliminary extrusion testing was performed to see what the minimum storage modulus required to form a self-supporting filament when extruded from a needle. It was empirically concluded that only MDP inks with storage moduli greater than 1 KPa were able to form robust fibers (Figure S3, Supporting Information). Based on this and the previous rheological findings, larger batches of MDP inks were made at 4% K2, 4% E2, and 3% K2.
Unlike most materials used as 3D printing inks, MDPs rely solely on supramolecular forces for assembly and do not require covalent crosslinking for stabilization. This also means that MDP nanofibers exist in a dynamic state. Thus, batch-to-batch differences were screened along with the effect of temperature and time on rheological properties. Frequency sweeps on 3 replicates of each ink showed reproducible rheological properties that matched closely to initial rheological testing (Figure 3i). In addition, the MDP inks showed stable G′ values between 4 – 37°C, with the two 4% inks averaging just a ~5% change over the temperature sweep (Figure 3j). Thus, MDP inks avoid the need for temperature control during 3D printing, which is a necessary complication for many 3D printing inks that have temperature-dependent properties. In addition, the introduction of cells in vitro or implantation of printed constructs in vivo will undoubtedly necessitate gel stability at 37°C, which is demonstrated here for MDP inks. The MDP inks also showed long term stability when stored at 4°C (Figure 3k). Frequency sweeps on the different inks after being stored at 4°C for up to 2.5 months revealed a negligible change in their rheological properties. In contrast to a time sensitive ink, MDP inks offer much more flexibility and the ability for on-demand use, such as what might be necessary in a clinical setting. In total, these tests revealed a high level of stability for the supramolecular MDP inks.
Fiber formation tests were performed on 4% K2 and 4% E2 inks paired with 25G, 27G, and 30G needles to determine the finest needle that could be used to form uniform fibers for 3D printing (Figure S3, Supporting Information). 4% K2 inks formed uniform fibers when extruded through 25G, 27G, and 30G needles. 4% E2 inks only formed uniform fibers when extruded through a 25G needle, and fiber “flaring” was evident when extruded from 27G and 30G needles. Thus, 25G needles were used for the remainder of the study to have an equal comparison between the K2 and E2 inks.
3D printing of MDPs was performed on an Allevi 3 bioprinter. The same print optimization process was completed for the 4% K2, 4% E2 and 3% K2 inks. First, the minimum pressure needed for ink extrusion through a 25G needle was determined by increasing pressure in 0.5 PSI increments until filament flow occurred. Using this pressure, a series of calibration lines at varying printhead speeds were then printed. These lines were designed in a U shape to ensure that initial under or over extrusion did not influence the width of the lines measured (Figure S4, Supporting Information). The width of calibration lines was measured as a function of printhead speed (Figure 4a). The calibration curves for the three inks followed an exponential decay curve with R2 values of 0.96, 0.79, and 0.81 for 4% K2, 4% E2, and 3% K2, respectively. At 300 mm/min, 4% K2, 4% E2, and 3% K2 had an average line width of 543 μm, 426 μm, and 590 μm, respectively. A filament extruded from a 25G needle has an ideal height and width of 250 μm, or the inner diameter of the needle. Gravity, though, deforms a printed fiber and leads to one with a smaller height than width.[63] Because of this, MDP filaments were not observed to have widths = 250 μm; instead, fibers fractured at high print speeds. A print speed of 300 mm/min was used for the remainder of the study, as this was the first point on each calibration curve where the data started to form an asymptote and had heights ~250 μm. Using the same process outlined here, an optimized printing speed can be experimentally determined for a range of MDPs using different gauge needles.
Figure 4.
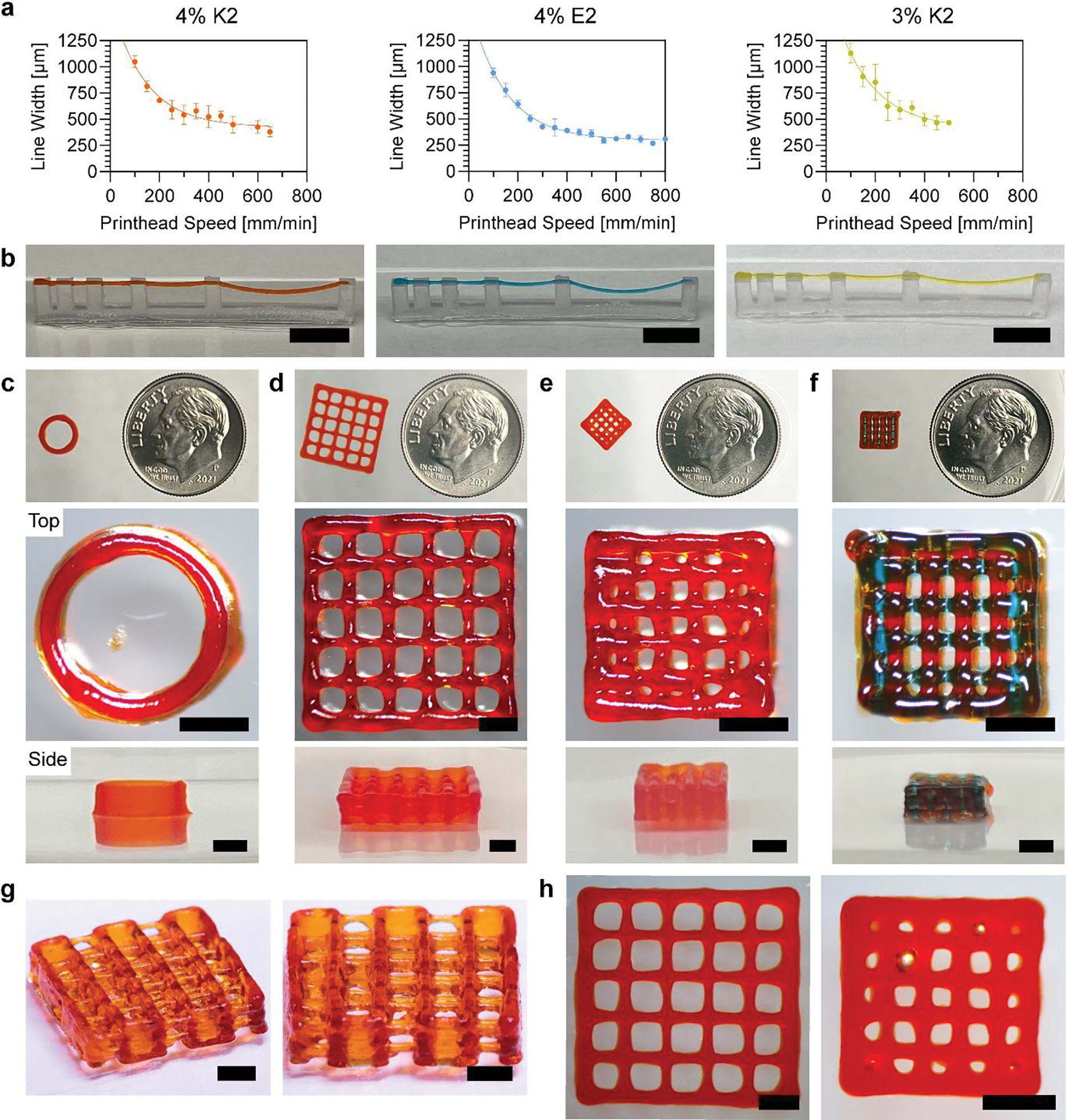
MDP 3D printing optimization and printed structures. a) Printhead speed calibration curves for 4% K2, 4% E2, and 3% K2 MDP inks (n=3 – 9). b) Overhang tests for MDP inks, where orange, blue, and yellow correspond to 4% K2, 4% E2, and 3% K2 MDP inks (Scale bars = 8 mm). c) Cylinder, d) 2×2 log pile, e) 1×1 log pile, and f) multimaterial 1×1 log pile. (Top to bottom) 3D printed structures imaged next to a dime for scale, top views, and side views for each of the constructs. The first three structures were printed with 4% K2 alone and the fourth had alternating 4% K2 and 4% E2 at each layer (Scale bars = 2 mm). g) Modified 2×2 log pile with visible overhanging layers (Scale bars = 2 mm). h) Top view images of 2×2 log pile and 1×1 log pile after being incubated in HBSS for 1 day.
An “overhang test” was performed as previously described[64] to demonstrate the printability of all three MDP inks. The overhang test structure was designed with overhang lengths of 1, 2, 4, 8, and 16 mm and itself was 3D printed on a Form 3 printer (Figure S5, Supporting Information). Using optimized pressures and print speeds, all three inks were successfully printed over the entire overhang length (Figure 4b and Videos 1 – 3, Supporting Information). Minor deflection of the filament was only observed at overhangs of 8 and 16 mm. These represent the first time SAPs have been printed into an overhanging structure with the use of a monoaxial printing strategy. In addition, it shows that noncovalent assemblies can be strong enough to self-support, avoiding the need for covalent crosslinking.
To extend the proof-of-concept test prints, a series of 3D structures with increasing complexity were printed using MDPs. 4% K2 was found to be the best performing ink and therefore was used as the primary ink moving forward. G-code was manually written ignoring the thickness of printed filaments, so the X/Y dimensions of printed constructs were expected to be offset from the designed dimensions by the width of the filaments. In addition, the layer height of all constructs was set as 250 μm (the inner diameter of a 25G needle). First, a 10-layer tall cylinder was designed to have a diameter of 6 mm and height of 2.5 mm (Figure S6, Supporting Information). Although a cylinder is a relatively simple stacking structure without overhangs, it is one of the most difficult 3D design that has been successfully printed with SAPs in previous publications.[45,46] A cylinder was successfully printed with 4% K2 with smooth sides and no observable defects (Figure 4c).
To increase complexity, the log pile design was used, which has been used in multiple 3D printability studies and is difficult to print due to having overhangs at each layer and internal porosity.[37,63] A 10-layer tall log pile was designed to have a length and width of 10 mm, a height of 2.5 mm, and to contain 2 × 2 mm pores (Figure S7, Supporting Information). The 2×2 log pile was successfully printed with 4% K2 and had all 25 pores unobstructed (Figure 4d). To assess print quality, the designed pore dimensions were compared to those of the printed construct. Since the G-code contained 2 × 2 mm pores, the expected pore dimensions when accounting for the filament width is 1457 × 1457 μm (2000 μm - 543 μm from the 4% K2 calibration curve). The inner 9 pores were measured to have an average length and width of 1528 μm, which represents just a 4.9% error between design and print.
Following this success, a 10-layer tall log pile was designed to have a length and width of 5 mm, a height of 2.5 mm, and to contain 1 × 1 mm pores (Figure S8, Supporting Information). The 1×1 log pile was printed with 4% K2 and had all 9 inner pores unobstructed (Figure 4e). The expected pore length and width for this design is 457 × 457 μm (1000 μm - 543 μm from the 4% K2 calibration curve). The inner 9 pores were measured to have an average length and width of 595 μm, which represents a 30.1% error between design and print. This high error is attributed to hardware limitations as opposed to a limitation of the MDP ink, as the printer had a slight delay in movement during turns that caused over extrusion and negatively impacted the accuracy of the 1×1 log pile.
In addition to printing with 4% K2 alone, a multi-MDP structure was successfully fabricated. A 1×1 log pile was designed to have 4% K2 and 4% E2 alternating at each layer (Figure 4f). The pore dimensions were measured separately, with pore width being used to assess K2 print quality and pore length being used to assess E2 print quality. The expected pore width is 457 μm (1000 μm – 543 μm from the 4% K2 calibration curve) and the expected pore length is 574 μm (1000 μm – 426 μm from the 4% E2 calibration curve). The measured pore width and length were an average of 517 and 644 μm, and represented errors of 13.1% and 12.2%, respectively. Together, there was a 12.7% error in the K2/E2 combined 1×1 pore print.
To better visualize that overhangs could be printed within a complex structure, a modified 2×2 log pile that alternated layer direction every 2 layers was designed (Figure S9, Supporting Information). The structure was successfully printed with the 4% K2 ink and overhangs throughout this structure were clearly visible (Figure 4g and Figure S10, Supporting Information).
Attempts to print log pile structures with 4% E2 alone were not as successful as those created with 4% K2 alone or the combined K2/E2 constructs. Although 4% E2 filaments were robust enough to self-support, excessive stacking, such as what is required to print 10-layer log piles, led to sagging of the bulk structure and failed prints (Figure S11, Supporting Information). Because the height of subsequent layers did not match with where previous layers had been deposited, we observed various printing errors.
Structure drying was not observed to be a significant issue, as all the constructs printed were relatively small and had short print times. Still, multiple overhang test lines were left out to dry to observe the potential effects. Within 15 minutes, the hydrogel fibers were observed drying into straight, rigid fibers (Figure S12 and Video 4, Supporting Information). While interesting, the long-term stability and swelling of MDP constructs in a hydrated environment was more relevant for in vitro use. Thus, all complex hydrogel constructs were incubated in HBSS directly after printing. The successfully printed 2×2 and 1×1 log piles were stored in HBSS for 24 hours and maintained their structure and internal porosity (Figure 4h). There were no statistically significant changes in the pore dimensions in either the 1×1 or 2×2 log pile structures for up to 10 days (Figure S13, Supporting Information). In addition, the constructs could be manipulated with a spatula without causing damage (Video 5, Supporting Information).
This work represents the first time a SAP has been used to 3D print structures containing overhangs and porosity. Using MDPs, log pile structures were fabricated with pores as small as 600 μm. In addition, this work represents the first high quality, multimaterial print accomplished using SAPs. The ability to print layered structures without any covalent crosslinks, and the stability of these constructs over time in HBSS both serve as a proof that MDPs are excellent hydrogel candidates for 3D printing. Frequently used crosslinking chemistries can lead to the foreign body response in vivo, so non-covalent self-assembly chemistries may lend to better biocompatibility.[65] In addition, the ability to print with multiple MDPs allows for chemical complexity to be coupled with the three dimensional complexity achieved through printing. Because MDP inks primarily consist of HBSS (>95%), they are inherently cell-friendly in terms of osmolarity and pH. In addition, incubation in cell friendly solutions (such as HBSS) does not negatively affect the structural integrity of 3D printed MDP constructs, showing their viability for long-term in vitro use.
2.3. In Vitro Studies
The charge of a substrate has a large effect on cells both in vitro and in vivo.[66,67] Thus, in vitro studies were conducted to assess how the charge of 3D printed MDPs affected C2C12 cells, a myoblast cell line frequently used in studies of skeletal muscle.[68,69] The ability to 3D print MDPs allowed for the creation of structurally similar, yet oppositely charged 3D constructs with macroscopic porosity. 2-layer tall 1×1 log pile patterns were printed with only 4% K2, only 4% E2, and a combination where the first layer was K2 and the second was E2. Cells were then seeded onto the constructs, and cell viability and morphology were analyzed over time. Live/dead staining at 1 day post seeding revealed that cells adhered to all three constructs and that few dead cells were observed across groups (Figure 5a). After 1 day of culture, K2, E2, and the combined structure supported 97, 92, and 95% cell viability, respectively (Figure 5c). At days 3, 5, and 10, the viabilities remained high for K2 (89, 77, 79%), E2 (88, 74, 79%), and K2/E2 (81, 90, 95%), and there were no statistically significant changes in viability within inks from days 3 – 10. Although MDP charge did not significantly influence viability, more cells adhered to the cationic K2 MDP compared to the anionic E2 MDP (Figure 5d). At days 1, 3, and 5, the average number of cells observed was 79, 143, and 377 cells/field on K2 and 12, 24, and 27 cells/field on E2. In addition to higher seeding, K2 led to faster cell proliferation compared to E2. Using the average number of viable cells measured at day 1 and 5, the doubling times on K2 and E2 were calculated to be 1.8 and 3.4 days, respectively. Regardless, both MDPs supported a significantly higher number of cells by day 5 of culture compared to the number observed on day 1, which suggests that the poorly adhesive nature of E2 was not enough to stop proliferation, but only slow it down. Combined K2/E2 constructs shared this high cell viability up to 10 days in culture, which confirms that combining these oppositely charged MDPs does not cause cytotoxicity.
Figure 5.
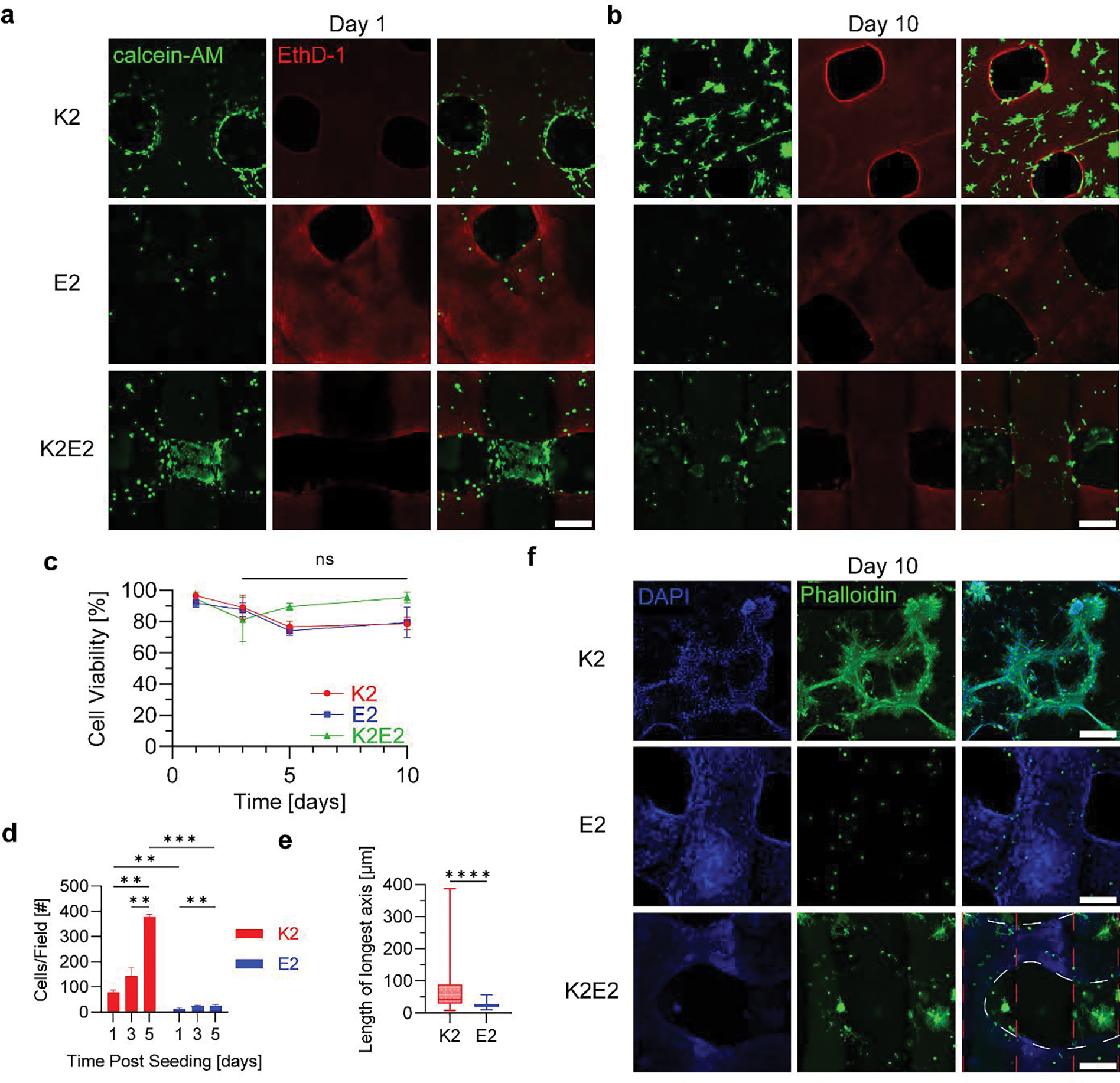
In vitro characterization of 3D printed MDP log pile hydrogels with differing charge. Live/dead staining of C2C12 cells seeded onto 3D printed K2, E2, and K2/E2 hydrogels after a) 1 day and b) 10 days of culture (Scale bars = 300 μm). All images are maximum intensity projections of 300 μm z-stacks. c) Cell viability of cells seeded onto 3D printed MDPs over 10 days (n = 3). The statistical analyses used were multiple one-way ANOVAs with Tukey’s multiple comparisons test between time points of the same group. d) Number of cells that adhered to and grew on 3D printed MDPs over 5 days (n=3). The statistical analyses used were multiple two-way ANOVAs with Tukey’s and Sidak’s multiple comparisons tests within gels at each timepoint and across gels for each timepoint, respectively. e) Length of longest axis of all live cells on each 3D printed MDP after 10 days in culture (n=3). The statistical analysis used was an unpaired t test and the whiskers represent min to max values. f) Immunostaining of cells on 3D printed MDPs after 10 days of culture (Scales bar = 300 μm). The red dashed lines represent regions where K2 was printed, while the white dashed lines represent where E2 gel was printed over the K2. Significance is represented as: * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Cells were qualitatively observed to adopt different morphologies on K2 compared to E2, which was most clearly seen on day 10 of culture (Figure 5b). Differences in cell spreading were also visible on K2/E2 combined constructs, where the inner K2 region had high confluency of extended cells, whereas the outer E2 regions showed less confluent and more spherical cells (Figure 5a, b). To quantify this phenomenon, the length of the longest axis of each cell volume was calculated and compared between constructs, which is a metric that has been previously used to quantify cell spreading.[70] Cells adhered to K2 were observed to spread significantly more than those adhered to E2 after 10 days in culture, with average lengths of 66 and 24 μm, respectively (Figure 5e).
Immunostaining of actin filaments was performed to get a higher quality look at cell morphology (Figure 5f). Myoblasts were observed fusing into myotubes on the K2 gels and extending in multiple directions. In contrast, cells on the E2 constructs were much smaller in size and very few were seen expanding actin filaments at all. Interestingly, cells on the K2/E2 constructs seemed to adopt a morphology dictated by which gel they were directly on, while also being influenced by the oppositely charged peptide in close proximity. The red dashed lines represent regions where K2 was printed, while the white dashed lines represent where E2 gel was printed over the K2 (Figure 5F Bottom Right). These images are maximum intensity projections through the full thickness of the constructs. The actin filaments of cells within the K2 region looked spread out and there seemed to be noticeable nuclei clumping. In contrast, cells on the E2 region were primarily spherical, but some actin filaments could be observed protruding outward from these cells. This was not observed in the E2 only constructs. Overall, the results suggest that K2 and E2 may be strategically used together to dictate cellular behavior in vitro.
In addition to charge, the physical properties of a substrate have a large effect on cells both in vitro and in vivo.[67] Different cell types have been found to respond differently to hydrogel mechanical properties, and there have been numerous studies characterizing differences among stem cells,[71–73] neural cells,[74,75] and myoblasts.[76,77] Using mechanical actuation has even been proposed to aid in muscle generation by modifying the internal cellular environment of muscle cells.[78] The work presented here can hopefully expand this avenue of research to gain an understanding of the interplay between mechanical properties, charge, and their combined effects on cells.
Multiple other aspects of MDPs make them a good candidate for further development for in vitro testing. Because they are synthesized one amino acid at a time, MDPs are tunable and can easily be modified with bioactive peptide sequences to influence cell fate and behavior. Future work will include the printing of more complex geometries as well as exploring the behavior of other cell types when cultured onto MDPs. Although preliminary printing of cell laden MDPs was demonstrated (Figure S15, Supporting Information), further work is needed to fully characterize their use as a bioink.
3. Conclusion
In this study, we establish MDPs as a new class of 3D printable hydrogels. Following characterization of their supramolecular assembly, MDPs are shown to possess the necessary rheological properties to be used for extrusion 3D printing. Printing optimization is then undertaken and utilized to create complex, overhanging structures and multi-material prints. This work represents one of very few 3D printing papers involving self-assembling peptides and surpasses the level of printing complexity that has been previously accomplished. In addition, in vitro characterization of cationic and anionic MDP constructs revealed charge-dependent differences in cell morphology and charge-independent high cell viability. Further research should be undertaken to expand the number of MDPs printed, increase printing resolution, and explore the strategic use of MDP charge to modulate cell behavior.
4. Experimental Section
Materials:
Low loading Rink Amide MBHA Resin and FMOC protected amino acids were purchased from EMD Millipore (Burlington, MA). O-(7-Azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) was purchased from P3 BioSystems (Louisville, KY). Dichloromethane (DCM), N,N-Dimethylformamide (DMF), Acetic Anhydride, and Diethyl Ether were purchased from Thermo Fisher Scientific (Waltham, MA). Dimethyl sulfoxide (DMSO), Piperidine, N,N-Diisopropylethylamine (DiEA), Trifluoroacetic acid (TFA), Triisopropylsilane (TIPS), and Anisole were purchased from Sigma Aldrich (St. Louis, MO).
Peptide Synthesis:
All peptides were synthesized via Solid Phase Peptide Synthesis. For each FMOC deprotection, 25% Piperidine in 50% DMF/ 50% DMSO was added to the reaction vessel for 10 minutes. For each coupling, 4 equivalents of amino acid, 4 equivalents of HATU, and 6 equivalents of DiEA were dissolved in 50% DMF/ 50% DMSO and mixed in the reaction vessel for 20 minutes. Acetylation of the N-terminus was completed with an excess of DiEA and Acetic Anhydride in DCM. Peptides were cleaved from the resin with TFA for 3 hours in the presence of Milli-Q water, TIPS and Anisole, which acted as scavengers. TFA was then evaporated with Nitrogen, and the crude peptide was triturated in cold Diethyl Ether. Centrifugation and resuspension of the crude peptide was repeated 3 times to dissolve cleavage scavengers, before the crude peptide was left to dry overnight.
Peptide Purification:
Crude peptides were dissolved at 10 mg/mL and dialyzed for 4 days against Milli-Q water in 100 – 500 Dalton Spectra/Por® Biotech Cellulose Ester Dialysis Membranes (Spectrum Laboratories Inc. Rancho Dominguez, CA). Peptides were then passed through 0.2 μm Cellulose Acetate Sterile Syringe Filters (VWR International, Radnor, PA) under sterile conditions, frozen, lyophilized, and stored at −20°C. Peptide mass was confirmed to be correct using a Bruker AutoFlex Speed MALDI ToF (Bruker Instruments, Billerica, MA).
Circular Dichroism (CD):
10 μL of peptide in Hank’s Balanced Salt Solution (HBSS, Corning Inc., Corning, NY) at 10 mg/mL was pipetted into a 0.01 mm cuvette and loaded into a Jasco J-810 spectropolarimeter (JASCO Corporation, Tokyo, Japan). A wavelength scan from 180 to 250 nm was performed for 5 accumulations with a pitch of 0.1 nm and a scanning speed of 50 nm/minute.
Attenuated Total Reflectance Fourier Transform Infrared (ATR-FTIR) Spectroscopy:
10 μL of peptide in HBSS at 10 mg/mL was pipetted onto a Golden Gate diamond window within a Nicolet iS20 FT/IR spectrometer (Thermo Scientific, Waltham, MA) and dried with nitrogen. An infrared scan was performed for 30 accumulations at a resolution of 4 cm−1 and background subtraction was performed on the spectra.
Scanning Electron Microscopy (SEM):
Samples were immersed in a series of ethanol dilutions (30%, 50%, 60%, 70%, 80%, 90%, 2 × 100%) for 10 minutes each to dehydrate the gels and then dried using a Leica EM CPD300 Critical Point Dryer (Leica Biosystems, Deer Park, IL. Samples were then transferred to a Denton Desk V Sputter System (Denton Vacuum, Moorestown, NJ) and coated with 10 nm of gold. Samples were then imaged on a Helios NanoLab 660 Scanning Electron Microscope (FEI Company, Hillsboro, OR) at 1 kV and 25 pA.
Rheology:
Rheology tests were performed on an AR-G2 rheometer (TA Instruments, New Castle, DE) with a 12 mm parallel plate. 100 μL of peptide was extruded onto the stage and the parallel plate was lowered to a gap of 550 μm. Any excess gel was scraped away, before the gap was lowered to 500 μm and oil was applied to prevent evaporation during testing. Before each test, an hour-long equilibration period was performed at 1 rad/s and 1% strain. The parameters for each test were as follows: Frequency sweep: 0.1 – 100 rad/s at 1% strain. Shear sweep: 0.01 to 100 s−1. Strain sweep: 0.1 to 100% strain at 1 rad/s. Shear recovery: A minute each at high (500%), low (1%), high (500%), low (1%) strain at 1 rad/s. Temperature sweep: 4 – 39°C at 5°C increments, with 5 minutes at 1 rad/s and 1% strain at each temperature.
Ink Preparation:
Pure peptides were dissolved in HBSS at either 30 or 40 mg/mL. Sonication was used to dissolve all the peptide, while centrifugation was used to remove any bubbles. For the 3 wt% K2, 4 wt% K2, and 4 wt% E2 peptide bioinks, Tartrazine, Allura red, and Brilliant green (Sigma Aldrich, St. Louis, MO) were dissolved in HBSS at 0.1, 0.1 and 0.01 mg/mL, respectively, prior to the addition of peptide. After being fully dissolved, the peptide bioinks were drawn into a 3mL syringe, centrifuged to remove any bubbles, and then loaded into an Allevi plastic syringe (Allevi by 3D Systems, Philadelphia, PA) via a female-to-female syringe coupler. All procedures were done under sterile conditions.
3D Printing:
3D printing was performed on an Allevi 3 (Allevi by 3D Systems, Philadelphia, PA). Repetier-Host (Hot-World GmbH & Co. KG.) was used to manually write G-Code, which was then uploaded to Bioprint Essential (Allevi by 3D Systems, Philadelphia, PA) to be read by the 3D printer. The calibration line print was designed to have three U-shaped patterns, each with a length of 8 mm and width of 4 mm (Figure S4, Supporting Information). Only the final line of the U was used for measurements to account for any differences in preflow. Each print contained 3 U shapes, each being 50 mm/min greater than the previous one. The filament collapse test platform STL file was downloaded from a previous publication and contained overhangs of 1, 2, 4, 8, and 16 mm (Figure S5, Supporting Information).[64] The platform was printed using a Form 3+ (Formlabs, Boston, MA) and glued to a standard glass slide. Constructs were imaged using an OMAX 18 MP USB 3.0 Digital Camera (OMAX Microscope) and analyzed with ToupView (ToupTek Photonics, Hangzhou, China).
Cell Seeding:
C2C12 myoblasts (ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA) supplemented with high glucose (4500mg/L), L-glutamine (4mM), pyruvate (1mM), penicillin-streptomycin (1%), and Fetal Bovine Serum (10%). Nunc Non-Treated 6-well plates (Thermo Fisher Scientific, Waltham, MA) were loaded on the Allevi 3 stage and printed onto directly. Printed gels were briefly submerged in HBSS to prevent evaporation, which was subsequently aspirated off prior to cell seeding. Cells were trypsinized, resuspended in medium, and 100,000 cells were pipetted onto each printed gel. 2mL of medium was added to each well and changed daily for the duration of the experiment. Live/Dead staining was performed using Calcein AM and Ethidium homodimer (Thermo Fisher Scientific, Waltham, MA) according to manufacturer protocols. For immunostaining, gels were washed with PBS, fixed in 4% Paraformaldehyde (Thermo Fisher Scientific, Waltham, MA) for 30 minutes, washed with PBS, quenched with 100mM glycine (Thermo Fisher Scientific, Waltham, MA), permeabilized with 0.2% Triton X-100 (Thermo Fisher Scientific, Waltham, MA) in PBS for 30 minutes, blocked with 1% BSA (Genetex, Irvine, CA) in 0.2% Triton X in PBS for 1 hour, incubated with Alexa Fluor 488 Phalloidin (1:20) (Thermo Fisher Scientific, Waltham, MA) for 1–2 hours, washed with PBS, counterstained with Dapi (1:500) (Invitrogen, Carlsbad, CA) for 10 minutes, washed with PBS and cleared in 88% Glycerol for at least 30 minutes prior to imaging. All imaging was performed on a Nikon A1 Confocal Laser Microscope (Nikon Corporation, Tokyo, Japan) and all images were 300 μm z-stacks and shown visually as maximum intensity projections. Quantification for live/dead staining and cell spreading were performed in image J using the “3D objects counter” plugin. Doubling time was calculated using the following formula: .
Statistical Analysis:
Data were not preprocessed in any way. All results were graphed as the mean +/− standard deviation aside from Figure 5e, which contains box and whiskers representing min and max values. Sample sizes were noted in figure captions along with the statistical tests applied. GraphPad Prism 9 was used to carry out analyses. Significance was represented as: * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Supplementary Material
Acknowledgements
This work was supported by funding from the National Institute of Health (NIH) (R01 DE021798). We gratefully acknowledge additional funding from the National Science Foundation Graduate Research Fellowship Program that supports A.C.F and B.H.P. We also acknowledge the support of Rice University’s Biomaterials Lab.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Adam C. Farsheed, Department of Bioengineering, Rice University, Houston, TX, 77005, USA
Adam J. Thomas, Department of Chemistry, Rice University, Houston, TX, 77005, USA
Brett H. Pogostin, Department of Bioengineering, Rice University, Houston, TX, 77005, USA
Jeffrey D. Hartgerink, Department of Bioengineering, Rice University, Houston, TX, 77005, USA Department of Chemistry, Rice University, Houston, TX, 77005, USA.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, Feinberg AW, Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- [2].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Johansson F, Randles A, Rosenkrantz JE, Louis-Rosenberg JD, Galie PA, Stevens KR, Miller JS, Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Daly AC, Prendergast ME, Hughes AJ, Burdick JA, Cell 2021, 184, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bedell ML, Navara AM, Du Y, Zhang S, Mikos AG, Chem. Rev. 2020, 120, 10744. [DOI] [PubMed] [Google Scholar]
- [5].Patil P, Russo KA, McCune JT, Pollins AC, Cottam MA, Dollinger BR, DeJulius CR, Gupta MK, D’Arcy R, Colazo JM, Yu F, Bezold MG, Martin JR, Cardwell NL, Davidson JM, Thompson CM, Barbul A, Hasty AH, Guelcher SA, Duvall CL, Sci. Transl. Med. 2022, 14, eabm6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ataie Z, Kheirabadi S, Zhang JW, Kedzierski A, Petrosky C, Jiang R, Vollberg C, Sheikhi A, Small 2022, 18, 2202390. [DOI] [PubMed] [Google Scholar]
- [7].Yi S, Liu Q, Luo Z, He JJ, Ma H-L, Li W, Wang D, Zhou C, Garciamendez CE, Hou L, Zhang J, Zhang YS, Small 2022, 18, 2106357. [DOI] [PubMed] [Google Scholar]
- [8].Ahrens JH, Uzel SGM, Skylar-Scott M, Mata MM, Lu A, Kroll KT, Lewis JA, Adv. Mater. 2022, 34, 2200217. [DOI] [PubMed] [Google Scholar]
- [9].Boularaoui S, Shanti A, Lanotte M, Luo S, Bawazir S, Lee S, Christoforou N, Khan KA, Stefanini C, ACS Biomater. Sci. Eng. 2021, 7, 5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Soliman BG, Major GS, Atienza-Roca P, Murphy CA, Longoni A, Alcala-Orozco CR, Rnjak-Kovacina J, Gawlitta D, Woodfield TBF, Lim KS, Adv. Healthc. Mater. 2022, 11, 2101873. [DOI] [PubMed] [Google Scholar]
- [11].Lee S, Sani ES, Spencer AR, Guan Y, Weiss AS, Annabi N, Adv. Mater. 2020, 32, 2003915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, Adv. Mater. 2014, 26, 3124. [DOI] [PubMed] [Google Scholar]
- [13].Ding A, Jeon O, Cleveland D, Gasvoda KL, Wells D, Lee SJ, Alsberg E, Adv. Mater. 2022, 34, 2109394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kiseleva M, Omar MM, Boisselier É, V Selivanova S, Fortin MA, ACS Biomater. Sci. Eng. 2022, 8, 1200. [DOI] [PubMed] [Google Scholar]
- [15].Williams AH, Roh S, Jacob AR, Stoyanov SD, Hsiao L, Velev OD, Nat. Commun. 2021, 12, 2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kajtez J, Wesseler MF, Birtele M, Khorasgani FR, Ottosson DR, Heiskanen A, Kamperman T, Leijten J, Martínez-Serrano A, Larsen NB, Angelini TE, Parmar M, Lind JU, Emnéus J, Kajtez J, Birtele M, Ottosson DR, Parmar M, Adv. Sci. 2022, 9, 2201392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dhand AP, Davidson MD, Galarraga JH, Qazi TH, Locke RC, Mauck RL, Burdick JA, Adv. Mater. 2022, 34, 2202261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Daly AC, Davidson MD, Burdick JA, Nat. Commun. 2021, 12, 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Davidson MD, Prendergast ME, Ban E, Xu KL, Mickel G, Mensah P, Dhand A, Janmey PA, Shenoy VB, Burdick JA, Sci. Adv. 2021, 7, 8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Highley CB, Rodell CB, Burdick JA, Adv. Mater. 2015, 27, 5075. [DOI] [PubMed] [Google Scholar]
- [21].Szklanny AA, Machour M, Redenski I, Chochola V, Goldfracht I, Kaplan B, Epshtein M, Yameen HS, Merdler U, Feinberg A, Seliktar D, Korin N, Jaroš J, Levenberg S, Adv. Mater. 2021, 33, 2102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brassard JA, Nikolaev M, Hübscher T, Hofer M, Lutolf MP, Nat. Mater. 2020, 20, 22. [DOI] [PubMed] [Google Scholar]
- [23].Huo Y, Xu Y, Wu X, Gao E, Zhan A, Chen Y, Zhang Y, Hua Y, Swieszkowski W, Zhang YS, Zhou G, Adv. Sci. 2022, 9, 2202181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Behre A, Tashman JW, Dikyol C, Shiwarski DJ, Crum RJ, Johnson SA, Kommeri R, Hussey GS, Badylak SF, Feinberg AW, Adv. Healthc. Mater. 2022, 2200866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pati F, Jang J, Ha DH, Kim SW, Rhie JW, Shim JH, Kim DH, Cho DW, Nat. Commun. 2014, 5, 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schmid R, Schmidt SK, Detsch R, Horder H, Blunk T, Schrüfer S, Schubert DW, Fischer L, Thievessen I, Heltmann-Meyer S, Steiner D, Schneidereit D, Friedrich O, Grüneboom A, Amouei H, Wajant H, Horch RE, Bosserhoff AK, Arkudas A, Kengelbach-Weigand A, Adv. Funct. Mater. 2022, 32, 2107993. [Google Scholar]
- [27].Sun Y, You Y, Jiang W, Wang B, Wu Q, Dai K, Sci. Adv. 2020, 6, eaay1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Skylar-Scott MA, Uzel SGM, Nam LL, Ahrens JH, Truby RL, Damaraju S, Lewis JA, Sci. Adv. 2019, 5, eaaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ouyang L, Highley CB, Sun W, Burdick JA, Adv. Mater. 2017, 29, 1604983. [DOI] [PubMed] [Google Scholar]
- [30].Liu W, Zhang YS, Heinrich MA, De Ferrari F, Jang HL, Bakht SM, Alvarez MM, Yang J, Li YC, Trujillo-de Santiago G, Miri AK, Zhu K, Khoshakhlagh P, Prakash G, Cheng H, Guan X, Zhong Z, Ju J, Zhu GH, Jin X, Shin SR, Dokmeci MR, Khademhosseini A, Adv. Mater. 2017, 29, 1604630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A, Adv. Mater. 2016, 28, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol. 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [33].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Proc. Natl. Acad. Sci. 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rutz AL, Hyland KE, Jakus AE, Burghardt WR, Shah RN, Adv. Mater. 2015, 27, 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue HJ, Ramadan MH, Hudson AR, Feinberg AW, Sci. Adv. 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ouyang L, Armstrong JPK, Lin Y, Wojciechowski JP, Lee-Reeves C, Hachim D, Zhou K, Burdick JA, Stevens MM, Sci. Adv. 2020, 6, eabc5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hull SM, Lindsay CD, Brunel LG, Shiwarski DJ, Tashman JW, Roth JG, Myung D, Feinberg AW, Heilshorn SC, Adv. Funct. Mater. 2021, 31, 2007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lou J, Mooney DJ, Nat. Rev. Chem. 2022 2022, 6, 726. [DOI] [PubMed] [Google Scholar]
- [39].Sinha NJ, Langenstein MG, Pochan DJ, Kloxin CJ, Saven JG, Chem. Rev. 2021, 121, 13915. [DOI] [PubMed] [Google Scholar]
- [40].Du X, Zhou J, Shi J, Xu B, Chem. Rev. 2015, 115, 13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cui H, Webber MJ, Stupp SI, Pept. Sci. 2010, 94, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Majumder P, Singh A, Wang Z, Dutta K, Pahwa R, Liang C, Andrews C, Patel NL, Shi J, de Val N, Walsh STR, Jeon AB, Karim B, Hoang CD, Schneider JP, Nat. Nanotechnol. 2021 1611 2021, 16, 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yan C, Pochan DJ, Chem. Soc. Rev. 2010, 39, 3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Loo Y, Lakshmanan A, Ni M, Toh LL, Wang S, Hauser CAE, Nano Lett. 2015, 15, 6919. [DOI] [PubMed] [Google Scholar]
- [45].Susapto HH, Alhattab D, Abdelrahman S, Khan Z, Alshehri S, Kahin K, Ge R, Moretti M, Emwas AH, Hauser CAE, Nano Lett. 2021, 21, 2719. [DOI] [PubMed] [Google Scholar]
- [46].Sather NA, Sai H, Sasselli IR, Sato K, Ji W, Synatschke CV, Zambrotta RT, Edelbrock JF, Kohlmeyer RR, Hardin JO, Berrigan JD, Durstock MF, Mirau P, Stupp SI, Small 2021, 17, 2005743. [DOI] [PubMed] [Google Scholar]
- [47].Nolan MC, Caparrós AMF, Dietrich B, Barrow M, Cross ER, Bleuel M, King SM, Adams DJ, Soft Matter 2017, 13, 8426. [DOI] [PubMed] [Google Scholar]
- [48].Raphael B, Khalil T, Workman VL, Smith A, Brown CP, Streuli C, Saiani A, Domingos M, Mater. Lett. 2017, 190, 103. [Google Scholar]
- [49].Jian H, Wang M, Dong Q, Li J, Wang A, Li X, Ren P, Bai S, ACS Appl. Mater. Interfaces 2019, 11, 46419. [DOI] [PubMed] [Google Scholar]
- [50].Dong H, Paramonov SE, Aulisa L, Bakota EL, Hartgerink JD, J. Am. Chem. Soc. 2007, 129, 12468. [DOI] [PubMed] [Google Scholar]
- [51].Aulisa L, Dong H, Hartgerink JD, Biomacromolecules 2009, 10, 2694. [DOI] [PubMed] [Google Scholar]
- [52].Bakota EL, Wang Y, Danesh FR, Hartgerink JD, Biomacromolecules 2011, 12, 1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lopez-Silva TL, Leach DG, Azares A, Li IC, Woodside DG, Hartgerink JD, Biomaterials 2020, 231, 119667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Moore AN, Lopez Silva TL, Carrejo NC, Origel Marmolejo CA, Li IC, Hartgerink JD, Biomaterials 2018, 161, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lopez-Silva TL, Cristobal CD, Lai CSE, Leyva-Aranda V, Lee HK, Hartgerink JD, Biomaterials 2020, 265, 120401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lai CSE, Leyva-Aranda V, Kong VH, Lopez-Silva TL, Farsheed AC, Cristobal CD, Swain JWR, Lee HK, Hartgerink JD, ACS Appl. Bio Mater. 2022, 5, 4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Leach DG, Dharmaraj N, Piotrowski SL, Lopez-Silva TL, Lei YL, Sikora AG, Young S, Hartgerink JD, Biomaterials 2018, 163, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Leach DG, Newton JM, Florez MA, Lopez-Silva TL, Jones AA, Young S, Sikora AG, Hartgerink JD, ACS Biomater. Sci. Eng. 2019, 5, 6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Leach DG, Dharmaraj N, Lopez-Silva TL, Venzor JR, Pogostin BH, Sikora AG, Hartgerink JD, Young S, ACS Biomater. Sci. Eng. 2021, 7, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Pogostin BH, Yu MH, Azares AR, Euliano EM, Lai CSE, Saenz G, Wu SX, Farsheed AC, Melhorn SM, Graf TP, Woodside DG, Hartgerink JD, Mchugh KJ, Biomater. Sci. 2022, 10, 6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Paxton N, Smolan W, Böck T, Melchels F, Groll J, Jungst T, Biofabrication 2017, 9, 044107. [DOI] [PubMed] [Google Scholar]
- [62].Chen MH, Wang LL, Chung JJ, Kim YH, Atluri P, Burdick JA, ACS Biomater. Sci. Eng. 2017, 3, 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA, ACS Biomater. Sci. Eng. 2016, 2, 1743. [DOI] [PubMed] [Google Scholar]
- [64].Ribeiro A, Blokzijl MM, Levato R, Visser CW, Castilho M, Hennink WE, Vermonden T, Malda J, Biofabrication 2017, 10, 014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Delgado LM, Bayon Y, Pandit A, Zeugolis DI, Tissue Eng. Part B Rev. 2015, 21, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Metwally S, Stachewicz U, Mater. Sci. Eng. C 2019, 104, 109883. [DOI] [PubMed] [Google Scholar]
- [67].Huang G, Li F, Zhao X, Ma Y, Li Y, Lin M, Jin G, Lu TJ, Genin GM, Xu F, Chem. Rev. 2017, 117, 12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gilbert-Honick J, Iyer SR, Somers SM, Takasuka H, Lovering RM, Wagner KR, Mao HQ, Grayson WL, Biomaterials 2020, 255, 120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kim W, Lee H, Lee J, Atala A, Yoo JJ, Lee SJ, Kim GH, Biomaterials 2020, 230, 119632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee HP, Lippens E, Duda GN, Mooney DJ, Nat. Mater. 2016, 15, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Engler AJ, Sen S, Sweeney HL, Discher DE, Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- [72].Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ, Nat. Mater. 2014, 13, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Marklein RA, Burdick JA, Soft Matter 2009, 6, 136. [Google Scholar]
- [74].Leipzig ND, Shoichet MS, Biomaterials 2009, 30, 6867. [DOI] [PubMed] [Google Scholar]
- [75].Banerjee A, Arha M, Choudhary S, Ashton RS, Bhatia SR, Schaffer DV, Kane RS, Biomaterials 2009, 30, 4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bauer A, Gu L, Kwee B, Li WA, Dellacherie M, Celiz AD, Mooney DJ, Acta Biomater. 2017, 62, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Silver JS, Günay KA, Cutler AA, Vogler TO, Brown TE, Pawlikowski BT, Bednarski OJ, Bannister KL, Rogowski CJ, McKay AG, DelRio FW, Olwin BB, Anseth KS, Sci. Adv. 2021, 7, eabe4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Seo BR, Payne CJ, Mcnamara SL, Freedman BR, Kwee BJ, Nam S, de Lázaro I, Darnell M, Alvarez JT, Dellacherie MO, Vandenburgh HH, Walsh CJ, Mooney DJ, Sci. Transl. Med 2021, 13, eabe8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


