Abstract
Recent work in structural biology is shedding light on how many of the enzymes of intermediary metabolism are self- and co-assembling into large, filamentous polymers or agglomerates to organize and regulate the complex and essential biochemical pathways in cells. Filament assembly provides an additional layer of regulation by modulating the intrinsic allostery of the enzyme protomers which tunes activity in response to a variety of environmental cues. Enzyme filaments dynamically assemble and disassemble in response to changes in metabolite levels and environmental cues, shifting metabolic flux on a more rapid timescale than transcriptional or translational reprogramming. Here we present recent examples of high-resolution structures of filaments from proteins in intermediary metabolism and we discuss how filament assembly modulates the activities of these and other proteins.
Keywords: Filament assembly, Metabolic enzymes, Apolar filaments, Allostery, Cryo Electron Microscopy
Introduction
Large assemblies of purified enzymes from across intermediary metabolism have been documented since their initial purifications in the 1960s. Early examples of enzymes that reversibly assemble in large filaments include glutamine synthetase [1], acetyl CoA carboxylase [2,3], glutaminase [4], and phosphoribosyl pyrophosphate synthetase [5,6] (for a comprehensive review, see Park & Horton, 2019 [7]). In each case, assembly was modulated by the presence of specific ligands known to affect enzyme activity, suggesting that assembly might play a role in regulation. These early examples of enzyme filaments were characterized using analytical centrifugation, size exclusion chromatography, and transmission electron microscopy, and in some cases changes in activity could be correlated with assembly states. Due to the technical challenges associated with studying large, dynamic polymers at the time, however, the structural underpinnings of the enzyme assemblies and the biochemical and physiological effects of assembly remained uncharacterized.
Advances in light microscopy and subsequent identification of cellular protein assemblies containing enzymes from intermediary metabolism have supported physiological roles for the large polymers. Screens using fluorescently labeled proteins have identified numerous enzyme assemblies in budding yeast, with some paralogs or enzymes within the same pathways co-localizing within the cell [8–11]. Recent studies have begun to define the conditions under which the punctate and filamentous assemblies appear in cells, demonstrating that the macrostructures are dynamic and assemble reversibly [12–19]. The assemblies often appear at times of cell stress, including starvation, cell division, and proliferation. Many of the assemblies tend to contain proteins that sit at regulatory nodes within metabolic networks, suggesting that assembly provides another layer of enzyme regulation within the cell and driving the exploration of the molecular mechanisms and functional consequences of polymerization [10].
The uncovering of physiological roles for large-scale assemblies prompted the exploration of their structural organization. Cryo-electron microscopy, which can provide high-resolution structures of polymers spanning tens of nanometers in length, has greatly expanded our understanding of the molecular underpinnings of the protein assemblies observed in cells. This technique, paired with other structural and biochemical tools, has made it possible to dissect the functional consequences of polymerization [20]. Mutagenesis studies targeted to the filament interface show that filament assembly serves as a platform for allosteric regulation [21–24], where mutations lead to changes in catalytic activity. Here, we reflect on recent work, describing patterns in the filaments assembled from enzymes in intermediary metabolism. We discuss the allosteric regulatory mechanisms made possible by filament formation, and we contemplate the next steps in the field.
Filaments in intermediary metabolism: leveraging symmetry
Filaments from intermediary metabolism share some traits with the better-understood dynamic cytoskeletal polymers. For example, actin filaments and microtubules are extremely well conserved and found in all eukaryotes, and both have homologues in prokaryotes. Both are dynamic and change assembly states in response to changes in cellular conditions. However, both actin filaments and microtubules are directional (polar) and assemble from monomers (actin) (Figure 1A) or asymmetric dimers (tubulin) into helical or pseudo-helical filaments [25].
Figure 1. Many enzymes from intermediary metabolism form nondirectional filaments with dihedral symmetry.
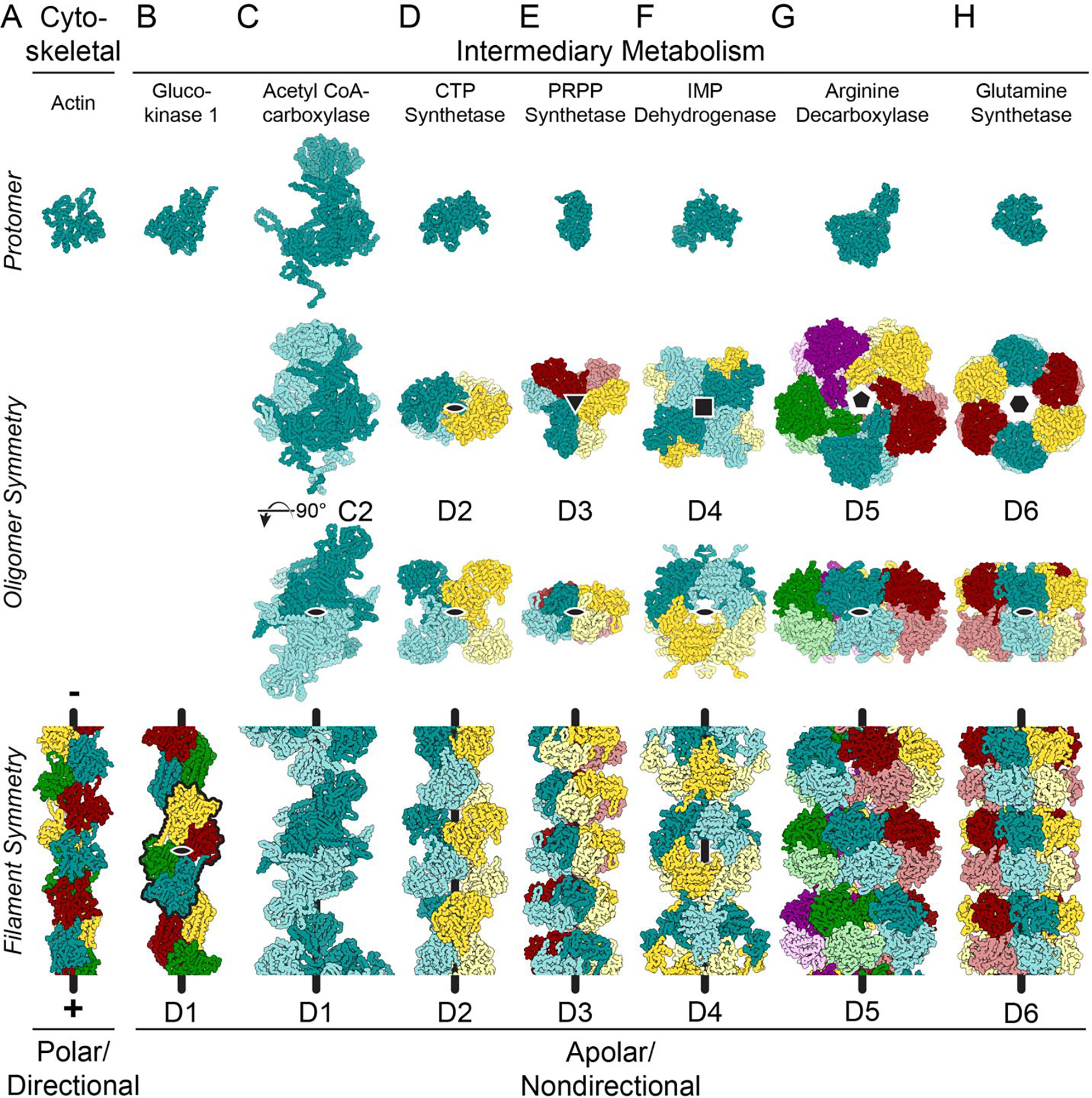
Images show mainchain C, N, and Cα atoms only. The symmetry for the helical axes is shown in the oligomer (black shapes, second row); one, two-fold axis is shown on the oligomer (black oval, third row); and the helical axis is shown as a black line on the filaments (bottom row). A. Cytoskeletal proteins, like actin (shown - PDB ID 6DJM [60]) and tubulin assemble from monomers or asymmetrical dimers, which produces a directional filament. The filaments grow and shrink from a defined pole of the filament structure, and the longitudinal interfaces within the filaments are comprised of residues found on opposite sides of the protein. B. While glucokinase 1 filaments (6PDT) assemble from monomers, the repeating units in the filament (black outline) assemble with C2 symmetry around an axis (black oval) perpendicular to the helical axis, generating a filament with overall D1 symmetry. C. Symmetric C2 dimers can also assemble into filaments with D1 symmetry (Acetyl CoA-carboxylase: 6G2D). The symmetry axis perpendicular to the helical axis is shown in both the oligomer and the filament (black oval). D-H. Enzymes found in intermediary metabolism have dihedral symmetry within the oligomer which is propagated in the filament form of the enzyme (CTP synthase: 6PK4; PRPP synthetase: 8DBF; IMP dehydrogenase: 7RGL/7RGM; arginine decarboxylase: 7PK6; glutamine synthetase: 7W85).
Unlike actin and tubulin, metabolic enzymes that form polymers generally assemble into helical filaments that lack directionality (i.e. apolar filaments) (Figure 1B–E), even when the assembling protomers are monomers. For example, glucokinase 1 from Saccharomyces cerevisiae, a distant actin homolog that catalyzes the first step in glycolysis, assembles from monomers into filaments [26]. The repeating unit in the filament of glucokinase 1 (black outline in Figure 1B, bottom row) contains twofold symmetry, which adds a symmetry axis perpendicular to the helical axis. This axial symmetry coupled with the helical symmetry places the filament in dihedral filament symmetry group D1 [27] (Figure 1B), where a 180° rotation around the axis perpendicular to the helical axis will superimpose the filament on itself.
Most other metabolic enzyme filaments assemble from protomers that are themselves oligomers with closed point group symmetry. In most cases, the defined oligomerization is required for activity or allosteric regulation. Panels C through H in Figure 1 illustrate several examples of nondirectional filaments formed from oligomers of increasing symmetry. Acetyl CoA-carboxylase forms a symmetric dimer in solution, and these dimers stack to form filaments with D1 symmetry (Figure 1C) [28]. Higher dihedral symmetries are also observed (Figure 1D–H) [16,21–23,29–32], and oligomers with D3 symmetry or greater tend to stack with their highest symmetry axes coincident with the helical axis of the filament. Oligomers composed of multiple proteins or protein isoforms with pseudo dihedral filament symmetry have been identified and characterized [33,34].
A filament interface containing dihedral symmetry becomes much more sensitive to a single amino acid change at the protomer level and can more rapidly evolve towards or away from filament assembly (Figure 2) [35]. When a mutated amino acid is located on the outer surface of the oligomer, the change can result in a “self-interacting patch” on the protein, triggering formation of filaments [36]. The propensity for assembly into filaments can be calculated [36] and increases with higher-order symmetries due to avidity effects. Negative selection also occurs at potential assembly interfaces on dihedral oligomers, with specific surface residues preventing filament formation [37]. The degree to which the filament interfaces are evolutionarily constrained varies by protein and suggests polymerization can evolve and be selected. For example, there is evidence for widespread conservation of PRPP synthetase filament interfaces [16,23], but CTP synthase forms different interfaces in animals, fungi, and prokaryotes, implying that polymerization has arisen multiple times in evolution [22,29].
Figure 2. Increasing symmetry at filament interfaces increases the impact of single amino acid changes within the interface.
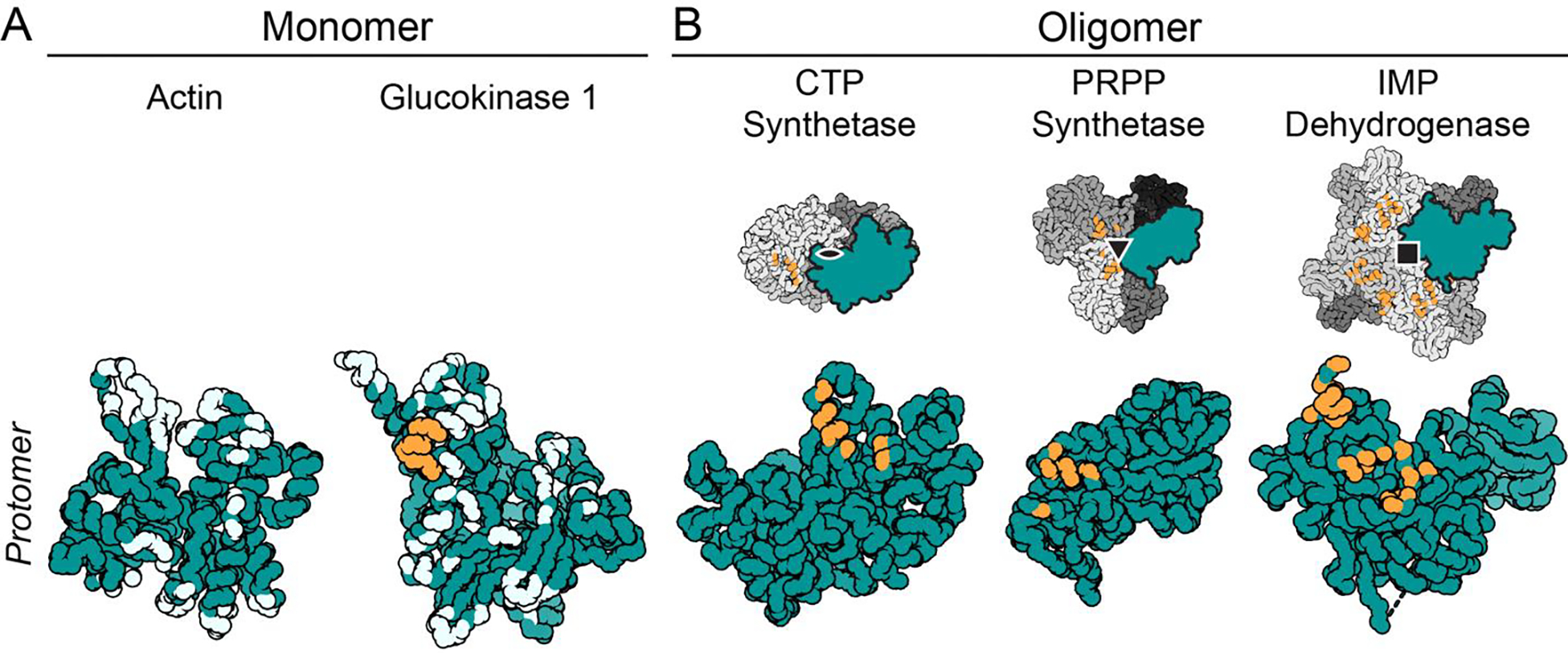
Images show mainchain C, N, and Cα atoms only, with residues within 4 Å of the filament interface shown in orange (self-interacting patch) or light blue (non self-interacting patch). A. Each monomer in a filament composed of monomers has many interface residues spread across the protein. No amino acids on actin self-interact within the filament (PDB ID 6DJM). In contrast, the glucokinase 1 interface does contain one self-interacting patch (orange residues) (PDB ID 6PDT). B. Filaments composed of symmetrical oligomers contain filament interfaces composed of self-interacting patches. In addition, each protomer of the oligomer only contains a small portion of the filament interface, narrowly localized on the protomer (PDB IDs 6PK4, 8DBF, 7RGL/7RGM).
Individual filaments and their larger organization within the cell can be described in terms of their quinary structures, or their quasi-regular assembly into agglomerates via weak interactions [38,39]. Individual filaments like those in Figure 1 can be defined as a level of quinary structure, as they are not bound by a specific protomer number; they are theoretically infinite, and their assembly is dependent on their environment. Quinary structural assemblies build upon the characteristics generated by quaternary assembly by reversibly tuning protein behavior in a variety of ways [40–43]. Quinary assembly can stabilize or destabilize protein structure (Figure 3A, PRPP synthetase [23]), modulate protein activity (Figure 3B, IMP dehydrogenase [30]), and localize proteins to specific regions within a cell [37], all in response to changes in the cellular environment.
Figure 3: Examples of different assembly-based mechanisms for tuning allosteric regulation.
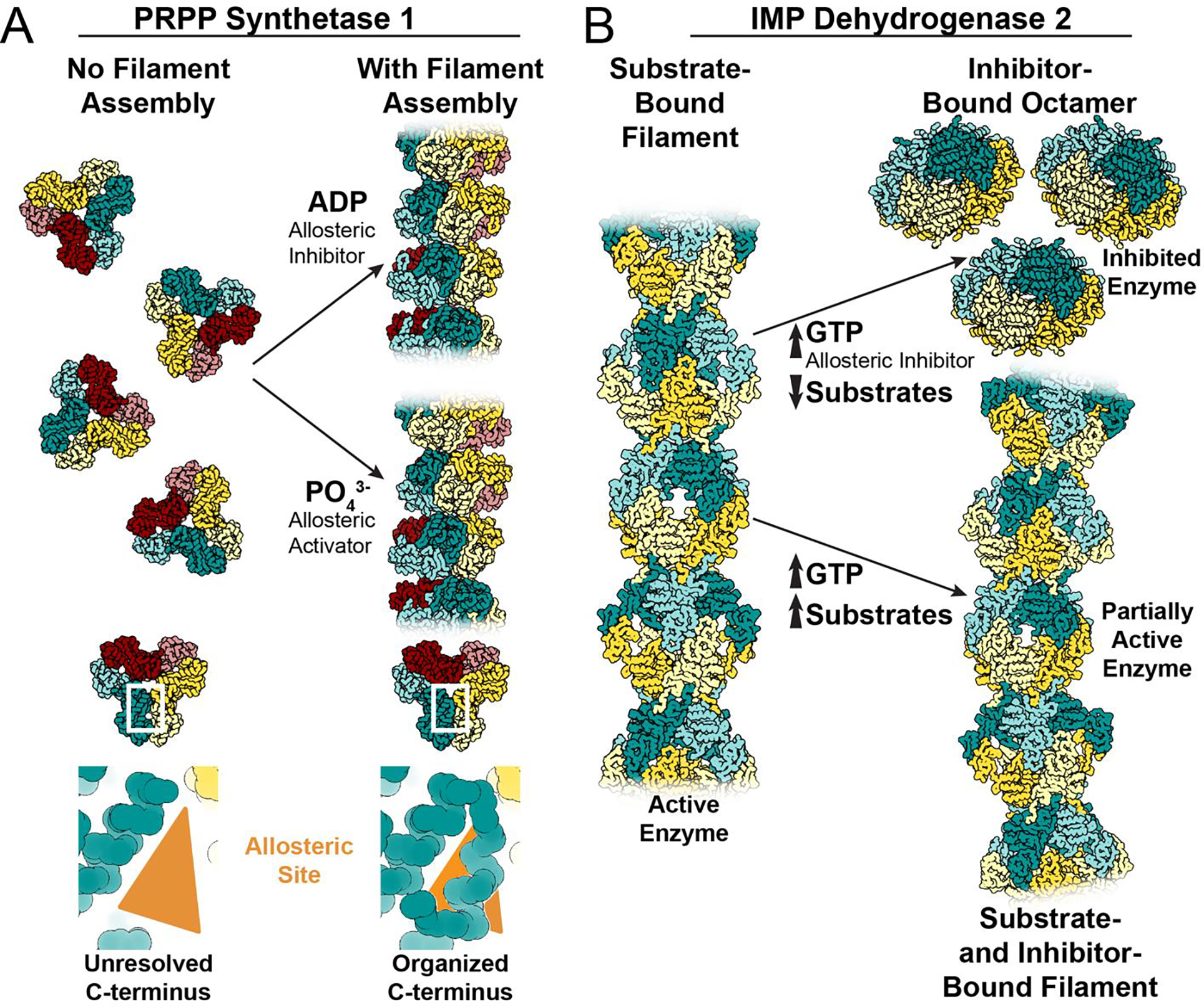
A. Without filament formation, the C-terminus of PRPP synthetase 1 is unresolved. Filament assembly organizes the amino acids in the C-terminus, stabilizing the allosteric site to increase affinity for effectors, increasing sensitivity to both positive and negative regulation (PDB IDs 8DBD and 8DBF). B. IMP dehydrogenase 2 assembles filaments in the active, extended conformation when bound to substrates (PDB ID 6U8E/6N8N). The allosteric inhibitor GTP stabilizes an inactive, compressed octamer conformation when substrate concentrations are limiting (PDB ID 6UAJ). When substrate and inhibitor concentrations are high, filament assembly contacts resist the GTP-induced compression to retain activity in the filament at GTP concentrations which completely inhibit free octamers (PDB ID 6U8R/6U8S). The partially active, bent filaments merge the competing signals that arise in an environment with high concentrations of both substrate and inhibitor.
Many filaments are hypothesized to form larger lateral arrays in cells, with notable examples in the literature [9,22,44,45]. While some of these assemblies may consist of quasi-ordered filament arrays without a strictly ordered lattice, some filaments have been shown to assemble with defined geometry. Both CTP synthase isoforms from S. cerevisiae assemble reversibly into multi-filament bundles with well-defined inter-filament assembly contacts [22]. Bundles of IMP dehydrogenase filaments have been observed in vivo via ultrathin section EM [9] and bundles can be induced by crowding in vitro [45,46].
Enzyme regulation by filament assembly
Enzyme filament structures are intimately linked to their effects on biochemical function, which are surprisingly varied among different enzymes. Filament assembly can act as a mechanism of allosteric control, either by directly stabilizing active or inactive states of an enzyme or by influencing the affinity for other allosteric effectors (Figure 3). Some filaments accommodate conformational changes between different functional states, influencing the equilibria between active and inhibited conformations. In other cases, filaments act as an integration node, where signals from binding partners and phosphorylation can be merged [28]. One advantage of such assembly-based regulation is that it can happen rapidly in response to sudden changes in metabolic demand, while other forms of control like transcriptional reprogramming or degradation require longer response times. Below, we discuss several recent examples of assembly-based control of enzyme activity (Figures 3 and 4) and suggest other possible mechanisms.
Figure 4. Filaments exploit many regulation modalities.
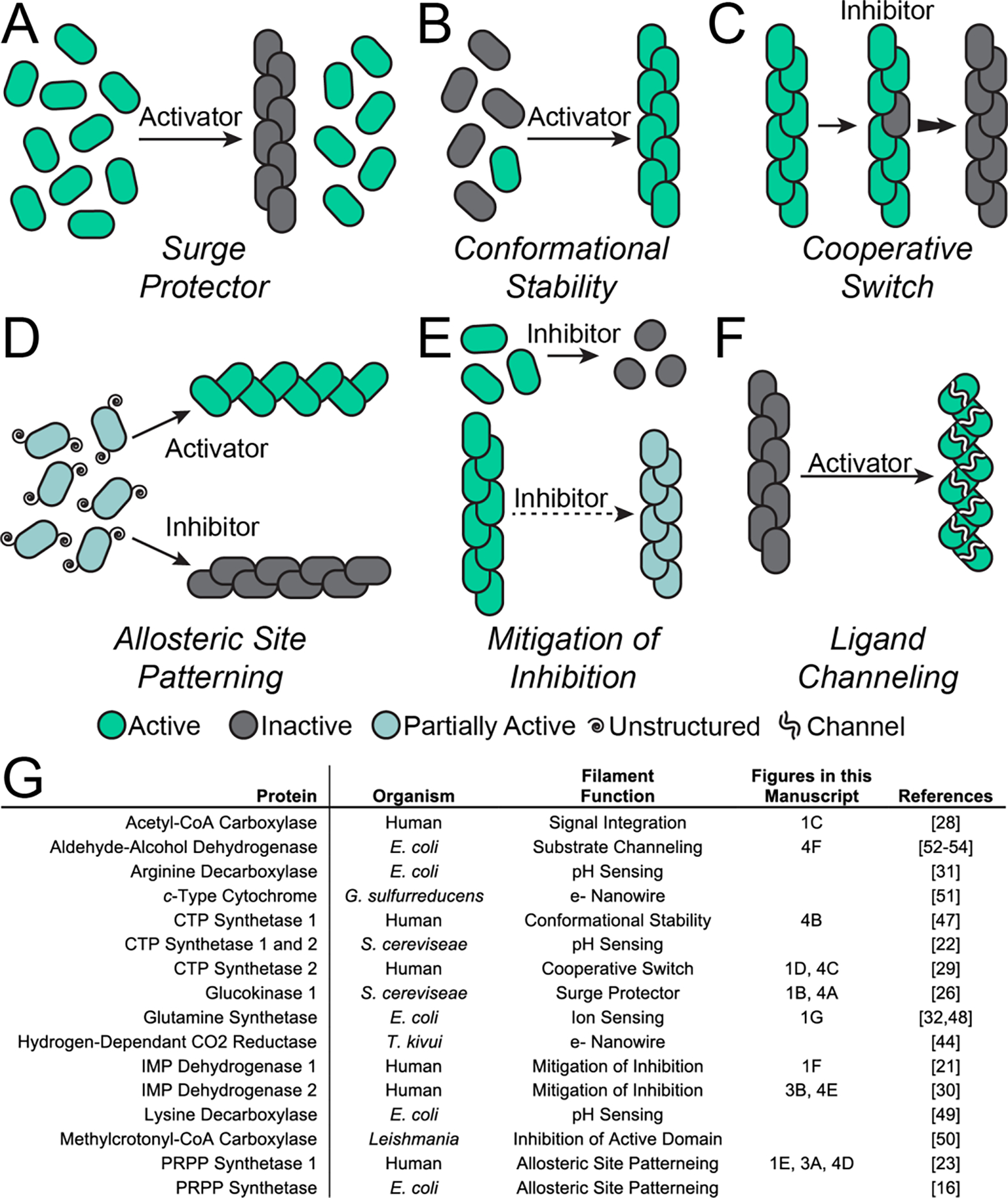
A. A spike in substrate concentration shifts protomers into filaments, inactivating the enzyme and preventing an overconsumption of substrate and cellular toxicity (glucokinase 1). B. Filaments stabilize the active form of the enzyme (CTP synthase 1). C. Filaments act as a cooperative switch, flipping the entire filament into the active or inactive conformation upon local binding of the activator or inhibitor, respectively (CTP synthase 2). D. Filaments stabilize the allosteric site of a protein, supporting the active or inactive conformations of the enzyme (PRPP synthetase 1). E. In the presence of high feedback inhibitor, filaments mitigate inhibition, allowing for sustained enzyme activity (IMP dehydrogenase). F. Filament assembly facilitates substrate channeling between the active sites in different domains within the enzyme (aldehyde-alcohol dehydrogenase). G. List of filament-forming proteins used as examples in this manuscript.
Filaments can act as a mechanism of inhibition and “protein buffering” within a cell. For example, glucokinase 1 (Glk1), which regulates entry into glycolysis, is active as a monomer. Glk1 has a high affinity for glucose, and is typically expressed when glucose levels are low, where it remains monomeric and active. However, when glucose levels spike Glk1 assembles into filaments in which it is prevented from turning over substrate, so that only the small pool of unassembled protein at the critical concentration remains active (Figure 4A) [26]. Assembly-based inactivation of Glk1 prevents toxic over commitment to glycolysis, acting as a “molecular surge protector” to prevent overactivity due to sudden sharp changes in metabolic state.
Enzyme filaments, however, are not all inactive. For example, assembly into filaments stabilizes the active conformation of human CTP synthase 1, which upregulates its activity (Figure 4B) [47]. CTP synthase 2 from humans assembles into filaments that can dynamically shift from the inhibited state to the active state and vice versa [29]. Physical coupling of CTP synthase 2 protomers within a filament results in highly cooperative transitions between low- and high-activity conformations via propagation along the filament (Figure 4C). The cooperativity of CTP synthase 2 filaments makes for a conformational switch that is exquisitely sensitive to the balance of substrates and allosteric inhibitor, allowing cells to quickly adapt to changes in nucleotide demand.
Assembly of filaments can also act to stabilize ligand binding sites, promoting activity, inhibition, or both. For example, human PRPP synthetase 1 assembles into filaments that stabilize a critical allosteric site that binds either the activator phosphate or the inhibitor ADP [23]. In free hexamers, the short helix that forms the allosteric site is disordered, making PRPP synthetase 1 insensitive to activation by phosphate. But in the filament, assembly contacts constrain the short helix and pre-patterns the allosteric pocket (Figure 3A). This stabilizes binding of either phosphate or ADP, and the filaments then adopt either the active or inhibited conformation of the enzyme (Figure 4D). Similar filaments and regulatory mechanisms have also been observed in the E. coli PRPP synthetase [16], suggesting this mode of regulation is broadly conserved.
Filaments can also tune the response of enzymes to allosteric effectors. Assembly of IMP dehydrogenase 2 into filaments decreases its response to the allosteric inhibitor GTP. Binding of GTP compresses and allosterically inhibits free octamers of IMP dehydrogenase 2, but filament contacts stabilize a conformation of the catalytic core that resists this GTP-induced compression, making the filaments less sensitive to GTP inhibition (Figure 3B and 4E) [30]. Filaments formed by IMP dehydrogenase 1 also modulate the enzyme’s response to GTP, and inhibition is further tuned by N- and C-terminal extensions that are a part of the retinal splice variants of the protein [21]. This ability to remain functional at high GTP levels is especially important in cells that have a high nucleotide demand, like those in the retina, where the enzyme must maintain activity in the presence of large pools of a feedback inhibitor.
Many other regulatory mechanisms have been associated with filament formation in enzymes. Filament assembly can occur as a direct response to environmental cues, such as divalent cations, as is the case in glutamine synthetase [32,48], or pH, as for CTP synthase from S. cerevisiae [22] and arginine and lysine decarboxylases in bacteria [31,49]. Assembly can provide a platform for integration of regulatory signals; filament formation of acetyl-CoA carboxylase allows for the integration of signals from binding partners and phosphorylation [28]. Filament formation can stabilize the domains located at the filament interface, inhibiting the activity of those domains, as happens in methylcrotonyl-CoA carboxylase [50]. Filaments can create a cofactor “nanowire” required for the transfer of electrons [44,51]. Recent work on aldehyde-alcohol dehydrogenase has demonstrated that “spirosome” filament assembly is not only critical for its activity, but that changes in the filament architecture facilitate substrate channeling between different active sites in the enzyme (Figure 4F) [52–54].
We can postulate additional mechanisms of assembly-based regulation, beyond those observed to date. Like the allosteric site stabilization in filaments of PRPP synthetase, assembly of filaments could form emergent allosteric sites not found in the enzyme protomers, making filaments required for activity or inhibition. Filament formation might also accomplish the opposite affect; by assembling into a filament, the active or allosteric sites of an enzyme could be occluded, preventing activity or regulation. Filaments might impose hysteresis on an enzyme; once the filament has assembled in either the active or inactive form, a higher effector concentration might be required to switch the enzyme into the opposing conformation than is required when the enzyme is in its monomeric or stoichiometric oligomer form. This modality would buffer enzyme activity or inhibition against short or shallow spikes in ligand concentration, preventing the cell from shifting into a new regime prematurely. Whether any such mechanisms exist in nature remains to be seen, and considering these alternatives may be useful for efforts to design novel filamentous assemblies to control biochemical activities [55–57].
Conclusions: where are filaments extending next?
Recent work has highlighted the many ways enzyme filament assembly provides regulatory control, yet the field remains young. Many metabolic proteins have multiple isoforms and splice variants, which often contain conserved oligomer and filament interface residues. As many of these isoforms and variants are expressed within the same cell type in multicellular organisms, it is very likely that these proteins can co-assemble in heteromeric oligomers and filaments. However, very little work has been done to characterize the emergent biochemical and structural properties that arise from mixed assembly [58]. In a similar vein, many proteins in intermediary metabolism are post-translationally modified, but how these modifications affect filament assembly states is only beginning to be explored [59]. Likely, additional cellular factors modulate enzyme assembly, and there will be many impactful developments related to this multilayered regulation in the coming years, including the development of pharmaceutical therapies that directly target specific isoforms of metabolic enzymes filaments [58,60].
In addition, much work remains to be done at the intermediate scale of protein assembly. While cell biologists have identified many large protein assemblies and structural biologists have defined the interfaces within many enzyme filaments, questions remain where these two fields meet. For example, how do individual filaments bundle into the larger structures observed in cells? How is this assembly regulated? Do polymers of different enzymes co-assemble, and if so, are these interactions defined and specific or more loosely associated agglomerations? What do these large assemblies look like at the angstrom and nanometer scale and how can researchers define the content of puncta and filaments in cells? Identifying and defining the structural underpinnings to these processes in cells will provide valuable insight into cellular organization and function.
Acknowledgements
This work was supported by the National Institutes of Health (R01 GM118396 to J.M.K. and F32 AI145111 to K.L.H.)
Footnotes
Declarations of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Valentine RC, Shapiro BM, Stadtman ER: Regulation of Glutamine Synthetase. XII. Electron Microscopy of the Enzyme from Escherichia coli. Biochemistry 1968, 7:2143–2151. [DOI] [PubMed] [Google Scholar]
- 2.Kleinschmidt AK, Moss J, Daniel Lane M: Acetyl Coenzyme A Carboxylase: Filamentous Nature of the Animal Enzymes. Science (1979) 1969, 166:1276–1278. [DOI] [PubMed] [Google Scholar]
- 3.Meredith MJ, Lane MD: Acetyl-CoA carboxylase. Evidence for polymeric filament to protomer transition in the intact avian liver cell. Journal of Biological Chemistry 1978, 253:3381–3383. [PubMed] [Google Scholar]
- 4.Olsen BR, Svenneby G, Kvamme E, Tveit B, Eskeland T: Formation and Ultrastructure of Enzymically Active Polymers of Pig Renal Glutaminase. J Mol Biol 1970, 52:239–245. [DOI] [PubMed] [Google Scholar]
- 5.Roth DG, Shelton E, Deuel TF: Purification and Properties of Phosphoribosyl Pyrophosphate Synthetase from Rat Liver. J Biol Chem 1974, 249:291–296. [PubMed] [Google Scholar]
- 6.Meyer LJ, Becker MA: Human Erythrocyte Phosphoribosylpyrophosphate Synthetase: Dependance of Activity on State of Subunit Association. J Biol Chem 1977, 252:3919–3925. [PubMed] [Google Scholar]
- 7.Park CK, Horton NC: Structures, functions, and mechanisms of filament forming enzymes: a renaissance of enzyme filamentation. Biophys Rev 2019, 11:927–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen QJ, Kassim H, Huang Y, Li H, Zhang J, Li G, Wang PY, Yan J, Ye F, Liu JL: Filamentation of Metabolic Enzymes in Saccharomyces cerevisiae. Journal of Genetics and Genomics 2016, 43:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiavon CR, Griffin ME, Pirozzi M, Parashuraman R, Zhou W, Jinnah HA, Reines D, Kahn RA: Compositional complexity of rods and rings. Mol Biol Cell 2018, 29:2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noree C, Begovich K, Samilo D, Broyer R, Monfort E, Wilhelm JE: A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Mol Biol Cell 2019, 30:2721–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’connell JD, Mirrielees J, Ellington AD, Marcotte EM: Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. The Proceedings of the National Academy of Sciences 2009, 106:10147–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begovich K, Yelon D, Wilhelm JE: PRPS polymerization influences lens fiber organization in zebrafish. Developmental Dynamics 2020, 249:1018–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marini G, Nöske E, Leng W, Alberti S, Pigino G: Reorganization of budding yeast cytoplasm upon energy depletion. Mol Biol Cell 2020, 31:1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simonet JC, Foster MJ, Lynch EM, Kollman JM, Nicholas E, O’Reilly AM, Peterson JR: CTP synthase polymerization in germline cells of the developing Drosophila egg supports egg production. Biol Open 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleghorn WM, Burrell AL, Giarmarco MM, Brock DC, Wang Y, Chambers ZS, Du J, Kollman JM, Brockerhoff SE: A highly conserved zebrafish IMPDH retinal isoform produces the majority of guanine and forms dynamic protein filaments in photoreceptor cells. Journal of Biological Chemistry 2022, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu HH, Lu GM, Chang CC, Li Y, Zhong J, Guo CJ, Zhou X, Yin B, Zhang T, Liu JL: Filamentation modulates allosteric regulation of PRPS. Elife 2022, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang B, Zhang J, Zhao G, Liu M, Hu J, Lin F, Wang J, Zhao W, Ma H, Zhang C, et al. : Filamentous GLS1 promotes ROS-induced apoptosis upon glutamine deprivation via insufficient asparagine synthesis. Mol Cell 2022, 82:1821–1835.e6. ** This paper shows that Glutaminase 1 can assemble into large, super-active filaments within cells during glutamine deprivation. These super-active filaments facilitate apoptosis during glutamine deprivation and constitutive filaments suppress cancer cell growth.
- 18.Wang PY, Lin WC, Tsai YC, Cheng ML, Lin YH, Tseng SH, Chakraborty A, Pai LM: Regulation of CTP synthase filament formation during DNA endoreplication in Drosophila. Genetics 2015, 201:1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noree C, Sato BK, Broyer RM, Wilhelm JE: Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. Journal of Cell Biology 2010, 190:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziegler SJ, Mallinson SJB, John PC, Bomble YJ: Advances in integrative structural biology: Towards understanding protein complexes in their cellular context. Comput Struct Biotechnol J 2021, 19:214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burrell AL, Nie C, Said M, Simonet JC, Fernández-Justel D, Johnson MC, Quispe J, Buey RM, Peterson JR, Kollman JM: IMPDH1 retinal variants control filament architecture to tune allosteric regulation. Nat Struct Mol Biol 2022, 29:47–58. * This paper describes the structure and biochemical properties of IMP dehydrogenase 1 from humans and demonstrates how splice variants in the retina alter both the assembly properties and the biochemical behavior of the protein. The paper also links disease mutations to changes in filament assembly or filament conformation.
- 22. Hansen JM, Horowitz A, Lynch EM, Farrell DP, Quispe J, Dimaio F, Kollman JM: Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation. 2021, 10. * This paper describes how the pH sensitivity of CTP synthase from S. cerevisiae intersects with filament formation and shows that filaments slow substrate turnover. Point mutations of CTP synthase engineered into S. cerevisiae illustrate that filament dysregulation slows the growth of S. cerevisiae.
- 23. Hvorecny KL, Hargett KA, Quispe J, Kollman JM: Human PRPS1 filaments stabilize allosteric sites to regulate activity. bioRxiv 2022, doi: 10.1101/2022.06.27.496812. * This paper describes the filament structure and biochemical effects of filaments of human PRPP synthetase 1. Formation of filaments stabilizes the active or the inhibited conformations of the enzyme via stabilization of the allosteric site in complex with the ligand.
- 24.Zhang M, Zhang L, Guo R, Xiao C, Yin J, Zhang S, Yang M: Structural basis for the catalytic activity of filamentous human serine beta-lactamase-like protein LACTB. Structure 2022, 30:685–696.e5. [DOI] [PubMed] [Google Scholar]
- 25.Pollard TD, Goldman RD: Overview of the cytoskeleton from an evolutionary perspective. Cold Spring Harb Perspect Biol 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stoddard PR, Lynch EM, Farrell DP, Dosey AM, Dimaio F, Williams TA, Kollman JM, Murray AW, Garner EC: Polymerization in the actin ATPase clan regulates hexokinase activity in yeast. Science (1979) 2020, 367:1039–1032. * This paper dissects the effect filament formation has on the activity of Glucokinase 1 from S. cerevisiae, identifying filament formation as molecular surge protector within the cell.
- 27.He S, Scheres SHW: Helical reconstruction in RELION. J Struct Biol 2017, 198:163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunkeler M, Hagmann A, Stuttfeld E, Chami M, Guri Y, Stahlberg H, Maier T: Structural basis for regulation of human acetyl-CoA carboxylase. Nature 2018, 558:470–474. [DOI] [PubMed] [Google Scholar]
- 29.Lynch EM, Kollman JM: Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Nat Struct Mol Biol 2020, 27:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson MC, Kollman JM: Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jessop M, Huard K, Desfosses A, Tetreau G, Carriel D, Bacia-Verloop M, Mas C, Mas P, Fraudeau A, Colletier JP, et al. : Structural and biochemical characterisation of the Providencia stuartii arginine decarboxylase shows distinct polymerisation and regulation. Commun Biol 2022, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang PC, Chen SK, Chiang WH, Ho MR, Wu KP: Structural basis for the helical filament formation of Escherichia coli glutamine synthetase. Protein Science 2022, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chicano TM, Dietrich L, de Almeida NM, Akram M, Hartmann E, Leidreiter F, Leopoldus D, Mueller M, Sánchez R, Nuijten GHL, et al. : Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Nat Microbiol 2021, 6:1129–1139. ** This paper solves the tubule structure of Nitrite Oxidoreductase multiprotein complex found in a subset of bacteria, which produces almost all nitrate found in nature. Using a combination of X-ray crystallography, cryo electron tomography, and biochemical assays, the study reveals that a previously unknown, but related, protein triggers tubule formation and shows that these tubules are also found in cells.
- 34.Dey S, Levy ED: PDB-wide identification of physiological hetero-oligomeric assemblies based on conserved quaternary structure geometry. Structure 2021, 29:1303–1311.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Seisdedos H, Villegas JA, Levy ED: Infinite Assembly of Folded Proteins in Evolution, Disease, and Engineering. Angewandte Chemie 2019, 131:5568–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Seisdedos H, Empereur-Mot C, Elad N, Levy ED: Proteins evolve on the edge of supramolecular self-assembly. Nature 2017, 548:244–247. [DOI] [PubMed] [Google Scholar]
- 37. Garcia-Seisdedos H, Levin T, Shapira G, Freud S, Levy ED: Mutant libraries reveal negative design shielding proteins from supramolecular self-assembly and relocalization in cells. The Proceedings of the National Academy of Sciences 2022, 119:1–8. ** This paper demonstrates how mutations changing the surface chemistry of a protein can alter the protein’s ability to form supramolecular assemblies. Additionally, these same mutations can also affect the localization of the protein or protein assembly within the cell.
- 38.Edelstein SJ: Patterns in the quinary structures of proteins. Plasticity and inequivalence of individual molecules in helical arrays of sickle cell hemoglobin and tubulin. Biophys J 1980, 32:347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McConkey EH: Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc Natl Acad Sci USA 1982, 79:3236–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang N, An LL, Li J, Liu Z, Yao L: Quinary interactions weaken the electric field generated by protein side-chain charges in the cell-like environment. J Am Chem Soc 2017, 139:647–654. [DOI] [PubMed] [Google Scholar]
- 41.Khan T, Kandola TS, Wu J, Venkatesan S, Ketter E, Lange JJ, Rodríguez Gama A, Box A, Unruh JR, Cook M, et al. : Quantifying Nucleation In Vivo Reveals the Physical Basis of Prion-like Phase Behavior. Mol Cell 2018, 71:155–168.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gnutt D, Timr S, Ahlers J, König B, Manderfeld E, Heyden M, Sterpone F, Ebbinghaus S: Stability Effect of Quinary Interactions Reversed by Single Point Mutations. J Am Chem Soc 2019, 141:4660–4669. [DOI] [PubMed] [Google Scholar]
- 43.Ribeiro S, Ebbinghaus S, Marcos JC: Protein folding and quinary interactions: creating cellular organisation through functional disorder. FEBS Lett 2018, 592:3040–3053. [DOI] [PubMed] [Google Scholar]
- 44. Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Trischler R, Schuller SK, Wagner J, Schwarz FM, Engel BD, Müller V, et al. : Membrane-anchored HDCR nanowires drive hydrogen-powered CO2 fixation. Nature 2022, 607:823–830. * This paper describes the structure of the specialized metabolic subcompartment responsible for carbon dioxide fixation in some anaerobic, autotrophic bacteria. The authors trace the path of electron transfer through the filamentous assembly and demonstrate that filaments facilitate rapid electron transfer.
- 45.Fernández-Justel D, Núñez R, Martín-Benito J, Jimeno D, González-López A, Soriano EM, Revuelta JL, Buey RM: A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity. J Mol Biol 2019, 431:956–969. [DOI] [PubMed] [Google Scholar]
- 46.Chang CC, Peng M, Zhong J, Zhang Z, Keppeke GD, Sung LY, Liu JL: Molecular crowding facilitates bundling of IMPDH polymers and cytoophidium formation. Cellular and Molecular Life Sciences 2022, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch EM, Hicks DR, Shepherd M, Endrizzi JA, Maker A, Hansen JM, Barry RM, Gitai Z, Baldwin EP, Kollman JM: Human CTP synthase filament structure reveals the active enzyme conformation. Nat Struct Mol Biol 2017, 24:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrovska I, Nüske E, Munder MC, Kulasegaran G, Malinovska L, Kroschwald S, Richter D, Fahmy K, Gibson K, Verbavatz JM, et al. : Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. Elife 2014, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jessop M, Liesche C, Felix J, Desfosses A, Baulard M, Adam V, Fraudeau A, Huard K, Effantin G, Kleman J-P, et al. : Supramolecular assembly of the Escherichia coli LdcI upon acid stress. 2022, doi: 10.1073/pnas.2014383118/. [DOI] [PMC free article] [PubMed]
- 50.Hu JJ, Lee JKJ, Liu Y-T, Yu C, Huang L, Aphasizheva I, Aphasizhev R, Hong Zhou Z: Discovery, Structure, and Function of Filamentous 3-Methylcrotonyl-CoA Carboxylase. bioRxiv 2022, doi: 10.1101/2022.08.19.504621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F, Mustafa K, Suciu V, Joshi K, Chan CH, Choi S, Su Z, Si D, Hochbaum AI, Egelman EH, et al. : Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing. Nat Microbiol 2022, 7:1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim G, Azmi L, Jang S, Jung T, Hebert H, Roe AJ, Byron O, Song JJ: Aldehyde-alcohol dehydrogenase forms a high-order spirosome architecture critical for its activity. Nat Commun 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim G, Yang J, Jang J, Choi JS, Roe AJ, Byron O, Seok C, Song JJ: Aldehyde-alcohol dehydrogenase undergoes structural transition to form extended spirosomes for substrate channeling. Commun Biol 2020, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pony P, Rapisarda C, Terradot L, Marza E, Fronzes R: Filamentation of the bacterial bi-functional alcohol/aldehyde dehydrogenase AdhE is essential for substrate channeling and enzymatic regulation. Nat Commun 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen H, Fallas JA, Lynch E, Sheffler W, Parry B, Jannetty N, Decarreau J, Wagenbach M, Vicente JJ, Chen J, et al. : De novo design of self-assembling helical protein filaments. Science (1979) 2018, 362:705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linghu C, An B, Shpokayte M, Celiker OT, Shmoel N, Zhang C, Park WM, Ramirez S, Boyden ES: Recording of cellular physiological histories along optically readable self-assembling protein chains. [date unknown], doi: 10.1101/2021.10.13.464006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin D, Xiuyuan †, Li T, Park P, Tang, Shen H, Grimm JB, Falco N, Baker D, Lavis LD, et al. : Time-tagged ticker tapes for intracellular recordings. bioRxiv 2021, doi: 10.1101/2021.10.13.463862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amara N, Cooper MP, Voronkova MA, Webb BA, Lynch EM, Kollman JM, Ma T, Yu K, Lai Z, Sangaraju D, et al. : Selective activation of PFKL suppresses the phagocytic oxidative burst. Cell 2021, 184:4480–4494.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burrell AL, Kollman JM: IMPDH dysregulation in disease: A mini review. Biochem Soc Trans 2022, 50:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lynch EM, Dimattia MA, Albanese S, van Zundert GCP, Hansen JM, Quispe JD, Kennedy MA, Verras A, Borrelli K, Toms A v, et al. : Structural basis for isoform-specific inhibition of human CTPS1. The Proceedings of the National Academy of Sciences 2021, 118:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


