Abstract
Salmonella enterica is one of the most widespread bacterial pathogens found worldwide, resulting in approximately 100 million infections and over 200,000 deaths per year. Salmonella isolates, termed “serovars”, can largely be classified as either non-typhoidal or typhoidal Salmonella, which differ in regard to disease manifestation and host tropism. Non-typhoidal Salmonella causes gastroenteritis in many hosts, while typhoidal Salmonella is human-restricted and causes typhoid fever, a systemic disease with a mortality rate of up to 30% without treatment. There has been considerable interest in understanding how different Salmonella serovars cause different diseases, but the molecular details that underlie these infections have not yet been fully characterized, especially in the case of typhoidal Salmonella. In this review, we highlight the current state of research into understanding the pathogenesis of both non-typhoidal and typhoidal Salmonella, with a specific interest in serovar-specific traits that allow human-adapted strains of Salmonella to cause enteric fever. Overall, a more detailed molecular understanding of how different Salmonella isolates infect humans will provide critical insights into how we can eradicate these dangerous enteric pathogens.
Keywords: enteric fever, diarrhea, salmonella, Evolution, virulence, pseudogenes, pathogenesis, persistence, granulomas, macrophages, macrophage polarization
1. Introduction to typhoidal Salmonella
Salmonella enterica subspecies enterica is comprised of over 2,400 serovars that infect animal hosts via the fecal-oral route, resulting in a range of disease states ranging from self-limiting gastroenteritis to lethal systemic infection [1]. Typhoidal Salmonella serovars S. Typhi and S. Paratyphi A are clinically important, causing over 200,000 deaths world-wide and a significant burden on developing, under-resourced healthcare systems [2]. The H58 lineage of S. Typhi is particularly problematic, as it is strongly associated with antimicrobial resistance and is rapidly becoming the most prevalent typhoidal lineage across the world [3]. Most individuals infected by typhoid fever live in South and Southeast Asia [4], and the emergence of an extensively drug-resistant (XDR) strain in these densely populated areas is extremely worrisome [5]. Infections with Typhi and Paratyphi A, which are human-restricted pathogens, usually begin with ingestion of contaminated water or food. The pathogens invade the gastrointestinal mucosa, translocate to the lymphoid follicles where they survive and replicate within macrophages, and then disseminate to the intestinal lymph nodes, liver, spleen, bone marrow and gallbladder [4]. Vaccinations against typhoid fever are available but differ in regard to the protection they offer: the live oral Ty21a vaccines elicits some level of cross-protection against multiple strains of Salmonella [6], while the typhoid conjugate vaccines do not provide protection against paratyphoid fever, a cause of one in five cases of enteric fever in some areas of Asia [7]. Thus, an increase in paratyphoid fever cases in endemic areas is predicted to occur [8].
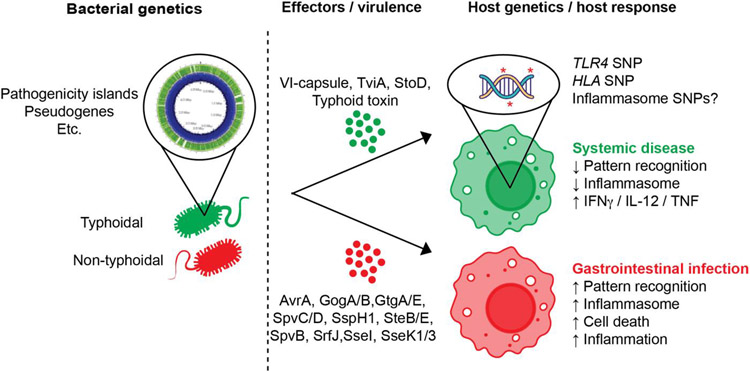
The restriction of Typhi and Paratyphi A to human hosts limits experimental approaches available to study the pathogenesis of enteric fever caused by these serovars. Typhimurium infection in mice is commonly considered a model system for studying the pathogenesis of typhoid and paratyphoid fever, but important differences among these serovars exist. For example, while both non-typhoidal and typhoidal Salmonella encode two type 3 secretion systems (T3SSs) on Salmonella pathogenicity islands (SPI) 1 and 2, which are required for translocating effector proteins into mammalian cells, prior studies have shown that the SPI-2 T3SS is not required for the intracellular survival and replication of typhoidal Salmonella in the human macrophage (MΦ) cell line THP-1 [9,10] or for infection in a humanized mouse model [11]. In addition, many T3SS effector genes are either absent or pseudogenes in Typhi and Paratyphi A, and thus the functions of T3SS-1 and −2 and their effectors may be absent or altered in Typhi and Paratyphi A [1]. In this review we will discuss the genetic basis for divergent pathogenicity between serovars, key bacterial virulence factors underlying typhoid and paratyphoid infection, and the human immune basis for heterogeneous clinical outcomes (Fig. 1).
Figure 1. Summary figure highlighting some key known differences in the pathogenesis between typhoidal and non-typhoidal Salmonella.
Typhoidal and non-typhoidal serovars of Salmonella are genetically similar but have distinct features that lead to divergent disease phenotypes. For example, typhoidal and non-typhoidal serovars genomes differ in the repertoire of pathogenicity islands and pseudogenes that they contain. In addition, these genetic variations give rise to key differences in secreted effectors and virulence factors that influence the intracellular survival of Salmonella serovars. Finally, on the host side, SNPs in key genes that influence pattern recognition and antigen presentation may help typhoidal serovars evade local responses in the gastrointestinal tract and spread to systemic sites, triggering either enteric fever and/or chronic persistent infections.
2. Evolution of human-restricted Salmonella serovars: divergence of typhoidal serovars from generalist serovars
The genetic basis underlying host-restriction of divergent Salmonella enterica serovars is a topic of considerable interest. While typhoidal and non-typhoidal Salmonella share >90% of genes, these serovars also encode hundreds of unique genes that may underpin differences in disease manifestations [12]. Many serovar-specific genes are within integrated prophage regions, which represent important sites of genetic diversity across bacterial genomes (Table 1). Prophages often encode “cargo” genes that can significantly impact microbial behavior and virulence. For example, multiple genes encoding type III secretion system (T3SS) effectors are contained on prophage regions across Salmonella serovars, including sopE, sspH1, sseI, and sopE2. The importance of T3SSs during typhoidal Salmonella virulence is discussed in more detail in Section 3 of this review. Other serovar-specific genes are found on Salmonella Pathogenicity Islands (SPIs), which are large, horizontally acquired genetic cassettes that often encode virulence genes (Table 1). For example, SPI-7,−15,−17 and −18 are unique to typhoidal serovars, while SPI-14 is only found in non-typhoidal isolates. SPI-7 is an especially important region of the S. Typhi genome that encodes the Vi antigen, a capsular polysaccharide that coats the surface of the bacterium and contributes to virulence. More specifically, the Vi capsule prevents complement binding [13] and masks lipopolysaccharide (LPS) from TLR-4 recognition [14], thereby avoiding immune recognition. The Vi capsule has also recently been found to promote phagocytosis of S. Typhi by human macrophages while simultaneously allowing S. Typhi to evade phagocytosis by human neutrophils [15]. TviA, the transcriptional activator of Vi production, is contained within SPI-7 and can also suppress the expression of flagellin and T3SS-1, thereby allowing S. Typhi to evade TLR-5 and NAIP-mediated inflammatory responses [16]. Together, these findings suggest that Vi capsule production contributes to a “stealth” S. Typhi phenotype, which may allow S. Typhi to disseminate to systemic sites without eliciting strong inflammatory immune responses. Interestingly, S. Paratyphi A also causes enteric fever but does not encode the Vi capsule. Instead, S. Paratyphi A encodes fepE, which leads to the generation of very long O-antigen chains that help this pathogen escape the immune response by limiting inflammasome activation and subsequent cell death [17]. While non-typhoidal strains of Salmonella also encode fepE, previous studies have shown that S. Paratyphi A expresses fepE at significantly higher levels compared to S. Typhimurium [17]. Modification of surface antigens may therefore be a convergently evolved mechanism of Typhi and Paratyphi A to evade the human immune system.
Table 1.
Horizontally Acquired Regions of the S. Typhi Ty2 Chromosome
| Name | Length | Key Virulence Features |
|---|---|---|
| SPI-1 | 40,278 bp | T3SS-1; invasion, host cell modulation |
| SPI-2 | 39,633 bp | T3SS-2; intracellular replication, host cell modulation |
| SPI-3 | 36,845 bp | Magnesium uptake genes, many pseudogenes in S. Typhi |
| SPI-4 | 23,392 bp | SiiE adhesin & T1SS |
| SPI-5 | 7,408 bp | SopB, PipA, PipB |
| SPI-6 | 57,634 bp | T6SS; gut colonization & unique 10 kb in Typhi |
| SPI-7 | 131,750 bp | Unique to Typhi; capsule biosynthesis, SopEφ, type IVB pilus |
| SPI-8 | - | (Unique 8kb part of SPI-13) |
| SPI-9 | 15,696 bp | T1SS, putative adhesin |
| SPI-10 | 33,675 bp | Unique to Typhi; contains ST46 |
| SPI-11 | 10,080 bp | PhoP-activated genes, typhoid toxin |
| SPI-12 | 6,439 bp | SspH2 |
| SPI-13 | 24,963 bp | An 8 kb portion is different between Typhi and Typhimurium |
| SPI-15 | 6,009 bp | Absent form Typhimurium, present in Typhi |
| SPI-16 | 4,087 bp | Three ORFs with a high level of identity with P22 phage genes |
| SPI-17 | 4,939 bp | Absent from Typhimurium, present in Typhi |
| SPI-18 | 1,678 bp | Absent from Typhimurium, present in Typhi; ClyA & TaiA |
| ST10 | 42,759 bp | Absent from Typhimurium, present in Typhi (prophage) |
| ST15 | 32,588 bp | Absent from Typhimurium, present in Typhi (prophage) |
| ST27 | 22,785 bp | Salmochelin iron acquisition operon (iroCDEN), PipB2, VirK |
| ST2-27 | 10,268 bp | Absent from Typhimurium, present in Typhi (prophage) |
| ST35 | 35,194 bp | Absent from Typhimurium, present in Typhi (prophage) |
| ST46 | 7,055 bp | Absent from Typhimurium, present in Typhi (prophage) |
| CS54 | 24,674 bp | RatA, SivI |
| SopEφ (within SPI-7) | 33,971 bp | Present in some Typhimurium strains, present in Typhi Ty2 strain |
Several pathogenicity islands are also shared between typhoidal and non-typhoidal Salmonella, but there are many serovar-specific genes encoded within these regions. For example, SPI-11 is present in both non-typhoidal and typhoidal serovars, but this island contains genes that encode the typhoid toxin in S. Typhi and S. Paratyphi A, including cdtB and pltAB [18]. While the molecular details of how typhoid toxin benefits Salmonella in the host are still being investigated, previous work has shown that the typhoid toxin causes DNA damage and host cell cycle arrest in cultured intestinal epithelial cells [19] and in human primary gallbladder cells [20]. There are also observations linking the toxin to gallbladder cancer through a mechanism where the CdtB subunit causes DNA damage and subsequent activation of the p53/P21 axis [21], and indeed previous work has shown that patients who are carriers of typhoidal Salmonella are significantly more likely to develop carcinomas in the gallbladder compared to non-carriers [22]. In addition, researchers have shown that introducing typhoid toxin-encoding genes to S. Typhimurium increases the frequency at which asymptomatic carriers emerge in long-term murine models of Salmonella infection [23], while another study assessing a two-day infection of S. Typhi in humanized mice did not observe a typhoid toxin-dependent effect [11]. In agreement with this mouse study, one recent study in humans has suggested that the typhoid toxin is not required for virulence at the acute stage of typhoid fever [24]. Together, these studies suggest that this toxin may play a role in systemic virulence and long-term persistent infection, but may not be necessary during early stages of typhoid fever.
Another distinguishing feature of typhoidal serovars is the large proportion of pseudogenes that are present in their genomes, which is a hallmark of genome degradation. Approximately 5% of the genome in sequenced typhoidal isolates are predicted to be pseudogenes, which arise either from single base mutations that result in frameshift mutations, or from the appearance of early stop codons [12]. These pseudogenes include loci that encode many different functions, including metabolic genes (ie, narW, wcaK), transporters (i.e., fhuA, mglA), chemotaxis genes (i.e., trg), and cell-wall related genes (i.e., dacD) [25,26]. One study has shown that the loss of the loss of the ydiQRSTD operon in S. Typhi, which is responsible for the utilization of butyrate by non-typhoidal Salmonella, may have contributed to the transition of S. Typhi from a gastrointestinal to an extraintestinal pathogen [27]. Interestingly, there are multiple virulence genes in S. Typhimurium that are pseudogenes in S. Typhi and/or S. Paratyphi A, including several SPI-1 and SPI-2 T3SS effectors (i.e., sopA, sseI, steB, sopE2, sseK2, and sopD2) [25]. sseI is also a pseudogene in non-typhoidal Salmonella belonging to the ST313 lineage, which causes systemic bacteremia in humans [28]; these invasive non-typhoidal Salmonella infections have been reviewed elsewhere extensively [29-32]. Previous work has shown that the loss of SseI contributes to the ability of ST313 Salmonella to hyperdisseminate from the gut to systemic sites [33] and thus the pseudogenization of sseI may contribute to the ability of ST313 Salmonella to cause systemic bacteremia. In addition, many genes encoded by non-typhoidal Salmonella that contribute to colonization of the gut are pseudogenes in S. Typhi and S. Paratyphi A, including the autotransporter misL, the adhesin-encoding gene siiE, and the type VI secretion system (T6SS) genes sciI and sciS [12]. It is likely that the loci mutated in S. Typhi and S. Paratyphi A are not necessary for systemic disease in humans. In agreement with this hypothesis, many of these virulence-related pseudogenes have been shown in S. Typhimurium to promote intestinal persistence, which is not a hallmark of typhoidal serovars [12]. More generally, gene loss has been found to result in fundamental changes in the behavior of numerous bacterial pathogens and can even enhance virulence phenotypes [34-38]. In the case of S. Typhi, it has been reported that the loss of the fepE gene enhances immune evasion mediated by the Vi capsule [39].
Although both S. Typhi and S. Paratyphi A encode many pseudogenes, there is little overlap in pseudogenes between these typhoidal serovars; for example, less than 20% of pseudogenes in S. Paratyphi A 9150 A are also pseudogenes in S. Typhi CT18 [25]. This observation strongly suggests that S. Typhi and S. Paratyphi A have evolved independently to converge on the same disease phenotype. Based on the high number of pseudogenes in these serovars, it is likely that both S. Typhi and S. Paratyphi A primarily evolved via reductive evolution from a non-typhoidal ancestor, losing genes that are no longer necessary for survival or intracellular replication during the progression of typhoid fever [40]. Interestingly, reductive evolution appears to be a feature of other human-restricted pathogens including Mycobacterium leprae, Shigella flexneri, and human-restricted strains of Bordetella spp [41-43]. In addition, recent work has suggested that some bioinformatically predicted pseudogenes in typhoidal Salmonella may produce functional proteins, although this finding should be confirmed experimentally by other groups [44]. Thus, a more careful analysis of pseudogenes in the above pathogens may yield deeper insights into the molecular mechanisms by which human-restricted pathogens evolve to cause disease.
3. Molecular mechanisms underlying typhoid and paratyphoid pathogenesis
As previously mentioned, Salmonella encodes two T3SS, which are conserved between non-typhoidal and typhoidal Salmonella [45]. However, the repertoire of T3SS effectors differ between these serovars (Table 2). The roles that T3SS-secreted effectors play are best described for Typhimurium infections – ranging from manipulation of host cytoskeleton to immune evasion, intracellular trafficking and cell survival [45-47]. Differences in expression of T3SS and the effector repertoire between typhoidal and non-typhoidal serovars will be highlighted here.
Table 2.
T3SS-Dependent Effectors in the Typhi Ty2 Genome
| Effector | Ty2 Locus | CT18 Locus | STm LT2 Locus | Paratyphi A 9150 Locus |
Locus Location |
T3SS-1 | T3SS- 2 |
|---|---|---|---|---|---|---|---|
| AvrA | ⊘ | ⊘ | STM2865 | ⊘ | SPI-1 | ✔ | ✔ |
| GogB | ⊘ | ⊘ | STM2584 | ⊘ | Gifsy-1 | ✔ | ✔ |
| GtgA | ⊘ | ⊘ | STM1026 | ⊘ | Gifsy-2 | ✔ | ✔ |
| GtgE | ⊘ | ⊘ | STM1055 | ⊘ | Gifsy-2 | ✔ | ✔ |
| PipB2 | T_RS13590 | STY2897 | STM2780 | SPA_RS13395 | ST27 | ✔ | ✔ |
| SlrP | T_RS10620 | STY0833 | STM0800 | SPA_RS09800 | - | ✔ | ✔ |
| SpvC | ⊘ | ⊘ | PSLT038 | ⊘ | pSLT | ✔ | ✔ |
| SpvD | ⊘ | ⊘ | PSLT037 | ⊘ | pSLT | ✔ | ✔ |
| SspH1 | ⊘ | ⊘ | STM14_RS07010 | ⊘ | Gifsy-3 | ✔ | ✔ |
| SteA | T_RS07605 | STY1482 | STM1583 | SPA_RS06440 | - | ✔ | ✔ |
| SteB | ⊘ | ⊘ | STM1629 | ⊘ | - | ✔ | ✔ |
| SteE (SarA) | ⊘ | ⊘ | STM2585 | ⊘ | Gifsy-1 | ✔ | ✔ |
| SipA | T_RS14125 | STY3005 | STM2882 | SPA_RS13925 | SPI-1 | ✔ | |
| SipB | T_RS14140 | STY3008 | STM2885 | SPA_RS13940 | SPI-1 | ✔ | |
| SipC | T_RS14135 | STY3007 | STM2884 | SPA_RS13935 | SPI-1 | ✔ | |
| SipD | T_RS14130 | STY3006 | STM2883 | SPA_RS13930 | SPI-1 | ✔ | |
| SopA | T_RS04065 | STY2275 | STM2066 | SPA_RS04025 | - | ✔ | |
| SopB | T_RS09305 | STY1121 | STM1091 | SPA_RS08830 | SPI-5 | ✔ | |
| SopD | T_RS14435 | STY3073 | STM2945 | SPA_RS14235 | - | ✔ | |
| SopE | T_RS21905 | STY4609 | SL1344_RS13925 | SPA_RS12970 | SopEφ | ✔ | |
| SopE2 | T_RS05225 | STY1987 | STM1855 | SPA_RS05115 | - | ✔ | |
| SopF | T_RS05655 | STY1893 | STM1239 | SPA_RS08080 | SPI-11 | ✔ | |
| SptP* | T_RS14105 | STY3001 | STM2878 | SPA_RS13905 | SPI-1 | ✔ | |
| StoD | T_RS09505 | STY1076 | ⊘ | ⊘ | ST10 | ✔ | |
| GogA | ⊘ | ⊘ | STM2614 | ⊘ | Gifsy-1 | ✔ | |
| CigR | T_RS19110 | STY4024 | STM3762 | SPA_RS18310 | SPI-3 | ✔ | |
| PipA | T_RS09330 | STY1115 | STM1087 | SPA_RS08855 | SPI-5 | ✔ | |
| PipB | T_RS09325 | STY1117 | STM1088 | SPA_RS08850 | SPI-5 | ✔ | |
| SifA | T_RS08635 | STY1264 | STM1224 | SPA_RS08155 | - | ✔ | |
| SifB | T_RS07700 | STY1462 | STM1602 | SPA_RS06345 | - | ✔ | |
| SopD2 | T_RS09990 | STY0971 | STM0972 | SPA_RS09170 | - | ✔ | |
| SpiC | T_RS06440 | STY1727 | SL1344_RS06920 | SPA_RS07330 | SPI-2 | ✔ | |
| SpvB | ⊘ | ⊘ | PSLT039 | ⊘ | pSLT | ✔ | |
| SrfA | T_RS07655 | STY1472 | STM1593 | SPA_RS06390 | - | Pred. | |
| SrfB | T_RS07660 | STY1471 | STM1594 | SPA_RS06385 | - | Pred. | |
| SrfC | T_RS07665 | STY1470 | STM1595 | SPA_RS06380 | - | Pred. | |
| SrfJ | ⊘ | ⊘ | STM4426 | ⊘ | - | ✔ | |
| SseB | T_RS06465 | STY1722 | STM1398 | SPA_RS07305 | SPI-2 | ✔ | |
| SseC | T_RS06475 | STY1720 | STM1400 | SPA_RS07295 | SPI-2 | ✔ | |
| SseD | T_RS06480 | STY1719 | STM1401 | SPA_RS07290 | SPI-2 | ✔ | |
| SseF | T_RS06495 | STY1716 | STM1404 | SPA_RS07275 | SPI-2 | ✔ | |
| SseG | T_RS06500 | STY1715 | STM1405 | SPA_RS07270 | SPI-2 | ✔ | |
| SseI (srfH) | ⊘ | ⊘ | STM1051 | ⊘ | Gifsy-2 | ✔ | |
| SseJ | T_RS25885 | STY_RS06750 | STM1631 | ⊘ | - | ✔ | |
| SseK1 | ⊘ | ⊘ | STM4157 | ⊘ | - | ✔ | |
| SseK2 | T_RS23900 | STY_RS11125 | SL1344_RS10980 | SPA_RS23070 | - | ✔ | |
| SseK3 | ⊘ | ⊘ | SL1344_RS10000 | ⊘ | ST64B | ✔ | |
| SseL | T_RS02915 | STY2517 | SL1344_RS11745 | SPA_RS02885 | - | ✔ | |
| SspH2 | T_RS03160 | STY2467 | STM2241 | ⊘ | SPI-12 | ✔ | |
| SteC | T_RS08195 | STY1353 | STM1698 | SPA_RS05905 | - | ✔ | |
| SteD | T_RS03630 | STY2367 | STM2139 | SPA_RS03565 | - | ✔ |
Grey box = Gene has a frameshift or early truncation in S. Typhi relative to S. Typhimurium, assumed to be non-functional
= The sptP gene is mutated and non-functional in S. Typhi
⊘ = Gene is absent from this serovar
✔ = S. Typhimurium translocation of this effector is dependent on T3SS-1, −2 or both. “Pred.” means predicted but not yet directly demonstrated.
If there is no homolog in S. Typhimurium str. LT2, homolog in strain SL1344 or 14028 given instead.
T3SS-1, encoded in the SPI-1 locus, enables Salmonella species to gain a foothold in the gastrointestinal tract by triggering Salmonella uptake by non-phagocytic cells, including epithelial cells. Previous work has shown that the regulation of T3SS-1 dependent invasion may differ between non-typhoidal and typhoidal Salmonella. For example, Winter et. al. found that TviA, a transcriptional regulator that is only present in S. Typhi, positively regulates Vi capsule production and represses T3SS-1 expression [48]. In addition, S. Typhi up-regulates T3SS-1 and invasion of epithelial cells in response to bile, which is present in the gallbladder, whereas S. Typhimurium does not [49], which may partly explain why the gallbladder is a site of chronic S. Typhi infections in humans [50].
In addition to differences in T3SS regulation, the effectors encoded on these virulence loci also differ between non-typhoidal and typhoidal Salmonella (Table 2). For example, previous work has shown that many T3SS-1 effectors contribute to intestinal inflammation in S. Typhimurium [51], but some of these effectors are pseudogenes in S. Typhi and S. Paratyphi A. In turn, one hypothesis for why typhoidal serovars do not cause acute intestinal inflammation is genetic degradation of the specific T3SS-1 dependent effectors. For example, six T3SS-1 effectors, SipA, SopA, SopB, SopD, SopE, and SopE2, elicit neutrophil infiltration and fluid accumulation during S. Typhimurium infection of bovine illeal loops [52], but SopA and SopE2 are pseudogenized in S. Typhi [53]. Another potential hypothesis for reduced intestinal inflammation is that typhoidal serovars encode unique genes to specifically dampen inflammatory responses during intestinal infection. Interestingly, a T3SS-1 dependent effector present in S. Typhi, named StoD, is an E3/4 ubiquitin ligase that likely causes degradation of host targets and may play a role in dampening immune responses [54].
After adhering to the surface of epithelial cells, T3SS-1 injects effector proteins into the host cell cytoplasm to facilitate internalization. S. Typhi and S. Typhimurium both induce T3SS-1 dependent ruffling of epithelial cell surfaces upon invasion, indicating a conserved “trigger” mechanism of invasion caused by cytoskeletal rearrangement. The injected effectors SipA and SipC directly nucleate actin [55], while SopE and SopE2 initiate actin polymerization through host GTPases Cdc42 and Rac1[56]. Notably, SipA, SipC and SopE are conserved in typhoidal serovars while SopE2 is pseudogenized [53]. The T3SS-1 dependent effector SptP is deployed by S. Typhimurium to antagonize SopE-mediated cytoskeletal rearrangements by degrading Cdc42 & Rac1 and return the cytoskeleton to homeostasis after internalization occurs [57]. However, the sptP ORF within S. Typhi contains a point mutation rendering the protein non-functional [58]. Therefore, the mechanism by which S. Typhi resolves host cytoskeletal rearrangements post-invasion is currently unclear.
While T3SS-1 dependent invasion is required to enter nonphagocytic cells, macrophages naturally phagocytose pathogens, and thus SPI-1 mediated invasion is not required for Salmonella internalization [59]. Paradoxically, macrophages, which typically kill phagocytosed pathogens, are a major replicative niche for Salmonella during systemic infection [60]. Salmonella uses multiple molecular mechanisms to facilitate intra-macrophage replication. Effector proteins deployed through the T3SS-2 are required for S. Typhimurium intra-macrophage replication within the Salmonella-containing vacuole (SCV) [61]. However, T3SS-2 is not strictly required for S. Typhi intracellular survival and/or replication in THP-1 macrophages [9,10]. The molecular mechanisms underlying this discrepancy between non-typhoidal and typhoidal serovars remain unclear.
Most of the mechanisms by which Salmonella survive and replicate within a vacuole inside of macrophages has been performed with S. Typhimurium [62]. For example, previous studies have shown that two T3SS-2 dependent effectors, SseF and SseG, play an important role in intracellular trafficking of the SCV [63], and these effectors are conserved in typhoidal Salmonella. It has also been shown in S. Typhimurium that multiple effectors, including SifA, PipB2, SopD2 and SseJ, contribute to growth and elongation of the SCV membrane to form Salmonella-induced filaments (Sifs) [64]. However, these Sif-inducing effectors are not universally conserved; for example, SseJ, which normally localizes to the SCV surface and recruits SifA in Typhimurium, is a pseudogene in typhoidal Salmonella [65]. Similarly, the gene that encodes SopD2, which plays a role in limiting Sif extension and ensuring optimal membrane dynamics, is also a pseudogene in typhoidal Salmonella [26]. Whether S. Typhi and S. Paratyphi A genomes contain different effectors that function similarly to SseJ and SopD2 is currently unclear. In S. Typhimurium, SopD2 also works cooperatively with another effector protein, GtgE, which is absent in typhoidal serovars [66]. GtgE degrades various Rab GTPases, which typically regulate many components of membrane trafficking in eukaryotic cells and may deliver antimicrobial factors to the SCV during Salmonella infection [67]. Previous work has shown that the presence of Rab32 leads to the death of S. Typhi in mouse macrophages, which may partly explain why typhoidal Salmonella cannot infect mice [66]. In agreement with this model, trans-expression of GtgE from S. Typhimurium allows S. Typhi to overcome host restriction by removing Rab32 from the SCV surface and replicate in murine macrophages [66]. While typhoidal Salmonella does not encode GtgE, one recent study has suggested that S. Typhi utilizes its SPI-1 T3SS to evade Rab32-mediated killing in human macrophages [68]. The molecular mechanisms by which S. Typhi can overcome the same Rab-mediated killing in human macrophages but not in murine macrophages are currently unclear.
While both typhoidal and non-typhoidal Salmonella survive and thrive within macrophages, the fate of intracellular Salmonella in human macrophages are heterogeneous, and include host-killed, non-replicating, persisting, and actively replicating intracellular bacteria [69]. As demonstrated with S. Typhimurium, macrophages with host-killed bacteria and bystander cells share a transcriptomic phenotype that is characterized by an M1 polarization state [70]. In contrast, macrophages that allow for intracellular replication and non-replicating persisting bacteria tend to have a transcriptomic profile dominated by M2 polarization genes [70]. Thus, the state of individual macrophages influences the ability of intracellular Salmonella to replicate and manipulating macrophage activation states can alter the overall outcome of Salmonella infection. Whether or not these bacterial populations and/or macrophage polarization states are different during typhoidal vs. non-typhoidal Salmonella infections is currently an ongoing area of investigation. Determining the relative sizes of each of these populations, their role in maintaining persistent infection, and characterizing the molecular mechanisms underlying each state may reveal additional serovar-specific differences during macrophage infection.
4. Host factors that influence the outcome of disease
While multiple differences have been identified between non-typhoidal and typhoidal Salmonella that may influence disease manifestation, as highlighted in the above sections, there are also numerous studies investigating differences in host factors and genetic predispositions that may influence disease severity in the infected host. A variety of pathogen-associated molecular patterns (PAMPs) in both typhoidal and non-typhoidal Salmonella interact with extracellular and intracellular pattern recognition receptors (PRRs). For example, TLRs including TLR2, TLR4 and TLR5 recognizes CsgA, LPS and FliC, respectively [71-73]. As these receptors are engaged, the adaptor proteins TIRAP/MyD88 or TRIF/TRAM are phosphorylated and initiate distinct signaling cascades that lead to cytokine and chemokine production, which recruit and activate myeloid and lymphoid cells necessary to clear the infection [74]. Consequently, Tlr2−/−, Tlr4−/− and Myd88−/− mice have distinct disease phenotypes and succumb to infection within days of S. Typhimurium infection [75]. TLRs are highly involved in inducing inflammasome formation and the subsequent processing and release of IL-1 superfamily proteins which is the hallmark of pyroptosis [76]. As such, inflammasome deficient mice, including Casp1−/−, Il18−/− and Il1b−/− are more susceptible to S. Typhimurium infection than wild type control mice [77]. In addition, one study has shown that TLR11 is responsible for host restriction of S. Typhi in mice [78], although other groups have failed to replicate this finding [79].
Other factors beyond PRRs have also been shown to influence host susceptibility to Salmonella. For example, a point mutation in the Nramp1 allele, which regulates iron-availability in macrophages, has also been shown to increase the susceptibility of conventional C57BL/6J and BALB/C mice to Salmonella infection, and C57BL/6 mice complemented with the wild type Nramp1 allele have increased survival compared to WT controls [80]. In addition, MHC-II deficient mice that fail to present Salmonella antigens to host immune cells are more susceptible to infection than wild type mice [81]. Some of these afore-mentioned genes have further been implicated to play a role in the susceptibility to typhoid fever in humans, suggesting that there are shared immunological pathways that are important for causing more severe disease in both humans and animals. For example, SNPs in both TLR4 [82] and HLA genes [83] has been shown to influence typhoid susceptibility. In contrast, there are some genes that may influence infection outcomes differently in different hosts. For example, while Nramp1 is important for control of non-typhoidal Salmonella infections in mice [80,84], there are no reports that SNPs in the human homologue SLC11A1 influence typhoid susceptibility [85]. In addition, SNPs in IL4 [86], PARK2 [87], PACRG [87], CFTR [88] and VAC14 [89] have been implicated to increase the susceptibility to typhoidal Salmonella infection in humans, but they have not been shown to have effects in murine models. Studies have also shown that defects in the IFNγ-IL12 axis are risk factors for invasive non-typhoidal Salmonella infections, but not for typhoid fever [90-92].
While mouse models are commonly used to study non-typhoidal Salmonella infections in the laboratory, there are critical differences between human and murine cells that are important to consider. For example, Salmonella infection activates the NAIP/NLRC4 inflammasome in both human and murine macrophages, but these responses are distinctly different. Mice encode multiple NAIPs which are constitutively expressed by macrophages to sense bacterial ligands in the cytosol while humans encode a single NAIP [93]. The NLRC4/NAIP inflammasome recognizes SPI-1 and SPI-2 associated proteins (PrgI, PrgJ and SsaG) as well as flagellin [94]. For human NAIP to recognize flagellin, a full-length isoform is needed which is expressed in hMDMs but not in monocytic/macrophage cell lines such as THP-1 or U937 cells [95]. On the pathogen side, Salmonella serovars have evolved different mechanisms to counteract host recognition and inflammasome activation. For example, non-typhoidal strains of Salmonella within the ST313 lineage trigger less inflammasome activation compared to serovars belonging to the ST19 lineage [28]. Differences in these responses have been attributed to lower expression of the SPI-1 effector SopE2 and the flagellin FliC in ST313 strains compared to ST19 isolates during host cell infection [28]. Typhoidal Salmonella also triggers lower amounts of inflammasome activation compared to non-typhoidal Salmonella, in part due to the presence of the TviA transcriptional regulator, which is uniquely found in typhoidal Salmonella. TviA decreases the expression of flagellin during host cell infections by S. Typhi, in turn suppressing inflammasome activation and subsequent IL-1β and pyroptosis responses [16].
One hallmark of typhoid fever is that ~5-10% of people carry Salmonella persistently for more than 1 year and thus serve as a reservoir of infection for this disease [96]. Although our understanding of the mechanisms that underlie typhoidal persistence remains limited, it has been shown that typhoidal Salmonella can persist in the gallbladders of chronic carriers [97]. Intriguingly, bile, which is found at high concentrations in the gallbladder, has been shown to trigger increased biofilm formation in typhoidal Salmonella [98], which may partly explain how this pathogen is able to persist in the gallbladder. It has also been reported that not all chronic carriers of typhoidal Salmonella have gallstones [99], strongly suggesting that there are other factors that contribute to the persistent stage of this infection. However, given the human-restricted nature of typhoidal Salmonella, the chronic phase of this disease has remained difficult to study. Instead, researchers have used S. Typhimurium infections of the 129x1/svJ mouse model to study persistent infections, as S. Typhimurium can persist in tissues of these mice for at least one year [84]. Data from long term studies in 129x1/svJ mice indicate that there is a shift from TH1 driven response at early time points, characterized by IL-1β, TNFα, IFNγ and IL-12 production, to TH2 driven responses during persistent infection [100]. Although there are no proteomic studies on the cytokine profile in human carriers of typhoidal Salmonella, several studies have demonstrated that acute typhoid fever is also accompanied by high serum levels of pro-inflammatory cytokines including IFNγ, IL-1, IL-6 and IL-8 [101-104]. While the cytokine response during the persistent phase of typhoid fever is currently unclear, some case reports suggest that children with prolonged Mycobacterium and Salmonella co-infection have reduced IFN-receptor- or IL-12 receptor expression in isolated blood cells, suggesting that functional interferon signaling is essential for the host to counteract long-term Salmonella infection [105-107].
One key feature of persistent Salmonella infections is the formation of granulomas, a structure formed to contain macrophages infected by Salmonella that cannot be cleared by the host. Salmonella driven formation of granulomas has been found in the MLN, liver and spleen in humans infected with typhoid fever, and thus granulomas may represent another mechanism by which the host influences Salmonella persistence [108-110]. These granuloma structures have also been observed during S. Typhimurium infection of 129x1/SvJ mice, and in this chronic infection model, granulomas form rapidly, with observable structures found by 1 week post infection [111]. Splenic granulomas are characterized by a lack of TH1 cells, which allows Salmonella to persist and replicate in the activated macrophages [112]. Recent studies have also shown that S. Typhimurium facilitates the use of the effector protein SteE to polarize macrophages into an M2-like, anti-inflammatory phenotype within splenic granulomas, allowing Salmonella to persist within these structures during long-term infection [111,113]. Intriguingly, there is no homolog of steE encoded in the genomes of typhoidal Salmonella, and thus more research is needed to identify typhoidal-specific effectors that allow typhoidal Salmonella to persist within human granulomas.
5. Conclusion
As extensively antibiotic-resistant outbreaks of typhoid and paratyphoid fever increase in frequency and magnitude, a deeper understanding of human typhoidal disease is critical for the development of new therapies and vaccines. Identifying specific molecular mechanisms unique to typhoidal pathogenesis is key to developing anti-virulence strategies for targeted treatment. While much progress has been made in characterizing the molecular mechanisms that underlie Salmonella infections, especially in the context of non-typhoidal Salmonella, many questions remain. For example, while many genetic differences have been identified between non-typhoidal and typhoidal Salmonella, how do these changes in genotypes relate to differences in phenotype or disease manifestation? How do typhoidal strains of Salmonella replicate within human macrophages, and are there typhoid-specific virulence factors that play a role in these infections? What are the specific molecular determinants that allow typhoidal Salmonella to persist in human hosts for years, and are there better ways that we can model these human-adapted infections? Overall, by studying human-restricted typhoidal Salmonella in a variety of human-associated model systems, including human macrophages, human-derived intestinal organoids, and humanized mice, researchers may uncover human-specific mechanisms of virulence. In addition, understanding the human immune response to these “stealth” pathogens will likely reveal novel mechanisms of immune modulation.
Highlights.
Salmonella enterica is a bacterial pathogen that is comprised of non-typhoidal and typhoidal isolates, aka “serovars”.
While non-typhoidal Salmonella causes self-limiting gastroenteritis, typhoidal Salmonella causes typhoid fever.
The molecular mechanisms that underlie different disease manifestations are still being explored.
In this review, we focus on the mechanisms that underlie the pathogenesis of typhoidal Salmonella.
We highlight specific genetic traits that may contribute to typhoid fever, including serovar-specific virulence factors.
We discuss how the host responds to infections by typhoidal vs. non-typhoidal Salmonella.
Acknowledgements
We thank members of the Monack group for insightful conversations. Research reported in this publication was supported by grants R01-AI116059 and R01-AI095396 from the National Institute of Allergy and Infectious Diseases, United States (D.M.M.), Paul Allen Stanford Discovery Center on Systems Modeling of Infection (to D.M.M.), Gates Grand Challenge Grant from Bill & Melinda Gates Foundation (to D.M.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Gal-Mor O, Boyle EC, Grassl GA: Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol 2014, 5:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crump JA, Luby SP, Mintz ED: The global burden of typhoid fever. Bull World Health Organ 2004, 82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 3.Thanh DP, Karkey A, Dongol S, Thi NH, Thompson CN, Rabaa MA, Arjyal A, Holt KE, Wong V, Thieu NTV, et al. : A novel ciprofloxacin-resistant subclade of h58. Salmonella typhi is associated with fluoroquinolone treatment failure. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dougan G, Baker S: Salmonella enterica Serovar Typhi and the Pathogenesis of Typhoid Fever. http://dx.doi.org/101146/annurev-micro-091313-103739 2014, 68:317–336. [DOI] [PubMed] [Google Scholar]
- 5.Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, et al. : Emergence of an extensively drug-resistant Salmonella enterica serovar typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine MM, Ferreccio C, Black RE, Lagos R, San Martin O, Blackwelder WC: Ty21a live oral typhoid vaccine and prevention of paratyphoid fever caused by Salmonella enterica Serovar Paratyphi B. Clin Infect Dis 2007, 45 Suppl 1. [DOI] [PubMed] [Google Scholar]
- 7. Garrett DO, Longley AT, Aiemjoy K, Yousafzai MT, Hemlock C, Yu AT, Vaidya K, Tamrakar D, Saha S, Bogoch II, et al. : Incidence of typhoid and paratyphoid fever in Bangladesh, Nepal, and Pakistan: results of the Surveillance for Enteric Fever in Asia Project. Lancet Glob Health 2022, 10:e978–e988. The authors establish the Surveillance for Enteric Fever in Asia Project (SEAP) and find that there is a high burden of enteric fever in Southeast Asia, with an average incidence rate of over 100 infections per 100,000 people.
- 8.Ochiai RL, Wang X, von Seidlein L, Yang J, Bhutta ZA, Bhattacharya SK, Agtini M, Deen JL, Wain J, Kim DR, et al. : Salmonella Paratyphi A Rates, Asia. Emerg Infect Dis 2005, 11:1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabbagh SC, Lepage C, McClelland M, Daigle F: Selection of Salmonella enterica Serovar Typhi Genes Involved during Interaction with Human Macrophages by Screening of a Transposon Mutant Library. PLoS One 2012, 7:36643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forest CG, Ferraro E, Sabbagh SC, Daigle F: Intracellular survival of Salmonella enterica serovar Typhi in human macrophages is independent of Salmonella pathogenicity island (SPI)-2. Microbiology (Reading) 2010, 156:3689–3698. [DOI] [PubMed] [Google Scholar]
- 11.Karlinsey JE, Stepien TA, Mayho M, Singletary LA, Bingham-Ramos LK, Brehm MA, Greiner DL, Shultz LD, Gallagher LA, Bawn M, et al. : Genome-wide Analysis of Salmonella enterica serovar Typhi in Humanized Mice Reveals Key Virulence Features. Cell Host Microbe 2019, 26:426–434.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F: So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett 2010, 305:1–13. [DOI] [PubMed] [Google Scholar]
- 13.Wilson RP, Winter SE, Spees AM, Winter MG, Nishimori JH, Sanchez JF, Nuccio SP, Crawford RW, Tükel Ç, Bäumler AJ: The Vi Capsular Polysaccharide Prevents Complement Receptor 3-Mediated Clearance of Salmonella enterica Serotype Typhi. Infect Immun 2011, 79:830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson RP, Raffatellu M, Chessa D, Winter SE, Tükel Ç, Bäumler AJ: The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol 2008, 10:876–890. [DOI] [PubMed] [Google Scholar]
- 15.Zhang LF, Lepenies B, Nakamae S, Young BM, Santos RL, Raffatellu M, Cobb BA, Hiyoshi H, Bäumler AJ: The Vi Capsular Polysaccharide of Salmonella Typhi Promotes Macrophage Phagocytosis by Binding the Human C-Type Lectin DC-SIGN. mBio 2022, doi: 10.1128/MBIO.02733-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winter SE, Winter MG, Atluri V, Poon V, Romão EL, Tsolis RM, Bäumler AJ: The Flagellar Regulator TviA Reduces Pyroptosis by Salmonella enterica Serovar Typhi. Infect Immun 2015, 83:1546–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mylona E, Sanchez-Garrido J, Hoang Thu TN, Dongol S, Karkey A, Baker S, Shenoy AR, Frankel G: Very long O-antigen chains of Salmonella Paratyphi A inhibit inflammasome activation and pyroptotic cell death. Cell Microbiol 2021, 23. The authors show that S. Paratyphi A encodes a very long O-antigen that plays a similar role to the Vi capsule in S. Typhi by inhibiting inflammasome activation and suppressing immune response.
- 18.Spanò S, Ugalde JE, Galán JE: Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 2008, 3:30–38. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Gao X, Galán JE: Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature 2013 499:7458 2013, 499:350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sepe LP, Hartl K, Iftekhar A, Berger H, Kumar N, Goosmann C, Chopra S, Schmidt SC, Gurumurthy RK, Meyer TF, et al. : Genotoxic effect of salmonella paratyphi a infection on human primary gallbladder cells. mBio 2020, 11:1–39. The authors show that S. Paratyphi A uses its typhoid toxin to cause DNA damage in gallbladder cells, which may have implications for why enteric fever is associated with increased risk of gallbladder cancer.
- 21.di Domenico EG, Cavallo I, Pontone M, Toma L, Ensoli F: Biofilm Producing Salmonella Typhi: Chronic Colonization and Development of Gallbladder Cancer. Int J Mol Sci 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla VK, Singh H, Pandey M, Upadhyay SK, Nath G: Carcinoma of the Gallbladder—Is It a Sequel of Typhoid? Digestive Diseases and Sciences 2000 45:5 2000, 45:900–903. [DOI] [PubMed] [Google Scholar]
- 23.del Bel Belluz L, Guidi R, Pateras IS, Levi L, Mihaljevic B, Rouf SF, Wrande M, Candela M, Turroni S, Nastasi C, et al. : The Typhoid Toxin Promotes Host Survival and the Establishment of a Persistent Asymptomatic Infection. PLoS Pathog 2016, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibani MM, Jones E, Barton A, Jin C, Meek J, Camara S, Galal U, Heinz E, Rosenberg-Hasson Y, Obermoser G, et al. : Investigation of the role of typhoid toxin in acute typhoid fever in a human challenge model. [date unknown], doi: 10.1038/s41591-019-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, Mclellan M, et al. : Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. 2004, 36. [DOI] [PubMed] [Google Scholar]
- 26.Holt KE, Thomson NR, Wain J, Langridge GC, Hasan R, Bhutta ZA, Quail MA, Norbertczak H, Walker D, Simmonds M, et al. : Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 2009, 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bronner DN, Faber F, Olsan EE, Byndloss MX, Sayed NA, Xu G, Yoo W, Kim D, Ryu S, Lebrilla CB, et al. : Genetic Ablation of Butyrate Utilization Attenuates Gastrointestinal Salmonella Disease. Cell Host Microbe 2018, 23:266–273.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carden S, Okoro C, Dougan G, Monack D: Non-typhoidal Salmonella Typhimurium ST313 isolates that cause bacteremia in humans stimulate less inflammasome activation than ST19 isolates associated with gastroenteritis. Pathog Dis 2015, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchello CS, Birkhold M, Crump JA, Martin LB, Ansah MO, Breghi G, Canals R, Fiorino F, Gordon MA, Kim JH, et al. : Complications and mortality of non-typhoidal salmonella invasive disease: a global systematic review and meta-analysis. Lancet Infect Dis 2022, 22:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haselbeck AH, Panzner U, Im J, Baker S, Meyer CG, Marks F: Current perspectives on invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis 2017, 30:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uche IV, MacLennan CA, Saul A: A Systematic Review of the Incidence, Risk Factors and Case Fatality Rates of Invasive Nontyphoidal Salmonella (iNTS) Disease in Africa (1966 to 2014). PLoS Negl Trop Dis 2017, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fierer J: Invasive Non-typhoidal Salmonella (iNTS) Infections. 2022, doi: 10.1093/cid/ciac035. [DOI] [PubMed] [Google Scholar]
- 33.Carden SE, Walker GT, Honeycutt J, Lugo K, Pham T, Jacobson A, Bouley D, Idoyaga J, Tsolis RM, Monack D: Pseudogenization of the secreted effector gene sseI confers rapid systemic dissemination of S. Typhimurium ST313 within migratory dendritic cells. Cell Host Microbe 2017, 21:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albalat R, Cañestro C: Evolution by gene loss. 2016, doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 35.Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, Tavazoie S: Bacterial adaptation through loss of function. PLoS Genet 2013, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurelli AT: Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol Lett 2007, 267:1–8. [DOI] [PubMed] [Google Scholar]
- 37.Moore RA, Reckseidler-Zenteno S, Kim H, Nierman W, Yu Y, Tuanyok A, Warawa J, DeShazer D, Woods DE: Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect Immun 2004, 72:4172–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurelli AT, Fernández fernández RE, Bloch CA, Rode CK, Fasano A: “Black holes” and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. 1998, 95:3943–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford RW, Wangdi T, Spees AM, Xavier MN, Tsolis RM, Bäumler AJ: Loss of very-long O-antigen chains optimizes capsule-mediated immune evasion by Salmonella enterica serovar Typhi. mBio 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson SM, Rajachandran V, Majumdar S, Saha S, Das S, Chattopadhyay S: Distinct Potentially Adaptive Accumulation of Truncation Mutations in Salmonella enterica serovar Typhi and Salmonella enterica serovar Paratyphi A. Microbiol Spectr 2022, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gómez-Valero L, Rocha EPC, Latorre A, Silva FJ: Reconstructing the ancestor of Mycobacterium leprae: The dynamics of gene loss and genome reduction. Genome Res 2007, 17:1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mai SNT, Bodhidatta L, Turner P, Wangchuk S, Thanh TH, Vinh PV, Pham DT, Rabaa MA, Thwaites GE, Thomson NR, et al. : The evolutionary history of Shigella flexneri serotype 6 in Asia. Microb Genom 2021, 7:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Octavia S, Maharjan RP, Sintchenko V, Stevenson G, Reeves PR, Gilbert GL, Lan R: Insight into Evolution of Bordetella pertussis from Comparative Genomic Analysis: Evidence of Vaccine-Driven Selection. Mol Biol Evol 2011, 28:707–715. [DOI] [PubMed] [Google Scholar]
- 44. Feng Y, Wang Z, Chien KY, Chen HL, Liang YH, Hua X, Chiu CH: “Pseudo-pseudogenes” in bacterial genomes: Proteogenomics reveals a wide but low protein expression of pseudogenes in Salmonella enterica. Nucleic Acids Res 2022, 50:5158–5170. The authors show that many pseudogenes in typhoidal Salmonella may be functional, which raises the possibility that they play novel roles during infection. However, this finding requires more experimental validation in the future.
- 45.Johnson R, Mylona E, Frankel G: Typhoidal Salmonella: Distinctive virulence factors and pathogenesis. Cell Microbiol 2018, 20:e12939. [DOI] [PubMed] [Google Scholar]
- 46.Ramos-Morales F: Impact of Salmonella enterica Type III Secretion System Effectors on the Eukaryotic Host Cell. International Scholarly Research Network ISRN Cell Biology 2012, 2012:36. [Google Scholar]
- 47.Lou L, Zhang P, Piao R, Wang Y: Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network. Front Cell Infect Microbiol 2019, 9:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Rüssmann H, Bäumler AJ: The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol 2009, 74:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson R, Ravenhall M, Pickard D, Dougan G, Byrne A, Frankel G: Comparison of Salmonella enterica Serovars Typhi and Typhimurium Reveals Typhoidal Serovar-Specific Responses to Bile. Infect Immun 2018, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basnyat B, Baker S: Typhoid carriage in the gallbladder. The Lancet 2015, 386:1074. [DOI] [PubMed] [Google Scholar]
- 51.Coburn B, Sekirov I, Finlay BB: Type III Secretion Systems and Disease. Clin Microbiol Rev 2007, 20:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG: The Salmonella enterica Serotype Typhimurium Effector Proteins SipA, SopA, SopB, SopD, and SopE2 Act in Concert To Induce Diarrhea in Calves. Infect Immun 2002, 70:3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valenzuela LM, Hidalgo AA, Rodríguez L, Urrutia IM, Ortega AP, Villagra NA, Paredes-Sabja D, Calderón IL, Gil F, Saavedra CP, et al. : Pseudogenization of sopA and sopE2 is functionally linked and contributes to virulence of Salmonella enterica serovar Typhi. Infect Genet Evol 2015, 33:131–142. [DOI] [PubMed] [Google Scholar]
- 54.McDowell MA, Byrne AMP, Mylona E, Johnson R, Sagfors A, Crepin VF, Lea S, Frankel G: The S. Typhi effector StoD is an E3/E4 ubiquitin ligase which binds K48- and K63-linked diubiquitin. Life Sci Alliance 2019, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGhie EJ, Hayward RD, Koronakis V: Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J 2001, 20:2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friebel A, Ilchmann H, Aepfelbacher M, Ehrbar K, Machleidt W, Hardt W-D: SopE and SopE2 from Salmonella typhimurium Activate Different Sets of RhoGTPases of the Host Cell*. 2001, doi: 10.1074/jbc.M100609200. [DOI] [PubMed] [Google Scholar]
- 57.Fu Y, Galán JE: A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 1999, 401:293–297. [DOI] [PubMed] [Google Scholar]
- 58.Johnson R, Byrne A, Berger CN, Klemm E, Crepin VF, Dougan G, Frankel G: The Type III Secretion System Effector SptP of Salmonella enterica Serovar Typhi. J Bacteriol 2017, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Velden AWM, Lindgren SW, Worley MJ, Heffron F: Salmonella Pathogenicity Island 1-Independent Induction of Apoptosis in Infected Macrophages by Salmonella enterica Serotype Typhimurium. Infect Immun 2000, 68:5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang L, Wang P, Song X, Zhang H, Ma S, Wang J, Li W, Lv R, Liu X, Ma S, et al. : Salmonella Typhimurium reprograms macrophage metabolism via T3SS effector SopE2 to promote intracellular replication and virulence. Nature Communications 2021 12:1 2021, 12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Figueira R, Watson KG, Holden DW, Helaine S: Identification of Salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar typhimurium: Implications for rational vaccine design. mBio 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jennings E, Thurston TLM, Holden DW: Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms And Physiological Consequences. Cell Host Microbe 2017, 22:217–231. [DOI] [PubMed] [Google Scholar]
- 63.Yu XJ, Liu M, Holden DW: Salmonella effectors SseF and SseG interact with mammalian protein ACBD3 (GCP60) to anchor salmonella-containing vacuoles at the Golgi network. mBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knuff K, Finlay BB: What the SIF Is Happening—The Role of Intracellular Salmonella-Induced Filaments. Front Cell Infect Microbiol 2017, 7:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trombert AN, Berrocal L, Fuentes JA, Mora GC: S. Typhimurium sseJ gene decreases the S. Typhi cytotoxicity toward cultured epithelial cells. BMC Microbiol 2010, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spanò S, Galán JE: A Rab32-dependent pathway contributes to Salmonella typhi host restriction. Science 2012, 338:960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spanò S: Host restriction in Salmonella: insights from Rab GTPases. Cell Microbiol 2014, 16:1321–1328. [DOI] [PubMed] [Google Scholar]
- 68. Baldassarre M, Solano-Collado V, Balci A, Colamarino RA, Dambuza IM, Reid DM, Wilson HM, Brown GD, Mukhopadhyay S, Dougan G, et al. : The Rab32/BLOC-3–dependent pathway mediates host defense against different pathogens in human macrophages. Sci Adv 2021, 7. This paper shows that S. Typhi has evolved mechanisms to target human Rab-32, which may help explain why S. Typhi can survive in human macrophages but not in murine macrophages.
- 69.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW: Internalization of Salmonella by Macrophages Induces Formation of Nonreplicating Persisters. Science 2014, 343:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saliba AE, Li L, Westermann AJ, Appenzeller S, Stapels DAC, Schulte LN, Helaine S, Vogel J: Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat Microbiol 2016, 2. [DOI] [PubMed] [Google Scholar]
- 71.Steiner TS: How Flagellin and Toll-Like Receptor 5 Contribute to Enteric Infection. Infect Immun 2007, 75:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aldapa-Vega G, Moreno-Eutimio MA, Berlanga-Taylor AJ, Jiménez-Uribe AP, Nieto-Velazquez G, López-Ortega O, Mancilla-Herrera I, Cortés-Malagón EM, Gunn JS, Isibasi A, et al. : Structural variants of Salmonella Typhimurium lipopolysaccharide induce less dimerization of TLR4/MD-2 and reduced pro-inflammatory cytokine production in human monocytes. Mol Immunol 2019, 111:43–52. [DOI] [PubMed] [Google Scholar]
- 73.Tükel Ç, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akçelik M, et al. : CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol 2005, 58:289–304. [DOI] [PubMed] [Google Scholar]
- 74.Kawasaki T, Kawai T: Toll-like receptor signaling pathways. Front Immunol 2014, 5:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss DS, Raupach B, Takeda K, Akira S, Zychlinsky A: Toll-Like Receptors Are Temporally Involved in Host Defense. The Journal of Immunology 2004, 172:4463–4469. [DOI] [PubMed] [Google Scholar]
- 76.Nyström S, Antoine DJ, Lundbäck P, Lock JG, Nita AF, Högstrand K, Grandien A, Erlandsson-Harris H, Andersson U, Applequist SE: TLR activation regulates damage-associated molecular pattern isoforms released during pyroptosis. EMBO J 2013, 32:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raupach B, Peuschel S-K, Monack DM, Zychlinsky A: Caspase-1-mediated activation of interleukin-1beta (IL-1beta) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect Immun 2006, 74:4922–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mathur R, Oh H, Zhang D, Park SG, Seo J, Koblansky A, Hayden MS, Ghosh S: A mouse model of salmonella typhi infection. Cell 2012, 151:590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song J, Wilhelm CL, Wangdi T, Maira-Litran T, Lee SJ, Raetz M, Sturge CR, Mirpuri J, Pei J, Grishin N v., et al. : Absence of TLR11 in Mice Does Not Confer Susceptibility to Salmonella Typhi. Cell 2016, 164:827–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P: The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1 Gly169 allele. Infect Immun 1996, 64:2923–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chapes SK, Beharka AA: Salmonella infections in the absence of the major histocompatibility complex II. J Leukoc Biol 1998, 63:297–304. [DOI] [PubMed] [Google Scholar]
- 82.Hue NT, Lanh MN, Phuong LT, Vinh H, Chinh NT, Hien TT, Hieu NT, Farrar JJ, Dunstan SJ: Toll-Like Receptor 4 (TLR4) and Typhoid Fever in Vietnam. PLoS One 2009, 4:e4800–e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunstan SJ, Hue NT, Han B, Li Z, Tram TTB, Sim KS, Parry CM, Chinh NT, Vinh H, Lan NPH, et al. : Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat Genet 2014, 46:1333–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Monack DM, Bouley DM, Falkow S: Salmonella typhimurium Persists within Macrophages in the Mesenteric Lymph Nodes of Chronically Infected Nramp1+/+ Mice and Can Be Reactivated by IFNγ Neutralization. Journal of Experimental Medicine 2004, 199:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunstan SJ, Ho VA, Duc CM, Lanh MN, Phuong CXT, Luxemburger C, Wain J, Dudbridge F, Peacock CS, House D, et al. : Typhoid Fever and Genetic Polymorphisms at the Natural Resistance–Associated Macrophage Protein 1. J Infect Dis 2001, 183:1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fadl MA, Aydarous MA, Maom C, Yasmeen A: An association of VNTR polymorphism in intron3 of IL-4 gene with susceptibility to typhoid fever in Khartoum State, Sudan. Kuwait J Sci 2016, 43:185–192. [Google Scholar]
- 87.Ali S, Vollaard AM, Widjaja S, Surjadi C, van de Vosse E, van Dissel JT: PARK2/PACRG polymorphisms and susceptibility to typhoid and paratyphoid fever. Clin Exp Immunol 2006, 144:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de van Vosse E, de Visser AW, Al-Attar S, Vossen R, Ali S, van Dissel JT: Distribution of CFTR Variations in an Indonesian Enteric Fever Cohort. Clinical Infectious Diseases 2010, 50:1231–1237. [DOI] [PubMed] [Google Scholar]
- 89.MI Alvarez, Glover LC, Luo P, Wang L, Theusch E, Oehlers SH, Walton EM, Tram TTB, Kuang Y-L, Rotter JI, et al. : Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol. Proceedings of the National Academy of Sciences 2017, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feasey NA, Archer BN, Heyderman RS, Sooka A, Dennis B, Gordon MA, Keddy KH: Typhoid Fever and Invasive Nontyphoid Salmonellosis, Malawi and South Africa. Emerg Infect Dis 2010, 16:1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ali S, Vollaard AM, Kremer D, de Visser AW, Martina CAE, Widjaja S, Surjadi C, Slagboom E, van de Vosse E, van Dissel JT: Polymorphisms in Proinflammatory Genes and Susceptibility to Typhoid Fever and Paratyphoid Fever. https://home.liebertpub.com/jir 2007, 27:271–279. [DOI] [PubMed] [Google Scholar]
- 92.Gordon MA: Salmonella infections in immunocompromised adults. J Infect 2008, 56:413–422. [DOI] [PubMed] [Google Scholar]
- 93.Yang J, Zhao Y, Shi J, Shao F: Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A 2013, 110:14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Naseer N, Egan MS, Reyes Ruiz VM, Scott WP, Hunter EN, Demissie T, Rauch I, Brodsky IE, Shin S: Human NAIP/NLRC4 and NLRP3 inflammasomes detect Salmonella type III secretion system activities to restrict intracellular bacterial replication. PLoS Pathog 2022, 18:e1009718–e1009718. The authors find new roles for human NAIP during macrophage infection, and they show that human NAIP recognizes the SPI-2 needle protein during Salmonella infection.
- 95.Kortmann J, Brubaker SW, Monack DM: Cutting Edge: Inflammasome Activation in Primary Human Macrophages Is Dependent on Flagellin. The Journal of Immunology 2015, 195:815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Srinivasan M, Sindhu KN, Giri S, Kumar N, Mohan VR, Grassly NC, Kang G: Salmonella Typhi Shedding and Household Transmission by Children With Blood Culture-Confirmed Typhoid Fever in Vellore, South India. J Infect Dis 2021, 224:S593–S600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gonzalez-Escobedo G, Marshall JM, Gunn JS: Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol 2011, 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Prouty AM, Schwesinger WH, Gunn JS: Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun 2002, 70:2640–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hoffman SA, Desai SN, Sikorski MJ, Fatupaito G, Tupua S, Thomsen RE, Rambocus S, Nimarota-Brown S, Punimata LL, Sialeipata M, et al. : Point-of-Care Ultrasound by Nonexpert Operators Demonstrates High Sensitivity and Specificity in Detecting Gallstones: Data from the Samoa Typhoid Fever Control Program. Am J Trop Med Hyg 2022, 106:798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eisele NA, Ruby T, Jacobson A, Manzanillo PS, Cox JS, Lam L, Mukundan L, Chawla A, Monack DM: Salmonella Require the Fatty Acid Regulator PPARδ for the Establishment of a Metabolic Environment Essential for Long-Term Persistence. Cell Host Microbe 2013, 14:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sheikh A, Khanam F, Sayeed MA, Rahman T, Pacek M, Hu Y, Rollins A, Bhuiyan MS, Rollins S, Kalsy A, et al. : Interferon-γ and proliferation responses to Salmonella enterica Serotype Typhi proteins in patients with S. Typhi Bacteremia in Dhaka, Bangladesh. PLoS Negl Trop Dis 2011, 5:e1193–e1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gasem MH, Keuter M, Dolmans WM v, van der Ven-Jongekrijg J, Djokomoeljanto R, van der Meer JWM: Persistence of Salmonellae in Blood and Bone Marrow: Randomized Controlled Trial Comparing Ciprofloxacin and Chloramphenicol Treatments against Enteric Fever. Antimicrob Agents Chemother 2003, 47:1727–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Butler T, Ho M, Acharya G, Tiwari M, Gallati H: Interleukin-6, gamma interferon, and tumor necrosis factor receptors in typhoid fever related to outcome of antimicrobial therapy. Antimicrob Agents Chemother 1993, 37:2418–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gal-Mor O, Suez J, Elhadad D, Porwollik S, Leshem E, Valinsky L, McClelland M, Schwartz E, Rahav G: Molecular and Cellular Characterization of a Salmonella enterica Serovar Paratyphi A Outbreak Strain and the Human Immune Response to Infection. Clinical and Vaccine Immunology 2012, 19:146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Jong R, Altare F, Haagen I-A, Elferink DG, de Boer T, van Breda Vriesman PJC, Kabel PJ, Draaisma JMT, van Dissel JT, Kroon FP, et al. : Severe Mycobacterial and Salmonella Infections in Interleukin-12 Receptor-Deficient Patients. Science (1979) 1998, 280:1435–1438. [DOI] [PubMed] [Google Scholar]
- 106.Jouanguy E, Lamhamedi-Cherradi S, Altare F, Fondanèche MC, Tuerlinckx D, Blanche S, Emile JF, Gaillard JL, Schreiber R, Levin M, et al. : Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette-Guérin infection and a sibling with clinical tuberculosis. Journal of Clinical Investigation 1997, 100:2658–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Altare F, Lammas D, Revy P, Jouanguy E, Döffinger R, Lamhamedi S, Drysdale P, Scheel-Toellner D, Girdlestone J, Darbyshire P, et al. : Inherited interleukin 12 deficiency in a child with bacille Calmette-Guérin and Salmonella enteritidis disseminated infection. Journal of Clinical Investigation 1998, 102:2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bharadwaj S, Anim JT, Ebrahim F, Aldahham A: Granulomatous inflammatory response in a case of typhoid fever. Med Princ Pract 2009, 18:239–241. [DOI] [PubMed] [Google Scholar]
- 109.Mert A, Tabak F, Ozaras R, Ozturk R, Aki H, Aktuglu Y: Typhoid fever as a rare cause of hepatic, splenic, and bone marrow granulomas. Intern Med 2004, 43:436–439. [DOI] [PubMed] [Google Scholar]
- 110.Narechania S, Duran M, Karivedu V, Gopalakrishna K v: A case of typhoid Fever with hepatic granulomas and enteritis. Case Rep Pathol 2015, 2015:745461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pham THM, Brewer SM, Thurston T, Massis LM, Honeycutt J, Lugo K, Jacobson AR, Vilches-Moure JG, Hamblin M, Helaine S, et al. : Salmonella-Driven Polarization of Granuloma Macrophages Antagonizes TNF-Mediated Pathogen Restriction during Persistent Infection. Cell Host Microbe 2020, 27:54–67.e5. This paper explains how Salmonella drive macrophages into an M2 phenotype, altering their permissiveness in splenic granulomas while simultaneously evading TNF-mediated restriction.
- 112.Goldberg MF, Roeske EK, Ward LN, Pengo T, Dileepan T, Kotov DI, Jenkins MK: Salmonella Persist in Activated Macrophages in T Cell-Sparse Granulomas but Are Contained by Surrounding CXCR3 Ligand-Positioned Th1 Cells. Immunity 2018, 49:1090–1102.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Panagi I, Jennings E, Zeng J, Günster RA, Stones CD, Mak H, Jin E, Stapels DAC, Subari NZ, Pham THM, et al. : Salmonella Effector SteE Converts the Mammalian Serine/Threonine Kinase GSK3 into a Tyrosine Kinase to Direct Macrophage Polarization. Cell Host Microbe 2020, 27:41–53.e6. This paper shows that the Salmonella effector SteE causes macrophages to convert to an anti-inflammatory state, which is beneficial for Salmonella survival.